Open Access Article
Open Access ArticleComplexation and conjugation between phenolic compounds and proteins: mechanisms, characterisation and applications as novel encapsulants
Bo
Wang
 *a,
Loc B.
Pham
b and
Benu
Adhikari
*a,
Loc B.
Pham
b and
Benu
Adhikari
 *b
*b
aSchool of Behavioural and Health Science, Australian Catholic University, Sydney, NSW 2060, Australia. E-mail: bo.wang@acu.edu.au; Tel: +61 298392744
bSchool of Science, RMIT University, Melbourne, VIC 3028, Australia. E-mail: benu.adhikari@rmit.edu.au; Tel: +61 399259940
First published on 22nd July 2024
Abstract
Food phenolic compounds (PCs) and proteins interact and react via non-covalent and covalent routes to form phenolic compound–protein (PCP) complexes and conjugates. In the last decade, increasing research has focused on protein modification based on these interactions in various food systems. This review provides the mechanism of PCP complexation and conjugation and relevant analytical techniques for detection and quantification purposes. Moreover, key functional properties of PCP complexes and conjugates, including solubility, emulsifying property, antioxidant activity, thermal stability, anti-microbial activity and digestibility, are discussed. The applications of the complexes and conjugates as novel encapsulants to stabilise bioactive but sensitive as novel food ingredients are also overviewed. It is worth noting that the correlation between PCP complexation and conjugation and these functional properties are not fully understood. There is still research paucity exploring the applications of PCP complexes and conjugates as promising encapsulants. Future research is required to advance the science in this area and facilitate application.
Sustainability spotlightThe contemporary food industry does not favour the interactions (non-covalent complexation) and reactions (covalent conjugation) between food proteins and phenolic compounds. As such, separating phenolic compounds from food materials before processing significantly increases the fabrication cost of the final product. However, an increasing amount of research indicates improved protein techno-functional properties based on the optimisation of phenolic compound-protein complexation and conjugation. Various produced complexes and conjugates have shown improved antioxidation activities and resistance to in vitro enzymatic digestion in a simulated gastric environment. Therefore, they are promising food materials to stabilise susceptible food compounds to extend their shelf life to achieve the Sustainable Development Goals (goal 12, responsible production). This approach will also ensure the successful delivery of sensitive food compounds to the target site to provide biological benefits for better human health (Sustainable Development Goals, goal 3, good health and well-being). In this context, this manuscript overviews relevant studies published in the last decade (2013–2023). It discusses the mechanism of complexation and conjugation, characterisation methods, tailored protein techno-functional properties and their tested applications to stabilise sensitive food compounds. It highlights the technological pathways to design, monitor, characterise and apply phenolic compound–protein complexes and conjugates as novel encapsulants. Therefore, it meets the Sustainable Development Goals established by the UN. |
1 Introduction
Plants synthesise phenolic compounds (PCs) as secondary metabolites against abiotic and biotic factors.1 These compounds chemically comprise an aromatic ring(s) with at least one attached hydroxyl group.2 Recently, many health benefits associated with the dietary intake of PCs, including cardio-protection as well as anticarcinogenic, antioxidant, anti-apoptotic, and anti-inflammatory activities, have been reported.3–6 Ingested PCs exhibit these biological benefits via various mechanisms such as scavenging free radicals, eliminating radical precursors, chelating metal ions and inhibiting the enzymes involved in reactive oxygen species (ROS) production.7–9Owing to their highly reactive nature, PCs readily interact or react with other food compounds during food processing, storage and digestion. Conventionally, phenolic compound–protein (PCP) interactions or reactions are considered negative because they alter the properties of food. For instance, these interactions or reactions may cause the formation of insoluble compounds, which compromises the appearance of beverages and wine and the digestibility of proteins.10–12 As such, PCs are usually separated from the food system using multiple techniques such as solvent extraction, ultrasound- or microwave-assisted separation and supercritical fluid extraction before further processing.13–17 Although these methods gained some success to different extents, many resources and expenses such as energy, chemicals and advanced equipment are required.
Furthermore, food bioactive compounds such as polyunsaturated lipids, probiotics and essential oils have health benefits. However, these compounds are chemically unstable and readily degraded during food processing storage when exposed to environmental stresses such as light, heat, and oxygen, thereby reducing their bioavailability.18 In this context, proteins have been commonly used as foaming agents, colloid stabilisers, biodegradable film-forming materials and encapsulants to stabilise susceptible compounds. In general, the efficacy and effectiveness of food proteins as encapsulants are closely related to their physicochemical properties, such as structure, solubility as well as swelling, foaming, emulsifying, gelling and oil-binding abilities.19,20 There is a demand for developing novel protein-based encapsulants with improved functional properties. To date, the effect of chemical (e.g. via acetylation, succinylation, phosphorylation, lyophilisation, glycosylation, thiolation or crosslinking),21–23 physical (e.g. via high pressure, radiation or ultrasound)24–26 and enzymatic (e.g. via endopeptidases, transglutaminase and oxidase)27–29 treatments on the protein structure and property change have been studied by the researchers. The PCP interaction and reaction between plant-based proteins and phenolic compounds have been overviewed in some recent papers.30,31 However, most of these existing studies only focused on PCP interaction or reaction mechanism and changed the physicochemical properties.32–36 There is a lack of research investigating the effect of interaction or reaction on the protein property change and exploring the potential applications of the derived products as potential encapsulants.
Therefore, this review paper provides an overview of the interaction and reaction between PCs and proteins, the PCP complexation and conjugation mechanism, the characterisation method, the effect of complexation and conjugation on protein properties, and the end product applications as encapsulants. We aim to enlighten future researchers to develop novel food materials with improved functional properties by strategically modifying protein structure and property using interaction or reaction.
2 Literature search and result analysis
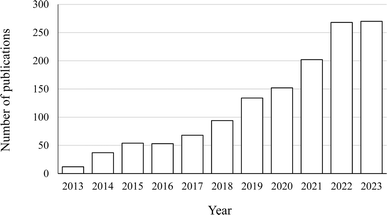
To overview the most recently published studies in this area, the following search criteria were used to source the journal articles published in Scopus from 2013 to 2023.(ALL (protein–phenolic AND complex) OR ALL (protein–phenolic AND interaction) OR ALL (protein–phenol AND complex) OR ALL (protein–phenol AND interaction) OR TITLE-ABS-KEY (protein–phenolic AND conjugate) OR TITLE-ABS-KEY (protein–phenolic AND reaction) OR TITLE-ABS-KEY (protein–phenol AND conjugate) OR TITLE-ABS-KEY (protein–phenol AND conjugate))
Subsequently, review papers and book chapters were excluded, as this review only discusses the original research. The scope was further limited to research papers written in English and published in the areas of food, biochemistry, bioengineering, and biomaterials. The literature search result is shown in Fig. 1.
3 Mechanism of PCP complexation and conjugation
From a chemical point of view, PCs can either non-covalently interact or covalently react with proteins.20,37–39 In the following sections of this paper, the interaction and reaction are referred to as PCP complexation and conjugation, with the end products as complex and conjugates, respectively.3.1 PCP complexation
The mechanism of PCP complexation is shown in Fig. 2. This complexation is reversible due to the weaker binding between PCs and proteins. This phenomenon is mainly driven by hydrophobic interaction, van der Waals association and hydrogen bonding.40–43 Ultimately, soluble or insoluble PCP complexes are formed.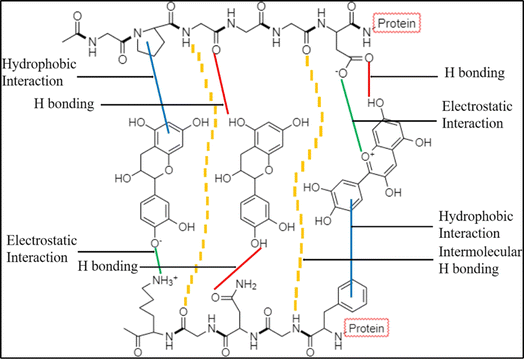 | ||
| Fig. 2 Mechanism of PCP complexation (adapted from Shahidi and Dissanayak's study,40 under the Creative Commons Attribution 4.0 International License). | ||
Hydrogen bonding is one of the main drivers to bind PCs with proteins in a PCP complex. The hydrogen atom in the hydroxyl group acts as a hydrogen donor and forms hydrogen bonds via its interaction with the oxygen atom in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the amide group, oxygen or nitrogen on the side chain of amino acid residues such as –OH and –NH2 groups.44 In Guo and Jarregi's study, resveratrol and β-lactoglobulin were complexed via hydrogen bonds to form nanoparticles around 181.8 nm.45 Jiang et al. induced PCP complexation between chalconoids and alpha-actalubin.46 The authors found that Trp 118, Glu 11 and Lys 5 in the protein molecule formed hydrogen bonds with hydroxyl safflower yellow A.
O of the amide group, oxygen or nitrogen on the side chain of amino acid residues such as –OH and –NH2 groups.44 In Guo and Jarregi's study, resveratrol and β-lactoglobulin were complexed via hydrogen bonds to form nanoparticles around 181.8 nm.45 Jiang et al. induced PCP complexation between chalconoids and alpha-actalubin.46 The authors found that Trp 118, Glu 11 and Lys 5 in the protein molecule formed hydrogen bonds with hydroxyl safflower yellow A.
Hydrophobic interactions also facilitate the PCP complexation. It involves the nonpolar aromatic ring of the PCs and hydrophobic amino acid residues in the protein molecule chain.47 In Lu et al.'s study, PCP complexation between whey protein and rosmarinic acid was investigated.48 The authors proved that the hydrophobic pocket formed by Ser 191, Arg 198, Leu 237, His 241, Leu 259, Ile 263, His 287, Ala 290 and Glu 291 amino acid residues in the protein chain “entrapped” rosmarinic acid via hydrophobic interactions. A similar result was also reported in myosin–curcumin complexes.49
The van der Waals association is a weak force that attracts neutral molecules to one another.50 For example, Wu et al. reported that green tea (−)-epicatechin gallate (EGCG) complexed with bovine-lactoglobulin (BLG) spontaneously via van der Waals interactions and hydrogen bonding.51 Similarly, the complexation between β-lactoglobulin and PCs including ferulic acid, quercetin and vanillic acid was driven by hydrogen bonds, van der Waals association and hydrophobic forces.52
3.2 PCP conjugation
The covalent PCP conjugation is an irreversible reaction due to the nature of covalent bonds between the PCs and proteins. Compared to the PCP complexation, this conjugation is preferable from a food processing perspective due to its intrinsic strength. Briefly, the PCP conjugation starts from the oxidation of PCs by enzymatic and non-enzymatic methods, among which the alkaline treatment is the most used approach.
Fig. 3 presents the reaction scheme of PCP conjugation. Briefly, PCs are oxidised to form quinone or semiquinone radical intermediates. In this step, the formation of these radical intermediates depends on the structural configuration of PCs. For example, quinone radical intermediates are usually formed from PCs such as caffeic acid, gallic acid, quercetin and myricetin, which have catechol in their structure,53,54 while semiquinone radical intermediates are produced from ferulic acid, p-coumaric acid, and sinapic acid, which have monophenols in their structure. Subsequently, the formed quinone or semiquinone radical intermediates react with amino groups in the protein chain via Schiff-base (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and Michael addition mechanisms (C–NH), as shown in Fig. 3(a). Meanwhile, the radical attack by hydroxyl radicals on the protein molecular chain may induce the formation of protein radicals, which further conjugate with PCs. These protein radicals may also covalently bind with semiquinone radical intermediates, as shown in Fig. 3(b).
N) and Michael addition mechanisms (C–NH), as shown in Fig. 3(a). Meanwhile, the radical attack by hydroxyl radicals on the protein molecular chain may induce the formation of protein radicals, which further conjugate with PCs. These protein radicals may also covalently bind with semiquinone radical intermediates, as shown in Fig. 3(b).
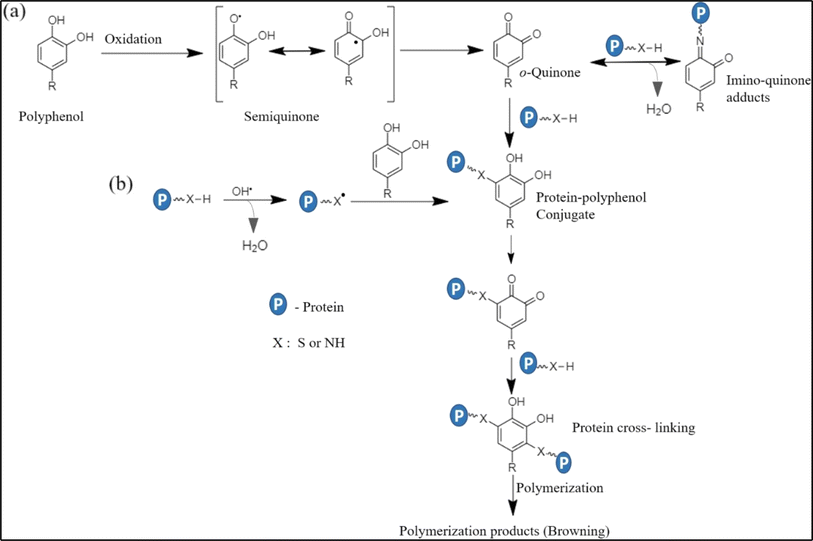 | ||
| Fig. 3 Mechanism of PCP conjugation via (a) oxidation process-Michael addition and amino-quinone adducts and (b) the free radical-mediated process (adapted from Shahidi and Dissanayak's study,40 under the Creative Commons Attribution 4.0 International License). | ||
To date, most published studies on PCP conjugation focused on animal-derived proteins including myoglobin,55 gelatin,54 and dairy protein.56 Recently, there have been an increasing number of publications on conjugation between PCs and plant-based proteins. For example, Jia et al. mixed EGCG and whey protein isolate in an alkaline condition at pH 9 for 12 h to induce PCP conjugation.57 The PCP conjugate formation was confirmed via several physicochemical measurements. Similarly, Parolia et al. induced PCP conjugation between lentil protein and various PCs including quercetin, rutin and ellagic acid under alkaline conditions.58 Xu et al. also prepared zein–chlorogenic acid, zein–garlic acid and zein–carreic acid PCP conjugates.59
Similar to the above-mentioned PCP complexation, the PCP conjugation is also affected significantly by their nature and structure. Sui et al. investigated the PCP complexation and conjugation between anthocyanins from black rice and soybean protein isolate.60 The authors reported that the anthocyanins were more likely to form conjugates than complexes. The attachment of the anthocyanins to the soy protein molecular chain changed the protein structure and some of the functional properties. In Pham et al.'s study, hydroxytyrosol, flaxseed polyphenols and ferulic acid were used to form conjugation with flaxseed protein isolates.61 The authors observed that only hydroxytyrosol crosslinked multiple protein chains, while ferulic acid and flaxseed polyphenols only attached themselves on single protein molecules.
