Open Access Article
Open Access ArticleAnti-diabetic properties of brewer's spent yeast peptides. In vitro, in silico and ex vivo study after simulated gastrointestinal digestion
Marilin E.
Aquino
a,
Silvina R.
Drago
 a,
Fermín
Sánchez de Medina
b,
Olga
Martínez-Augustin
a,
Fermín
Sánchez de Medina
b,
Olga
Martínez-Augustin
 *c and
Raúl E.
Cian
*c and
Raúl E.
Cian
 a
a
aInstituto de Tecnología de Alimentos, CONICET, FIQ – UNL, 1° de Mayo 3250, (3000) Santa Fe, Argentina
bDepartment of Pharmacology, CIBERehd, School of Pharmacy, Instituto de Investigación Biosanitaria ibs.GRANADA, University of Granada, Granada, Spain
cDepartment of Biochemistry and Molecular Biology II, CIBERehd, School of Pharmacy, Instituto de Investigación Biosanitaria ibs.GRANADA, Instituto de Nutrición y Tecnología de los Alimentos José Mataix, University of Granada, Granada, Spain. E-mail: omartine@ugr.es; Fax: +34 958 248960; Tel: +34 958 241305
First published on 7th March 2024
Abstract
Brewer's spent yeast (BSY) hydrolysates are a source of antidiabetic peptides. Nevertheless, the impact of in vitro gastrointestinal digestion of BSY derived peptides on diabetes has not been assessed. In this study, two BSY hydrolysates were obtained (H1 and H2) using β-glucanase and alkaline protease, with either 1 h or 2 h hydrolysis time for H1 and H2, respectively. These hydrolysates were then subjected to simulated gastrointestinal digestion (SGID), obtaining dialysates D1 and D2, respectively. BSY hydrolysates inhibited the activity of α-glucosidase and dipeptidyl peptidase IV (DPP-IV) enzymes. Moreover, although D2 was inactive against these enzymes, D1 IC50 value was lower than those found for the hydrolysates. Interestingly, after electrophoretic separation, D1 mannose-linked peptides showed the highest α-glucosidase inhibitory activity, while non-glycosylated peptides had the highest DPP-IV inhibitory activity. Kinetic analyses showed a non-competitive mechanism in both cases. After peptide identification, GILFVGSGVSGGEEGAR and IINEPTAAAIAYGLDK showed the highest in silico anti-diabetic activities among mannose-linked and non-glycosylated peptides, respectively (AntiDMPpred score: 0.70 and 0.77). Molecular docking also indicated that these peptides act as non-competitive inhibitors. Finally, an ex vivo model of mouse jejunum organoids was used to study the effect of D1 on the expression of intestinal epithelial genes related to diabetes. The reduction of the expression of genes that codify lactase, sucrase-isomaltase and glucose transporter 2 was observed, as well as an increase in the expression of Gip (glucose-dependent insulinotropic peptide) and Glp1 (glucagon-like peptide 1). This is the first report to evaluate the anti-diabetic effect of BSY peptides in mouse jejunum organoids.
1. Introduction
Brewer's spent yeast (BSY) is the second largest by-product of the brewing industry, representing around 15% of the total by-products.1 BSY is generally recognized as a safe raw material and is essentially constituted by yeast cells.2,3 Yeast cells are predominantly composed of carbohydrates and proteins such as β-glucans (35–45% dry basis) and mannoproteins (35–60% dry basis), respectively.4 These proteins are bound to mannose residues (glycoproteins) and are found mainly in the yeast cell wall.5Different bioactive peptides such as antioxidant, anti-hypertensive and antimicrobial peptides have been obtained from BSY.2 Nevertheless, there are few works focused on studying the anti-diabetic properties of BSY peptides. In this sense, Marson et al.6 reported that low molecular weight peptides obtained from BSY with Brauzyn®, Protamex® and Alcalase® had in vitro α-amylase and α-glucosidase inhibitory properties. However, there are very few studies on the inhibition of dipeptidyl peptidase IV (DPP-IV) by BSY peptides. This enzyme acts to inactivate the incretin hormones glucagon-like peptide 1 (GLP-1) and glucose-dependent insulinotropic peptide (GIP) that enhance insulin secretion. Therefore, DPP-IV is a molecular target in diabetes treatment.7 In this regard, DPP-IV inhibitors have been used as drugs to control postprandial glycemia in type 2 diabetes mellitus.8 The epithelial intestine is responsible for the absorption of glucose and produces incretin hormones that regulate glucose metabolism.9 Their main effects include the induction of insulin secretion by the pancreas and the decrease of blood glucose levels.10
Physiological effects of bioactive peptides largely depend on their structural stability in the gastrointestinal environment. Many bioactive peptides can lose their bioactivity after gastrointestinal digestion due to proteolysis, or they can increase it due to the generation of more active peptides.11 In this regard, simulation of physiological digestion is a useful tool to evaluate in vitro bioactive peptide stability against digestive enzymes and weigh the remaining bioactivity.12 However, there is no research about in vitro α-glucosidase and DPP-IV inhibitory properties of BSY peptides after simulated gastrointestinal digestion, nor on the effect of these peptides on the intestinal epithelium, a key component of the glucose absorption and incretin hormone production. In general, intestinal cell lines have been used to assess effects of food components on intestinal epithelium, nevertheless these models present several limitations; in general they are 2D cultures of immortalized cell lines containing only one cell type and no stem cells. In our study we have used intestinal organoids that constitute a powerful research tool that overcomes these limitations, including stem cells and their niche, as well as several intestinal epithelial cell types, partially recapitulating the heterogeneity and cell behaviour of the original tissue.13 The aims of this work were: (i) to obtain bioactive peptides from BSY using β-glucanase and alkaline protease; (ii) to evaluate the α-glucosidase and DPP-IV inhibitory activity of BSY peptides before and after simulated gastrointestinal digestion; (iii) to identify the peptides responsible of α-glucosidase and DPP-IV inhibitory activity using tandem mass spectrometry and in silico analysis; and (iv) to evaluate the anti-diabetic effect of BSY peptides after simulated gastrointestinal digestion on mouse jejunum organoids.
2. Material and methods
2.1 Raw materials and reagents
BSY was supplied by Okcidenta (Santa Fe, Santa Fe, Argentina). LAMINEX® MaxFlow 4G (β-glucanase) and alkaline protease-Protex 6L® were provided by Danisco Argentina S.A (Buenos Aires, Argentina) and DuPont (Buenos Aires, Argentina), respectively. DPP-IV (D4943), α-glucosidase enzyme (G5003), α-amylase enzyme (A3306), 4-nitrophenyl α-D-glucopyranoside (N1377), Gly-Pro-p-nitroanilide (G0513), 3,5-dinitrosalicylic acid (D0550), pepsin from porcine gastric mucosa (P-7000), pancreatin from porcine pancreas (P-1750), peptide N glycosidase F (PNGase F) (P7367), and o-phthaldialdehyde (P1378) were obtained from Sigma Chemical Co. (St Louis, MO, USA). Molecular weight standards conalbumin, carbonic anhydrase, cytochrome C, aprotinin, bacitracin, cytidine, and glycine were obtained from Pharmacia Fine Chemicals (Piscataway, NJ, USA). Trifluoroacetic acid (TFA) and acetonitrile HPLC grade were obtained from Merk (Buenos Aires, Argentina). Soluble starch, sodium hydroxide, hydrochloric acid, sulfuric acid, and other reagents were of analytical grade and obtained from Cicarelli Laboratorios (San Lorenzo, Santa Fe, Argentina).2.2 Brewers’ spent yeast hydrolysates
In order to obtain bioactive peptides, BSY was dispersed in phosphate buffer (50 mmol L−1, pH 5.0) during 4 h at 90 °C according to Li and Karboune.14 The dispersion was then centrifuged at 3000g for 30 min at 4 °C and the residue was used as substrate for β-glucanase hydrolysis. For hydrolysis with β-glucanase, two enzymatic conditions were used: (a) temperature 70 °C, pH 5.0, enzyme/substrate ratio 5 g per 100 g, time: 1 h; and (b) temperature 70 °C, pH 5.0, enzyme/substrate (E/S) ratio 5 g per 100 g, time: 2 h. The hydrolysates were then centrifuged at 3000g for 30 min at 4 °C and the residues were used as substrate for alkaline protease. The residues obtained after 1 h and 2 h of hydrolysis with β-glucanase were named R1 and R2, respectively.Glucose and mannose content of β-glucanase hydrolysates and residues were measured using Megazyme kit (MANGL 04/20) after acid hydrolysis.15 Protein content of β-glucanase hydrolysates and residues was measured according to Lowry et al.16 Also, the mannose/protein ratio content was calculated. The degree of hydrolysis (DH) of β-glucans was calculated as:
| DH-β-glucanase = [(G − G0)/Gt] × 100 |
The residues obtained from β-glucanase hydrolysis (R1 and R2) were hydrolyzed with alkaline protease-Protex 6L® during 2 h at 55 °C and pH 9.5. The enzyme/substrate (E/S) ratio was 5 g per 100 g. The hydrolysates were then centrifuged at 3000g for 30 min at 4 °C and the supernatants were named H1 and H2.
2.3 Simulated gastrointestinal digestion
Simulated gastrointestinal digestion of BSY hydrolysates (H1 and H2) with gastric and intestinal phases was performed according to Van de Velde et al.17 Moreover, the intestinal phase of the digestive process was simulated using dialysis bags. After this process, bag contents corresponding to dialysates of the in vitro intestinal phase were transferred to flasks, weighted and frozen at −20 °C until analysis. H1 and H2 dialysates from the gastrointestinal digestion process were named D1 and D2, respectively.2.4 Characterization of hydrolysates and dialysates
Free amino groups of H1, H2, D1 and D2 were measured using o-phthaldialdehyde, according to Nielsen et al.18 and the degree of hydrolysis (DH) was calculated as:| DH = [(h − h0)/htot] × 100 |
Protein and mannose content of H1, H2, D1 and D2 were measured as mentioned above. The proportion of mannose-linked peptides in dialysates was estimated by affinity chromatography (Glycoprotein Isolation Kit-ConA 89804, Thermo Scientific). Peptide analysis of H1, H2, D1 and D2 was performed by RP-HPLC according to Garzón et al.19 A Shimadzu LC-20AT pump, with an SPD-M20A diode detector, and a Phenomenex column (Gemini 110A C18, 250 mm × 4.6 mm, 5 μm particle size), were used. Fractions with absorbance peaks at 280 and 220 nm were analyzed. The retention times and the areas of peptides in samples were registered. Moreover, the ratio between peaks area and total area of chromatogram was performed.
Fast protein liquid chromatography (FPLC) of H1, H2, D1 and D2 was performed according to Cian et al.20 using an AKTA Prime system equipped with a Superdex 75 (GE Life Sciences, Piscataway, NJ, USA). Molecular mass of proteins fractions was estimated using molecular weight standards (Pharmacia Fine Chemicals, Piscataway, NJ, USA). Moreover, the ratio between peaks area and total area of chromatogram was performed.
2.5 In vitro α-glucosidase and DPP-IV inhibitory activity
The evaluation of α-glucosidase inhibitory activity of H1, H2, D1 and D2 was performed according to Donkor et al.21 The DPP-IV inhibition assay was made according to Wang et al.22The inhibition rate of α-glucosidase and DPP-IV enzymes was calculated as follows:
 | (1) |
The concentration of sample needed to inhibit 50% of enzyme activity was defined as IC50 value. To determine the IC50 value of α-glucosidase and DPP-IV enzymes, serial dilutions of samples from 0 to 10 mg mL−1 of protein were prepared. Moreover, the IC50 value was calculated according to Cian et al.8 All determinations were performed by triplicate.
2.6 Fractionation and characterization of dialysate peptides
The most bioactive sample was selected to characterize and evaluate the peptide fraction responsible of in vitro α-glucosidase and DPP-IV inhibitory activity. For this, D1 was fractionated using affinity chromatography (Glycoprotein Isolation Kit-ConA 89804, Thermo Scientific). The fractions obtained from D1 by affinity chromatography were named as D1–F1 and D1–F2, corresponding to peptides without glycosylation or mannose-linked peptides respectively. These fractions were subjected to the α-glucosidase and DPP-IV inhibition assays as mentioned before. Also, the IC50 value was determined and serial dilutions of samples from 0 to 10 mg mL−1 of protein were prepared. Moreover, the kinetic analysis of enzymes was performed using Michaelis–Menten equation. For α-glucosidase, different substrate concentrations (0.5–10 mmol L−1p-nitrophenyl α-D-glucopyranoside) were incubated with enzyme solution with and without D1–F2 peptide fraction at their IC50 concentration (0.26 mg protein per mL). For DPP-IV, different substrate concentrations (0.8–10 mmol L−1 Gly-Pro-p-nitroanilide) were incubated with enzyme solution with and without D1–F1 peptide fraction at their IC50 concentration (1.45 mg protein per mL). Then, the experimental data were fitted with GraphPad Prism software version 6.07 (GraphPad Software, La Jolla, San Diego, CA, USA) using the Michaelis–Menten equation. Vmax and Km parameters were provided by the same software taking into account all the experimental plots. All determinations were performed by triplicate.To determine the presence of O-glycosidic bonds in D1–F2, a reductive β-elimination assay of peptide fraction was performed according to Cavallero et al.23 For N-glycosidic bonds study, D1–F2 was subjected to PGNase F digestion.23 The analysis of the sugar composition from β-elimination assay or PGNase F digestion of D1–F2 was performed by high performance anion exchange chromatography according to Cavallero et al.23 Sugar analysis was performed in a DX-3000 Dionex BioLC system (Dionex Corp.) with a pulse amperometric detector. The β-elimination products were separated using a Carbopack P-20 column equipped with a P-20 pre-column (Dionex), while the PGNase F products were separated using a Carbopack P-100 microbore column equipped with a P-100 pre-column (Dionex).
Peptide identification from H1, D1–F1, and D1–F2 was performed by a nanoLC 1000 coupled to an EASY-SPRAY Q Exactive Mass Spectrometer (Thermo Scientific) with a high collision dissociation and an Orbitrap analyzer. An Easy Spray PepMap RSLC C18 column (50 μm × 150 mm, particle size 2.0 μm, pore size: 100 Å) at 40 °C was used for separation. Separation was performed according to Cavallero et al.23 Data were manually evaluated. Automatic search of peptides was assisted by Sequest HT on Proteome Discoverer 1.4 (Thermo Fisher Sc.). The mass accuracy tolerance was set to 10 ppm for precursor ions. The static modification was carbamidomethylation in the Cys residues. Deconvolution was assisted by Xtract on Thermo Xcalibur 3.0.63. Peptide identifications were accepted if they were statistically significant (p < 0.05).
2.7 In silico analysis of identified peptides from dialysate fractions
Studies in silico of the peptide obtained were conducted. The potential anti-diabetic property of the identified peptides was predicted using AntiDMPpred tool (https://i.uestc.edu.cn/AntiDMPpred/cgi-bin/AntiDMPpred.pl). Peptides are scored from 0 to 1. A score higher than 0.5 indicate high probability to be anti-diabetic.24 Moreover, peptide hydrophobicity was predicted by PepDraw tool, and the glycosylation site of peptides was obtained from Saccharomyces genome database.2.8 Molecular docking
The identified peptides from D1–F1 and D1–F2 with the highest anti-diabetic score were used for molecular docking assays. HPEPDOCK server (https://huanglab.phys.hust.edu.cn/hpepdock/) was used to perform flexible protein-peptide docking. The α-glucosidase and DPP-IV structure were selected from https://www.rcsb.org/in PDB format (PDB entry code: 3 l4y and 5J3J, for α-glucosidase and DPP-IV respectively). The best model of each protein–peptide interaction was selected according to the most negative docking score. The structures obtained were used for the second stage of refinement to find high-resolution modeling of protein–peptide interactions using the FlexPepDock server (https://flexpepdock.furmanlab.cs.huji.ac.il/). The enzyme–peptide interactions were analyzed with FlexPepDock server.25 Visual analyses to obtain the interaction points were performed using the PyMol 4.60 software.2.9 Anti-diabetic effect of dialysate on mouse jejunum organoids
Organoid culture was performed according to Arredondo-Amador et al.26 Briefly, jejunum crypts were cultured in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dome of Matrigel® Growth Factor Reduced Basement Membrane Matrix, Phenol Red-Free (Corning, New York, USA) and IntestiCult™ Organoid Growth Medium (Mouse) (Stemcell, Vancouver, Canada) following the manufacturer's instructions. When Matrigel® was set, 500 μL of IntestiCult™ were added to each well. Experiments were conducted after two to three organoid passages.
1 dome of Matrigel® Growth Factor Reduced Basement Membrane Matrix, Phenol Red-Free (Corning, New York, USA) and IntestiCult™ Organoid Growth Medium (Mouse) (Stemcell, Vancouver, Canada) following the manufacturer's instructions. When Matrigel® was set, 500 μL of IntestiCult™ were added to each well. Experiments were conducted after two to three organoid passages.
To evaluate the regulation of gene expression, organoids were treated with D1 (0.1 g protein per L) in basal and in TNF (10 ng mL−1) (eBioscience, Inc., San Diego, California, USA) plus fetal bovine serum (FBS) 5% (v/v) stimulated conditions. After 24 h of incubation, organoids underwent RNA extraction.
Total RNA from tissue was isolated with the RNeasy minikit (Qiagen, CA, USA), whereas organoid RNA was obtained with QIAzol Lysis Reagent (Qiagen, California, USA), 1 μg of RNA per sample was retrotranscribed using iScript Select cDNA Synthesis kit (Biorad Laboratories, California, USA). Specific DNA sequences were amplified with a Biorad CFX connect real-time PCR device (Alcobendas, Madrid, Spain). Primers used are shown in Table 1. Results are expressed as 2−ddCt using Ppib, Hprt and 18S as reference genes. Please note that GLP-1 is generated by postranslational processing of proglucagon; for clarity we refer to Glp1 to the PCR amplification product.
| Gene | Forward 5′–3′ | Reverse 3′–5′ |
|---|---|---|
| 18s | TGGTGGAGCGATTTGTCTGG | ACGCTGAGCCAGTCAGTGTACG |
| Gip | CATCAGTGATTACAGCATCG | TTTCCAGTCACTCTTCTTCC |
| Glp-1 | ACCTTTACCAGTGATGTGAG | GTGGCAAGATTATCCAGAATG |
| Hprt | AGGGATTTGAATCACGTTTG | TTTACTGGCAACATCAACAG |
| Lct | TTCCTATCAGGTTGAAGGTG | GTCATTCCCAATCTTCAGTG |
| Ppib | CAAATCCTTTCTCTCCTGTAG | TGGAGATGAATCTGTAGGAC |
| Slc2a2 | TTGTGCTGCTGGATAAATTC | AAATTCAGCAACCATGAACC |
| Sis | GAAGATAACTCTGGCAAGTC | GTCCAATGAGCTCTTGATATTG |
2.10 Statistical analysis
Results were expressed as the mean ± standard deviation and were analyzed by one-way analysis of variance. The statistical differences among samples were determined using the least significant difference test with a level of signification α: 0.05, using the STATGRAPHICS Centurion XV 15.2.06 (StatPoint Technologies, Inc., Warrenton, Virginia, USA).GraphPad Prism 6 software was used to analyze the RT-qPCR data. RT-qPCR data are given as relative expression (fold change) versus the control, which is assigned a mean value of 1. Differences among means were tested for statistical significance by one-way analysis of variance and Fisher's least significant difference tests to determine differences between samples (p < 0.05).
3. Results and discussion
3.1 Characterization and in vitro anti-diabetic activity of hydrolysates and dialysates
As shown in Fig. 1A, the degree of hydrolysis of β-glucans increased with the reaction time. Moreover, the mannose/protein ratio of supernatant obtained after 2 h of hydrolysis with β-glucanase was higher than that found for 1 h (4.0 ± 0.2 vs. 2.5 ± 0.1 mg mannose per 100 g protein, respectively). Similar results were found by Li et al.27 for β-glucanase hydrolysis of brewer's yeast. These authors reported that mannose/protein ratio obtained during 20 h of hydrolysis with Zymolyase® was higher than that found for 4 h (16.4 vs. 13.1%, respectively). As it is known, the β-glucanase enzyme hydrolyzes β-glucans at β-1,3-linkages, which promotes cell wall rupture and the release of covalently bound mannoproteins.28The DH of protein hydrolysates was 17.8 ± 1.2 and 25.4 ± 1.3% for H1 and H2, respectively. The protein profile of hydrolysates shows that the proportion of 10–0.05 kDa peak area in H2 was significantly higher than that obtained for H1 (Fig. 1B), confirming the lower proteolysis of the latter. Moreover, H2 showed lower proportion of intermediate molecular weight species (83–10 kDa) than that found for H1 (Fig. 1B). Furthermore, the mannose/protein ratio of H2 was lower than that obtained for H1 (42.3 ± 0.9 vs. 143.1 ± 4.3 mg mannose g per protein, respectively). These results suggest that the protease produces different kinds of peptides depending on the starting substrate (R1 or R2). Thus, the hydrolysis with β-glucanase for 2 h promoted the release of mannoproteins, decreasing their content in R2, which was evidenced as a lower mannose/protein ratio in H2.
On the other hand, the degree of hydrolysis of dialysates (42.8 ± 1.3 and 35.7 ± 2.1 for D1 and D2, respectively) was higher than that obtained for hydrolysates. Therefore, the simulated gastrointestinal digestion (SGID) promoted the generation of new peptides. In agreement with these results, Cian et al.20 reported that the DH of brewers’ spent grain hydrolysate increased 5 times after SGID due to pepsin and pancreatin enzymes activity. Moreover, the degree of hydrolysis of D1 was higher than that found for D2 (p < 0.05), indicating that gastrointestinal enzymes were more effective on the proteins and peptides of H1. Besides, D1 showed a higher proportion of intermediate and low molecular weight peptides (2.6–0.6 kDa and 643–57 Da) than those found for D2 (Fig. 1C). Also, the mannose/protein ratio of D1 was higher than that obtained for D2 (49.9 ± 1.5 vs. 16.1 ± 0.6 mg mannose g per protein, respectively). In agreement with these results, D1 showed higher proportion of mannose-linked peptides than that obtained for D2 (74.7 ± 1.9 vs. 30.1 ± 1.4%, respectively).
Fig. 1D shows the hydrolysates and dialysates’ peptide profile obtained by RP-HPLC. At low retention times (0–20 min), the peak area of dialysates was higher than that obtained for the hydrolysates. In contrast, the dialysates showed the lowest peak area at high retention times (36–52 min). Garzón et al.19 reported that the most hydrophilic peptides elute at low retention times during RP-HPLC, while the hydrophobic ones elute at high retention times. Thus, the dialysates had higher content of hydrophilic peptides than the hydrolysates, probably due to production of low molecular weight peptides during digestive hydrolysis. Note that the SGID promotes the release of highly hydrophilic small peptides.29 In agreement with this result, D1 showed higher content of hydrophilic peptides than that found for D2 (Fig. 1D), which is consistent with the content of mannose-linked peptides of the dialysates.
As shown in Table 2, both BSY hydrolysates inhibited the α-glucosidase enzyme. The IC50 value obtained by D1 was lower than that found for hydrolysates. However, D2 did not show α-glucosidase inhibitory activity. Similar results were obtained for the DPP-IV enzyme, where D1 showed the lowest IC50 value and therefore the highest potency (Table 2). These results suggest that pepsin and pancreatin digestion promoted the generation of new more active peptides from H1. However, the digestives enzymes degraded the α-glucosidase and DPP-IV inhibitory peptides present in H2. As mentioned above, the SGID can increase, maintain or reduce peptide bioactivity.11 Changes in bioactive properties may be due to new molecular characteristics of peptides generated after SGID, such as molecular weight, hydrophobicity, net charge, and sequence.30 Zhang et al.31 reported that the α-glucosidase inhibitory activity of peptides is molecular weight dependent. Moreover, it was reported that α-glucosidase inhibitory peptides have molecular weights lower than 2000 Da.32 Instead, Wang et al.22 found that most DPP-IV inhibitory peptides had low molecular weight. Thus, the high inhibitory activity of D1 against α-glucosidase and DPP-IV enzymes could be partly due to the high content of low molecular weight peptides (Fig. 1B). Moreover, D1 had higher content of mannose-linked peptides than D2. It was reported that low molecular weight glycopeptides generated after SGID from glycosylated α-lactalbumin showed stronger α-glucosidase inhibitory activity.33 In this sense, D1–F2 fraction showed a lower IC50 value than that obtained for D1 (Table 2), indicating that mannose-linked peptides were responsible for inhibiting the α-glucosidase enzyme. Note that D1–F1 fraction did not inhibit this enzyme. Glycosylation analysis of the D1–F2 fraction showed that mannose is linked to the peptide sequence through both O- and N-glycosidic bonds (retention time ≈18.6 min) (Fig. 2A and B), which is evidenced by the release of mannose after β-elimination assay and PGNase F digestion (retention time ≈18.6 min).23 In addition, the mannose-linked peptides had other carbohydrates in their structure, such as glucose, whose retention time was 20 min. Note that glucose was only present in PGNase F products (Fig. 2B), indicating that this carbohydrate is linked to the peptide sequence by N-glycosidic bonds.
| IC50-α-glucosidase (mg protein per mL) | IC50-DPP-V (mg protein per mL) | |
|---|---|---|
| Results are expressed as mean value ± standard deviation. Different letters in the same column mean significant differences between samples (p < 0.05) according to least significant difference (LSD) test. | ||
| H1 | 4.78 ± 0.34c | 4.81 ± 0.17c |
| H2 | 6.95 ± 0.28d | 5.83 ± 0.21d |
| D1 | 3.04 ± 0.13b | 2.99 ± 0.19b |
| D2 | — | — |
| D1–F1 | — | 1.55 ± 0.14a |
| D1–F2 | 0.26 ± 0.07a | 3.12 ± 0.11b |
Regarding DPP-IV inhibition, the D1–F1 fraction showed a lower IC50 value than D1 and D1–F2 fractions (Table 2). Thus, the non-glycosylated peptides were the main responsible species for inhibiting this enzyme.
3.2 Enzyme kinetic analysis, peptide identification and in silico study
The enzyme kinetic analysis of DPP-IV was performed using the Michaelis–Menten model (Fig. 3A). As shown in Table 3, there was no significant difference in Kappm without inhibitor or in the presence of D1–F1 (p > 0.05). However, the Vappmax value for DPP-IV activity was lower in the presence of peptides (p < 0.05), indicating that the type of inhibition of DPP-IV by D1–F1 is non-competitive. Non-competitive inhibitors bind to a site different from the active center of the enzyme (allosteric site), which induces conformational changes in the DPP-IV, reducing its activity.34 Similar inhibition mode on DPP-IV enzyme was found for peptide fractions obtained from silver carp swim bladder hydrolysates.35| Peptide concentration (mg protein mL−1) | K appm (mmol L−1) | V appmax (μmol min−1 L−1) | |
|---|---|---|---|
| K appm: Michaelis constant. Vappmax: maximum reaction velocity. Results are expressed as mean value ± standard deviation. Different letters in the same column for each enzyme mean significant differences between samples (p < 0.05) according to least significant difference (LSD) test. | |||
| DPP-IV | 0 | 1.102 ± 0.091a | 1.304 ± 0.028b |
| DPP-IV + D1–F1 fraction | 1.4536 | 1.125 ± 0.102a | 0.877 ± 0.02a |
| α-Glucosidase | 0 | 5.254 ± 0.278a | 3.486 ± 0.156b |
| α-Glucosidase + D1–F2 fraction | 0.2598 | 5.352 ± 0.229a | 2.165 ± 0.115a |
Regarding the kinetic analysis of α-glucosidase inhibition, it was observed that mannose-linked peptides inhibited the enzyme by non-competitive mode (Fig. 3B). As shown in Table 3, there was no significant difference in Kappm without inhibitor or in the presence of D1–F2, while the Vappmax value for α-glucosidase activity was reduced with the addition of the glycopeptides (p < 0.05). Similar results were reported by Hu et al.,32 who isolated the non-competitive peptide GLLGY from fermented rice bran. As mentioned before, non-competitive inhibitors bind to an essential group of the enzyme which differ from the active site.31 Thus, the mannose-linked peptides would bind to functional groups of the α-glucosidase (allosteric site), causing its inhibition.
In order to characterize the molecular mass and amino acid sequence of D1–F1 and D1–F2 peptide fractions, analysis by tandem mass spectrometry was performed. A search for MS/MS fragments allowed identifying the peptides shown in Table 4. The peptides identified were statistically significant (p < 0.05), thereby confirming their identity with 95% confidence.
| Fractions | Peptide sequencea | Mass (MH+, Da) | Hydrophobicity (kcal mol−1)b | Glycosidic bonda | ADPc |
|---|---|---|---|---|---|
| a Bold letters indicate glycosylation site according to Saccharomyces genome database (SGD). b Otained with PepDraw program. c Obtained with AntiDMPpred tool. The potential anti-diabetic property was estimated with a score higher than 0.5. | |||||
| D1–F1 | IINEPTAAAIAYGLDK | 1658.9 | +17.04 | — | 0.70 |
| VEVEEKDGK | 1031.5 | +28.26 | — | 0.68 | |
| DAGTIAGLNVLR | 1198.7 | +13.67 | — | 0.66 | |
| HLTGEFEK | 959.5 | +18.73 | — | 0.56 | |
| FAGDDAPR | 847.4 | +17.57 | — | 0.52 | |
| KVEKPLS | 799.5 | +16.02 | — | 0.5 | |
| TTPSYVAFTDTER | 1486.7 | +16.20 | — | 0.49 | |
| HFSVEGQLEFR | 1347.7 | +16.55 | — | 0.48 | |
| EVRNSNLD | 945.5 | +17.43 | — | 0.43 | |
| IWHHTFYNELR | 1514.7 | +12.22 | — | 0.30 | |
| LLIKKIS | 813.6 | +9.22 | — | 0.26 | |
| ISFSTWNAFR | 1227.6 | +5.60 | — | 0.18 | |
| NVPNWHR | 921.5 | +11.33 | — | 0.18 | |
| YLRIRRPLK | 1213.8 | +11.94 | — | 0.09 | |
| D1–F2 | GILFVGSGVSGGEEGAR | 1590.8 | +20.29 | O-Glycosidic | 0.77 |
| NMSVIAHVDHGKS | 1393.7 | +19.71 | O-Glycosidic | 0.69 | |
| SYELPDGQVITIGNER | 1789.9 | +20.72 | O-Glycosidic | 0.64 | |
| VPTVDVSVVDLTVK | 1469.8 | +15.07 | O-Glycosidic | 0.52 | |
| AYLPVNESFGFTGELR | 1798.9 | +14.38 | O-Glycosidic | 0.49 | |
| VVNETIQDK | 1044.5 | +17.80 | N-Glycosidic | 0.47 | |
| PTVGNQRIPS | 1067.6 | +11.89 | O-Glycosidic | 0.40 | |
As mentioned above, the fraction with the highest DPP-IV inhibitory activity was D1–F1. Fourteen peptides were identified in this fraction with molecular weight ranging from 799.5 to 1658.9 Da. Prados et al.36 reported that the DPP-IV inhibitory peptides had a size ranging from 700 to 1500 Da. Moreover, Garzón et al.25 identified eleven peptides from the most active fraction against the DPP-IV enzyme, whose molecular weight ranged from 516.2 to 2012 Da. Thus, the identified peptides had a molecular weight within the range expected. All the identified peptides had positively charged residues, such as Arg, His and Lys. In this regard, different identified peptides with DPP-IV inhibitory activity have basic amino acid residues in their sequences.37 Moreover, eight peptides identified from D1–F1 showed Pro or Ala in their structure. In this sense, it was reported that peptides containing hydrophobic residues such as Pro or Ala could act as potent DPP-IV inhibitors.38,39 The in silico hydrophobicity of the identified peptides ranged from +5.6 to +28.3 kcal mol−1 (Table 4). Similar hydrophobicity results were reported by Garzón et al.25 for different DPP-IV inhibitory peptides obtained from sorghum spent hydrolysate. Moreover, the most active anti-diabetic peptide identified from D1–F1 was IINEPTAAAIAYGLDK (AntiDMPpred score: 0.70), which has 62.5% hydrophobic residues in their sequence. This peptide had four alanine residues and lysine at C-terminal position, which could influence inhibitory activity against DPP-IV.
For D1–F2, seven peptides were identified with molecular weight ranging from 1044.5 to 1798.9 Da (Table 4). As mentioned above, α-glucosidase inhibitory peptides have a molecular weight <2000 Da.32 Thus, the identified peptides had a molecular weight within the expected range. All D1–F2 peptides had amino acids with a hydroxyl or basic group near to N-terminal position. In this regard, it was reported that Ser, Thr, Tyr, Lys, Arg and Asn at the N-terminal position of peptide sequence increase the α-glucosidase inhibitory activity.40 Moreover, it was observed that the presence of Glu, Gly and Leu in the peptide sequence is crucial to have good α-glucosidase inhibitory activity.41 Except for VVNETIQDK, all the identified peptides had one of these amino acids in their sequence. Moreover, hydrophobic residues such as Pro, Gly and Leu play a key role in α-glucosidase inhibition.40,42 According to Saccharomyces genome database, six of the seven peptides identified from D1–F2 present O-glycosylation sites (Table 4). Kröger et al.43 reported that O-glycosylated amino acids act as inhibitors of galactosidases. In this regard, the most active anti-diabetic peptide identified from D1–F2 was GILFVGSGVSGGEEGAR (AntiDMPpred score: 0.77), which has two O-glycosylation sites in the peptide sequence (Ser7 and Ser10). Besides the glycosylation type, the inhibitory activity of glycopeptides against α-glucosidase enzyme depends on the peptide sequence.44 Thus, the presence of Ala, Leu, and Gly in the peptide sequence of GILFVGSGVSGGEEGAR could influence their inhibitory activity against α-glucosidase.
In order to evaluate the effect of simulated gastrointestinal digestion on H1 peptides and to estimate the bioaccessibility of D1 peptides, the molecular mass and amino acid sequence analysis of H1 peptides by tandem mass spectrometry was performed. A search for MS/MS fragments allowed the identification of 223 peptides. As shown in Table 5, the main peptides identified were 49. Peptide identification was significant at p < 0.05. Sixteen identified peptides have encrypted peptide sequences (peptide number: 1, 4, 6, 14, 17, 22, 30, 31, 33, 35, 38, 40, 41, 42, 44, and 46) which were found in D1. These results indicate that digestive proteases cleave some peptides present in H1, allowing the release of new peptides, which dialyze across the membrane and are present in D1. Moreover, some of these peptides were glycosylated. Thus, the peptide bioaccessibility was not affected by glycosylation. The presence of some peptides in D1 which do not have a precursor in H1 may be due to the proteolytic action of digestive enzymes on other peptide precursors with high MW present in the hydrolysate.
| Peptide number | Peptide sequencea | Mass (MH+, Da) |
|---|---|---|
| a Bold letters indicate encrypted peptides that are released after simulated gastrointestinal digestion and were identified in D1 LC-MS/MS spectrum. | ||
| 1 | ADREVRNSNLDYTIL | 1777.9 |
| 2 | CK | 249.1 |
| 3 | DCDIITVHSLHGPKVNTEGQPLVIINHR | 3104.6 |
| 4 | DDMEKIWHHTFYNELR | 2132.9 |
| 5 | DPLLK | 584.4 |
| 6 | DTDTIISLLIKKISYDCRYYNY | 2699.3 |
| 7 | DVHQMNK | 870.4 |
| 8 | EISSPGTR | 845.4 |
| 9 | FK | 293.2 |
| 10 | FPVTKILVFDK | 1305.8 |
| 11 | GEK | 332.2 |
| 12 | GKPELRK | 826.5 |
| 13 | GLKGALLR | 826.5 |
| 14 | GMCKAGFAGDDAPRA | 1465.6 |
| 15 | GSGSRLNR | 845.4 |
| 16 | HQYPSSK | 845.4 |
| 17 | IINEPTAAAIAYGLDKKS | 1874.0 |
| 18 | ILVFDK | 733.4 |
| 19 | ITADTQAVTHAAFLSMGSAWAKIK | 2517.3 |
| 20 | KGEHDFTTTLTLSSDGSLTTTTSTHTTHK | 3104.5 |
| 21 | KHK | 411.3 |
| 22 | KKKGILFVGSGVSGGEEGARY | 2138.1 |
| 23 | KNFLDK | 763.4 |
| 24 | KNSSANNK | 861.4 |
| 25 | LAAKSFK | 763.5 |
| 26 | LLLQRR | 797.5 |
| 27 | MPPGIPR | 766.4 |
| 28 | NIESKQK | 845.6 |
| 29 | NSAIHITYRISLR | 1542.9 |
| 30 | NVARVVNETIQDKSSAGA | 1857.9 |
| 31 | VPTVDVSVVDLTVKLEKEATY | 2304.2 |
| 32 | QCLSLPSKVEFYPEPPSSSVPAR | 2517.3 |
| 33 | QRQATKDAGTIAGLNVLR | 1911.1 |
| 34 | RHASQK | 725.4 |
| 35 | RITYKNVPNWHRDLVR | 2066.1 |
| 36 | SK | 233.1 |
| 37 | SPQAVDFLSQRVTTSMTPLSKPK | 2517.3 |
| 38 | SYELPDGQVITIGNERFRAPEAL | 2574.3 |
| 39 | TSPANTSSIFEDHHITPCKPGGQLKFHR | 3104.5 |
| 40 | VARISFSTWNAFRRCDIN | 2155.1 |
| 41 | VEIIANDQGNRTTPSYVAFTDTERL | 2809.4 |
| 42 | VEVEEKDGKTQTKLTQHRF | 2272.1 |
| 43 | VFNILK | 732.5 |
| 44 | VGDGGTGKTTFVKRHLTGEFEKKY | 2654.4 |
| 45 | VMPFEK | 749.4 |
| 46 | VRNMSVIAHVDHGKS | 1648.8 |
| 47 | VYPESR | 749.4 |
| 48 | WK | 332.2 |
| 49 | YINAFKGLLFHVCLHFCSIHR | 2517.3 |
3.3 Molecular docking
According to the AntiDMPpred tool, the most active anti-diabetic peptides identified from D1–F1 and D1–F2 were IINEPTAAAIAYGLDK and GILFVGSGVSGGEEGAR, respectively. Moreover, D1–F1 showed the highest DPP-IV inhibitory activity, while D1–F2 peptides had the highest α-glucosidase inhibitory activity. Thus, the probable mode of interaction of the IINEPTAAAIAYGLDK peptide with DPP-IV and GILFVGSGVSGGEEGAR peptide with α-glucosidase was studied using the HPEPDOCK server.Diprotin A (IPI) has been reported as the most potent DPP-IV inhibitory peptide,45 having a docking score of −101.9. Thus, this tripeptide can be used as control. In this regard, the docking score obtained for IINEPTAAAIAYGLDK peptide was −231.5, indicating a favorable interaction between IINEPTAAAIAYGLDK and DPP-IV. Similar docking scores were reported by Garzón et al.25 for peptides obtained from sorghum spent grain hydrolysate (docking score range from −154.837 to −235.975). Moreover, Rosetta score and interface energies of IINEPTAAAIAYGLDK were −939.8 and −19.745 kcal mol−1, respectively. These results confirm that the interaction between IINEPTAAAIAYGLDK and DPP-IV was possible and favorable. As shown in Fig. 3C, the peptide IINEPTAAAIAYGLDK is within the pocket of DPP-IV. However, there were no interactions lower than 3.0 Å with the active site of enzyme. This result is consistent with non-competitive inhibition mode found for D1–F1 and DPP-IV.
Acarbose is a competitive inhibitor of α-glucosidase enzyme and is used as a positive control in molecular docking.31 The docking binding energy of acarbose is – 7.4 kcal mol−1.32 In this regard, the interface energy obtained for GILFVGSGVSGGEEGAR was −38.038 kcal mol−1, indicating that the interaction between GILFVGSGVSGGEEGAR and α-glucosidase was possible and favorable. Moreover, the docking and Rosetta score were −169.4 and −1606.8, respectively. However, the peptide GILFVGSGVSGGEEGAR was not within the active site of the enzyme. Moreover, the binding site of peptide to the α-glucosidase was very distant from the catalytic center (Fig. 3D). This result agrees with the non-competitive inhibition mechanism found for D1–F2.
3.4 Anti-diabetic effect on mouse jejunum organoids
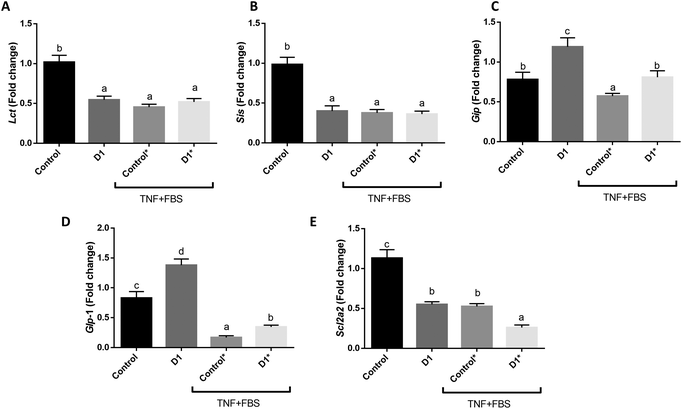
As mentioned before, D1 showed the highest α-glucosidase and DPP-IV inhibitory activity. Thus, it was selected to evaluate the anti-diabetic effect on mouse jejunum organoids.The intestinal epithelium contains different cell types that are present in intestinal organoids.13 Therefore, when compared to cultured intestinal cell lines, the organoid system is more complex and valuable to study the effects of nutrients on the intestinal epithelium. Note that intestinal organoids also contain pluripotent crypt cells.13 Given that the intestinal epithelium is in charge of carbohydrate absorption and produces hormones such as incretins involved in carbohydrate metabolism, organoids are a suitable in vitro model to study the regulation of gene expression of the intestinal epithelium related to carbohydrate metabolism. Therefore, we decided used mouse jejunum organoids to study how the peptides derived from the in vitro digestion of BSY hydrolysates affect the expression of enzymes that digest carbohydrates (sucrase-isomaltase and lactase), the glucose transporter GLUT2 (encoded by Slc2a2), and the expression of incretins (GLP-1 and GIP). As shown in Fig. 4A and B, D1 reduced the gene expression of lactase and sucrase-isomaltase enzymes, which are encoded by the Lct and Sis genes, respectively. These disaccharidases found along the brush border of the small intestine break down sugars into monosaccharides, allowing their absorption.46 Moreover, sucrase-isomaltase belongs to the α-glucosidase family.47 In this regard, the down-regulation exerted by D1 on the Lct and Sis genes was biologically consistent with the in vitro α-glucosidase inhibitory activity. Moreover, D1 increased the expression of the Glp1 and Gip incretin genes in basal conditions (Fig. 4C and D). The stimulation with TNF inhibited the expression of these genes and D1 partially counteracted the effect on Glp1. As mentioned before, the DPP-IV enzyme degrades the incretin hormones (GLP-1 and GIP), reducing insulin secretion.7 Thus, D1 could exert an anti-diabetic effect not only through the inhibition of the DPP-IV enzyme, but also increasing the expression of incretin hormones. Additionally, D1 inhibited the gene expression of Slc2a2 in basal and stimulated conditions (TNF) (Fig. 4E). This gene codes for the glucose transporter 2 (GLUT2), which is the main glucose transporter in the apical membrane of the intestinal epithelium.48 Therefore, D1 not only regulated the gene expression of intestinal enzymes involved in the carbohydrate digestion, but could also modulate glucose uptake on mouse jejunum organoids. Similar results were reported by Mojica et al.49 for peptides isolated from black bean protein hydrolysates. These authors found that peptides AKSPLF, ATNPLF, FEELN, and LSVSVL effectively inhibited the GLUT2 and sodium-dependent glucose transporter 1 (SGLT1) in hyperglycemic rat model, which was evidenced by the reduction of the blood glucose level.
4. Conclusions
Peptides with in vitro α-glucosidase and DPP-IV inhibitory activities from BSY were obtained. The simulated gastrointestinal digestion significantly modified the anti-diabetic activity of these peptides since only one hydrolysate dialysate maintained this activity, which even presented lower α-glucosidase and DPP-IV inhibitory IC50 values than before digestion, indicating a higher anti-diabetic potential. This dialysate presented a high content of peptides linked to mannose through O- and N-glycosidic bonds. The mannose-linked peptides were responsible for inhibiting the α-glucosidase enzyme, while the non-glycosylated fraction strongly inhibited the DPP-IV enzyme. This is the first report where the role of glycosylated BSY peptides in anti-diabetic properties was studied. On the other hand, the inhibition of these enzymes by mannose-linked peptides and the non-glycosylated fraction was non-competitive. This was confirmed by tandem mass spectrometry and molecular docking analysis. In addition, dialysate peptides downregulated the expression of genes related to carbohydrate catabolism and glucose uptake in a mouse jejunal organoid model. Thus, our data indicate BSY may be a good source of anti-diabetic peptides that regulate the intestinal epithelium glucose metabolism after simulated gastrointestinal digestion. However, in vivo studies are needed for confirm these effects.Abbreviations
| BSY | Brewer's spent yeast |
| D1 | Dialysate obtained from H1 after SGID |
| D2 | Dialysate obtained from H2 after SGID |
| D1–F1 | Non-glycosylated peptides obtained from D1 by affinity chromatography |
| D1–F2 | Mannose-linked peptides obtained from D1 by affinity chromatography |
| DH | Degree of hydrolysis |
| DPP-IV | Dipeptidyl peptidase IV enzyme |
| FBS | Fetal bovine serum |
| GLUT2 | Glucose transporter 2 |
| H1 | Hydrolysate obtained from R1 |
| H2 | Hydrolysate obtained from R2 |
| GIP | Glucose-dependent insulinotropic peptide |
| GLP-1 | Glucagon-like peptide 1 |
| R1 | Residue obtained after hydrolysis with β-glucanase for 1 h |
| R2 | Residue obtained after hydrolysis with β-glucanase for 2 h |
| SGID | Simulated gastrointestinal digestion |
| SGLT1 | Sodium-dependent glucose transporter 1. |
Author contributions
Marilin E. Aquino and Raúl E. Cian: formal analysis; investigation; methodology. Silvina R. Drago, Fermín Sanchez de Medina, Olga Martínez-Augustín and Raúl E. Cian: data curation; investigation; methodology; validation; writing – review & editing. Raúl E. Cian, Fermín Sánchez de Medina and Olga Martínez-Augustin: resources and funding acquisition. All authors read and approved the final manuscript.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
The authors are grateful to Dr Tena-garitaonaindia's assistance. This work was supported by ANPCyT – Argentina (Project PICT-2020-Serie A-1985) and Agencia Santafesina de Ciencia Tecnología e Innovación – Argentina (Res. 132/23-Project PEICID-2022-133), by grants PID2020-112768RB-I00 and PI21/00952 funded by MICINN and by Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Spain, respectively, and by grants A-AGR-468-UGR20 and P20-00695 funded by Junta de Andalucía and FEDER. Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas (CIBERehd) is funded by Instituto de Salud Carlos III, Spain.References
- A. S. Oliveira, C. Ferreira, J. O. Pereira, M. E. Pintado and A. P. Carvalho, Valorisation of protein-rich extracts from spent brewer's yeast (Saccharomyces cerevisiae): an overview, Biomass Convers. Biorefin., 2022 DOI:10.1007/s13399-022-02636-5.
- R. Ribeiro-Oliveira, Z. E. Martins, J. B. Sousa, I. M. P. L. V. O. Ferreira and C. Diniz, The health-promoting potential of peptides from brewing by-products: An up-to-date review, Trends Food Sci. Technol., 2021, 118(Part A), 143–153 CrossRef CAS.
- G. V. Marson, R. J. S. de Castro, M. P. Belleville and M. D. Hubinger, Spent brewer's yeast as a source of high added value molecules: a systematic review on its characteristics, processing and potential applications, World J. Microbiol. Biotechnol., 2020, 36, 95 CrossRef PubMed.
- M. Amorim, J. O. Pereira, D. Gomes, C. D. Pereira, H. Pinheiro and M. Pintado, Nutritional ingredients from spent brewer's yeast obtained by hydrolysis and selective membrane filtration integrated in a pilot process, J. Food Eng., 2016, 185, 42–47 CrossRef CAS.
- P. G. Spontón, R. Spinelli, S. R. Drago, G. G. Tonarelli and A. C. Simonetta, Acetylcholinesterase-inhibitor hydrolysates obtained from ‘in vitro’ enzymatic hydrolysis of mannoproteins extracted from different strains of yeasts, Int. J. Food Sci. Technol., 2016, 51, 300–308 CrossRef.
- G. V. Marson, S. Lacour, M. D. Hubinger and M. P. Belleville, Serial fractionation of spent brewer's yeast protein hydrolysate by ultrafiltration: A peptide-rich product with low RNA content, J. Food Eng., 2022, 312, 110737 CrossRef CAS.
- J. Yan, J. Zhao, R. Yang and W. Zhao, Bioactive peptides with antidiabetic properties: A review, Int. J. Food Sci. Technol., 2019, 54, 1909–1191 CrossRef CAS.
- R. E. Cian, A. E. Nardo, A. G. Garzón, M. C. Añon and S. R. Drago, Identification and in silico study of a novel dipeptidyl peptidase IV inhibitory peptide derived from green seaweed Ulva spp. hydrolysates, LWT–Food Sci. Technol., 2022, 154, 112738 CrossRef CAS.
- T. Zietek, E. Rath, D. Haller and H. Daniel, Intestinal organoids for assessing nutrient transport, sensing and incretin secretion, Sci. Rep., 2015, 5, 16831 CrossRef CAS PubMed.
- J. J. Holst, L. S. Gasbjerg and M. M. Rosenkilde, The Role of Incretins on Insulin Function and Glucose Homeostasis, Endocrinology, 2021, 162, bqab065 CrossRef PubMed.
- R. E. Cian, A. Campos-Soldini, L. Chel-Guerrero, S. R. Drago and D. Betancur-Ancona, Bioactive Phaseolus lunatus peptides release from maltodextrin/gum arabic microcapsules obtained by spray drying after simulated gastrointestinal digestion, Int. J. Food Sci. Technol., 2019, 54, 2002–2009 CrossRef CAS.
- R. E. Cian, A. G. Garzón, D. Betancur-Ancona, L. Chel-Guerrero and S. R. Drago, Hydrolyzates from Pyropia columbina seaweed have antiplatelet aggregation, antioxidant and ACE I inhibitory peptides which maintain bioactivity after simulated gastrointestinal digestion, LWT–Food Sci. Technol., 2015, 64, 881–888 CrossRef CAS.
- L. Szabó, A. C. Seubert and K. Kretzschmar, Modelling adult stem cells and their niche in health and disease with epithelial organoids, Semin. Cell Dev. Biol., 2023, 144, 20–30 CrossRef PubMed.
- J. Li and S. Karboune, A comparative study for the isolation and characterization of mannoproteins from Saccharomyces cerevisiae yeast cell wall, Int. J. Biol. Macromol., 2018, 119, 654–661 CrossRef CAS PubMed.
- Y. Zhang and W. Gu, Determination of mannose in yeast by ultraviolet spectrometry, Food Ferment. Ind., 1999, 25, 32–36 CAS.
- O. Lowry, N. Rosebrough, L. Farr and R. Randall, Proteins measurement with the folin phenol reagent, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS PubMed.
- F. Van de Velde, M. E. Pirovani and S. R. Drago, Bioaccessibility analysis of anthocyanins and ellagitannins from blackberry at simulated gastrointestinal and colonic levels, J. Food Compos. Anal., 2018, 72, 22–31 CrossRef CAS.
- P. Nielsen, D. Petersen and C. Dambmann, Improved method for determining food protein degree of hydrolysis, J. Food Sci., 2001, 66, 642–646 CrossRef CAS.
- A. G. Garzón, R. E. Cian, M. A. Aquino and S. R. Drago, Isolation and identification of cholesterol esterase and pancreatic lipase inhibitory peptides from brewer's spent grain by consecutive chromatography and mass spectrometry, Food Funct., 2020, 11, 4994–5003 RSC.
- R. E. Cian, P. R. Salgado, A. N. Mauri and S. R. Drago, Pyropia columbina phycocolloids as microencapsulating material improve bioaccessibility of brewers’ spent grain peptides with ACE-I inhibitory activity, Int. J. Food Sci. Technol., 2020, 55, 1311–1317 CrossRef CAS.
- O. Donkor, L. Stojanovska, P. Ginn, J. Ashton and T. Vasiljevic, Germinated grains – Sources of bioactive compounds, Food Chem., 2012, 135, 950–959 CrossRef CAS PubMed.
- T. Wang, C. Hsieh, C. Hung, C. Jao, P. Lin, Y. Hsieh and K. Hsu, A study to evaluate the potential of an in silico approach for predicting dipeptidyl peptidase-IV inhibitory activity in vitro of protein hydrolysates, Food Chem., 2017, 234, 431–438 CrossRef CAS PubMed.
- G. J. Cavallero, M. Malamud, A. C. Casabuono, M. L.Á. Serradell and A. S. Couto, A glycoproteomic approach reveals that the S-layer glycoprotein of Lactobacillus kefiri CIDCA 83111 is O- and N-glycosylated, J. Proteomics, 2017, 162, 20–29 CrossRef CAS PubMed.
- X. Chen, J. Huang and B. He, B. AntiDMPpred: a web service for identifying anti-diabetic peptides, Peer J, 2022, 10, e13581 CrossRef PubMed.
- A. G. Garzón, F. F. Veras, A. Brandelli and S. R. Drago, Purification, identification and in silico studies of antioxidant, antidiabetogenic and antibacterial peptides obtained from sorghum spent grain hydrolysate, LWT–Food Sci. Technol., 2022, 153, 112414 CrossRef.
- M. Arredondo-Amador, C. J. Aranda, B. Ocón, R. González, O. Martínez-Augustin O and F. Sánchez de Medina, Epithelial deletion of the glucocorticoid receptor (Nr3c1) protects the mouse intestine against experimental inflammation, Br. J. Pharmacol., 2021, 178, 2482–2495 CrossRef CAS PubMed.
- J. Li, S. Karboune, J. Sedman and A. Ismail, Characterization of the structural properties of mannoproteins isolated from selected yeast-based products upon the enzymatic treatment, LWT–Food Sci. Technol., 2020, 131, 109596 CrossRef CAS.
- J. Li, S. Karboune and A. Asehraou, Mannoproteins from inactivated whole cells of baker's and brewer's yeasts as functional food ingredients: Isolation and optimization, J. Food Sci., 2020, 85, 1438–1449 CrossRef CAS PubMed.
- F. B. Pimentel, R. C. Alves, P. A. Harnedy and R. J. FitzGerald, Macroalgal-derived protein hydrolysates and bioactive peptides: Enzymatic release and potential health enhancing properties, Trends Food Sci. Technol., 2019, 93, 106–124 CrossRef CAS.
- T. Ahmed, X. Sun and C. C. Udenigwe, Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: A systematic review, Trends Food Sci. Technol., 2022, 120, 265–273 CrossRef CAS.
- Y. Zhang, F. Wu, Z. He, X. Fang and X. Liu, Optimization and Molecular Mechanism of Novel Glucosidase Inhibitory Peptides Derived from Camellia Seed Cake through Enzymatic Hydrolysis, Foods, 2023, 12, 393 CrossRef CAS PubMed.
- J. Hu, X. Lai, X. Wu, H. Wang, N. Weng, J. Lu, M. Lyu and S. Wang, Isolation of a Novel Anti-Diabetic-Glucosidase Oligo-Peptide Inhibitor from Fermented Rice Bran, Foods, 2023, 12, 183 CrossRef CAS PubMed.
- Y. Ma, Y. Liu, H. Yu, S. Mu, H. Li, X. Liu, M. Zhang, Z. Jiang and J. Hou, Biological activities and in vitro digestion characteristics of glycosylated α-lactalbumin prepared by microwave heating: Impacts of ultrasonication, LWT–Food Sci. Technol., 2022, 158, 113141 CrossRef CAS.
- N. T. P. Nong, Y. K. Chen, W. L. Shih and J. L. Hsu, Characterization of Novel Dipeptidyl Peptidase-IV Inhibitory Peptides from Soft-Shelled Turtle Yolk Hydrolysate Using Orthogonal Bioassay-Guided Fractionations Coupled with In Vitro and In Silico Study, Pharmaceuticals, 2020, 13, 308 CrossRef CAS PubMed.
- H. Hong, Y. Zheng, S. Song, Y. Zhang, C. Zhang, J. Liu and Y. Luo, Identification and characterization of DPP-IV inhibitory peptides from silver carp swim bladder hydrolysates, Food Biosci., 2020, 38, 100748 CrossRef CAS.
- I. Prados, M. Marina and M. García, Isolation and identification by high resolution liquid chromatography tandem mass spectrometry of novel peptides with multifunctional lipid lowering capacity, Food Res. Int., 2018, 111, 77–86 CrossRef CAS PubMed.
- X. Kong, L. Zhang, W. Song, C. Zhang, Y. Hua, Y. Chen and X. Li, Separation, identification and molecular binding mechanism of dipeptidyl peptidase IV inhibitory peptides derived from walnut (Juglans regia L.) protein, Food Chem., 2021, 47, 129062 CrossRef PubMed.
- M. Cermeño, J. Stack, P. Tobin, M. O'Keeffe, P. Harnedy, D. B. Stengel and R. J. FitzGerald, Peptide identificatiob from a Porphyra dioica protein hydrolysate with antioxidant, angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory peptides, Food Funct., 2019, 10, 3421–3429 RSC.
- A. B. Nongonierma and R. J. FitzGerald, Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins, J. Food Biochem., 2017, 43, e12451 CrossRef PubMed.
- S. Abbasi, M. Moslehishad and M. Salami, Antioxidant and alpha-glucosidase enzyme inhibitory properties of hydrolyzed protein and bioactive peptides of quinoa, Int. J. Biol. Macromol., 2022, 213, 602–609 CrossRef CAS PubMed.
- P. Pramai, N. A. A. Hamid, A. Mediani, M. Maulidiani, F. Abas and S. Jiamyangyuen, Metabolite profiling, antioxidant, and α-glucosidase inhibitory activities of germinated rice: nuclear-magnetic-resonance-based metabolomics study, J. Food Drug Anal., 2018, 26, 47–57 CrossRef CAS PubMed.
- X. Zheng, H. Chi, S. Ma, L. Zhao and S. Cai, Identification of novel α-glucosidase inhibitory peptides in rice wine and their antioxidant activities using in silico and in vitro analyses, LWT–Food Sci. Technol., 2023, 178, 114629 CrossRef CAS.
- L. Kröger, D. Henkensmeier, A. Schäfer and J. Thiem, Novel O-glycosyl amino acid mimetics as building blocks for O-glycopeptides act as inhibitors of galactosidases, Bioorg. Med. Chem. Lett., 2004, 14, 73–75 CrossRef PubMed.
- G. Wei, Q. Zhao, D. Wang, Y. Fan, Y. Shi and A. Huang, Novel ACE inhibitory, antioxidant and α-glucosidase inhibitory peptides identified from fermented rubing cheese through peptidomic and molecular docking, LWT–Food Sci. Technol., 2022, 159, 113196 CrossRef CAS.
- F. Rivero-Pino, F. J. Espejo-Carpio and E. M. Guadix, Production and identification of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from discarded Sardine pilchardus protein, Food Chem., 202, 328, 127096 CrossRef PubMed.
- L. Viswanathan and S. S. Rao, Intestinal Disaccharidase Deficiency in Adults: Evaluation and Treatment, Curr. Gastroenterol. Rep., 2023, 25, 134–139 CrossRef PubMed.
- B. Gericke, N. Schecker, M. Amiri and H. Y. Naim, Structure-function analysis of human sucrase-isomaltase identifies key residues required for catalytic activity, J. Biol. Chem., 2017, 292, 11070–11078 CrossRef CAS PubMed.
- P. Solverson, Anthocyanin Bioactivity in Obesity and Diabetes: The Essential Role of Glucose Transporters in the Gut and Periphery, Cells, 2020, 9, 2515 CrossRef CAS PubMed.
- L. Mojica, E. Gonzalez de Mejia, M.Á. Granados-Silvestre and M. Menjivar, Evaluation of the hypoglycemic potential of a black bean hydrolyzed protein isolate and its pure peptides using in silico, in vitro and in vivo approaches, J. Funct. Foods, 2017, 31, 274–286 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |