Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biotransformation of camu–camu galloylated ellagitannins by Lactiplantibacillus plantarum with extracellular tannase activity†
Elena C.
Pulido-Mateos
 ab,
Jacob
Lessard-Lord
ab,
Jacob
Lessard-Lord
 a,
Yves
Desjardins
a,
Yves
Desjardins
 a and
Denis
Roy
a and
Denis
Roy
 *ab
*ab
aInstitut sur la nutrition et les aliments fonctionnels de l'Université Laval, Faculté des sciences de l'agriculture et de l'alimentation, Université Laval, Quebec, QC, Canada. E-mail: Denis.Roy@fsaa.ulaval.ca
bLaboratoire de génomique microbienne, Département des sciences des aliments, Faculté des sciences de l'agriculture et de l'alimentation, Université Laval, Quebec, QC, Canada
First published on 19th June 2024
Abstract
Some strains of Lactiplantibacillus plantarum produce specific tannases that could enable the metabolism of ellagitannins into more bioavailable phenolic metabolites, thereby promoting the health effects of these polyphenols. However, the metabolic ability of these strains remains poorly understood. In this study, we analyzed the ability of broad esterase-producing (Est_1092+) and extracellular tannase-producing (TanA+) strains to convert a wide assortment of ellagitannins from camu–camu (Myrciaria dubia) fruit. To this end, forty-three strains were screened to identify and sequence (WGS) those producing Est_1092. In addition, six previously reported TanA+ strains were included in the study. Each strain (Est_1092+ or TanA+) was inoculated into a minimal culture medium supplemented with an aqueous camu–camu extract. After fermentation, supernatants were collected for semi-quantification of ellagitannins and their metabolites by mass spectrometry. For analysis, the strains were grouped according to their enzyme type and compared with an Est_1092 and TanA-lacking strain. Out of the forty-three isolates, three showed Est_1092 activity. Of the Est_1092+ and TanA+ strains, only the latter hydrolyzed the tri-galloyl-HHDP-glucose and various isomers of HHDP-galloyl-glucose, releasing HHDP-glucose and gallic acid. TanA+ strains also transformed three isomers of di-HHDP-galloyl-glucose, liberating di-HHDP-glucose and gallic acid. Overall, TanA+ strains released 3.6–4.9 times more gallic acid than the lacking strain. In addition, those exhibiting gallate decarboxylase activity pursued gallic acid metabolism to release pyrogallol. Neither Est_1092+ nor TanA+ strains transformed ellagitannin-core structures. In summary, TanA+ L. plantarum strains have the unique ability to hydrolyze a wide range of galloylated ellagitannins, releasing phenolic metabolites with additional health benefits.
1. Introduction
Camu–camu (Myrciaria dubia) is a tropical berry from the Amazon region that has been shown to improve glucose and lipid homeostasis and prevent weight gain, fat accumulation, low-grade inflammation, endotoxemia, and hepatic steatosis.1,2 These benefits are attributed in part to the phenolic content of this fruit, including proanthocyanidins, flavonoids, ellagitannins and ellagic acid derivatives.3 However, increasing evidence suggests that the health impact of some of these (poly)phenols may be influenced by the metabolic capacity of the gut microbiota.4 This is the case for certain (poly)phenols, such as ellagitannins, which, due to their size, are not absorbed but are instead converted into more bioaccessible phenolic metabolites (e.g., urolithins) by the colonic microbiota.5 These resulting microbial phenolic metabolites have lower molecular weights, allowing them to cross membranes, reach target tissues, and exert local benefits, such as anti-neuroinflammatory activities.6 However, inter-individual variations in the diversity of bacteria and strains comprising the colonic microbiota caused by aging, lifestyle, diet, and other factors may affect the abundance of ellagitannin-converting species and, therefore, the production of these beneficial phenolic metabolites.5 Strategies to promote the metabolism of these compounds are therefore being sought but are currently confined by the limited knowledge of ellagitannin-converting species.Presumably, tannase-producing bacteria are responsible for the hydrolysis of the ester bonds of the ellagitannin-core structures (the hexahydroxydiphenoyl [HHDP] moieties esterified with glucose or galloyl molecules) and the subsequent release of ellagic acid.5 However, this cannot be generalized to all microbial tannases, as these enzymes vary widely in their substrate specificity.7 Furthermore, there is a limited understanding of the hydrolytic capacity of bacterial tannases, as the existing reports on ellagitannin biotransformation are scarce and almost exclusively restricted to fungal species.8,9 A deeper understanding of ellagitannin-transforming bacteria is crucial, as they may play a key role in driving ellagitannin metabolism toward the production of bioactive phenolic metabolites.
Among tannase-producing species, Lactiplantibacillus plantarum is a potential probiotic bacterium that attracts particular attention for its outstanding repertoire of tannases and other (poly)phenol-associated enzymes (PAZymes).4,10 Certain enzymes are present in most L. plantarum strains, such as TanB, an intracellular tannase,11 and gallate decarboxylase (GD), which converts gallic acid to pyrogallol.12 Others are strain-specific, such as Est_1092, an intracellular broad esterase, and TanA, an extracellular tannase.11,13
Specifically, the isolated form of Est_1092 has shown both feruloyl esterase and tannase activities, acting on a wide range of polyphenols.13 However, its capacity to metabolize small ellagitannins (<600 Da) that could enter the microbial cell remains unexplored. In contrast, TanA is known for its specific ability to hydrolyze gallotannins (gallate polyesters) into gallic acid, as these molecules cannot enter the microbial cell. In addition to gallotannins, L. plantarum strains with TanA activity (TanA+) may also act on galloylated ellagitannins,11 such as those found in camu–camu, as these molecules are also surrounded by ester-linked galloyl units. Although it is still questionable whether TanA+ strains can continue the transformation of the ellagitannin-core molecules, as in a previous study, these strains failed to transform punicalagin, a monomeric and non-galloylated ellagitannin from pomegranate.12 However, other ellagitannins with simpler and smaller structures, such as the HHDP-glucose (482 Da) found in camu–camu,3 may be more easily hydrolyzed by TanA+ or Est_1092-producing (Est_1092+) strains. In fact, the different chemical complexity of camu–camu ellagitannins makes this fruit an attractive model to verify and explore the metabolic capacity of L. plantarum.
The aim of this study was to determine if the L. plantarum strains with TanA or Est_1092 activity enable the metabolism of camu–camu ellagitannins. For this, we developed a camu–camu ellagitannin-rich extract and characterized its phenolic content. We screened L. plantarum strains for those producing Est_1092 and sequenced their genome. In addition, the PAZymes genomic features of these Est_1092+ strains were analyzed together with those of six previously reported TanA+ strains.12 Finally, we examined the metabolic ability of the selected Est_1092+ and TanA+ L. plantarum strains towards the wide range of ellagitannins present in camu–camu.
2. Materials and methods
2.1 Strains and growth conditions
WCFS1 and ATCC 14917 L. plantarum strains were purchased from the American Type Culture Collection (ATCC) and were used as reference strains. The L. plantarum WCFS1, which only produces TanB (TanB+), was used as a negative control of TanA and Est_1092 enzymatic activities, and the L. plantarum ATCC 14917 (TanB+ and TanA+) was used as a reference strain for TanA activity.11,13 The L. plantarum ATCC 8014 (TanB+ and Est_1092+), kindly provided by Probi AB (Lund, Sweden), was used as a reference strain for Est_1092 activity.13 In addition, six previously reported TanA+ L. plantarum strains12 (PROBI S204, PROBI S126, RKG 1-473, RKG 1-500, RKG 2-219, and RKG 2-690) were included in this study, constituting the TanA+ L. plantarum group. To form the Est_1092+ L. plantarum group, thirty-seven L. plantarum isolates provided by Probi AB and the six TanA+ strains were screened for Est_1092 activity, as explained further. The identity of the L. plantarum isolates was previously confirmed by Pulido-Mateos et al.12 A stock culture of each isolate was stored at −80 °C in the Man-Rogosa-Sharpe (MRS) medium supplemented with glycerol (20%). For each experiment, the strains were reactivated 24 h in MRS medium, and the third sub-culture was used for inoculation.The basal medium developed by Rozès and Peres14 was chosen to evaluate the metabolizing capacity of the strains in the presence of the camu–camu extract with some modifications (RP-M).12 Glucose was replaced with galactose to avoid a possible carbon catabolite repression. The medium was supplemented with 1% of DMSO to facilitate the dissolution of camu–camu (poly)phenols and 1.9% of β-glycerophosphate disodium salt hydrate to improve its buffering capacity. The pH was adjusted to 5.0 to prevent ellagitannin degradation15 and sterilized by filtration.
2.2 Selection of Est_1092+ L. plantarum strains
Forty-three L. plantarum isolates were screened for the presence of the est_1092 gene, encoding the broad esterase enzyme. For this, their chromosomal DNA was obtained as previously described.16 For the PCR assay, the primers proposed by Esteban-Torres et al.13 (5′-atgatatcaaaagaattgagtcggt and 5′ ggccatatgttcctgcaaaaagcg) targeting the est_1092 gene in L. plantarum, were used. The resulting 900-pb amplicon was visualized on 2% (w/v) agarose gels after electrophoresis.Feruloyl esterase activity was confirmed in the resulting est_1092-harboring isolates using the method proposed by Donaghy et al.17 Briefly, an agar MRS medium lacking glucose was supplemented with a filter-sterilized solution of ethyl ferulate (0.1% in ethanol). After agar solidification, tiny wells were created with the help of a sterile toothpick. Then, one colony of each L. plantarum strain grown during 48 h in MRS agar medium was transferred to the ethyl-ferulate supplemented MRS agar plates. After 72 h, the feruloyl esterase activity of Est_1092+ L. plantarum strains was evidenced by a surrounding clear halo (Fig. S1†). The strain L. plantarum ATCC 8014 was used as a positive control for Est_1092 activity, and the strain WCFS1 as a negative control.13
Est_1092+ isolates were sequenced to confirm that each represented a different strain. For this, the chromosomal DNA was extracted, as previously described, and sent for whole genome sequencing to the IBIS genomic analysis platform (Université Laval, https://www.ibis.ulaval.ca/). Sequencing was performed using the Illumina MiSeq platform, which can generate 300 bp pair-ended reads. Raw reads were assembled using Unicycler18 in the Bacterial and Viral Bioinformatics Resource Center (BV-BRC, https://www.bv-brc.org/, formerly PATRIC).19 Genome scaffolding was completed using MEDUSA scaffolder,20 with the L. plantarum WCFS1 genome as the reference. Genome functions were annotated with the RAST tool kit20 in the BV-BRC. Est_1092 producing strains genomes were compared with the pairwise genome comparison tool21 available at https://jspecies.ribohost.com/jspeciesws/.
2.3 Analysis of PAZymes genomic features in Est_1092+ and TanA+ strains
The BLAST feature and the multiple alignments tool, available in the BV-BRC, were employed to identify and compare the PAZymes genomic features in the Est_1092+ and TanA+ strains. For this, the ATCC 8014 Est_1092 amino acid sequence (GenBank accession number, ATQ33180.1) and the WCFS1 LpB and LpdC amino acid sequences (GenBank accession numbers, CCC77798.1 and CCC80016.1), encoding the gallate decarboxylase enzyme, were used as queries.13,212.4 Extraction and purification of camu–camu ellagitannins and ellagic acid derivatives
Freeze-dried camu–camu (Myrciaria dubia) raw fruit powder was kindly provided by Symrise (Diana Food Canada Inc.). The powder was extracted twice with aqueous ethanol (70% v/v) at 60 °C for 15 min. The obtained extract was filtered with Whatman® qualitative filter paper (Grade 1) and then concentrated with a rotary evaporator (Buchi, New Castle, DE) at 45 °C to remove the ethanol. The dry crude extract was obtained after freeze-drying the remaining concentrate (Fig. 1).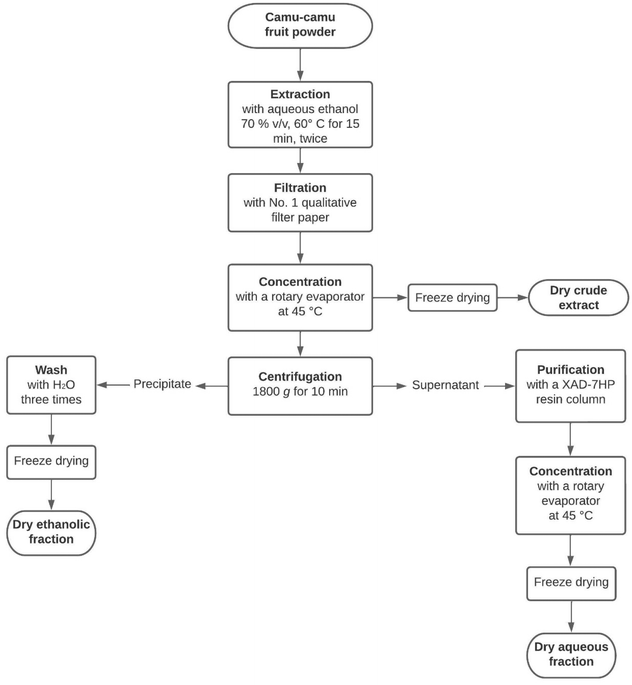 | ||
| Fig. 1 The extraction process to obtain three camu–camu products: the crude extract, the aqueous fraction and the ethanolic fraction. | ||
To purify the ellagitannins and ellagic acid derivatives, the concentrate was separated into two fractions (Fig. 1) by centrifugation at 1800g for 10 min. Then, the supernatant was transferred into a column containing XAD-7HP resin to remove sugars and small organic acids, such as vitamin C, as described by Dufour et al.22 Finally, the purified eluate was concentrated by rotary evaporation at 45 °C and freeze-dried to obtain a dried solid extract. For the ethanolic fraction, the precipitate was washed three times with water before being freeze-dried. All the dried extracts (crude extract, aqueous fraction and ethanolic fraction) were kept at −20 °C until analysis.