4 Measurement of PCP complexation and conjugation
To date, there is still limited information about the mechanism of PCP complexation and conjugation at the molecular level. However, the formation of complexes and conjugates significantly changes the protein structure, molecular weight, solubility, antioxidative activity, etc. Therefore, some analytical methods that have been used to characterise the protein properties can be employed to assess and quantify the PCP complexation and conjugation.4.1 Fluorescence spectrophotometry
Fluorescence quenching is a useful technique, providing information on the protein change when it associates with PCs.62 The measurement of fluorescence parameters, in terms of fluorescence emission, fluorescence anisotropy and fluorescence lifetime, is carried out for this purpose.63–65 Briefly, proteins have an excitation wavelength of 240–280 nm due to the presence of aromatic amino acids. This causes their fluorescence emission at 340–350 nm. Once proteins form complex or conjugates with PCs, their fluorescence emission intensity and wavelength in this range significantly change.12,52In Li et al.'s study, the PCP complexation between four kinds of PCs (apigenin, naringenin, kaempferol, genistein) and β-lactoglobulin was investigated.66 The authors observed a decreased protein fluorescence intensity with the increased flavonoid concentration in the PCP system. This result indicated strong PCP complexation via fluorescence quenching.
The fluorescence emission spectrum is also used to study the PCP conjugation. The protein intrinsic fluorescence emission spectrum is usually dominated by tryptophan and may be altered by PCP conjugation. Wang et al. observed a significant decrease in a-La when the protein was conjugated with CA or EGCG.67 This indicated that the indole ring of tryptophan was most likely involved in the covalent reaction. Similarly, PCP conjugation between whey protein isolates and EGCG caused the change in fluorescence emission wavelength of protein from 336 to 339 nm.57 This result suggested that the major fluorophore tryptophan was exposed to a more hydrophilic environment after conjugation.
4.2 Protein structure
PCP complexation and conjugation change the conformation of proteins. Particularly, it involves reducing relevant groups and introducing new functional groups on protein backbones. Fourier transform infrared (FT-IR) spectroscopy and circular dichroism (CD) spectroscopy are commonly used to detect and quantify these structural changes, as shown in Table 1.| Protein and PCs | Protein structure and test method | Result | References |
|---|---|---|---|
| a Abbreviations in the table: circular dichroism: CD; Fourier transform infrared spectroscopy: FTIR; epigallocatechin-3-gallate: EGCG; β-lactoglobulin: β-LG; tryptophan: Trp; tyrosine: Tyr. | |||
| PCP complexation | |||
| α-Lactalbumin, caffeic acid | CD | • α-Lactalbumin–caffeic acid complex increased the α-helical structure with increased caffeic acid content | Al-Shabib et al.68 |
| Tertiary structure via fluorescence spectroscopy | • Complexation altered the tertiary structure of α-lactalbumin | ||
| β-Lactoglobulin, ferulic acid | Secondary structure via CD and FTIR | • PCP complexation occurred in both the monomer and dimer forms | Abdollahi et al.69 |
| • The form (dimer/monomer) of the protein played an important role in the effect that ligand binding has on the secondary structure of β-lactoglobulin | |||
| • At pH 7.3, FTIR result suggested that the PCP complex decreased the β-sheet content but increased the unordered structure. This trend was consistent with the CD result | |||
| β-Lactoglobulin, vanillic acid | Secondary structure via CD, FTIR | • The complexation increased random coil and β-turns of protein, whereas decreasing trends were observed for β-sheet and α-helix contents | Abdollahi et al.70 |
| • Complexation opened the protein structure and facilitated electrostatic and hydrogen bond interactions with negatively charged Pcs | |||
| Ovalbumin, ferulic acid | Secondary structure via CD | • The complexation with ferulic acid with ovalbumin decreased the α-helix content while increasing contents of β-sheet, β-turn and random contents | Chen et al.71 |
| • PCP complexation did not weaken the ordered structure but merely rearranged the secondary structural pattern | |||
| Ovalbumin, PCs extracted from the leaves of Campomanesia xanthocarpa (Mart.) O. Berg | Secondary structure via FTIR | • PCP complexation decreased the β-sheet content of protein | Castanha et al.72 |
| • The A and B amide bands increased and decreased based on complexation. It indicated that the complexation was driven by hydrogen bonds | |||
| Rice glutelin, gallic acid | Secondary structure via CD | • There was a reduction in the α-helix structure and an increase in the β-sheet structure present in protein after the interaction | Dai et al.73 |
| • PCP complexation did not affect the protein random coil content | |||
| Complexation promoted the unfolding of the polypeptide chain, disruption of hydrogen bonding networks, and induced rearrangement of hydrogen bond in the protein molecule | |||
| Tertiary structure via fluorescence emission spectrum | • Complexation altered the tertiary protein tertiary structure | ||
| α-Lactalbumin, EGCG | Secondary structure via CD, FTIR | • Both CD and FTIR results suggested complexation between α-lactalbumin and EGCG decreased protein α-helix and increased β-sheet contents, while the random coil content remained unchanged | Al-Hanish et al.74 |
| • PCP complexation did not reduce the ordered structure but only rearrange pattern of secondary structures | |||
| Tertiary structure via fluorescence quenching analysis | • Complexation distorted the tertiary structure of α-lactalbumin | ||
| β-LG, 3,4-dihydroxybenzoic acid, gallic acid, syringic acid, caffeic acid, ferulic acid, chlorogenic acid | Secondary structure via CD | • The selected PCs induced secondary structure transition of β-LG from β-sheet to β-turn and α-helix | Li et al.75 |
| • PCP complexation decreased the β-sheet content of protein but increased β-turn and α-helical contents | |||
| • Original structure of β-LG was partial destabilised by binding to PCs within the hydrophobic pocket, located in the interior of the β-barrel | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| PCP conjugation | |||
| Whey protein, gallic acid | Secondary structure via CD | • Crosslinking of whey protein using gallic acid decreased α-helix content and increased the irregular curl content of WP | Fei et al.76 |
| • Conjugation induced structural loosening of protein | |||
| Soy protein, black rice anthocyanins | Secondary structure via FTIR | • Conjugation decreased α-helix and β-sheet contents in the protein | Jiang et al.77 |
| • There was a significant increase in β-turn and random structure contents in the conjugate | |||
| Tertiary structure via three-dimensional fluorescence spectroscopy | • The fluorescence intensity of SPI at Peak 1 decreased, due to the covalent interactions between Trp or Tyr residues of soy protein and quinones | ||
| • The fluorescence intensity at Peak 2 of soy protein decreased after conjugation, which indicates that the unfolding of polypeptide chain in the protein | |||
| Bovine bone protein, EGCG | Secondary structure via FTIR | • Conjugation decreased α-helix and β-sheet contents of the protein | Pan et al.78 |
| Tertiary structure via fluorescence spectroscopy | • The maximum fluorescence intensity of protein decreased significantly after conjugation, due to the quenching of aromatic amino acids in the protein by EGCG | ||
| Whey protein isolate, EGCG | Secondary structure via FTIR | • Conjugation increased β-turn but decreased β-sheet content of the protein | Han et al.79 |
| • The random coil remained unchanged | |||
Wu et al. studied the PCP complexation between (−)-epigallocatechin (EGC) and β-lactoglobulin.51 The result suggests an amide I band shift from 1639.37 to 1641.30 cm−1 and an amide II band shift from 1541.01 to 1542.93 cm−1, respectively. This indicated a significant protein structure change by the complexation. A similar result was reported in Chen et al.'s study, where ovalbumin formed a PCP complex with tannic acid.80 The original protein and PCs have typical characteristic bands at 1538 and 1450 cm−1, respectively, due to vibrations of secondary N–H bonds in the protein and symmetrical stretching vibrations of the carboxyl groups in the PCs. The PCP complexation shifted the peak of N–H bending to 1547 cm−1, with the one for carboxyl groups to 1466 cm−1. Wang et al. detected the reduced amplitude of amide A, and I and II bands of a-La after it complexed with EGCG at pH 8.0. Particularly, the PCP complexation significantly shifted the protein band at 1692 cm−1, which was contributed by a structure of turns and bends, towards a larger wave number range, suggesting a structural change.81
The shift of characteristic band in the protein spectra is also observed when it forms conjugates with Pcs. In Pham et al.'s study, conjugation was induced between a flaxseed protein isolate and various phenolic compounds.61 Due to the conjugation, the band of amide II was shifted towards a higher wavelength number. Liu et al. induced the PCP conjugation between lactoferrin and EGCG.82 The authors found the secondary structure of the protein (11.5% α-helix, 9.5% β-sheet, 54.1% β-turn and 24.9% random coil) was changed (18.1% α-helix, 15.2% β-sheet, 43.5% β-turn and 23.1% random coil).
In Al-Hanish et al.'s study, PCP complexation between bovine α-lactalbumin and green tea polyphenol, EGCG, was induced.74 This complexation decreased the α-helix content of protein from 31.7 to 24.1%, while increasing its β-sheet content from 38.9 to 46.7%. The random coil content of the protein remained unchanged. Therefore, the authors suggested that the EGCG binding does not reduce ordered α-lactalbumin structure but only rearranges the secondary structure pattern. Korpela et al. induced the complexation between procyanidin B2 and transglutaminase-crosslinked oat globulin. The PCP complexation significantly reduced α-helix content in the protein, most likely because procyanidin B2 destroyed the protein's hydrogen bonding network by inserting itself into the hydrophobic surface of OG molecules.83 The decreased α-helix content during the PCP complexation was also observed by other researchers.84,85 However, a different trend was observed during the PCP complexation between ovalbumin and ellagic acid.86 In this study, when the protein and phenolic compounds interacted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, the overall structure of ovalbumin did not change significantly. The α-helix content slightly increased from 40.0 to 42.8%, while the β-turn content decreased by 3.5%.
30, the overall structure of ovalbumin did not change significantly. The α-helix content slightly increased from 40.0 to 42.8%, while the β-turn content decreased by 3.5%.
CD can detect the conformation changes of protein caused by PCP conjugation. In Yi et al.'s study, α-lactalbumin was conjugated with catechin.87 The authors reported that the conjugation reduced the α-helix content from 29.8 to 25.1% and increased the β-sheet content from 14.6 to 18.8% in protein. A decreased α-helix content in the protein was also observed in PCP conjugation formed by bovine serum albumin and chlorogenic acid.12 These results indicate the partial unfolding of the protein during the reaction. However, the protein conformational changes caused by PCP conjugation are somewhat affected by polyphenol type. In Liu et al.'s study, zein formed conjugate with quercetagetin.88 The authors observed a decreased α-helix and a β-sheet while increasing the β-turn and random coil contents in the protein secondary structure.
Furthermore, PCP conjugation between chlorogenic acid and EGCG and zein did not induce any protein secondary structure. Similarly, Pham et al. reported the increased α-helix and β-sheet of flaxseed protein isolate when it was conjugated with flaxseed polyphenol.61 However, increased α-helix and β-turn contents of the same protein were detected when it was conjugated with ferulic acid.
4.3 Molecule docking
Molecular docking has been used as an effective computational tool to predict the intermolecular interaction or reaction between multiple molecules. Several studies investigated the possible binding sites of dairy proteins and different polyphenols via PCP complexation. It is worth noting that during the molecular docking of the PCP complex, the proteins are often considered as rigid molecules while the phenolic compounds are molecules with flexibility to some extent. However, the molecular docking approach is based on classical physics. Therefore, it is more suitable for PCP complexation than conjugation.The molecular docking result of some example PCP complexation is presented in Table 2. In Li et al.'s study, the complexation of four PCs, namely, apigenin, naringenin, kaempferol and genistein with β-lactoglobulin was investigated.100 Among the binding sites, the most suitable result was the one with the highest −CDOCKER energy (−ECD) and highest −CDOCKER interaction energy (−IECD). According to the result of computational docking simulation, the site II of the protein molecular chain, with residues of Trp19, Tyr20, Tyr42, Gln44, Gln59, Gln68, Leu156, Glu157, Glu158 and His161, was found to be the best suitable binding sites for PCP complexation. Similarly, Gholami and Bordbar induced the PCP complexation between naringenin and bovine β-lactoglobulin.101 The molecular docking result indicated that naringenin interacted with some hydrophobic residues such as Val, Phe, Ala, and Leu and some polar and electrically charged residues such as Thr, Gln, Cys, and Lys in the outer surface of the protein. Kaur et al. mixed β-casein and p-coumaric acid solutions and heated the mixture at 145 °C for 8 s.102 The molecule docking result indicated at least one hydrogen bond between p-coumaric acid and the peptide backbone of isoleucine (Ile27).
| Protein and PCs | Complexation mechanism | References |
|---|---|---|
| a Abbreviations in the table: β-lactoglobulin: β-lg; tryptophan: Trp; lysine: Lys; glutamine: Gln; arginine: Arg; tyrosine: Tyr; serine: Ser; aspartic acid: Asp; leucine: Leu; threonine: Thr; glycine: Gly; epigallocatechin-3-gallate: EGCG; epicatechin 3-gallate: ECG; epigallocatechin: EGC. | ||
| Bovine and camel dairy proteins, date leaf polyphenolic extracts, ferulic acid | • PCP complexation depends on the molecular structure of PCs and protein molecules | Baba et al.89 |
| • PCs with lower polarity were more suitable for complexation with camel dairy proteins | ||
| • The major driving force for camel casein–date leaf polyphenolic extracts complexation may be van der Waal forces | ||
| • In bovine casein–date leaf polyphenolic extracts complex, β-lactoglobulin showed a higher number of hydrogen bonds and hydrophobic interactions than α-lactalbumin as well as total contacts with ferulic acid | ||
| β-Lactoglobulin, luteolin, apigenin, eriodictyol | • Apigenin binds in the internal cavity, luteolin binds at the mouth of the cavity, and eriodictyol binds outside the cavity of the protein | Baruah et al.90 |
| • Eriodictyol and apigenin exhibit better binding interactions with the protein than luteolin | ||
| • For eriodictyol and luteolin, van der Waals, hydrophobic, and hydrogen bonding interactions are the main interacting forces, whereas for apigenin, hydrophobic and van der Waals interactions play major roles | ||
| Pea protein, (Z)-2-penten-1-ol, hexanal, (E)-2-octenal | • Hydrophobic interactions were dominant in the PCP complexation between (E)-2-octenal/(Z)-2-penten-1-ol and pea protein | Bi et al.91 |
| • Hydrogen bonding was dominant in the PCP complexation between hexanal and pea protein | ||
| • The Lys357 residue in pea protein provided the main contribution to the formation of hydrophobic interactions with (Z)-2-penten-1-ol | ||
| • The Leu12 and Lys39 residues in pea protein contributed to hydrophobic interactions with hexanal, as well as a hydrogen bond between the Gln40 residue of protein and the oxygen atom in the PCs | ||
| • The Lys482 and Thr449 residues in pea protein made major contributions to the formation of hydrophobic interactions with (E)-2-octenal | ||
| Soy protein isolates, young apple polyphenols | • Young apple polyphenols physically bound to soy protein through hydrogen bonds and hydrophobic interactions | Chao et al.92 |
| • The four main PCs in young apple polyphenols were all more likely to interact with 7S protein than with 11S protein fractions | ||
| • Among the major PCs in the YAP, chlorogenic acid exhibited the strongest binding ability to both 7S and 11S proteins among PCs in the YAP. It formed three hydrogen bonds and 14 main hydrophobic interactions with the SPI 7S protein and interacted with the SPI 11S protein through five hydrogen bonds and seven hydrophobic interactions | ||
| Bovine β-lg, 28 flavonoids | • Thermodynamic analysis showed that hydrogen bonds and van der Waals force played the main roles in all the PCP complexations | Geng et al.93 |
| • Among 28 flavonoids, myricetin had the highest affinity with β-lg | ||
| • β-lg interacted with myricetin with Trp19 of β-lg, causing the entrapment of the PCs in hydrophobic cavity of the protein | ||
| Whey protein isolate, galangin, genistein | • Both galangin and genistein were bound into the β-lg in whey protein isolate via very similar binding site, i.e., hydrophobic pockets of the β-lg | Ma and Zhao94 |
| • Binding sites between β-lg and PCs (galangin and genistein) were different (Lys-77 vs. Gln-13) | ||
| • The α-lactalbumin in whey protein isolate had lower interaction energy than β-lg when forming PCP complexes with both PCs | ||
| Bovine lactoferrin, dihydromyricetin, myricetin | • Thermodynamic parameters revealed that hydrogen bond and van der Waals force were major forces in the formation of lactoferrin-dihydromyricetin complex, while hydrophobic interactions played major roles in the formation of the complex | Huang et al.95 |
| • Dihydromyricetin did not enter hydrophobic chamber of lactoferrin. Dihydromyricetin molecule formed six hydrogen bonds with amino acid residues of Arg121, Tyr192, Ser193, Asp297, Leu299, Thr58, and one hydrophobic bonds formed with Arg 296 | ||
| • Myricetinmolecule formed eight hydrogen bonds with residues Arg121, Ser185, Tyr192, Ser193, Arg296, Asp297, Glu15, Thr58, Leu59, and three hydrogen bonds with residues of Gly191 and Arg296 | ||
| Quinoa protein, quercetin, curcumin, luteolin, resveratrol | • The strongest hydrogen bonding interaction occurred between quinoa protein and quercetin, due its most hydroxyl groups | Liu et al.96 |
| • All the flavonoids were successfully assembled into the hydrophobic region of quinoa protein nanomicelles | ||
| • The hydrophobic interaction and hydrogen bonding were the main interaction driving forces between quinoa protein and hydrophobic flavonoids | ||
| Scallop (Patinopecten yessoensis) gonad protein isolates, EGCG, ECG, EGC, catechin | • The EGCG was bound into the hydrophobic central of vitellogenin or β-actin in the protein and surrounded by 17 and 20 amino acid residues on the vitellogenin and β-actin | Han et al.97 |
| • Hydrogen bond and van der Waals dominated the interaction process between SGPIs and EGCG | ||
| • Two hydrophobic interaction forces (π-alkyl) were found between aromatics rings of EGCG and alkyl groups of amino acid residues (Leu 80, Leu 821) of the protein, indicating the hydrophobic forces in the PCP complex | ||
| Myofibrillar protein, gallic acid, quercetin, EGCG | • Gallic acid formed hydrogen bonds with myofibrillar protein mainly through phenolic hydroxyl groups | Li et al.98 |
| • Quercetin had hydrophobic and electrostatic interactions with myofibrillar protein, leading to an increase in protein surface non-polarity, depending on the hydroxyl and flavonol structures | ||
| • EGCG had hydrogen bonds and hydrophobic interactions with myofibrillar protein | ||
| Casein, α-casein, β-casein, gallic acid, quercetin quercitrin, EGC, EGCG, tannic acid, bovine serum albumin | • Bonds, hydrophobic interactions and van der Waals were found in the interactions between PCs and α-casein and β-casein | Chen et al.99 |
| • PCs approached the hydrophobic cavities of α-casein and β-casein, and the nonpolar aromatic rings of phenolic compounds could interact with hydrophobic amino acids (such as proline, alanine and valine) residues through hydrophobic interaction | ||
| • The binding affinity of gallic acid, EGC and EGCG to casein gradually increased, due to the increased molecular weight and phenolic hydroxyl groups | ||
4.4 UV-vis spectrophotometer
UV-vis spectroscopy is used to provide insights into the impact of polyphenol type and the nature of the interactions on the protein molecular structure. Phenolic acids often exhibit UV absorption maxima at 270–280 nm and 305–330 nm, while flavonoids show UV absorption maxima at 270–280 nm and 310–350 nm. Furthermore, proteins exhibit UV absorption maxima at 214 nm (absorption of peptide bond) and 280 nm (absorption of aromatic amino acids). Once the phenolic compounds are conjugated with proteins, the UV absorption maxima of proteins shift, but this shift depends on the type of phenolic compounds and proteins and the reaction conditions.Liu et al. found that the zein–EGCG PCP conjugate had an appreciably higher absorbance than their physical mixture.88 Based on these results, the authors suggested that the formation of covalent bonds changed the structure of zein. The conjugate formation (at pH 9.0) may lead to structural changes in the protein, resulting in exposure of more tyrosine and tryptophan residues to the surrounding solvent. In Yan et al.'s study, the authors observed that the maximum absorption peak in the UV-vis spectra of the native protein isolate from Cinnamomum camphora seed kernels was around 280 nm, possibly attributed to the π → π* transition of Phe, Trp and Tyr residues.39 After this protein was conjugated with PCs, the intensity of the maximum absorption peak at approximately 280 nm of PI was progressively increased with the increase in PPE concentration. This result suggested that the conjugation might change the microenvironment of the Tyr and Trp residues of protein. In addition, the PCP complexes exhibited distinct broad peaks at around 315 nm, which may be mainly caused by the addition of PPE (with an absorbance peak at 315 nm).
4.5 Molecular weight
Based on the attachment of PCs on the protein molecular chain via PCP conjugation, the protein molecular weight is changed permanently. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) technique has been commonly used to detect this molecular weight change. Briefly, the SDS interacts with proteins to form an SDS-protein complex, thus disrupting non-covalent interactions in the sample such as hydrophobic interaction, ionic bonds and hydrogen bonds.103–106 Therefore, only the molecular weight change caused by PCP conjugation can be detected and quantified.In Sui et al.'s study, a PCP conjugate between black rice anthocyanins and a soybean protein isolate was produced.60 The SDS-PAGE result suggested that the intensity of β-conglycinin and glycinin subunits (<100 kDa) decreased, with the formation of a new band at a molecular weight of approximately 180 kDa. It was due to the crosslinking of the proteins by the PCs. Gu et al. also reported the molecular weight increase of egg white protein via PCP conjugation with catechin.107
It is worth noting that multiple advanced chromatographic and mass spectrometric techniques such as size exclusion chromatography (SEC), electrospray ionisation-mass spectrometry (ESI-MS) or matrix-assisted laser desorption/ionisation time-of-light mass spectrometry (MALDI-TOF-MS) have been further used to quantify the molecular weight of PCP conjugates. In Jia et al.'s study, the PCP conjugation between the whey protein isolate and EGCG was performed.57 The authors observed the formation of a new molecular weight band at approximately 36 kDa using SDS-PAGE under reducing conditions due to the crosslinking of the protein. Then, the authors used SEC measurement to confirm the protein crosslinking by PCs. Yi et al. observed a slightly increased molecular weight of α-lactalbumin via its conjugation with catechin using SDS-PAGE.87 Subsequently, ESI-MS was used to quantify a molecular weight increase of 272.6 Da, which is the molecular weight of catechin. This suggested that at least one catechin molecule was adducted to the protein molecular chain.
Similarly, Pham et al. reported the PCP conjugation between the flaxseed protein isolate and various phenolic compounds including flaxseed polyphenols, ferulic acid and hydroxytyrosol, respectively.61 Although the SDS-PAGE result did not show any significant change in the molecular weight of flaxseed protein isolate-flaxseed polyphenols and flaxseed protein isolate-ferulic acid conjugates, a slight increase in the molecular weight by several hundred Dalton was successfully detected by MALDI-TOF-MS. This indicated that these phenolic compounds simply formed the semiquinone and then attached onto the side chain of FPI. These authors also confirmed the crosslinking of the flaxseed protein moleculars by hydroxytyrosol based on SDS-PAGE and MALDI-TOF-MS analyses.
5 Characteristics of PCP complexes and conjugates
PCP complexation and conjugation can change the functional properties of both polymers, particularly in terms of solubility, emulsifying property, thermal stability, antioxidant activity, anti-microbial activity and digestibility, as shown in Table 3.| Protein and PCs | Functional properties | Result | References |
|---|---|---|---|
| a Abbreviations in the table: epigallocatechin-3-gallate: EGCG; β-Lactoglobulin: β-lg; Immunoglobulin E: IgE; scanning electron microscope: SEM; epigallocatechin: EGC; epicatechin: EC. | |||
| PCP complex | |||
| Dairy proteins, sea buckthorn polyphenols | In vitro antioxidant capacity, stability, bioaccessibility | • PCP conjugation reduced the in vitro antioxidant capacity of polyphenols but enhanced its stability during simulated in vitro digestion | Ashwar and Gani108 |
| • Compared with original PCs, the PCP conjugates had increased bioaccessibility (40.01% vs. 60.97–64.97%) | |||
| • The particle size of casein–polyphenol and whey protein–polyphenol complexes was in the nanosize region (650–983 nm), with narrow particle size distribution | |||
| Pea protein, quercetin, quercitrin, rutin | Surface hydrophobicity, surface tension, foaming and emulsifying properties | • Complexation decreased surface hydrophobicity of protein and caused a looser structure | Fu et al.109 |
| • Complex showed improved foaming and emulsifying properties, compared with protein | |||
| Whey protein isolate (WPI), EGCG, caffeic acid | In vitro digestion protein structural change antioxidant property | • Reducing capacity and FRAP values increased as the phenolic compound concentration increased in the PCP complex, while oxygen radical absorbance capacity values remained unchanged | de Morais et al.110 |
• At a protein-to-PCs molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, PCP complexation did not affect the protein digestibility (83%)., except for whey protein isolate–caffeic acid complex at pH 7 (73%) 0.5, PCP complexation did not affect the protein digestibility (83%)., except for whey protein isolate–caffeic acid complex at pH 7 (73%) |
|||
| • The pH of complexation and type of phenolic affected the protein cleavage pattern | |||
| • Complexation protected PCs during the in vitro digestion | |||
| Myofibrillar protein, mulberry PCs | Emulsifying property, gelling capacity, antioxidant capacity, water-holding capacity, rheological properties | • Mulberry PCs enhanced the inoxidizability of myofibrillar protein emulsion but decreased its emulsifying property | Cheng et al.111 |
| • Mulberry PCs at intermediate concentrations (5–20 mM) improved the elasticity, strength, and water-holding capacity of PCP complex-based emulsion gel | |||
| • Mulberry PCs at a high concentrations (40 mM) destroyed the emulsion gel, resulting in an “oil leakage” | |||
| Dairy and plant proteins, blueberry polyphenols | Protein digestion, binding affinity | • The hemp and pea proteins showed greater PCs binding affinities than whey | Chima et al.112 |
| • PCP complexation did not affect the digestion of any protein studied | |||
| • Solution affected the formation of PCP complex | |||
| Pea protein, EGCG, chlorogenic acid, resveratrol | Protein free sulfhydryl groups, protein surface hydrophobicity, emulsifying and foaming property, in vitro digestibility | • Free sulfhydryl groups and surface hydrophobicity of protein significantly decreased after complexing with PCs | Hao et al.113 |
| • EGCG, chlorogenic acid and resveratrol enhanced the emulsification, foaming and in vitro digestibility of pea protein | |||
| Lactoferrin, procyanidin | Surface hydrophobicity, foaming property | • PCP complexation reduced the surface hydrophobicity of the lactoferrin, suggesting procyanidin bound to nonpolar patches on lactoferrin's surfaces | Li et al.114 |
| • The binding of the procyanidin to the lactoferrin enhanced its foaming properties | |||
| Myofibrillar protein, gallic acid, quercetin, EGCG | Oxidative stability, rheological properties gelling property, protein surface hydrophobicity | • PCP complexation improved the oxidative stability of myofibrillar protein but decreased its emulsification properties | Li et al.98 |
| • PCs improved the rheological properties and gelling behavior of myofibrillar protein | |||
| • Gallic acid, quercetin and EGCG reduce the surface hydrophobicity of protein | |||
| • QT slightly increased protein surface hydrophobicity | |||
| Whey protein isolate, gallic acid chlorogenic acid, EGCG | Protein free sulfhydryl groups, protein surface hydrophobicity, solubility, foaming property, emulsifying capacity | • PCP complexation decreased free sulfhydryl groups and surface hydrophobicity of whey protein isolate | Meng et al.115 |
| • Compared with original protein, PCP complex showed enhanced solubility, foaming and emulsifying capacities | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| PCP conjugation | |||
| β-lg, caffeic acid | Protein thermal stability, protein solubility and antioxidant activity | • PCP conjugate showed increased antioxidant activity with the increased CA content attached to the protein | Abd El-Maksoud et al.116 |
| • PCP conjugates formed by β-lg and caffeic acid displayed comparable or superior water solubility than native protein and PCP mixture | |||
| • Conjugate significantly enhanced β-lg thermal stability | |||
| Ovalbumin, EGCG | Antioxidant activity, in vitro digestibility, emulsifying and foaming properties, thermal stability, IgE-binding capacity | • After conjugation with EGCG, ovalbumin showed increased digestibility, antioxidant activity, and emulsifying and foaming properties | He et al.117 |
| • Ovalbumin–EGCG conjugated had reduced thermal stability than the original protein | |||
| • IgE-binding capacities of ovalbumin were decreased with EGCG conjugation, indicating a reduced antigenicity | |||
| Beef myofibrillar protein, tea polyphenols | Solubility, surface hydrophobicity and emulsion activity index | • PCP conjugation decreased solubility, surface hydrophobicity and emulsion activity index of MPs but did not affect myofibrillar protein-stabilised emulsion stability | Hu et al.118 |
| Surface morphological | • Tea polyphenol addition was harmful to the formation process of MPs gels but increased surface complexity of the gels | ||
| • The degree of myofibrillar protein aggregation was related to how much polyphenols were added to myofibrillar protein suspension | |||
| • PCP conjugation caused the aggregation of myofibrillar protein | |||
| Whey protein isolate, chlorogenic acid, rosmarinic acid, quercetin | Protein isoelectric point, antioxidant property | • PCP conjugation increased the antioxidant capacity of proteins about 2.7 to 3.4 folds | Ali et al.119 |
| • Conjugation caused very slight shifts in the isoelectric points compared to the unmodified protein | |||
| Rice protein concentrate, legume PCs | Size, surface charge, crystallinity, SEM, protein thermal stability, sensory evaluation | • PCP conjugation reduced protein size (178.4 nm) and surface charge (−19.5 mV) | Chawla et al.120 |
| • Compared with original protein, conjugate showed a decreased crystallinity and smooth and continuous surface | |||
| • Conjugation enhanced the protein thermal stability | |||
| • PCP conjugated added fruit-based smoothie exhibited acceptable sensory profile | |||
| Pseudosciaena crocea roe protein isolate, EGCG | Protein molecular weight, thermal stability, antioxidant capacity | • PCP conjugation increased protein molecular weight from 86.9 to 215.3 kDa | Du et al.121 |
| • The protein–EGCG conjugates exhibited higher thermal stability than native protein | |||
| • The radical scavenging and reducing power of native protein were increased by 2.0–2.5- and 1.4-fold, respectively, after the EGCG-grafting reaction | |||
| Zein, adzuki bean seed coat polyphenol | Interfacial property, rheological property, antioxidant property, in vitro digestibility, colour profile | • PCP conjugate could be absorbed on the water–oil surface and stabilised ZAE, which presented as a non-Newtonian fluid state with good rheological properties | Ge et al.122 |
| • Conjugate exhibited stronger antioxidant property than original protein | |||
| • In vitro gastrointestinal digestion result suggested increased PCs content reduced the free fatty acids release rate of oil in the emulsion | |||
| • PCP conjugate exhibited a strong red–yellow color | |||
| Soy protein, tannic acid | Antioxidant property, gelling property, gel texture and structure, gel rheological behaviour | • PCP conjugate antioxidant activity and gelation characteristics soy protein | Guo et al.123 |
| • Gel prepared using PCP conjugate produced at pH 11 showed best hardness and elasticity | |||
| • Storage (G′) and loss modulus (G′′) of gel increased with increased tannic acid concentrations in the conjugate | |||
| • Increase in tannic acid concentration in the PCP conjugate increased the compactness of the gel | |||
| Soybean protein isolate, EGCG, EGC, EC | Fluorescence quenching functional property of SPI | • All the PCP conjugates showed improved the emulsifying activity, emulsion stability and antioxidant property, compared with the original protein | Han et al.124 |
| • Soybean protein isolate–EGCG conjugate exhibited best functional properties among the PCP conjugates | |||
| Whey protein isolate, EGCG, quercetin, apigenin, naringenin | Particle size, structure, polyphenols antioxidant properties and thermal stabilities | • Compared with the protein, all the PCP conjugates showed increased particle size | Liu et al.125 |
| • PCP conjugates had enhanced antioxidant property and thermal stability than PCs | |||
| • Conjugate reduced surface hydrophobicity of PCs | |||
| • Among the PCP conjugates, whey protein isolate-EGCG had the best functional properties, followed by whey protein isolate- quercetin, whey protein isolate-apigenin, and whey protein isolate-naringenin | |||
| Ovalbumin, chlorogenic acid | Allergenic capacity, antioxidant property | • Ovalbumin–chlorogenic acid conjugate reduced the IgE binding capacity of the protein | Lu et al.126 |
| • The ability of the ovalbumin–chlorogenic acid conjugate to activate histamine release was reduced | |||
| • Ovalbumin–chlorogenic acid conjugate showed high antioxidant activity | |||
| Sunflower protein, chlorogenic acid | Solubility, gelling ability, colour profile | • PCP conjugation increased protein solubility | Jia et al.127 |
• Covalently modified samples showed colour changes at a protein-to-PCs molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and higher 1 and higher |
|||
• Maximum gel strength was obtained when the conjugate was prepared at a protein-to-PCs molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
|||
| Casein, quercetin | Improved foaming activity, emulsifying activity and antioxidant activity, solubility, secondary structural | • Conjugate showed a decreased surface hydrophobicity and enhanced solubility, compared with the original protein | Ke et al.128 |
| • Conjugation improved foaming activity, emulsifying activity and antioxidant activity of casein | |||
| • Conjugation decreased foaming stability and emulsifying stability of casein | |||
5.1 Solubility
Generally, both PCP complexation and conjugation usually cause protein aggregation and precipitation, reducing protein solubility. Cano et al. incorporated tannin into caseinate- and gelatin-based films. The authors observed a reduced water solubility of both proteins with increased tannin content.129 In a recent study by Qian et al., a lentil protein isolate/cyanidin-3-O-glucoside complex was formed. It reduced the solubility and increased the particle size and turbidity of the protein.130 This trend was also reported in gluten- and gelatin-based films.131 This change was due to the disruption of hydrogen bonds among the protein chains and new hydrogen interactions between hydroxyl groups of the tannin molecule and polar groups of protein (amide carbonyl of the peptide backbone) and hydrophobic interaction.131,132 In von Staszewski et al.'s study, insoluble PCP complexes between green tea polyphenols and β-lactoglobulin/caseinomacropeptide were formed immediately once the protein and PCs were mixed.133 However, Prigent et al. did not find any significant effect on the solubility of BSA and α-lactalbumin when they formed a PCP complex binding with chlorogenic acid.134Santos et al. developed a zein-based film with lower water vapour permeability via its PCP conjugation with oxidised tannic acid.135 The phenolic compound crosslinked the protein and reduced its solubility. A similar trend was also observed in flaxseed protein–ferulic acid conjugates,61 flaxseed protein–flaxseed polyphenolic conjugates,61 egg white protein–EGCG conjugates136 and some myoglobin-based conjugates.46 In Strauch and Lila's study, cranberry polyphenols were conjugated with a pea protein isolate. The conjugation reduced protein solubility by 75%.137 However, the solubility of the PCP conjugate varies depending on the nature of the PCs and proteins. Abd El-Maksoud et al. reported the comparable or even superior water solubility of the β-lactoglobulin/caffeic conjugate over the original protein.138 In Pham et al.'s study, the PCP conjugate formed by the flaxseed protein and hydroxytyrosol showed a reduced hydrophobicity compared with the native protein, due to the exposure of hydrophilic groups during the PCP conjugation.61
5.2 Emulsifying properties
Due to their amphiphilic nature, proteins exhibit an emulsifying property to participate in forming and stabilising the emulsion. It is one of the most important protein functional properties for their applications as food ingredients. The emulsifying property of protein is significantly affected by its solubility. Thus, both PCP complexation and conjugation may affect these functional properties. The degree of change depends on the nature of the protein and PCs and the reaction nature.Once ovalbumin formed PCP complexes with tannic acid, a decreased emulsifying property of the protein was observed, due to the sheltering of the hydrophobic groups of protein.80 In another study, although the emulsion droplet stabilised by β-lactoglobulin/caffeic acid complexes exhibited a similar size to the ones coated by the native protein, the creaming index of the emulsion significantly increased with the incorporation of PCs in the system.138 However, different trends were reported in other studies. For example, the emulsifying property of the soy protein isolate–resveratrol PCP complex to produce an oil-in-water emulsion was not significantly different, compared with the native soy protein.139 In another study, the PCP complexation between the soy protein isolate and curcumin did not change the emulsifying and foaming properties of the protein.140 In Jiang et al.'s study, the complexation between α-lactalbumin and chalconoids improved the emulsion stability of the protein.141 The authors suggested that it might be due to the increased electrostatic repulsive force between emulsion particles due to the PCP complexation.
PCs have a predominantly positive influence on the emulsifying properties of proteins through conjugation. The emulsion droplets coated with the lactoferrin–chlorogenic acid conjugate had a smaller droplet size than the one stabilised by the protein alone.88 This was possibly due to the increased electrostatic and steric repulsion when the phenolic compounds were adducted on the protein molecular chain. Similar results were also reported in α-lactalbumin/EGCG and α-lactalbumin/chlorogenic acid conjugates,67 whey protein isolate/chlorogenic acid conjugates142 and β-lactoglobulin/coffee phenolics,143etc. However, different trends were reported, too. The gelatin–tannic acid conjugate–stabilised oil-in-water emulsion had a slightly bigger droplet size than the emulsion with native gelatin as an emulsifier.144 It might be due to the decreased surface hydrophobicity of the conjugate, which hindered the migration of gelatin to the oil/water interface. Similarly, decreased emulsifying activity and emulsion stability were found in emulsions emulsified by flaxseed protein isolate-flaxseed polyphenol and flaxseed protein isolate-hydroxytyrosol conjugates, compared with the ones with native protein as emulsifiers.145
5.3 Antioxidant activity
The antioxidant activities of PCs have been well reported.146–148 As such, both PCP complexes and conjugates exhibited enhanced antioxidant activity to a certain extent, compared with the native protein. However, it is worth noting that the antioxidant activity of the complex and conjugate is usually less powerful than that of the native PCs. Technically, the antioxidant activity of the PCP complex and conjugate can be evaluated by various in vitro tests, such as reducing power, lipid peroxidation and low density lipoprotein oxidation inhibition assays, 2,20-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging assays and oxygen radical absorbance capacity (ORAC) assay.Many studies reported the reduced antioxidant activity of PCs due to their complexation with dairy proteins, as a result of some dietary behaviours (e.g., drinking tea or coffee with milk). In Sharma et al.'s study, the effect of added milk and sugar on the antioxidant activity of black tea was evaluated using 1,1-diphenyl-2-picrylhydrazyl (DPPH) and β-carotene–linoleic acid model systems.149 The result suggested the radical scavenging and antioxidant activities of the systems were in the order of plain black tea > black tea with sugar > black tea with milk. This was because the presence of the dairy protein might hinder the radical scavengers from reaching their optimum scavenging capacity. The PCP complex between proteins such as caseinate and gelatin and tannins from different sources, including white peel grape, red peel grape and oak bark, showed decreased antioxidant activity than the phenolics, although the complexes were more antioxidative than the native protein.129 In Wan et al.'s study, the soy protein isolate was complexed with resveratrol.139 The product exhibited significantly improved oxidant activity than the native protein, in terms of DPPH radical scavenging activity, ORAC and reducing power, due to the presence of resveratrol in the complex. Additionally, this antioxidant activity was further improved by thermal processing at 90 °C for 30 min.
The antioxidant activity of the PCP conjugate was observed in gelatine–tannic acid, gelatine–caffeic acid and gelatine–ferulic acid conjugates,144 lactoferrin–EGCG conjugates150 and BLG–CLA conjugates.143 The PCP conjugate formed by α-lactalbumin and ECCG exhibited significantly higher DPPH and ABTS radical scavenging activities than those of the protein, due to the covalent incorporation of the PCs.20,81 In Yin et al.'s study, phosphorylated egg white protein was conjugated with EGCG and the conjugation was found to be responsible for its significantly improved antioxidant activity, measured using ABTS+ free radical scavenging activity and oxygen radical antioxidant capacity assay.136 Geng et al. used an ultrasound technique to facilitate the PCP conjugation between the soy protein isolate and EGCG. Among the natural protein, PCP complex and PCP conjugate, the conjugate had the highest DPPH (84.84 ± 1.34%) and ABTS (88.89 ± 1.23%) values.151 Interestingly, the increased antioxidant activity of phenolic compounds due to conjugation was also reported in zein–EGCG, zein-Q and zein–CA conjugate.88 Several recent studies reported the stronger antioxidant activity of the PCP conjugate than the complex. Wei et al. measured the antioxidant activity of the EGCG-protein conjugate in terms of DPPH, ABTS and FRAP assays and compared these capacities with the complex.152 The authors confirmed the superiority of EGCG–protein conjugates due to the relatively strong binding by the covalent conjugation. Similar results were reported in the catechin polymer-egg white protein and caffeic acid–β-lactoglobulin conjugate and complex.107,116
5.4 Thermal stability
The thermal stability of the protein depends on its structure. As such, PCP complexation and conjugation have significant effects on this functional property. The investigation can be performed by detecting the protein coagulation during heating or using a thermodynamic technique such as differential scanning calorimetry (DSC) and isothermal titration calorimetry (ITC).Generally, the thermal property of the proteins is enhanced via PCP complexation and conjugation.153–156 However, some contradictory results were also observed. O'Connell and fox and O'Connell et al. investigated the effect of phenolic compound addition on the thermal stability of milk proteins, based on the detection of protein coagulation at 120 and 140 °C.157,158 These authors observed an increased stability of milk proteins after they were complexed with ferulic acid and vanillic acid. However, a contrary phenomenon was observed when tannic and quinic acid were added to the milk. In Ojha et al.'s study, the PCP complexation between bovine serum albumin (BSA) and ferulic acid (FA) was induced at pH 7.4.159 The binding parameters determined using ITC indicated an increased thermal stability of the protein. Similarly, the enhanced thermal stability of whey protein was observed once it formed a PCP complex with coffee phenolics.143
The PCP conjugates usually exhibited enhanced thermal stability, compared with native protein and even the PCP complexes, due to the presence of covalent bonds.138,160,161 For example, Liu et al. reported that the conjugation between lactoferrin and various PCs, including EGCG, chlorogenic acid and gallic acid, significantly improved the protein thermal stability.82 Similarly, soy glycinin exhibited a higher thermal denaturation temperature after it formed conjugates with phenolic acids and flavonoids.162 In Liu et al.'s study, complexation and conjugation between zein and EGCG were induced in ethanol–water solutions. The authors observed that the denaturation temperature value of the zein–EGCG conjugate was higher than that of the control zein and zein–EGCG complex.88
5.5 Anti-microbial activity
In recent years, the use of natural phenolic compounds as anti-microbial agents to preserve food has gained research interest. Based on PCP complexation and conjugation, food proteins may also exhibit an inhibitory effect on some foodborne pathogens. However, studies on the effect of PCP complexation or conjugation on the anti-microbial activity of the products are still limited. Furthermore, most research focused on PCP conjugates, possibly due to the weak binding between the polymers in the PCP complex.In Fu et al.'s study, the PCP conjugate formed chlorogenic acid and gelatin was produced via the formation of covalent bonds between the carboxyl group of chlorogenic acid and free amino groups of gelatin.163 The anti-microbial activity of this conjugate against selected Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa) and Gram-positive bacteria (Listeria monocytogenes and Staphylococcus aureus) was further investigated. The result showed that the inhibitory activity of the conjugate was quite close to that of free chlorogenic acid against the test pathogenic microbe. In Novakovic et al.'s study, the authors induced the PCP conjugation between the lysozyme and the avarone. The formed conjugate exhibited strong anti-microbial activity against multiple bacteria.164 However, contradictory results were reported by Keppler et al.165 These authors prepared a rosmarinic acid quinone-BLG conjugate by an alkaline-induced method. This conjugate showed no significant anti-microbial effect on Staphylococcus aureus.
5.6 Digestibility
The PCP complexation and conjugation may reduce protein digestibility, due to the inhibition of digestive enzymes to access the active sites. In Stojadinovic et al.'s study, PCs from tea, coffee and cocoa were complexed with β-lactoglobulin.63 The in vitro digestibility of the resulting PCP complex was studied using the enzymatic method. The result suggested that complexation significantly hindered the protein digestion by protecting its secondary structure at pH 1.2. Additionally, a positive correlation was found between the strength of PCP complexation and the inhibitory effect against enzyme hydrolysis under simulated gastric conditions. Similarly, the sorghum bran proanthocyanidins and onion skin phenolic compounds significantly decreased the digestibility of wheat gluten after the PCP complex was formed.166,167 It is worth noting that the complexation between PCs and protein may affect the digestibility of protein differently in the different stages of digestion. For example, Shen et al. induced the PCP complexation between egg white protein and tea polyphenol. The complexation significantly improved the digestibility of egg white protein in the pepsin solution at pH 1.2 but inhibited its digestion in the pancreatin solution at pH 7.5.64 It was due to the changed secondary structure of ovalbumin and lysozyme.The PCP conjugation also usually reduces the protein bioavailability of essential amino acids. It can be because the phenolic compound/protein conjugation sterically blocks cleavage sites for the enzymes.168 Jiang et al. induced the conjugation between the soy protein isolate and black rice anthocyanins and investigated the in vitro hydrolysis of the conjugate.77 The authors found that the conjugate had a higher hydrolysis degree than the native protein due to polypeptide chains unfolding during conjugation. However, this conjugation reduced the peptide permeabilities across Caco-2 cell monolayers, indicating the potentially compromised bioavailability of the conjugate. The same phenomenon was also observed in Rawel et al.'s study, where the chlorogenic acid–bovine serum albumin conjugates exhibited a lower bioavailability than native BSA based on exposure to trypsin groups.12 Strauch and Lila compared the in vitro digestibility of the pea protein isolate and pea protein isolate/cranberry polyphenol conjugate. These authors observed that the PCP conjugation slowed gastric (pepsin) digestion by 25% and intestinal (pancreatin) digestion by 35%.169
Interestingly, the reduced protein digestibility by conjugation with PCs may help decrease their allergenicity since the attached phenolic compounds may bind with the IgE recognition epitope.112 This phenomenon was reported in whey protein isolate-chlorogenic acid, β-lactoglobulin/EGCG and β-lactoglobulin/chlorogenic acid conjugates.38,142
6 Application of PCP complexes and conjugates as novel encapsulants
Due to their amphiphilic, non-toxic and biocompatible nature, proteins are effective encapsulants for developing emulsion-based encapsulation systems. Bioactive compounds can be protected against degradation by avoiding reacting with other food ingredients and environmental stresses. In general, the efficacy of proteins for this application depends on their structure, as shown in Table 4. The key characteristics of the encapsulation system, such as particle size, shape, structure, surface properties and anti-microbial properties, are affected by the PCP complexation and conjugation.| Encapsulant component | Core material | Result | References |
|---|---|---|---|
| a Abbreviations in the table: epigallocatechin-3-gallate: EGCG; thiobarbituric acid reactive substance: TBARS. | |||
| PCP complex | |||
| Porcine myofibrillar protein, EGCG | Soy oil | • The results showed EGCG at all levels (0, 50, 100, 200, 500, and 1000 mg L−1) effectively suppressed lipid oxidation in emulsion during the entire chill storage (at 4 °C for 0, 3, or 7 days) | Cao et al.170 |
| • The incorporation of EGCG at higher concentrations (>100 mg L−1) promoted the loss of sulfhydryls, reduction of surface hydrophobicity, and aggregation and crosslinking of MP | |||
| • High concentrations of EGCG (500 and 1000 mg L−1) hampered emulsification and gel formation of MP | |||
| • EGCG at lower concentrations (50–200 mg L−1) improved the oxidative stability of emulsions without jeopardising the textural stability | |||
| Coconut globulin, coffee polyphenols | Soybean oil | • PCP complexation of changed the lipophilicity of protein and facilitated the formation of a dense and thick interfacial film at the oil–water interface | Chen et al.171 |
| • Furthermore, the emulsion stabilised by coconut globulin–coffee polyphenols complex showed excellent stability after storage, centrifugation, pH, and salt treatment | |||
| Pea proteins (PP), grape seed proanthocyanidins (GSP) | Flaxseed oil | • PCP complexation slightly decreased the isoelectric point, thermostability, and salt stability of the emulsions, compared to the ones with protein alone | Dai et al.172 |
| • PCP complexation increased emulsion storage stability | |||
| Whey protein isolate, lignin | Lactobacillus reuteri KUB-AC5 | • The results showed that microencapsulation using whey protein isolate–lignin complexes at 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio offered the best protection to probiotics 1 ratio offered the best protection to probiotics |
Diêp et al.173 |
| • Lignin improved survival of spray-dried probiotics through improving the structure and antioxidant properties of whey protein isolate | |||
| Sodium caseinate, resveratrol | Walnut oil | • Walnut oil emulsions stabilised by the complex exhibited excellent physical stability at 55 °C for 12 days or at room temperature for 10 months | Gong et al.174 |
| • More than 90% of resveratrol was loaded at the interface | |||
| • Lipid hydroperoxides and TBARS were reduced by ∼30% and 20% in the sodium caseinate–walnut oil emulsions containing 6 mM resveratrol than the control | |||
| Sunflower protein isolate, chlorogenic acid | Sunflower oil | • Emulsion droplets stabilised by the complexes exhibited high stability against coalescence | Karefyllakis et al.175 |
| Rice protein isolate, ferulic acid | Corn oil | • Emulsion stabilised by rice protein isolate-ferulic acid complex could decrease the concentration of hydroperoxide, TBARS, and hexanal, thereby effectively restraining fat oxidation degradation | Jia et al.176 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| PCP conjugate | |||
| Walnut protein isolate, ellagic acid, EGCG | Medium-chain triglyceride oil | • PCP conjugation resulted in decreased mean particle sizes and increased surface charges of the walnut protein-covered oil droplets | Huang et al.177 |
| • The emulsions stabilised by PCP conjugates were greater storage stability than those stabilised by unmodified protein | |||
| β-lg, polymeric units containing caffeic acid | Fish oil | • Compared to native protein-based emulsions, PCP conjugate-stablised one had larger oil droplet sizes, stronger negative zeta potentials (−69.9 mv), narrower size distributions (PDI: 0.22) and less creaming index | El-Maksoud et al.178 |
| • In the PCP conjugate-based emulsion with 50% fish oil, lipid oxidation was reduced by 4.3- and 1.8 folds, compared to the ones with protein and PCP mixture as the emulsifiers, respectively | |||
| Scallop (Patinopecten yessoensis) gonad protein isolate, EGCG | Tuna oil | • Compared with emulsion stabilised by protein, tuna oil emulsions emulsified by the PCP conjugate exhibited a smaller particle size and better storage stability | Han et al.179 |
| • The PCP conjugate inhibited lipid and fatty acid oxidation during storage more significantly in tuna oil emulsions than the protein due to its higher interfacial accumulation and antioxidant activities | |||
| Rice bran protein, catechin | Corn oil | • PCP conjugation reduced particle size, enhanced electrostatic repulsion, and improved emulsion stability | Li et al.180 |
| • The emulsions stabilised by conjugate exhibited superior rheological properties and enhanced oxidation stability compared to the one with protein as emulsifier | |||
| β-lg, gentisic acid | Hemp oil | • Under high gentisic acid grafting number (protein-to-PCs molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150), the soluble PCP conjugate exhibits a similar capacity as raw β-lg in physically stabilising emulsions, while exerting greater antioxidant activity against emulsion oxidation 150), the soluble PCP conjugate exhibits a similar capacity as raw β-lg in physically stabilising emulsions, while exerting greater antioxidant activity against emulsion oxidation |
Li et al.181 |
6.1 PCP complex
PCP complexation may improve the physicochemical stability of the encapsulation systems. Chen et al. reported that the oil-in-water emulsion stability was increased when the ovalbumin–tannic acid complex was used as an emulsifier, compared with the one stabilised by the native protein.80 Although the PCP complex exhibited a lower surface charge than that of the original protein, the presence of hydrogen bonds and hydrophobic interaction due to PCP complexation contributed to this increased stability. Similarly, the soy protein isolate–anthocyanin complex exhibited a higher emulsifying activity index than the protein. The complex-stabilised emulsion had a reduced particle size and increased emulsion stability index, compared with the one emulsified by proteins.60 However, a different trend was observed in Wan et al.'s study.139 The authors found that the complexation between the soy protein isolate and resveratrol did not affect the oil-in-water emulsion droplet sizes, although the accumulation of resveratrol at the oil/water interface significantly improved the oxidative stability of the emulsion. Baba et al. induced complexation between palm leaf polyphenols and bovine and camel dairy proteins.89 The formed complexes were then used to encapsulate curcumin in nanoemulsions. The result suggested that the thermal stability, photostability and bioaccessibility of curcumin were significantly improved when the complex was used as the encapsulant, compared to the ones stabilised by the original proteins. In Diêp et al.'s study, lignin formed a complex with the whey protein isolate. This PCP complex was then used to microencapsulate Lactobacillus reuteri KUB-AC5 via spray drying.173 The authors reported an increased probiotic survival rate with the increase in lignin content in the PCP complex. Gong et al.174 developed an oil-in-water emulsion with a sodium caseinate/resveratrol complex as an encapsulant to stabilise walnut oil. The presence of resveratrol in this emulsion system significantly reduced lipid oxidation by approximately 30 and 20% in terms of hydroperoxides and TBARS, compared with the emulsion with only proteins as emulsifiers. It is worth noting that although some PCP complexes are promising encapsulants, there is still research paucity in this area. It might be possibly due to the unstable nature of the complex.6.2 PCP conjugate
The PCP conjugate has been more studied than the complex for encapsulation applications, due to the stronger binding by covalent bonds. In Jia et al.'s study, the oil-in-water emulsion stabilised by the WPI-EGCG conjugate exhibited significantly reduced droplet size, compared with the one prepared using native WPI, due to conformational changes.57 Similarly, the physical stability of the α-lactalbumin-based emulsion containing β-carotene was improved once the protein was conjugated with EGCG. The use of these PCP conjugates as encapsulants significantly improved the chemical stability of β-carotene, possibly due to the enhanced emulsifying capacity and antioxidant activity of protein.67 Wang et al. induced the PCP conjugation between the glycosylated black bean protein isolate and EGCG, followed by preparing an oil-in-water emulsion using the conjugate as the stabiliser. This emulsion showed considerable stability against storage, oxidation, thermal treatments and freeze–thaw. It was because the PCP conjugation significantly increased protein adsorption at the oil/water interface, reduced the droplet size and produced a compact layer.182 In Fan et al.'s study, resveratrol-enriched zein nanoparticles were successfully encapsulated in the system stabilised by the bovine serum albumin–caffeic acid conjugate.183 The albumin–caffeic acid conjugation significantly increased the encapsulation efficiency of resveratrol from 91.5 to 96.3%. Similarly, beta-lactoglobulin and chlorogenic acid were conjugated to form nanoparticles to enhance the stability of EGCG, as reported by Fan et al.184 This PCP conjugate also significantly protected these tea PCs in the simulated gastrointestinal digestion fluid.Moreover, the thermal stability and UV stability of resveratrol were also increased due to the antioxidant activity of the conjugate. Wei et al. fabricated the gallic acid-ovotransferrin conjugate and used this novel material to prepare a Pickering emulsion in the presence of carboxymethyl dextran to deliver curcumin.185 The authors observed improved bio-accessibility and lipolysis of curcumin. Yi et al. developed a nanoencapsulation system to deliver beta-carotene by using catechin and β-lactoglobulin conjugates as encapsulants.186 Compared with the emulsion with the original protein as the emulsifier, the retention of beta-carotene in the conjugate-based emulsion was significantly improved. It was possible due to the high affinity between the catechin residual and beta-carotene. Similarly, the enhanced stability of fish oil was also observed in the nanoemulsion with catechin–egg white protein, catechin–α-lactalbumin and catechin–ovalbumin as emulsifiers, compared to the oils in the protein-based emulsions.187 However, it is worth noting that different results are also reported. Abd El-Maksoud et al. did not observe a clear correlation between the fish oil-in-water emulsion creaming index and the number of caffeic acids conjugated with β-lactoglobulin, although the oxidative stability of the stabilised fish oil increased.138
PCP conjugates were also trialled as a part of the encapsulant. The studies by Pham et al.'s group can be referred to as an example with the schematic diagram shown in Fig. 4. First, the flaxseed protein isolate was conjugated with various phenolic compounds such as flaxseed polyphenol and hydroxytyrosol.61 The formed PCP conjugate was used to stabilise flaxseed oil in an oil-in-water emulsion. During the 32 days storage at 20 and 40 °C, the PCP conjugate provided more effective protection to the sensitive flaxseed oil against lipid oxidation than protein alone.188 Subsequently, the authors induced complex coacervation between these PCP conjugates and flaxseed gum to maximise the microencapsulation efficiency and protection of the lipids. The complex coacervates produced using the PCP conjugate and flaxseed gum were more effective in protecting the sensitive oil against oxidation than the one formed by protein and gum during 4 weeks storage at 40 °C.189 In the in vitro digestion test, these PCP conjugate–gum complex coacervate-based microcapsules kept their integrity during the gastric stage but successfully digested in the intestinal stage to release the stabilised oil, suggesting the PCP conjugate-based complex coacervates as promising encapsulants.190
 | ||
| Fig. 4 Microencapsulation of flaxseed oil using flaxseed protein isolate/phenolic compound/flaxseed gum complex coacervates as encapsulants.61,188–190 | ||
7 Conclusion and future perspectives
There have been an increasing number of studies focusing on the modification of proteins to facilitate their food applications. Recently, complexation and conjugation between PCs and food proteins have been explored as promising methods for this purpose. In this context, this review provided an overview of the complexation and conjugation between PCs and food proteins, as well as the effect of these interactions or reactions on the functional properties of the resulting compounds. Generally, PCs and food proteins can form complexes via hydrophobic interactions, hydrogen bonding and van der Waals forces. However, the complex products are susceptible to environmental stresses and are easily disrupted. As such, PCP conjugates with permanent covalent bonds between PCs and proteins may be more suitable for applications. This review also discussed the potential application of the produced complex and conjugate as novel encapsulants to stabilise bioactive compounds.It is worth noting that the development of novel encapsulants via PCP complexation or conjugation is still in its early stages. Although various studies have proved that PCP complexation or conjugation is affected by the properties of PCs and proteins, their amount, and their interaction or reaction nature, the synthetic mechanism of PCP complexation or conjugation is still unclear. The correlation between PCP complexation or conjugation and protein/phenolic with structure types also remains unknown. The existing PCP complexation and conjugation studies are based on the changed functional properties of the protein, as well as the control of the PCP complexation and conjugation to produce end products with desired functional properties. Most of the existing studies only focused on the complexation or conjugation between proteins and simple phenols. The PCP complexation or conjugation between proteins and polyphenols has not been well investigated. Moreover, the impact of PCP complex- or conjugate-based encapsulation systems on quality attributes of actual food products has not been studied. Future research is required in these areas. Furthermore, there are quite few studies focusing on the biological activities of PCP complexes and conjugates. To facilitate their food applications, some biological properties such as toxicity, anti-diabetic, anti-allergic, anti-inflammatory and antimutagenic activities should be prioritised.
Author contributions
Bo Wang: conceptualization, supervision, writing – original draft, reviewing and editing. Loc B. Pham: writing – original draft, Benu Adhikari: conceptualization, supervision, writing – reviewing and Editing.Conflicts of interest
We have no conflict of interest to declare.References
- D. Šamec, E. Karalija, I. Šola, V. Vujčić Bok and B. Salopek-Sondi, Plants, 2021, 10, 118 CrossRef PubMed.
- M. Vuolo, V. S. Lima and M. R. Maróstica Junior, in Bioactive Compounds, ed. M. R. S. Campos, Woodhead Publishing, 2019, pp. 33–50, DOI:10.1016/B978-0-12-814774-0.00002-5.
- M. Lutz, E. Fuentes, F. Ávila, M. Alarcón and I. Palomo, Molecules, 2019, 24, 366 CrossRef PubMed.
- F. Jiang and G. J. Dusting, Curr. Vasc. Pharmacol., 2003, 1, 135–156 CrossRef CAS PubMed.
- M. Mauramo, T. Onali, W. Wahbi, J. Vasara, A. Lampinen, E. Mauramo, A. Kivimäki, S. Martens, H. Häggman, M. Sutinen and T. Salo, Antioxidants, 2021, 10, 1319 CrossRef CAS PubMed.
- S. Yıkmış, E. Bozgeyik and M. A. Şimşek, J. Food Sci. Technol., 2020, 57, 3445–3456 CrossRef PubMed.
- D. Villaño, M. S. Fernández-Pachón, M. L. Moyá, A. M. Troncoso and M. C. García-Parrilla, Talanta, 2007, 71, 230–235 CrossRef PubMed.
- T. Wang, R. Jónsdóttir and G. Ólafsdóttir, Food Chem., 2009, 116, 240–248 CrossRef CAS.
- E. Oskoueian, N. Abdullah, R. Hendra and E. Karimi, Int. J. Mol. Sci., 2011, 12, 8610–8625 CrossRef CAS PubMed.
- D. Zeng, G. Xiao, Y. Xu, B. Zou, J. Wu and Y. Yu, Food Sci. Biotechnol., 2019, 28, 945–953 CrossRef CAS PubMed.
- M. Esteruelas, N. Kontoudakis, M. Gil, M. F. Fort, J. M. Canals and F. Zamora, Food Res. Int., 2011, 44, 77–83 CrossRef CAS.
- H. M. Rawel, S. Rohn, H.-P. Kruse and J. Kroll, Food Chem., 2002, 78, 443–455 CrossRef CAS.
- N. D'Alvise, C. Lesueur-Lambert, B. Fertin, P. Dhulster and D. Guillochon, Sep. Sci. Technol., 2000, 35, 2453–2472 CrossRef.
- V. Cheynier, Phytochem. Rev., 2012, 11, 153–177 CrossRef CAS.
- M. Saifullah, R. McCullum, A. McCluskey and Q. Vuong, Heliyon, 2020, 6, e03666 CrossRef PubMed.
- L. L. Marques, G. P. Panizzon, B. A. Aguiar, A. S. Simionato, L. Cardozo-Filho, G. Andrade, A. G. de Oliveira, T. A. Guedes and J. C. Mello, Food Chem., 2016, 212, 703–711 CrossRef CAS PubMed.
- Z. Liu, J. Jia, F. Chen, F. Yang, Y. Zu and L. Yang, Molecules, 2014, 19, 19471–19490 CrossRef PubMed.
- L. Mao, Y. Lu, M. Cui, S. Miao and Y. Gao, Crit. Rev. Food Sci., 2020, 60, 1651–1666 CrossRef CAS PubMed.
- I. D. Nwachukwu and R. E. Aluko, in Food Proteins and Peptides: Emerging Biofunctions, Food and Biomaterial Applications, The Royal Society of Chemistry, 2021, pp. 1–33, 10.1039/9781839163425-00001.
- M. P. Almajano, M. E. Delgado and M. H. Gordon, Food Chem., 2007, 101, 126–130 CrossRef CAS.
- J. Miedzianka, A. Zambrowicz, M. Zielińska-Dawidziak, W. Drożdż and A. Nemś, Molecules, 2021, 26, 1575 CrossRef CAS PubMed.
- O. S. Lawal and K. O. Adebowale, Nahrung, 2004, 48, 129–136 CrossRef CAS PubMed.
- I. J. Boggione Santos, H. L. Hernandez Hernandez, M. H. Cardoso Costa, J. A. de Queiroz Lafetá and J. S. Dos Reis Coimbra, Food Res. Int., 2019, 492–498 CrossRef CAS PubMed.
- S. N. Sawali, V. K. Mukund and S. Deepti, Food Chem. Adv., 2023, 3, 100546 CrossRef.
- L. Gordonand and A. M. R. Pilosof, Food Biophys., 2010, 5, 203–210 CrossRef.
- M. Gulzar, S. Bouhallab, R. Jeantet, P. Schuck and T. Croguennec, Food Chem., 2011, 129, 110–116 CrossRef CAS.
- M. Vogelsang-O'Dwyer, A. W. Sahin, E. K. Arendt and E. Zannini, Foods, 2022, 11, 1307 CrossRef PubMed.
- X. Wu, Y. Luo, Q. Liu, S. Jiang and G. Mu, J. Sci. Food Agric., 2019, 99, 4942–4951 CrossRef CAS PubMed.
- I. A. Rasiah, K. H. Sutton, F. L. Low, H.-M. Lin and J. A. Gerrard, Food Chem., 2005, 89, 325–332 CrossRef CAS.
- D. J. M. Lorenzo and E. Capanoglu, Food Res. Int., 2023, 173, 113269 CrossRef PubMed.
- X. Yan, Z. Zeng, D. J. McClements, X. Gong, P. Yu, J. Xia and D. Gong, Compr. Rev. Food Sci. Food Saf., 2023, 22, 1312–1336 CrossRef CAS PubMed.
- E. Castanha, R. L. kavalek, R. B. Hoff, M. V. Dacoreggio, B. A. P. D. Jesus, M. D. L. B. Magalhães, G. F. D. Silva, A. S. D. Silva and A. P. Kempka, Food Chem. Adv., 2023, 2, 100303 CrossRef.
- D. Li, R. Wang, Y. Ma and D. Yu, Int. J. Biol. Macromol., 2023, 249, 126003 CrossRef CAS PubMed.
- M. Fu, Q. Geng, J. Chen, X. He, X. He, T. Li, C. Liu and T. Dai, J. Mol. Liq., 2023, 386, 122487 CrossRef CAS.
- Z. C. Song, H. Zhang, P. F. Niu, L. S. Shi, X. Y. Yang, Y. H. Meng, X. Y. Wang, T. Gong and Y. R. Guo, Food Chem., 2023, 420, 136110 CrossRef PubMed.
- M. Manzoor, Z. F. N. Tchameni, Z. F. BhatA, K. Jaiswal and S. Jaglan, Food Bioprocess Technol., 2023, 1–24 Search PubMed.
- S. Dubeau, G. Samson and H.-A. Tajmir-Riahi, Food Chem., 2010, 122, 539–545 CrossRef CAS.
- X. Wu, Y. Lu, H. Xu, D. Lin, Z. He, H. Wu, L. Liu and Z. Wang, Food Chem., 2018, 256, 427–434 CrossRef CAS PubMed.
- X. Yan, Y. Gao, S. Liu, G. Zhang, J. Zhao, D. Cheng, Z. Zeng, X. Gong, P. Yu and D. GOng, Food Chem., 2021, 352, 129377 CrossRef CAS PubMed.
- F. Shahidi and C. S. Dissanayaka, Food Prod., Process. Nutr., 2023, 5, 2 CrossRef CAS.
- A. J. Charlton, N. J. Baxter, M. L. Khan, A. J. G. Moir, E. Haslam, A. P. Davies and M. P. Williamson, J. Agric. Food Chem., 2002, 50(6), 1593–1601 CrossRef CAS PubMed.
- S. V. E. Prigent, H. Gruppen, A. J. W. G. Visser, G. A. Van Koningsveld, G. A. H. De Jong and A. G. J. Voragen, J. Agric. Food Chem., 2003, 51(17), 5088–5095 CrossRef CAS PubMed.
- T. Richard, X. Vitrac, J. M. Merillon and J. P. Monti, Biochim. Biophys. Acta, Gen. Subj., 2005, 1726(3), 238–243 CrossRef CAS PubMed.
- Y. Feng, X. Feng, S. Liu, H. Zhang and J. Wang, J. Sci. Food Agric., 2022, 102, 7387–7396 CrossRef CAS PubMed.
- Y. Guo and P. Jauregi, Food Chem., 2018, 266, 101–109 CrossRef CAS PubMed.
- Z. Jiang, T. Li, L. Ma, W. Chen, H. Yu, Q. Abdul, J. Hou and B. Tian, Food Res. Int., 2020, 131, 109006 CrossRef CAS PubMed.
- W. N. Baba, D. J. McClements and S. Maqsood, Food Chem., 2021, 365, 130455 CrossRef CAS PubMed.
- Y. Lu, R. Zhao, C. Wang, X. Zhang and C. Wang, Food Hydrocolloids, 2022, 132, 107895 CrossRef CAS.
- L. Zhang, P. Wang, Z. Yang, F. Du, Z. Li, C. Wu, A. Fang, X. Xu and G. Zhou, Food Hydrocolloids, 2020, 101, 105455 CrossRef CAS.
- A. K. Singh, in Engineered Nanoparticles, ed. A. K. Singh, Academic Press, 2016, pp. 19–76 Search PubMed.
- X. Wu, H. WU, M. Liu, Z. Liu, H. Xu and F. Lai, Spectrochim. Acta, Part A, 2011, 82, 164 CrossRef CAS PubMed.
- S. Zhang, X. Li, B. Ai, L. Zheng, X. Zheng, Y. Yang, D. Xiao and Z. Sheng, Food Chem.: X, 2022, 15, 100369 CAS.
- H. M. Rawel, D. Czajka, S. Rohn and J. Kroll, Int. J. Biol. Macromol., 2002, 30, 137–150 CrossRef CAS PubMed.
- G. Strauss and S. M. Gibson, Food Hydrocolloids, 2004, 18, 81–89 CrossRef CAS.
- J. Kroll, H. M. Rawel and N. Seidelmann, J. Agric. Food Chem., 2000, 48, 1580–1587 CrossRef CAS PubMed.
- H. M. Rawel, J. Kroll and U. C. Hohl, Food/Nahrung, 2001, 45, 72–81 CrossRef CAS PubMed.
- Z. Jia, M. Zheng, F. Tao, W. CHen, G. HUang and J. Jiang, LWT–Food Sci. Technol., 2016, 66, 305–310 CrossRef CAS.
- S. Parolia, J. Maley, R. Sammynaiken, R. Green, M. Nickerson and S. Ghosh, Food Chem., 2022, 367, 130603 CrossRef CAS PubMed.
- Y. Xu, Z. Wei, C. Xue and Q. Huang, J. Food Sci., 2022, 87, 2965–2979 CrossRef CAS PubMed.
- X. Sui, H. Sun, B. Qi, M. Zhang, Y. Li and L. Jiang, Food Chem., 2018, 245, 871–878 CrossRef CAS PubMed.
- L. B. Pham, B. Wang, B. Zisu and B. Adhikari, Food Chem., 2019, 293, 463–471 CrossRef CAS PubMed.
- C. Ribeiro, E. Brandão, C. Bravo, R. Dias, V. de Freitas, S. Soares and R. Pérez-Gregorio, in Extraction, Characterization, and Functional Assessment of Bioactive Compounds, ed. M. F. González, P. R. Rodríguez, and E. M. Carballo, Humana, New York, NY, 2024, DOI:10.1007/978-1-0716-3942-9_6.
- M. Stojadinovic, J. Radosavljevic, J. Ognjenovic, J. Vesic, I. Prodic, D. Stanic-Vucinic and T. Cirkovic Velickovic, Food Chem., 2013, 136, 1263–1271 CrossRef CAS PubMed.
- F. Shen, F. Niu, J. Li, Y. Su, Y. Liu and Y. Yang, Food Res. Int., 2014, 59, 100–107 CrossRef CAS.
- Z. He, B. Yuan, M. Zeng, G. Tao and J. Chen, Food Chem., 2015, 175, 457–464 CrossRef CAS PubMed.
- T. Li, P. Hu, T. Dai, P. Li, X. Ye, J. Chen and C. Liu, Spectrochim. Acta, Part A, 2018, 201, 197–206 CrossRef CAS PubMed.
- X. Wang, F. Liu, L. Liu, Z. Wei, F. Yuan and Y. Gao, Food Chem., 2015, 173, 564–568 CrossRef CAS PubMed.
- N. A. Al-Shabib, J. M. Khan, A. M. Al-Amri, A. Malik, F. M. Husain, P. Sharma, A. Emerson, V. Kumar and P. Sen, ACS Omega, 2023, 8, 19853–19861 CrossRef CAS PubMed.
- K. Abdollahi, C. Ince, L. Condict, A. Hung and S. Kasapis, Food Hydrocolloids, 2020, 101 Search PubMed.
- K. Abdollahi, L. Condict, A. Hung and S. Kasapis, Food Chem., 2022, 367, 130655 CrossRef CAS PubMed.
- L. Chen, M. Zhu, X. Hu, J. Pan and G. Zhang, J. Sci. Food Agric., 2022, 102, 3835–3846 CrossRef CAS PubMed.
- E. Castanha, R. L. Kavalek, R. B. Hoff, M. V. Dacoreggio, B. A. P. D. Jesus, M. D. L. B. MagalhÃEs, G. F. D. Silva, A. S. D. Silva and A. P. Kempka, Food Chem. Adv., 2023, 2, 100303 CrossRef.
- T. Dai, X. Yan, Q. Li, T. Li, C. Liu, D. J. McClements and J. Chen, Food Res. Int., 2017, 102, 274–281 CrossRef CAS PubMed.
- A. Al-Hanish, D. Stanic-Vucinic, J. Mihailovic, I. Prodic, S. Minic, M. Stojadinovic, M. Radibratovic, M. Milcic and T. Cirkovic Velickovic, Food Hydrocolloids, 2016, 61, 241–250 CrossRef CAS.
- X. Li, T. Dai, P. Hu, C. Zhang, J. Chen, C. Liu and T. Li, Food Hydrocolloids, 2020, 105 Search PubMed.
- X. Fei, Y. Yan, L. Wang, Z. Huang, D. Gong and G. Zhang, Food Res. Int., 2023, 170, 113000 CrossRef CAS PubMed.
- L. Jiang, Y. Liu, L. Li, B. Qi, M. Ju, Y. Xu, Y. Zhang and X. Sui, Food Res. Int., 2019, 120, 603–609 CrossRef CAS PubMed.
- L. Pan, Q. Li, J. Chen, Z. Qi, J. Jin, W. Shi, S. Lu, J. Dong and Q. Wang, Colloids Surf., A, 2023, 675 CrossRef CAS.
- X. Han, Z. Liang, S. Tian, L. Liu and S. Wang, Food Res. Int., 2022, 158 Search PubMed.
- Y. Chen, J. Hu, X. Yi, B. Ding, W. Sun, F. Yan, S. Wei and Z. Li, LWT, 2018, 95, 282–288 CrossRef CAS.
- X. Wang, J. Zhang, F. Lei, C. Liang, F. Yuan and Y. Gao, Food Chem., 2014, 150, 341–347 CrossRef CAS PubMed.
- F. Liu, C. Sun, W. Yang, F. Yuan and Y. Gao, RSC Adv., 2015, 5, 15641–15651 RSC.
- B. Korpela, L. Pitkänen and M. Heinonen, Food Chem., 2022, 395, 133568 CrossRef CAS PubMed.
- H. Zhang, D. Yu, J. Sun, H. Guo, Q. Ding, R. Liu and F. Ren, J. Mol. Struct., 2014, 1058, 228–233 CrossRef CAS.
- K. S. Ghosh, B. K. Sahoo and S. Dasgupta, Chem. Phys. Lett., 2008, 452, 193–197 CrossRef CAS.
- C. Yan and Z. Zhou, Food Hydrocolloids, 2020, 100, 105408 CrossRef CAS.
- J. Yi, Y. Fan, Y. Zhang and L. Zhao, Food Chem., 2016, 205, 73–80 CrossRef CAS PubMed.
- F. Liu, C. Ma, D. J. McClements and Y. Gao, Food Hydrocolloids, 2017, 63, 625–634 CrossRef CAS.
- W. N. Baba, R. Abdelrahman and S. Maqsood, Food Biosci., 2023, 53, 102690 CrossRef CAS.
- I. Baruah, C. Kashyap, A. K. Guha and G. Borgohain, ACS Omega, 2022, 7, 23083–23095 CrossRef CAS PubMed.
- S. Bi, X. Pan, W. Zhang, Z. Ma, F. Lao, Q. Shen and J. Wu, Food Chem., 2022, 389, 133044 CrossRef CAS PubMed.
- Z. Chao Song, H. Zhang, P. Fei Niu, L. S. Shi, X. Yan Yang, Y. Hong Meng, X. Yu Wang, T. Gong and Y. Rong Guo, Food Chem., 2023, 420, 133044 CrossRef PubMed.
- S. Geng, Z. Jiang, H. Ma, Y. Wang, B. Liu and G. Liang, Food Chem., 2020, 312, 126066 CrossRef CAS PubMed.
- C. M. Ma and X. H. Zhao, Foods, 2019, 8, 360 CrossRef CAS PubMed.
- J. Huang, Z. He, R. Cheng, Z. Cheng, S. Wang, X. Wu, B. Niu, G. X. Shen and X. Liao, Spectrochim. Acta, Part A, 2020, 243, 118731 CrossRef CAS PubMed.
- K. Liu, X. Q. Zha, Q. M. Li, L. H. Pan and J. P. Luo, Food Hydrocolloids, 2021, 118 Search PubMed.
- J. Han, Y. Du, J. Yan, X. Jiang, H. Wu and B. Zhu, Food Chem., 2021, 357, 129690 CrossRef CAS PubMed.
- N. Li, K. Zhang, X. Dong, Y. Xu, Z. Tan, G. Cao, X. Liu, D. Zhou and D. Li, Food Biosci., 2023, 53, 102676 CrossRef CAS.
- L. Chen, N. Chen, Q. He, Q. Sun, M. R. Gao and W. C. Zeng, Food Res. Int., 2022, 162, 112110 CrossRef CAS PubMed.
- T. Li, P. Hu, T. Dai, P. Li, X. Ye, J. Chen and C. Liu, Spectrochim. Acta, Part A, 2018, 201, 197–206 CrossRef CAS PubMed.
- S. Gholami and A.-K. Bordbar, Biophys. Chem., 2014, 187–188, 33–42 CrossRef CAS PubMed.
- J. Kaur, L. Katopo, A. Hung, J. Ashton and S. Kasapis, Food Chem., 2018, 252, 163–170 CrossRef CAS PubMed.
- J. M. Jung, G. Savin, M. Pouzot, C. Schmitt and R. Mezzenga, Biomacromolecules, 2008, 9, 2477–2486 CrossRef CAS PubMed.
- R. Jain, D. Sharma, R. Kumar and R. Kumar, J. Biochem., 2019, 165, 125–137 CrossRef CAS PubMed.
- E. L. Gelamo and M. Tabak, Spectrochim. Acta, Part A, 2000, 56A, 2255–2271 CrossRef CAS PubMed.
- D. Otzen, Biochim. Biophys. Acta, 2011, 1814, 562–591 CrossRef CAS PubMed.
- L. Gu, N. Peng, C. Chang, D. J. McClements, Y. Su and Y. Yang, Food Biophys., 2017, 12, 198–210 CrossRef.
- B. A. Ashwar and A. Gani, ACS Food Sci. Technol., 2021, 1, 1206–1214 CrossRef CAS.
- M. Fu, Q. Geng, J. Chen, X. He, X. He, T. Li, C. Liu and T. Dai, J. Mol. Liq., 2023, 386 Search PubMed.
- F. P. R. de Morais, T. B. Pessato, E. Rodrigues, L. Peixoto Mallmann, L. R. B. Mariutti and F. M. Netto, Food Res. Int., 2020, 133, 109104 CrossRef CAS PubMed.
- J. Cheng, D. Tang, H. Yang, X. Wang, M. Zhu and X. Liu, Food Chem., 2021, 360 Search PubMed.
- B. Chima, P. Mathews, S. Morgan, S. A. Johnson and C. B. Van Buiten, Foods, 2022, 11, 2846 CrossRef CAS PubMed.
- L. Hao, J. Sun, M. Pei, G. Zhang, C. Li, C. Li, X. Ma, S. He and L. Liu, Food Chem., 2022, 383, 132623 CrossRef CAS PubMed.
- C. Li, T. Dai, J. Chen, X. Li, T. Li, C. Liu and D. J. McClements, Food Chem., 2021, 339, 128145 CrossRef CAS PubMed.
- Y. Meng and C. Li, Food Chem., 2021, 364, 129622 CrossRef CAS PubMed.
- A. A. Abd El-Maksoud, I. H. Abd El-Ghany, H. S. El-Beltagi, S. Anankanbil, C. Banerijee, S. V. Petersen, B. Pérez and Z. Guo, Food Chem., 2018, 241, 281–289 CrossRef CAS PubMed.
- W. He, H. Xu, Y. Lu, T. Zhang, S. Li, X. Lin, B. Xu and X. Wu, J. Funct. Foods, 2019, 61, 103490 CrossRef CAS.
- Y. Hu, Y. Gao, I. Solangi, S. Liu and J. Zhu, LWT, 2022, 154 Search PubMed.
- M. Ali, J. Food Meas. Charact., 2019, 13, 2970–2979 CrossRef.
- P. Chawla, K. Sridhar and A. Bains, Food Res. Int., 2023, 171, 113075 CrossRef PubMed.
- Y. N. Du, J. R. Han, Z. K. Yin, J. N. Yan, X. Y. Jiang and H. T. Wu, J. Sci. Food Agric., 2021, 101, 5948–5955 CrossRef CAS PubMed.
- S. Ge, R. Jia, W. Liu, J. Xie, M. Liu, D. Cai, M. Zheng, H. Liu and J. Liu, Food Chem., 2022, 386 Search PubMed.
- Y. Guo, Y. H. Bao, K. F. Sun, C. Chang and W. F. Liu, Food Hydrocolloids, 2021, 112 Search PubMed.
- L. Han, X. Peng, Y. Cheng, Y. Zhu, Y. Huang, S. Zhang and B. Qi, LWT, 2023, 173 Search PubMed.
- X. Liu, Q. Song, X. Li, Y. Chen, C. Liu, X. Zhu, J. Liu, D. Granato, Y. Wang and J. Huang, Food Chem., 2021, 361, 130071 CrossRef CAS PubMed.
- Y. Lu, S. Li, H. Xu, T. Zhang, X. Lin and X. Wu, J. Agric. Food Chem., 2018, 66, 9794–9800 CrossRef CAS PubMed.
- W. Jia, D. S. Sethi, A. J. van der Goot and J. K. Keppler, Food Hydrocolloids, 2022, 131 Search PubMed.
- C. Ke, B. Liu, O. E. Dudu, S. Zhang, L. Meng, Y. Wang, W. Wei, J. Cheng and T. Yan, Food Hydrocolloids, 2023, 137, 108394 CrossRef CAS.
- A. Cano, M. Andres, A. Chiralt and C. González-Martinez, Food Hydrocolloids, 2020, 100, 105443 CrossRef CAS.
- H. Qian, F. Guo, H. Xiong, H. Zhang, L. Jiang and Y. Sun, Foods, 2022, 12, 104 CrossRef PubMed.
- C. Peña, K. de la Caba, A. Eceiza, R. Ruseckaite and I. Mondragon, Bioresour. Technol., 2010, 101, 6836–6842 CrossRef PubMed.
- K. Yi, G. Cheng and F. Xing, J. Appl. Polym. Sci., 2006, 101, 3125–3130 CrossRef CAS.
- M. von Staszewski, F. L. Jara, A. L. T. G. Ruiz, R. J. Jagus, J. E. Carvalho and A. M. R. Pilosof, J. Funct. Foods, 2012, 4, 800–809 CrossRef CAS.
- S. V. Prigent, H. Gruppen, A. J. Visser, G. A. Van Koningsveld, G. A. De Jong and A. G. Voragen, J. Agric. Food Chem., 2003, 51, 5088–5095 CrossRef CAS PubMed.
- T. M. Santos, M. d. S. M. Souza Filho, C. R. Muniz, J. P. S. Morais, L. R. V. Kotzebue, A. L. S. Pereira and H. M. Azeredo, J. Sci. Food Agric., 2017, 97, 4580–4587 CrossRef CAS PubMed.
- C. Yin, L. Yang, H. Zhao and C. P. Li, Food Res. Int., 2014, 64, 855–863 CrossRef CAS PubMed.
- R. C. Strauch and M. A. Lila, Food Sci. Nutr., 2021, 9, 3740–3751 CrossRef CAS PubMed.
- A. A. Abd El-Maksoud, I. H. Abd El-Ghany, H. S. El-Beltagi, S. Anankanbil, C. Banerjee, S. V. Petersen, B. Pérez and Z. Guo, Food Chem., 2018, 241, 281–289 CrossRef CAS PubMed.
- Z. L. Wan, J. M. Wang, L. Y. Wang, Y. Yuan and X. Q. Yang, Food Chem., 2014, 161, 324–331 CrossRef CAS PubMed.
- A. Tapal and P. K. Tiku, Food Chem., 2012, 130, 960–965 CrossRef CAS.
- Z. Jiang, T. Li, L. Ma, W. Chen, H. Yu, Q. Abdul, J. Hou and B. Tian, Food Res. Int., 2020, 131, 109006 CrossRef CAS PubMed.
- H. Xu, T. Zhang, Y. Lu, X. Lin, X. Hu, L. Liu, Z. He and X. Wu, Food Chem., 2019, 298, 125024 CrossRef CAS PubMed.
- M. Ali, T. Homann, M. Khalil, H.-P. Kruse and H. Rawel, J. Agric. Food Chem., 2013, 61, 6911–6920 CrossRef CAS PubMed.
- T. Aewsiri, S. Benjakul, W. Visessanguan, J.-B. Eun, P. A. Wierenga and H. Gruppen, Food Chem., 2009, 117, 160–168 CrossRef CAS.
- L. B. Pham, B. Wang, B. Zisu and B. Adhikari, Food Hydrocolloids, 2019, 94, 20–29 CrossRef CAS.
- G. Lin, L. Xian, X. Zhou, S. Wang, Z. H. Shah, S. A. Edwards and Y. Gao, ACS Appl. Mater. Interfaces, 2020, 12(44), 50152–50160, DOI:10.1021/acsami.0c14802.
- E. G. Wrigglesworth and J. H. Johnston, RSC Adv., 2017, 7, 45757–45762 RSC.
- B. Mukhtar, M. Mushtaq, S. Akram and A. Adnan, Anal. Methods, 2018, 10, 4310–4319 RSC.
- V. Sharma, H. Vijay Kumar and L. Jagan Mohan Rao, Food Res. Int., 2008, 41, 124–129 CrossRef CAS.
- F. Liu, D. Wang, C. Ma and Y. Gao, Food Hydrocolloids, 2016, 58, 49–59 CrossRef CAS.
- M. Geng, X. Feng, H. Yang, X. Wu, L. Li, Y. Li and F. Teng, Comparison of soy protein isolate-(–)-epigallocatechin gallate complexes prepared by mixing, chemical polymerization, and ultrasound treatment, Ultrason. Sonochem., 2022, 90, 106172 CrossRef CAS PubMed.
- Z. Wei, W. Yang, R. Fan, F. Yuan and Y. Gao, Food Hydrocolloids, 2015, 45, 337–350 CrossRef CAS.
- H. Ojha, K. Mishra, M. I. Hassan and N. K. Chaudhury, Thermochim. Acta, 2012, 548, 56–64 CrossRef CAS.
- M. Yan, B. Li, X. Zhao and J. Yi, Food Hydrocolloids, 2011, 25(5), 907–914 CrossRef CAS.
- P.-J. Tsai and P.- J She, J. Agric. Food Chem., 2006, 54(22), 8491–8494 CrossRef CAS PubMed.
- A. A. Abd El-Maksoud, I. H. Abd El-Ghany, H. S. El-Beltagi, S. Anankanbil, C. Banerjee, S. V. Petersen, B. Pérez and Z. Guo, Food Chem., 2018, 241, 281–289 CrossRef CAS PubMed.
- J. O'Connell and P. fox, J. Dairy Res., 1999, 66, 399–407 CrossRef.
- J. E. O'Connell, P. F. Fox, R. Tan-Kintia and P. F. Fox, Int. Dairy J., 1998, 8, 689–693 CrossRef.
- H. Ojha, K. Mishra, M. I. Hassan and N. K. Chaudhury, Thermochim. Acta, 2012, 548, 56–64 CrossRef CAS.
- M. Ishtikhar, E. Ahmad, Z. Siddiqui, S. Ahmad, M. V. Khan, M. Zaman, M. K. siddiqi, S. Nusrat, T. I. Chandel, M. R. Ajmal and R. H. Khan, Int. J. Biol. Macromol., 2018, 107, 2450–2464 CrossRef CAS PubMed.
- S. Wu, Y. Zhang, F. Ren, Y. Qin, J. Liu, J. Liu, Q. Wang and H. Zhang, Food Chem., 2018, 245, 613–619 CrossRef CAS PubMed.
- H. M. Rawel, D. Czajka, S. Rohn and J. Kroll, Int. J. Biol. Macromol., 2002, 30, 137–150 CrossRef CAS PubMed.
- S. Fu, C. Wu, T. Wu, H. Yu, S. Yang and Y. Hu, Food Chem., 2017, 221, 657–663 CrossRef CAS PubMed.
- I. Novaković, U. Anđelković, M. Zlatović, M. J. Gašić and D. Sladić, Bioconjug. Chem., 2012, 23, 57–65 CrossRef PubMed.
- J. K. Keppler, K. Schwarz and A. J. van der Goot, Trends Food Sci. Technol., 2020, 101, 38–49 CrossRef CAS.
- M. Świeca, U. Gawlik-Dziki, D. Dziki, B. Baraniak and J. Czyż, Food Chem., 2013, 141, 451–458 CrossRef PubMed.
- K. L. Dunn, L. Yang, A. Girard, S. Bean and J. M. Awika, J. Agric. Food Chem., 2015, 63, 1234–1241 CrossRef CAS PubMed.
- T. D. Cirkovic Velickovic and D. J. Stanic-Vucinic, Compr. Rev. Food Sci. Food Saf., 2018, 17, 82–103 CrossRef CAS PubMed.
- R. C. Strauch and M. A. Lila, Food Sci. Nutr., 2021, 9, 3740–3751 CrossRef CAS PubMed.
- Y. Cao, N. Ai, A. D. True and Y. L. Xiong, Food Chem., 2018, 245, 439–445 CrossRef CAS PubMed.
- Y. Chen, Y. Chen, L. Jiang, Z. Huang, W. Zhang and Y. Yun, Food Chem.: X, 2023, 20, 100954 CAS.
- T. Dai, T. Li, R. Li, H. Zhou, C. Liu, J. Chen and D. J. McClements, Food Chem., 2020, 329 Search PubMed.
- P. Diêp Huy Vũ, A. Rodklongtan and P. Chitprasert, LWT, 2021, 148 Search PubMed.
- T. Gong, B. Chen, C. Y. Hu, Y. R. Guo, Y. H. Shen and Y. H. Meng, Food Res. Int., 2022, 158 Search PubMed.
- D. Karefyllakis, S. Altunkaya, C. C. Berton-Carabin, A. J. van der Goot and C. V. Nikiforidis, Food Hydrocolloids, 2017, 73, 326–334 CrossRef CAS.
- X. Jia, M. Zhao, N. Xia, J. Teng, C. Jia, B. Wei, L. Huang and D. Chen, J. Cereal Sci., 2019, 89, 102818 CrossRef CAS.
- X. Huang, C. Yan, M. Lin, C. He, Y. Xu, Y. Huang and Z. Zhou, Food Res. Int., 2022, 161, 111910 CrossRef CAS PubMed.
- A. A. A. El-Maksoud, S. Anankanbil, Y. Zhou, I. H. A. El-Ghany, H. S. El-Beltagi, C. Banerjee, S. V. Petersen and Z. Guo, Food Chem., 2019, 301 Search PubMed.
- J. Han, Y. Du, W. Shang, J. Yan, H. Wu, B. Zhu and H. Xiao, Food Funct., 2019, 10, 6752–6766 RSC.
- D. Li, R. Wang, Y. Ma and D. Yu, Int. J. Biol. Macromol., 2023, 249, 126003 CrossRef CAS PubMed.
- H. Li, Y. Pan, Z. Yang, J. Rao and B. Chen, Food Hydrocolloids, 2022, 128, 107578 CrossRef CAS.
- J. Wang, H. Zheng, S. Zhang, J. Li, X. Zhu, H. Jin and J. Xu, RSC Adv., 2021, 11, 2546–2555 RSC.
- Y. Fan, Y. Liu, L. Gao, Y. Zhang and J. Yi, Food Chem., 2018, 261, 283–291 CrossRef CAS PubMed.
- Y. Fan, Y. Zhang, W. Yokoyama and J. Yang, RSC Adv., 2017, 7, 21366–21374 RSC.
- Z. Wei, H. Zhang and Q. Huang, Food Funct., 2019, 10, 4911–4923 RSC.
- J. Yi, Y. Zhang, R. Liang, F. Zhong and J. Ma, J. Agric. Food Chem., 2015, 63, 297–303 CrossRef CAS PubMed.
- T. H. Quan and S. Benjakul, Colloids Surf., A, 2019, 579, 123711 CrossRef CAS.
- L. B. Pham, B. Wang, B. Zisu and B. Adhikari, Food Hydrocolloids, 2019, 94, 20–29 CrossRef CAS.
- L. B. Pham, B. Wang, B. Zisu, T. Truong and B. Adhikari, Food Hydrocolloids, 2020, 107, 105944 CrossRef CAS.
- L. B. Pham, B. Wang, B. Zisu, T. Truong and B. Adhikari, Food Hydrocolloids, 2021, 112, 106325 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |