Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A type 4 resistant potato starch alters the cecal microbiome and gene expression in mice fed a western diet based on NHANES data†
Allen D.
Smith
 *a,
Celine
Chen
a,
Lumei
Cheung
a,
Robert E.
Ward
b,
B. Sky
Jones
b,
Elizabeth A.
Pletsch
a and
Harry D.
Dawson
a
*a,
Celine
Chen
a,
Lumei
Cheung
a,
Robert E.
Ward
b,
B. Sky
Jones
b,
Elizabeth A.
Pletsch
a and
Harry D.
Dawson
a
aDiet, Genomics, and Immunology Laboratory, Beltsville Human Nutrition Research Center, Agricultural Research Service, United States Department of Agriculture, Rm. 228, Bldg. 307C, BARC-East, 10, 300 Baltimore Ave., Beltsville, MD 20705, USA. E-mail: allen.smith@usda.gov; Tel: +(301) 504-8577 ext. 280
bDepartment of Nutrition, Dietetics and Food Sciences, Utah State University, USA
First published on 5th March 2024
Abstract
Four major types of resistant starch (RS1–4) are present in foods, all of which can alter the microbiome and are fermented in the cecum and colon to produce short-chain fatty acids (SCFAs). Type 4 RSs are chemically modified starches, not normally found in foods, but have become a popular food additive as their addition increases fiber content. Multiple studies, in humans and rodents, have explored how different RS4 affect post-prandial glucose metabolism, but fewer studies have examined the effects of RS4 consumption on the microbiome. In addition, many RS studies conducted in rodents use high-fat diets that do not approximate what is typically consumed by humans. To address this, mice were fed a Total Western Diet (TWD), based on National Health and Nutrition Examination Survey (NHANES) data that mimics the macro and micronutrient composition of a typical American diet, for six weeks, and then supplemented with 0, 2, 5, or 10% of the RS4, Versafibe 1490™ (VF), a phosphorylated and cross-linked potato starch, for an additional three weeks. The cecal contents were analyzed for SCFA content and microbiota composition. Butyrate production was increased while branched chain SCFA production decreased. The alpha-diversity of the microbiome decreased in mice fed the TWD with 10% VF 1490 added while the beta-diversity plot showed that the 5% and 10% VF groups were distinct from mice fed the TWD. Similarly, the largest changes in relative abundance of various genera were greatest in mice fed the 10% VF diet. To examine the effect of VF consumption on tissue gene expression, cecal and distal colon tissue mRNA abundance were analyzed by RNASeq. Gene expression changes were more prevalent in the cecum than the colon and in mice fed the 10% VF diet, but the number of changes was substantially lower than we previously observed in mice fed the TWD supplemented with native potato starch (RPS). These results provide additional evidence that the structure of the RS is a major factor determining its effects on the microbiome and gene expression in the cecum and colon.
Introduction
Dietary fiber (DF) is found in plant- and fruit-based foods and includes a wide variety of compounds including polysaccharides, oligosaccharides, lignin, β-glucan, pectin, cellulose, resistant starches (RS) and other associated plant-derived substances. Four major types of RS (RS1–4) have been defined based upon their physical and chemical properties.1 Daily consumption of 15–20 g d−1 of RS is recommended but most people eating a western diet consume on average 4.9 g day−1.1,2 Consumption of RS alters the microbiome and SCFA levels by microbial fermentation in the cecum and colon in rodents,3–6 pigs7–9 and humans10,11 that has been linked to health benefits.12 Different fibers and RS differentially alter microbial composition and bacterial metabolites13 and even members of the same RS type can have differential effects on the microbiome and bacterial metabolites due to differences in their fine structure that may favor certain bacteria strains over others.14,15Multiple rodent studies have looked at the effect of RS consumption in mice fed a “Western” high fat diet (WD).16–19 In general, these studies used modified AIN-93 diets with 45–60% of the calories coming from lard or milkfat and 8–19% from sucrose, however, the diets used in these studies do not reflect a typical American diet. We and others20–22 have used the TWD developed using the 50th percentile daily intake levels for macro and micronutrients from NHANES including fewer calories from protein and carbohydrates, double the fat with more saturated and monounsaturated fats, less polyunsaturated fat, fewer complex carbohydrates, and twice the level of simple sugars compared to the AIN-93 diet.23 In previous studies, we demonstrated that the addition of RPS to the TWD induced significant changes to the intestinal microbiome and transcriptome24 and unexpectantly found that higher levels of RPS (10%) increased the efficiency of infection and colon pathology induced by Citrobacter rodentium,25 a bacterium that naturally infects mice and shares 67% of its genes with enteropathogenic Escherichia coli strains.26–28
Type 4 RSs (RS4) are chemically modified starches not normally found in nature but have become a popular food additive as their addition increases fiber content, as defined by the FDA. There have been multiple studies in both humans and rodents exploring how different RS4 affect postprandial glucose metabolism.29–33 Studies with VF, a type 4 phosphorylated and crosslinked potato starch, have provided conflicting results with some studies showing a positive effect and others no effect on postprandial glucose metabolism.34–36 Fewer studies exploring the effects of RS4 consumption on the microbiome have been undertaken. Deehan et al. (2020) demonstrated that RS4 derived from maize and tapioca altered the microbiome and SCFA production in human subjects while VF did not.37 Similarly, a study comparing feeding different types of RS4 to rats found that VF did not alter SCFA production or cecal content pH as compared to other RS4s suggesting that VF was not being fermented.38 These results contrast with an in vitro culture study which found that VF was fermented by the microbiomes of human donors.39 To follow up on our previous investigation of a RS2 potato starch (RPS, PenPure 10, Ingredion),24 and further explore the effects of feeding VF on the microbiome and gene expression, we fed different levels of VF to mice and looked for changes in the cecal microbiota and SCFA production, and gene expression in the cecum and colon.
Materials and methods
Animals and diet
C57BL/6 mice originally purchased from Charles River (Frederick, MD) were bred in-house. Breeding pairs were fed rodent chow (Teklad 2020X, Frederick, MD). Ventilated filter-top cages were used for housing the mice at the USDA BHNRC animal facility under 12 h light/dark cycle. Timed breedings were set up and offspring were weaned at 3–4 weeks of age. After weaning, male mice were group housed (4–5 per cage) and placed on the TWD [ESI Table 1†].23 After feeding mice the TWD for 6–7 weeks, mice were divided into 4 dietary treatment groups and fed for an additional 3 weeks with TWD or TWD with 2%, 5%, or 10% w/w VF (Ingredion, Westchester, IL) substituted for an equivalent amount of corn starch. VF is a phosphorylated and crosslinked potato starch that contains 85% fiber as measured by the AOAC 991.43 method (Ingredion Technical Specification sheet). Body weights and food consumption were recorded weekly. All experiments were approved by the USDA-ARS Beltsville Institutional Animal Care and Use Committee.Sample collection and processing
Samples for various analyses were obtained from three replicate experiments due to limitations on the amount of material, especially cecal contents, available for analysis. Cecal samples for 16S sequencing (n = 9–10), cecum and distal colon RNASeq (n = 5) were obtained from experiment 1, while body weights (n = 20) and samples for SCFA analysis were obtained from experiment 3 (10 of 20 samples). For other analyses, samples from all three experiments were used (n = 37–38). Fecal samples collected during the last 5 d of feeding were flash frozen and stored at −80° C. After approximately three weeks on the four dietary treatments mice were then euthanized and tissues collected. The colon was excised, fecal pellets removed, and the terminal six cm weighed and one cm segments from the distal and proximal colon (PC) were collected and snap frozen for RNA isolation. The cecum was excised, weighed and the cecal contents collected, the empty cecum reweighed, a section collected for RNA isolation. Fecal pellets were weighed and homogenized in 10 volumes of water, centrifuged to remove debris and the pH of the supernatant measured.Short-chain fatty acid analysis
SCFA analysis was performed according to the method of Ward et al., with slight modifications.22 Cecal contents were weighed. Distilled water and a metaphosphoric acid solution (250 g L−1) containing 1 g L−1 of ethyl butyric acid as an internal standard were added according to the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (sample
2 (sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water
water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acid/internal standard). After vortexing for 5 m, tubes were centrifuged at 14
acid/internal standard). After vortexing for 5 m, tubes were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 m. Approximately 200 μl of the supernatant was transferred to an insert in a gas chromatography vial. An eight-point calibration standard was prepared using a mix of six acids. Acetic, propionic, and butyric acids were present from 10 to 0.08 mM, and isobutyric, isovaleric and valeric acids were present from 5 to 0.04 mM. To prepare calibrations, 1 ml of standard was mixed with 0.2 ml of the metaphosphoric/ethyl butyric acid solution. Samples were analyzed by gas chromatography on an Agilent 6890 equipped with a ZB-FFAP column (30 mm × 0.52 mm ID × 1.0 μm film thickness; Phenomenex, Torrance, CA) and a flame ionization detector. The injector was maintained at 200 °C, and 1 μl was injected in splitless mode. The initial column temperature was 60 °C which was held for 1 m. The column was heated at 17 °C m−1 to 260 °C and held for 8 m. Peaks were identified according to retention time of individually run acids. For both external standards and the samples, raw peak areas were normalized to the ethyl butyric acid area. Peak area ratios were converted to concentration using regression equations for each acid. The overall concentration of acid in the sample was determined by multiplying the volume of water added by the concentration derived from the regression equation and dividing by the sample mass.
000g for 20 m. Approximately 200 μl of the supernatant was transferred to an insert in a gas chromatography vial. An eight-point calibration standard was prepared using a mix of six acids. Acetic, propionic, and butyric acids were present from 10 to 0.08 mM, and isobutyric, isovaleric and valeric acids were present from 5 to 0.04 mM. To prepare calibrations, 1 ml of standard was mixed with 0.2 ml of the metaphosphoric/ethyl butyric acid solution. Samples were analyzed by gas chromatography on an Agilent 6890 equipped with a ZB-FFAP column (30 mm × 0.52 mm ID × 1.0 μm film thickness; Phenomenex, Torrance, CA) and a flame ionization detector. The injector was maintained at 200 °C, and 1 μl was injected in splitless mode. The initial column temperature was 60 °C which was held for 1 m. The column was heated at 17 °C m−1 to 260 °C and held for 8 m. Peaks were identified according to retention time of individually run acids. For both external standards and the samples, raw peak areas were normalized to the ethyl butyric acid area. Peak area ratios were converted to concentration using regression equations for each acid. The overall concentration of acid in the sample was determined by multiplying the volume of water added by the concentration derived from the regression equation and dividing by the sample mass.
16S sequencing of cecal contents
DNA was isolated from cecal contents using a Quick-DNA Fecal/Soil Microbe Microprep Kit (Zymo, Irvine, CA) following the manufacturer's instructions and were further purified using the DNA Clean and Concentration kit (Zymo, Irvine, CA). The Quant-it PicoGreen dsDNA kit (Invitrogen, Waltham, MA) was used to determine the DNA concentration of the samples. DNA was submitted to the Michigan State University RTSF Genomics Core for targeted amplicon library preparation and sequencing. The V3–V4 hypervariable regions of the 16S rRNA gene was amplified using indexed, Illumina compatible primers 341f/806r essentially as described40 except that the V3 flanking primer 341f was substituted for the V4 primer 515f as described in the paper. The pooled libraries were loaded into an Illumina MiSeq v2 500 cycle reagent cartridge. Custom sequencing and index primers complementary to the 341f/806r target specific sequences were added to appropriate wells as described in ref. 40. The FASTQ files with raw data were submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under Bioproject number PRJNA1019368.The 16S rRNA tag data curation and processing were performed using the CLC Microbial Genomics Module (QIAGEN Bioinformatics, Redwood City CA) following its standard OTU clustering workflow. Paired reads were first merged into contigs, followed by removal of the adapters, nucleotides below Q30, reads containing more than two ambiguous nucleotides or shorter than five. Samples were then filtered by minimum reads of 100 or the minimum 50% from the median times the median number of reads across all samples and the aligned contigs were mapped to the SILVA SSU database from release v138.1.41 Chimeras were detected with k-mer search and removed from further processing and analysis. Sequences were then clustered into Operational Taxonomic Units (OTUs) at 97% similarity. Alpha and beta diversity were estimated using OTUs aligned with the MUSCLE tool42 to reconstruct the phylogenetic tree by a Maximum Likelihood approach. PERMANOVA43 was used to measure the effect size and significance of beta diversity. OTU count abundance was further collapsed into different taxon levels for downstream analysis. Successively, principal component analysis (PCA) and hierarchical clustering were performed with the OTU or taxon-specific abundance profiles to examine changes induced by the VF treatments. Changes in data distribution induced by the VF treatments were visualized by PCA analysis in JMP Genomics 10 (JMP Statistical Discovery LLC, Cary, NC) using default settings. To identify RPS-specific biomarkers at multiple taxonomical levels, the abundance profiles were subjected to linear discriminant analysis effect size (LEfSe) analysis.44 The differential expression analysis for all taxa and OTUs were performed with the Bioconductor package Deseq2 (version 3.14; run on Rstudio, version 4.0.3, Boston, MA).45 Relative abundance heatmaps were generated using log2 (composition × 100 + 1) transformed data in JMP Genomics 10.
RNASeq analysis of cecum and distal colon tissue
RNA from the cecum and distal colon (DC) was isolated using Tri-Reagent (Zymo, Irvine, CA) and Purelink RNA kits (Invitrogen, Carlsbad, CA). RNA was further purified using RNA Clean and Concentrate columns (Zymo, Irvine, CA). Five cecum samples from each group with a RIN scores of 7.9 or higher (Agilent Bioanalyzer) and their matching distal colon samples were chosen for further analysis. RNA was submitted to the Michigan State University RTSF Genomics Core facility for sequencing. Libraries were prepared using Illumina Stranded mRNA Prep, Ligation kit with IDT for Illumina Unique Dual Indexes following manufacturer's recommendations by the Michigan State University RTSF Genomics Core facility. Completed libraries were QC'd and quantified using a combination of Qubit dsDNA HS and Agilent 4200 TapeStation HS DNA1000 assays. The libraries were pooled in equimolar amounts for multiplexed sequencing, and the pool quantified using the Invitrogen Collibri Quantification qPCR kit. This pool was loaded onto one (1) lane of an Illumina NovaSeq S4 flow cell and sequencing was performed in 2 × 150 bp single read format using a NovaSeq 6000 v1.5 300 cycle reagent kit. Base calling was done by Illumina Real Time Analysis (RTA) v3.4.4 and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.20.0. The FASTQ files with raw data and the gene expression profiles were submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under Bioproject number PRJNA1019368.The resulting sequences were processed to determine gene expression levels as follows.