2.5 Characterization of the camu–camu extract by liquid chromatography coupled with an ultraviolet detector and quadrupole–time of flight mass spectrometer (UPLC-UV-QToF)
The method was adapted from Fracassetti et al.,3 Briefly, 50 mg of the dried extract was dissolved in 1 mL of methanol–water (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v) acidified with 1% (v/v) of formic acid. Then, the solution was vortexed for 2 min, sonicated for 15 minutes at 37 °C, re-vortexed for 2 min and passed through a 0.22 μm Nylon filter before analysis.
50 v/v) acidified with 1% (v/v) of formic acid. Then, the solution was vortexed for 2 min, sonicated for 15 minutes at 37 °C, re-vortexed for 2 min and passed through a 0.22 μm Nylon filter before analysis.
Chromatographic separation was achieved with an Acquity I-Class UPLC equipped with an ACQUITY UPLC® HSS T3 column (2.1 × 100 mm, 1.8 μm) protected with an ACQUITY UPLC® HSS T3 VanGuard pre-column (2.1 × 5 mm, 1.8 μm) (Waters, Milford, MA), which were maintained at 30 °C. Mobile phases were composed of water (A) and acetonitrile (B), both acidified with 0.1% (v/v) of formic acid, and the elution was carried out with the following gradient: 0–2 min: 1% B, 2–18.33 min: 1–48% B, 18.33–22.33 min: 48–95% B, 22.33–25.33 min: 95% B, 25.33–25.4 min: 95–1% B and 25.4–28 min: 1% B. The injection volume was 1 μL, and the flow rate was 0.3 mL min−1. UV data were collected from 200 to 500 nm. MS data were acquired using a Synapt G2-Si (Waters, Milford, MA) with the following source parameters: capillary voltage: −2.40 kV, source temperature: 120 °C, desolvation temperature: 600 °C, cone gas flow: 50 L h−1, and desolvation gas flow: 700 L h−1. The fast-DDA acquisition method was used in negative electrospray and resolution mode (≈25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) with an MS survey scan range (m/z) of 50 to 1200 and a scan time of 0.2 s. For each MS survey scan, 3 MS/MS were performed with a scan time of 0.1 s and a collision energy ranging from 15 to 45 V. Dynamic peak exclusion was used to exclude masses within 10 ppm during 3 s after MS/MS acquisition. Leucine-enkephaline (200 pg μL−1) was infused at a flow rate of 10 μL min−1 for internal mass correction.
000) with an MS survey scan range (m/z) of 50 to 1200 and a scan time of 0.2 s. For each MS survey scan, 3 MS/MS were performed with a scan time of 0.1 s and a collision energy ranging from 15 to 45 V. Dynamic peak exclusion was used to exclude masses within 10 ppm during 3 s after MS/MS acquisition. Leucine-enkephaline (200 pg μL−1) was infused at a flow rate of 10 μL min−1 for internal mass correction.
Gallic acid was quantified at 280 nm, ellagitannins at 240 nm and ellagic acid derivatives at 360 nm, using a calibration curve obtained for gallic acid (1–100 mg L−1), vescalagin (5–250 mg L−1) and ellagic acid (1–100 mg L−1), respectively. Each extract was injected in triplicate. UV data were processed using TargetLynx XS v4.2 software (Waters, Milford, MA), while MS data were analyzed with Progenesis QI 3.0 (Nonlinear Dynamics).
2.6 Analysis of the ellagitannin-transforming capacity of the TanA and Est_1092-producing L. plantarum strains
The capacity of TanA+ and Est_1092+ L. plantarum strains to transform the broad range of ellagitannins present in camu–camu was tested by cultivating the selected strains in RP-M medium supplemented with 0.15% (w/v) of the camu–camu aqueous extract. This dose was chosen since it allowed ellagitannin compounds to be accurately semi-quantified when lower doses showed considerably higher variations (data not shown). For the assay, the camu–camu supplemented medium was inoculated with 1% of a 13h MRS culture of each strain and incubated at 30 °C without agitation. At the end of the fermentation (10 d), the supernatants of each strain were collected and analyzed.Colony counts were performed in triplicate at the beginning and after days 2, 4, 6, 8, and 10 of fermentation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). Each function was collected with a scan range (m/z) of 50 to 1200 and a scan time of 0.2 s. In the high energy function, a collision energy ramp of 15 to 45 V. MS data were processed using Skyline 21.1
000). Each function was collected with a scan range (m/z) of 50 to 1200 and a scan time of 0.2 s. In the high energy function, a collision energy ramp of 15 to 45 V. MS data were processed using Skyline 21.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 for the semi-quantification of ellagitannins and ellagic acid derivatives.
23 for the semi-quantification of ellagitannins and ellagic acid derivatives.
Absolute quantification of pyrogallol and gallic acid was performed using UV data at 266 nm and 280 nm, respectively, with the appropriate calibration curve ranging from 1 to 100 mg L−1.
Ellagitannins and ellagic acid derivatives were only semi-quantified by MS as UV did not provide enough sensibility.
2.7 Statistical analysis
The results are expressed as the mean ± standard error of three independent experiments. For the analysis, the results from the strains were grouped into WCFS1 (Est_1092 and TanA-lacking reference strain), Est_1092+ strains (ATCC 8014, PROBI 56-12, PROBI 56-24, PROBI 59-12) and TanA+ strains (ATCC 14917, PROBI S204, PROBI S126, RKG 1-473, RKG 1-500, RKG 2-219, and RKG 2-690). When indicated, groups were further divided according to their capacity to produce the GD enzyme. Statistical significance was determined by a one-way ANOVA analysis with Tukey–Kramer multiple comparisons. If data were not distributed normally, the Kruskal–Wallis test by ranks was applied. For statistical inference, results were evaluated for normality by the Shapiro–Wilk test and by visually inspecting the distribution of residuals in quantile–quantile plots. All statistical analyses were performed in GraphPad Prism 9.4.1. Differences were considered significant at p < 0.05.3. Results
3.1 Phenolic characterization of camu–camu crude extract and its aqueous and ethanolic fractions
A dry crude extract and its aqueous and ethanolic fractions were obtained in order to find the most suitable source of ellagitannins. The phenolic compounds characterized in these three powders, their exact mass, and their mass accuracy are indicated in Table S1.† The crude extract and its aqueous and ethanolic fractions showed the presence of a wide variety of ellagitannins of different chemical complexity, such as vescalagin, castalagin, HHDP-galloyl-glucose, three isomers of di-HHDP-glucose, tri-galloyl-HHDP-glucose, and di-galloyl-HHDP-glucose (Table 1). Among the ellagitannins detected, castalagin stood out as the most abundant form, accounting for 56.4 ± 0.4% of the total ellagitannins of the crude extract, 62.7 ± 0.4% of those of the aqueous fraction, and 46.6 ± 0.6% of those of the ethanolic fraction (Table 1). The aqueous fraction showed the highest content of total ellagitannins, with 2.3 times more ellagitannins than the crude extract and 12.7 times more than the ethanolic fraction (Table 1). The aqueous fraction also showed the highest content of ellagic acid derivatives such as valoneic acid dilactone, ellagic acid hexoside, ellagic acid pentoside, ellagic acid desoxyhexoside, ellagic acid, two isomers of ellagic acid acetyl rhamnoside, and four isomers of ellagic acid glycosides (Table 1). Overall, the ethanolic fraction showed a lower abundance of the phenolic compounds analyzed. The aqueous fraction was selected for subsequent experiments as it consistently showed the highest quantities of all the different types of ellagitannins (Table 1).| Class | Compound | Crude extract (mg per 100 g) | Aqueous fraction (mg per 100 g) | Ethanolic fraction (mg per 100 g) |
|---|---|---|---|---|
| Gallic acid and derivatives | Gallic acid | 139.16 ± 0.83 | 22.11 ± 0.16 | 20.9 ± 0.10 |
| Ellagic acid and derivatives | Valoneic acid dilactone | 11.89 ± 0.13 | 37.66 ± 0.23 | 3.58 ± 0.06 |
| Ellagic acid hexoside | 28.53 ± 0.18 | 100.2 ± 0.39 | 7.76 ± 0.05 | |
| Ellagic acid pentoside | 54.37 ± 0.53 | 199.4 ± 1.43 | 13.89 ± 0.33 | |
| Ellagic acid desoxyhexoside | 49.67 ± 0.27 | 183.7 ± 0.53 | 12.81 ± 0.08 | |
| Ellagic acid | 28.41 ± 0.18 | 96.06 ± 0.48 | 10.84 ± 0.11 | |
| Ellagic acid acetyl rhamnoside – 1 | 11.78 ± 0.08 | 40.76 ± 0.18 | 4.063 ± 0.05 | |
| Ellagic acid acetyl rhamnoside – 2 | 11.72 ± 0.10 | 36.81 ± 0.12 | 4.13 ± 0.10 | |
| Ellagic acid glycoside – 1 | 2.39 ± 0.01 | 5.510 ± 0.01 | 1.8 ± 0.01 | |
| Ellagic acid glycoside – 2 | 4.14 ± 0.01 | 11.99 ± 0.02 | 2.33 ± 0.01 | |
| Ellagic acid glycoside – 3 | 3.22 ± 0.02 | 7.92 ± 0.04 | 2.13 ± 0.02 | |
| Ellagic acid glycoside – 4 | 3.36 ± 0.03 | 9.28 ± 0.04 | 2.02 ± 0.02 | |
| Ellagitannins | Vescalagin | 373.2 ± 2.21 | 384.5 ± 1.19 | 55.73 ± 0.33 |
| Castalagin | 860.6 ± 6.36 | 2198 ± 15.58 | 128.9 ± 1.63 | |
| HHDP-galloyl-glucose | 19.47 ± 0.28 | 75.92 ± 1.39 | 8.45 ± 0.03 | |
| Di-HHDP-glucose – 1 | 57.59 ± 0.59 | 129.2 ± 0.67 | 13.83 ± 0.12 | |
| Di-HHDP-glucose – 2 | 33.49 ± 0.28 | 110.7 ± 0.92 | 10.34 ± 0.07 | |
| Di_HHDP-galloyl-glucose – 1 | 66.08 ± 0.99 | 225.1 ± 0.98 | 17.33 ± 0.08 | |
| Di_HHDP-galloyl-glucose – 2 | 69.66 ± 1.03 | 251.1 ± 0.69 | 16.89 ± 0.28 | |
| Di_HHDP-galloyl-glucose – 3 | 24.17 ± 0.47 | 79.39 ± 0.23 | 9.93 ± 0.09 | |
| Tri-galloyl-HHDP-glucose | 9.62 ± 0.09 | 16.28 ± 0.12 | 7.40 ± 0.04 | |
| Di-galloyl-HHDP-glucose | 11.21 ± 0.12 | 35.48 ± 0.57 | 7.91 ± 0.03 | |
| Total ellagitannins | 1525 ± 12.01 | 3505 ± 16.16 | 276.7 ± 2.6 |
3.2 Selected L. plantarum strains and their PAZymes genomic features
Out of forty-three L. plantarum isolates, three contained the est_1092 gene encoding the broad esterase enzyme (Table S2†). These are L. plantarum PROBI 56-12, L. plantarum PROBI 56-24, and L. plantarum PROBI 59-12, isolated from fermented sorghum. The three est_1092-harboring strains showed Est_1092 activity, as they were able to hydrolyze the ethyl-ferulate added in the medium into ferulic acid.Pairwise genome comparisons of the novel Est_1092+ isolates showed an average nucleotide identity (ANI) ranging between 99.08% and 99.97% (Table S3†). Notably, PROBI 56-12, PROBI 56-24, and PROBI 59-12 isolates presented an ANI higher than 99.7%, indicating that these isolates are potential variants of the same strain (although there is no available consensus on the cut-off). The genome sequences of these Est_1092+ strains are available in the BV-BRC with the following IDs: 1590.3114, 1590.3115, 1590.3116.
The PAZymes genomic features of the Est_1092+ strains were analyzed to determine their potential to convert camu–camu phenolic compounds. A multiple sequence alignment of the Est_1092 genomic feature showed that the three producing strains have an identical 295-amino acid sequence (100% identity with the query reference sequence) (Table S4†). In addition, all Est_1092+ strains have both gallate decarboxylase genomic features, LpdB and LpdC (100% of identity with the query reference sequence) (Table S4†), indicating that these strains have the potential to convert gallic acid into pyrogallol.
