Open Access Article
Open Access ArticleTryptophan metabolites relieve intestinal Candida albicans infection by altering the gut microbiota to reduce IL-22 release from group 3 innate lymphoid cells of the colon lamina propria†
Ziyao
Peng
 a,
Jiali
Zhang
b,
Meng
Zhang
c,
Liping
Yin
a,
Ziyang
Zhou
a,
Cuiting
Lv
b,
Zetian
Wang
*a and
Jianguo
Tang
*a
a,
Jiali
Zhang
b,
Meng
Zhang
c,
Liping
Yin
a,
Ziyang
Zhou
a,
Cuiting
Lv
b,
Zetian
Wang
*a and
Jianguo
Tang
*a
aDepartment of Trauma-Emergency and Critical Care Medicine, Shanghai Fifth People's Hospital, Fudan University, Shanghai, China
bCentral Laboratory, Shanghai Fifth People's Hospital, Fudan University, Shanghai, China
cDepartment of Pulmonary and Critical Care Medicine, Shanghai Fifth People's Hospital, Fudan University, Shanghai, China
First published on 13th March 2024
Abstract
Invasive candidiasis may be caused by Candida albicans (C. albicans) colonization of the intestinal tract. Preventing intestinal damage caused by Candida albicans infection and protecting intestinal barrier function have become a critical issue. Integrated analyses of the microbiome with metabolome revealed a remarkable shift of the gut microbiota and tryptophan metabolites, kynurenic acid (KynA), and indolacrylic acid (IA) in mice infected with C. albicans. The transcriptome sequencing indicated that differentially expressed genes were significantly associated with innate immune responses and inflammatory responses. The results of this study suggest that KynA and IA (KI) can alleviate intestinal damage caused by Candida albicans infection in mice by reducing intestinal permeability, increasing intestinal firmness, alleviating intestinal inflammation, and reducing the secretion of interleukin-22 (IL-22) in the 3 groups of colon innate lymphoid cells (ILC3). We performed a fecal microbiota transplantation (FMT) experiment and found that the intestinal barrier function, inflammation, and IL-22 secretion of ILC3 in the colon lamina propria of the recipient mice subjected to C. albicans infection and KI treatment were consistent with the trends of the donor mice. Our results suggest that tryptophan metabolites may directly regulate colon lamina ILC3 to promote intestinal resistance to C. albicans invasion, or indirectly regulate the ILC3 secretion of IL-22 to play a protective role in the intestinal barrier by affecting intestinal microorganisms, which may become a potential target for alleviating intestine borne C. albicans infection.
Introduction
C. albicans is an opportunistic pathogen that is a normal component of the human gut microbiota. In recent years, with the extensive use of tumor chemotherapy, organ transplantation, glucocorticoids, immunosuppressants, and broad-spectrum antibiotics, the incidence of invasive candidiasis has been on the rise.1 Among them, candidemia and disseminated candidiasis are the most common clinical types of invasive candidiasis, and the prognosis is often poor. C. albicans and the host are in a state of homeostasis. When the balance is disturbed, the yeast is able to break through the barrier of the intestinal mucosa and cause invasive candidiasis and candidemia.2–4Candida albicans can be effectively prevented from penetrating the intestinal mucosa and entering deep tissues by the mechanical intestinal barrier, which is composed of intestinal epithelial cells, tight intercellular junctions, and intestinal peristalsis, the most important line of the intestinal mucosal barrier.5 In the neighboring cells of the intestinal epithelium, there is a selective permeability gap, known as the paracellular pathway, which regulates the size of the cellular gap using tight junctions and the interaction of associated proteins. Reduced expression of these proteins disrupts the physical barrier of the intestinal epithelial cells and increases their permeability, allowing pathogens, toxins, antigens, and others to enter the lamina propria. It triggers an inflammatory cascade and promotes intestinal damage.6,7Candida albicans is capable of sensing a variety of extracellular stimuli, inducing adaptive transformation from a yeast to a hyphal state, and increasing its ability to penetrate mucosal membranes. This morphological transformation is considered to be an important factor in its pathogenesis. Candidalysin is synthesized by the mycelium-forming gene Ece1 of Candida albicans, which can penetrate the epithelial cell membrane and disrupt the intestinal epithelial barrier.7 At the same time, there are proteins on the surface of Candida albicans that bind to host-specific ligands, called invasins, the most widely recognized of which are agglutinin-like sequence 3 (Als3) and stress seventy related protein-a (Ssa). At the same time, Candida albicans secretes a variety of proteolytic enzymes that promote invasiveness, including secreted aspartyl proteinase (Saps), phospholipase (Pl), and lipase (Lip).8,9 Therefore, the effective protection of the intestinal epithelial barrier, the resistance, and the control of the Candida albicans infection are an important problem in the field of severe infections.Several studies have shown that the gut microbiota may influence C. albicans colonization and invasion.10C. albicans in the gut can also cause bacteremia and imbalance in the gut microbiota.11,12 Therefore, the interaction of C. albicans with other gut bacteria is closely linked to symptomatic C. albicans infections.4,13 An increasing number of studies have suggested that the intestinal microbiota, metabolites, and intestinal barrier may influence C. albicans colonization and invasion. Earlier studies based on 16S rRNA sequencing have suggested that C. albicans from the gut may erode the gut lining by regulating the gut microbiota. In addition, there is a close relationship between the gut microbiota and metabolites. In addition, the effect of C. albicans on the intestinal microbiota causes changes in the microbiota's metabolic products, although little research has been conducted in this field.14–16
Tryptophan (Trp) is an essential amino acid used to build proteins. It is also a biochemical precursor of metabolites that has important effects on mammalian physiology, such as the gastrointestinal, immune, metabolic, and nervous systems. In the gastrointestinal tract, the metabolism of L-tryptophan can follow three main pathways, all of which are influenced by the gut microbiota: (i) the kynurenine (KP) route in both immune and epithelial cells, (ii) the serotonergic route in enterochromaffin cells (ECC), a specialized subtype of intestinal epithelium cells, and (iii) direct conversion of L-Trp into various molecules, including aryl hydrocarbon receptor (AhR) ligands.17 In humans, tryptophan (Trp) is an essential amino acid that can only be obtained through dietary sources. Intestinal flora plays an important role in tryptophan metabolism. Gut microbes directly convert tryptophan into various molecules, such as indole and its derivatives. Indole and its derivatives maintain intestinal homeostasis by regulating the expression of pro-inflammatory and anti-inflammatory cytokines.18 Metabolomic analysis has shown that gut bacteria influence host metabolism and immunity through a variety of chemically distinct metabolites, including amino acid metabolites. In particular, a lack of Trp in the diet impairs intestinal immunity in mice and alters the microbial community in the gut,19 suggesting that mucosal homeostasis is a multifactorial phenomenon in which Trp metabolism is an important regulatory component. Owing to its binding with AhR, kynurenic acid (KynA) is an important metabolite with potent immunoregulatory functions.20,21 Some species of Peptostreptococci contain a cluster of genes that enable the production of the tryptophan metabolite indole acrylic (IA), which enhances intestinal epithelial barrier function and reduces inflammatory responses.22 However, there are few studies on how Candida albicans infection disrupts intestinal microecology and its effects on tryptophan metabolism. Meanwhile, how tryptophan metabolites (such as kynurenic acid and indole acrylate (KI)) alleviate intestinal damage caused by Candida albicans infection remains to be studied.
Interleukin-22 (IL-22) is a cytokine of the IL-10 superfamily. It is involved in the innate immune response to bacterial pathogens and is mainly secreted by Th17, Th22, NK, and other immune cells, particularly in epithelial cells such as respiratory and intestinal epithelial cells.23 Studies have shown that midgut epithelial cells secrete antimicrobial peptides mainly by stimulating IL-22. Pathogenic bacteria promote the secretion of IL-22 by various innate immune cells and the secretion of antimicrobial peptides (mainly including lysozyme, regIIIγ, cryptate, and secreted phospholipase A2) by the IL-22Ra/IL-10Rp/Stat3 signaling pathway.24 Bacterial translocation, microbial imbalance, and other causes can lead to decreased secretion of IL-22, destruction of the intestinal immune barrier, decreased secretion of antimicrobial peptides, and altered intestinal homeostasis.25 In animals, the dynamic changes in IL-22 are regular in the process of repairing damage to the intestine, and this balance cannot be disturbed. On the one hand, when the intestinal epithelium is injured, the epithelial structure is severely damaged and the high expression of IL-22 is conducive to its role in promoting cell proliferation and supporting the reconstruction of the intestinal epithelium. However, the late stage of inflammation is also the time of the formation of a large number of terminally differentiated cells, so the sustained high expression of IL-22 does not promote the differentiation of intestinal epithelial cells. At this time, the body regulates the downregulation of IL-22 expression and promotes the repair of gut tissue damage.26 At present, the regulatory mechanism of IL-22 induced by tryptophan metabolites remains to be clarified.
In this study, we compared the intestinal microbiome and metabolic changes of mice infected with Candida albicans and control mice, and screened out tryptophan metabolites with significant changes in content to investigate the mechanism of their protective effect on intestinal damage caused by Candida albicans infection. We constructed a mouse model of DSS-induced Candida albicans intestinal infection to investigate the effects of KI on inflammation, intestinal microbiome, intestinal barrier, and immunity. In addition, transcriptome sequencing and 2bRAD-M sequencing revealed the related gene changes of KI in the mouse gut and the alteration of the gut microbiome. Since the relationship between the secretion of IL-22 regulated by tryptophan metabolites and intestinal flora has not been studied, we constructed a fecal microbiota transplantation (FMT) model to study the stool samples of germ-free (GF) mice (donor mice), healthy mice with intestinal Candida albicans infection and mice treated with KI. We found that the intestinal flora after Candida albicans infection had a destructive effect on the intestinal barrier of recipient mice, and transplantation of intestinal flora after KI treatment could reverse this effect. Meanwhile, the secretion of IL-22 and inflammation of ILC3 in recipient mice were similar to those in donor mice. Therefore, we inferred that the role of tryptophan metabolites in ILC3's secretion of IL-22 to protect the intestinal barrier is partly through the intestinal microbiota, and our results provide evidence for the regulatory role of KI in the intestinal microbiota and suggest that KI may be a promising therapeutic approach for the treatment of intestinal damage caused by intestinal Candida albicans infection.
Results
Changes in gut microbiota diversity and associated metabolites caused by Candida albicans infection
We established a mouse model of DSS-induced Candida albicans intestinal infection. We performed a joint analysis by metabolomic sequencing and 16s RNA microbiome sequencing. We found significant changes in tryptophan metabolites (KynA and IA), indicating that Candida albicans infection disrupts the intestinal flora. As shown in Fig. 1a and b, Shannon's and Simpson's diversity indices were significantly lower in the CA group than in the C group. A clear distinction between the CA and C groups was observed in the Bray–Curtis PCoA analysis (Fig. 1c). At the phylum level (Fig. 1d), Bacteroidetes and Verrucomicrobia ratios and relative abundances were similar between the CA and control groups. At the species level (Fig. 1e), Muribaculaceae_bacterium_Isolate_080_Janvier and Bacteroides_sp_NM69_E16B were more abundant in the CA group than in the C group. Muribaculaceae_bacterium_Isolate_080_Janvier was enriched in the CA group using linear discriminant analysis coupled with effect size measurements (LEfSe) (Fig. 1g).To elucidate the effects of C. albicans gut infection on microbial metabolites, LC-MS analysis was performed to detect the differentially expressed metabolites and relevant key pathways between the two groups. Partial least squares discriminant analysis (PLS-DA) score plots showed a clear separation of metabolites between the two groups (ESI Fig. 1a†), which were divided into two clusters based on concentration profiles. The tryptophan metabolites kynurenic acid and indoleacetic acid were significantly different between the C and CA groups when 46 different organoheterocyclic compounds were screened between the two groups (Fig. 1f). Our metabolome sequencing showed that the expression of indoleacrylic acid in the CA group was higher than that in the C group and the combined analysis of 16s and the metabolome showed that Akkermansia in the CA group was positively correlated with metabolites such as indoleacrylic acid, that is, Akkermansia was also elevated in the CA group. A. muciniphila decreases the secretion of several pro-inflammatory cytokines while increasing the production of IL-10 in different mouse models.27 Thus, it is found that our sequencing data support that the process of Candida albicans infection leads to an increase in the abundance of Akk and an increase in IA. This process is an anti-inflammatory process of the body towards infection, which is also in line with the inflammatory response pathway shown in Fig. 2c. These results suggest that the gut microbiota and metabolites can be effectively altered by C. albicans intestinal infection compared with the control group.
Candida albicans infection causes intestinal inflammatory responses and immune responses
To investigate the underlying mechanisms of intestinal barrier damage caused by C. albicans infection, colon tissues from the CA and C groups (n = 4 per group) were subjected to transcriptome sequencing. A total of 2123 differentially expressed genes were selected (Fig. 2a) using a cut-off of p < 0.05. To understand the biological functions potentially regulated by these genes, further investigation was performed using the Gene Ontology (GO) database. We found that these genes may play a role in immune function, innate immunity, and inflammation (Fig. 2b). Cytoscape software was then used to predict interactions between selected genes based on their corrected p-values and fold change values (Fig. 2c). To validate the transcriptomic sequences, additional PCR arrays for the mouse inflammatory response and immunity were used to profile the expression of genes involved in the C. albicans induced inflammatory response (n = 4 per group). The data showed that 15 candidate genes were upregulated in the CA group compared to those in the C group (Fig. 2d). One pathway involved in mucosal inflammation and immunity has been identified in the literature: ILC3 effector cytokines (IL-22, IL-17α, and IL-18). PCR analysis confirmed higher expression of IL-22 in the CA group than in the C group (ESI Fig. 1b†).Combining KynA and IA may reduce intestinal inflammation caused by Candida albicans infection and help protect the mucosal barrier
In the process of validating Kl's phenotype for intestinal barrier protection, we found that Kl can reduce intestinal permeability and inhibit intestinal inflammation, thereby improving the overall condition of the intestine after infection with Candida albicans. The CA group showed a higher incidence of colon shortening than the control group, whereas the KI group showed a higher incidence of colon lengthening than the CA group (Fig. 3a). In addition, the mRNA and protein levels of ZO-1 and occludin increased in the colons of the KI group compared to those in the CA group (Fig. 3b and c). Intestinal infection with C. albicans dramatically reduced the intestinal permeability of the mice, as measured with plasma FD-4 levels, and treatment with KI significantly reversed this trend (Fig. 3d). In the KI-treated mice, there was a decrease in the mRNA levels of pro-inflammatory cytokines, together with a significant up-regulation of mRNAs involved in anti-inflammatory function compared to the CA group (Fig. 3e). Studies have shown that butyrate is the main metabolite in the bacterial supernatant that causes an increase of TGF-β1.28 Metabolites associated with intestinal inflammation and IBD include tryptophan metabolites, short-chain fatty acids (SCFAs), and bile acid derivatives.29 Our study proved that tryptophan metabolites can also regulate the increase of TGF-β1, and in particular the expression of TGF-β1 in the FKI group transplanted with the feces of the tryptophan metabolite group was also increased. It can be inferred that the regulation of TGF-β1 by tryptophan metabolites plays a role through intestinal flora. Therefore, our study enriches our knowledge on the function and mechanism of these metabolites in intestinal inflammation models. Next, using immunofluorescence to detect barrier-related proteins in colonic tissues, we evaluated the effect of KI on the barrier function in C. albicans infected mice. KI caused a significant increase in the expression of occludin and ZO-1 in the intestinal epithelium compared with that in the CA group (Fig. 3f). HE staining showed that the mice in the KI group had significantly reduced histopathological damage and histological scoring in the large intestine compared to those in the CA group (Fig. 3g). Our results indicate that KI plays a role in both gut inflammation and barrier function.Effects of KynA and IA on mucosal inflammation and immunity in mice with intestinal infection with Candida albicans