Nucleotides below Q30 or reads containing more than two ambiguous nucleotides were removed before sequence alignments performed by the CLC Genomics Workbench version 20.01 (QIAGEN Bioinformatics, Redwood City CA). To calculate gene expression in count, reads were mapped to the Mus musculus genome assembly GRCm39. Transcriptomes built from the alignment results were subjected to differential expression analysis. The statistical analyses were carried out with Deseq2. Genes were considered differentially expressed with the thresholds of a false discovery rate (FDR) ≤0.05 and an absolute fold change ≥1.5. We functionally annotated differentially expressed genes using our Porcine Translational Research Database46 as previously described.47 The database serves to translate data found in rodents or pigs to human. Gene expression heatmaps were generated using log2 transformed data in JMP Genomics 10.
Statistical analyses
Data was analyzed using a Student's t-test or one-way ANOVA. Data was transformed as necessary to achieve equal variance and normality. For graphical purposes, the mean and SEM of untransformed data are shown.Results
Effect of VF on short-chain fatty acid production, tissue weights and fecal pH
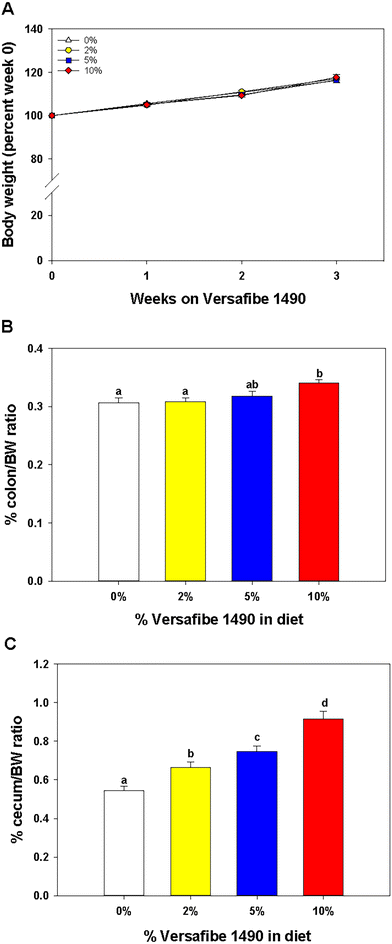
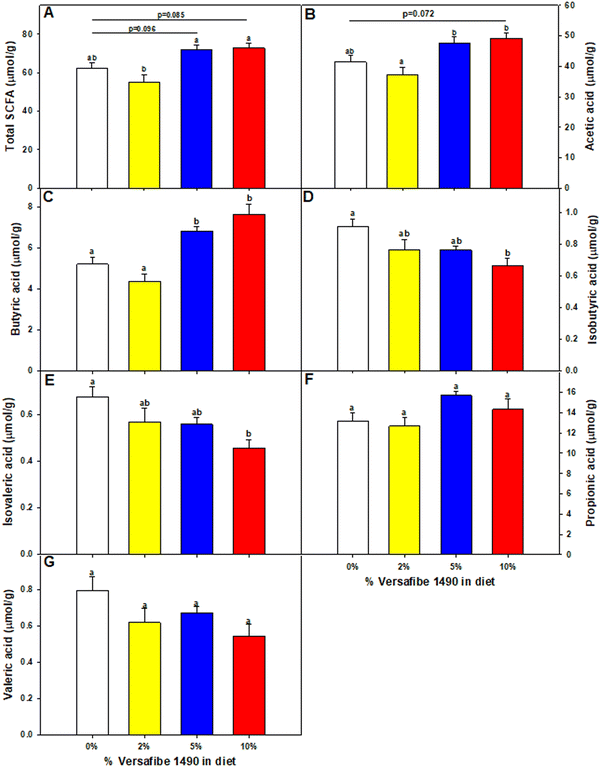
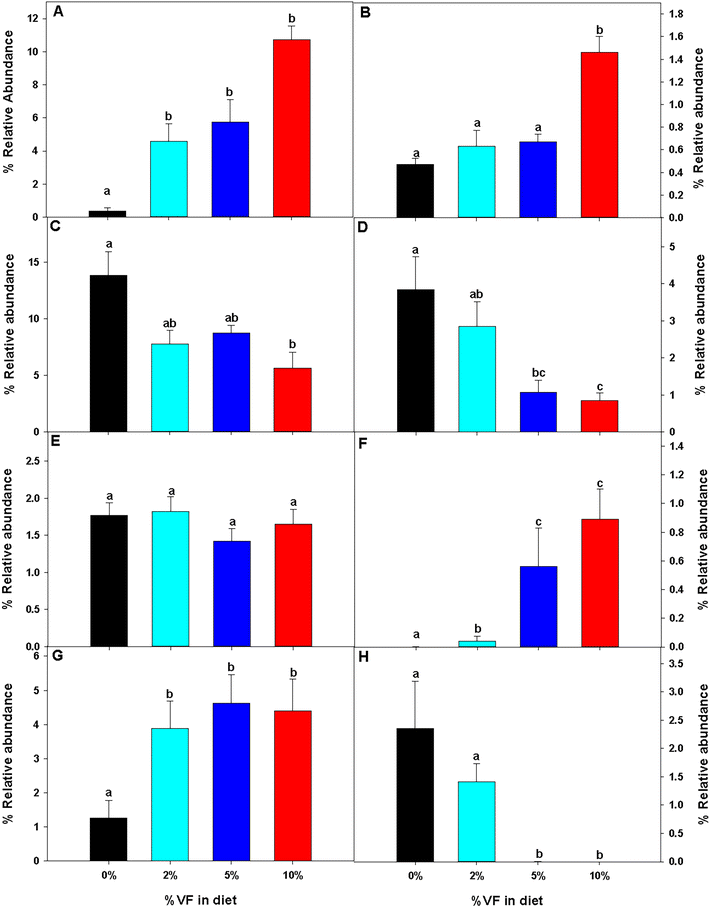
Mice were euthanized and tissues collected for analysis after feeding the TWD for six weeks followed by subsets being fed the TWD/VF containing diets for three weeks. Body weights were not affected by feeding the different levels of VF containing diets compared to mice fed only the TWD (Fig. 1A). Feeding mice, the 10% VF diet, but not the 2% or 5% VF diets led to a significant increase in the % colon/BW (Fig. 1B). There were, however, no differences in the mucosa depth in either the distal or proximal colon (data not shown). The % cecum/BW ratio increased with increasing levels of dietary VF suggesting increased fermentation and/or accumulation of undigested material in mice consuming VF-containing diets (Fig. 1C). Because fermentation of RSs has been shown to increase production of SCFAs,3–6 we measured SCFA production to assess whether consumption of VF was being fermented. Total SCFA and acetic acid levels were significantly lower in the 2% group versus the 5 an 10% groups (Fig. 2a and b). As we saw previously in mice fed RPS, a raw potato starch containing RS2,24 feeding VF resulted in increased butyrate production in mice fed the 5 or 10% VF diets (Fig. 2c). Propionic acid and valeric acid production was not significantly increased over levels observed in TWD fed mice (Fig. 2f and g). Both isobutyric and isovaleric acid levels decreased but were only significantly different in mice fed the 10% VF diet (Fig. 2d and e). We looked to see if fecal pH decreased with increased with consumption of VF. One experiment showed a significant decrease in pH with increasing dietary VF, but two replicate experiments showed no effect suggesting that VF consumption did not result in reproducible effects on fecal pH (data not shown).Effect of VF on the microbiome diversity and composition
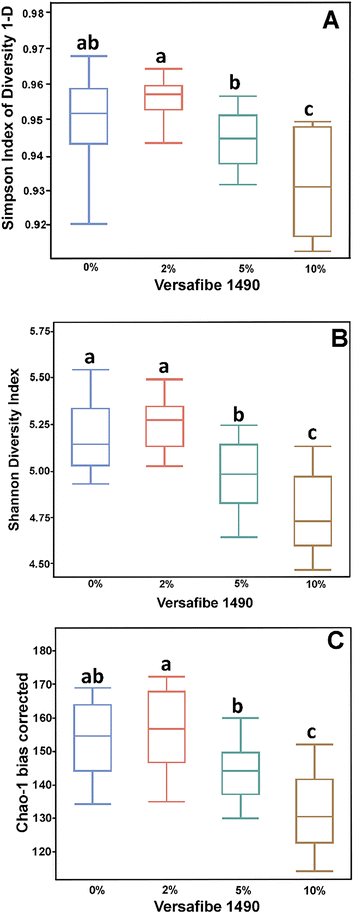
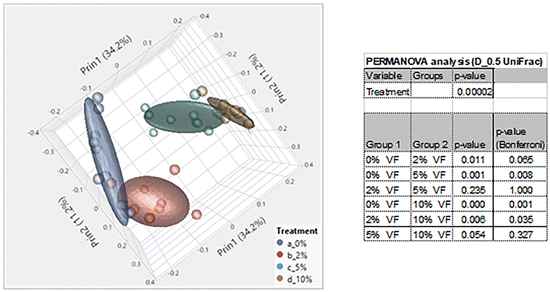
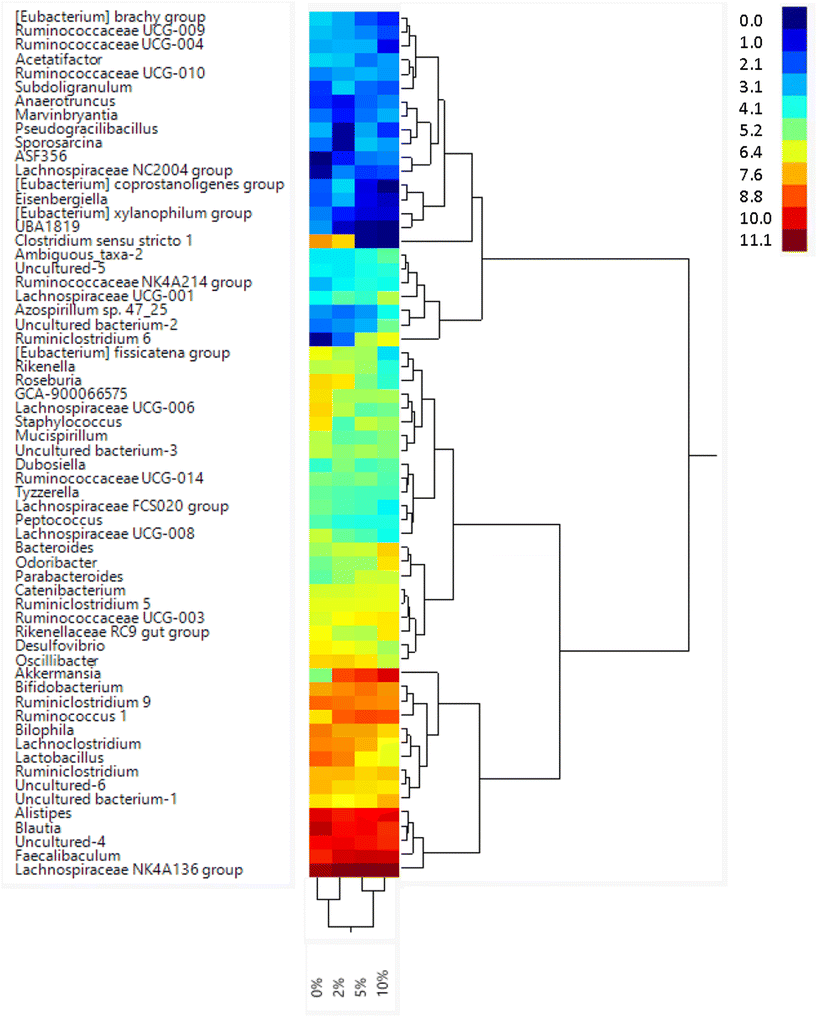
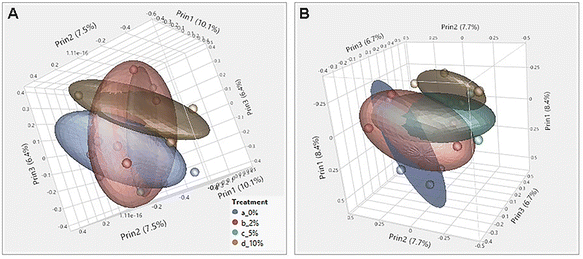
Consumption of DF and RSs can differentially alter the microbiome.10,48 We previously showed that feeding mice RPS had profound effects on the microbiome.24 To investigate the effect of consuming VF, a phosphorylated, cross-linked potato starch, on the microbiome, DNA isolated from cecal contents of mice fed the TWD or TWD/VF diets were subjected to 16S sequencing analysis. Microbiome alpha diversity and richness, as measured by the Shannon and the Chao-1 biased and Simpson indices, showed decreased diversity in mice fed diets containing 10% VF compared to mice fed the basal diet with only the Shannon index showing a significant decrease in alpha diversity for mice fed the 5% VF (Fig. 3); however, no effect of feeding a 2% VF diet on alpha diversity compared to the basal diet was observed. Beta-diversity was analyzed by PERMANOVA (0.5 UniFrac) and is illustrated by a PCA (Pearson product-moment correlation) plot (Fig. 4). The UniFrac analysis which also considers both abundance and phylogenic similarity indicated that the 5% and 10% VF groups clustering more closely together and significantly separated from mice fed the basal diet while mice fed the 2% VF diet were not significantly different from mice fed the basal diet.The relative abundance of various genera with a relative abundance of greater than 0.05% is summarized in a heat map plot shown in Fig. 5 and in ESI Table 2.†Lachnospirecia NK4A136 group was the genus with the highest relative abundance but was not substantially changed by consumption of increasing amounts of VF. Akkermansia levels were increased in mice fed all three levels of the VF diet compared to mice fed the basal diet (ESI Table 2,†Fig. 5). Bacteroides, Azospirillum sp. 47_25, ASF356, Faecalibaculum and Odoribacter were increased only in mice fed the 10% VF diet while Ruminiclostridium 6 and Ruminococcus 1 were increased in mice fed all three levels of VF suggesting that they were particularly sensitive to dietary VF levels (ESI Table 2,†Fig. 5). Bifidobacterium and Alistipes, both present at substantial levels, were unaffected by feeding VF. Numerous genera had significant decreases in relative abundance in response to feeding 10% VF including Bilophila, Blautia, Eisenbergiella, Lachnoclostridium, Lactobacillus, Oscillibacter, Rikenella, Roseburia, members of the Eubacterium and Clostridium senso stricto 1 being particularly sensitive to feeding of VF with the abundance decreasing from 2.4% to undetectable levels. Changes in select genera are shown in Fig. 6.