A BLAST search using the WCFS1 TanB amino acid sequence as a query revealed that both, Est_1092+ and TanA+ (selected in a previous study12) strains also contain the TanB genomic feature (intracellular tannase), showing 99% to 100% identity with the TanB query reference sequence (Table S5†).
3.3 Metabolism of camu–camu phenolic compounds by L. plantarum strains with different tannase ability
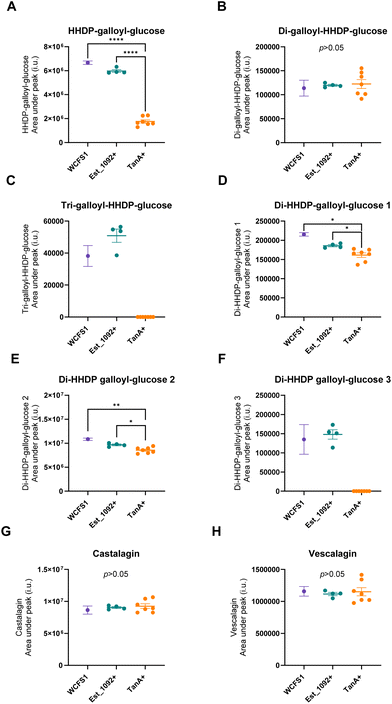
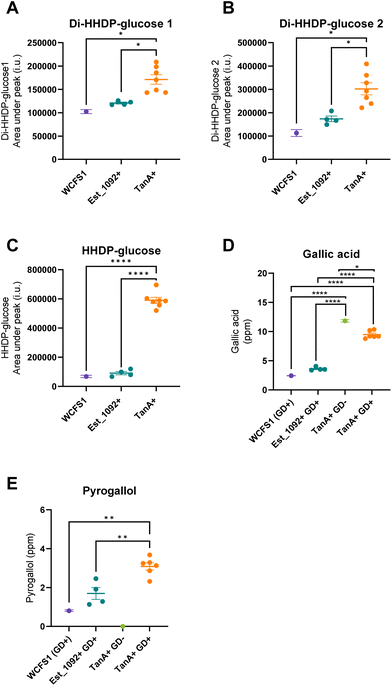
As a result of the hydrolysis of di-HHDP-galloyl-glucose isomers by TanA+ strains, two isomers of di-HHDP-glucose and gallic acid were released (p < 0.05, Fig. 3A, B and D). Similarly, HHDP-glucose and gallic acid were released from the hydrolysis of tri-galloyl-HHDP-glucose and HHDP-galloyl-glucose by TanA+ strains (p < 0.05, Fig. 3C and D). Overall, TanA+ L. plantarum strains released 3.6 to 4.9 times more gallic acid than the TanA-lacking strains (WCFS1 and Est_1092+ strains) (p < 0.0001) (Fig. 3D). Additionally, no statistical differences were found between the gallic acid released from WCFS1 or Est_1092+ strains and a blank (non-inoculated media) (p > 0.05, Fig. S2†). The strains with TanA and GD activity (PROBI S204, PROBI S126, RKG 1-473, RKG 2-219, RKG 2-690) followed the gallic acid metabolism until the release of pyrogallol. These strains released 2.8 to 4.5 times more pyrogallol than the WCFS1 strain (Fig. 3E).
In addition, all L. plantarum strains maintained most of their viability throughout the fermentation, with viable counts remaining greater than 6 log CFU mL−1. Indeed, only a reduction of 2.31 ± 0.32 log CFU mL−1 in the camu–camu supplemented media and 1.76 ± 0.38 log CFU mL−1 in the control media was observed (Fig. S4†).
4. Discussion
Lactobacilli tannases have been suggested as part of the putative ellagitannin gut-transforming microorganisms,5 but little is known about their ability to metabolize these substrates. In this study, we shed some light on this metabolism by investigating the ability of L. plantarum producing a strain-specific tannase (i.e., TanA) or a broad esterase (i.e., Est_1092) to transform camu–camu ellagitannins with chemical structures of varying complexity. Our results show that TanA+ L. plantarum are the only strains that hydrolyze camu–camu galloylated ellagitannins, releasing (di)HHDP-glucose, HHDP-glucose and gallic acid (Fig. 4).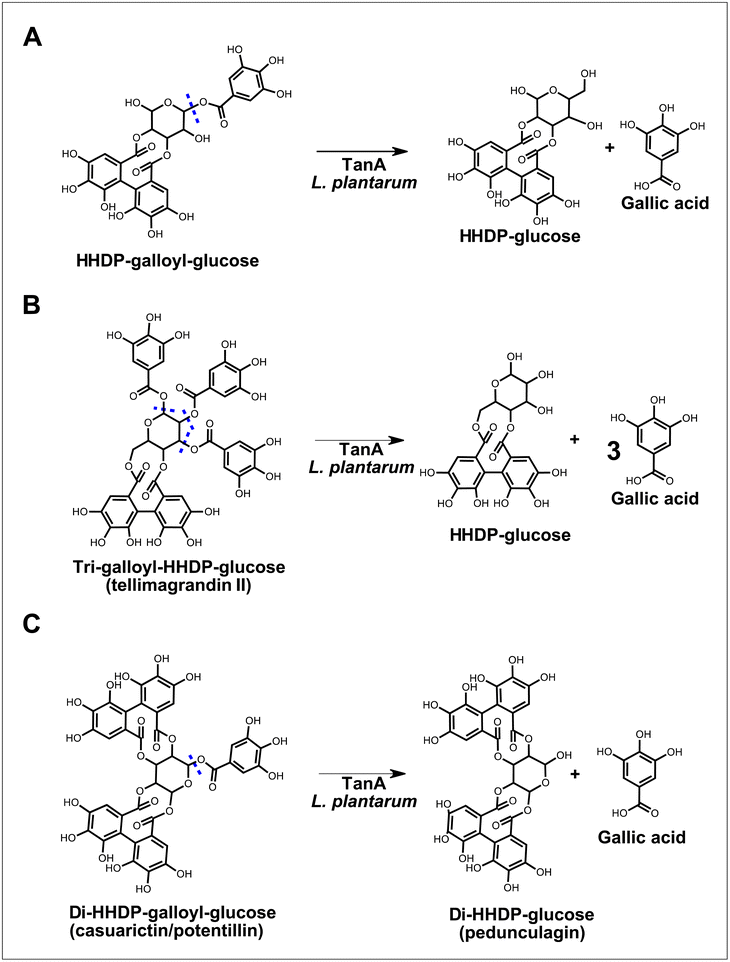 | ||
| Fig. 4 Proposed biotransformations of galloylated ellagitannins by L. plantarum strains with TanA activity. The predicted sites of hydrolysis are marked in blue dotted lines. | ||
In addition to their hydrolyzing capacity, the TanA+ strains showed great substrate versatility, acting on five (out of six) different galloylated ellagitannins found in camu–camu fruit. The only galloylated ellagitannin that remained unchanged during fermentation by this group of strains was the di-galloyl-HHDP glucose. We suggest that this is because, in our study, this molecule is both a substrate (as it is present in the extract) and an intermediate metabolite of the tri-galloyl-HHDP glucose (molecule formed after partial removal of a galloyl group), resulting in unaltered concentrations. We also observed that two galloylated ellagitannins, the di-HHDP-galloyl-glucose 3 and the tri-galloyl-HHDP-glucose, were completely transformed by the TanA+ L. plantarum strains, while the others were only partially metabolized. These differences are probably due to the availability of these compounds in the camu–camu aqueous extract, as those that were fully metabolized were present in lower amounts. Based on the observed hydrolytic versatility, it could be hypothesized that TanA+ strains act on other galloylated ellagitannins, such as sanguiin H-6 and fragariin from strawberry, and the lambertianin C from raspberry and cloudberry.15,24,25 Future studies will confirm whether this enzymatic robustness can be extended to other galloylated molecules, such as the galloylated proanthocyanidins (polymeric flavonoids) found in grapes, wine and in persimmon fruit.26,27
By hydrolyzing camu–camu galloylated ellagitannins, TanA+ L. plantarum strains favour the release of bioactive phenolic metabolites. Indeed, this study shows that these strains release almost four times more gallic acid and at least two and a half times more pyrogallol when compared to a non-producing reference strain (WCFS1). This is significant as these phenolic metabolites have been shown to have antidiabetic, antiobesity, neuroprotective, and anticancer properties,28–35 thus highlighting the potential of these strains to act synergistically to enhance the health benefits of camu–camu (poly)phenols. However, it is essential to note that gallic acid and other (poly)phenols under certain biological conditions may also exhibit pro-oxidant or pro-carcinogenic effects.36,37
In this study, we also observed that TanA+ L. plantarum released di-HHDP-glucose and HHDP-glucose. In the intestinal environment, this could favour the uptake of these molecules by yet unknown members of the gut microbiota, which convert ellagitannins-core structures to ellagic acid. The ellagic acid released could ultimately be used to produce urolithins either by probiotic species, such as Bifidobacterium pseudocatenulatum INIA P815, or by gut species, such as Gordonibacter, Ellagibacter and Enterocloster sp., which are being investigated as next-generation probiotics.38–40 Future studies using colonic digestion simulation systems may illustrate the contribution of TanA+ strains in the collaborative transformation of the diverse and complex structures of ellagitannins.
This study showed that neither TanA+ nor Est_1092+ L. plantarum strains transformed non-galloylated ellagitannins (i.e., castalagin and vescalagin) or ellagitannin-core structures, indicating that none of these strain-specific enzymes are involved in this type of hydrolysis. This result differs from that of Caballero et al.,41 who reported the biodegradation of punicalagin (a pomegranate ellagitannin) into ellagic acid by different lactic acid bacteria, including L. plantarum. Indeed, these authors observed a small release of ellagic acid (conversion rate of <5%) after incubation of each strain in a culture medium supplemented with a punicalagin-rich extract from pomegranate (31% of purity). However, as punicalagin was not quantified in the fermented medium, it is unclear whether the ellagic acid released resulted from the hydrolysis of this ellagitannin or if it arose from the metabolism of unidentified (poly)phenols from the extract (e.g., methyl-ellagic acid42).
The fact that Est_1092+ L. plantarum strains do not hydrolyze ellagitannins suggests that this trait is unlikely to be involved in tannin catabolism, as previously suggested by Esteban-Torres et al.13 Indeed, these authors observed an unexpected reduction in est_1092 gene expression after exposure of a producing strain to methyl gallate (an ester substrate hydrolyzed by tannases), suggesting that this species does not use this enzyme as a tannase. It is noteworthy that the Est_1092 feruloyl esterase activity allows the transformation of other phenolic substrates of interest (not covered in this study), such as the hydroxycinnamic acid esters that are abundant in plant cell walls.13 As previously suggested, the production of this enzyme is highly strain-specific, as only three L. plantarum strains out of forty-three isolates analyzed in this study showed this characteristic. Interestingly, the ANI value of these strains (an indicator of genomic similarity) revealed that these selected strains were highly genetically related since they had an ANI of more than 99.08%. Indeed, this value is close to those found between L. plantarum strains of the same clade (99.19% ± 0.22%).43 Curiously, the Est_1092+ strains selected in this study and those previously reported were mostly isolated from plant sources such as corn silage (ATCC 8014), grass silage (ATCC 14431, JDM1), fermented sorghum (PROBI 56-12, PROBI 56-24, PROBI 59-12), bread dough (DSM 1055), wine (RM-35, RM-73), and fermented bamboo shoot (EGD-AQ4),13,44 suggesting that the Est_1092 trait may facilitate the survival of this species in plant niches.12,37
Regardless of their type of enzyme (i.e. Est_1092 or TanA), all L. plantarum groups (WCFS1, Est_1092+, and TanA+) showed a similar growth response in the presence of the camu–camu aqueous extract. During the first four days of fermentation, they showed a slight reduction in viable counts compared to their growth in the (poly)phenol-free medium. The observed antimicrobial effect may be mainly due to the content of castalagin and vescalagin, the two most abundant forms of ellagitannins in the extract. Indeed, these ellagitannins have been observed to alter the normal assembly of the peptidoglycans located on the surface of Gram-positive bacteria, thus promoting cell disruption and death.45 Despite their slightly higher growth, all L. plantarum groups showed resistance to the presence of ellagitannins, remaining viable throughout the fermentation period.
Finally, it is essential to note that, despite its antimicrobial activity, the aqueous camu–camu fraction was a suitable substrate for the simultaneous study of the transformation of different types of ellagitannins. Overall, this fraction showed a higher and purer ellagitannin content compared to the crude extract and its ethanolic fraction. It also provided at least eight times more ellagitannins than a previously characterized dried camu–camu flour produced from the peel and seeds of this fruit.3 Nevertheless, the previously reported camu–camu flour and the aqueous fraction of this study showed a similar ellagitannin profile, with castalagin and vescalagin highlighted as the predominant forms. Ellagitannin-rich extracts, such as the reported in this study, along with emerging techniques using UPLC-MS/MS, are key tools for future studies investigating the hydrolytic capacity of microorganisms and purified enzymes towards ellagitannins.
5. Conclusions
The TanA enzyme is a strain-specific feature that enables L. plantarum strains to hydrolyze the esterified galloyl units of a wide range of galloylated ellagitannins. TanA+ L. plantarum strains convert HHDP-galloyl glucose, tri-galloyl-HHDP-glucose and different regioisomers of Di-HHDP-galloyl-glucose found in camu–camu and other dietary sources such as raspberry, cloudberry, tea, and walnut.24,46,47 This exceptional and versatile metabolic activity allows these strains to release bioactive phenolic metabolites, such as gallic acid and pyrogallol, as well as ellagitannin-core structures that can be further metabolized into beneficial metabolites by other members of the gut microbiota.Based on these findings, TanA+ strains can be considered for strategies to potentialize the production of bioactive phenolic metabolites from galloylated ellagitannins or for inclusion in synbiotic formulations containing camu–camu. Furthermore, in the context of personalized nutrition, these strains may be suitable as potential “precision probiotics”48 aiming to favour the metabolism of galloylated tannins in individuals with unfavourable metabotypes. Future research is warranted to explore the potential of TanA+ strains to release bioactive phenolic metabolites in the complex gut environment and to improve a range of health outcomes in in vivo models.
Author contributions
ECPM: conceptualization, methodology, validation, investigation formal analysis, and writing – original draft; JLL: conceptualization, methodology, validation, investigation, writing-review, and editing; YD: supervision, resources, funding, writing-review, and editing; DR: supervision, resources, funding, writing-review, and editing.Conflicts of interest
ECPM, JLL, DR, and YD research works are funded by a Collaborative Research and Development Grant (RDC), partly funded by Symrise (Diana Food Canada Inc.) and the Natural Sciences and Engineering Research Council of Canada (NSERC). YD holds an NSERC-DianaFood Industrial Chair on the prebiotic effects of fruit and vegetable polyphenols.Acknowledgements
This study was supported by a Collaborative Research & Development (CRD) program on the development of synergistic combinations of prebiotic polyphenols and probiotic bacteria (RDCPJ 518138 - 17) and by The NSERC Industrial Research Chair (IRC) on the prebiotic effect of polyphenols in fruits and vegetables. ECPM was a scholarship recipient from the Consejo Nacional de Humanidades, Ciencias y Tecnologías (Conahcyt) (Mexico). JLL was a scholarship recipient from the NSERC. The authors are also grateful to Ashraf Badr, who performed the aqueous extraction of camu–camu ellagitannins of this work, and D. Roy's team, for their technical and scientific, especially Alexandre J. Kennang Ouamba. We would also like to acknowledge the INAF platforms for providing access to the analytical instruments used in this work.References
- A. Abot, A. Brochot, N. Pomié, E. Wemelle, C. Druart, M. Régnier, N. M. Delzenne, W. M. de Vos, C. Knauf and P. D. Cani, Metabolites, 2022, 12, 301 CrossRef CAS.
- F. F. Anhê, V. Varin, J. Trottier, S. Dudonné, M. Le Barz, P. Feutry, G. Pilon, O. Barbier, Y. Desjardins, D. Roy and A. Marette, Gut, 2019, 68, 453–464 CrossRef.
- D. Fracassetti, C. Costa, L. Moulay and F. A. Tomás-Barberán, Food Chem., 2013, 139, 578–588 CrossRef CAS.
- M. C. Rodríguez-Daza, E. C. Pulido-Mateos, J. Lupien-Meilleur, D. Guyonnet, Y. Desjardins and D. Roy, Front. Nutr., 2021, 8, 347 Search PubMed.
- R. García-Villalba, J. A. Giménez-Bastida, A. Cortés-Martín, M.Á Ávila-Gálvez, F. A. Tomás-Barberán, M. V. Selma, J. C. Espín and A. González-Sarrías, Mol. Nutr. Food Res., 2022, 66, 2101019 CrossRef.
- D. Carregosa, R. Carecho, I. Figueira and C. N. Santos, J. Agric. Food Chem., 2020, 68, 1790–1807 CrossRef CAS.
- B. de las Rivas, H. Rodríguez, J. Anguita and R. Muñoz, Appl. Microbiol. Biotechnol., 2019, 103, 603–623 CrossRef CAS.
- J. A. Ascacio-Valdés, J. J. Buenrostro, R. De la Cruz, L. Sepúlveda, A. F. Aguilera, A. Prado, J. C. Contreras, R. Rodríguez and C. N. Aguilar, J. Basic Microbiol., 2014, 54, 28–34 CrossRef.
- C. Crestini and H. Lange, Microchem. J., 2015, 123, 139–147 CrossRef CAS.
- R. Muñoz, B. de Las Rivas, H. Rodríguez, M. Esteban-Torres, I. Reverón, L. Santamaría, J. M. Landete, L. Plaza-Vinuesa, A. Sánchez-Arroyo, N. Jiménez and J. A. Curiel, Int. J. Food Microbiol., 2024, 412, 110555 CrossRef . Top of Form.
- N. Jimenez, M. Esteban-Torres, J. M. Mancheno, B. de las Rivas and R. Munoz, Appl. Environ. Microbiol., 2014, 80, 2991–2997 CrossRef.
- E. C. Pulido-Mateos, J. Lessard-Lord, D. Guyonnet, Y. Desjardins and D. Roy, Sci. Rep., 2022, 12, 1–15 CrossRef.
- M. Esteban-Torres, J. M. Landete, I. Reverón, L. Santamaría, B. de las Rivas and R. Muñoz, Appl. Environ. Microbiol., 2015, 81, 3235–3242 CrossRef CAS.
- N. Rozès and C. Peres, Appl. Microbiol. Biotechnol., 1998, 49, 108–111 CrossRef.
- M. Sójka, M. Janowski and K. Grzelak-Błaszczyk, Eur. Food Res. Technol., 2018, 245, 1–10 Search PubMed.
- M. Gagnon, A. J. K. Ouamba, G. LaPointe, P. Y. Chouinard and D. Roy, J. Dairy Sci., 2020, 103, 5931–5946 CrossRef CAS.
- J. Donaghy, P. F. Kelly and A. M. McKay, Appl. Microbiol. Biotechnol., 1998, 50, 257–260 CrossRef CAS.
- R. R. Wick, L. M. Judd, C. L. Gorrie and K. E. Holt, PLoS Comput. Biol., 2017, 13(6), e1005595 CrossRef.
- J. J. Davis, A. R. Wattam, R. K. Aziz, T. Brettin, R. Butler, R. M. Butler, P. Chlenski, N. Conrad, A. Dickerman, E. M. Dietrich, J. L. Gabbard, S. Gerdes, A. Guard, R. W. Kenyon, D. MacHi, C. Mao, D. Murphy-Olson, M. Nguyen, E. K. Nordberg, G. J. Olsen, R. D. Olson, J. C. Overbeek, R. Overbeek, B. Parrello, G. D. Pusch, M. Shukla, C. Thomas, M. Vanoeffelen, V. Vonstein, A. S. Warren, F. Xia, D. Xie, H. Yoo and R. Stevens, Nucleic Acids Res., 2020, 48, D606 CAS.
- E. Bosi, B. Donati, M. Galardini, S. Brunetti, M. F. Sagot, P. Lió, P. Crescenzi, R. Fani and M. Fondi, Bioinformatics, 2015, 31, 2443–2451 CrossRef CAS.
- N. Jimenez, J. A. Curiel, I. Reveron, B. de las Rivas and R. Munoz, Appl. Environ. Microbiol., 2013, 79, 4253–4263 CrossRef CAS.
- C. Dufour, J. A. Villa-Rodriguez, C. Furger, J. Lessard-Lord, C. Gironde, M. Rigal, A. Badr, Y. Desjardins and D. Guyonnet, Antioxidants, 2022, 11, 1561 CrossRef CAS.
- K. J. Adams, B. Pratt, N. Bose, L. G. Dubois, L. St. John-Williams, K. M. Perrott, K. Ky, P. Kapahi, V. Sharma, M. J. Maccoss, M. A. Moseley, C. A. Colton, B. X. Maclean, B. Schilling and J. W. Thompson, J. Proteome Res., 2020, 19, 1447 CrossRef CAS.
- M. Kähkönen, P. Kylli, V. Ollilainen, J.-P. Salminen and M. Heinonen, J. Agric. Food Chem., 2012, 60, 1167–1174 CrossRef.
- E. Karlińska, Ł. Pecio, J. Macierzyński, A. Stochmal and M. Kosmala, Food Chem., 2019, 296, 109–115 CrossRef.
- C. Li, R. Leverence, J. D. Trombley, S. Xu, J. Yang, Y. Tian, J. D. Reed and A. E. Hagerman, J. Agric. Food Chem., 2010, 58, 9033–9042 CrossRef CAS.
- C. P. Passos, S. M. Cardoso, M. R. M. Domingues, P. Domingues, C. M. Silva and M. A. Coimbra, Food Chem., 2007, 105, 1457–1467 CrossRef CAS.
- S. S. Patel and R. K. Goyal, Pharmacogn. Res., 2011, 3, 239 CrossRef CAS.
- I. A. Adedara, S. E. Owumi, A. K. Oyelere and E. O. Farombi, J. Biochem. Mol. Toxicol., 2021, 35, e22684 CrossRef CAS.
- Y. Oi, I. C. Hou, H. Fujita and K. Yazawa, Phytother. Res., 2012, 26, 475–481 CrossRef CAS.
- M. T. Mansouri, Y. Farbood, M. J. Sameri, A. Sarkaki, B. Naghizadeh and M. Rafeirad, Food Chem., 2013, 138, 1028–1033 CrossRef CAS.
- C.-L. Hsu and G.-C. Yen, Br. J. Nutr., 2007, 98, 727–735 CrossRef CAS.
- D. H. Priscilla and P. S. M. Prince, Chem.-Biol. Interact., 2009, 179, 118–124 CrossRef CAS.
- A. Ahad, H. Ahsan, M. Mujeeb and W. A. Siddiqui, Chem.-Biol. Interact., 2015, 240, 292–303 CrossRef CAS.
- M. J. Nemec, H. Kim, A. B. Marciante, R. C. Barnes, S. T. Talcott and S. U. Mertens-Talcott, Food Funct., 2016, 7, 3825–3833 RSC.
- E. Kadosh, I. Snir-Alkalay, A. Venkatachalam, S. May, A. Lasry, E. Elyada, A. Zinger, M. Shaham, G. Vaalani, M. Mernberger, T. Stiewe, E. Pikarsky, M. Oren and Y. Ben-Neriah, Nature, 2020, 586, 133–138 CrossRef CAS.
- F. López de Felipe, B. de Las Rivas and R. Muñoz, Front. Microbiol., 2014, 5, 121517 Search PubMed.
- C. E. Iglesias-Aguirre, R. García-Villalba, D. Beltrán, M. D. Frutos-Lisón, J. C. Espín, F. A. Tomás-Barberán and M. V. Selma, J. Agric. Food Chem., 2023, 71(9), 4029–4035 CrossRef CAS.
- C. E. Iglesias-Aguirre, A. González-Sarrías, A. Cortés-Martín, M. Romo-Vaquero, L. Osuna-Galisteo, J. J. Cerón, J. C. Espín and M. V. Selma, Food Funct., 2023, 14(6), 2657–2667 RSC.
- P. Gaya, Á. Peirotén, M. Medina, I. Álvarez and J. M. Landete, J. Funct. Foods, 2018, 45, 95–99 CrossRef CAS.
- V. Caballero, M. Estévez, F. A. Tomás-Barberán, D. Morcuende, I. Martín and J. Delgado, J. Agric. Food Chem., 2022, 70(51), 16273–16285 CrossRef CAS.
- R. F. Wang, W. D. Xie, Z. Zhang, D. M. Xing, Y. Ding, W. Wang, C. Ma and L. J. Du, J. Nat. Prod., 2004, 67, 2096–2098 CrossRef CAS.
- K. Li, S. Wang, W. Liu, L. Y. Kwok, M. Bilige and W. Zhang, Food Microbiol., 2022, 104, 103989 CrossRef CAS.
- F. Dellaglio, V. Bottazzi and M. Vescovo, Int. J. Syst. Bacteriol., 1975, 25, 160–172 CrossRef.
- A. R. ArauÌjo, A. C. ArauÌjo, R. L. Reis and R. A. Pires, ACS Biomater. Sci. Eng., 2021, 7, 1022–1030 CrossRef.
- S. K. Chang, C. Alasalvar, B. W. Bolling and F. Shahidi, J. Funct. Foods, 2016, 26, 88–122 CrossRef CAS.
- X. Yang and F. A. Tomás-Barberán, J. Agric. Food Chem., 2019, 67, 5394–5404 CrossRef CAS.
- P. Veiga, J. Suez, M. Derrien and E. Elinav, Nat. Microbiol., 2020, 5, 878–880 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo00149d |
| This journal is © The Royal Society of Chemistry 2024 |