To investigate the mechanisms underlying KI-mediated C. albicans infection and barrier function changes, transcriptome sequencing was performed on colonic tissues from the KI and CA groups (n = 4 per group). The results showed that KI targeted inflammatory cytokines and ILC3 cells to reduce C. albicans infection-induced gut injury. Using a cutoff of p < 0.05, we selected 2226 differentially expressed genes (Fig. 4a). Further investigation using the Gene Ontology (GO) database was performed to understand the biological functions potentially regulated by these genes. We found that these genes could be involved in antimicrobial peptide-mediated humoral immunity, adaptive immunity, negative regulation of type 2 immunity, organic acid binding and organic acid metabolism (Fig. 4b). Using Cytoscape software, the genes selected by these functional pathways were predicted to interact with each other (Fig. 4c). ELISA showed significantly increased IL-22 levels in the gut and serum of the CA group compared to those in the C group, whereas IL-22 levels were significantly decreased in the KI group (Fig. 4d). In addition, immunofluorescence showed that the intestinal lamina propria of the CA group was enriched with a higher number of RORγt+IL-22+CD3− ILC3 cells than that in the C and KI groups (Fig. 4e and f).Effects on the intestinal microbiota in mice administered KynA and IA combined with Candida albicans infection
To understand the specific mechanism of KI targeting inflammatory cytokines and ILC3, we used 2bRAD-M sequencing to clarify the effect of KI on the gut microbiota of C. albicans mice. The KI group had lower Chao 1 and Shannon indices than the CA group, although this difference was not significant (Fig. 5a and b), whereas Adonis PCoA analysis showed differentiation between the two groups (Fig. 5c). At the phylum level (Fig. 5d), the abundance of Proteobacteria in the KI group was significantly higher than that in the CA group. At the genomic level (Fig. 5e), Roseburia and Akkermansia were enriched in KI and CA groups, respectively. Two sample clusters representing the KI and CA groups were identified on a heat map based on the binary Jaccard distance (Fig. 5f). Furthermore, the relative abundance of Faecalibaculum, Aminipila, Subdoligranulum, and Ruthenibacter was lower in the KI group than in the CA group (Fig. 5g). Finally, GO pathway analysis suggested that the CA microbiota was associated with an uncharacterized protein and ABC polysaccharide transport system, a permease component, and Asp/Glu/hydantoin racemase components, whereas the KI microbiota was associated with an uncharacterized protein and ABC polysaccharide transport system and an Asp/Glu/hydantoin racemase component (Fig. 5h).Effects of KI-treated FMT on the inflammatory response and mucosal barrier function in germ-free mice with Candida albican-induced enteritis
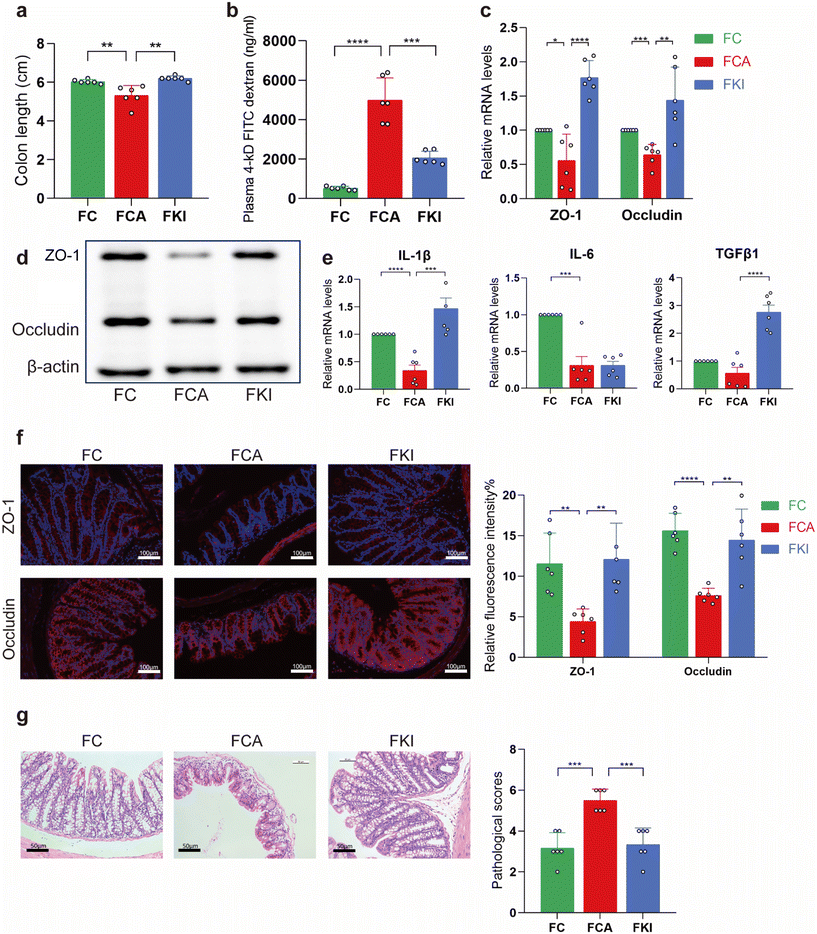
To further confirm that the effect of KI was dependent on the gut microbiota, we built an FMT model, which was used on the control, C. albicans infected, and KI-treated mice (FC, FCA, and FKI) (Fig. 7a). We demonstrated that FMT from mice infected with C. albicans affected the intestinal barrier and intestinal inflammation. Furthermore, the analysis of alpha and beta diversity showed that the gut microbial diversity of ABX-treated mice (receiver group) was significantly lower than that of the donor group mice and was able to cluster completely (Fig. 7b and c). The frequency of colonic shortening was higher in the FCA group than in the FC and FKI groups (Fig. 6a). In addition, the FKI group showed dramatically reduced intestinal permeability compared with the FCA group, as measured by plasma FD-4 levels (Fig. 6b). The expression of the barrier molecules of occludin and ZO-1 was significantly higher in the FC and FKI groups than that in the FCA group (Fig. 6c and d, f). IL-1β and IL-6 mRNA levels were decreased in the FCA group and TGFβ1 was significantly upregulated in the FKI group (Fig. 6e). Compared to the FCA group, the mice in the FKI group showed markedly reduced histopathological and histological values in the large intestine, as observed by HE staining (Fig. 6g).Effects of KI-treated FMT on mucosal immunity in germ-free mice with Candida albican-induced enteritis
Next, we investigated whether FMT from mice infected with C. albicans affected mucosal immunity. Immunofluorescence showed that the intestinal lamina propria of the FCA group was enriched with more RORγt+IL-22+CD3− ILC3 cells than those of the FC and FKI groups (Fig. 7d and e). IL-22 mRNA levels were higher in colonic tissue from the FKI group than those from the FCA group, as determined by qRT-PCR (Fig. 7f). Fecal microbial profiles confirmed significant differences between the genera present in the microbiota of all three recipient groups, with Bacteroides, Akkermansia and Klebsiella being predominant in the FC, FCA, and FKI groups, respectively (Fig. 7g).Discussion
This study found that C. albicans altered specific gut microbiota, metabolite composition, and function. Following C. albicans infection, kynurenic acid and indoleacetic acid significantly decreased inflammatory reactions, protected gut barrier function, and decreased IL-22 production from colonic lamina propria ILC3 and expression of associated inflammatory cytokines. Furthermore, FMT revealed that the inhibitory effect of KI on inflammation depended on the alteration of the gut microbiota. The changes in intestinal flora may be related to the secretion of IL-22 by ILC3 in the lamina propria, but the overall IL-22 level may not be regulated. This is the first study to show that tryptophan metabolites regulate the secretion of IL-22 by ILC3 in the lamina propria of the colon by the gut microbiota, and these findings suggest that KI may be a promising therapeutic strategy for the treatment of C. albicans infections in the gut (Fig. 7f).A key regulator of epithelial homeostasis is the cytokine interleukin-22. It is involved in many aspects of epithelial barrier function, including the regulation of epithelial cell growth and permeability, production of mucous and antimicrobials (AMP), and complement production.30 Two recent studies have suggested that the mechanism by which IL-22 promotes regeneration in vivo and organoid growth in vitro is through the stimulation of transit-amplifying cells, whereas IL-22 inhibits the expansion of ISCs by activating the Wnt and Notch pathways.31,32 In a series of elegant experiments, Gronke et al.33 showed that selective depletion of IL-22RA1 from IECs leads to the suppression of DNA damage-induced apoptosis and consequently to increased tumor formation in an inflammation-driven tumor model. Through STAT3-dependent signaling, IL-22 directly induces the expression of mucin genes in mucosal epithelial cells.34 IL-22 treatment also stimulates goblet cell hyperplasia, a response to worm infection, which is why IL-22 null animals show less goblet cell hyperplasia.35 Stimulation of IL-22 leads to the up-regulation of epithelial claudin-2, a key regulatory subunit of epithelial tight junctions, and may thereby induce water efflux, diarrhea, and the clearance of pathogens in the gut.36 IL-22 also functions in the clearance of pathogens that have already managed to penetrate the barrier. A separate group described IL-22-deficient mice with less bacterial diversity and decreased abundance of Lactobacillus, but increased abundance of Escherichia, Salmonella, and Helicobacter, which showed increased susceptibility to dextran sulfate sodium (DSS) colitis in cohoused wild-type animals, indicating that IL-22-deficient animals can harbor microbiota with transferable effects on barrier function.37 IL-22 and IDO1 are crucial in balancing resistance with tolerance to Candida, their deficiencies are risk factors for RVVC, and targeting tolerance via therapeutic kynurenines may benefit patients with RVVC.38 Studies have shown that Minapuram, a metabolite of intestinal flora, enhances the tolerance of intestinal I/R injury by regulating AHR/ILC3/IL-22 signaling, providing a new therapeutic strategy for intestinal I/R injury and enteric-borne sepsis.39 IL-22 is produced by innate lymphoid cells (ILCs) and CD4+ T cells and plays an important role in host defense and mucosal homeostasis. IL-21 and AhR signaling controls IL-22 production in T cells and the development of DSS induced colitis in ILC-deficient mice.40 Tryptophan metabolites are known to be ligands of AHR, so our study further demonstrates the certainty of its mechanism of action.
Intestinal infection with C. albicans is associated with changes in several genes involved in immune system processes, innate immune responses, and inflammatory responses. Unexpectedly, we found that C. albicans increased the expression of IL-22 and the number of ILC-3 cells in the intestinal mucosa. This is consistent with the finding that IL-22 is strongly upregulated in the inflamed colonic mucosal tissue and serum of patients.41 This process may be linked to chemotactic cytokines, which can stimulate leukocytes to migrate to areas of inflammation and make them activate the immune cells.42 In this study, we found that KI reduced IL-22 expression and ILC3 cell numbers in the inflamed colon epithelium after C. albicans infection, and FMT indicated that this effect was associated with KI-altered gut microbiota. Recently, it was been shown that ILC3 is involved in the maintenance of GI mucosal homeostasis and the progression of IBD.43 Previous mechanistic studies have shown that ILC3 cells promote IL-22 production to exacerbate IBD development by damaging the intestinal mucosal barrier, directly supporting our array results showing reduced levels in KI-treated mouse gut tissue.44 In addition, the composition of the microbiota in infectious or inflammatory diseases is a critical factor affecting ILC3 cells, according to a newly published study.45 Thus, we hypothesized that the anti-inflammatory effect of KI may be mediated, in part, by a reduction in ILC3 cell production. Future studies are needed to clarify which KI-associated bacterial species are predominantly responsible for the production of ILC3 in the inflammatory microenvironment.
This study had several limitations. On the one hand, no significant decrease in the number of C. albicans was detected in the 2bRAD-M sequencing after KI treatment, which suggests that KI may not have a specific anti-C. albicans activity but may act as an anti-inflammatory and protective intestinal barrier. On the other hand, based on more accurate detection techniques, such as metagenomics, future work is still needed to confirm the fungal diversity of KI. Second, we did not confirm the function of IL-22 after its knockout in mice by rescue experiments. To determine whether IL-22−/− mice can restore gut barrier function and anti-inflammatory effects, we plan to perform further experiments. Third, the host–microbe interplay might be of interest for understanding the impact of their interplay with epithelial cells on gut homeostasis and gut disease. Finally, the actual role of KI in humans, particularly in those at risk of C. albicans infection, remains unknown, and our study was based entirely on mouse models. Well-designed clinical trials with ethical approval are required to address this limitation. For example, tryptophan metabolites with appropriate concentrations were added to the oral medication of patients with Candida albicans infection, and the load of Candida albicans in patients’ blood and faeces, as well as the intestinal barrier and inflammation were observed to verify the clinical efficacy and significance of tryptophan metabolites in patients with Candida albicans infection.
Materials and methods
Animals and C. albicans culture
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of East China Normal University and approved by the Animal Ethics Committee of East China Normal University (Approval number: R20211202). Male C57BL/6 mice were acquired at about 8 weeks of age. The animals were housed at the East China Normal University Animal Center (Shanghai, China). The mice were given unlimited food and given a week to adjust to their surroundings (24 ± 2 °C, 60 ± 5% relative humidity, and a 12 h day−1 light/dark cycle). C. albicans (strain SC5314) (CGMCC) was purchased from the China General Microbiological Culture Collection Center. Then, it was cultivated in a liquid medium composed of yeast extract, peptone, and dextrose (YEPD). A loop was injected and streaked on a chromogenic medium to detect C. albicans (CHROMagar Company, France). Thereafter, a single colony was plated on YEPD agar, incubated for 25 h at 35 °C, and then its mass spectrometric identity was determined (Shanghai Fifth People's Hospital, Laboratory Department). A prepared inoculum of 1.0 × 106C. albicans cells in 0.3 ml of phosphate-buffered saline (PBS, pH 7.4) (Servicebio, Wuhan, China) was used.46Mouse model of DSS induces intestinal C. albicans infection
Mouse models of C. albicans infection of the gut have been established. Three groups were created: a normal control group (C), a C. albicans infection group (CA), and a KynA/IA (Sigma-Aldrich, USA) treatment group (KI). Group C received 0.2 ml of PBS daily. Dextran sulfate (DSS; 2% w/v, 40 kDa, Sigma-Aldrich, USA) was added to the drinking water of the CA and KI groups to induce colonic injury.46 To promote C. albicans colonization in the CA and KI groups, PBS yeast suspension (5 × 107 CFU kg−1) was administered orally daily. In the KI group, a mixed solution of KynA (2.5 mg kg−1) and IA (20 mg kg−1) was injected intraperitoneally daily (Fig. 7a). Feces were collected from all mice and stored at −80 °C after 14 days of treatment. The mice were then sacrificed by cervical dislocation and their colons were collected for blood sampling by retro-orbital puncture.FMT experiments
Recipient mice (C57BL/6J male mice between the ages of 6–8 weeks) were fed in a sterile environment and given antibiotics (ABX) (vancomycin 100 mg kg−1, neomycin sulfate 200 mg kg−1, metronidazole 200 mg kg−1, and ampicillin 200 mg kg−1) once a day for one week. In the second week, both donor (C, CA, and KI groups) and recipient mice (FC, FCA, and FKI groups) began to exhibit enteric-borne Candida albicans infection models, which lasted for 2 weeks. Starting from the third week, feces of donor mice were collected every day, and resuspended in PBS at 0.125 g ml−1. An amount of 0.1 ml of the solution was administered to recipient mice intragastrically once each day for 1 week.47 All the mice had unrestricted access to food and drinks while being gavaged in a sterile setting (Fig. 7a).2bRAD-M sequencing
With a few minor alterations, the initial procedure developed by Wang et al.48 for the creation of the 2bRAD-M library was followed in theory. 4 U BcgI enzyme (NEB) was used to digest DNA (1 pg–200 ng) for 3 h at 37 °C. The adaptors were then ligated to the DNA fragments. Five liters of digested DNA were combined with ten liters of a ligation master mix that contained 800 U of T4 DNA ligase and 0.2 M of each of the two adaptors (NEB). The ligation product was amplified and the PCR results were run on an 8% polyacrylamide gel. DNA was released from the gel over 12 hours in nuclease-free water at 4 °C. The 100 bp bands were removed from the polyacrylamide gel. To add sample-specific barcodes, PCR was performed in conjunction with platform-specific primers. Each 20 l PCR comprised 25 ng of gel-extracted PCR product, 0.2 M of each primer, 0.3 M dNTP, 1 U of Phusion High-Fidelity DNA Polymerase, and 1 U of Phusion HF buffer (NEB). A QIAquick PCR Purification Kit (Qiagen) was used to purify the PCR products before sequencing on an Illumina Nova PE150 platform.Initially, 173![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 165 microbial genomes were downloaded from the NCBI RefSeq database (including bacteria, fungi, and archaea). Next, using built-in Perl scripts to sample restriction fragments from microbial genomes with each of the 16 type 2B restriction enzymes, a sizable 2bRAD microbial genome database was produced. Each genome collection of 2bRAD tags was assigned a GCF number and taxonomy description that corresponded to the complete genome. Finally, the 2bRAD tags from every GCF were compared with those from every other GCF. These 2bRAD tags were created as species-specific 2bRAD markers and collectively constitute a database of 2bRAD markers.49
165 microbial genomes were downloaded from the NCBI RefSeq database (including bacteria, fungi, and archaea). Next, using built-in Perl scripts to sample restriction fragments from microbial genomes with each of the 16 type 2B restriction enzymes, a sizable 2bRAD microbial genome database was produced. Each genome collection of 2bRAD tags was assigned a GCF number and taxonomy description that corresponded to the complete genome. Finally, the 2bRAD tags from every GCF were compared with those from every other GCF. These 2bRAD tags were created as species-specific 2bRAD markers and collectively constitute a database of 2bRAD markers.49
Fecal metabolome analysis
In brief, 500 μl of solvent and 60 mg of feces were combined, followed by crushing, vortexing, and centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C. For LC-MS analysis, the supernatants were then filtered through a 0.22 μm filter and kept at −80 °C. A quality control (QC) group was created by combining equal quantities of the supernatant from each sample to determine whether the mass spectrometric platform of the system remained stable throughout the experiment. Using an AB TripleTOF 6600 mass spectroscopy system (AB-Siex), metabolic profiles were examined using a mix of ESI sources in both positive and negative ion scanning modes. High-performance liquid chromatography (HPLC)–grade reagents were utilized throughout the experiments. Metabolites were identified using Progenesis QI data processing software after LC-MS data from fecal pellets were processed using Progenesis QI software (Waters Corporation, Milford, USA). Using the ropls package in R, Principal Component Analysis (PCA) and Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) were performed to visualize the normalized data. Ellipses in PCA and OPLS-DA plots were utilized with a 95% confidence threshold to characterize metabolic perturbations between groups in a Hotelling's T2 area. The OPLS-DA model was used to determine the variable importance in projection (VIP), which was used to identify significant metabolites with VIP > 1.0 and P-value = 0.05.50
000 rpm for 15 min at 4 °C. For LC-MS analysis, the supernatants were then filtered through a 0.22 μm filter and kept at −80 °C. A quality control (QC) group was created by combining equal quantities of the supernatant from each sample to determine whether the mass spectrometric platform of the system remained stable throughout the experiment. Using an AB TripleTOF 6600 mass spectroscopy system (AB-Siex), metabolic profiles were examined using a mix of ESI sources in both positive and negative ion scanning modes. High-performance liquid chromatography (HPLC)–grade reagents were utilized throughout the experiments. Metabolites were identified using Progenesis QI data processing software after LC-MS data from fecal pellets were processed using Progenesis QI software (Waters Corporation, Milford, USA). Using the ropls package in R, Principal Component Analysis (PCA) and Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) were performed to visualize the normalized data. Ellipses in PCA and OPLS-DA plots were utilized with a 95% confidence threshold to characterize metabolic perturbations between groups in a Hotelling's T2 area. The OPLS-DA model was used to determine the variable importance in projection (VIP), which was used to identify significant metabolites with VIP > 1.0 and P-value = 0.05.50
Transcriptomics
RNA sequences and differentially expressed genes were analysed. Libraries were sequenced to produce 150 bp paired-end reads using the Illumina Novaseq 6000 platform. Approximately 50 million raw readings were obtained for each sample. Fastp51 was first used to handle fastq format raw readings. Low-quality reads were eliminated to produce a clean one. Each sample had a total of 49.2 million clean readings that were stored for further processing. HISAT2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 was used to map clean reads to the reference genome (GRCm39). Using HTSeqCount,53 the read counts of each gene and the FPKMs54 of each gene were computed. PCA analysis was carried out using R to evaluate the biological duplication of the samples (v 3.2.0).
52 was used to map clean reads to the reference genome (GRCm39). Using HTSeqCount,53 the read counts of each gene and the FPKMs54 of each gene were computed. PCA analysis was carried out using R to evaluate the biological duplication of the samples (v 3.2.0).
DESeq2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 was used to analyze differential expression. Significantly Differentially Expressed Genes (DEGs) were defined as those with a fold change >2 and a P value of 0.05. R was used to perform the hierarchical cluster analysis of DEGs to display gene expression patterns in various groups and samples (v 3.2.0). The radar map of the top 30 genes was created using the R package ggradar to display the expression of up- or downregulated DEGs.
55 was used to analyze differential expression. Significantly Differentially Expressed Genes (DEGs) were defined as those with a fold change >2 and a P value of 0.05. R was used to perform the hierarchical cluster analysis of DEGs to display gene expression patterns in various groups and samples (v 3.2.0). The radar map of the top 30 genes was created using the R package ggradar to display the expression of up- or downregulated DEGs.
GO,56 KEGG,57 Reactome, and WikiPathways enrichment analyses were carried out based on the hypergeometric distribution. We searched for important enrichment terms using R (version 3.2.0): R (v 3.2.0) for the chordal and blister plots, as well as for the important enrichment terms.
16S rRNA analysis of the microbial community of the sample
Feces were collected from the ABX and control groups of mice. For microbiome analysis, six fecal pellets were randomly selected. Briefly, a standard DNA extraction kit (QIAGEN) was used to extract DNA from feces. The quality and amount of DNA were verified by agarose gel electrophoresis. The quality of amplification of the 16S rRNA genes’ V3–V4 regions was examined. The V3–V4 gene amplicons were sequenced using Illumina MiSeq technology. Filtering and elimination of clean tags from the raw data produced valid tags that were used to create OTUs, which were then categorized using Vsearch software (version 2.4.2) with a threshold of 97% sequence similarity.58 Afterward, the past was used to compare the OTUs (v0.1). This software was used for the phylogenetic analysis. The diversity and makeup of the gut microbiota were identified using an enriched OTU table. The alpha diversity indices of fecal samples were obtained from a uniform depth normalized OTU table. Beta diversity indices were generated using the Bray–Curtis algorithm and unweighted UniFrac distance to determine whether significant gut microbiota differences existed between groups, and were also determined by Principal Component Analysis (PCA).46Western blot analysis
Western blot analysis was performed using standard methods to analyze protein expression in colon tissue. The antibodies used were anti-occludin (Proteintech, 13409-1-AP, Wuhan, China), anti-ZO-1 (Affinity, AF5145, USA), and anti-β-actin (Cell Signaling Technology, CST-3077, USA). The secondary antibody anti-rabbit IgG (111-035-003) was purchased from Jackson ImmunoResearch. Protein bands were visualized using an ECL chemiluminescence imaging system. The ImageJ software was used for quantification, which calculates the ratios of IntDen (target protein)/IntDen (β-actin).Measurement of FITC–dextran leakage
Gut permeability was assessed by measuring FITC–dextran leakage. After approximately 8 h of overnight starvation, the mice were administered 25 mg ml−1 FITC-Dixon (4 kDa, Sigma-Aldrich, USA) dissolved in PBS. FITC–dextran at 0.6 mg FITC–dextran per gram of body weight was administered via gavage. After 4 h, 400 ml of blood was collected through a retro-orbital puncture and centrifuged with PBS. A multimode reader (excitation: 485 nm, emission: 528 nm, and bandwidth: 20 nm) was used to detect the fluorescence intensity of the diluted serum (100 ml) from each sample. A standard curve was used to calculate the FITC concentration.59qRT-PCR
A reverse transcription enzyme was used by the manufacturer's instructions to create cDNA (RR036A, Takara Bio Inc., Takara, Japan). The PCR was carried out using an ABI Q6 PCR machine and SYBR Green detection technique (RK21203, AB-Clonal, China). Target gene expression in mice was normalized to that of the housekeeping gene 18S using the 2-CT technique. The target gene primers are listed in Table 1.| Genotyping | Sequence 5′-3′ | |
|---|---|---|
| ZO-1 | Forward primer | CCAGCAACTTTCAGACCACC |
| Reverse primer | TTGTGTACGGCTTTGGTGTG | |
| Occludin | Forward primer | TAAGAGCTTACAGGCAGAACTAG |
| Reverse primer | CTGTCATAATCTCCCACCATC | |
| IL-1β | Forward primer | AAGGTCCACGGGAAAGACAC |
| Reverse primer | AGCTTCAGGCAGGCAGTATC | |
| IL-6 | Forward primer | GAGGATACCACTCCCAACAGACC |
| Reverse primer | AAGTGCATCATCGTTGTTCATACA | |
| TGFβ1 | Forward primer | TGATACGCCTGAGTGGCTGTCT |
| Reverse primer | CACAAGAGCAGTGAGCGCTGAA | |
| IL-10 | Forward primer | CGGGAAGACAATAACTGCACCC |
| Reverse primer | CGGTTAGCAGTATGTTGTCCAGC | |
| IL-22 | Forward primer | CATGCAGGAGGTGGTACCTT |
| Reverse primer | CAGACGCAAGCATTTCTCAG | |
Enzyme-linked immunosorbent assays
The mice's blood was drawn, and the serum was extracted by centrifugation at 3000 rpm for 10 min at 4 °C. The colonic tissues were ground nine times in a homogenization medium. Centrifugation was performed to separate the supernatants for 10 min at 4 °C and 3000 rpm. All serum and tissue supernatants were stored at −80 °C for subsequent simultaneous detection. Following the manufacturer's instructions, IL-22 levels were measured using a mouse ELISA kit (A106907-96T, Fusheng, Shanghai, China).Histomorphological analysis
Colon specimens were embedded in paraffin, dried, and preserved in a 4% neutral paraformaldehyde solution before being sectioned at a thickness of 5 μm. Hematoxylin and eosin (H&E) staining was used for observation. Two skilled pathologists, who were blinded simultaneously, rated the histological alterations according to a grading system47 and averaged the results.Immunofluorescence
Paraffin-embedded samples were blocked for 1 h and incubated overnight at 4 °C with anti-ZO-1 antibodies (Servicebio, China) and anti-occludin antibodies (Servicebio, China). Images were captured using fluorescence microscopy (Olympus, Japan) after the colon tissues were washed and stained with DAPI for 10 min. Six randomly selected ×200 fields of each sample were used to quantify the fluorescence intensity of ZO-1 and occludin staining of small intestinal tissues by automated image analysis.60 The samples of paraffin sections were blocked for 1 h and incubated with an anti-CD3 antibody (Servicebio, Wuhan, China), anti-RORγt antibody (Bioss, Beijing, China), and anti-IL-22 antibody (Servicebio, Wuhan, China) at 4 °C overnight.47 Images were captured using fluorescence microscopy (Olympus, Japan) after the colon tissues were washed and stained with DAPI for 10 min. Six randomly selected ×400 fields of each sample were quantified using automated image analysis.Statistical analysis
All analyses were conducted using GraphPad Prism 8 software, except for microbial data, which were subjected to multivariable and advanced statistical analysis as previously mentioned. All data were calculated from at least three repetitions and are presented as mean ± SEM. Multigroup comparisons were performed by one-way analysis of variance (ANOVA) followed by post-hoc Duncan's multiple-comparison test. Wilcoxon test was used to analyze the difference between two unpaired groups. Statistical significance has been established at p < 0.05.Author contributions
Conceptualization: Ziyao Peng and Zetian Wang. Investigation: Ziyao Peng and Meng Zhang. Formal Analysis: Jiali Zhang. Data curation: Liping Yin and Ziyang Zhou. Resources: Cuiting Lv. Supervision: Jianguo Tang. Writing – original draft: Ziyao Peng. Writing – review & editing: Jianguo Tang. All authors approved the final version of the manuscript.Abbreviations
| ABX | Antibiotic |
| ANOVA | Analysis of variance |
| C | Control |
| CA | Candida albicans |
| FD-4 | FITC-dextran 4 |
| FMT | Fecal microbiota transplantation |
| FC | FMT-control |
| FCA | FMT-C. albicans infection |
| FKI | FMT-kynurenic acid and indoleacrylic acid treatment |
| GF | Germ-free |
| HE | Hematoxylin–eosin |
| ILC3 | Group 3 innate lymphoid cell |
| KI | Kynurenic acid and indoleacrylic acid |
| LEfSe | Linear discriminant effect size |
| PBS | Phosphate-buffered saline |
| PCoA | Principal coordinates analysis |
| PCR | Polymerase chain reaction |
Ethics approval and consent to participate
All animal experiments were approved by the Experimental Animal Ethical Review Committee, East China Normal University (Shanghai, China).Data availability
The raw sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA957306) under accession numbers PRJNA957306 and PRJNA778766. All other data associated with this study are presented in the paper or ESI.†Conflicts of interest
All authors declare that they have no competing interests.Acknowledgements
We would like to thank the Central Laboratory, Fifth People's Hospital of Shanghai, and Fudan University, for their support and assistance. This study was supported by the Shanghai Science and Technology Commission (Grant Number 22ZR1448800).References
- A. A. Mohamed, X.-l. Lu and F. A. Mounmin, Diagnosis and Treatment of Esophageal Candidiasis: Current Updates, Can. J. Gastroenterol. Hepatol., 2019, 2019, 1–6 CrossRef PubMed.
- L. Yan, C. Yang and J. Tang, Disruption of the intestinal mucosal barrier in Candida albicans infections, Microbiol. Res., 2013, 168, 389–395 CrossRef CAS PubMed.
- L. Basmaciyan, F. Bon, T. Paradis, P. Lapaquette and F. Dalle, Candida Albicans Interactions With The Host: Crossing The Intestinal Epithelial Barrier, Tissue Barriers, 2019, 7, 1612661 CrossRef PubMed.
- C. A. Kumamoto, M. S. Gresnigt and B. Hube, The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine, Curr. Opin. Microbiol., 2020, 56, 7–15 CrossRef CAS PubMed.
- J. Kinchen, H. H. Chen, K. Parikh, A. Antanaviciute, M. Jagielowicz, D. Fawkner-Corbett, N. Ashley, L. Cubitt, E. Mellado-Gomez, M. Attar, E. Sharma, Q. Wills, R. Bowden, F. C. Richter, D. Ahern, K. D. Puri, J. Henault, F. Gervais, H. Koohy and A. Simmons, Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease, Cell, 2018, 175, 372–386 CrossRef CAS PubMed.
- J. H. Hageman, M. C. Heinz, K. Kretzschmar, J. van der Vaart, H. Clevers and H. J. G. Snippert, Intestinal Regeneration: Regulation by the Microenvironment, Dev. Cell, 2020, 54, 435–446 CrossRef CAS PubMed.
- Y. Fang, C. Wu, Q. Wang and J. Tang, Farnesol contributes to intestinal epithelial barrier function by enhancing tight junctions via the JAK/STAT3 signaling pathway in differentiated Caco-2 cells, J. Bioenerg. Biomembr., 2019, 51, 403–412 CrossRef PubMed.
- N. O. Ponde, L. Lortal, G. Ramage, J. R. Naglik and J. P. Richardson, Candida albicans biofilms and polymicrobial interactions, Crit. Rev. Microbiol., 2021, 47, 91–111 CrossRef CAS PubMed.
- P. E. Sudbery, Growth of Candida albicans hyphae, Nat. Rev. Microbiol., 2011, 9, 737–748 CrossRef CAS PubMed.
- P. G. Pappas, M. S. Lionakis, M. C. Arendrup, L. Ostrosky-Zeichner and B. J. Kullberg, Invasive candidiasis, Nat. Rev. Dis. Primers, 2018, 4, 18026 CrossRef PubMed.
- M. Valentine, E. Benade, M. Mouton, W. Khan and A. Botha, Binary interactions between the yeast Candida albicans and two gut-associated Bacteroides species, Microb. Pathog., 2019, 135, 103619 CrossRef CAS PubMed.
- P. Hiengrach, W. Panpetch, N. Worasilchai, A. Chindamporn, S. Tumwasorn, T. Jaroonwitchawan, A. Wilantho, P. Chatthanathon, N. Somboonna and A. Leelahavanichkul, Administration of Candida Albicans to Dextran Sulfate Solution Treated Mice Causes Intestinal Dysbiosis, Emergence and Dissemination of Intestinal Pseudomonas Aeruginosa and Lethal Sepsis, Shock, 2020, 53, 189–198 CrossRef CAS PubMed.
- Z. Peng and J. Tang, Intestinal Infection of Candida albicans: Preventing the Formation of Biofilm by C. albicans and Protecting the Intestinal Epithelial Barrier, Front. Microbiol., 2021, 12, 783010 CrossRef PubMed.
- L. Romani, T. Zelante, M. Palmieri, V. Napolioni, M. Picciolini, A. Velardi, F. Aversa and P. Puccetti, The cross-talk between opportunistic fungi and the mammalian host via microbiota's metabolism, Semin. Immunopathol., 2015, 37, 163–171 CrossRef CAS PubMed.
- A. Bessede, M. Gargaro, M. T. Pallotta, D. Matino, G. Servillo, C. Brunacci, S. Bicciato, E. M. Mazza, A. Macchiarulo, C. Vacca, R. Iannitti, L. Tissi, C. Volpi, M. L. Belladonna, C. Orabona, R. Bianchi, T. V. Lanz, M. Platten, M. A. Della Fazia, D. Piobbico, T. Zelante, H. Funakoshi, T. Nakamura, D. Gilot, M. S. Denison, G. J. Guillemin, J. B. DuHadaway, G. C. Prendergast, R. Metz, M. Geffard, L. Boon, M. Pirro, A. Iorio, B. Veyret, L. Romani, U. Grohmann, F. Fallarino and P. Puccetti, Aryl hydrocarbon receptor control of a disease tolerance defence pathway, Nature, 2014, 511, 184–190 CrossRef CAS PubMed.
- T. Zelante, R. G. Iannitti, C. Cunha, A. De Luca, G. Giovannini, G. Pieraccini, R. Zecchi, C. D'Angelo, C. Massi-Benedetti, F. Fallarino, A. Carvalho, P. Puccetti and L. Romani, Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22, Immunity, 2013, 39, 372–385 CrossRef CAS PubMed.
- M. Modoux, N. Rolhion, S. Mani and H. Sokol, Tryptophan Metabolism as a Pharmacological Target, Trends Pharmacol. Sci., 2021, 42, 60–73 CrossRef CAS PubMed.
- C. Xue, G. Li, Q. Zheng, X. Gu, Q. Shi, Y. Su, Q. Chu, X. Yuan, Z. Bao, J. Lu and L. Li, Tryptophan metabolism in health and disease, Cell Metab., 2023, 35, 1304–1326 CrossRef CAS PubMed.
- T. Hashimoto, T. Perlot, A. Rehman, J. Trichereau, H. Ishiguro, M. Paolino, V. Sigl, T. Hanada, R. Hanada, S. Lipinski, B. Wild, S. M. Camargo, D. Singer, A. Richter, K. Kuba, A. Fukamizu, S. Schreiber, H. Clevers, F. Verrey, P. Rosenstiel and J. M. Penninger, ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation, Nature, 2012, 487, 477–481 CrossRef CAS PubMed.
- H. Lemos, L. Huang, G. C. Prendergast and A. L. Mellor, Immune control by amino acid catabolism during tumorigenesis and therapy, Nat. Rev. Cancer, 2019, 19, 162–175 CrossRef CAS PubMed.
- Y. Liu, X. Liang, W. Dong, Y. Fang, J. Lv, T. Zhang, R. Fiskesund, J. Xie, J. Liu, X. Yin, X. Jin, D. Chen, K. Tang, J. Ma, H. Zhang, J. Yu, J. Yan, H. Liang, S. Mo, F. Cheng, Y. Zhou, H. Zhang, J. Wang, J. Li, Y. Chen, B. Cui, Z. W. Hu, X. Cao, F. Xiao-Feng Qin and B. Huang, Tumor-Repopulating Cells Induce PD-1 Expression in CD8(+) T Cells by Transferring Kynurenine and AhR Activation, Cancer Cell, 2018, 33, 480–494 CrossRef CAS PubMed.
- M. Wlodarska, C. Luo, R. Kolde, E. d'Hennezel, J. W. Annand, C. E. Heim, P. Krastel, E. K. Schmitt, A. S. Omar, E. A. Creasey, A. L. Garner, S. Mohammadi, D. J. O'Connell, S. Abubucker, T. D. Arthur, E. A. Franzosa, C. Huttenhower, L. O. Murphy, H. J. Haiser, H. Vlamakis, J. A. Porter and R. J. Xavier, Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation, Cell Host Microbe, 2017, 22, 25–37 CrossRef CAS PubMed.
- J. A. Dudakov, A. M. Hanash and M. R. M. van den Brink, Interleukin-22: Immunobiology and Pathology, Annu. Rev. Immunol., 2015, 33, 747–785 CrossRef CAS PubMed.
- Q. Zhang, Y. Pan, R. Yan, B. Zeng, H. Wang, X. Zhang, W. Li, H. Wei and Z. Liu, Commensal bacteria direct selective cargo sorting to promote symbiosis, Nat. Immunol., 2015, 16, 918–926 CrossRef CAS PubMed.
- T. Ito, K. Hirose and H. Nakajima, Bidirectional roles of IL-22 in the pathogenesis of allergic airway inflammation, Allergol. Int., 2019, 68, 4–8 CrossRef CAS PubMed.
- D. M. Ponce, A. M. Alousi, R. Nakamura, J. Slingerland, M. Calafiore, K. S. Sandhu, J. N. Barker, S. Devlin, J. Shia, S. Giralt, M.-A. Perales, G. Moore, S. Fatmi, C. Soto, A. Gomes, P. Giardina, L. Marcello, X. Yan, T. Tang, K. Dreyer, J. Chen, W. L. Daley, J. U. Peled, M. R. M. van den Brink and A. M. Hanash, A phase 2 study of interleukin-22 and systemic corticosteroids as initial treatment for acute GVHD of the lower GI tract, Blood, 2023, 141, 1389–1401 CrossRef CAS PubMed.
- P. D. Cani, C. Depommier, M. Derrien, A. Everard and W. M. de Vos, Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms, Nat. Rev. Gastroenterol. Hepatol., 2022, 19, 625–637 CrossRef PubMed.
- C. Martin-Gallausiaux, F. Beguet-Crespel, L. Marinelli, A. Jamet, F. Ledue, H. M. Blottiere and N. Lapaque, Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells, Sci. Rep., 2018, 8, 9742 CrossRef PubMed.
- A. Lavelle and H. Sokol, Gut microbiota-derived metabolites as key actors in inflammatory bowel disease, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 223–237 CrossRef PubMed.
- M. Keir, T. Yi, T. Lu and N. Ghilardi, The role of IL-22 in intestinal health and disease, J. Exp. Med., 2020, 217, e20192195 CrossRef PubMed.
- J.-M. Zha, H.-S. Li, Q. Lin, W.-T. Kuo, Z.-H. Jiang, P.-Y. Tsai, N. Ding, J. Wu, S.-F. Xu, Y.-T. Wang, J. Pan, X.-M. Zhou, K. Chen, M. Tao, M. A. Odenwald, A. Tamura, S. Tsukita, J. R. Turner and W.-Q. He, Interleukin 22 Expands Transit-Amplifying Cells While Depleting Lgr5+ Stem Cells via Inhibition of Wnt and Notch Signaling, Cell. Mol. Gastroenterol. Hepatol., 2019, 7, 255–274 CrossRef PubMed.
- B. Zwarycz, A. D. Gracz, K. R. Rivera, I. A. Williamson, L. A. Samsa, J. Starmer, M. A. Daniele, L. Salter-Cid, Q. Zhao and S. T. Magness, IL22 Inhibits Epithelial Stem Cell Expansion in an Ileal Organoid Model, Cell. Mol. Gastroenterol. Hepatol., 2019, 7, 1–17 CrossRef PubMed.
- K. Gronke, P. P. Hernández, J. Zimmermann, C. S. N. Klose, M. Kofoed-Branzk, F. Guendel, M. Witkowski, C. Tizian, L. Amann, F. Schumacher, H. Glatt, A. Triantafyllopoulou and A. Diefenbach, Interleukin-22 protects intestinal stem cells against genotoxic stress, Nature, 2019, 566, 249–253 CrossRef CAS PubMed.
- K. Sugimoto, A. Ogawa, E. Mizoguchi, Y. Shimomura, A. Andoh, A. K. Bhan, R. S. Blumberg, R. J. Xavier and A. Mizoguchi, IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis, J. Clin. Invest., 2008, 118, 534–544 CAS.
- J.-E. Turner, B. Stockinger and H. Helmby, IL-22 Mediates Goblet Cell Hyperplasia and Worm Expulsion in Intestinal Helminth Infection, PLoS Pathog., 2013, 9, e1003698 CrossRef PubMed.
- P.-Y. Tsai, B. Zhang, W.-Q. He, J.-M. Zha, M. A. Odenwald, G. Singh, A. Tamura, L. Shen, A. Sailer, S. Yeruva, W.-T. Kuo, Y.-X. Fu, S. Tsukita and J. R. Turner, IL-22 Upregulates Epithelial Claudin-2 to Drive Diarrhea and Enteric Pathogen Clearance, Cell Host Microbe, 2017, 21, 671–681 CrossRef CAS PubMed.
- L. A. Zenewicz, X. Yin, G. Wang, E. Elinav, L. Hao, L. Zhao and R. A. Flavell, IL-22 Deficiency Alters Colonic Microbiota To Be Transmissible and Colitogenic, J. Immunol., 2013, 190, 5306–5312 CrossRef CAS PubMed.
- A. De Luca, A. Carvalho, C. Cunha, R. G. Iannitti, L. Pitzurra, G. Giovannini, A. Mencacci, L. Bartolommei, S. Moretti, C. Massi-Benedetti, D. Fuchs, F. De Bernardis, P. Puccetti and L. Romani, IL-22 and IDO1 Affect Immunity and Tolerance to Murine and Human Vaginal Candidiasis, PLoS Pathog., 2013, 9, e1003486 CrossRef CAS PubMed.
- F. Deng, J. J. Hu, Z. B. Lin, Q. S. Sun, Y. Min, B. C. Zhao, Z. B. Huang, W. J. Zhang, W. K. Huang, W. F. Liu, C. Li and K. X. Liu, Gut microbe-derived milnacipran enhances tolerance to gut ischemia/reperfusion injury, Cell Rep. Med., 2023, 4, 100979 CrossRef CAS PubMed.
- A. Yeste, I. D. Mascanfroni, M. Nadeau, E. J. Burns, A. M. Tukpah, A. Santiago, C. Wu, B. Patel, D. Kumar and F. J. Quintana, IL-21 induces IL-22 production in CD4+ T cells, Nat. Commun., 2014, 5, 3753 CrossRef CAS PubMed.
- A. Andoh, Z. Zhang, O. Inatomi, S. Fujino, Y. Deguchi, Y. Araki, T. Tsujikawa, K. Kitoh, S. Kim-Mitsuyama, A. Takayanagi, N. Shimizu and Y. Fujiyama, Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts, Gastroenterology, 2005, 129, 969–984 CrossRef CAS PubMed.
- S. Danese, F. Furfaro and S. Vetrano, Targeting S1P in Inflammatory Bowel Disease: New Avenues for Modulating Intestinal Leukocyte Migration, J. Crohns Colitis., 2018, 12, S678–S686 CrossRef PubMed.
- B. Zeng, S. Shi, G. Ashworth, C. Dong, J. Liu and F. Xing, ILC3 function as a double-edged sword in inflammatory bowel diseases, Cell Death Dis., 2019, 10, 315 CrossRef PubMed.
- P. Goncalves and J. P. Di Santo, An Intestinal Inflammasome - The ILC3-Cytokine Tango, Trends Mol. Med., 2016, 22, 269–271 CrossRef CAS PubMed.
- F. M. Lehmann, N. von Burg, R. Ivanek, C. Teufel, E. Horvath, A. Peter, G. Turchinovich, D. Staehli, T. Eichlisberger, M. Gomez de Aguero, M. Coto-Llerena, M. Prchal-Murphy, V. Sexl, M. Bentires-Alj, C. Mueller and D. Finke, Microbiota-induced tissue signals regulate ILC3-mediated antigen presentation, Nat. Commun., 2020, 11, 1794 CrossRef CAS PubMed.
- W. Hu, L. Huang, Z. Zhou, L. Yin and J. Tang, Diallyl Disulfide (DADS) Ameliorates Intestinal Candida albicans Infection by Modulating the Gut microbiota and Metabolites and Providing Intestinal Protection in Mice, Front. Cell. Infect. Microbiol., 2021, 11, 743454 CrossRef CAS PubMed.
- C. Kong, X. Yan, Y. Liu, L. Huang, Y. Zhu, J. He, R. Gao, M. F. Kalady, A. Goel, H. Qin and Y. Ma, Ketogenic diet alleviates colitis by reduction of colonic group 3 innate lymphoid cells through altering gut microbiome, Signal Transduction Targeted Ther., 2021, 6, 154 CrossRef CAS PubMed.
- S. Wang, E. Meyer, J. K. McKay and M. V. Matz, 2b-RAD: a simple and flexible method for genome-wide genotyping, Nat. Methods, 2012, 9, 808–810 CrossRef CAS PubMed.
- Z. Sun, S. Huang, P. Zhu, L. Tzehau, H. Zhao, J. Lv, R. Zhang, L. Zhou, Q. Niu, X. Wang, M. Zhang, G. Jing, Z. Bao, J. Liu, S. Wang and J. Xu, Species-resolved sequencing of low-biomass or degraded microbiomes using 2bRAD-M, Genome Biol., 2022, 23, 36 CrossRef CAS PubMed.
- Z. Liu, F. Liu, G. Li, X. Chi, Y. Wang, H. Wang, L. Ma, K. Han, G. Zhao, X. Guo and B. Xu, Metabolite Support of Long-Term Storage of Sperm in the Spermatheca of Honeybee (Apis mellifera) Queens, Front. Physiol., 2020, 11, 574856 CrossRef PubMed.
- S. Chen, Y. Zhou, Y. Chen and J. Gu, fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, 2018, 34, i884–i890 CrossRef PubMed.
- D. Kim, B. Langmead and S. L. Salzberg, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods, 2015, 12, 357–360 CrossRef CAS PubMed.
- S. Anders, P. T. Pyl and W. Huber, HTSeq–a Python framework to work with high-throughput sequencing data, Bioinformatics, 2015, 31, 166–169 CrossRef CAS PubMed.
- A. Roberts, C. Trapnell, J. Donaghey, J. L. Rinn and L. Pachter, Improving RNA-Seq expression estimates by correcting for fragment bias, Genome Biol., 2011, 12, R22 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- C. The Gene Ontology, The Gene Ontology Resource: 20 years and still GOing strong, Nucleic Acids Res., 2019, 47, D330–D338 CrossRef PubMed.
- M. Kanehisa, M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda, T. Tokimatsu and Y. Yamanishi, KEGG for linking genomes to life and the environment, Nucleic Acids Res., 2008, 36, D480–D484 CrossRef CAS PubMed.
- T. Rognes, T. Flouri, B. Nichols, C. Quince and F. Mahé, VSEARCH: a versatile open source tool for metagenomics, PeerJ, 2016, 4, e2584 CrossRef PubMed.
- D. J. Brayden, S. Maher, B. Bahar and E. Walsh, Sodium caprate-induced increases in intestinal permeability and epithelial damage are prevented by misoprostol, Eur. J. Pharm. Biopharm., 2015, 94, 194–206 CrossRef CAS PubMed.
- J. Hu, F. Deng, B. Zhao, Z. Lin, Q. Sun, X. Yang, M. Wu, S. Qiu, Y. Chen, Z. Yan, S. Luo, J. Zhao, W. Liu, C. Li and K. X. Liu, Lactobacillus murinus alleviate intestinal ischemia/reperfusion injury through promoting the release of interleukin-10 from M2 macrophages via Toll-like receptor 2 signaling, Microbiome, 2022, 10, 38 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo00432a |
| This journal is © The Royal Society of Chemistry 2024 |