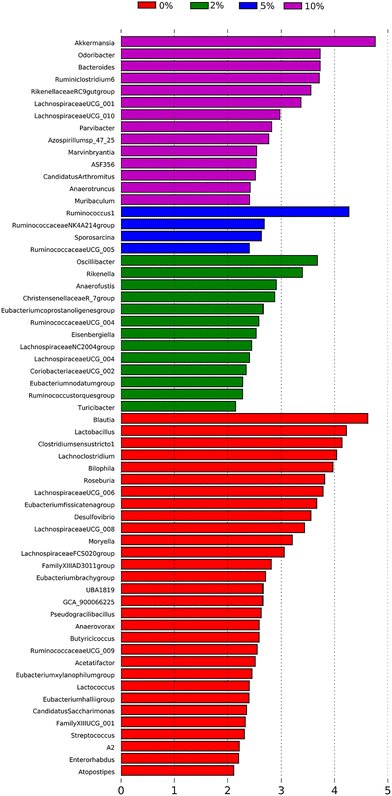
To identify bacterial genera that discriminate between consumption of different levels of dietary VF, a Lefse plot was generated for Genera taxa (Fig. 7). The all-group LefSe comparison showed that Akkermansia and Odoribacter and Bacteroides, all significantly increased by feeding 10% VF, were the top three discriminating genera. Only four genera were discriminating for mice fed a 5% VF diet and both Ruminococcus 1 and Ruminococcaceae NK4A214 group were significantly increased in mice fed either the 5% or 10% VF diets. For mice fed the 2% VF diet, Oscillibacter and Rikenella were discriminating, and their relative abundance was unchanged compared to mice fed the basal diet but was decreased in mice fed the 10% VF diet. Blautia, Lactobacillus, Clostridium senso stricto 1, Lachnoclostridium, Bilophila and Roseburia were discriminating for mice fed the basal diet and all were significantly decreased in mice fed the 10% VF diet.
Effect of VF on gene expression in the cecum and DC
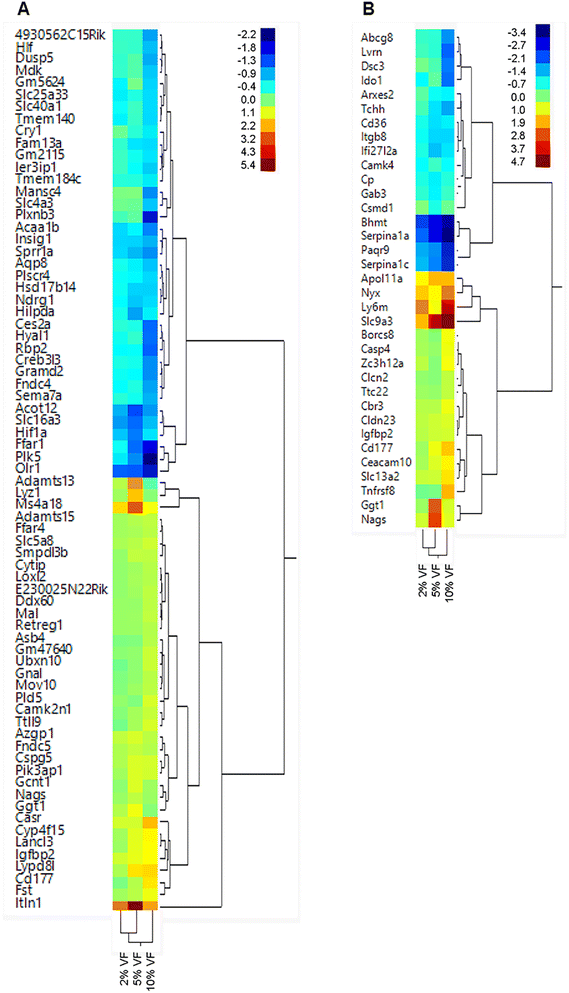
Feeding increasing levels of VF did not lead to significant separation of the dietary groups in a PCA plot for the cecum or distal colon Fig. 8. However, in the cecum of VF fed mice, we observed a significant change (FDR adjusted p < 0.05, fold change <−1.5 or >+1.5) in the expression of 2, 24 and 65 genes in the 2, 5 and 10% VF groups versus control, respectively (Fig. 9A, ESI Tables 3 and 4†). In the 2% group, both genes were downregulated. In the 5% group 10 genes were downregulated and 14 were upregulated. The most highly upregulated gene was Intelectin 1 (Itln1), an Il-13 induced adipocytokine and pattern recognition receptor.49 Lysozyme 1 (Lyz1), an enzyme involved in antibacterial responses, was induced 3.9-fold. N-Acetylglutamate synthase (Nags), a protein involved in arginine metabolism50 was also upregulated 1.6-fold. Among the downregulated genes were, solute carrier family 16, member 3 (Slc16a3), a lactate and pyruvate transporter51 and aquaporin 8 (Aqp8), a water transporter.52 In the 10% group, 36 genes were downregulated and 29 upregulated. Among the upregulated genes were 3 interferon-induced genes (myelin and lymphocyte protein); T-cell differentiation protein (Mal), Mov10 RNA helicase (Mov10), DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (Ddx60), involved in antiviral responses.53,54In the distal colon we observed a significant change (FDR adjusted p < 0.05, fold change <−1.5 or >+1.5) in the expression of 0, 9 and 33 genes in the 2, 5 and 10% VF groups versus control, respectively (Fig. 9B, ESI Tables 3 and 5†). In the 5% group, 6 genes were downregulated and 3 were upregulated. The most highly upregulated gene was Nags (7.7-fold). The most highly downregulated (−1.9-fold) gene was ceruloplasmin (Cp), a copper transporter and positive acute phase reactant.55 Another downregulated gene was Interferon, alpha-inducible protein 27 like 2A (Ifi27l2a), an interferon induced, adipocytokine.56,57 In the 10% group, 15 genes were downregulated and 16 upregulated. The most highly upregulated gene (25.6-fold) was solute carrier family 9 (sodium/hydrogen exchanger), member 3 (Slc9a3) an interleukin-13 induced,58 chloride ion transporter59 involved in the production of mucus.60 Solute carrier family 13 (sodium-dependent citrate transporter), member (Slc13a2), a transporter for citrate and succinate61 was modestly induced (2.7-fold). Another gene that was modestly induced was Claudin 23 (Cldn23) (1.8-fold) a member of the Claudin family and has previously shown to be upregulated by butyrate in mice and humans.62 The most interesting gene that was down regulated was indoleamine–pyrrole 2;3 dioxygenase (Ido1) a macrophage expressed, interferon gamma-induced protein63 that is anti-inflammatory due to its ability to degrade tryptophan. Several genes involved in lipid transport or metabolism (ATP-binding cassette sub-family G member 8 (Abcg8), CD36 blood group (Cd36), apolipoprotein B, Apob) and several serine protease inhibitor 1 paralogs (Serpina1a, Serpina1c) were also among the downregulated genes. The human ortholog of Serpina1a, SERPINA1, is a positive acute phase reactant and in mice has broad anti-inflammatory activities.64
Discussion
The results presented here demonstrate that mice fed VF, a phosphorylated and crosslinked potato starch, in the context of a basal diet that emulates a typical American diet for both macro and micronutrients,23 had a significant effect on the microbiome. This approach differs from many RS studies that employ unrealistic high fat, high sucrose diets that do not mimic a typical American diet. The levels of RS4 consumed by the mice is approximately equivalent to a human consuming between 4 and 20 g of RS per day based on the energy needs conversion factor for mice to humans and average feed consumption for mice consuming between 2–10% RPS.65 Alpha-diversity was significantly decreased in mice fed the 10% VF diet, but the 5% VF group was only significantly different from the basal group in the Shannon diversity plot and the 2% group was not different from the basal diet in all three alpha diversity plots. We and others have also shown that feeding RS can decrease alpha diversity in rodents.6,17,24,66–68 Feeding pigs diets containing RS2 including RPS also showed decreased alpha-diversity in the colon but not the cecum.69,70 Similar results were observed in pigs fed a diet containing RS3.71 However, our results with VF only partially recapitulated what we found with RPS, where decreased alpha diversity was more pronounced and observed at the 2%, 5%, and 10% levels. Similarly, VF had a less pronounced effect on beta diversity compared to RPS with differences only observed in mice fed the 5% and 10% VF diets compared to the basal diet. The 2% VF diet had no effect on beta diversity whereas the 2% RPS diet caused a significant change in beta-diversity compared to the basal diet. Together these results suggest that phosphorylating and crosslinking the potato starch altered its ability to affect changes to the diversity of the microbiome as compared to RPS.SCFA production was altered in VF fed mice. As was observed in our previous study using RPS,24 there were no differences in total, or propionic acid levels but butyrate levels were increased in the 5% and 10% VF groups. Changes in isobutyric and isovaleric acid were less pronounced in mice fed the VF diets than we observed with RPS with only the 10% VF group significantly reduced compared to the basal diet but are consistent with a shift to saccharolytic fermentation and a decrease in proteolytic fermentation (branched-chain SCFAs).72,73 In contrast to the previously observed dose dependent RPS induced decrease in valeric acid production, VF had no effect on valeric acid levels. These results indicate that phosphorylating and crosslinking potato starch changed but did not eliminate its ability to be fermented by commensal bacteria and alter SCFA production. Others have also reported increased butyrate production in response to feeding RS to rodents.6,74,75 These results are different than what was observed in rats38 and humans37 fed VF where no changes in SCFA production was observed. The reasons for these differences are not clear but it is known that the composition of the basal commensal microbiota can affect its ability to digest RS and other compounds.76
Consumption of resistant starches has also been associated with a decrease in fecal pH6,75 which can favor butyrate forming taxa.77 Feeding RPS to mice resulted in a dose dependent decrease in fecal pH.24 This was not reproducibly observed in mice fed VF with only one of three experiments showing decreasing fecal pH with increasing dietary VF. A lack of fecal pH change was also seen in rats fed VF38 and contrasts with results obtained with other RS where decreased fecal or cecal pH was observed.6,38,75 We and others have reported that feeding RPS to mice can result in an increase in colon and cecum weight.24,78,79 Similar effects were observed with VF. A modest increase in colon/body weight ratio was observed only in mice fed the 10% VF diet but no effect was observed on mucosa depth (data not shown). Cecum weight increased with increasing dietary VF reflecting the increased levels of undigested material reaching the cecum for subsequent fermentation by commensal bacteria. Other have also observed decreased fecal pH6,75 and enlargement of the cecum in rodents fed RS5,16,17,66 including VF.38
Feeding VF led to specific changes to the microbiota at the genus level. These changes were mostly observed in mice fed 5% or 10% VF diets. About equal numbers of genera increased or decreased in relative abundance at each dietary level of VF. This contrasts to what we observed with RPS where only a few genera increased in abundance in response to feeding RPS. There were about 4× and 2× as many changes in mice fed the 10% VF diet than was seen in mice fed the 2% or 5% VF diets, respectively. Of particular note, we previously observed that the Lachnospiraceae NK4A136 group went from approximately 10% to 50% relative abundance in a graded manner in response to increasing dietary RPS,24 but this large increase was not observed when mice were fed VF suggesting that this genus cannot utilize VF to the same extent as RPS. Also of note, there was an increase in Akkermansia in response to feeding any level of VF. Bifidobacterium, considered a beneficial commensal, did not increase in response to feeding VF although there was a significant increase in Bacteroides. There were multiple genera that decreased in abundance including Bilophila, Blautia, Clostridium sensu stricto 1, Lactobacillus, Rikenella, and Roseburia that were also reduced in mice fed RPS.24 While Ruminococcus 1, Ruminiclostridium 5 and 9, and Alistipes decreased in response to feeding RPS, Ruminococcus 1 and Ruminiclostridium 6 increased but other Ruminiclostridium and Ruminococcaceae genera decreased or remained unchanged in mice fed VF. In toto, these results suggest that commensal bacteria responded very differently to feeding a phosphorylated, crosslinked potato starch compared to our previous results using an unmodified potato starch.24
The Lachnospiraceae and Ruminococcaceae families contain the largest number of butyrate producers.80 Other genera can also contribute to butyrate production including Bacteroides, Faecalibaculum, and Ruminococcus 1 all of which increased in response to feeding VF. Akkermansia is a known mucin degrader81 that can produce metabolites that cross-feed other commensal strains leading to increased SCFA production82 that may have also contributed to the increased butyrate production observed with feeding VF. Ruminiclostridium 6 expression was extremely low in TWD fed mice and increased markedly upon feeding VF (but not RPS) suggesting that that members of this genus can metabolize the phosphorylated cross-linked starch and have a growth advantage. Members of this genus are known to digest a variety of substrates including cellulose, hemicellulose and other complex carbohydrates.83Ruminococcus 1 abundance was also increased by feeding VF and is another genus associated with degradation of starches.84,85 Another member of this genus, Ruminococcus bromii is considered to be a primary RS degrader in humans.86R. bromii contain enzyme complexes called amylosomes, that can bind to RS granules via carbohydrate binding domains and digest the RS releasing products that can subsequently be acted upon by secondary degrader such as B. thetaiotaomicron and Eubacterium rectale.87
RS4s have been produced from a variety of starting materials including corn, wheat, tapioca and potatoes and by different methods. The ability of these different RS4s to modulate SCFA production and the microbiota differs. Deehan et al., (2020)37 demonstrated that crystalline maize and cross-linked tapioca RS4s were able to affect SCFA production and the microbiota in humans while crosslinked potato starch (VF) did not, suggesting that VF was not a fermentable substrate. This contrasts with a recent study where VF altered the microbiome of human subjects88 and results from in vitro human fecal cultures where VF as well as crystalline maize and cross-linked tapioca were all fermented to varying degrees.39 Our results indicate that feeding VF to mice fed a western-style diet altered both SCFA production and the microbiome while feeding rats VF did not alter SCFA production or change cecal pH while crystalline maize and cross-linked tapioca RS4s did.38 Other RS4s have also been tested. An in vitro study using human feces showed fermentation of RS4s derived from wheat (Fibersym RW™) and potatoes (Fibersym 80ST).89 There are, however, conflicting results for human studies using the RS4, Fibersym RW, with one study finding no effect of Fibersym RW on the metaproteome90 while two studies showed significant changes to the microbiome of subjects fed Fibersym RW91,92 as did a study in mice.93 Together these results suggest that the host response to feeding VF and other RS4s can vary substantially. These conflicting results could be due to differences in the basal microbiome composition. Interindividual differences in human microbiota are well documented and treatment induced changes to the microbiome can often exceed intraindividual differences due to treatment.94 SCFA production in humans in response to RS consumption can depend on whether specific bacterial strains are present.76 Additional studies are necessary to gain a better understanding of how the basal microbiome affects subsequent responses to feeding different types of RS.
Global gene expression in the cecum and colon were analyzed. PCA plots of the cecum and colon did show separation of mice fed the basal vs. the 10% VF diet but there was not clear separation of the different VF groups as we had observed with mice fed RPS.24 There was a dramatic decrease in the total number of gene expression changes in mice fed VF in both the cecum and colon compared to our results with RPS. While, in general, mice fed 10% RPS diet had more genes upregulated in the cecum than downregulated,24 the number of up- and downregulated genes in mice fed the 5% or 10% VF diet were similar. A number of genes associated with the immune response were altered by consuming VF as was observed with RPS. The reasons for the differences in gene expression between this study and our previous study with RPS are not clear but may be related to the observed differences in the effect of RPS vs. VF on the microbiome and/or their metabolites. Future studies comparing the effect of consumption of RPS vs. VF on the metabolome may shed light on this. Alternatively, different RS may have other non-microbial dependent effects on gene expression.
Conclusions
Previous studies by others have provided mixed results on the ability of various RS4s to be fermented. Of note were studies where feeding VF to humans, rats or in vitro culture systems gave conflicting results as to the fermentability of VF. Here we showed that mice are capable of fermenting VF as demonstrated by increased butyrate and reduced branch-chain SCFA production, as well as changes to the cecal microbiome and gene expression in the cecum and distal colon. Gene expression changes were more prevalent in the cecum and in mice fed the 10% VF diet. The number of changes, however, were substantially lower than we previously observed in mice fed RPS. VF induced changes to the microbiome were also different than we observed with RPS providing additional evidence that the structure of the RS is a major factor determining its effects on the microbiome and gene expression in the cecum and colon.Abbreviations
| DC | Distal colon |
| DF | Dietary fiber |
| EC | Epithelial cell |
| HFD | High fat diet |
| HAMS | High-amylose maize starch |
| LEfSe | Linear discriminant analysis effect size |
| Nags | N-Acetylglutamate synthase |
| NHANES | National Health and Nutrition Examination Survey |
| PC | Proximal colon |
| RS | Resistant starch |
| RPS | Resistant potato starch |
| SCFA | Short chain fatty acids |
| Serpina | Serpin family A member |
| Slc | Solute carrier family |
| TWD | Total western diet |
| VF | Versafibe 1490™ |
Institutional review board statement
The animal study protocol was approved by the by the USDA/ARS Beltsville Area Institutional Animal Care and Use Committee (Protocol 22-04).Author contributions
AS and HD conceived and designed this experiment and supervised all experimental works. AS, CC, LC, RW, BSJ, EP and HD performed experiments. AS, CC, RW, EP and HD performed data analysis. AS, CC, and HD drafted the manuscript. AS, CC, RW, and HD helped to revise the manuscript. All authors have read and approved the final manuscript.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by USDA ARS project 8040-53000-021-000D.References
- M. G. Sajilata, R. S. Singhal and P. R. Kulkarni, Resistant starch - A review, Compr. Rev. Food Sci. Food Saf., 2006, 5, 1–17 CrossRef CAS PubMed.
- M. M. Murphy, J. S. Douglass and A. Birkett, Resistant starch intakes in the United States, J. Am. Diet. Assoc., 2008, 108, 67–78 CrossRef PubMed.
- K. Lange, F. Hugenholtz, M. C. Jonathan, H. A. Schols, M. Kleerebezem, H. Smidt, M. Müller and G. J. Hooiveld, Comparison of the effects of five dietary fibers on mucosal transcriptional profiles, and luminal microbiota composition and SCFA concentrations in murine colon, Mol. Nutr. Food Res., 2015, 59, 1590–1602 CrossRef CAS PubMed.
- A. Kaur, T. Chen, S. J. Green, E. Mutlu, B. R. Martin, P. Rumpagaporn, J. A. Patterson, A. Keshavarzian and B. R. Hamaker, Physical Inaccessibility of a Resistant Starch Shifts Mouse Gut Microbiota to Butyrogenic Firmicutes, Mol. Nutr. Food Res., 2019, 63, e1801012 CrossRef PubMed.
- R. Nagata, N. Innami, S. Pelpolage, K. Shimada, H. Koaze, M. Tani, K. H. Han and M. Fukushima, Effects of Raw Potato Starch with High Resistant Starch Levels on Cecal Fermentation Properties in Rats, J. Nutr. Sci. Vitaminol., 2019, 65, S192–s195 CrossRef PubMed.
- Y. Hu, R. K. Le Leu, C. T. Christophersen, R. Somashekar, M. A. Conlon, X. Q. Meng, J. M. Winter, R. J. Woodman, R. McKinnon and G. P. Young, Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats, Carcinogenesis, 2016, 37, 366–375 CrossRef CAS PubMed.
- J. M. Heo, A. K. Agyekum, Y. L. Yin, T. C. Rideout and C. M. Nyachoti, Feeding a diet containing resistant potato starch influences gastrointestinal tract traits and growth performance of weaned pigs, J. Anim. Sci., 2014, 92, 3906–3913 CrossRef CAS PubMed.
- D. Haenen, J. Zhang, C. Souza da Silva, G. Bosch, I. M. van der Meer, J. van Arkel, J. J. van den Borne, O. Pérez Gutiérrez, H. Smidt, B. Kemp, M. Müller and G. J. Hooiveld, A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine, J. Nutr., 2013, 143, 274–283 CrossRef CAS PubMed.
- M. S. Hedemann and K. E. Bach Knudsen, Resistant starch for weaning pigs—Effect on concentration of short chain fatty acids in digesta and intestinal morphology, Livest. Sci., 2007, 108, 175–177 CrossRef.
- X. Yang, K. O. Darko, Y. Huang, C. He, H. Yang, S. He, J. Li, J. Li, B. Hocher and Y. Yin, Resistant Starch Regulates Gut Microbiota: Structure, Biochemistry and Cell Signalling, Cell. Physiol. Biochem., 2017, 42, 306–318 CrossRef CAS PubMed.
- R. L. Hughes, W. H. Horn, P. Finnegan, J. W. Newman, M. L. Marco, N. L. Keim and M. E. Kable, Resistant Starch Type 2 from Wheat Reduces Postprandial Glycemic Response with Concurrent Alterations in Gut Microbiota Composition, Nutrients, 2021, 13, 645–664 CrossRef CAS PubMed.
- J. H. Cummings, E. W. Pomare, W. J. Branch, C. P. Naylor and G. T. Macfarlane, Short chain fatty acids in human large intestine, portal, hepatic and venous blood, Gut, 1987, 28, 1221–1227 CrossRef CAS PubMed.
- J. M. Wong, R. de Souza, C. W. Kendall, A. Emam and D. J. Jenkins, Colonic health: fermentation and short chain fatty acids, J. Clin. Gastroenterol., 2006, 40, 235–243 CrossRef CAS PubMed.
- M. Benmoussa, K. A. Moldenhauer and B. R. Hamaker, Rice amylopectin fine structure variability affects starch digestion properties, J. Agric. Food Chem., 2007, 55, 1475–1479 CrossRef CAS PubMed.
- B. R. Hamaker and Y. E. Tuncil, A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota, J. Mol. Biol., 2014, 426, 3838–3850 CrossRef CAS PubMed.
- D. A. Kieffer, B. D. Piccolo, M. L. Marco, E. B. Kim, M. L. Goodson, M. J. Keenan, T. N. Dunn, K. E. Knudsen, R. J. Martin and S. H. Adams, Mice Fed a High-Fat Diet Supplemented with Resistant Starch Display Marked Shifts in the Liver Metabolome Concurrent with Altered Gut Bacteria, J. Nutr., 2016, 146, 2476–2490 CrossRef CAS PubMed.
- J. Barouei, Z. Bendiks, A. Martinic, D. Mishchuk, D. Heeney, Y. H. Hsieh, D. Kieffer, J. Zaragoza, R. Martin, C. Slupsky and M. L. Marco, Microbiota, metabolome, and immune alterations in obese mice fed a high-fat diet containing type 2 resistant starch, Mol. Nutr. Food Res., 2017, 61 DOI:10.1002/mnfr.201700184.
- D. Chang, X. Hu and Z. Ma, Pea-Resistant Starch with Different Multi-scale Structural Features Attenuates the Obesity-Related Physiological Changes in High-Fat Diet Mice, J. Agric. Food Chem., 2022, 70, 11377–11390 CrossRef CAS PubMed.
- C. P. Rosado, V. Rosa, B. Martins, A. Soares, A. Almo, E. Monteiro, A. Mulder, N. Moura-Nunes and J. B. Daleprane, Green banana flour supplementation improves obesity-associated systemic inflammation and regulates gut microbiota profile in high-fat diet-fed mice, Appl. Physiol., Nutr., Metab., 2021, 46, 1469–1475 CrossRef CAS PubMed.
- A. D. Benninghoff, K. J. Hintze, S. P. Monsanto, D. M. Rodriguez, A. H. Hunter, S. Phatak, J. J. Pestka, A. J. V. Wettere and R. E. Ward, Consumption of the Total Western Diet Promotes Colitis and Inflammation-Associated Colorectal Cancer in Mice, Nutrients, 2020, 12, 544–578 CrossRef CAS PubMed.
- D. M. Rodriguez, K. J. Hintze, G. Rompato, A. J. V. Wettere, R. E. Ward, S. Phatak, C. Neal, T. Armbrust, E. C. Stewart, A. J. Thomas and A. D. Benninghoff, Dietary Supplementation with Black Raspberries Altered the Gut Microbiome Composition in a Mouse Model of Colitis-Associated Colorectal Cancer, although with Differing Effects for a Healthy versus a Western Basal Diet, Nutrients, 2022, 14, 5270–5303 CrossRef CAS PubMed.
- R. E. Ward, A. D. Benninghoff, B. J. Healy, M. Li, B. Vagu and K. J. Hintze, Consumption of the total Western diet differentially affects the response to green tea in rodent models of chronic disease compared to the AIN93G diet, Mol. Nutr. Food Res., 2017, 61 DOI:10.1002/mnfr.201600720.
- K. J. Hintze, A. D. Benninghoff and R. E. Ward, Formulation of the Total Western Diet (TWD) as a basal diet for rodent cancer studies, J. Agric. Food Chem., 2012, 60, 6736–6742 CrossRef CAS PubMed.
- A. D. Smith, C. Chen, L. Cheung, R. Ward, K. J. Hintze and H. D. Dawson, Resistant Potato Starch Alters the Cecal Microbiome and Gene Expression in Mice Fed a Western Diet Based on NHANES Data, Front. Nutr., 2022, 9, 782667 CrossRef PubMed.
- A. D. Smith, C. Chen, L. Cheung and H. D. Dawson, Raw potato starch alters the microbiome, colon and cecal gene expression, and resistance to Citrobacter rodentium infection in mice fed a Western diet, Front. Nutr., 2022, 9, 1057318 CrossRef PubMed.
- N. K. Petty, R. Bulgin, V. F. Crepin, A. M. Cerdeno-Tarraga, G. N. Schroeder, M. A. Quail, N. Lennard, C. Corton, A. Barron, L. Clark, A. L. Toribio, J. Parkhill, G. Dougan, G. Frankel and N. R. Thomson, The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli, J. Bacteriol., 2010, 192, 525–538 CrossRef CAS PubMed.
- A. P. Gobert, K. T. Wilson and C. Martin, Cellular responses to attaching and effacing bacteria: activation and implication of the innate immune system, Arch. Immunol. Ther. Exp., 2005, 53, 234–244 CAS.
- J. W. Collins, K. M. Keeney, V. F. Crepin, V. A. Rathinam, K. A. Fitzgerald, B. B. Finlay and G. Frankel, Citrobacter rodentium: infection, inflammation and the microbiota, Nat. Rev. Microbiol., 2014, 12, 612–623 CrossRef CAS PubMed.
- M. L. Stewart, M. L. Wilcox, M. Bell, M. A. Buggia and K. C. Maki, Type-4 Resistant Starch in Substitution for Available Carbohydrate Reduces Postprandial Glycemic Response and Hunger in Acute, Randomized, Double-Blind, Controlled Study, Nutrients, 2018, 10, 129–140 CrossRef PubMed.
- T. J. Steele, C. C. Steele, C. C. Maningat, P. A. Seib, M. D. Haub and S. K. Rosenkranz, Glycemic and Insulinemic Responses of Healthy Humans to a Nutrition Bar with or without Added Fibersym(®) RW, a Cross-Linked Phosphorylated RS4-Type Resistant Wheat Starch, Int. J. Environ. Res. Public Health, 2022, 19, 13804–13814 CrossRef CAS PubMed.
- C. Li, W. Yu and R. G. Gilbert, The Effects of Starch Molecular Fine Structure on Thermal and Digestion Properties of Rice Starch, Foods, 2022, 11, 4012–4028 CrossRef CAS PubMed.
- E. Mah, V. Garcia-Campayo and D. Liska, Substitution of Corn Starch with Resistant Starch Type 4 in a Breakfast Bar Decreases Postprandial Glucose and Insulin Responses: A Randomized, Controlled, Crossover Study, Curr. Dev. Nutr., 2018, 2, nzy066 CrossRef PubMed.
- T. J. Steele, C. C. Maningat, P. A. Seib, M. D. Haub and S. K. Rosenkranz, Metabolic Responses to Native Wheat Starch (Midsol(TM) 50) versus Resistant Wheat Starch Type 4 (Fibersym(®) RW): Standard versus Marketplace Testing Protocols, Curr. Dev. Nutr., 2021, 5, nzab011 CrossRef CAS PubMed.
- V. Gourineni, M. L. Stewart, M. L. Wilcox and K. C. Maki, Nutritional Bar with Potato-Based Resistant Starch Attenuated Post-Prandial Glucose and Insulin Response in Healthy Adults, Foods, 2020, 9, 1679 CrossRef CAS PubMed.
- M. L. Stewart and J. P. Zimmer, A High Fiber Cookie Made with Resistant Starch Type 4 Reduces Post-Prandial Glucose and Insulin Responses in Healthy Adults, Nutrients, 2017, 9, 237–253 CrossRef PubMed.
- Y. Du, Y. Wu, D. Xiao, G. Guzman, M. L. Stewart, V. Gourineni, B. Burton-Freeman and I. Edirisinghe, Food prototype containing resistant starch type 4 on postprandial glycemic response in healthy adults, Food Funct., 2020, 11, 2231–2237 RSC.
- E. C. Deehan, C. Yang, M. E. Perez-Munoz, N. K. Nguyen, C. C. Cheng, L. Triador, Z. Zhang, J. A. Bakal and J. Walter, Precision Microbiome Modulation with Discrete Dietary Fiber Structures Directs Short-Chain Fatty Acid Production, Cell Host Microbe, 2020, 27, 389–404 CrossRef CAS PubMed.
- D. B. Coulon, R. Page, A. M. Raggio, J. Guice, B. Marx, V. Gourineni, M. L. Stewart and M. J. Keenan, Novel Resistant Starch Type 4 Products of Different Starch Origins, Production Methods, and Amounts Are Not Equally Fermented when Fed to Sprague-Dawley Rats, Mol. Nutr. Food Res., 2020, 64, e1900901 CrossRef PubMed.
- J. M. Erickson, J. L. Carlson, M. L. Stewart and J. L. Slavin, Fermentability of Novel Type-4 Resistant Starches in In Vitro System, Foods, 2018, 7, 18–31 CrossRef PubMed.
- J. G. Caporaso, C. L. Lauber, W. A. Walters, D. Berg-Lyons, C. A. Lozupone, P. J. Turnbaugh, N. Fierer and R. Knight, Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(Suppl 1), 4516–4522 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2012, 41, D590–D596 CrossRef PubMed.
- R. C. Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 2004, 32, 1792–1797 CrossRef CAS PubMed.
- M. J. Anderson, A new method for non–parametric multivariate analysis of variance, Aust. Ecol., 2001, 26, 32–46 Search PubMed.
- N. Segata, J. Izard, L. Waldron, D. Gevers, L. Miropolsky, W. S. Garrett and C. Huttenhower, Metagenomic biomarker discovery and explanation, Genome Biol., 2011, 12, R60 CrossRef PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- H. D. Dawson, C. Chen, B. Gaynor, J. Shao and J. F. Urban Jr., The porcine translational research database: a manually curated, genomics and proteomics-based research resource, BMC Genomics, 2017, 18, 643 CrossRef PubMed.
- H. D. Dawson, C. Chen, R. W. Li, L. N. Bell, T. Shea-Donohue, H. Kringel, E. Beshah, D. E. Hill and J. F. Urban Jr., Molecular and metabolomic changes in the proximal colon of pigs infected with Trichuris suis, Sci. Rep., 2020, 10, 12853 CrossRef CAS PubMed.
- Z. A. Bendiks, K. E. B. Knudsen, M. J. Keenan and M. L. Marco, Conserved and variable responses of the gut microbiome to resistant starch type 2, Nutr. Res., 2020, 77, 12–28 CrossRef CAS PubMed.
- R. Datta, M. L. deSchoolmeester, C. Hedeler, N. W. Paton, A. M. Brass and K. J. Else, Identification of novel genes in intestinal tissue that are regulated after infection with an intestinal nematode parasite, Infect. Immun., 2005, 73, 4025–4033 CrossRef CAS PubMed.
- M. Geng, T. Li, X. Kong, X. Song, W. Chu, R. Huang, Y. Yin and G. Wu, Reduced expression of intestinal N-acetylglutamate synthase in suckling piglets: a novel molecular mechanism for arginine as a nutritionally essential amino acid for neonates, Amino Acids, 2011, 40, 1513–1522 CrossRef CAS PubMed.
- A. P. Halestrap and D. Meredith, The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond, Pfluegers Arch., 2004, 447, 619–628 CrossRef CAS PubMed.
- M. Agemark, J. Kowal, W. Kukulski, K. Nordén, N. Gustavsson, U. Johanson, A. Engel and P. Kjellbom, Reconstitution of water channel function and 2D-crystallization of human aquaporin 8, Biochim. Biophys. Acta, 2012, 1818, 839–850 CrossRef CAS PubMed.
- J. Zhang, F. Huang, L. Tan, C. Bai, B. Chen, J. Liu, J. Liang, C. Liu, S. Zhang, G. Lu, Y. Chen and H. Zhang, Host Protein Moloney Leukemia Virus 10 (MOV10) Acts as a Restriction Factor of Influenza A Virus by Inhibiting the Nuclear Import of the Viral Nucleoprotein, J. Virol., 2016, 90, 3966–3980 CrossRef CAS PubMed.
- A. Fullam and M. Schröder, DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication, Biochim. Biophys. Acta, 2013, 1829, 854–865 CrossRef CAS PubMed.
- P. D. Eckersall, P. K. Saini and C. McComb, The acute phase response of acid soluble glycoprotein, alpha(1)-acid glycoprotein, ceruloplasmin, haptoglobin and C-reactive protein, in the pig, Vet. Immunol. Immunopathol., 1996, 51, 377–385 CrossRef CAS PubMed.
- N. Parker and A. C. Porter, Identification of a novel gene family that includes the interferon-inducible human genes 6–16 and ISG12, BMC Genomics, 2004, 5, 8 CrossRef PubMed.
- S. Ohgaki, K. Iida, T. Yokoo, K. Watanabe, R. Kihara, H. Suzuki, H. Shimano, H. Toyoshima and N. Yamada, Identification of ISG12b as a putative interferon-inducible adipocytokine which is highly expressed in white adipose tissue, J. Atheroscler. Thromb., 2007, 14, 179–184 CrossRef CAS PubMed.
- C. Zeng, S. Vanoni, D. Wu, J. M. Caldwell, J. C. Wheeler, K. Arora, T. K. Noah, L. Waggoner, J. A. Besse, A. N. Yamani, J. Uddin, M. Rochman, T. Wen, M. Chehade, M. H. Collins, V. A. Mukkada, P. E. Putnam, A. P. Naren, M. E. Rothenberg and S. P. Hogan, Solute carrier family 9, subfamily A, member 3 (SLC9A3)/sodium-hydrogen exchanger member 3 (NHE3) dysregulation and dilated intercellular spaces in patients with eosinophilic esophagitis, J. Allergy Clin. Immunol., 2018, 142, 1843–1855 CrossRef CAS PubMed.
- G. Lamprecht, J. Schaefer, K. Dietz and M. Gregor, Chloride and bicarbonate have similar affinities to the intestinal anion exchanger DRA (down regulated in adenoma), Pfluegers Arch., 2006, 452, 307–315 CrossRef CAS PubMed.
- F. Xiao, Q. Yu, J. Li, M. E. Johansson, A. K. Singh, W. Xia, B. Riederer, R. Engelhardt, M. Montrose, M. Soleimani, D. A. Tian, G. Xu, G. C. Hansson and U. Seidler, Slc26a3 deficiency is associated with loss of colonic HCO3 (-) secretion, absence of a firm mucus layer and barrier impairment in mice, Acta Physiol., 2014, 211, 161–175 CrossRef CAS PubMed.
- A. M. Pajor and N. N. Sun, Single nucleotide polymorphisms in the human Na+-dicarboxylate cotransporter affect transport activity and protein expression, Am. J. Physiol.: Renal Physiol., 2010, 299, F704–F711 CrossRef CAS PubMed.
- W. Xu, Y. Ishii, D. M. Rini, Y. Yamamoto and T. Suzuki, Microbial metabolite n-butyrate upregulates intestinal claudin-23 expression through SP1 and AMPK pathways in mouse colon and human intestinal Caco-2 cells, Life Sci., 2023, 329, 121952 CrossRef CAS PubMed.
- W. Dai and S. L. Gupta, Molecular cloning, sequencing and expression of human interferon-gamma-inducible indoleamine 2,3-dioxygenase cDNA, Biochem. Biophys. Res. Commun., 1990, 168, 1–8 CrossRef CAS PubMed.
- S. H. Meghadri, B. Martinez-Delgado, L. Ostermann, G. Gomez-Mariano, S. Perez-Luz, S. Tumpara, S. Wrenger, D. S. DeLuca, U. A. Maus, T. Welte and S. Janciauskiene, Loss of Serpina1 in Mice Leads to Altered Gene Expression in Inflammatory and Metabolic Pathways, Int. J. Mol. Sci., 2022, 23, 10425–10435 CrossRef CAS PubMed.
- J. Whelan and J. Whelan, Conversion of dietary polyunsaturated fats between humans and rodents: A review of allometric scaling models, Prostaglandins, Leukotrienes Essent. Fatty Acids, 2020, 158, 102094 CrossRef CAS PubMed.
- F. Goldsmith, J. Guice, R. Page, D. A. Welsh, C. M. Taylor, E. E. Blanchard, M. Luo, A. M. Raggio, R. W. Stout, D. Carvajal-Aldaz, A. Gaither, C. Pelkman, J. Ye, R. J. Martin, J. Geaghan, H. A. Durham, D. Coulon and M. J. Keenan, Obese ZDF rats fermented resistant starch with effects on gut microbiota but no reduction in abdominal fat, Mol. Nutr. Food Res., 2017, 61, 1501025 CrossRef PubMed.
- T. S. Nielsen, Z. Bendiks, B. Thomsen, M. E. Wright, P. K. Theil, B. L. Scherer and M. L. Marco, High-Amylose Maize, Potato, and Butyrylated Starch Modulate Large Intestinal Fermentation, Microbial Composition, and Oncogenic miRNA Expression in Rats Fed A High-Protein Meat Diet, Int. J. Mol. Sci., 2019, 20, 2137–2155 CrossRef CAS PubMed.
- Y. Bai, Y. Li, T. Marion, Y. Tong, M. M. Zaiss, Z. Tang, Q. Zhang, Y. Liu and Y. Luo, Resistant starch intake alleviates collagen-induced arthritis in mice by modulating gut microbiota and promoting concomitant propionate production, J. Autoimmun., 2021, 116, 102564 CrossRef CAS PubMed.
- Y. Sun, L. Zhou, L. Fang, Y. Su and W. Zhu, Responses in colonic microbial community and gene expression of pigs to a long-term high resistant starch diet, Front. Microbiol., 2015, 6, 877 Search PubMed.
- Y. Sun, Y. Su and W. Zhu, Microbiome-Metabolome Responses in the Cecum and Colon of Pig to a High Resistant Starch Diet, Front. Microbiol., 2016, 7, 779 Search PubMed.
- O. C. Umu, J. A. Frank, J. U. Fangel, M. Oostindjer, C. S. da Silva, E. J. Bolhuis, G. Bosch, W. G. Willats, P. B. Pope and D. B. Diep, Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations, Microbiome, 2015, 3, 16 CrossRef PubMed.
- H. M. Hamer, V. De Preter, K. Windey and K. Verbeke, Functional analysis of colonic bacterial metabolism: relevant to health?, Am. J. Physiol.: Gastrointest. Liver Physiol., 2012, 302, G1–G9 CrossRef CAS PubMed.
- L. A. David, C. F. Maurice, R. N. Carmody, D. B. Gootenberg, J. E. Button, B. E. Wolfe, A. V. Ling, A. S. Devlin, Y. Varma, M. A. Fischbach, S. B. Biddinger, R. J. Dutton and P. J. Turnbaugh, Diet rapidly and reproducibly alters the human gut microbiome, Nature, 2014, 505, 559–563 CrossRef CAS PubMed.
- Y. Zhang, L. Chen, M. Hu, J. J. Kim, R. Lin, J. Xu, L. Fan, Y. Qi, L. Wang, W. Liu, Y. Deng, J. Si and S. Chen, Dietary type 2 resistant starch improves systemic inflammation and intestinal permeability by modulating microbiota and metabolites in aged mice on high-fat diet, Aging, 2020, 12, 9173–9187 CrossRef CAS PubMed.
- R. Nagata, N. Innami, S. Pelpolage, K. Shimada, H. Koaze, M. Tani, K.-H. Han and M. Fukushima, Effects of Raw Potato Starch with High Resistant Starch Levels on Cecal Fermentation Properties in Rats, J. Nutr. Sci. Vitaminol., 2019, 65, S192–S195 CrossRef PubMed.
- A. Venkataraman, J. R. Sieber, A. W. Schmidt, C. Waldron, K. R. Theis and T. M. Schmidt, Variable responses of human microbiomes to dietary supplementation with resistant starch, Microbiome, 2016, 4, 33 CrossRef CAS PubMed.
- P. Louis and H. J. Flint, Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine, FEMS Microbiol. Lett., 2009, 294, 1–8 CrossRef CAS PubMed.
- S.-J. Bang, E.-S. Lee, E.-J. Song, Y.-D. Nam, M.-J. Seo, H.-J. Kim, C.-S. Park, M. Y. Lim and D.-H. Seo, Effect of raw potato starch on the gut microbiome and metabolome in mice, Int. J. Biol. Macromol., 2019, 133, 37–43 CrossRef CAS PubMed.
- R. J. Calvert, M. Otsuka and S. Satchithanandam, Consumption of raw potato starch alters intestinal function and colonic cell proliferation in the rat, J. Nutr., 1989, 119, 1610–1616 CrossRef CAS PubMed.
- D. Parada Venegas, M. K. De la Fuente, G. Landskron, M. J. González, R. Quera, G. Dijkstra, H. J. M. Harmsen, K. N. Faber and M. A. Hermoso, Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases, Front. Immunol., 2019, 10, 277–292 CrossRef PubMed.
- M. Derrien, E. E. Vaughan, C. M. Plugge and W. M. de Vos, Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium, Int. J. Syst. Evol. Microbiol., 2004, 54, 1469–1476 CrossRef CAS PubMed.
- A. Pellegrino, G. Coppola, F. Santopaolo, A. Gasbarrini and F. R. Ponziani, Role of Akkermansia in Human Diseases: From Causation to Therapeutic Properties, Nutrients, 2023, 15, 1815–1839 CrossRef CAS PubMed.
- A. Fosses, M. Maté, N. Franche, N. Liu, Y. Denis, R. Borne, P. de Philip, H. P. Fierobe and S. Perret, A seven-gene cluster in Ruminiclostridium cellulolyticum is essential for signalization, uptake and catabolism of the degradation products of cellulose hydrolysis, Biotechnol. Biofuels, 2017, 10, 250 CrossRef PubMed.
- F. M. Cerqueira, A. L. Photenhauer, H. L. Doden, A. N. Brown, A. M. Abdel-Hamid, S. Moraïs, E. A. Bayer, Z. Wawrzak, I. Cann, J. M. Ridlon, J. B. Hopkins and N. M. Koropatkin, Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome, J. Biol. Chem., 2022, 298, 101896 CrossRef CAS PubMed.
- A. A. Rangarajan, H. E. Chia, C. A. Azaldegui, M. H. Olszewski, G. V. Pereira, N. M. Koropatkin and J. S. Biteen, Ruminococcus bromii enables the growth of proximal Bacteroides thetaiotaomicron by releasing glucose during starch degradation, Microbiology, 2022, 168 DOI:10.1099/mic.0.001180.
- X. Ze, S. H. Duncan, P. Louis and H. J. Flint, Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon, ISME J., 2012, 6, 1535–1543 CrossRef CAS PubMed.
- D. H. Jung and C. S. Park, Resistant starch utilization by Bifidobacterium, the beneficial human gut bacteria, Food Sci. Biotechnol., 2023, 32, 441–452 CrossRef CAS PubMed.
- S. L. S. Devarakonda, D. K. Superdock, J. Ren, L. M. Johnson, A. P. Loinard-Gonzalez and A. C. Poole, Gut microbial features and dietary fiber intake predict gut microbiota response to resistant starch supplementation, medRxiv, 2023, DOI:10.1101/2023.03.24.23287665.
- L. U. Thompson, C. C. Maningat, K. Woo and P. A. Seib, In Vitro Digestion of RS4-Type Resistant Wheat and Potato Starches, and Fermentation of Indigestible Fractions, Cereal Chem., 2011, 88, 72–79 CrossRef CAS.
- L. Li, J. Ryan, Z. Ning, X. Zhang, J. Mayne, M. Lavallée-Adam, A. Stintzi and D. Figeys, A functional ecological network based on metaproteomics responses of individual gut microbiomes to resistant starches, Comput. Struct. Biotechnol. J., 2020, 18, 3833–3842 CrossRef CAS PubMed.
- I. Martínez, J. Kim, P. R. Duffy, V. L. Schlegel and J. Walter, Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects, PLoS One, 2010, 5, e15046 CrossRef PubMed.
- B. Upadhyaya, L. McCormack, A. R. Fardin-Kia, R. Juenemann, S. Nichenametla, J. Clapper, B. Specker and M. Dey, Impact of dietary resistant starch type 4 on human gut microbiota and immunometabolic functions, Sci. Rep., 2016, 6, 28797 CrossRef CAS PubMed.
- L. B. Bindels, R. R. Segura Munoz, J. C. Gomes-Neto, V. Mutemberezi, I. Martínez, N. Salazar, E. A. Cody, M. I. Quintero-Villegas, H. Kittana, C. G. de Los Reyes-Gavilán, R. J. Schmaltz, G. G. Muccioli, J. Walter and A. E. Ramer-Tait, Resistant starch can improve insulin sensitivity independently of the gut microbiota, Microbiome, 2017, 5, 12 CrossRef PubMed.
- G. R. Healey, R. Murphy, L. Brough, C. A. Butts and J. Coad, Interindividual variability in gut microbiota and host response to dietary interventions, Nutr. Rev., 2017, 75, 1059–1080 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table 1. Composition of the basal TWD. Table 2. Differential expression of genera, Table 3. Gene expression changes, Table 4. RNASeq data for the cecum. Table 5. RNASeq data for the distal colon. See DOI: https://doi.org/10.1039/d3fo04512a |
| This journal is © The Royal Society of Chemistry 2024 |