Open Access Article
Open Access Article(Poly)phenol-related gut metabotypes and human health: an update
Jiaying
Hu
a,
Robin
Mesnage
ab,
Kieran
Tuohy
 c,
Christian
Heiss
c,
Christian
Heiss
 d and
Ana
Rodriguez-Mateos
d and
Ana
Rodriguez-Mateos
 *a
*a
aDepartment of Nutritional Sciences, School of Life Course and Population Sciences, Faculty of Life Sciences and Medicine, King's College London, London, UK. E-mail: ana.rodriguez-mateos@kcl.ac.uk
bBuchinger Wilhelmi Clinic, Überlingen, Germany
cSchool of Food Science and Nutrition, Faculty of Environment, University of Leeds, Leeds, UK
dDepartment of Clinical and Experimental Medicine, Faculty of Health and Medical Sciences, University of Surrey, Surrey, UK
First published on 19th February 2024
Abstract
Dietary (poly)phenols have received great interest due to their potential role in the prevention and management of non-communicable diseases. In recent years, a high inter-individual variability in the biological response to (poly)phenols has been demonstrated, which could be related to the high variability in (poly)phenol gut microbial metabolism existing within individuals. An interplay between (poly)phenols and the gut microbiota exists, with (poly)phenols being metabolised by the gut microbiota and their metabolites modulating gut microbiota diversity and composition. A number of (poly)phenol metabolising phenotypes or metabotypes have been proposed, however, potential metabotypes for most (poly)phenols have not been investigated, and the relationship between metabotypes and human health remains ambiguous. This review presents updated knowledge on the reciprocal interaction between (poly)phenols and the gut microbiome, associated gut metabotypes, and subsequent impact on human health.
Introduction
Dietary (poly)phenols are plant-derived secondary metabolites that are abundant in many fruits and vegetables, tea, coffee, cocoa, soy products, olive oil, and red wine.1 Extensive preclinical studies have demonstrated promising biological activities of individual (poly)phenols, including anti-inflammatory, antioxidant, and anti-proliferative.2–7 However, the physiological relevance of such studies, in particular the evidence from in vitro studies, has been largely questioned.8,9 In recent years, increasing evidence from observational studies and randomized controlled trials (RCT) has demonstrated an inverse association between (poly)phenol consumption and the risk of various non-communicable diseases, such as cardiometabolic and neurodegenerative diseases.10–15 However, inconsistent results also exist and a high inter-individual variability in response to (poly)phenols has been reported.16,17 In addition to heterogenous study designs and differences in the physical and genetic make-up of individuals, the widely documented high variability in bioavailability and metabolism of (poly)phenols is likely to play an important role in explaining the observed variability in response.18,19While a limited amount of the ingested (poly)phenols are absorbed in the small intestine, a large proportion (∼95%) reach the colon where they are metabolised by gut microorganisms into smaller phenolic compounds for further uptake.20,21 These compounds can then undergo phase II metabolism, generating conjugated metabolites that may have very different biological activities from their parent compounds.22 In this context, genetic differences, such as single nucleotide polymorphisms in transporters and enzymes, may account for some of the individual variability in the absorption and metabolism of (poly)phenols.23,24 However, the well-known variability in gut microbiota diversity, composition and functionality between individuals, points to the gut microbiota metabolising capacity as a key factor to explain the individual differences on the biological responses to (poly)phenol consumption. (Poly)phenol metabolising phenotypes, or metabotypes, have been proposed, with isoflavone and ellagitannin related metabotypes being the most widely studied so far.25,26
When investigating the effects of dietary (poly)phenols on human health, it is important to consider the two-way interaction between the gut microbiota and (poly)phenols: (poly)phenols are transformed into metabolites via the enzymatic activity of gut microbes, and these metabolites may in turn modulate the gut microbial community.27
In this review, we summarise the current evidence on inter-individual differences in circulating (poly)phenol gut microbial metabolites, (poly)phenol-related gut metabotypes, associated gut microbiota and effects on human health.
Gut microbial metabolism of (poly)phenols
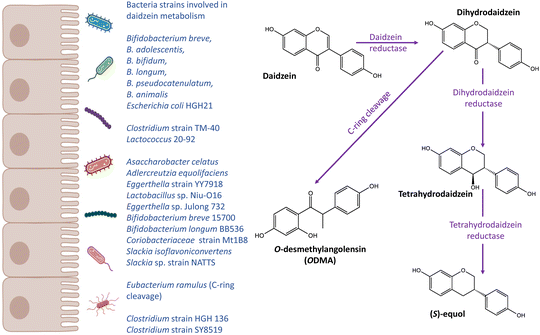
The gut microbiota transforms (poly)phenols into more bioavailable metabolites of lower molecular weight through several catabolic pathways, including hydrolysis, cleavage, and reduction.28 To date, only a few gut microbial species have been identified to participate in the metabolism of specific (poly)phenols, while most species involved still remain unknown. Moreover, whilst the traditional approach has been tried to identify individual microbial species involved in particular transformations, it is highly likely that the transformation of complex (poly)phenols derives from the concerted action of multiple species working together and also on local pH. Deeper investigation into the composition of multi-species consortia awaits wider application of high-throughput microbiome co-culture or mixed culture methodologies. In Table 1 we summarise the main species that have been identified to date. Lactobacillus and Bifidobacterium strains have been found to deconjugate flavonoid rhamnoglucosides through rhamnosidase activity, and many species within Bifidobacteriaceae, Bacteroidaceae, Porphyromonadaceae, and Enterococcaceae phyla exert O-deglycosylation activity in flavanones, isoflavones and flavan-3-ols.29 Cleavage of C–C bonds in flavonoids is also a characterised gut microbial activity. For instance, the isoflavone daidzein has two distinct metabolites, a reduction product equol and a ring cleavage product O-demethylangolensin.30Eubacterium ramulus and Strain SY8519 have been reported to catalyse the C-ring cleavage of isoflavones (Table 1).31,32Flavonifractor plautii also showed similar activity for flavonols.29 Gut bacteria also catalyse reduction, hydrogenation of double bonds and dehydroxylation reactions of (poly)phenols. In the metabolism of isoflavones, Eggerthella sp. YY7918, Slackia equolifaciens, Slackia isoflavoniconvertens, and Lactococcus garvieae have been reported to show reduction activity, thus generating equol (Table 1).33–36 An example of dehydroxylation is the biotransformation of ellagic acid to urolithins. After hydrolysis and reduction of carboxylic acid, dehydroxylation occurs at the p-hydroxy group of a semi-hydroquinone, to form urolithin C and A.37Gordonibacter urolithinfaciens and Ellagibacter isourolithinifaciens are involved in this process (Table 1).38 They also metabolise catechol dehydroxylation and reduction.38 Importantly, the gut microbiota can also influence host metabolism. For example, (poly)phenol phase II metabolites that are excreted via the bile into the small intestine can be converted back to aglycones in the colon by gut microbes with glucuronidase and sulfatase activity, and re-absorbed.37 Therefore, the gut microbiota is a key factor affecting (poly)phenol bioavailability and metabolism in multiple ways.| Phenolic class | Species/strain | Substrate(s) | Reaction | Ref. |
|---|---|---|---|---|
| Anthocyanins | Bifidobacterium lactis | Anthocyanin | β-Glucosidase | 39 |
| Lactobacillus acidophilus | Anthocyanin | β-Glucosidase | 39 | |
| Lactobacillus casei | Anthocyanin | β-Glucosidase | 39 | |
| Lactobacillus.plantarum | Anthocyanin | β-Glucosidase | 39 | |
| Ellagitannins | Bifidobacterium pseudocatenulatum | Ellagic acid | Metabolise ellagic acid to urolithin A and B | 40 |
| Clostridium coccoides members | Ellagic acid | Metabolise ellagic acid to urolithins | 41 | |
| Ellagibacter isourolithinifaciens | Ellagic acid | Metabolise ellagic acid to isourolithin A | 42 | |
| Enterococcus faecium FUA027 | Ellagic acid | Metabolise ellagic acid to urolithin A | 43 | |
| Gordonibacter pamelaeae | Ellagic acid | Metabolise ellagic acid to urolithins | 44 | |
| Gordonibacter urolithinfaciens | Ellagic acid | Metabolise ellagic acid to urolithins | 45 | |
| Lactococcus garvieae FUA009 | Ellagic acid | Metabolise ellagic acid to urolithin A | 46 | |
| Streptococcus thermophilus FUA329 | Ellagic acid | Metabolise ellagic acid to urolithin A | 47 | |
| Flavanones | Bacteroides distasonis | Eriocitrin | Hydrolysis | 48 |
| Bacteroides uniformis | Eriocitrin | Hydrolysis | 48 | |
| Bifidobacterium catenulatum | Hesperidin | Hydrolysis | 49 | |
| Bifidobacterium pseudocatenultum | Hesperidin | Hydrolysis | 49 | |
| Clostridium butyricum | Eriocitrin | C-ring cleavage | 48 | |
| Flavan-3-ols | Adlercreutzia equolifaciens JCM 14793 | (−)-Epigallocatechin, (−)-gallocatechin | Dihydroxylation | 50 |
| Asaccharobacter celatus JCM 14811 | (−)-Epigallocatechin, (−)-gallocatechin | C-ring cleavage | 50 | |
| Eggerthella lenta | (−)-Epicatechin, (+)-catechin | C-ring cleavage | 51 | |
| Slackia equolifaciens JCM 16059 | (−)-Epigallocatechin, (−)-gallocatechin | C-ring cleavage | 50 | |
| Flavones | Blautia sp. MRG-PMF1 | Apigetrin | O-Glucose hydrolysis | 52 |
| Eubacterium cellulosolvens | Homoorientin, isovitexin | Deglycosylation of C- and O-glucosides | 53 | |
| Flavonols | Bacillus subtilis | Quercetin | C-ring cleavage | 54 |
| Bacteroides distasonis | Robinin | Hydrolyse robinin to kaempferol | 55 | |
| Bacteroides ovatus | Rutin | β-Glucosidase, hydrolyse rutin to quercetin | 55 | |
| Bacteroides uniformis | Rutin | β-Glucosidase, hydrolyse rutin to quercetin | 55 | |
| Bifidobacterium adolescentis | Kaempferol 3-O-glucoside | β-Glucosidase | 56 | |
| Bifidobacterium bifidum | Kaempferol 3-O-glucoside | β-Glucosidase | 56 | |
| Bifidobacterium breve | Kaempferol 3-O-glucoside | β-Glucosidase | 56 | |
| Bifidobacterium catenulatum | Kaempferol 3-O-glucoside | Hydrolysis, β-Glucosidase | 56 | |
| Bifidobacterium dentium | Rutin, poncirin | Hydrolysis | 57 | |
| Bifidobacterium infantis | Kaempferol 3-O-glucoside | β-Glucosidase | 56 | |
| Bifidobacterium longum | Kaempferol 3-O-glucoside | β-Glucosidase | 56 | |
| Bifidobacterium pseudocatenulatum | Kaempferol 3-O-glucoside | Hydrolysis, β-glucosidase | 56 | |
| Blautia sp. MRG-PMF1 | Hesperidin, Poylmethoxyflavones | O-Rutinose hydrolysis, demethylation, degylcosylation | 52 | |
| Clostridium orbiscindens | Quercetin, taxifolin, luteolin, apigenin, naringenin, phloretin | C-ring cleavage | 58 | |
| Enterococcus avium | Rutin | O-Deglycosylation | 59,60 | |
| Enterococcus casseliflavus | Quercetin-3-glucoside | Hydrolysis | 61 | |
| Eubacterium ramulus | Rutin, quercetin, kaempferol, taxifolin, luteolin, quercetin-3-glucoside | C-ring cleavage | 61–63 | |
| Flavonifractor plautii | Quercetin | C-ring cleavage | 64 | |
| Lactobacillus acidophilus | Rutin, nicotiflorin, narirutin | α-Rhamnosidase | 65 | |
| Lactobacillus plantarum | Rutin, nicotiflorin, narirutin | α-Rhamnosidase | 65 | |
| Isoflavones | Adlercreutzia equolifaciens | Daidzein | Bioconversion of daidzein to equol | 66 |
| Asaccharobacter celatus | Daidzein | Bioconversion of daidzein to equol | 67 | |
| Bifidobacterium adolescentis | Daidzein | Hydrolysis | 68 | |
| Bifidobacterium animalis | Daidzein | Hydrolysis | 69 | |
| Bifidobacterium bifidum | Daidzein | Hydrolysis | 68 | |
| Bifidobacterium breve | Daidzein | Hydrolysis | 68 | |
| Bifidobacterium longum | Daidzein | Hydrolysis | 69,70 | |
| Bifidobacterium pseudocatenulatum | Daidzein | Hydrolysis | 69,70 | |
| Blautia sp. MRG-PMF1 | Daidzein, genistin, glycitin | Hydrolysis, O-glucose & O-methyl hydrolysis | 52 | |
| Clostridium strain HGH 136 | Daidzein | Bioconversion of daidzein to ODMA | 71 | |
| Clostridium strain SY8519 | Daidzein | Bioconversion of daidzein to ODMA | 32 | |
| Clostridium strain TM-40 | Daidzein | Bioconversion of daidzein to dihydrodaidzein | 72 | |
| Coriobacteriaceae strain Mt1B8 | Daidzein | Bioconversion of daidzein to equol | 73 | |
| Eggerthella strain YY7918 | Daidzein | Bioconversion of daidzein to equol | 74 | |
| Eggerthella sp. Julong 732 | Daidzein | Bioconversion of daidzein to equol | 75 | |
| Enterococcus sp. MRG-IFC-2 | Puerarin | O-Glycosidase | 76 | |
| Escherichia coli HGH21 | Daidzein, genistin | β-Glucosidase | 77 | |
| Eubacterium ramulus | Daidzein, genistin | C-ring cleavage | 78 | |
| Lachnospiraceae strain CG19-1 | Puerarin | Deglycosylation | 79 | |
| Lactobacillus sp. Niu-O16 | Daidzein | Bioconversion of daidzein to equol | 75 | |
| Lactococcus sp. MRG-IFC-1 | Puerarin | O-Glycosidase | 76 | |
| Lactococcus 20-92 | Daidzein | Bioconversion of daidzein to dihydrodaidzein | 36 | |
| Slackia isoflavoniconvertens | Daidzein | Bioconversion of daidzein to equol | 80 | |
| Slackia sp. strain NATTS | Daidzein | Bioconversion of daidzein to equol | 81 | |
| Lignans | Bacteroides distasonis DSM 20701T | Secoisolariciresinol (SECO) | Deglycosylation | 82 |
| Bacteroides fragilis DIfE-05 | SECO | Deglycosylation | 82 | |
| Bacteroides fragilis SDG-Mt85-4C, B. fragilis SDG-Mt85-5B | SECO | Deglycosylation | 82 | |
| Bacteroides methylotrophicum DSM 3468T | SECO | Demethylation | 82 | |
| Bifidobacterium bifidum INIA P466 | SECO | Metabolise SECO to enterodiol | 83 | |
| Bifidobacterium catenulatum INIA P732 | SECO | Metabolise SECO to enterodiol | 83 | |
| Bifidobacterium pseudocatenulatum INIA P946 | SDG | Deglycosylation of SDG to SECO | 83 | |
| Bifidobacterium pseudolongum INIA P2 | SECO | Metabolise SECO to enterodiol | 83 | |
| Blautia producta DSM 3507 | SECO | Demethylation | 84 | |
| Clostridium cocleatum | Secoisolariciresinol diglucoside (SDG) | Deglycosylation | 82 | |
| Clostridium ramosum | SDG | Deglycosylation | 82 | |
| Eggerthella lenta DSM 2243 | Pinoresinol, lariciresinol | Reduction | 84 | |
| Eubacterium callanderi DSM 3662T | SECO | Demethylation | 82 | |
| Eubacterium limosum DSM 20543T | SECO | Demethylation | 82 | |
| Gordonibacter pamelaeae | Didemethyl-SECO | dehydroxylation | 84 | |
| Lactobacillus gasseri INIA P508, | SECO | Metabolise SECO to enterolignans | 83 | |
| Lactobacillus salivarius INIA P183, Lactobacillus salivarius INIA P448 | SECO | Metabolise SECO to enterolignans | 83 | |
| Lactonifactor longoviformis DSM 17459 | Enterodiol | Lactonization, bioconversion of enterodiol to enterolactone | 84 | |
| Gordonibacter pamelaeae | Didemethyl-SECO | dehydroxylation | 84 | |
| Peptostreptococcus productus DSM 2950T, Peptostreptococcus productus DSM 3507 | SECO | Demethylation | 82 | |
| Stilbenes | Adlercreutzia equolifaciens | Resveratrol | Metabolism resveratrol into dihydroresveratrol | 85 |
| Slackia equolifaciens | Resveratrol | Metabolism resveratrol into dihydroresveratrol | 85 | |
| Xanthohumol | Eubacterium ramulus | Xanthohumol | Hydrogenation | 86 |
| Eubacterium limosum | Isoxanthohumol | O-Demethylation | 86 |
(Poly)phenols as modulators of the gut microbiota
While dietary (poly)phenols are bio-transformed into absorbable metabolites by the intestinal microbiota, these metabolites are capable of modulating gut microbial communities. Both in vitro assays and in vivo studies have revealed that (poly)phenols exhibit prebiotic activities through promoting the growth of beneficial bacteria such as Lactobacillus and Bifidobacterium,87 and through inhibiting colonies of pathogenic bacteria such as Escherichia coli, Clostridium perfringens and Helicobacter pylori.88,89 For example, an RCT conducted with 10 healthy male participants showed that 4-week consumption of red wine (poly)phenols significantly enhanced the growth of Enterococcus, Prevotella, Bacteroides, Bifidobacterium, Bacteroides uniformis, Eggerthella lenta and Blautia coccoides–Eubacterium rectale groups.90 Quercetin has been demonstrated to inhibit the growth of E. coli, Pseudomonas aeruginosa and Staphylococcus aureus in in vitro assays.91,92 It has also been suggested that the altered gut microbial ecology may prevent against metabolic diseases through several physiological actions, including increasing production of short-chain fatty acids (SCFAs), decreasing adipogenesis and lipogenesis, or alleviating systemic inflammation.93 Zhang et al.94 reported that oolong tea flavan-3-ols are able to promote the growth of Bifidobacterium and Lactobacillus/Enterococcus groups while inhibiting the growth of Bacteroides–Prevotella, Clostridium histolyticum, and Eubacterium–Clostridium groups in vitro, and further increase the concentration of SCFAs. In mouse models, grape (poly)phenols significantly increased Akkermansia muciniphila abundance and decreased the Firmicutes to Bacteroidetes ratio, paralleled with attenuation of high-fat-diet-induced inflammation.95 An RCT conducted among 22 healthy volunteers found that cocoa flavan-3-ols promoted the beneficial bifidobacterial and lactobacilli populations, and reduced plasma triacylglycerol and C-reactive protein concentrations.96 However, the evidence from human intervention studies, especially for (poly)phenol extracts or pure (poly)phenol compounds is limited, and the relationship between gut bacteria species, (poly)phenol gut microbial metabolites and health outcomes is still unclear. Some evidence from in vitro studies suggest direct biological activities of (poly)phenol gut microbial metabolites, however whether they are the major bioactive compounds responsible for the health benefits or whether they act as biomarkers of a healthy intestinal microbial community remains unknown.97 The answer to this question is likely to explain at least in part the differential individual biological responses to (poly)phenols observed in clinical trials and their interaction with other putative bioactives in whole plant foods, such as dietary fibre. Nevertheless, what has become clear is that (poly)phenol consumption can modulate gut microbiota diversity, composition and function and this may have important implications for human health.Variability in (poly)phenol gut microbial metabolism: the concept of (poly)phenol metabotypes
