Exploring immunoregulatory properties of a phenolic-enriched maple syrup extract through integrated proteomics and in vitro assays†
Tess
Puopolo
 a,
Ying
Chen
a,
Ying
Chen
 ab,
Hang
Ma
ab,
Hang
Ma
 a,
Chang
Liu
a,
Chang
Liu
 *a and
Navindra P.
Seeram
*a and
Navindra P.
Seeram
 *a
*a
aDepartment of Biomedical and Pharmaceutical Sciences, College of Pharmacy, University of Rhode Island, Kingston, RI 02881, USA. E-mail: hichang813@uri.edu; nseeram@uri.edu
bDepartment of Obstetrics and Gynecology, The Second Affiliated Hospital of Soochow University, Suzhou, 215004, China
First published on 21st November 2023
Abstract
Our laboratory has established a comprehensive program to investigate the phytochemical composition and nutritional/medicinal properties of phenolic-enriched maple syrup extract (MSX). Previous studies support MSX's therapeutic potential in diverse disease models, primarily through its anti-inflammatory effects. We recently demonstrated MSX's ability to regulate inflammatory signaling pathways and modulate inflammatory markers and proteins in a lipopolysaccharide (LPS)-induced peritonitis mouse model. However, MSX's immunoregulatory properties remain unknown. Herein, we investigated MSX's immunoregulatory properties for the first time using an integrated approach, combining data-dependent acquisition (DDA) and data-independent acquisition (DIA) strategies in a proteomic analysis of spleen tissue collected from the aforementioned peritonitis mouse model. Additionally, we conducted immune cell activation assays using macrophages and T lymphocytes. The DIA analysis unveiled a distinctive expression pattern involving three proteins—Krt83, Thoc2, and Vps16—which were present in both the control and MSX-treated groups but absent in the LPS-induced model group. Furthermore, proteins Ppih and Dpp9 exhibited significant reductions in the MSX-treated group. Ingenuity pathway analysis indicated that MSX may modulate several critical signaling pathways, exerting a suppressive effect on immune responses in various cell types involved in both innate and adaptive immunity. Our in vitro cell assays supported findings from the proteomics, revealing that MSX significantly reduced the levels of interleukin-1 beta (IL-1β) and tumor necrosis factor-alpha (TNF-α) in LPS-stimulated human macrophage cells, as well as the levels of IL-2 in anti-CD3/anti-CD28-induced Jurkat T cells. Taken together, our investigations provide evidence that MSX exerts immune regulatory effects that impact both innate and adaptive immunity, which adds to the data supporting MSX's development as a functional food.
Introduction
Maple syrup, a natural sweeter derived from the sap of maple trees, serves as an alternative diet option to refined sugars.1,2 Native to Canada and the United States, maple syrup is collected from trees of the Acer species, predominantly the sugar maple (Acer saccharum).1 Our previous research reported the phytochemical composition of maple syrup, revealing a profile of diverse compounds including minerals, vitamins, amino acids, organic acids, phytohormones, and a substantial presence of phenolic compounds (such as lignans, phenolic acids, stilbenes, coumarins, and various flavonoid subclasses).3–6 These constituents have been recognized for their potential human health benefits.3,7 In addition to our phytochemical compositional investigations, our laboratory has been dedicated to developing maple constituents for their applications as functional foods, culminating in the development of a novel food-grade phenolic-enriched maple syrup extract (MSX).3 Over the past decade, our studies have supported the nutraceutical potential of MSX. These studies included in-depth analyses of the phytochemical composition where we characterized MSX phenolic content (primarily lignans), in comparison to maple sugar, maple syrup, and maple water.6,8 These findings are comparable to other phenolic-rich foods such as chocolate, pomegranate and cranberry.6 We have also conducted safety assessment on MSX and reported that there were no overt signs of toxicity with daily doses of up to 1000 mg kg−1 in rats.3 Further, our studies have encompassed an examination of MSX's biological effects, spanning diseases and/or biological functions including diabetes, neurodegenerative disorders, inflammation, wound healing, and oxidative stress.3,9–13Inflammation stands as a predominant factor contributing to more than half of global mortality, owing to its pervasive involvement in various disease conditions, including heart disease, kidney disease, diabetes, neurodegenerative disorders, and autoimmune diseases.14 While acute and chronic inflammation can affect multiple body tissues, the spleen emerges as a promising target for therapeutics aimed at mitigating inflammation.15 The spleen, structurally composed of distinct red pulp and white pulp regions, assumes critical roles in the immune response.16 Specifically, the spleen orchestrates both innate and adaptive immune processes, facilitating pathogen clearance and cytokine secretion.17,18 The spleen accommodates both leukocytes with innate immune functions and T and B cells crucial for adaptive immunity, playing a central role in various immune functions.16
Systemic inflammation can be triggered by lipopolysaccharide (LPS), a bacterial endotoxin and pathogen-associated molecular pattern.19 While LPS primarily modulates the innate immune system, it has also been shown to affect adaptive immunity.20 In the spleen, LPS administration leads to neutrophil migration to the T cell zone and the redistribution of T cells in mice.20 Furthermore, LPS induces a reduction of T cells in the red pulp, accompanied by the reorganization of immune effectors, including cytokines and chemokines.20 Splenic macrophages are activated by LPS to express high levels of proinflammatory cytokines such as IL-6, IL-12, and TNF-α.21 Additionally, intraperitoneal injections of LPS are eliminated from systemic circulation by macrophages, monocytes, and neutrophils, as well as by resident macrophages in the spleen.22 Further, the spleen, with its dual role in the innate and adaptive immune systems, plays a crucial role in counteracting endotoxemia.22 Given our previous findings of MSX's therapeutic potential in the treatment of various diseases, primarily due to its anti-inflammatory properties, it is important to conduct a comprehensive investigation into the impact of MSX on immune cells and immune function within spleen tissue.
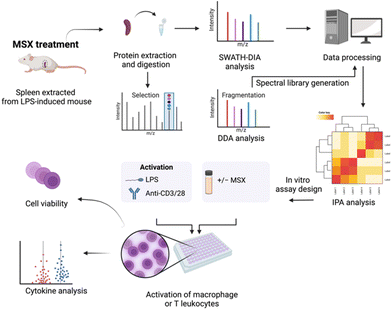
Recently, our group elucidated the anti-inflammatory mechanisms of MSX in an LPS-induced peritonitis mouse model, utilizing data-independent acquisition (DIA) proteomics to gain a comprehensive understanding of its action.23 Building upon the foundation of that study, the current research aims to evaluate MSX's potential immunoregulatory effects in the context of splenic inflammation induced by LPS. To facilitate this investigation, we have taken several strategic steps. First, we constructed a spectral library using data-dependent acquisition (DDA) proteomics. Subsequently, we conducted DIA proteomics methods to investigate the molecular changes caused by the administration of LPS and MSX (Fig. 1). To validate our proteomic findings, we conducted in vitro assays to assess MSX's impact on both innate and adaptive immunity. These experiments involved in vitro cell activation models, including differentiated monocytes (THP-1 cells stimulated by LPS) and T cells (Jurkat cells activated by anti-CD3/CD28), as depicted in Fig. 1.
Experimental
MSX preparation and characterization
A food-grade MSX was prepared at the facility of Silicycle Inc. (Quebec, Canada) at a gram-scale and its phytochemical profile was characterized by our laboratory using previously published protocols.3Chemicals and reagents
Lipopolysaccharide (LPS) from Escherichia coli was obtained from the Cayman Chemical Company (Ann Arbor, MI, USA). The Pierce BCA Protein Assay kit and the CyQUANT™ MTT Cell Viability Assay kit were purchased from Thermo Fisher Scientific (Waltham, MA, USA). The RIPA lysis buffer (10×), methanol, formic acid, 1,4-dithiothreitol (DTT), triethyl ammonium bicarbonate (TEAB), ammonium bicarbonate, urea buffer, sodium deoxycholate (DOC), iodoacetamide (IAA) and phorbol myristate acetate (PMA) were purchased from Sigma-Aldrich (St Louis, MO, USA). TPCK-treated trypsin was purchased from SCIEX (Framingham, MA, USA). Purified anti-CD3 and anti-CD28 antibodies were purchased from BioLegend Inc. (San Diego, CA, USA). Fetal bovine serum (FBS) and Roswell Park Memorial Institute (RPMI) 1640 medium were purchased from Gibco Life Technologies (Gaithersburg, MD, USA). The kits of ELISA Max Deluxe human IL-1β, IL-2, and TNF-α were purchased from BioLegend (San Diego, CA, USA). A Cell Counting kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies, Inc. (Rockville, MD, USA).Animals
The spleen tissues used in this study were obtained from an animal study we previously reported.23 Specifically, we utilized an LPS-induced peritonitis model in mice where the optimal MSX dose (0.05%) was selected in a dose-finding study, followed by a DIA proteomic analysis of the kidney and liver. The study was conducted in strict adherence to the guidelines outlined in the “Guide for the Care and Use of Laboratory Animals,” 8th Edition, as released by the National Institutes of Health (NIH), the “Public Health Service Policy on Humane Care and Use of Laboratory Animals” (PHS Policy), and the “Animal Welfare Regulations” (AWRs) set forth by the United States Department of Agriculture (USDA). Additionally, all procedures were executed in strict accordance with the approved protocol number AN1819-011, as authorized by the University of Rhode Island Institutional Animal Care and Use Committee (IACUC).Briefly, five-week-old male C57BL/6 mice were procured from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained under Specific-pathogen-free (SPF) conditions at a controlled temperature of 21 °C ± 2 and humidity ranging from 30–70%. The mice were provided a 2020X Teklad Global Soy Protein-Free Extruded Rodent Diet as we previously reported.23 All experimental procedures undertaken in this study align with those detailed in our earlier publications.13,23 In detail, three groups including the control group, model, and MSX groups were employed in this study. For the control group, mice were administered an intraperitoneal (i.p.) injection of PBS. For the model (LPS) group, mice were subjected to an i.p. injection of 1 mg kg−1 LPS. For the MSX group, the mice received an oral gavage of 60 mg kg−1 (0.05% MSX per b.w.) daily for a consecutive duration of 10 days. On the 10th day, precisely one hour after the final oral gavage of MSX, the mice underwent an i.p. injection of 1 mg kg−1 LPS.
Collection of spleen tissue samples from LPS-induced inflammation mouse model
The LPS-induced peritonitis model was established as we previously described.23 After a one-week acclimation period, the mice were stratified into three groups. In the control group, mice received an intraperitoneal (i.p.) injection of PBS. In the model (LPS) group, mice were subjected to an i.p. injection of 1 mg kg−1 LPS. For the MSX group, mice were administered 60 mg kg−1 (0.05%) MSX per b.w. via oral gavage for a duration of 10 consecutive days. On the 10th day, precisely one hour following the oral gavage of MSX, mice underwent an i.p. injection of 1 mg kg−1 LPS. Subsequent to a four-hour interval following the LPS stimulation, all mice were humanely euthanized, and specimens of spleen (n = 4) were harvested for subsequent analyses.Chloroform–methanol protein extraction for proteomics analysis
Spleen tissues were retrieved from frozen storage and prepared in a homogenization buffer (8 M urea & 50 mM TEAB) at a concentration of 50 mg mL−1. Chloroform–methanol protein extraction was performed as previously described.23 For a more comprehensive description, please refer to our earlier publication.23LC-MS/MS
Both data-dependent acquisition (DDA) and data-independent acquisition (DIA) were conducted on an AB Sciex TripleTOF5600 mass spectrometer using a DuoSpray™ ion source (SCIEX, Framingham, MA, USA) coupled to an Acquity H Class HPLC system (Waters Corp., Milford, MA, USA). An Acquity UPLC Peptide BEH C18 column (2.1 Å–150 mm2, 300 Å, 1.7 μm) with VanGuard pre-column (2.1 Å–5 mm2, 300 Å, 1.7 μm) was used for the sample separation. The column temperature was set to 50 °C and the autosampler was set to 10 °C. Sample separation was employed within a 180 min gradient method of 10 μL min−1 flow rate. Mobile phase A was water containing 0.1% w/v formic acid, and mobile phase B was acetonitrile containing 0.1% w/v formic acid. A solvent composition scheme was described as follows: 98% A, 0–5 min; 98% to 70% A, 5–155 min; 70% to 50% A, 155–160 min; 50% to 5% A, 160–170 min; and 5% to 98% A held from 170–175 min. The gradient was returned to initial conditions from 170 to 175 min to equilibrate the column between samples. β-Galactosidase (SCIEX, Framingham, MA, USA) was injected every 4 samples for TripleTOF5600 mass calibration. Data was acquired using analyst TF 1.7.1.For DDA experiments, positive ionization mode via the DuoSpray™ ion source was employed. The ion source gas 1 (GS1), ion source gas 2 (GS2), and curtain gas (CUR) were set to 55, 60, and 25 psi, respectively. The source temperature (TEM) was maintained at 450 °C, and ion spray voltage floating (ISVF) was set to 5500 V. Parameters for declustering potential (DP), collision energy (CE), and collision energy spread (CES) were specified as 100, 10, and 15, respectively. Ions within a mass range of m/z 300–1250, surpassing 25 cps, were selected for MS/MS analysis during survey scans. In DIA utilizing SWATH-MS experiments, the ion mode was set to positive. The ESI source parameters included: GS1, 55; GS2, 60; CUR, 25 psi; ISVF, 5500 V; TEM, 450 °C; DP, 100; CE, 10; and CES, 15. SWATH data was acquired within the mass range of m/z 400–1100, utilizing 70 SWATH windows per cycle, each with a window size of m/z 10.
Data-independent acquisition analysis
DIA analysis was conducted on spleen tissue samples. Spectral libraries were generated using DDA experiments in conjunction with the mouse FASTA file (UP000000589_10090) sourced from UNIPROT. The Trypsin/P enzymatic cleavage rule was employed for digestion, allowing for a maximum of 2 missed cleavages. Peptides ranging from 7 to 52 amino acids in length were selected for analysis. This resulted in the creation of a distinct spectral library for spleen tissue.For the MSX spleen samples, a total of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568 precursor ions were produced across 12 DDA runs (three groups consisting of four biological replicates). Subsequent to spectral library construction, DIA was carried out using three groups, each comprising four biological replicates. Default settings were employed in Spectronaut17, where Trypsin-P was chosen for protease specificity, and carbamidomethylation was designated as a fixed modification. Variable modifications included acetylation at protein N-termini and oxidation of methionine (M). False Discovery Rate (FDR) was set at 0.01 for peptide, protein, and peptide-spectrum match (PSM) levels. The XIC Retention Time (RT) window was dynamically configured, with optimal tolerances determined by Spectronaut17, and the correction factor was established as per system default. Spectronaut17 was responsible for normalizing protein intensities, utilizing a local normalization strategy. The total protein approach (TPA) was used to quantify proteins where quantities were obtained from raw intensity values using the following equation: Protein (pmol per mg protein) = (total intensity/(MW (g mol−1) × total protein intensity)) × 109.24,25
568 precursor ions were produced across 12 DDA runs (three groups consisting of four biological replicates). Subsequent to spectral library construction, DIA was carried out using three groups, each comprising four biological replicates. Default settings were employed in Spectronaut17, where Trypsin-P was chosen for protease specificity, and carbamidomethylation was designated as a fixed modification. Variable modifications included acetylation at protein N-termini and oxidation of methionine (M). False Discovery Rate (FDR) was set at 0.01 for peptide, protein, and peptide-spectrum match (PSM) levels. The XIC Retention Time (RT) window was dynamically configured, with optimal tolerances determined by Spectronaut17, and the correction factor was established as per system default. Spectronaut17 was responsible for normalizing protein intensities, utilizing a local normalization strategy. The total protein approach (TPA) was used to quantify proteins where quantities were obtained from raw intensity values using the following equation: Protein (pmol per mg protein) = (total intensity/(MW (g mol−1) × total protein intensity)) × 109.24,25
Bioinformatic analysis
The application of Ingenuity Pathway Analysis (IPA), specifically the release version 62089861 from 2021 (Qiagen), was undertaken to analyze the proteins displaying differential expression. In this analysis, the total protein concentrations within each experimental group were transformed into expression ratios relative to the control group. These derived ratios, in conjunction with corresponding p-values (pertaining to Model versus Control and MSX versus Model comparisons), were input into IPA to facilitate the construction of the IPA database. The input file for the IPA analysis has been included as ESI,† ensuring the reproducibility of the data.To generate canonical pathways, disease functions, and upstream regulators, Fisher's Exact Test was employed. This statistical test evaluated associations between input data and established annotations within the IPA database. The outcome of this analysis provided insights into potential pathways, regulators, and disease functions relevant to the dataset.
Projections of pathways, regulators, and disease functions were ascertained by contrasting the directionality of expression changes observed in the dataset with anticipated expression patterns derived from pertinent literature annotations. The assignment of z-scores allowed for the determination of statistical significance regarding these predictions.
For the 35 proteins uniquely expressed by LPS administration, we conducted an analysis utilizing the STRING database (Version 12), to map proteins to specific gene ontology biological processes (https://string-db.org/). Specifically, the 35 protein identifiers were input into the STRING analysis with the organism selected as Mus musculus. Basic settings included selecting full STRING network as the network type, selecting evidence under meaning of network edges, selecting active interaction sources including textmining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence, and the minimum required interaction score set to medium confidence (0.0400). Advanced settings included the network display mode set to interactive svg. Significant functional enrichments corresponding to gene ontology biological processes were reported and enriched networks were exported.
Cell line and cell culture
Human THP-1 monocytes and Jurkat T cells were purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). The cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS at 5% CO2 and 37 °C as recommended by ATCC.THP-1 cell differentiation and activation
The differentiation of THP-1 monocytes into macrophages was conducted as we previously reported.26 The cells were seeded at a density of 5 × 104 cells per well in a 48-well plate and differentiated with PMA (25 nM) for 48 h. PMA was then removed, and cells were grown in PMA-free media for an additional 24 h. Control cells received no treatment, and the model group received LPS (100 ng mL−1) to induce inflammation. To determine the effect of MSX on LPS-induced THP-1 cell activation, differentiated THP-1 monocytes were treated with MSX (0.156, 0.312, 0.625, and 1.25 μg mL−1) for 1 h, followed by the addition of LPS (100 ng mL−1) for 24 h.Jurkat T cell activation
Jurkat T-cells were seeded at 5 × 104 per well in a 48-well plate. Cells received no treatment (control) or were activated with the addition of anti-CD3 (100 ng mL−1)/anti-CD28 antibodies (100 ng mL−1) according to an established protocol with minor modifications.27 To determine the effect of MSX against T-cell activation, cells were treated with MSX (0.001, 0.01, 0.1, and 1 μg mL−1) for 1 h, followed by the addition of anti-CD3 (100 ng mL−1)/anti-CD28 (100 ng mL−1) antibodies for 24 h.Cell viability assays
To determine THP-1 cell viability an MTT assay was performed where THP-1 monocytes were seeded at 4 × 104 in a 96-well plate and differentiated with PMA (50 nM) for 48 h. PMA was then removed and cells were grown in PMA-free media for an additional 24 h. Control cells received no treatment, and the model group received LPS (1 μg mL−1). To determine the cytotoxic effect of MSX on cells, THP-1 cells were treated with MSX (0.001, 0.01, 0.1, 1, 10 and 100 μg mL−1) for 1 h, followed by the addition of LPS (1 μg mL−1) for 24 h. The medium was removed and 100 μL fresh medium was added to each well followed by MTT stock solution (12 mM; 10 μL). A negative control was employed using MTT stock solution (10 μL) to 100 μL of medium. The plate was incubated for four hrs and all medium was removed. Next, DMSO (100 μL) was added to each well with a subsequent incubation at 37 °C for 10 min. The absorbance at 540 nm was read with a microplate reader (SpectraMax M2, Molecular Devices Corp.).Jurkat T-cells cell viability was evaluated using a CCK-8 assay where T-cells were seeded at 1 × 104 cells per well in a 96 well plate. Cells received no treatment (control), anti-CD3/anti-CD28 antibodies (T-cell activation model), MSX (0.001–100 μg mL−1) 1 h + 24 h no additional treatment, or MSX 0.001–100 μg mL−1 1 h + anti-CD3 (100 ng mL−1)/anti-CD28 (100 ng mL−1) antibodies for 24 h. Each group also included a corresponding blank with the same treatment and absence of cells. After 24 h, CCK-8 (10 μL) solution was added to each well and incubated at 37 °C for 4 h. Absorbance was read at 450 nm with a microplate reader (SpectraMax M2, Molecular Devices Corp.).
ELISAs
The levels of IL-1β and TNF-α were obtained from collected differentiated THP-1 cell supernatants using ELISA kits (Biolegend, Inc., San Diego, CA, USA). IL-2 was quantified from Jurkat T cell supernatants using an ELISA kit (Biolegend, Inc., San Diego, CA, USA).Statistical analysis
To assess statistical difference across multiple groups, a one-way analysis of variance (ANOVA) was performed, followed by Tukey's multiple comparison. For the analysis utilizing IPA, the Fisher's Exact Test was employed as the statistical method of choice for the generation of canonical pathways, disease functions, and upstream regulators. The inclusion of z-scores played a pivotal role in the evaluation of the statistical significance pertaining to predictions related to pathways, regulators, and disease functions.Data availability
The raw mass spectrometry data and corresponding quantification results, along with the Spectranaut sne format file, have been deposited in the MassIVE repository (http://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the project identifier: MSV000093293 (FTP Download link: ftp://massive.ucsd.edu/v02/MSV000093293). Additionally, the protein quantification results, disease and function comparisons, and upstream regulation data have been included in the ESI† to ensure transparency and accessibility. The IPA input data has also been provided as ESI to support data reproducibility. Complete IPA results are available upon request.Results
Impact of MSX treatment on protein profiles during LPS-induced inflammation
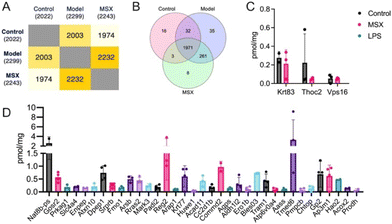
As shown in Fig. 2, the proteomics analysis identified a total of 2022 proteins in the control group, 2299 proteins in the LPS-induced model group, and 2243 proteins in the MSX-treated group (Fig. 2A). A Venn diagram analysis revealed that a core set of 1971 proteins were consistently expressed across all three groups (Fig. 2B). Of particular interest, three proteins—namely, Krt83, Thoc2, and Vps16—demonstrated a unique expression pattern. These proteins were present in both the control and MSX-treated groups but absent in the LPS-induced model group (Fig. 2C).Furthermore, our investigation revealed 35 proteins exclusive in the LPS-induced model group, with no detectable expression in neither the control nor MSX-treated groups (Fig. 2D). Specifically, these proteins included Nat8b-ps, Gosr2, Prkag1, Slc4a4, Enpep, Atxn10, Dpep1, Srprb, Fmo1, Arsb, Rras2, Mark3, Padi2, Reg2, Arap1, Krt77, Huwe1, Acad11, Cc2d1b, Commd2, Agps, Aldh1/2, Ero1b, Bles03, Tram1, Atp6v0a4, Aass, Tmed6, Pmpcb, Chtop, Gp2, Ap3m1, Hao2, Acox2, and Prodh (Fig. 2D). This characterization of protein expression profiles highlights distinct molecular responses associated with LPS-induced inflammation and MSX treatment.
Treatment of MSX modulated protein expressions in LPS-induced inflammation
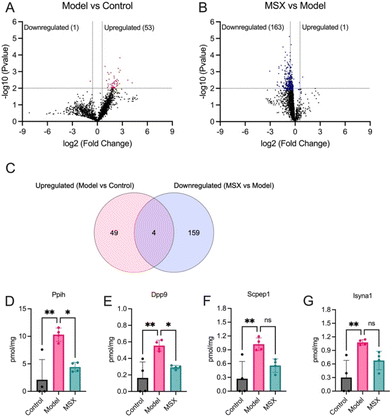
Subsequently, we conducted a comprehensive analysis of the protein expression alterations within the shared set of 1971 proteins utilizing the TPA. As illustrated in Fig. 3, our examination revealed a significant increase in the expression of 53 proteins in response to LPS-induction when compared to the control group (Fig. 3A). Intriguingly, upon comparing the MSX-treated and LPS-induction groups, we observed a substantial decrease in the expression levels of 163 proteins (Fig. 3B). The specific proteins upregulated or downregulated in the LPS-induction group versus the control group, and the MSX-treated versus the LPS-induction group are shown in details in Table 1.| Model vs. control | MSX vs. model | ||
|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated |
| Fxr1, G3bp1, Denr, Lamtor1, Chil3, Slc4a1, Bpgm, Hbb-bs, Epb42, Irgm1, Ca2, Col4a1, Ifit3b, Hba, Hp, Ltf, Ube2 h, Tmem33, Wdfy4, Nagk, Abhd12, Tuba3b, Ppp1r8, Eif3e, Ngp, Slfn5, Ppih, Carhsp1, Sptb, Ube2l3, S100a10, Dynll2, Isyna1, Vps25, Arl1, Ca1, Clu, Ahsg, Abcf1, Atp6v0c, Dpp9, Tsg101, Stk4, Pon3, Serpina3 m, Sec24b, Pgls, Arl8b, Lnpep, Eif3i, Scpep1, RtcA, Rad23b | Alad | Dnll1 | Mat2b, Sqor, Dpm1, Ndufa5, Isyna1, Pmpca, Pgm2, Rps16, Gnas, Ppig, Adpgk, Cavin1, Wars1, Qki, Ndufa9, Gmds, Usp9x, Copa, Itpa,Ndufs1, Dcn, Gmfb, Aifm1, Naxd, Ankfy1, Sfxn1, Aprt,Mccc2, Nceh1, Slc25a1, Glrx, Igkv6-23, Plec, Immt, Hsd17b8, Atp5f1e, Uggt1, Anxa11, Pdlim7, Aimp2,Ctsa, Tst, Drg1, Ndufs7, Dpysl3, Ndufs2, Copb2, Dync1i2, Sf3b3, Ddx46, Acsl5, Cbr1, Rel, Cad, Pgrmc2, Btk, Pycr3, Pds5b, Trap1, Add3, Arhgap1, Ak3, Ykt6, Hibch, Preb, Shmt2, Eif2s3x, Psma3, Mtnd4, Atp5 mg, Cope, Phpt1, Rala, Tomm40, Eif5b, Mtco2, Qars1, Emc1, Rab9a, Tecr, Pcyox1,Ndufa2, Pccb, Uqcrq, Rab8a, Gusb, Dpp9,Ctsd, Ppih, Pcca, Ganab, Chmp1a, Pdia4, Pitpna, Myo18a |
Upon further examination of these proteins that exhibited significant alterations, we identified a subset of four proteins including Ppih, Dpp9, Scpep1, and Isyna1, demonstrating a distinctive response pattern (Fig. 3C). These proteins exhibited a significant increase in expression following LPS-induction, while their levels were reduced upon MSX treatment (Fig. 3D–G). Specifically, the protein expression of Ppih was increased by 8.18 pmol mg−1 by LPS administration as compared to the control but was lowered by 5.89 pmol mg−1 with MSX treatment as compared to the model (Fig. 3D). The protein Dpp9 increased by 0.389 pmol mg−1 in the LPS group compared to the control, which was reduced by 0.261 pmol mg−1 in the MSX group (Fig. 3E). Scpep1 was upregulated by 0.758 pmol mg−1 as compared to the control, which was counteracted by MSX by 0.463 pmol mg−1 (Fig. 3F). Further, the protein Isyna1 was increased by 0.776 pmol mg−1 as compared to the control and lowered by 0.399 pmol mg−1 with MSX administration (Fig. 3G). Notably, subsequent one-way ANOVA analysis, followed by Tukey's multiple comparison post-hoc test, confirmed that only Ppih (Fig. 3D) and Dpp9 (Fig. 3E) displayed statistically significant reductions in the MSX-treated group compared to the model group. This approach was designed to ensure a robust examination of protein expression changes, involving an initial unpaired T-test followed by a subsequent one-way ANOVA analysis and Tukey's multiple comparison test, enhancing the reliability of our findings.
Treatment of MSX altered signaling pathways, canonical pathways, and upstream regulators in LPS-induced inflammation
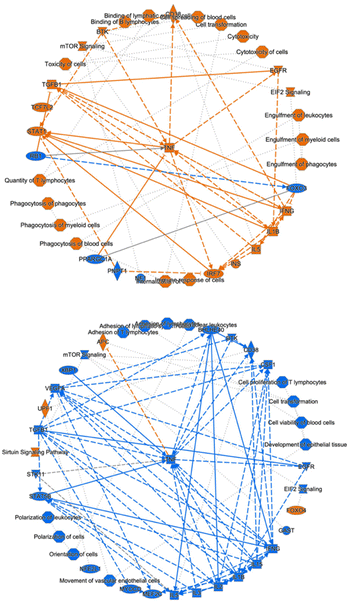
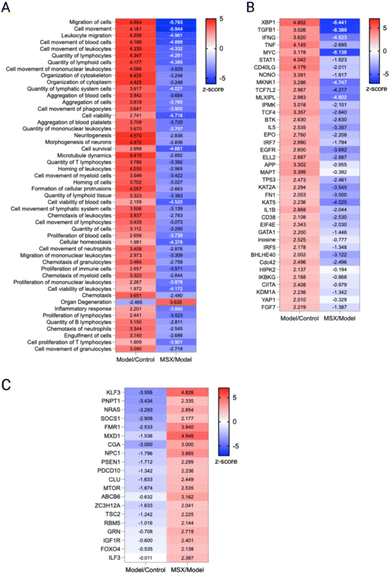
Next, we conducted an ingenuity pathway analysis (IPA) based on the observed alterations in protein expression profiles. Remarkably, our analysis revealed that LPS-induced inflammation elicited significant perturbations in several critical cellular processes (Fig. 4; top). These included heightened immune responses in various cell types, augmented binding of lymphocytes, increased engulfment of leukocytes, phagocytes, and myeloid cells, elevated quantities of T lymphocytes, enhanced phagocytic activity of phagocytes, as well as increased presence of myeloid cells and blood cells. Furthermore, LPS administration induced cellular cytotoxicity. Additionally, this inflammatory response was associated with the activation of key signaling pathways, notably EIF2 signaling, mTOR signaling, activation of BTK, and STAT1 signaling, along with the upregulation of regulatory factors such as IFNG, TNF, IL1B, and IL5.Notably, our investigation unveiled that MSX treatment exerted a substantial counteracting effect on these observed cellular events, signaling pathways, and regulatory factors involved with the inflammatory response induced by LPS (Fig. 4; bottom). Specifically, MSX treatment significantly attenuated the activation, adhesion, and proliferation of T cells, dampened the polarization of leukocytes and cells, and reduced cell viability in blood cells. Furthermore, MSX treatment suppressed the activation of pivotal signaling pathways, including EIF2 signaling and mTOR signaling. Moreover, MSX notably downregulated the expression of TNF, IL1B, and IL-2. Interestingly, MSX increased the expression of the transcription factor, Foxo4, the sirtuin signaling pathway, RNA helicase, UPF1, and the tumor suppressor, APC. These comprehensive analyses collectively suggest that MSX treatment exerts a substantial suppressive effect on inflammation through immune-regulatory mechanisms that impact both innate and adaptive immunity.
As we observed that the suppression of MSX on inflammation was associated with both innate and adaptive immunity, we further analyzed the cellular biofunctions as well as upstream regulators (Fig. 5). In the IPA analysis, we focused on the changes in disease and biological functions. Overall, we identified 708 events that were associated with the change of 1937 proteins in comparison to all three groups. Herein, we selected the top 50 events where many were identified as immune-related processes. In the LPS-induced groups, we observed the upregulation of leukocyte quantities, movement, homing, chemotaxis, proliferation, and viability (Fig. 5A). Further, the movement, quantity, and proliferation of T and B lymphocytes was increased (Fig. 5A). LPS also increased the cellular movement of myeloid cells (Fig. 5A). All of the noted processes induced by LPS were countered by MSX, where multiple immune functions were suppressed by MSX treatment (Fig. 5A). Further, several proteins, signaling molecules, and transcription factors of immune origins (for example, IFNG, TNF, and IL5) were found to be upregulated by LPS, and downregulated by MSX (Fig. 5B). Lastly, a subset of proteins were also observed to be downregulated by LPS, such as FOXO4, which was increased in the MSX group (Fig. 5C).
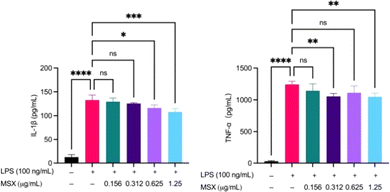
MSX reduces IL-1β and TNF-α induced by LPS in differentiated THP-1 cells
To validate the findings from the in vivo proteomics analysis, we established two in vitro immune cell activation models, namely, LPS-induced THP-1 cell activation and anti-CD3/anti-CD28-induced T cell activation models. MSX (0.01–100 μg mL−1) demonstrated no significant toxicity in THP-1 cells, whether induced with LPS or not (ESI Fig. S1†). Further, in a model of LPS-induced inflammation in THP-1 monocytes, MSX induced significant reductions in the pro-inflammatory cytokines, IL-1β and TNF-α (Fig. 6). LPS administration induced a significant increase in IL-1β by 120.2 pg mL−1 and TNF-α by 1213 pg mL−1 (Fig. 6). MSX pretreatment at 0.625 μg mL−1 and 1.25 μg mL−1 significantly reduced IL-1β by 16.46 pg mL−1 and 24.85 pg mL−1, respectively (Fig. 6; left). Further, MSX pretreatment at 0.312 and 1.25 pg mL−1 significantly reduced TNF-α by 187.7 and 193.9 pg mL−1, respectively (Fig. 6; right).MSX reduces IL-2 secretions prior to T-cell activation in Jurkat T-cells
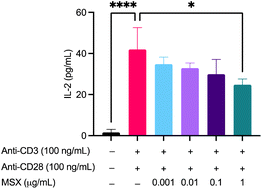
In a model of T cell activation, Jurkat cells were treated with MSX followed by the addition of anti-CD3 (100 ng mL−1)/anti-CD28 (100 ng mL−1) and assayed for IL-2 concentration. First of all, MSX (0.01–100 μg mL−1) demonstrated no significant toxicity in Jurkat T cells whether induced with anti-CD3/CD28 or not (ESI Fig. S2†). Therefore, we chose the MSX concentration as 0.01–1 μg mL−1. We found that treatment with anti-CD3 (100 ng mL−1)/anti-CD28 (100 ng mL−1) resulted in a significant increase of IL-2 by 40.39 pg mL−1 (Fig. 7). MSX (1 μg mL−1) pre-treatment significantly decreased IL-2 levels by 17.16 pg mL−1 as compared to the model group (Fig. 7).Discussion
In this study, we employed two distinct bottom-up proteomics acquisition strategies including DDA and DIA proteomics. The DDA approach selects a peptide based on its signal intensity at a specific chromatographic time point and subsequently fragments the peptide.28,29 The DIA proteomics selects peptide ions using a mass/charge window at a specific chromatographic time point and then fragments these ions.28 It is worth noting that DIA proteomics offers superior reproducibility and a wider dynamic range compared to DDA.30,31 In a previous study, we exclusively utilized the DIA strategy for the discovery of proteomics.23 However, a notable limitation of DIA workflows is the multiplexed spectra within each MS2 scan, making accurate peptide identification challenging.32 Herein, we strategically integrated the DDA proteomics into the DIA analysis process. Subsequently, we analyzed the results from DDA and generated a spectral library, which proved valuable for the analysis of the DIA data. These resulting spectral libraries tend to be more concise and include only peptides observable in complex, non-fractionated samples. Our combined approach allowed us to identify a total of 2022 proteins in the control group, 2299 proteins in the LPS-induced model group, and 2243 proteins in the MSX-treated group.We selected the spleen as our focus for several reasons. Firstly, the spleen plays a central role in both innate and adaptive immune responses, housing a diverse array of immune cells, including macrophages, dendritic cells, T cells, and B cells, facilitating a comprehensive exploration of immune mechanisms.33 Additionally, the spleen is responsive to inflammatory signals, such as those induced by LPS, resulting in significant alterations in cell populations, cytokine expression, and immune cell migration, offering valuable insights into immune responses.33 Our findings in this study corroborated the inflammatory impact of LPS administration, a phenomenon commonly observed in both in vitro and in vivo models, owing to the similarities with the inflammatory response observed in humans.34–36 Moreover, the spleen acts as a reservoir for immune cells, particularly monocytes and macrophages, enabling the study of their mobilization during inflammation.37 In this study, we observed various immune responses, including increased leukocyte quantities, movement, homing, chemotaxis, proliferation, and viability, as well as enhanced T and B lymphocyte activity and myeloid cell movement in spleen tissue from LPS-induced inflammation. Besides, LPS-induced inflammation led to notable perturbations in several cellular processes, such as heightened immune responses, enhanced lymphocyte binding, increased leukocyte and myeloid cell engulfment, and elevated T lymphocyte numbers in spleen tissues. Interestingly, our investigation revealed that the administration of MSX exerted a counteractive effect on the aforementioned observed cellular events. Furthermore, we observed modulation in the signaling pathways associated with these immune responses, shedding light on the intricate interplay between MSX and immune processes.
Our proteomics analysis provided critical information by identifying 35 uniquely expressed proteins in response to LPS administration (Fig. 2D). Notably, these proteins were absent in both the control and MSX-treated groups, suggesting they are potential targets for LPS stimulation. Nevertheless, it is important to corroborate these findings through further assays, such as Western Blotting or selected reaction monitoring. In this group of 35 proteins, a STRING analysis mapped two significantly enriched protein clusters to gene ontology biological processes where six corresponded to the carboxylic acid catabolic process, and nine mapped to the organic acid metabolic process (ESI Fig. S3†). Furthermore, our investigation showed that the administration of MSX, followed by immune challenge with LPS, triggered the upregulation of three distinct proteins, namely, Krt83, Thoc2, and Vps16. Notably, these proteins were expressed in the control group but absent in the LPS-induced group. This observation suggested a potential influence of MSX pretreatment on the expression of these three proteins, as they were present in the control group and absent in the LPS-induced group. Notably, pretreatment with MSX upregulated these three proteins.
Among the 1971 shared proteins across all three groups (control, model, and MSX), we observed that LPS upregulated the expression of 53 proteins. Of particular interest, within this shared protein pool, MSX pretreatment exerted a counteractive effect by downregulating 163 proteins to mitigate the effects of LPS-induced inflammation. Notably, our analysis unveiled four proteins—Ppih, Dpp9, Scpep1, and Isyna1—that exhibited upregulation in response to LPS and subsequent downregulation following MSX administration, with Ppih and Dpp9 exhibiting significant alterations. In particular, Dpp9 is known for its involvement in immune regulation through the inhibition of the NLRP1 inflammasome.38
The further analysis conducted using IPA has provided us with a comprehensive overview of MSX's immune regulatory effects on inflammation. Our findings underscore that the immune processes initially activated by LPS were subsequently downregulated by MSX. This regulatory effect of MSX was observed on various immune cell types, including lymphocytes, leukocytes, and myeloid cells, as well as on key signaling pathways, particularly EIF2 and mTOR. Furthermore, MSX exhibited a notable impact on the upregulation of specific transcription factors, such as Foxo4, and the sirtuin pathway. Foxo transcription factors are known to be expressed in immune cells and may play a pivotal role in maintaining immune system homeostasis, as well as influencing lymphocyte and T-cell selection.39 Other molecular pathways may also be involved in the modulation of the immunity by MSX, for instance, sirtuins have been recognized for their anti-inflammatory properties in response to LPS administration.40 Specifically, sirtuins achieve this by suppressing inflammasome activation in macrophages and endothelial cells and by inhibiting the secretion of cytokines such as IL-1β, IL-6, IL-8, and TNF-α.40 Of particular interest, our analysis revealed that MSX treatment had a regulatory effect on several immune mediators, including TNF, IL1B, and IL-2. TNF and IL1B are known for their pivotal roles in innate immunity, where they contribute to inflammation and immune responses.41 Conversely, IL-2 is primarily associated with adaptive immunity, mainly due to its central role in T cell activation and proliferation.42
Previous studies have reported the anti-inflammatory effects of MSX as demonstrated in murine macrophages, microglial and neuronal cells, and a mouse model of Alzheimer's disease.10,11,13 In addition, we also previously reported the immune suppressive effects of MSX (0.0125%–0.05% b.w.), the equivalence of daily human (60 kg) consumption of 1.2–4.8 mg, in a mouse acute inflammation model.23 However, in our current study, our IPA findings have shed new light on the broader immunomodulatory effects of MSX, which encompass both innate and adaptive immunity. This expanded understanding is corroborated by MSX's regulatory influence on key immune mediators, including TNF, IL-1β, and IL-2. To further validate these findings, we conducted two cell activation assays, utilizing LPS-induced THP-1 cells and anti-CD3/CD28-induced Jurkat T cells. In the case of THP-1 monocytes, which were differentiated into macrophages and treated with MSX prior to LPS administration, we observed a significant reduction in the pro-inflammatory cytokines IL-1β and TNF-α. This observation aligns with the proteomic data obtained from the spleen, where both of these cytokines were upregulated by LPS but downregulated by MSX. This cell model reflects the role of macrophages in the spleen, acting as intermediaries between the innate and adaptive immune systems, and their ability to release cytokines.43,44 Our IPA analysis suggested that MSX could modulate T cell function, hinting at a potential impact on adaptive immunity. We further employed a Jurkat T cell model, applying MSX treatment prior to LPS exposure. We then quantified the concentration of IL-2, a cytokine found to be decreased by MSX in our proteomics results. Our experiments confirmed a reduction in IL-2 levels in Jurkat T cells. Since T cell activation is crucial in adaptive immunity and is closely linked to IL-2 secretion, our data provides additional evidence supporting the notion that MSX exerts a modulatory effect on adaptive immunity, in addition to its well-established effects on innate immunity.45
Conclusions
In summary, our study highlights the potential of MSX as a promising nutraceutical with disease-modifying capabilities in inflammation-related conditions. Leveraging advanced proteomics, we observed MSX's ability to counteract both innate and adaptive immune responses, impacting immune cells, signaling pathways, and key immune mediators such as TNF, IL-1β, and IL-2. To validate these findings, we conducted cell activation assays using differentiated THP-1 cells and Jurkat T-cells, revealing that MSX effectively dampens pro-inflammatory cytokine production and impacts T-cell activation. These comprehensive insights contribute to our understanding of MSX's multifaceted role in immune regulation, supporting its potential as a promising nutraceutical for further exploration in the field of immune-related disorders.Author contributions
Tess Puopolo: investigation, formal analysis, writing – original draft, visualization; Ying Chen: investigation; Hang Ma: writing – reviewing and editing; Chang Liu: conceptualization, investigation, methodology, formal analysis, writing – reviewing and editing, supervision; Navindra P Seeram: supervision, resources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the Quebec Maple Syrup Producers for their support of this project. Food-grade MSX for this study was produced by Silicycle Inc. (Quebec, Canada). The authors also acknowledge the RI-INBRE core facility at the University of Rhode Island for granting access to the equipment, which was supported by Grant P20GM103430 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). The authors also acknowledge the proteomics facility at the College of Pharmacy, University of Rhode Island, for their invaluable data acquisition and analysis services.References
- M. F. Ramadan, H. A. Gad and M. A. Farag, Chemistry, processing, and functionality of maple food products: An updated comprehensive review, J. Food Biochem., 2021, 45(8), e13832 CrossRef CAS.
- A. Saraiva, et al., Maple Syrup: Chemical Analysis and Nutritional Profile, Health Impacts, Safety and Quality Control, and Food Industry Applications, Int. J. Environ. Res. Public Health, 2022, 19, 13684 CrossRef CAS.
- Y. Zhang, et al., Chemical Compositional, Biological, and Safety Studies of a Novel Maple Syrup Derived Extract for Nutraceutical Applications, J. Agric. Food Chem., 2014, 62, 6687–6698 CrossRef CAS.
- J. Sun, H. Ma, N. P. Seeram and D. C. Rowley, Detection of Inulin, a Prebiotic Polysaccharide, in Maple Syrup, J. Agric. Food Chem., 2016, 64, 7142–7147 CrossRef CAS.
- J. Sheng, et al., Phenolic–enriched maple syrup extract protects human keratinocytes against hydrogen peroxide and methylglyoxal induced cytotoxicity, Dermatol. Ther., 2020, 33(3), e13426 CrossRef CAS.
- Y. Liu, H. Ma and N. P. Seeram, Development and UFLC-MS/MS Characterization of a Product-Specific Standard for Phenolic Quantification of Maple-Derived Foods, J. Agric. Food Chem., 2016, 64, 3311–3317 CrossRef CAS.
- L. Li and N. P. Seeram, Maple Syrup Phytochemicals Include Lignans, Coumarins, a Stilbene, and Other Previously Unreported Antioxidant Phenolic Compounds, J. Agric. Food Chem., 2010, 58, 11673–11679 CrossRef CAS.
- L. Li and N. P. Seeram, Further Investigation into Maple Syrup Yields 3 New Lignans, a New Phenylpropanoid, and 26 Other Phytochemicals, J. Agric. Food Chem., 2011, 59, 7708–7716 CrossRef CAS PubMed.
- T. Toyoda, A. Kamei, T. Ishijima, K. Abe and S. Okada, A maple syrup extract alters lipid metabolism in obese type 2 diabetic model mice, Nutr. Metab., 2019, 16, 84 CrossRef.
- P. P. Nahar, M. V. Driscoll, L. Li, A. L. Slitt and N. P. Seeram, Phenolic mediated anti-inflammatory properties of a maple syrup extract in RAW 264.7 murine macrophages, J. Funct. Foods, 2014, 6, 126–136 CrossRef CAS.
- H. Ma, et al., Effects of a Standardized Phenolic-Enriched Maple Syrup Extract on β-Amyloid Aggregation, Neuroinflammation in Microglial and Neuronal Cells, and β-Amyloid Induced Neurotoxicity in Caenorhabditis elegans, Neurochem. Res., 2016, 41, 2836–2847 CrossRef CAS.
- W. Liu, et al., Anti-glycation and anti-oxidative effects of a phenolic-enriched maple syrup extract and its protective effects on normal human colon cells, Food Funct., 2017, 8, 757–766 RSC.
- K. N. Rose, et al., Anti-neuroinflammatory effects of a food-grade phenolic-enriched maple syrup extract in a mouse model of Alzheimer's disease, Nutr. Neurosci., 2021, 24, 710–719 CrossRef CAS.
- D. Furman, et al., Chronic inflammation in the etiology of disease across the life span, Nat. Med., 2019, 25, 1822–1832 CrossRef CAS.
- V. Bronte and M. J. Pittet, The Spleen in Local and Systemic Regulation of Immunity, Immunity, 2013, 39, 806–818 CrossRef CAS.
- S. M. Lewis, A. Williams and S. C. Eisenbarth, Structure and function of the immune system in the spleen, Sci. Immunol., 2019, 4(33), eaau6085 CrossRef CAS PubMed.
- C. M. D. Mota and C. J. Madden, Neural control of the spleen as an effector of immune responses to inflammation: mechanisms and treatments, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 2022, 323, R375–R384 CrossRef CAS PubMed.
- M. Aliyu, F. Zohora and A. Akbar Saboor-Yaraghi, Spleen in innate and, adaptive immunity regulation, AIMS Allergy Immunol., 2021, 5, 1–17 CAS.
- D. T. Skelly, E. Hennessy, M.-A. Dansereau and C. Cunningham, A Systematic Analysis of the Peripheral and CNS Effects of Systemic LPS, IL-1B, TNF-α and IL-6 Challenges in C57BL/6 Mice, PLoS One, 2013, 8, e69123 CrossRef CAS.
- I. M. Lalić, et al., Lipopolysaccharide induces tumor necrosis factor receptor-1 independent relocation of lymphocytes from the red pulp of the mouse spleen, Ann. Anat., 2018, 216, 125–134 CrossRef.
- C. Wang, et al., Characterization of murine macrophages from bone marrow, spleen and peritoneum, BMC Immunol., 2013, 14, 6 CrossRef CAS.
- E. Dervishi, D. Hailemariam, S. A. Goldansaz and B. N. Ametaj, Early-Life Exposure to Lipopolysaccharide Induces Persistent Changes in Gene Expression Profiles in the Liver and Spleen of Female FVB/N Mice, Vet. Sci., 2023, 10, 445 CrossRef PubMed.
- T. Puopolo, et al., Uncovering the anti-inflammatory mechanisms of phenolic-enriched maple syrup extract in lipopolysaccharide-induced peritonitis in mice: insights from data-independent acquisition proteomics analysis, Food Funct., 2023, 14, 6690–6706 RSC.
- J. R. Wiśniewski and M. Mann, A Proteomics Approach to the Protein Normalization Problem: Selection of Unvarying Proteins for MS-Based Proteomics and Western Blotting, J. Proteome Res., 2016, 15, 2321–2326 CrossRef.
- J. R. Wiśniewski and D. Rakus, Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome, J. Proteomics, 2014, 109, 322–331 CrossRef PubMed.
- C. Liu, H. Ma, A. L. Slitt and N. P. Seeram, Inhibitory Effect of Cannabidiol on the Activation of NLRP3 Inflammasome Is Associated with Its Modulation of the P2X7 Receptor in Human Monocytes, J. Nat. Prod., 2020, 83, 2025–2029 CrossRef CAS.
- J. W. Zagorski, et al., The Nrf2 Activator, tBHQ, Differentially Affects Early Events Following Stimulation of Jurkat Cells, Toxicol. Sci., 2013, 136, 63–71 CrossRef CAS PubMed.
- A. Sinha and M. Mann, A beginner's guide to mass spectrometry–based proteomics, Biochem, 2020, 42, 64–69 CAS.
- T. Guo and R. Aebersold, Recent advances of data–independent acquisition mass spectrometry–based proteomics, Proteomics, 2023, 23(7–8), 2200011 CrossRef CAS.
- L. Krasny and P. H. Huang, Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology, Mol. Omics, 2021, 17, 29–42 RSC.
- A. Hu, W. S. Noble and A. Wolf-Yadlin, Technical advances in proteomics: new developments in data-independent acquisition, F1000Research, 2016, 5, 419 Search PubMed.
- L. K. Pino, S. C. Just, M. J. MacCoss and B. C. Searle, Acquiring and Analyzing Data Independent Acquisition Proteomics Experiments without Spectrum Libraries, Mol. Cell. Proteomics, 2020, 19, 1088–1103 CrossRef.
- E. Semaeva, et al., Access to the Spleen Microenvironment through Lymph Shows Local Cytokine Production, Increased Cell Flux, and Altered Signaling of Immune Cells during Lipopolysaccharide-Induced Acute Inflammation, J. Immunol., 2010, 184, 4547–4556 CrossRef CAS.
- W. Lee, E.-J. Yang, S.-K. Ku, K.-S. Song and J.-S. Bae, Anti-inflammatory Effects of Oleanolic Acid on LPS-Induced Inflammation In Vitro and In Vivo, Inflammation, 2013, 36, 94–102 CrossRef CAS PubMed.
- S. Seemann, F. Zohles and A. Lupp, Comprehensive comparison of three different animal models for systemic inflammation, J. Biomed. Sci., 2017, 24, 60 CrossRef PubMed.
- S. Copeland, H. S. Warren, S. F. Lowry, S. E. Calvano and D. Remick, Acute Inflammatory Response to Endotoxin in Mice and Humans, Clin. Vaccine Immunol., 2005, 12, 60–67 CrossRef CAS.
- E. Liverani, et al., LPS-induced systemic inflammation is more severe in P2Y12 null mice, J. Leukocyte Biol., 2013, 95, 313–323 CrossRef.
- M. Huang, et al., Structural and biochemical mechanisms of NLRP1 inhibition by DPP9, Nature, 2021, 592, 773–777 CrossRef CAS.
- K. U. Birkenkamp and P. J. Coffer, FOXO Transcription Factors as Regulators of Immune Homeostasis: Molecules to Die for?, J. Immunol., 2003, 171, 1623–1629 CrossRef CAS.
- Z. Pan, H. Dong, N. Huang and J. Fang, Oxidative stress and inflammation regulation of sirtuins: New insights into common oral diseases, Front. Physiol., 2022, 13, 953078 CrossRef.
- K. Newton and V. M. Dixit, Signaling in Innate Immunity and Inflammation, Cold Spring Harbor Perspect. Biol., 2012, 4, a006049–a006049 Search PubMed.
- M. F. Bachmann and A. Oxenius, Interleukin 2: from immunostimulation to immunoregulation and back again, EMBO Rep., 2007, 8, 1142–1148 CrossRef CAS PubMed.
- N. A-Gonzalez and A. Castrillo, Origin and specialization of splenic macrophages, Cell. Immunol., 2018, 330, 151–158 CrossRef CAS.
- J. M. M. den Haan and G. Kraal, Innate Immune Functions of Macrophage Subpopulations in the Spleen, J. Innate Immun., 2012, 4, 437–445 CrossRef.
- K. T. Bashour, et al., CD28 and CD3 have complementary roles in T-cell traction forces, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2241–2246 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo04026g |
| This journal is © The Royal Society of Chemistry 2024 |