The concept of (poly)phenol metabolising phenotypes or metabotypes was first proposed by Bolca and colleagues as “clusters of gut microbial communities with similar metabolic profiles”.98 More recently, Espín et al.28 defined gut (poly)phenol metabotypes as “metabolic phenotypes defined by specific gut microbial metabolites and their associated microbial ecology in terms of composition and functionality”. This is comparable to the notion of enterotype, a classification of gut microbiome composition profiles which is proposed to support the development of personalised nutrition strategies.99Currently, there is a lack of consensus regarding whether the concept of (poly)phenol metabotypes should be used exclusively to differentiate between producers and non-producers of specific (poly)phenol gut microbial metabolites, or in a broader sense to distinguish individuals with different metabolising capacities, such as low vs. high producers (Fig. 1). One of the main arguments proposed by Iglesias-Aguirre et al. for the definition of metabotypes as an exclusive qualitative (i.e. presence or absence of unique (poly)phenol gut microbial metabolites) but not quantitative criteria is that the production gradient could be affected by external factors, such as food matrix, sample collection time or diet, and that the cut-off to consider an individual from one or another metabotype will be arbitrary and will depend on each cohort considered.97 While we fully agree with these points, we argue that these issues may also apply to the definition of producers vs. non-producers. For example, differences in sensitivity between analytical devices used to determine metabotypes (typically HPLC-UV or more commonly, LC-MS), could lead to low producers being classified as non-producers in some studies. This can make comparisons between studies conducted with different instruments and sensitivities difficult. In addition, the cut-off for the definition of non-producers is also arbitrary. For example, for the definition of equol non-producer metabotype, some researchers have used the Setchell method,100–102 which is a classification based on a cut-off (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ≥ 1.75) which leads to non-producers and very low producers being classified as non-producers. Other researchers have used different cut-off concentrations, or the limit of detection of their analytical device.103–105 The type of food matrix, sample collection time, or background diet could also affect this classification in a similar manner. We argue that the definition of metabotypes does not have to be an absolute criterion but a relative one, and can be used to classify individuals within the same cohort into groups with different (poly)phenol metabolising capacities. The effects of external factors will be minimised, as factors such as food matrix or sample collection time will be homogenous for each defined cohort, therefore unlikely to affect a relative cut-off. For example, tertiles or quintiles of excretion, depending on the sample size of the cohort can be used to classify individuals within the same cohorts into low, medium and high producers, as it has already been done for some (poly)phenols, such as flavanones or lignans.106,107 Another point to consider is that for some (poly)phenols, individuals classified as non-producers are either non-existent or are present in very low number (i.e. 2–10% of the population investigated), which limits the conclusions that can be made and comparisons between groups due to low sample sizes. In some cases therefore, clustering non-producers and low producers in one group may be more accurate than having small groups with not enough power to detect differences in health outcomes or gut microbiome diversity and composition.
10 ≥ 1.75) which leads to non-producers and very low producers being classified as non-producers. Other researchers have used different cut-off concentrations, or the limit of detection of their analytical device.103–105 The type of food matrix, sample collection time, or background diet could also affect this classification in a similar manner. We argue that the definition of metabotypes does not have to be an absolute criterion but a relative one, and can be used to classify individuals within the same cohort into groups with different (poly)phenol metabolising capacities. The effects of external factors will be minimised, as factors such as food matrix or sample collection time will be homogenous for each defined cohort, therefore unlikely to affect a relative cut-off. For example, tertiles or quintiles of excretion, depending on the sample size of the cohort can be used to classify individuals within the same cohorts into low, medium and high producers, as it has already been done for some (poly)phenols, such as flavanones or lignans.106,107 Another point to consider is that for some (poly)phenols, individuals classified as non-producers are either non-existent or are present in very low number (i.e. 2–10% of the population investigated), which limits the conclusions that can be made and comparisons between groups due to low sample sizes. In some cases therefore, clustering non-producers and low producers in one group may be more accurate than having small groups with not enough power to detect differences in health outcomes or gut microbiome diversity and composition.
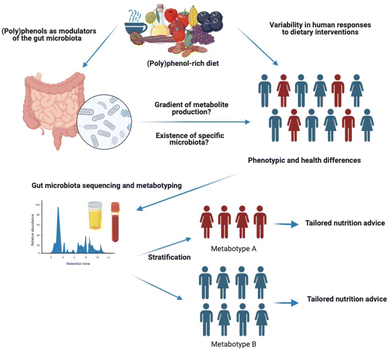 | ||
| Fig. 1 Inter-individual variability in gut metabolism of (poly)phenols due to distinct gut microbiome composition. | ||
(Poly)phenol metabotypes and human health: what we know so far
The most widely studied metabotypes are the ones related to the gut microbial metabolites of the isoflavone daidzein, equol and ODMA. More recently, ellagitannin and resveratrol related metabotypes have also been described.108Equol and ODMA metabotypes
Isoflavones are a class of phytoestrogens which are found mainly in soy products. They exist as glycoside conjugates in plants.109 After ingested by humans, isoflavones are hydrolysed by gut bacteria into bioactive aglycones, including daidzein, genistein and glycitein. Soy consumption is generally high in Asian countries whereas low in Western population.110–112Equol and ODMA are gut microbial metabolites of daidzein (Fig. 2), which have been related to health effects. To date, equol- and ODMA-producer metabotypes have been identified.113 It is also suggested that the capacity of an individual to produce equol is not influenced by the capacity to produce ODMA.114 Several distinct metabolic steps requiring specific intestinal bacteria species exist for the production of equol and ODMA, leading to the stratification of equol- and ODMA-producer metabotypes. For example, bacteria responsible for the C-ring cleavage is required to transform daidzein to ODMA but not needed for equol production.113
There are approximately 30%–50% and 80%–90% of Caucasian population being identified as equol- and ODMA-producers respectively following soy consumption.115–117 In Asian populations, the prevalence of ODMA-producers is slightly lower than that in Western population, where the prevalence of equol-producers reaches 50%–60%.117,118 Due to the stability in the long term, metabotypes are regarded as a biomarker for intestinal ecology and potential disease risks.116 A number of studies have investigated demographic, anthropometric and dietary factors (i.e. race, ethnicity, age, BMI, etc.) associated with daidzein-related metabotypes to describe the features of equol- and ODMA- producers. However, observed results are often inconsistent and no clear associations can be demonstrated.113,119 One of the reasons could be the inconsistent classification of metabotypes and arbitrary cut-offs used in different studies, as previously discussed.
The gut bacteria involved in the biotransformation of equol and ODMA are distinct from each other. Adlercreutzia, Asaccharobacter, Eggerthella, Bifidobacterium and Clostridium are gut bacteria that are associated with the production of equol from daidzein (Table 1).66–68,72,120 Importantly, the role of bifidobacterial in the biotransformation of isoflavones in soy milk has been well established.121–123 However, less is known about the bacteria population responsible for the production of ODMA, except for Eubacterium ramulus which is capable of C-ring cleavage activity.78Eubacterium ramulus also plays a role in the metabolism of other (poly)phenols, such as quercetin, xanthohumol, 8-prenylnaringenin and other flavonoids.86,124,125 This indicates that the bio-conversion of daidzein into ODMA share some metabolic steps with other (poly)phenols, and most individuals are likely to have the ability to produce ODMA. Further studies are warranted to investigate the gut microbiome composition and biological characteristics of equol- and ODMA-producer metabotypes, and their relationship with human health.
A 3-day cross-sectional study conducted among 99 Chinese participants found that equol-producers had higher abundance of Adlercreutzia equolifaciens and Bifidobacterium bifidum compared with non-producers (77.5% vs. 22.5%; 72.0% vs. 28.0%, respectively).126 The prevalence of dyslipidemia was significantly lower in equol-producers (27% vs. 50%). However, there was no significant difference in microbiome richness between equol-producers and non-producers.126 In contrast, an US study with 80 healthy females observed that equol-producers had lower gut microbiome diversity and beneficial bacteria taxa, such as Bacteroides spp., Faecalibacterium spp., and Butyrivibrium spp.127 In non-producers, a higher dominance of Akkermansia spp., Prevotella 9, and Megasphera elsdenii was presented. The authors also showed that even among individuals with the same metabotype, the consumption of soy or not would result in different gut microbiota composition.127 Considering the amount of regular soy intake and distribution of daidzein-related metabotypes are different between Asian and Western population, it is likely that the inconsistent results are attributable to sociodemographic factors and dietary patterns.
So far, although studies have suggested that equol-and ODMA producers may have lower cardiometabolic risk than non-producers, mixed results also exist and very few studies have investigated ODMA metabotypes. Miller et al. observed an association between ODMA non-producers and obesity in both peri- and post-menopausal women, whereas no significant association was found for equol non-producers.128 This is in line with the findings by Frankenfeld et al.,129 showing that among 297 adults participants, obesity was associated with being ODMA non-producers (OR: 2.8 [95% CI: 1.2, 6.2]). Cohort studies of Japanese men suggested a lower risk of coronary artery calcification (OR: 0.1 [95% CI: 0.01, 0.90, p < 0.04]) in equol-producers compared to equol non-producers.130,131 On the contrary, Usui et al. reported significant improvements in cardiometabolic risk parameters upon 12-week equol supplementation only in female equol non-producers.132 Interestingly, Hazim et al.100 reported acute benefits on vascular function in equol-producers after isoflavone consumption, and administration of commercially-made equol to non-producers did not cause any change in vascular function despite of increased plasma equol concentrations. Although this study only investigated acute effects and did not test the effect of equol in equol-producers, the study provided an insight that the health benefits might be attributed to the individual capacity of producing equol, i.e. the existence of gut microbiome responsible for the biotransformation of daidzein into equol. This would support the idea of the equol metabotype use as a means of defining microbiome health effects or at least in this case, a microbiome profile associated with improved vascular function. Future studies are therefore needed to link the composition and functionality of metabotype-related gut microbiota to health effects, and investigate the metabolic reactions for equol. Clear criteria for stratification of equol-producer, ODMA-producer and non-producer is also required. Furthermore, most daidzein-related studies are conducted in Asian population, and more research in Western countries are warranted.
In addition to the classic daidzein equol and ODMA metabotypes, a recent paper including 60 postmenopausal women investigated novel daidzein and genistein related gut metabotypes.133 After 12-week daily consumption of a soy isoflavone extract, the authors defined 5 metabotypes according to hierarchical cluster analysis. Cluster 1 and 2 shared similar characteristics in terms of high equol production, while cluster 2 showed higher 4-ethylphenol (4EP) production and lower genistein production than cluster 1; cluster 3 produced the highest proportions of 4EP but no or very small proportions of equol; cluster 4, in which most women were included, had the highest proportions of daidzein and genistein; while cluster 5 exhibited high proportions of dihydrodaidzein and dihydrogenistein. This study is unprecedented since the majority of isoflavone studies only include daidzein-related metabotypes, and highlights the complexity of (poly)phenol metabolism and the need for more research beyond the “classic” metabotypes.
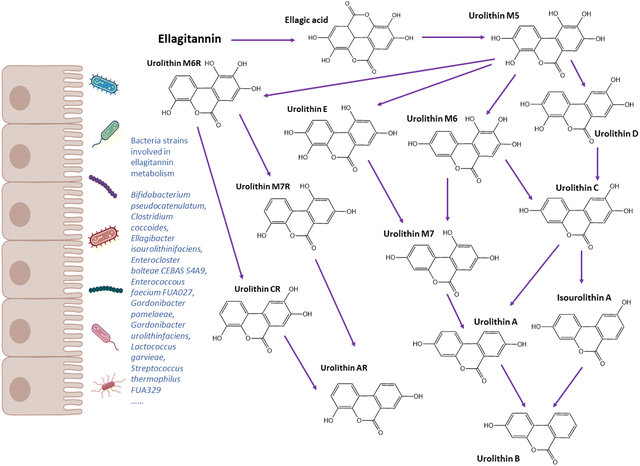
Urolithin metabotypes
Ellagitannins are rich in some berries, such as strawberries and raspberries, other fruits such as pomegranate and nuts, such as walnuts. In vitro and in vivo studies have demonstrated anti-inflammatory, antioxidant, antimicrobial, and anti-tumour activities of ellagitannins.134–136 Urolithins, which are dibenzopyran-6-one derivatives metabolised from ellagitannins (Fig. 3), have also shown anti-inflammatory properties.137,138Three urolithin-related metabotypes (UM) have been described for ellagitannin gut metabolism.26,139 Urolithin metabotype A (UMA), which is characterised by the production of urolithin A derivatives; urolithin metabotype B (UMB), which produces urolithin B and isourolithin A in addition to urolithin A derivatives; and urolithin metabotype zero (UM0), with no production of urolithin metabolites.26 The distribution of the three metabotypes was demonstrated in a relatively large Spanish cohort (n = 839) by Cortés-Martín et al.140 Approximately 50–80% of the population were UMA, 10–40% were UMB and 10% were UM0. The authors also suggested that the metabotype frequency was age-dependent, with the percentage of UMA decreasing and UMB increasing with age, while UM0 prevalence remained constant.140 However, the stability of urolithin metabotype over time in a given individual has yet to be measured in longitudinal studies. A recent study in healthy Chinese youth reported the approximate prevalence of 55% UMA, 30% UMB and 15% UM0.141 However, the sample size was too small (n = 35), therefore more studies conducted in Asian countries are required. Studies with large cohorts in different geographic locations are warranted to detect the distribution of metabotypes in different ethnicities and populations, as most of the evidence currently existing comes from Spanish cohorts.
Few small studies suggest that urolithin metabotypes can be potential biomarkers for gut microbiome balance and a healthy intestinal ecology. Tomás-Barberán et al.139 reported a higher prevalence of UMB among patients with metabolic syndrome or colorectal cancer. Selma et al.142 also found that UMB prevailed in individuals overweight or with obesity, and UMB metabolites were positively correlated with cardiometabolic disease biomarkers including total cholesterol, LDL, oxidised LDL, VLDL and apolipoprotein B, while urinary urolithin A was positively correlated with HDL and apolipoprotein A-I. However, this study was small (n = 69), and the correlation between UM and BMI was not confirmed in the larger cohort of 839 individuals conducted by the same research team.140 Regarding UM0, a study conducted among 52 Parkinson's disease (PD) patients and 117 healthy participants showed a significant higher proportion of UM0 in PD patients compared with healthy volunteers (27% vs. 9%, p = 0.004), and the proportion increased as the disease became more severe.143 The gut microbiota of UM0 patients displayed an increased pro-inflammatory Enterobacteriaceae and reduced protective butyrate-producing bacteria. This provides an insight that as the research about metabotype-associated gut microbiota goes further, disease risks might be predicted based on the features of distinct metabotypes.
The family Coriobacteriaceae has been found to display a strong correlation with urolithin production.44,45,144Gordonibacter urolithinfaciens and Gordonibacter pamelaeae, which are members of Coriobacteriaceae family, were identified to metabolise ellagitannins into pentahydroxy urolithin M5 and tetrahydroxy urolithins (D, E, and M6) through lactone-ring opening, decarboxylation, and dehydroxylation reactions.44,45Ellagibacter isourolithinifaciens sp. nov., a member of the family Eggerthellaceae, was isolated from human faeces and found to be capable of producing isourolithin A (Table 1).42 Romo-Vaquero et al.144 analysed gut microbiome composition from 249 healthy metabotyped participants. Olsenella, Senegalismassilia, and Slackia were positively correlated with isourolithin A and urolithin B production, while Gordonibacter and Eggerthella were positively correlated with urolithin A. A recent study by Iglesias-Aguirre et al.145 reported a novel bacterial strain Enterocloster bolteae CEBAS S4A9 that could convert Uro-C to Uro-A anaerobically. Very few studies have reported strains involved in the metabolism of ellagic acid to urolithin A, including Bifidobacterium pseudocatenulatum INIA P815, Lactococcus garvieae FUA009, Enterococcus faecium FUA027 and Streptococcus thermophilus FUA329.40,43,46,47 Among three metabotypes, UM0 showed the lowest diversity and richness of intestinal bacteria, while UMB had the highest richness at phylum and family level.144 The higher richness of gut microbiome may explain why UMB produces more types of metabolites than UMA. The authors also observed a positive correlation between Slackia and cardiometabolic risk factors, such as total cholesterol, LDL, apolipoprotein B and non-HDL.144 Overall, the gut bacteria species or consortia responsible for the production of urolithin metabolites and their relationship with disease risk need further investigation.
Very little evidence exists from randomised controlled trials on whether the response to ellagitannin consumption differs between urolithin metabotypes. A small RCT in healthy men (n = 10), which was not stratified into metabotypes, indicated significant increases in flow-mediated dilation (FMD) at 2 h and 24 h post-consumption of 200 g and 400 g raspberries, and these improvements were correlated with plasma ellagic acid and urolithin A metabolites.146 An RCT with 49 individuals with overweight or obesity indicated that after consumption of ellagitannins for 24 weeks, only UMB participants had a significant improvement in the blood lipid profile, while no effects were found in UMA individuals.147 However, it is important to point out that UMB individuals had a less favourable blood lipid profile to start with, so this could be a reason why only this group responded to the intervention. Supporting these findings, a 8-week crossover RCT with 51 older adults reported that a (poly)phenol-rich diet significantly reduced intestinal permeability in UMB but not in UMA individuals.148 Compared with UMA participants, UMB individuals showed a 2-fold higher improvement in zonulin levels, a marker of intestinal permeability, and an increase in HDL-cholesterol. Fatty acid oxidation was also upregulated in UMB participants after the treatment. Cortés-Martín et al.149 found that changes of gut microbiome and anthropometric metrics of post-partum mothers differed between UMs. During 1 year after delivery, UMB mothers showed a more robust gut microbial ecology that was resistant to changes, while UMA mothers had altered gut microbiota correlated with decreased waist circumference. The same authors later investigated associations between obesity prevalence and other factors including UM in a cohort of children and adolescents (n = 415).150 The ordinal logistic model showed that the prevalence of overweight-obesity was related to being a UMB or UM0 young boy, low adherence to Mediterranean diet and high contribution of 24 obesity-related single-nucleotide polymorphisms. The potential to modulate UM is therefore worthy to explore. Recently, Iglesias-Aguirre et al.151 conducted an in vivo animal study to transfer urolithin-producing bacterial consortia with the aim to convert UM0 to UMA and UMB. Urolithin-producing bacterial strains, Gordonibacter and Ellagibacter, successfully colonised the rats’ gut and replicated the ability to produce urolithins. This provides insights into the potential use of probiotics to convert non-producers into producers to benefit from (poly)phenol consumption. In addition to the investigation into bacterial strains, there are also some studies supporting the benefits of urolithin A supplements without gut metabotypes linked.152–154 To clearly demonstrate the health benefits of different metabotypes, larger cohort studies with pre-intervention stratification and balanced focus on the three metabotypes should be considered when designing protocols. Pre- and probiotic use to reproduce health-favouring gut ecology is also a novel area waiting further investigation.
Lunularin metabotypes
Resveratrol is a type of stilbenes that are mainly present in grapes, berries, peanuts and wines.155 Many preclinical studies have shown a wide range of biological properties, including anti-inflammation, anti-obesity, cardioprotective and neuroprotective effects.156–161 However, it is important to point out that all the studies showing resveratrol health benefits were conducted using amounts that are not achievable within a normal diet, and the use of resveratrol-enriched foods or supplements is needed.The research of resveratrol gut metabotypes is just at its infancy period. Bode et al.85 in 2013 discovered 2 novel metabolites of resveratrol besides dihydroresveratrol (DHR): 3,4′-dihydroxy-trans-stilbene (DHST) and 3,4′-dihydroxybibenzyl (lunularin, LUN) (Fig. 4). Among 9 subjects, only 3 produced LUN. Gut microbiome analysis showed that LUN-producers had higher abundances of Bacteroidetes, Actinobacteria, Verrucomicrobia, Cyanobacteria, Enterobacteriaceae, and Coriobacteriaceae.85Slackia equolifaciens and Adlercreutzia equolifaciens, were identified to be involved in the production of DHR (Table 1). Though only 9 participants were included, this was the first time to identify LUN and DHST.
Recently, Iglesias-Aguirre et al.162 reported a novel dehydroxylated product of LUN at 3-position, 4-hydroxydibenzyl (4HDB). This metabolite was found in urine samples from 41 participants (n = 59). The same authors then described the metabolic activities of resveratrol by the human gut microbiome and related metabotypes (i.e. LUN-producers vs. LUN non-producers).108 The gut microbiota first converts resveratrol into DHR through hydrogenation,163 then DHR undergoes two metabolic pathways: the major one in which DHR is dehydroxylated at the 5-position to yield LUN, and LUN then might be transformed into 4HDB via dihydroxylation at the 3-position; in the minor pathway, DHR is directly transformed into DHST.108 The distribution of gut metabotypes was also estimated. Among 159 healthy volunteers, there were 74.4% of LUN-producers and 25.6% of LUN non-producers. The distribution varied from geographic locations. Further analysis suggested a significant association between distribution and sex (p = 0.037), with more female being LUN non-producers.108 This study builds a foundation for future research about resveratrol-related gut metabotypes. Large sample size is necessary for the establishment of metabotypes and description of distributions, and the association with sex needs further validation.
While no studies investigating the link between resveratrol gut metabotypes and health have been conducted, a few preclinical studies have investigated the effects of DHR, LUN and DHST on different outcomes. LUN was found to inhibit E-selectin and IL-8 expression in vitro,164 and DHST was found to increase glucose uptake and induce AMPK phosphorylation independently of insulin.165 Li et al.166 reported that DHR and LUN exerted stronger anti-inflammatory and anti-cancer effects at the concentrations in mouse tissues. However, other studies found that DHR and LUN exhibited lower biological activities than resveratrol in animal models, including caloric restriction mimetics and inhibition of pancreatic oxidative damage.167,168 Therefore, it is important to conduct more research, including but not limited to in vitro and in vivo studies, to assess the bioactivity and health-related effects of resveratrol gut metabolites. Besides metabolites, resveratrol gut metabotypes, associated gut microbiota, and consequent health benefits also deserve in-depth studies, which will contribute to the understanding of variations in individual responses to resveratrol intake.
Low vs. high producer metabotypes
For many of the (poly)phenols investigated so far, a specific non-producer metabotype has not been reported, but a high inter-individual variability in their gut microbial metabolism. In this context, the variations are indicated by a gradient of metabolite production which stratifies individuals into low or high producers. With the limitations previously discussed regarding how such classification can be standardised, such gradients can be a marker of particular microbial consortia, and stratification may be useful to explain the high variability in response observed after (poly)phenol consumption. In this section we will discuss some examples of relevant (poly)phenols with very specific and unique gut microbial metabolites. Many other abundant classes of (poly)phenols, such as phenolic acids, anthocyanins or flavonols, have common gut microbial metabolites such as catechol, benzoic acids or hippuric acid derivatives which may come from multiple dietary sources and not exclusively from (poly)phenols, and are therefore not specific enough to stratify individuals into different metabotypes easily. However, the circulating levels of those common and abundant metabolites could be used as overall markers of (poly)phenol-rich food consumption and diet quality, and therefore their relationships with health outcomes and gut microbiota diversity, composition, and functionality are of great interest to investigate individual responses and mechanistic aspects.Lignans
Lignans are widely studied phenolic compounds that are rich in oilseeds, such as flaxseed, sesame, or sunflower seeds. Whole grains, legumes, fruits, and vegetables also contain low concentrations of lignans.169 Lignans have a similar structure to 17β-estradiol and are able to bind estrogen receptors, thus activating downstream signalling and exhibiting estrogenic or anti-estrogenic effects.170 Intervention and epidemiological studies have showed that lignans have a protective effect on cardiovascular diseases,171 whilst the effects on other chronic diseases, such as breast cancer, have not been unequivocally confirmed.172 The enterolignans enterodiol (ED) and enterolactone (EL) are the main gut microbial metabolites specific to lignans (Fig. 5). Despite studies suggest that these metabolites are produced by all participants, inter-individual variations in gut microbiome lead to the presence of high vs. low enterolignan excreters.173,174 Considering lignans undergo extensive phase-II metabolism, both the gut bacteria and phase-II enzymes play roles in varied metabolising capacity of lignans among individuals.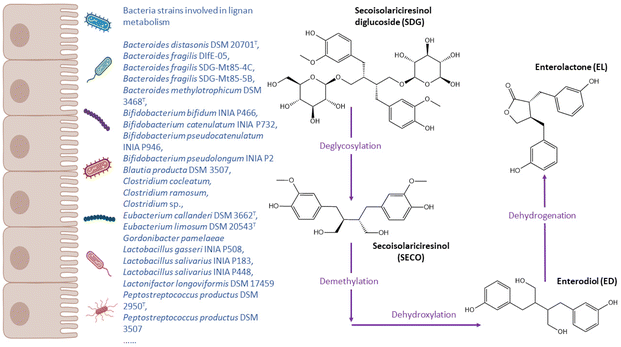 | ||
| Fig. 5 Metabolic pathways of lignan (secoisolariciresinol diglucoside) and bacteria related to enterolignan metabolism. | ||
Upon consumption, gut microorganism is responsible for the deglycosylation of lignan secoisolariciresinol diglucoside (SDG), leading to the production of secoisolariciresinol (SECO). Enterodiol is generated by the demethylation and dihydroxylation of SECO, and it then might be transformed into enterolactone through dehydrogenation reactions.175 Therefore, total of four reactions are involved in the bio-transformation of SDG into EL: deglycosylation, demethylation, dehydroxylation, and dehydrogenation. Clavel et al.176 demonstrated that the genera of Bacteroides and Clostridium were able to catalyse the first reaction, and Eubacterium limosum and Peptostreptococcus productus were involved in the demethylation step (Table 1). Peirotén et al.83 reported that some Bifidobacterium and Lactobacillus strains were capable of produce ED and EL from SECO but not other lignans (i.e. matairesinol).
The wide inter-individual variability observed in human studies is likely attributed to the biotransformation from ED to EL. When faecal samples from high and low EL producers were used to ferment flaxseed extract, 200 μM and 20 μM of EL were produced respectively, whereas ED production was similar between samples.177
In a prospective cohort study (NHS II) of 121![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 US women, higher urinary lignan metabolites, especially ED (median in the highest quartile, 342.8 nmol g−1 creatinine; median in the lowest quartile, 16.4 nmol g−1 creatinine), was associated with less weight gain (95% CI: 0.12, 0.41; p < 0.01).178 Another 10-year cohort study found that urinary EL levels (quartiles of urinary EL (ng ml−1): Q1: ≤ 127; Q2: 127–420; Q3: 420–979; Q4: > 979) were inversely associated with obesity (OR 0.30, 95% CI 0.17, 0.54; p < 0·001) and other cardiometabolic risk markers, including triglycerides, fasting glucose concentrations, and fasting insulin levels in men aged 20–60 years old.179 Two studies also suggested a positive association between plasma and urinary EL levels and diversity of gut microbiota.180,181 In the Men's Lifestyle Validation Study (MLVS) including 303 male participants, the relative abundance of Faecalibacterium prausnitzii, Alistipes shahii, Butyrivibrio crossotus, and Methanobrevibacter smithii was significantly associated with higher plasma EL levels (low EL = 4.4 nM, high EL = 22.9 nM).181 Among 115 premenopausal US women, EL production was significantly associated with alpha-diversity, and higher EL excretion (mean (μg mg−1 creatinine): 1st tertile = 0.46, 2nd tertile = 1.86, 3rd tertile = 5.66) was associated with gut microbial composition rich in Moryella spp., Acetanaerobacterium spp., Fastidiosipila spp., and Streptobacillus spp.180 The overall evidence favours enterolignan high producers as a beneficial phenotype. However, results from cohort studies should be taken with caution since many factors (diet, lifestyle, smoking habits, health status, drug use, etc.) could interfere with the results, and most observational studies did not stratify volunteers into low vs. high producers, therefore the associated links with health outcomes may simply be related to lignan intake rather than the metabolising capacity of the gut microbiota. Therefore, in the absence of stratified randomised controlled trials into low and high EL and ED producers, whether the variability in gut microbial metabolism can explain the variability in response to lignan consumption remains unknown.
700 US women, higher urinary lignan metabolites, especially ED (median in the highest quartile, 342.8 nmol g−1 creatinine; median in the lowest quartile, 16.4 nmol g−1 creatinine), was associated with less weight gain (95% CI: 0.12, 0.41; p < 0.01).178 Another 10-year cohort study found that urinary EL levels (quartiles of urinary EL (ng ml−1): Q1: ≤ 127; Q2: 127–420; Q3: 420–979; Q4: > 979) were inversely associated with obesity (OR 0.30, 95% CI 0.17, 0.54; p < 0·001) and other cardiometabolic risk markers, including triglycerides, fasting glucose concentrations, and fasting insulin levels in men aged 20–60 years old.179 Two studies also suggested a positive association between plasma and urinary EL levels and diversity of gut microbiota.180,181 In the Men's Lifestyle Validation Study (MLVS) including 303 male participants, the relative abundance of Faecalibacterium prausnitzii, Alistipes shahii, Butyrivibrio crossotus, and Methanobrevibacter smithii was significantly associated with higher plasma EL levels (low EL = 4.4 nM, high EL = 22.9 nM).181 Among 115 premenopausal US women, EL production was significantly associated with alpha-diversity, and higher EL excretion (mean (μg mg−1 creatinine): 1st tertile = 0.46, 2nd tertile = 1.86, 3rd tertile = 5.66) was associated with gut microbial composition rich in Moryella spp., Acetanaerobacterium spp., Fastidiosipila spp., and Streptobacillus spp.180 The overall evidence favours enterolignan high producers as a beneficial phenotype. However, results from cohort studies should be taken with caution since many factors (diet, lifestyle, smoking habits, health status, drug use, etc.) could interfere with the results, and most observational studies did not stratify volunteers into low vs. high producers, therefore the associated links with health outcomes may simply be related to lignan intake rather than the metabolising capacity of the gut microbiota. Therefore, in the absence of stratified randomised controlled trials into low and high EL and ED producers, whether the variability in gut microbial metabolism can explain the variability in response to lignan consumption remains unknown.
Flavanones
Citrus fruits are rich food sources of flavanones, in which hesperidin (hesperetin-7-O-rutinoside) represents a great amount of total flavanones in oranges and orange juices. After consumed by humans, a small fraction of flavanones is absorbed in the small intestine, the rest of hesperidin is cleaved by microorganism in the colon releasing hesperetin, then undergo phase-II metabolism.182 In this process, gut microbiome, including Bifidobacterium, Clostridium, Bacteroides, Lactobacillus and Eubacterium, plays a role in the deglycosylation of rhamnoglucoside moiety of hesperidin.29,183A large inter-individual variability has been observed in the metabolism and excretion of flavanones. Tomás-Navarro et al.184 defined high flavanone excretors as those with flavanone excretion values >10% of the ingested flavanones; medium excretors as those with excretion values between 5 and 10%, and low excretors those with excretion values <5%. Vallejo et al.185 used instead tertiles to stratify producers. Despite the different classification strategies, both studies reported that high hesperetin excretors produced nearly 5–6 times more urinary flavanones than low hesperetin excretors.184,185 The rhamnosidase activity of the gut microbiome is suggested to be one of the factors accounting for the variations. The urinary metabolites detected were mostly phase-II metabolites, such as hesperetin glucuronides and hesperetin sulfates.185,186 A cross-over study (n = 21) by Nishioka et al.187 was unable to find a significant correlation between excretion gradient and gut microbiome composition at the genus and/or species level. Moreover, the solubility of the food matrix also affects hesperidin metabolism.188 The gut microbiome associated to flavanone metabotypes therefore remains inconclusive. Important to note that flavanones share other common and abundant gut metabolites with other (poly)phenols, including hydroxyphenylacetic, hydroxybenzoic and hydroxyphenylpropionic acids.189 However, it is challenging to use them as metabotyping tools, due to their low specificity. This issue also exists for other (poly)phenols, such as flavan-3-ols.
Flavan-3-ols
Flavan-3-ol monomers (i.e. catechin and epicatechin), oligomers and polymers (also known as condensed tannins or proanthocyanidins) are among the most consumed (poly)phenols. In the human diet, the main sources are tea, pome fruits, berries and cocoa products.190 Phenyl-γ-valerolactones (PVL) and phenylvaleric acids (PVA) have been identified as main gut microbial metabolites of flavan-3-ols. The colonic metabolites might be further catabolised by intestinal bacteria into low molecular weight phenolic compounds or conjugated by phase-II enzymes, then excreted in urine.191 With respect to the involved gut microbiome, some bacteria capable of carrying out specific actions have been linked to the gut metabolism of flavan-3-ols: Adlercreutzia equolifaciens and Eggerthella lenta are able to stimulate C-ring cleavage of (epi)catechins, and Eggerthella lenta also performes 4′-dehydroxylation of ring fission product.192Flavonifractor plautii has been showed to convert phenyl-γ-valerolactones into 3-(phenyl)propionic acids.51 However, information regarding gut microbiome species involved in the biotransformation of flavan-3-ols into PVL and PVA is still limited.A large inter-individual variability in flavan-3-ol metabolism has been reported in studies using in vitro faecal fermentation and human interventions.193–197 Mena et al.196,198 described potential gut metabotypes related with flavan-3-ols: one with high production of tri- and di-hydroxyphenyl-γ-valerolactones and a reduced excretion of 3-(hydroxyphenyl)propionic acid; another is characterised by a low production of phenyl-γ-valerolactones but high amounts of 3-(hydroxyphenyl)propionic acid. Besides these two metabotypes, Cortés-Martín et al.199 proposed two additional clusters, one with high excretion of all the PVL and PVA derivatives and the other with low excretion of all of them. Considering that these colonic metabolites are not final products and might be further broken down by bacteria into other common (poly)phenol metabolites, such as phenylpropionic, phenylacetic, and benzoic acids, which are widely abundant in the diet and coming from multiple dietary and non-dietary sources, variations in the amount of metabolite excreted should be taken with caution when defining metabotypes in these studies. More recently, Tosi et al.200 proposed new additional metabotypes for cranberry (poly)phenols using data from a 12-week RCT among 60 healthy older adults. Metabotype 1 was characterized by a higher excretion of 5-(3′,4′-dihydroxyphenyl)-γ-valerolactones (3′,4′-diHPVLs) and 5-(3′-hydroxyphenyl)-γ-valerolactones (3′-HPVLs), metabotype 2 had a higher excretion of 3′-hydroxycinnamic acids (3′-HCAs), 3- (hydroxyphenyl)propanoic acids (HPPAs) and 3-hydroxybenzoic acids (3-HBAs), while metabotype 3 was characterized by a low excretion of all metabolites. The proposed metabotypes were not specific to flavan-3-ols, instead, the authors aimed to apply a quali-quantitative approach including various classes of (poly)phenols with different metabolic pathways. Using clustering techniques to establish metabotypes with multiple gut microbial metabolites is an useful approach, and more research in this area is needed,201 in particular for non-specific gut microbial metabolites such benzoic, catechol or phenylacetic acid metabolites, which are some of the most abundant (poly)phenol gut microbial metabolites, and the final products for most (poly)phenols.
Hop prenylflavonoids
Hop-derived prenylflavonoids, including xanthohumol, isoxanthohumol and 8-prenylnaringenin (8-PN), are not commonly found in most foods but are present in beer and hop-containing dietary supplements.202 They have been found to possess antioxidant, anti-proliferative, anti-inflammatory, estrogenic and immune-regulatory properties.202–206In vitro and animal studies have shown that isoxanthohumol can be converted into 8-PN by the action of Eubacterium limosum.207 In this regard, inter-individual differenced have been observed for the conversion, which is attributed to the biotransformation capacity of the gut bacteria.208,209 An intervention study with 50 healthy post-menopausal women found a considerable variation in the urinary recovery of 8-PN between low, moderate and high 8-PN producers.209 The stratification was based on the ratio 8-PN:(8-PN + isoxanthohumol), and the amount of 8-PN excreted in high excretors was nearly 5-fold than that in low excretors. An inverse relationship between antibiotics use and 8-PN production was also reported.209 These results supports the notion that gut microbiota is involved in the production of 8-PN. However, 8-PN can be further metabolised by liver microsomes, specifically in the prenyl group and the flavanone skeleton.210 Evidence regarding the metabolite production gradient of prenylflavonoids is limited, leading to the ambiguous demonstration of gut metabotypes for 8-PN.Metabotype clustering: is there a common metabotype for multiple (poly)phenol subclasses?
An important question that remains unknown is whether a (poly)phenol “superproducer” metabotype exist, or if same individuals share the same capacity to produce different (poly)phenol gut microbial metabolites. Very recently, Iglesias-Aguirre et al.211 investigated gut metabotype clusters (MC) for resveratrol, ellagic acid and daidzein in 127 individuals. To our knowledge, this is the first study investigating (poly)phenol gut metabotype clusters for different (poly)phenols, which would be more applicable to real life settings since multiple (poly)phenols are consumed together within the same foods and certain gut bacteria species might be involved in the metabolism of more than one (poly)phenol class. A total of 10 metabotype clusters were proposed, being the 5 more prevalent the following: MC1 (equol-non-producer (ENP) + UMB + LUN- producer (LP)) and MC2 (ENP + UMA + LP) were the most abundant, followed by MC3 (equol-producer (EP) + UMA + LP), MC4 (EP + UMB + LP) and MC5 (ENP + UMA + lunularin-non-producer (LNP)). The association between gut microbiome and metabotype clusters was explored, and for instance, Gordonibacter and Eggerthella were positively associated with urolithin A while inversely associated with isourolithin A and urolithin B, indicated by the higher abundance of these two genera in UMA-including MCs (i.e. MC2, MC3, and MC5). Akkermansia was positively associated with equol, urolithin A and LUN, which was showed by a higher abundance in MC2 and MC3 with UMA and LP prioritized. Although the sample size, participants’ ethnicity and intervention duration of this trial were limited, the study provides useful guidance for addressing the complexity of analyzing different classes of (poly)phenol gut metabolites in real-world settings. Considering that different (poly)phenols share some common metabolic pathways and metabolites, future studies could investigate the role of gut bacteria in both specific and combined (poly)phenol metabolism and further confirm the prediction for the models on health outcomes through pre-stratified randomized controlled trials.Conclusion
Available evidence demonstrated a two-way interaction between dietary (poly)phenols and human gut microbiota, that is, (poly)phenols consumed are transformed by gut microbiota into smaller absorbable compounds, and these metabolites in turn modulate gut microbial population and subsequent health effects. Current research has noted that variations in gut microbiota and (poly)phenol gut metabolism exist among individuals, which may explain the differences observed in the biological responses to (poly)phenol consumption. We have examples of specific bacterial species mediating key chemical transformations, but much evidence remains correlative and there is an over reliance on the reductionist approach, with few studies examining the relevance of mixed microbial consortia or cooperation between bacterial species in (poly)phenol biotransformation. Therefore, (poly)phenol-related gut metabotypes have been proposed as a biomarker for intestinal microbial ecology and individual health status. Based on related scientific evidence, this could contribute to the development of tailored dietary recommendations, especially regarding the consumption of polyphenol-rich diets, which is a critical aspect of personalised nutrition.Research on (poly)phenol-related gut microbial metabotypes is still in its infancy. Many open questions still remain for the most widely investigated metabotypes, the daidzein related equol and ODMA producers and non-producers. Mixed results exist for observational studies linking equol and ODMA production with health outcomes, as well as for stratified studies investigating variability in response with metabotypes. The main factors driving the prevalence of metabotypes among populations are also unclear, but indications suggest age, sex and disease state can impact on metabotypes. Whether the background diet, and in particular habitual (poly)phenol consumption is an important factor in metabotype prevalence is unclear, and the lack of reliable and accurate methods for estimating (poly)phenol intake is likely a confounding factor in this matter.
Metabotypes could be seen from a microbial point of view as markers of microbiome health. Since microbial metabolic activities are modifiable, then the concentrations of the key microbial metabolites which define metabotypes might change with changes in the microbiota e.g. induced by age, diet, or disease, with a clear example on the urolithin metabotype being modulated by age. Metabotypes then become a marker or readout of microbial activity at a particular time (age) and space (nutritional or dietary space for example), and could be used to define how healthy a given diet:microbiome state is along the scale from healthy to dysbiotic.
The existence of potential metabotypes for other (poly)phenols remains ambiguous. In this regard, an integration of metagenomics and metabolomics could contribute to a better understanding of (poly)phenol metabolism and the role of gut microbiota. Using metabolomics allows determination of functional aspects which is often not captured by gut microbiome composition analyses. Large cohort studies are a suitable means to examine the distribution and determinants of metabotypes and also concentration ranges of (poly)phenol unique marker metabolites, but also common metabolites of multiple (poly)phenols. Application of artificial intelligence here would help identify new, less obvious metabotypes defined by concentration ranges of common (poly)phenol metabolites. Clinical intervention trials should also be carried out to identify the role of (poly)phenol metabolites in the modulation of health effects. Overall, the relationships between (poly)phenol metabolism, gut microbiota composition, and subsequent health effects deserve further research.
Author contributions
J. H. and A. R. M. wrote the first draft. All authors improved and critically revised the manuscript, figures, and tables. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts of interest to declare.References
- M. Leri, M. Scuto, M. L. Ontario, V. Calabrese, E. J. Calabrese, M. Bucciantini and M. Stefani, Healthy Effects of Plant Polyphenols: Molecular Mechanisms, Int. J. Mol. Sci., 2020, 21, 1250 CrossRef CAS PubMed.
- M. I. Khan, J. H. Shin, T. S. Shin, M. Y. Kim, N. J. Cho and J. D. Kim, Anthocyanins from Cornus kousa ethanolic extract attenuate obesity in association with anti-angiogenic activities in 3T3-L1 cells by down-regulating adipogeneses and lipogenesis, PLoS One, 2018, 13, e0208556 CrossRef PubMed.
- D. Esposito, A. Chen, M. H. Grace, S. Komarnytsky and M. A. Lila, Inhibitory Effects of Wild Blueberry Anthocyanins and Other Flavonoids on Biomarkers of Acute and Chronic Inflammation in Vitro, J. Agric. Food Chem., 2014, 62, 7022–7028 CrossRef CAS PubMed.
- M. M. Coman, A. M. Oancea, M. C. Verdenelli, C. Cecchini, G. E. Bahrim, C. Orpianesi, A. Cresci and S. Silvi, Polyphenol content and in vitro evaluation of antioxidant, antimicrobial and prebiotic properties of red fruit extracts, Eur. Food Res. Technol., 2018, 244, 735–745 CrossRef CAS.
- M. Inglés, J. Gambini, M. G. Miguel, V. Bonet-Costa, K. M. Abdelaziz, M. El Alami, J. Viña and C. Borrás, PTEN Mediates the Antioxidant Effect of Resveratrol at Nutritionally Relevant Concentrations, BioMed Res. Int., 2014, 2014, e580852 Search PubMed.
- M. Yousef, I. A. Vlachogiannis and E. Tsiani, Effects of Resveratrol against Lung Cancer: In Vitro and In Vivo Studies, Nutrients, 2017, 9, 1231 CrossRef PubMed.
- P. Basu and C. Maier, Phytoestrogens and breast cancer: In vitro anticancer activities of isoflavones, lignans, coumestans, stilbenes and their analogs and derivatives, Biomed. Pharmacother., 2018, 107, 1648–1666 CrossRef CAS PubMed.
- M. Á. Ávila-Gálvez, A. González-Sarrías and J. C. Espín, In Vitro Research on Dietary Polyphenols and Health: A Call of Caution and a Guide on How To Proceed, J. Agric. Food Chem., 2018, 66, 7857–7858 CrossRef PubMed.
- P. Mena and D. Del Rio, Gold Standards for Realistic (Poly)phenol Research, J. Agric. Food Chem., 2018, 66, 8221–8223 CrossRef CAS PubMed.
- A. Tresserra-Rimbau, E. B. Rimm, A. Medina-Remón, M. A. Martínez-González, R. de la Torre, D. Corella, J. Salas-Salvadó, E. Gómez-Gracia, J. Lapetra, F. Arós, M. Fiol, E. Ros, L. Serra-Majem, X. Pintó, G. T. Saez, J. Basora, J. V. Sorlí, J. A. Martínez, E. Vinyoles, V. Ruiz-Gutiérrez, R. Estruch and R. M. Lamuela-Raventós, Inverse association between habitual polyphenol intake and incidence of cardiovascular events in the PREDIMED study, Nutr., Metab. Cardiovasc. Dis., 2014, 24, 639–647 CrossRef CAS PubMed.
- A. A. Fallah, E. Sarmast and T. Jafari, Effect of dietary anthocyanins on biomarkers of glycemic control and glucose metabolism: A systematic review and meta-analysis of randomized clinical trials, Food Res. Int., 2020, 137, 109379 CrossRef CAS PubMed.
- M. Bonaccio, G. Pounis, C. Cerletti, M. B. Donati, L. Iacoviello, G. de Gaetano and on behalf of the M.-S. S. Investigators, Mediterranean diet, dietary polyphenols and low grade inflammation: results from the MOLI-SANI study, Br. J. Clin. Pharmacol., 2017, 83, 107–113 CrossRef CAS PubMed.
- D. J. Lamport, L. Dye, J. D. Wightman and C. L. Lawton, The effects of flavonoid and other polyphenol consumption on cognitive performance: A systematic research review of human experimental and epidemiological studies, Nutr. Aging, 2012, 1, 5–25 Search PubMed.
- C. Valls-Pedret, R. M. Lamuela-Raventós, A. Medina-Remón, M. Quintana, D. Corella, X. Pintó, M. Á. Martínez-González, R. Estruch and E. Ros, Polyphenol-Rich Foods in the Mediterranean Diet are Associated with Better Cognitive Function in Elderly Subjects at High Cardiovascular Risk, J. Alzheimer's Dis., 2012, 29, 773–782 CAS.
- L. T. Fike, H. Munro, D. Yu, Q. Dai and M. J. Shrubsole, Dietary polyphenols and the risk of colorectal cancer in the prospective Southern Community Cohort Study, Am. J. Clin. Nutr., 2022, 115, 1155–1165 CrossRef CAS PubMed.
- N. M. Pham, V. V. Do and A. H. Lee, Polyphenol-rich foods and risk of gestational diabetes: a systematic review and meta-analysis, Eur. J. Clin. Nutr., 2019, 73, 647–656 CrossRef CAS PubMed.
- F. Potì, D. Santi, G. Spaggiari, F. Zimetti and I. Zanotti, Polyphenol Health Effects on Cardiovascular and Neurodegenerative Disorders: A Review and Meta-Analysis, Int. J. Mol. Sci., 2019, 20, 351 CrossRef PubMed.
- E. R. Gibney, D. Milenkovic, E. Combet, T. Ruskovska, A. Greyling, A. González-Sarrías, B. de Roos, F. Tomás-Barberán, C. Morand and A. Rodriguez-Mateos, Factors influencing the cardiometabolic response to (poly)phenols and phytosterols: a review of the COST Action POSITIVe activities, Eur. J. Nutr., 2019, 58, 37–47 CrossRef CAS PubMed.
- B. de Roos, A.-M. Aura, M. Bronze, A. Cassidy, M.-T. G. Conesa, E. R. Gibney, A. Greyling, J. Kaput, Z. Kerem, N. Knežević, P. Kroon, R. Landberg, C. Manach, D. Milenkovic, A. Rodriguez-Mateos, F. A. Tomás-Barberán, T. van de Wiele and C. Morand, Targeting the delivery of dietary plant bioactives to those who would benefit most: from science to practical applications, Eur. J. Nutr., 2019, 58, 65–73 CrossRef PubMed.
- L. Chen, H. Cao and J. Xiao, in Polyphenols: Properties, Recovery, and Applications, ed. C. M. Galanakis, Woodhead Publishing, 2018, pp. 45–67 Search PubMed.
- S. Mithul Aravind, S. Wichienchot, R. Tsao, S. Ramakrishnan and S. Chakkaravarthi, Role of dietary polyphenols on gut microbiota, their metabolites and health benefits, Food Res Int, 2021, 142, 110189 CrossRef CAS PubMed.
- A. González-Sarrías, J. A. Giménez-Bastida, M. Á. Núñez-Sánchez, M. Larrosa, M. T. García-Conesa, F. A. Tomás-Barberán and J. C. Espín, Phase-II metabolism limits the antiproliferative activity of urolithins in human colon cancer cells, Eur. J. Nutr., 2014, 53, 853–864 CrossRef PubMed.
- L. Rubio, A. Macia and M.-J. Motilva, Impact of Various Factors on Pharmacokinetics of Bioactive Polyphenols: An Overview, Curr. Drug Metab., 2014, 15, 62–76 CrossRef CAS PubMed.
- T. Rao, Z. Tan, J. Peng, Y. Guo, Y. Chen, H. Zhou and D. Ouyang, The pharmacogenetics of natural products: A pharmacokinetic and pharmacodynamic perspective, Pharmacol. Res., 2019, 146, 104283 CrossRef CAS PubMed.
- C. L. Frankenfeld, O-Desmethylangolensin: The Importance of Equol’s Lesser Known Cousin to Human Health, Adv. Nutr., 2011, 2, 317–324 CrossRef CAS PubMed.
- F. A. Tomás-Barberán, A. González-Sarrías, R. García-Villalba, M. A. Núñez-Sánchez, M. V. Selma, M. T. García-Conesa and J. C. Espín, Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status, Mol. Nutr. Food Res., 2017, 61, 1500901 CrossRef PubMed.
- F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, Interactions of gut microbiota with dietary polyphenols and consequences to human health, Curr. Opin. Clin. Nutr. Metab. Care, 2016, 19, 471–476 CrossRef PubMed.
- J. C. Espín, A. González-Sarrías and F. A. Tomás-Barberán, The gut microbiota: A key factor in the therapeutic effects of (poly)phenols, Biochem. Pharmacol., 2017, 139, 82–93 CrossRef PubMed.
- A. Braune and M. Blaut, Bacterial species involved in the conversion of dietary flavonoids in the human gut, Gut Microbes, 2016, 7, 216–234 CrossRef CAS PubMed.
- X. Feng, Y. Li, M. Brobbey Oppong and F. Qiu, Insights into the intestinal bacterial metabolism of flavonoids and the bioactivities of their microbe-derived ring cleavage metabolites, Drug Metab. Rev., 2018, 50, 343–356 CrossRef CAS PubMed.
- X.-L. Wang, K.-T. Kim, J.-H. Lee, H.-G. Hur and S.-I. Kim, C-Ring Cleavage of Isoflavones Daidzein and Genistein by a Newly-Isolated Human Intestinal Bacterium Eubacterium ramulus Julong 601, J. Microbiol. Biotechnol., 2004, 14, 766–771 CAS.
- S. Yokoyama, T. Niwa, T. Osawa and T. Suzuki, Characterization of an O-desmethylangolensin-producing bacterium isolated from human feces, Arch. Microbiol., 2010, 192, 15–22 CrossRef CAS PubMed.
- S. Yokoyama and T. Suzuki, Isolation and characterization of a novel equol-producing bacterium from human feces, Biosci., Biotechnol., Biochem., 2008, 72, 2660–2666 CrossRef CAS PubMed.
- J.-S. Jin, T. Nishihata, N. Kakiuchi and M. Hattori, Biotransformation of C-glucosylisoflavone puerarin to estrogenic (3S)-equol in co-culture of two human intestinal bacteria, Biol. Pharm. Bull., 2008, 31, 1621–1625 CrossRef CAS PubMed.
- A. Matthies, M. Blaut and A. Braune, Isolation of a Human Intestinal Bacterium Capable of Daidzein and Genistein Conversion, Appl. Environ. Microbiol., 2009, 75, 1740–1744 CrossRef CAS PubMed.
- Y. Shimada, S. Yasuda, M. Takahashi, T. Hayashi, N. Miyazawa, I. Sato, Y. Abiru, S. Uchiyama and H. Hishigaki, Cloning and Expression of a Novel NADP(H)-Dependent Daidzein Reductase, an Enzyme Involved in the Metabolism of Daidzein, from Equol-Producing Lactococcus Strain 20-92, Appl. Environ. Microbiol., 2010, 76, 5892–5901 CrossRef CAS PubMed.
- J. F. Stevens and C. S. Maier, The chemistry of gut microbial metabolism of polyphenols, Phytochem. Rev., 2016, 15, 425–444 CrossRef CAS PubMed.
- R. García-Villalba, D. Beltrán, M. D. Frutos, M. V. Selma, J. C. Espín and F. A. Tomás-Barberán, Metabolism of different dietary phenolic compounds by the urolithin-producing human-gut bacteria Gordonibacter urolithinfaciens and Ellagibacter isourolithinifaciens, Food Funct., 2020, 11, 7012–7022 RSC.
- M. Ávila, M. Hidalgo, C. Sánchez-Moreno, C. Pelaez, T. Requena and S. de Pascual-Teresa, Bioconversion of anthocyanin glycosides by Bifidobacteria and Lactobacillus, Food Res. Int., 2009, 42, 1453–1461 CrossRef.
- P. Gaya, Á. Peirotén, M. Medina, I. Álvarez and J. M. Landete, Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins A and B from ellagic acid, J. Funct. Foods, 2018, 45, 95–99 CrossRef CAS.
- R. García-Villalba, D. Beltrán, J. C. Espín, M. V. Selma and F. A. Tomás-Barberán, Time Course Production of Urolithins from Ellagic Acid by Human Gut Microbiota, J. Agric. Food Chem., 2013, 61, 8797–8806 CrossRef PubMed.
- D. Beltrán, M. Romo-Vaquero, J. C. Espín, F. A. Tomás-Barberán and M. V. Selma, Ellagibacter isourolithinifaciens gen. nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut, Int. J. Syst. Evol. Microbiol., 2018, 68, 1707–1712 CrossRef PubMed.
- X. Zhang, Y. Fang, G. Yang, X. Hou, Y. Hai, M. Xia, F. He, Y. Zhao and S. Liu, Isolation and characterization of a novel human intestinal Enterococcus faecium FUA027 capable of producing urolithin A from ellagic acid, Front. Nutr., 2022, 9, 1039697 CrossRef PubMed.
- M. V. Selma, F. A. Tomás-Barberán, D. Beltrán, R. García-Villalba and J. C. Espín, Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut, Int. J. Syst. Evol. Microbiol., 2014, 64, 2346–2352 CrossRef CAS PubMed.
- M. V. Selma, D. Beltrán, R. García-Villalba, J. C. Espín and F. A. Tomás-Barberán, Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species, Food Funct., 2014, 5, 1779–1784 RSC.
- H. Mi, S. Liu, Y. Hai, G. Yang, J. Lu, F. He, Y. Zhao, M. Xia, X. Hou and Y. Fang, Lactococcus garvieae FUA009, a Novel Intestinal Bacterium Capable of Producing the Bioactive Metabolite Urolithin A from Ellagic Acid, Foods, 2022, 11, 2621 CrossRef CAS PubMed.
- Q. Liu, Y. Bian, S. Mu, M. Chen, S. Liu, G. Yang, Y. Huang, X. Hou and Y. Fang, Genomic and phenotypic-based safety assessment and probiotic properties of Streptococcus thermophilus FUA329, a urolithin A-producing bacterium of human milk origin, Genomics, 2023, 115, 110724 CrossRef CAS PubMed.
- Y. Miyake, K. Yamamoto and T. Osawa, Metabolism of Antioxidant in Lemon Fruit (Citrus limon BURM. f.) by Human Intestinal Bacteria, J. Agric. Food Chem., 1997, 45, 3738–3742 CrossRef CAS.
- A. Amaretti, S. Raimondi, A. Leonardi, A. Quartieri and M. Rossi, Hydrolysis of the Rutinose-Conjugates Flavonoids Rutin and Hesperidin by the Gut Microbiota and Bifidobacteria, Nutrients, 2015, 7, 2788–2800 CrossRef CAS PubMed.
- A. Takagaki and F. Nanjo, Biotransformation of (-)-epigallocatechin and (-)-gallocatechin by intestinal bacteria involved in isoflavone metabolism, Biol. Pharm. Bull., 2015, 38, 325–330 CrossRef CAS PubMed.
- M. Kutschera, W. Engst, M. Blaut and A. Braune, Isolation of catechin-converting human intestinal bacteria, J. Appl. Microbiol., 2011, 111, 165–175 CrossRef CAS PubMed.
- M. Kim, N. Kim and J. Han, Metabolism of Kaempferia parviflora Polymethoxyflavones by Human Intestinal Bacterium Bautia sp. MRG-PMF1, J. Agric. Food Chem., 2014, 62, 12377–12383 CrossRef CAS PubMed.
- A. Braune and M. Blaut, Intestinal Bacterium Eubacterium cellulosolvens Deglycosylates Flavonoid C- and O-Glucosides, Appl. Environ. Microbiol., 2012, 78, 8151–8153 CrossRef CAS PubMed.
- M. R. Schaab, B. M. Barney and W. A. Francisco, Kinetic and spectroscopic studies on the quercetin 2,3-dioxygenase from Bacillus subtilis, Biochemistry, 2006, 45, 1009–1016 CrossRef CAS PubMed.
- V. D. Bokkenheuser, C. H. Shackleton and J. Winter, Hydrolysis of dietary flavonoid glycosides by strains of intestinal Bacteroides from humans., Biochem. J., 1987, 248, 953–956 CrossRef CAS PubMed.
- I. Marotti, A. Bonetti, B. Biavati, P. Catizone and G. Dinelli, Biotransformation of Common Bean (Phaseolus vulgaris L.) Flavonoid Glycosides by Bifidobacterium Species from Human Intestinal Origin, J. Agric. Food Chem., 2007, 55, 3913–3919 CrossRef CAS PubMed.
- S.-H. Bang, Y.-J. Hyun, J. Shim, S.-W. Hong and D.-H. Kim, Metabolism of rutin and poncirin by human intestinal microbiota and cloning of their metabolizing α-L-rhamnosidase from Bifidobacterium dentium, J. Microbiol. Biotechnol., 2015, 25, 18–25 CrossRef CAS PubMed.
- L. Schoefer, R. Mohan, A. Schwiertz, A. Braune and M. Blaut, Anaerobic Degradation of Flavonoids by Clostridium orbiscindens, Appl. Environ. Microbiol., 2003, 69, 5849–5854 CrossRef CAS PubMed.
- Y. Liu, Y. Liu, Y. Dai, L. Xun and M. Hu, Enteric Disposition and Recycling of Flavonoids and Ginkgo Flavonoids, J. Altern. Complementary Med., 2003, 9, 631–640 CrossRef PubMed.
- N. R. Shin, J. S. Moon, S.-Y. Shin, L. Li, Y. B. Lee, T.-J. Kim and N. S. Han, Isolation and characterization of human intestinal Enterococcus avium EFEL009 converting rutin to quercetin, Lett. Appl. Microbiol., 2016, 62, 68–74 CrossRef CAS PubMed.
- H. Schneider, A. Schwiertz, M. D. Collins and M. Blaut, Anaerobic transformation of quercetin-3-glucoside by bacteria from the human intestinal tract, Arch. Microbiol., 1999, 171, 81–91 CrossRef CAS PubMed.
- H. Schneider and M. Blaut, Anaerobic degradation of flavonoids by Eubacterium ramulus, Arch. Microbiol., 2000, 173, 71–75 CrossRef CAS PubMed.
- A. Braune, M. Gütschow, W. Engst and M. Blaut, Degradation of Quercetin and Luteolin by Eubacterium ramulus, Appl. Environ. Microbiol., 2001, 67, 5558–5567 CrossRef CAS PubMed.
- K. Ulbrich, N. Reichardt, A. Braune, L. W. Kroh, M. Blaut and S. Rohn, The microbial degradation of onion flavonol glucosides and their roasting products by the human gut bacteria Eubacterium ramulus and Flavonifractor plautii, Food Res. Int., 2015, 67, 349–355 CrossRef CAS.
- J. Beekwilder, D. Marcozzi, S. Vecchi, R. de Vos, P. Janssen, C. Francke, J. van Hylckama Vlieg and R. D. Hall, Characterization of Rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus, Appl. Environ. Microbiol., 2009, 75, 3447–3454 CrossRef CAS PubMed.
- T. Maruo, M. Sakamoto, C. Ito, T. Toda and Y. Benno, Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella, Int. J. Syst. Evol. Microbiol., 2008, 58, 1221–1227 CrossRef CAS PubMed.
- K. Minamida, K. Ota, M. Nishimukai, M. Tanaka, A. Abe, T. Sone, F. Tomita, H. Hara and K. Asano, Asaccharobacter celatus gen. nov., sp. nov., isolated from rat caecum, Int. J. Syst. Evol. Microbiol., 2008, 58, 1238–1240 CrossRef CAS PubMed.
- S. Raimondi, L. Roncaglia, M. De Lucia, A. Amaretti, A. Leonardi, U. M. Pagnoni and M. Rossi, Bioconversion of soy isoflavones daidzin and daidzein by Bifidobacterium strains, Appl. Microbiol. Biotechnol., 2009, 81, 943–950 CrossRef CAS PubMed.
- D. Tsangalis, J. F. Ashton, A. E. J. Mcgill and N. P. Shah, Enzymic Transformation of Isoflavone Phytoestrogens in Soymilk by β-Glucosidase-Producing Bifidobacteria, J. Food Sci., 2002, 67, 3104–3113 CrossRef CAS.
- S. Raimondi, L. Roncaglia, M. De Lucia, A. Amaretti, A. Leonardi, U. M. Pagnoni and M. Rossi, Bioconversion of soy isoflavones daidzin and daidzein by Bifidobacterium strains, Appl. Microbiol. Biotechnol., 2009, 81, 943–950 CrossRef CAS PubMed.
- H.-G. Hur, R. D. Beger, T. M. Heinze, J. O. Lay, J. P. Freeman, J. Dore and F. Rafii, Isolation of an anaerobic intestinal bacterium capable of cleaving the C-ring of the isoflavonoid daidzein, Arch. Microbiol., 2002, 178, 8–12 CrossRef CAS PubMed.
- M. Tamura, T. Tsushida and K. Shinohara, Isolation of an isoflavone-metabolizing, Clostridium-like bacterium, strain TM-40, from human faeces, Anaerobe, 2007, 13, 32–35 CrossRef CAS PubMed.
- A. Matthies, T. Clavel, M. Gütschow, W. Engst, D. Haller, M. Blaut and A. Braune, Conversion of Daidzein and Genistein by an Anaerobic Bacterium Newly Isolated from the Mouse Intestine, Appl. Environ. Microbiol., 2008, 74, 4847–4852 CrossRef CAS PubMed.
- S. Yokoyama, K. Oshima, I. Nomura, M. Hattori and T. Suzuki, Complete Genomic Sequence of the Equol-Producing Bacterium Eggerthella sp. Strain YY7918, Isolated from Adult Human Intestine, J. Bacteriol., 2011, 193, 5570–5571 CrossRef CAS PubMed.
- X.-L. Wang, H.-J. Kim, S.-I. Kang, S.-I. Kim and H.-G. Hur, Production of phytoestrogen S-equol from daidzein in mixed culture of two anaerobic bacteria, Arch. Microbiol., 2007, 187, 155–160 CrossRef CAS PubMed.
- M. Kim, J. Lee and J. Han, Deglycosylation of isoflavone C-glycosides by newly isolated human intestinal bacteria, J. Sci. Food Agric., 2015, 95, 1925–1931 CrossRef CAS PubMed.
- H.-G. Hur, J. O. Lay Jr., R. D. Beger, J. P. Freeman and F. Rafii, Isolation of human intestinal bacteria metabolizing the natural isoflavone glycosides daidzin and genistin, Arch. Microbiol., 2000, 174, 422–428 CrossRef CAS PubMed.
- L. Schoefer, R. Mohan, A. Braune, M. Birringer and M. Blaut, Anaerobic C-ring cleavage of genistein and daidzein by Eubacterium ramulus, FEMS Microbiol. Lett., 2002, 208, 197–202 CrossRef CAS PubMed.
- A. Braune and M. Blaut, Deglycosylation of puerarin and other aromatic C-glucosides by a newly isolated human intestinal bacterium, Environ. Microbiol., 2011, 13, 482–494 CrossRef CAS PubMed.
- C. Schröder, A. Matthies, W. Engst, M. Blaut and A. Braune, Identification and Expression of Genes Involved in the Conversion of Daidzein and Genistein by the Equol-Forming Bacterium Slackia isoflavoniconvertens, Appl. Environ. Microbiol., 2013, 79, 3494–3502 CrossRef PubMed.
- H. Tsuji, K. Moriyama, K. Nomoto and H. Akaza, Identification of an Enzyme System for Daidzein-to-Equol Conversion in Slackia sp. Strain NATTS, Appl. Environ. Microbiol., 2012, 78, 1228–1236 CrossRef CAS PubMed.
- T. Clavel, G. Henderson, W. Engst, J. Doré and M. Blaut, Phylogeny of human intestinal bacteria that activate the dietary lignan secoisolariciresinol diglucoside, FEMS Microbiol. Ecol., 2006, 55, 471–478 CrossRef CAS PubMed.
- Á. Peirotén, P. Gaya, I. Álvarez, D. Bravo and J. M. Landete, Influence of different lignan compounds on enterolignan production by Bifidobacterium and Lactobacillus strains, Int. J. Food Microbiol., 2019, 289, 17–23 CrossRef PubMed.
- E. N. Bess, J. E. Bisanz, F. Yarza, A. Bustion, B. E. Rich, X. Li, S. Kitamura, E. Waligurski, Q. Y. Ang, D. L. Alba, P. Spanogiannopoulos, S. Nayfach, S. K. Koliwad, D. W. Wolan, A. A. Franke and P. J. Turnbaugh, Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria, Nat. Microbiol., 2020, 5(1), 56–66 CrossRef CAS PubMed.
- L. M. Bode, D. Bunzel, M. Huch, G.-S. Cho, D. Ruhland, M. Bunzel, A. Bub, C. M. Franz and S. E. Kulling, In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota, Am. J. Clin. Nutr., 2013, 97, 295–309 CrossRef CAS PubMed.
- I. L. Paraiso, L. S. Plagmann, L. Yang, R. Zielke, A. F. Gombart, C. S. Maier, A. E. Sikora, P. R. Blakemore and J. F. Stevens, Reductive metabolism of xanthohumol and 8-prenylnaringenin by the intestinal bacterium Eubacterium ramulus, Mol. Nutr. Food Res., 2019, 63, e1800923 CrossRef PubMed.
- M. Moorthy, U. Sundralingam and U. D. Palanisamy, Polyphenols as Prebiotics in the Management of High-Fat Diet-Induced Obesity: A Systematic Review of Animal Studies, Foods, 2021, 10, 299 CrossRef CAS PubMed.
- H. C. Lee, A. M. Jenner, C. S. Low and Y. K. Lee, Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota, Res. Microbiol., 2006, 157, 876–884 CrossRef CAS PubMed.
- A. Duda-Chodak, T. Tarko, P. Satora and P. Sroka, Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review, Eur. J. Nutr., 2015, 54, 325–341 CrossRef CAS PubMed.
- M. I. Queipo-Ortuño, M. Boto-Ordóñez, M. Murri, J. M. Gomez-Zumaquero, M. Clemente-Postigo, R. Estruch, F. Cardona Diaz, C. Andrés-Lacueva and F. J. Tinahones, Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers, Am. J. Clin. Nutr., 2012, 95, 1323–1334 CrossRef PubMed.
- J.-P. Rauha, S. Remes, M. Heinonen, A. Hopia, M. Kähkönen, T. Kujala, K. Pihlaja, H. Vuorela and P. Vuorela, Antimicrobial effects of Finnish plant extracts containing flavonoids and other phenolic compounds, Int. J. Food Microbiol., 2000, 56, 3–12 CrossRef CAS PubMed.
- M. J. R. Vaquero, M. R. Alberto and M. C. M. de Nadra, Antibacterial effect of phenolic compounds from different wines, Food Control, 2007, 18, 93–101 CrossRef.
- V. Gowd, N. Karim, M. R. I. Shishir, L. Xie and W. Chen, Dietary polyphenols to combat the metabolic diseases via altering gut microbiota, Trends Food Sci. Technol., 2019, 93, 81–93 CrossRef CAS.
- X. Zhang, X. Zhu, Y. Sun, B. Hu, Y. Sun, S. Jabbar and X. Zeng, Fermentation in vitro of EGCG, GCG and EGCG3′′Me isolated from Oolong tea by human intestinal microbiota, Food Res. Int., 2013, 54, 1589–1595 CrossRef CAS.
- D. E. Roopchand, R. N. Carmody, P. Kuhn, K. Moskal, P. Rojas-Silva, P. J. Turnbaugh and I. Raskin, Dietary Polyphenols Promote Growth of the Gut Bacterium Akkermansia muciniphila and Attenuate High-Fat Diet–Induced Metabolic Syndrome, Diabetes, 2015, 64, 2847–2858 CrossRef CAS PubMed.
- X. Tzounis, A. Rodriguez-Mateos, J. Vulevic, G. R. Gibson, C. Kwik-Uribe and J. P. E. Spencer, Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study, Am. J. Clin. Nutr., 2011, 93, 62–72 CrossRef CAS PubMed.
- C. E. Iglesias-Aguirre, A. Cortés-Martín, M. Á. Ávila-Gálvez, J. A. Giménez-Bastida, M. V. Selma, A. González-Sarrías and J. C. Espín, Main drivers of (poly)phenol effects on human health: metabolite production and/or gut microbiota-associated metabotypes?, Food Funct., 2021, 12, 10324–10355 RSC.
- S. Bolca, T. Van de Wiele and S. Possemiers, Gut metabotypes govern health effects of dietary polyphenols, Curr. Opin. Biotechnol., 2013, 24, 220–225 CrossRef CAS PubMed.
- J. Boekhorst, N. Venlet, N. Procházková, M. L. Hansen, C. B. Lieberoth, M. I. Bahl, L. Lauritzen, O. Pedersen, T. R. Licht, M. Kleerebezem and H. M. Roager, Stool energy density is positively correlated to intestinal transit time and related to microbial enterotypes, Microbiome, 2022, 10, 223 CrossRef CAS PubMed.
- S. Hazim, P. J. Curtis, M. Y. Schär, L. M. Ostertag, C. D. Kay, A.-M. Minihane and A. Cassidy, Acute benefits of the microbial-derived isoflavone metabolite equol on arterial stiffness in men prospectively recruited according to equol producer phenotype: a double-blind randomized controlled trial, Am. J. Clin. Nutr., 2016, 103, 694–702 CrossRef CAS PubMed.
- K. D. R. Setchell and S. J. Cole, Method of Defining Equol-Producer Status and Its Frequency among Vegetarians12, J. Nutr., 2006, 136, 2188–2193 CrossRef CAS PubMed.
- C. Iino, T. Shimoyama, K. Iino, Y. Yokoyama, D. Chinda, H. Sakuraba, S. Fukuda and S. Nakaji, Daidzein Intake Is Associated with Equol Producing Status through an Increase in the Intestinal Bacteria Responsible for Equol Production, Nutrients, 2019, 11, 433 CrossRef CAS PubMed.
- R. Yoshikata, K. Z. Myint, H. Ohta and Y. Ishigaki, Inter-relationship between diet, lifestyle habits, gut microflora, and the equol-producer phenotype: baseline findings from a placebo-controlled intervention trial, Menopause, 2019, 26, 273 CrossRef PubMed.
- M. Igase, K. Igase, Y. Tabara, Y. Ohyagi and K. Kohara, Cross-sectional study of equol producer status and cognitive impairment in older adults, Geriatr. Gerontol. Int., 2017, 17, 2103–2108 CrossRef PubMed.
- V. Ahuja, K. Miura, A. Vishnu, A. Fujiyoshi, R. Evans, M. Zaid, N. Miyagawa, T. Hisamatsu, A. Kadota, T. Okamura, H. Ueshima and A. Sekikawa, Significant inverse association of equol-producer status with coronary artery calcification but not dietary isoflavones in healthy Japanese men, Br. J. Nutr., 2017, 117, 260–266 CrossRef CAS PubMed.
- J. K. Aschoff, K. M. Riedl, J. L. Cooperstone, J. Högel, A. Bosy-Westphal, S. J. Schwartz, R. Carle and R. M. Schweiggert, Urinary excretion of Citrus flavanones and their major catabolites after consumption of fresh oranges and pasteurized orange juice: A randomized cross-over study, Mol. Nutr. Food Res., 2016, 60, 2602–2610 CrossRef CAS PubMed.
- M. K. Reger, T. W. Zollinger, Z. Liu, J. Jones and J. Zhang, Urinary phytoestrogens and cancer, cardiovascular, and all-cause mortality in the continuous National Health and Nutrition Examination Survey, Eur. J. Nutr., 2016, 55, 1029–1040 CrossRef CAS PubMed.
- C. E. Iglesias-Aguirre, F. Vallejo, D. Beltrán, E. Aguilar-Aguilar, J. Puigcerver, M. Alajarín, J. Berná, M. V. Selma and J. C. Espín, Lunularin Producers versus Non-producers: Novel Human Metabotypes Associated with the Metabolism of Resveratrol by the Gut Microbiota, J. Agric. Food Chem., 2022, 70, 10521–10531 CrossRef CAS PubMed.
- L. Pilšáková, I. Riečanský and F. Jagla, The physiological actions of isoflavone phytoestrogens, Physiol. Res., 2010, 59, 651–664 Search PubMed.
- Z. Liu, W. Li, J. Sun, C. Liu, Q. Zeng, J. Huang, B. Yu and J. Huo, Intake of soy foods and soy isoflavones by rural adult women in China, Asia Pac. J. Clin. Nutr., 2004, 13, 204–209 Search PubMed.
- S. Oba, C. Nagata, N. Shimizu, H. Shimizu, M. Kametani, N. Takeyama, T. Ohnuma and S. Matsushita, Soy product consumption and the risk of colon cancer: a prospective study in Takayama, Japan, Nutr. Cancer, 2007, 57, 151–157 CrossRef CAS PubMed.
- C. L. Frankenfeld, Dairy consumption is a significant correlate of urinary equol concentration in a representative sample of US adults, Am. J. Clin. Nutr., 2011, 93, 1109–1116 CrossRef CAS PubMed.
- C. L. Frankenfeld, Cardiometabolic risk and gut microbial phytoestrogen metabolite phenotypes, Mol. Nutr. Food Res., 2017, 61, 1500900 CrossRef PubMed.
- C. L. Frankenfeld, O-Desmethylangolensin: The Importance of Equol's Lesser Known Cousin to Human Health, Adv. Nutr., 2011, 2, 317–324 CrossRef CAS PubMed.
- C. Atkinson, C. L. Frankenfeld and J. W. Lampe, Gut Bacterial Metabolism of the Soy Isoflavone Daidzein: Exploring the Relevance to Human Health, Exp. Biol. Med., 2005, 230, 155–170 CrossRef CAS PubMed.
- C. L. Frankenfeld, C. Atkinson, W. K. Thomas, A. Gonzalez, T. Jokela, K. Wähälä, S. M. Schwartz, S. S. Li and J. W. Lampe, High concordance of daidzein-metabolizing phenotypes in individuals measured 1 to 3 years apart, Br. J. Nutr., 2005, 94, 873–876 CrossRef CAS PubMed.
- K. B. Song, C. Atkinson, C. L. Frankenfeld, T. Jokela, K. Wähälä, W. K. Thomas and J. W. Lampe, Prevalence of Daidzein-Metabolizing Phenotypes Differs between Caucasian and Korean American Women and Girls, J. Nutr., 2006, 136, 1347–1351 CrossRef CAS PubMed.
- N. Li, X. Wu, W. Zhuang, L. Xia, Y. Chen, R. Zhao, M. Yi, Q. Wan, L. Du and Y. Zhou, Soy and Isoflavone Consumption and Multiple Health Outcomes: Umbrella Review of Systematic Reviews and Meta-Analyses of Observational Studies and Randomized Trials in Humans, Mol. Nutr. Food Res., 2020, 64, 1900751 CrossRef CAS PubMed.
- C. Atkinson, K. M. Newton, E. J. A. Bowles, M. Yong and J. W. Lampe, Demographic, anthropometric, and lifestyle factors and dietary intakes in relation to daidzein-metabolizing phenotypes among premenopausal women in the United States, Am. J. Clin. Nutr., 2008, 87, 679–687 CrossRef CAS PubMed.
- S. Yokoyama and T. Suzuki, Isolation and characterization of a novel equol-producing bacterium from human feces, Biosci., Biotechnol., Biochem., 2008, 72, 2660–2666 CrossRef CAS PubMed.
- D. Tsangalis, G. Wilcox, N. P. Shah, A. E. J. McGill and L. Stojanovska, Urinary excretion of equol by postmenopausal women consuming soymilk fermented by probiotic bifidobacteria, Eur. J. Clin. Nutr., 2007, 61, 438–441 CrossRef CAS PubMed.
- Q.-K. Wei, T.-R. Chen and J.-T. Chen, Using of Lactobacillus and Bifidobacterium to product the isoflavone aglycones in fermented soymilk, Int. J. Food Microbiol., 2007, 117, 120–124 CrossRef CAS PubMed.
- T. T. Pham and N. P. Shah, Biotransformation of Isoflavone Glycosides by Bifidobacterium animalis in Soymilk Supplemented with Skim Milk Powder, J. Food Sci., 2007, 72, M316–M324 CrossRef CAS PubMed.
- G. P. Rodriguez-Castaño, M. R. Dorris, X. Liu, B. W. Bolling, A. Acosta-Gonzalez and F. E. Rey, Bacteroides thetaiotaomicron Starch Utilization Promotes Quercetin Degradation and Butyrate Production by Eubacterium ramulus, Front. Microbiol., 2019, 10, 1145 CrossRef PubMed.
- A. Braune, W. Engst, P. W. Elsinghorst, N. Furtmann, J. Bajorath, M. Gütschow and M. Blaut, Chalcone Isomerase from Eubacterium ramulus Catalyzes the Ring Contraction of Flavanonols, J. Bacteriol., 2016, 198, 2965–2974 CrossRef CAS PubMed.
- W. Zheng, Y. Ma, A. Zhao, T. He, N. Lyu, Z. Pan, G. Mao, Y. Liu, J. Li, P. Wang, J. Wang, B. Zhu and Y. Zhang, Compositional and functional differences in human gut microbiome with respect to equol production and its association with blood lipid level: a cross-sectional study, Gut Pathog., 2019, 11, 20 CrossRef PubMed.
- M. Y. Lacourt-Ventura, B. Vilanova-Cuevas, D. Rivera-Rodríguez, R. Rosario-Acevedo, C. Miranda, G. Maldonado-Martínez, J. Maysonet, D. Vargas, Y. Ruiz, R. Hunter-Mellado, L. A. Cubano, S. Dharmawardhane, J. W. Lampe, A. Baerga-Ortiz, F. Godoy-Vitorino and M. M. Martínez-Montemayor, Soy and Frequent Dairy Consumption with Subsequent Equol Production Reveals Decreased Gut Health in a Cohort of Healthy Puerto Rican Women, Int. J. Environ. Res. Public Health, 2021, 18, 8254 CrossRef CAS PubMed.
- L. M. Miller, J. W. Lampe, K. M. Newton, G. Gundersen, S. Fuller, S. D. Reed and C. L. Frankenfeld, Being overweight or obese is associated with harboring a gut microbial community not capable of metabolizing the soy isoflavone daidzein to O-desmethylangolensin in peri- and post-menopausal women, Maturitas, 2017, 99, 37–42 CrossRef CAS PubMed.
- C. L. Frankenfeld, C. Atkinson, K. Wähälä and J. W. Lampe, Obesity prevalence in relation to gut microbial environments capable of producing equol or O-desmethylangolensin from the isoflavone daidzein, Eur. J. Clin. Nutr., 2014, 68, 526–530 CrossRef CAS PubMed.
- V. Ahuja, K. Miura, A. Vishnu, A. Fujiyoshi, R. Evans, M. Zaid, N. Miyagawa, T. Hisamatsu, A. Kadota, T. Okamura, H. Ueshima and A. Sekikawa, Significant inverse association of equol-producer status with coronary artery calcification but not dietary isoflavones in healthy Japanese men, Br. J. Nutr., 2017, 117, 260–266 CrossRef CAS PubMed.
- X. Zhang, A. Fujiyoshi, V. Ahuja, A. Vishnu, E. Barinas-Mitchell, A. Kadota, K. Miura, D. Edmundowicz, H. Ueshima and A. Sekikawa, Association of equol producing status with aortic calcification in middle-aged Japanese men: The ERA JUMP study, Int. J. Cardiol., 2022, 352, 158–164 CrossRef PubMed.
- T. Usui, M. Tochiya, Y. Sasaki, K. Muranaka, H. Yamakage, A. Himeno, A. Shimatsu, A. Inaguma, T. Ueno, S. Uchiyama and N. Satoh-Asahara, Effects of natural S-equol supplements on overweight or obesity and metabolic syndrome in the Japanese, based on sex and equol status, Clin. Endocrinol., 2013, 78, 365–372 CrossRef CAS PubMed.
- S. T. Soukup, A. K. Engelbert, B. Watzl, A. Bub and S. E. Kulling, Microbial Metabolism of the Soy Isoflavones Daidzein and Genistein in Postmenopausal Women: Human Intervention Study Reveals New Metabotypes, Nutrients, 2023, 15, 2352 CrossRef CAS PubMed.
- M. Shukla, K. Gupta, Z. Rasheed, K. A. Khan and T. M. Haqqi, Consumption of Hydrolyzable Tannins Rich Pomegranate Extract (POMx) Suppresses Inflammation and Joint Damage In Rheumatoid Arthritis, Nutrition, 2008, 24, 733–743 CrossRef CAS PubMed.
- E. Barrajón-Catalán, S. Fernández-Arroyo, D. Saura, E. Guillén, A. Fernández-Gutiérrez, A. Segura-Carretero and V. Micol, Cistaceae aqueous extracts containing ellagitannins show antioxidant and antimicrobial capacity, and cytotoxic activity against human cancer cells, Food Chem. Toxicol., 2010, 48, 2273–2282 CrossRef PubMed.
- V.-I. Neli, S. Ivo, J. Remi, Q. Stephane and S. G. Angel, Antiviral activities of ellagitannins against bovine herpesvirus-1, suid alphaherpesvirus-1 and caprine herpesvirus-1, J. Vet. Med. Anim. Health, 2020, 12, 139–143 CrossRef.
- A. González-Sarrías, M. Larrosa, F. A. Tomás-Barberán, P. Dolara and J. C. Espín, NF-κB-dependent anti-inflammatory activity of urolithins, gut microbiota ellagic acid-derived metabolites, in human colonic fibroblasts, Br. J. Nutr., 2010, 104, 503–512 CrossRef PubMed.
- J. A. Giménez-Bastida, A. González-Sarrías, M. Larrosa, F. Tomás-Barberán, J. C. Espín and M.-T. García-Conesa, Ellagitannin metabolites, urolithin A glucuronide and its aglycone urolithin A, ameliorate TNF-α-induced inflammation and associated molecular markers in human aortic endothelial cells, Mol. Nutr. Food Res., 2012, 56, 784–796 CrossRef PubMed.
- F. A. Tomás-Barberán, R. García-Villalba, A. González-Sarrías, M. V. Selma and J. C. Espín, Ellagic Acid Metabolism by Human Gut Microbiota: Consistent Observation of Three Urolithin Phenotypes in Intervention Trials, Independent of Food Source, Age, and Health Status, J. Agric. Food Chem., 2014, 62, 6535–6538 CrossRef PubMed.
- A. Cortés-Martín, R. García-Villalba, A. González-Sarrías, M. Romo-Vaquero, V. Loria-Kohen, A. Ramírez-de-Molina, F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, The gut microbiota urolithin metabotypes revisited: the human metabolism of ellagic acid is mainly determined by aging, Food Funct., 2018, 9, 4100–4106 RSC.
- W. Xian, S. Yang, Y. Deng, Y. Yang, C. Chen, W. Li and R. Yang, Distribution of Urolithins Metabotypes in Healthy Chinese Youth: Difference in Gut Microbiota and Predicted Metabolic Pathways, J. Agric. Food Chem., 2021, 69, 13055–13065 CrossRef CAS PubMed.
- M. V. Selma, M. Romo-Vaquero, R. García-Villalba, A. González-Sarrías, F. A. Tomás-Barberán and J. C. Espín, The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism, Food Funct., 2016, 7, 1769–1774 RSC.
- M. Romo-Vaquero, E. Fernández-Villalba, A.-L. Gil-Martinez, L. Cuenca-Bermejo, J. C. Espín, M. T. Herrero and M. V. Selma, Urolithins: potential biomarkers of gut dysbiosis and disease stage in Parkinson’s patients, Food Funct., 2022, 13, 6306–6316 RSC.
- M. Romo-Vaquero, A. Cortés-Martín, V. Loria-Kohen, A. Ramírez-de-Molina, I. García-Mantrana, M. C. Collado, J. C. Espín and M. V. Selma, Deciphering the Human Gut Microbiome of Urolithin Metabotypes: Association with Enterotypes and Potential Cardiometabolic Health Implications, Mol. Nutr. Food Res., 2019, 63, 1800958 CrossRef PubMed.
- C. E. Iglesias-Aguirre, R. García-Villalba, D. Beltrán, M. D. Frutos-Lisón, J. C. Espín, F. A. Tomás-Barberán and M. V. Selma, Gut Bacteria Involved in Ellagic Acid Metabolism To Yield Human Urolithin Metabotypes Revealed, J. Agric. Food Chem., 2023, 71, 4029–4035 CrossRef CAS PubMed.
- G. Istas, R. P. Feliciano, T. Weber, R. Garcia-Villalba, F. Tomas-Barberan, C. Heiss and A. Rodriguez-Mateos, Plasma urolithin metabolites correlate with improvements in endothelial function after red raspberry consumption: A double-blind randomized controlled trial, Arch. Biochem. Biophys., 2018, 651, 43–51 CrossRef CAS PubMed.
- A. González-Sarrías, R. García-Villalba, M. Romo-Vaquero, C. Alasalvar, A. Örem, P. Zafrilla, F. A. Tomás-Barberán, M. V. Selma and J. C. Espín, Clustering according to urolithin metabotype explains the interindividual variability in the improvement of cardiovascular risk biomarkers in overweight-obese individuals consuming pomegranate: A randomized clinical trial, Mol. Nutr. Food Res., 2017, 61, 1600830 CrossRef PubMed.
- T. Meroño, G. Peron, G. Gargari, R. González-Domínguez, A. Miñarro, E. Vegas-Lozano, N. Hidalgo-Liberona, C. Del Bo’, S. Bernardi, P. A. Kroon, B. Carrieri, A. Cherubini, P. Riso, S. Guglielmetti and C. Andrés-Lacueva, The relevance of urolithins-based metabotyping for assessing the effects of a polyphenol-rich dietary intervention on intestinal permeability: A post-hoc analysis of the MaPLE trial, Food Res. Int., 2022, 159, 111632 CrossRef PubMed.
- A. Cortés-Martín, M. Romo-Vaquero, I. García-Mantrana, A. Rodríguez-Varela, M. C. Collado, J. C. Espín and M. V. Selma, Urolithin Metabotypes can Anticipate the Different Restoration of the Gut Microbiota and Anthropometric Profiles during the First Year Postpartum, Nutrients, 2019, 11, 2079 CrossRef PubMed.
- A. Cortés-Martín, G. Colmenarejo, M. V. Selma and J. C. Espín, Genetic Polymorphisms, Mediterranean Diet and Microbiota-Associated Urolithin Metabotypes can Predict Obesity in Childhood-Adolescence, Sci. Rep., 2020, 10, 1–13 CrossRef PubMed.
- C. E. Iglesias-Aguirre, A. González-Sarrías, A. Cortés-Martín, M. Romo-Vaquero, L. Osuna-Galisteo, J. J. Cerón, J. C. Espín and M. V. Selma, In vivo administration of gut bacterial consortia replicates urolithin metabotypes A and B in a non-urolithin-producing rat model, Food Funct., 2023, 14, 2657–2667 RSC.
- D. D'Amico, P. A. Andreux, P. Valdés, A. Singh, C. Rinsch and J. Auwerx, Impact of the Natural Compound Urolithin A on Health, Disease, and Aging, Trends Mol. Med., 2021, 27, 687–699 CrossRef PubMed.
- D. D'Amico, M. Olmer, A. M. Fouassier, P. Valdés, P. A. Andreux, C. Rinsch and M. Lotz, Urolithin A improves mitochondrial health, reduces cartilage degeneration, and alleviates pain in osteoarthritis, Aging Cell, 2022, 21, e13662 CrossRef PubMed.
- S. Liu, D. D'Amico, E. Shankland, S. Bhayana, J. M. Garcia, P. Aebischer, C. Rinsch, A. Singh and D. J. Marcinek, Effect of Urolithin A Supplementation on Muscle Endurance and Mitochondrial Health in Older Adults: A Randomized Clinical Trial, JAMA Netw. Open, 2022, 5, e2144279 CrossRef PubMed.
- R. Zamora-Ros, C. Andres-Lacueva, R. M. Lamuela-Raventós, T. Berenguer, P. Jakszyn, C. Martínez, M. J. Sánchez, C. Navarro, M. D. Chirlaque, M.-J. Tormo, J. R. Quirós, P. Amiano, M. Dorronsoro, N. Larrañaga, A. Barricarte, E. Ardanaz and C. A. González, Concentrations of resveratrol and derivatives in foods and estimation of dietary intake in a Spanish population: European Prospective Investigation into Cancer and Nutrition (EPIC)-Spain cohort, Br. J. Nutr., 2008, 100, 188–196 CrossRef CAS PubMed.
- P. Yin, L. Yang, Q. Xue, M. Yu, F. Yao, L. Sun and Y. Liu, Identification and inhibitory activities of ellagic acid- and kaempferol-derivatives from Mongolian oak cups against α-glucosidase, α-amylase and protein glycation linked to type II diabetes and its complications and their influence on HepG2 cells’ viability, Arabian J. Chem., 2018, 11, 1247–1259 CrossRef CAS.
- Y. Hou, K. Wang, W. Wan, Y. Cheng, X. Pu and X. Ye, Resveratrol provides neuroprotection by regulating the JAK2/STAT3/PI3K/AKT/mTOR pathway after stroke in rats, Genes Dis., 2018, 5, 245–255 CrossRef CAS PubMed.
- J. K. Bird, D. Raederstorff, P. Weber and R. E. Steinert, Cardiovascular and Antiobesity Effects of Resveratrol Mediated through the Gut Microbiota, Adv. Nutr., 2017, 8, 839–849 CrossRef CAS PubMed.
- B. N. M. Zordoky, I. M. Robertson and J. R. B. Dyck, Preclinical and clinical evidence for the role of resveratrol in the treatment of cardiovascular diseases, Biochim. Biophys. Acta, Mol. Basis Dis., 2015, 1852, 1155–1177 CrossRef CAS PubMed.
- X. Huang, Y. Dai, J. Cai, N. Zhong, H. Xiao, D. J. McClements and K. Hu, Resveratrol encapsulation in core-shell biopolymer nanoparticles: Impact on antioxidant and anticancer activities, Food Hydrocolloids, 2017, 64, 157–165 CrossRef CAS.
- M. Samsami-kor, N. E. Daryani, P. R. Asl and A. Hekmatdoost, Anti-Inflammatory Effects of Resveratrol in Patients with Ulcerative Colitis: A Randomized, Double-Blind, Placebo-controlled Pilot Study, Arch. Med. Res., 2015, 46, 280–285 CrossRef CAS PubMed.
- C. E. Iglesias-Aguirre, F. Vallejo, D. Beltrán, J. Berná, J. Puigcerver, M. Alajarín, M. V. Selma and J. C. Espín, 4-Hydroxydibenzyl: a novel metabolite from the human gut microbiota after consuming resveratrol, Food Funct., 2022, 13, 7487–7493 RSC.
- S. V. Luca, I. Macovei, A. Bujor, A. Miron, K. Skalicka-Woźniak, A. C. Aprotosoaie and A. Trifan, Bioactivity of dietary polyphenols: The role of metabolites, Crit. Rev. Food Sci. Nutr., 2020, 60, 626–659 CrossRef CAS PubMed.
- S. Vogl, A. G. Atanasov, M. Binder, M. Bulusu, M. Zehl, N. Fakhrudin, E. H. Heiss, P. Picker, C. Wawrosch, J. Saukel, G. Reznicek, E. Urban, V. Bochkov, V. M. Dirsch and B. Kopp, The Herbal Drug Melampyrum pratense L. (Koch): Isolation and Identification of Its Bioactive Compounds Targeting Mediators of Inflammation, Evid. Based Complement. Alternat. Med., 2013, 2013, 395316 CAS.
- T. Ito-Nagahata, C. Kurihara, M. Hasebe, A. Ishii, K. Yamashita, M. Iwabuchi, M. Sonoda, K. Fukuhara, R. Sawada, A. Matsuoka and Y. Fujiwara, Stilbene analogs of resveratrol improve insulin resistance through activation of AMPK, Biosci., Biotechnol., Biochem., 2013, 77, 1229–1235 CrossRef CAS PubMed.
- F. Li, Y. Han, X. Wu, X. Cao, Z. Gao, Y. Sun, M. Wang and H. Xiao, Gut Microbiota-Derived Resveratrol Metabolites, Dihydroresveratrol and Lunularin, Significantly Contribute to the Biological Activities of Resveratrol, Front. Nutr., 2022, 9, 912591 CrossRef PubMed.
- I. Günther, G. Rimbach, C. I. Mack, C. H. Weinert, N. Danylec, K. Lüersen, M. Birringer, F. Bracher, S. T. Soukup, S. E. Kulling and K. Pallauf, The Putative Caloric Restriction Mimetic Resveratrol has Moderate Impact on Insulin Sensitivity, Body Composition, and the Metabolome in Mice, Mol. Nutr. Food Res., 2020, 64, 1901116 CrossRef PubMed.
- S. W. Tsang, Y.-F. Guan, J. Wang, Z.-X. Bian and H.-J. Zhang, Inhibition of pancreatic oxidative damage by stilbene derivative dihydro-resveratrol: implication for treatment of acute pancreatitis, Sci. Rep., 2016, 6, 22859 CrossRef CAS PubMed.
- C. Rodríguez-García, C. Sánchez-Quesada, E. Toledo, M. Delgado-Rodríguez and J. J. Gaforio, Naturally Lignan-Rich Foods: A Dietary Tool for Health Promotion?, Molecules, 2019, 24, 917 CrossRef PubMed.
- R. Kiyama, Biological effects induced by estrogenic activity of lignans, Trends Food Sci. Technol., 2016, 54, 186–196 CrossRef CAS.
- J. Peterson, J. Dwyer, H. Adlercreutz, A. Scalbert, P. Jacques and M. L. McCullough, Dietary lignans: physiology and potential for cardiovascular disease risk reduction, Nutr. Rev., 2010, 68, 571–603 CrossRef PubMed.
- A. K. Zaineddin, K. Buck, A. Vrieling, J. Heinz, D. Flesch-Janys, J. Linseisen and J. Chang-Claude, The Association Between Dietary Lignans, Phytoestrogen-Rich Foods, and Fiber Intake and Postmenopausal Breast Cancer Risk: A German Case-Control Study, Nutr. Cancer, 2012, 64, 652–665 CrossRef CAS PubMed.
- F. L. Miles, S. L. Navarro, Y. Schwarz, H. Gu, D. Djukovic, T. W. Randolph, A. Shojaie, M. Kratz, M. A. J. Hullar, P. D. Lampe, M. L. Neuhouser, D. Raftery and J. W. Lampe, Plasma metabolite abundances are associated with urinary enterolactone excretion in healthy participants on controlled diets, Food Funct., 2017, 8, 3209–3218 RSC.
- J. M. Landete, Plant and mammalian lignans: A review of source, intake, metabolism, intestinal bacteria and health, Food Res. Int., 2012, 46, 410–424 CrossRef CAS.
- A. Senizza, G. Rocchetti, J. I. Mosele, V. Patrone, M. L. Callegari, L. Morelli and L. Lucini, Lignans and Gut Microbiota: An Interplay Revealing Potential Health Implications, Molecules, 2020, 25, 5709 CrossRef CAS PubMed.
- T. Clavel, J. Doré and M. Blaut, Bioavailability of lignans in human subjects, Nutr. Res. Rev., 2006, 19, 187–196 CrossRef CAS PubMed.
- E. Eeckhaut, K. Struijs, S. Possemiers, J.-P. Vincken, D. D. Keukeleire and W. Verstraete, Metabolism of the Lignan Macromolecule into Enterolignans in the Gastrointestinal Lumen As Determined in the Simulator of the Human Intestinal Microbial Ecosystem, J. Agric. Food Chem., 2008, 56, 4806–4812 CrossRef CAS PubMed.
- Y. Hu, Y. Song, A. A. Franke, F. B. Hu, R. M. van Dam and Q. Sun, A Prospective Investigation of the Association Between Urinary Excretion of Dietary Lignan Metabolites and Weight Change in US Women, Am. J. Epidemiol., 2015, 182, 503–511 CrossRef PubMed.
- C. Xu, Q. Liu, Q. Zhang, A. Gu and Z.-Y. Jiang, Urinary enterolactone is associated with obesity and metabolic alteration in men in the US National Health and Nutrition Examination Survey 2001–10, Br. J. Nutr., 2015, 113, 683–690 CrossRef CAS PubMed.
- M. A. J. Hullar, S. M. Lancaster, F. Li, E. Tseng, K. Beer, C. Atkinson, K. Wähälä, W. K. Copeland, T. W. Randolph, K. M. Newton and J. W. Lampe, Enterolignan-Producing Phenotypes Are Associated with Increased Gut Microbial Diversity and Altered Composition in Premenopausal Women in the United States, Cancer Epidemiol., Biomarkers Prev., 2015, 24, 546–554 CrossRef CAS PubMed.
- Y. Li, F. Wang, J. Li, K. L. Ivey, J. E. Wilkinson, D. D. Wang, R. Li, G. Liu, H. A. Eliassen, A. T. Chan, C. B. Clish, C. Huttenhower, F. B. Hu, Q. Sun and E. B. Rimm, Dietary lignans, plasma enterolactone levels, and metabolic risk in men: exploring the role of the gut microbiome, BMC Microbiol., 2022, 22, 82 CrossRef CAS PubMed.
- W. Mullen, M.-A. Archeveque, C. A. Edwards, H. Matsumoto and A. Crozier, Bioavailability and Metabolism of Orange Juice Flavanones in Humans: Impact of a Full-Fat Yogurt, J. Agric. Food Chem., 2008, 56, 11157–11164 CrossRef CAS PubMed.
- I. Najmanová, M. Vopršalová, L. Saso and P. Mladěnka, The pharmacokinetics of flavanones, Crit. Rev. Food Sci. Nutr., 2020, 60, 3155–3171 CrossRef PubMed.
- M. Tomás-Navarro, F. Vallejo, E. Sentandreu, J. L. Navarro and F. A. Tomás-Barberán, Volunteer Stratification Is More Relevant than Technological Treatment in Orange Juice Flavanone Bioavailability, J. Agric. Food Chem., 2014, 62, 24–27 CrossRef PubMed.
- F. Vallejo, M. Larrosa, E. Escudero, M. P. Zafrilla, B. Cerdá, J. Boza, M. T. García-Conesa, J. C. Espín and F. A. Tomás-Barberán, Concentration and Solubility of Flavanones in Orange Beverages Affect Their Bioavailability in Humans, J. Agric. Food Chem., 2010, 58, 6516–6524 CrossRef CAS PubMed.
- G. Pereira-Caro, G. Borges, J. van der Hooft, M. N. Clifford, D. Del Rio, M. E. Lean, S. A. Roberts, M. B. Kellerhals and A. Crozier, Orange juice (poly)phenols are highly bioavailable in humans, Am. J. Clin. Nutr., 2014, 100, 1378–1384 CrossRef CAS PubMed.
- A. Nishioka, E. de C. Tobaruela, L. N. Fraga, F. A. Tomás-Barberán, F. M. Lajolo and N. M. A. Hassimotto, Stratification of Volunteers According to Flavanone Metabolite Excretion and Phase II Metabolism Profile after Single Doses of ‘Pera’ Orange and ‘Moro’ Blood Orange Juices, Nutrients, 2021, 13, 473 CrossRef CAS PubMed.
- M. Á. Ávila-Gálvez, J. A. Giménez-Bastida, A. González-Sarrías and J. C. Espín, New Insights into the Metabolism of the Flavanones Eriocitrin and Hesperidin: A Comparative Human Pharmacokinetic Study, Antioxidants, 2021, 10, 435 CrossRef PubMed.
- W. Lin, W. Wang, H. Yang, D. Wang and W. Ling, Influence of Intestinal Microbiota on the Catabolism of Flavonoids in Mice, J. Food Sci., 2016, 81, H3026–H3034 CAS.
- A. Vogiatzoglou, A. A. Mulligan, R. N. Luben, M. A. H. Lentjes, C. Heiss, M. Kelm, M. W. Merx, J. P. E. Spencer, H. Schroeter and G. G. C. Kuhnle, Assessment of the dietary intake of total flavan-3-ols, monomeric flavan-3-ols, proanthocyanidins and theaflavins in the European Union, Br. J. Nutr., 2014, 111, 1463–1473 CrossRef CAS PubMed.
- P. Mena, L. Bresciani, N. Brindani, I. A. Ludwig, G. Pereira-Caro, D. Angelino, R. Llorach, L. Calani, F. Brighenti, M. N. Clifford, C. I. R. Gill, A. Crozier, C. Curti and D. D. Rio, Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: synthesis, analysis, bioavailability, and bioactivity, Nat. Prod. Rep., 2019, 36, 714–752 RSC.
- A. Takagaki and F. Nanjo, Bioconversion of (−)-Epicatechin, (+)-Epicatechin, (−)-Catechin, and (+)-Catechin by (−)-Epigallocatechin-Metabolizing Bacteria, Biol. Pharm. Bull., 2015, 38, 789–794 CrossRef CAS PubMed.
- S. Wiese, T. Esatbeyoglu, P. Winterhalter, H.-P. Kruse, S. Winkler, A. Bub and S. E. Kulling, Comparative biokinetics and metabolism of pure monomeric, dimeric, and polymeric flavan-3-ols: a randomized cross-over study in humans, Mol. Nutr. Food Res., 2015, 59, 610–621 CrossRef CAS PubMed.
- J. van Duynhoven, J. J. J. van der Hooft, F. A. van Dorsten, S. Peters, M. Foltz, V. Gomez-Roldan, J. Vervoort, R. C. H. de Vos and D. M. Jacobs, Rapid and sustained systemic circulation of conjugated gut microbial catabolites after single-dose black tea extract consumption, J. Proteome Res., 2014, 13, 2668–2678 CrossRef CAS PubMed.
- N. Brindani, P. Mena, L. Calani, I. Benzie, S.-W. Choi, F. Brighenti, F. Zanardi, C. Curti and D. Del Rio, Synthetic and analytical strategies for the quantification of phenyl-γ-valerolactone conjugated metabolites in human urine, Mol. Nutr. Food Res., 2017, 61, 9 CrossRef PubMed.
- P. Mena, I. A. Ludwig, V. B. Tomatis, A. Acharjee, L. Calani, A. Rosi, F. Brighenti, S. Ray, J. L. Griffin, L. J. Bluck and D. Del Rio, Inter-individual variability in the production of flavan-3-ol colonic metabolites: preliminary elucidation of urinary metabotypes, Eur. J. Nutr., 2019, 58, 1529–1543 CrossRef CAS PubMed.
- F. Sánchez-Patán, C. Cueva, M. Monagas, G. E. Walton, G. R. M. Gibson, J. E. Quintanilla-López, R. Lebrón-Aguilar, P. J. Martín-Álvarez, M. V. Moreno-Arribas and B. Bartolomé, In vitro fermentation of a red wine extract by human gut microbiota: changes in microbial groups and formation of phenolic metabolites, J. Agric. Food Chem., 2012, 60, 2136–2147 CrossRef PubMed.
- P. Mena, C. Favari, A. Acharjee, S. Chernbumroong, L. Bresciani, C. Curti, F. Brighenti, C. Heiss, A. Rodriguez-Mateos and D. Del Rio, Metabotypes of flavan-3-ol colonic metabolites after cranberry intake: elucidation and statistical approaches, Eur. J. Nutr., 2022, 61, 1299–1317 CrossRef CAS PubMed.
- A. Cortés-Martín, M. V. Selma, J. C. Espín and R. García-Villalba, The Human Metabolism of Nuts Proanthocyanidins does not Reveal Urinary Metabolites Consistent with Distinctive Gut Microbiota Metabotypes, Mol. Nutr. Food Res., 2019, 63, 1800819 CrossRef PubMed.
- N. Tosi, C. Favari, L. Bresciani, E. Flanagan, M. Hornberger, A. Narbad, D. Del Rio, D. Vauzour and P. Mena, Unravelling phenolic metabotypes in the frame of the COMBAT study, a randomized, controlled trial with cranberry supplementation, Food Res. Int., 2023, 172, 113187 CrossRef CAS PubMed.
- L. Narduzzi, V. Agulló, C. Favari, N. Tosi, C. Mignogna, A. Crozier, D. Del Rio and P. Mena, (Poly)phenolic compounds and gut microbiome: new opportunities for personalized nutrition, Microbiome Res. Rep., 2022, 1, 16 CrossRef CAS PubMed.
- C. Busch, S. Noor, C. Leischner, M. Burkard, U. M. Lauer and S. Venturelli, Anti-proliferative activity of hop-derived prenylflavonoids against human cancer cell lines, Wien. Med. Wochenschr., 2015, 165, 258–261 CrossRef PubMed.
- E. Sommella, G. Verna, M. Liso, E. Salviati, T. Esposito, D. Carbone, C. Pecoraro, M. Chieppa and P. Campiglia, Hop-derived fraction rich in beta acids and prenylflavonoids regulates the inflammatory response in dendritic cells differently from quercetin: unveiling metabolic changes by mass spectrometry-based metabolomics, Food Funct., 2021, 12, 12800–12811 RSC.
- B. Kontek, D. Jedrejek, W. Oleszek and B. Olas, Antiradical and antioxidant activity in vitro of hops-derived extracts rich in bitter acids and xanthohumol, Ind. Crops Prod., 2021, 161, 113208 CrossRef CAS.
- M. Rad, M. Hümpel, O. Schaefer, R. C. Schoemaker, W.-D. Schleuning, A. F. Cohen and J. Burggraaf, Pharmacokinetics and systemic endocrine effects of the phyto-oestrogen 8-prenylnaringenin after single oral doses to postmenopausal women, Br. J. Clin. Pharmacol., 2006, 62, 288–296 CrossRef CAS PubMed.
- L. A. Calvo-Castro, M. Burkard, N. Sus, G. Scheubeck, C. Leischner, U. M. Lauer, A. Bosy-Westphal, V. Hund, C. Busch, S. Venturelli and J. Frank, The Oral Bioavailability of 8-Prenylnaringenin from Hops (Humulus Lupulus L.) in Healthy Women and Men is Significantly Higher than that of its Positional Isomer 6-Prenylnaringenin in a Randomized Crossover Trial, Mol. Nutr. Food Res., 2018, 62, 1700838 CrossRef PubMed.
- S. Possemiers, S. Rabot, J. C. Espín, A. Bruneau, C. Philippe, A. González-Sarrías, A. Heyerick, F. A. Tomás-Barberán, D. De Keukeleire and W. Verstraete, Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8-prenylnaringenin in vitro and in rat intestine, J. Nutr., 2008, 138, 1310–1316 CrossRef CAS PubMed.
- J. Guo, D. Nikolic, L. R. Chadwick, G. F. Pauli and R. B. van Breemen, Identification of Human Hepatic Cytochrome P450 Enzymes Involved in the Metabolism of 8-Prenylnaringenin and Isoxanthohumol from Hops (humulus Lupulus L.), Drug Metab. Dispos., 2006, 34, 1152–1159 CrossRef CAS PubMed.
- S. Bolca, S. Possemiers, V. Maervoet, I. Huybrechts, A. Heyerick, S. Vervarcke, H. Depypere, D. D. Keukeleire, M. Bracke, S. D. Henauw, W. Verstraete and T. V. de Wiele, Microbial and dietary factors associated with the 8-prenylnaringenin producer phenotype: a dietary intervention trial with fifty healthy post-menopausal Caucasian women, Br. J. Nutr., 2007, 98, 950–959 CrossRef CAS PubMed.
- D. Nikolic, Y. Li, L. R. Chadwick, S. Grubjesic, P. Schwab, P. Metz and R. B. van Breemen, Metabolism of 8-prenylnaringenin, a potent phytoestrogen from hops (Humulus lupulus), by human liver microsomes, Drug Metab. Dispos., 2004, 32, 272–279 CrossRef CAS PubMed.
- C. E. Iglesias-Aguirre, M. V. Selma and J. C. Espín, Unveiling metabotype clustering in resveratrol, daidzein, and ellagic acid metabolism: Prevalence, associated gut microbiomes, and their distinctive microbial networks, Food Res. Int., 2023, 173, 113470 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |