Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Emerging investigator series: Inactivation of antibiotic resistant bacteria and inhibition of horizontal resistance gene transfer is more effective by 222 than 254 nm UV†
Yijing
Liu
a and
Natalie M.
Hull
 *ab
*ab
aCollege of Engineering, Department of Civil, Environmental and Geodetic Engineering, The Ohio State University, Columbus, OH 43210, USA. E-mail: hull.305@osu.edu; nataliehull@boisestate.edu
bThe Sustainability Institute, The Ohio State University, Columbus, OH 43210, USA
First published on 19th November 2024
Abstract
The presence of antibiotic resistant bacteria (ARB) and the horizontal gene transfer (HGT) of antibiotic resistant genes (ARGs) in water environments pose a large and increasing threat to human health. This work compares the treatment efficiency of different ultraviolet (UV) wavelengths (222 nm KrCl excimer lamp and 254 nm low pressure Hg lamp) for inactivating multidrug antibiotic resistant B. subtilis strain 1A189, damaging its intracellular and extracellular ARGs, and inhibiting HGT of ARGs into non-resistant strain 1A1. The 222 nm wavelength was more effective than 254 nm at inactivation (dose required for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction or D1 = 4.11 mJ cm−2 at 222 nm and 8.99 mJ cm−2 at 254 nm). ARG damage increased with dose and with increasing qPCR amplicon length for both UV wavelengths. Although extracellular ARG damage was similar between wavelengths, intracellular ARG damage was greater at 222 nm than 254 nm. Inhibition of HGT also increased with UV dose for both wavelengths, but was stronger at 222 nm for both extracted DNA (D1 8.57 mJ cm−2 at 222 nm and 50.23 mJ cm−2 at 254 nm) and intracellular DNA (D1 = 20.14 mJ cm−2 at 222 nm and 92.90 mJ cm−2 at 254 nm). When taking into account factors such as electrical efficiency and spectral absorbance that are less favorable for 222 nm, results showed that 222 nm was still more efficient at extracellular HGT inhibition, while 254 nm was more efficient for other assay endpoints. Overall, these comparisons demonstrate the superior mechanistic efficacy of 222 nm over 254 nm UV for disinfecting ARB and for inhibiting transfer of ARG despite similar ARG damage. This information will help inform and improve tools to address the global water challenge of antibiotic resistance to minimize risks to human health.
log reduction or D1 = 4.11 mJ cm−2 at 222 nm and 8.99 mJ cm−2 at 254 nm). ARG damage increased with dose and with increasing qPCR amplicon length for both UV wavelengths. Although extracellular ARG damage was similar between wavelengths, intracellular ARG damage was greater at 222 nm than 254 nm. Inhibition of HGT also increased with UV dose for both wavelengths, but was stronger at 222 nm for both extracted DNA (D1 8.57 mJ cm−2 at 222 nm and 50.23 mJ cm−2 at 254 nm) and intracellular DNA (D1 = 20.14 mJ cm−2 at 222 nm and 92.90 mJ cm−2 at 254 nm). When taking into account factors such as electrical efficiency and spectral absorbance that are less favorable for 222 nm, results showed that 222 nm was still more efficient at extracellular HGT inhibition, while 254 nm was more efficient for other assay endpoints. Overall, these comparisons demonstrate the superior mechanistic efficacy of 222 nm over 254 nm UV for disinfecting ARB and for inhibiting transfer of ARG despite similar ARG damage. This information will help inform and improve tools to address the global water challenge of antibiotic resistance to minimize risks to human health.
Water impactUltraviolet (UV) disinfection at 222 nm is more effective than 254 nm at inactivating antibiotic resistant bacteria and inhibiting transfer of resistance genes. Although the novel 222 nm technology is limited by electrical efficiency, the efficacy of inhibiting gene transfer was still more efficient than 254 nm. These results enhance understanding of ARB treatment mechanisms toward optimizing UV for safe water. |
1. Introduction
The presence of antibiotic resistant bacteria (ARB) and their antibiotic resistance genes (ARGs) jeopardize health by rendering antibiotics ineffective, thus posing risks for public health and ecosystems.1,2 ARB are a prominent public health challenge of the 21st century, causing more than 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in the United States (US) and around 25
000 deaths in the United States (US) and around 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in Europe annually in 2017 with enormous cost.3 Deaths in the US increased to 35
000 deaths in Europe annually in 2017 with enormous cost.3 Deaths in the US increased to 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in 2019.4 ARB and ARG have the potential to transmit between humans, animals and throughout ecosystems.5,6 ARB have been detected in a variety of environments including soils, streams, lakes, rivers, and coastlines.7,8 Concerns about the microbial quality of water and transmission of ARB in water environments resulting in human exposure9 have led to investigations of the presence and removal of ARB and ARG in engineered water systems.
000 in 2019.4 ARB and ARG have the potential to transmit between humans, animals and throughout ecosystems.5,6 ARB have been detected in a variety of environments including soils, streams, lakes, rivers, and coastlines.7,8 Concerns about the microbial quality of water and transmission of ARB in water environments resulting in human exposure9 have led to investigations of the presence and removal of ARB and ARG in engineered water systems.
ARB have been detected in wastewater treatment plant (WWTP) influents, activated sludge, effluents, and biofilms,10,11 and the transfer of ARGs among bacteria can occur in the environment even after disinfection.12–14 The selection pressure of engineered treatments, combined with the overuse of antibiotics, can promote the evolution of bacteria through both spontaneous mutation to confer resistant genes, and the spread of resistance genes among microbial communities through mechanisms such as horizontal gene transfer (HGT).15–17 Previous studies have indicated that the environment of WWTPs can be beneficial for the growth of ARB and can increase the likelihood of gene transfer.10,18 A review about the fate of ARB in WWTPs indicated that raw and treated wastewater exhibit a greater prevalence of ARB than surface water.18 In other studies, highly abundant ARB were detected in both influent and effluent of a WWTP after chlorine disinfection.19–21 ARB and ARGs have been detected in untreated drinking water sources, as well as in tap and bottled water.22,23 Water distribution systems can allow proliferation of ARB and dissemination of ARG, resulting in higher quantities of ARGs in tap water than in both finished water and source water.24
Despite the prevalent detection of ARB and ARGs, disinfection has shown some efficacy in reducing ARB and ARGs in water sources and can serve as a protective barrier to restrict the proliferation and dissemination of ARB and ARGs.13,25,26 However, field studies indicated that these methods cannot completely eliminate all resistant bacteria and genes, leading to potential dissemination into the environment.16 Thus, the mechanisms of HGT inhibition and disinfection of ARB needs to be further explored. Research findings indicate the effectiveness of different disinfection processes (chlorine, UV irradiation, and peracetic acid) and ARB with different resistances to disinfectants.27 Chlorination is one of the most widely used drinking water disinfection techniques. However, chlorination can result in the formation of toxic disinfection by-products (DBPs) such as trihalomethanes and haloacetic acids.28 Chlorination also may be selected for some ARGs and promote HGT from ARB to pathogens by inducing genetic mutations.12,22,29 Therefore, a deeper understanding of the dynamics between disinfectant exposure and the development of resistance is crucial for inhibiting the dissemination of ARGs, refining water treatment practices, and minimizing the risk of antibiotic resistance propagation.
Compared with chlorination, UV produces less potentially toxic DBPs, damages nucleic acids which can inhibit HGT of ARGs, and inhibits ARG expression.30,31 The germicidal effect of UV light on bacteria and viruses is primarily due to the formation of pyrimidine dimers in DNA.32 These DNA lesions disrupt the normal structure and function of the genetic material, interfering with essential cellular processes and inhibiting the microorganism's ability to replicate and survive.33 As a result, UV disinfection effectively deactivates microorganisms by damaging their DNA, making UV highly relevant for combatting genes that confer antibiotic resistance. For ten ARGs across various WWTPs, it was found that the WWTP utilizing UV light as a frequent disinfection method during tertiary treatment exhibited the lowest prevalence for all ten ARGs.34 However, other studies indicated low levels of removal of ARGs after UV disinfection.35,36 Previous studies also investigated the direct influence of disinfection on ARB and ARGs and revealed that UV disinfection can substantially reduce ARB and ARGs.2,19,37,38 A recent study evaluated the performance of a 254 nm low pressure (LP) Hg lamp to disinfect ARB and ARGs in swine wastewater and indicated variable removal efficiencies for different ARGs,39 indicating that more studies are needed to fully understand the impacts of UV on ARB and ARG.
LP Hg lamps emitting primarily 254 nm UV are commonly applied, but little is known about ARB disinfection, ARG damage, and HGT inhibition by other UV light sources and wavelengths. One alternative UV source is krypton-chloride excimer (KrCl) lamps, which have long lifetimes and mercury-free materials, and compared with LP Hg lamps they exhibit much fewer adverse effects on skin and eyes.40 222 nm UV is becoming highly relevant due to its enhanced efficacy for disinfection and general water treatment. For example, many organic micropollutants have higher photolysis rate constants and quantum yields at 222 nm than at 254 nm.41 Additionally, 222 nm has been shown to be more efficient than 254 nm in a study inactivating B. subtilis spores due to triplet energy transfer from dipicolinic acid (DPA) to thymine bases.42
This work was therefore undertaken to compare kinetic dose response parameters for two different UV wavelengths (254 nm and 222 nm) for disinfecting B. subtilis ARB, and for damaging and inhibiting HGT of their ARGs to inform optimal wavelength options to inactivate and prevent the spread of ARB and ARGs and therefore minimize risks. By understanding the UV wavelength specific impacts for disinfecting ARB and for damaging and inhibiting transfer of ARGs, UV-based disinfection in water treatment systems can be optimized to protect public and environmental health.
2. Materials and methods
The overall study design was adapted from a previous study that focused on LP UV and other water treatments43 to additionally compare 222 nm wavelength dependent dose responses for: disinfection of ARB using culture assays, damage of ARGs using molecular qPCR assays, and inhibition of HGT of ARGs using transformation assays. Additionally, the impact on ARG damage and HGT inhibition was compared for intracellular ARGs in cells versus extracellular ARGs in DNA extracted from cells before UV exposure. Finally, electrical efficiency was additionally considered in addition to mechanistic efficacy.2.1 Bacterial strains and growth
Bacillus subtilis, a Gram-positive and spore-producing bacterium, was selected due to its various antimicrobial resistance phenotypes and mechanisms, carriers for exchanging ARGs, ease of cultivation, and extensive availability of gene sequence data.43–45 Bacillus subtilis strain 1A189 with chromosomal ARG (blt) was selected to be explored because it is a common multidrug transporter with broad substrate specificity.43,46B. subtilis strains 1A1 (nonresistant) and 1A189 (multidrug-resistant) were obtained from the Bacillus Genetic Stock Center (BGSC; Ohio State University). B. subtilis 1A189 exhibits a point mutation with an A–T base-pair deletion in the promoter region of the blt gene. The mutation conveys its resistance to multiple drugs. For incubation, 0.01 mL of each bacterial 25% glycerol stock was added with 6 mL antibiotic medium 3 growth medium (BD Difco™ Catalog No. DF0243-17-8, at pH 7) into an individual Erlenmeyer flask for 50 hours to monitor the OD600 of the cell culture hourly, and then diluted in series with phosphate-buffered saline (PBS, Fisher BioReagents™ BP3991) to be plated in duplicate on nutrient agar (BD Difco™ DF0479-17-3) to determine its growth curve and timing of the exponential phase to be used for UV exposure (∼26 hours). The growth curves of B. subtilis 1A189 and B. subtilis 1A1 are shown in Fig. S1 and S2.† PBS was used as the negative control.2.2 DNA extraction
After culturing B. subtilis to the late exponential growth phase, DNA of B. subtilis strains 1A189 was extracted using a method with fewer steps and quicker turnaround than the protocol He et al. used,43 modifying a previous protocol47 using phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform
chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) isoamyl alcohol instead of phenol
isoamyl alcohol instead of phenol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chloroform. 500 μl exponential phase (∼108 CFU ml−1) bacterial culture was loaded to 20 DNA extraction tubes for extraction, and DNA pellets were resuspended in 200 μl Tris–EDTA solution and used for UV exposure in the extracellular DNA study. For the intracellular study, DNA was extracted from untreated and UV exposed cells by the same method.
chloroform. 500 μl exponential phase (∼108 CFU ml−1) bacterial culture was loaded to 20 DNA extraction tubes for extraction, and DNA pellets were resuspended in 200 μl Tris–EDTA solution and used for UV exposure in the extracellular DNA study. For the intracellular study, DNA was extracted from untreated and UV exposed cells by the same method.
2.3 Ultraviolet light sources
An LP Hg lamp emitting nearly monochromatic light at a peak wavelength of 254 nm and an unfiltered krypton-chloride excimer (KrCl) lamp exhibiting a dominant peak at 222 nm were used as UV sources. Emission spectra (Ocean Optics HDX UV-vis spectroradiometer) for both lamps are shown in Fig. 1.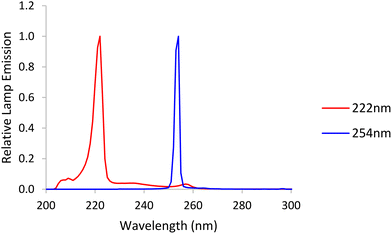 | ||
| Fig. 1 Emission spectra from a low-pressure (LP) mercury lamp, labelled 254 nm, and a krypton-chloride excimer (KrCl*) lamp, labelled 222 nm. | ||
2.4 UV dose determination and exposure
UV dose (mJ cm−2) is determined using two key parameters: wavelength and total UV. Standardized UV dose calculations described by Bolton and Linden (2003) include adjustment of variables such as measured incident irradiance (International Light ILT-5000 radiometer) and determination of Petri factor to account for non-uniformity across the sample surface, distance from light to the sample surface, sample depth, polychromatic sample absorbance (Agilent Cary 4000 UV-vis spectrophotometer), divergence of light throughout the sample depth, reflection factor at peak wavelength, and polychromatic lamp emission (Ocean Optics HDX UV-vis Spectroradiometer, NIST-traceable) and polychromatic radiometer response to calculate sample exposure times to achieve pre-determined UV doses.48,49 The protocol used is an open-source protocol.50 3 ml of exponential phase cells diluted to approximately 107 CFU ml−1, or their extracted DNA (1 ng μl−1), were exposed to fluences of 0–160 mJ cm−2. UV experiments were conducted fully in the dark environment and samples were covered with aluminum foil to avoid photo repair before transferring samples or plating. Log10 inactivation of cells, log10 gene damage, and log10 HGT inhibition were calculated using eqn (1).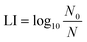 | (1) |
UV dose responses were modeled with a three-parameter log logistic model fitted to the disinfection data using the ‘drm’ function from the ‘drc’ package (Version 3.0-1) in R Studio51 using eqn (2).
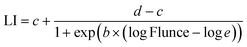 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) or 99% (2
log10) or 99% (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10) reduction, respectively, were calculated using modeled parameters and are summarized in Table S1.†
log10) reduction, respectively, were calculated using modeled parameters and are summarized in Table S1.†
2.5 Gene damage
Quantitative polymerase chain reaction (qPCR) was used to measure the damage of the target chromosomal blt gene with four different amplicon lengths (1017 bp, 870 bp, 832 bp and 266 bp) through loss of ability to amplify. All qPCR tests were performed in triplicate on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA) with a total volume of 20 μL, including 10 μL SsoFast EvaGreen PCR Master Mix (Applied Biosystems, USA), 10 μM forward and reverse primer, and 2 μL template DNA. The cycling protocol for qPCR follows 40 cycles of denaturation at 94 °C for 20 s, annealing for 20 s and extension at 72 °C for 40 s.43 Standard curves with gBlocks were used to calculate copy numbers. Sequences for each primer set and gBlocks were used without modification from a previous study.43 The ARG damage efficiency was calculated using eqn (3).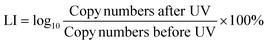 | (3) |
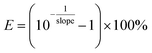 | (4) |
2.6 Horizontal gene transfer
Horizontal gene transfer of B. subtilis 1A189 DNA was quantified by natural transformation of the ARG into non-resistant B. subtilis 1A1 as described previously.43 Transformation frequency was calculated as transformant cell density (CFU mL−1) measured on selective media (with 4 mg L−1 acriflavine) over total recipient cell density (CFU mL−1) on nonselective media.2.7 Electrical efficiency
Electrical energy per order (EEO, kW h m−3 per order) represents the electric energy (kW h) required to remove the contaminant concentration per order of magnitude (90%) in 1 m3 water. EEO was calculated as previously52 according to the equation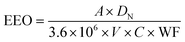 , where A is the irradiated surface area in cm2 calculated using the diameter of the Petri dish (9.62 cm2). V is the sample volume in liters (0.003 L). DN is the UV dose (mJ cm−2) required for obtaining a specific log reduction, which is the same as D1 or D2 for 1 or 2
, where A is the irradiated surface area in cm2 calculated using the diameter of the Petri dish (9.62 cm2). V is the sample volume in liters (0.003 L). DN is the UV dose (mJ cm−2) required for obtaining a specific log reduction, which is the same as D1 or D2 for 1 or 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log respectively, of cells or gene damage or HGT inhibition as summarized in Table S1.†C is the wall plug efficiency given by the manufacturer (0.35 for 254 nm (ref. 52) and 0.1 for 222 nm (ref. 53)). WF is the water factor, accounting for the UV absorbance and depth of the water, and we averaged the water factor from biological duplicate UV experiments. The factor 3.6 × 106 converts between hours and seconds, mW and kW, and L and m3.
log respectively, of cells or gene damage or HGT inhibition as summarized in Table S1.†C is the wall plug efficiency given by the manufacturer (0.35 for 254 nm (ref. 52) and 0.1 for 222 nm (ref. 53)). WF is the water factor, accounting for the UV absorbance and depth of the water, and we averaged the water factor from biological duplicate UV experiments. The factor 3.6 × 106 converts between hours and seconds, mW and kW, and L and m3.
3. Results and discussion
3.1 ARB inactivation
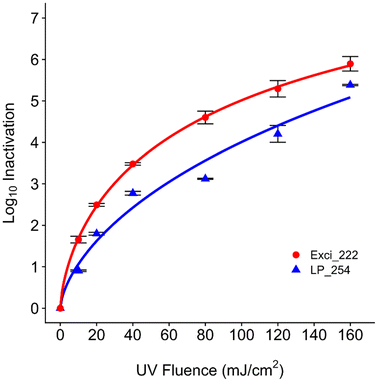
To explore the effect of disinfection dose and compare the effects of different wavelengths on ARB, we investigated the inactivation of multidrug antibiotic resistant B. subtilis 1A189 from a UV dose of 0–160 mJ cm−2. Both 222 nm and 254 nm UV wavelengths disinfected B. subtilis 1A189 (Fig. 2). Parameters of the fitted log logistic model are available in Table S1.† The log10 inactivation increased with increasing UV dose, but the 222 nm wavelength was more effective than 254 nm (D1 = 4.11 mJ cm−2 at 222 nm and 8.99 mJ cm−2 at 254 nm). The maximum observed inactivation was 5.9 ± 0.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log and 5.38 ± 0.01
log and 5.38 ± 0.01![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log at a dose of 160 mJ cm−2 for 222 nm and 254 nm, respectively, for the initial concentration of B. subtilis of ∼107 CFU mL−1. At the UV dose of 40 mJ cm−2 that is commonly applied in water treatment systems,54 3.48 ± 0.03 and 2.77 ± 0.05
log at a dose of 160 mJ cm−2 for 222 nm and 254 nm, respectively, for the initial concentration of B. subtilis of ∼107 CFU mL−1. At the UV dose of 40 mJ cm−2 that is commonly applied in water treatment systems,54 3.48 ± 0.03 and 2.77 ± 0.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log inactivation was observed for 222 nm and 254 nm, respectively.
log inactivation was observed for 222 nm and 254 nm, respectively.
In the study by He et al.,43 the rate constant of ARB inactivation was fitted to a first order reaction and tailing occurred at around 25 mJ cm−2; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction occurred around 5 mJ cm−2 by 254 nm, which was slightly lower than our observation of 9 mJ cm−2 required for 1
log reduction occurred around 5 mJ cm−2 by 254 nm, which was slightly lower than our observation of 9 mJ cm−2 required for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction at 254 nm and slightly higher than 4 mJ cm−2 required at 222 nm. Our findings align with previous research that demonstrated superiority of the KrCl excilamp over conventional Hg lamps for bactericidal efficacy against Bacillus spores55 and bacterial endospores.56 Similarly, a recent study applied 222 nm to disinfect waterborne pathogens and indicated the higher inactivation efficiency of MS2, E. coli, and S. aureus at 222 nm than 254 nm due to the damage of both proteins and nucleic acids.57 Another study also indicated the higher inactivation efficiency of 222 nm over 254 nm on vegetative B. subtilis.55 Differences in kinetics between wavelengths could be explained by previous studies which indicated higher inactivation of viruses at 222 nm over 254 nm due to more protein damage.58,59 Proteins are more abundant in cells than nucleic acids, while 222 nm is more intensively absorbed by proteins which can cause photooxidation and cross-linking.55,60 A study investigated different UV wavelengths (222 nm, 265 nm, and 285 nm) against chlorine-resistant bacteria and found 222 nm to promote the production of reactive oxygen species and cause cell membrane damage more than other wavelengths,61 which may have contributed to higher inactivation at 222 nm in our study. Another study additionally explored other UV mechanisms rather than the formation of DNA lesions, indicating the effects of reactive oxygen species formation which 254 nm altered greatly compared to 222 nm.60 However, in another study, higher disinfection performance at 254 nm than 222 nm on vegetative bacterial cells (E. coli K-12 and M. smegmatis) was observed in a thin-film aqueous solution,62 indicating potential differences across bacterial species and experimental setups and analyses or modeling that need to be further explored.
log reduction at 254 nm and slightly higher than 4 mJ cm−2 required at 222 nm. Our findings align with previous research that demonstrated superiority of the KrCl excilamp over conventional Hg lamps for bactericidal efficacy against Bacillus spores55 and bacterial endospores.56 Similarly, a recent study applied 222 nm to disinfect waterborne pathogens and indicated the higher inactivation efficiency of MS2, E. coli, and S. aureus at 222 nm than 254 nm due to the damage of both proteins and nucleic acids.57 Another study also indicated the higher inactivation efficiency of 222 nm over 254 nm on vegetative B. subtilis.55 Differences in kinetics between wavelengths could be explained by previous studies which indicated higher inactivation of viruses at 222 nm over 254 nm due to more protein damage.58,59 Proteins are more abundant in cells than nucleic acids, while 222 nm is more intensively absorbed by proteins which can cause photooxidation and cross-linking.55,60 A study investigated different UV wavelengths (222 nm, 265 nm, and 285 nm) against chlorine-resistant bacteria and found 222 nm to promote the production of reactive oxygen species and cause cell membrane damage more than other wavelengths,61 which may have contributed to higher inactivation at 222 nm in our study. Another study additionally explored other UV mechanisms rather than the formation of DNA lesions, indicating the effects of reactive oxygen species formation which 254 nm altered greatly compared to 222 nm.60 However, in another study, higher disinfection performance at 254 nm than 222 nm on vegetative bacterial cells (E. coli K-12 and M. smegmatis) was observed in a thin-film aqueous solution,62 indicating potential differences across bacterial species and experimental setups and analyses or modeling that need to be further explored.
To contextualize these 254 nm UV doses within engineered systems and other ARB, one study in a municipal WWTP found that only 5 mJ cm−2 was needed to obtain 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 inactivation of heterotrophic bacteria resistant to erythromycin and tetracycline,63 which is much lower than the doses studied here. 1
log10 inactivation of heterotrophic bacteria resistant to erythromycin and tetracycline,63 which is much lower than the doses studied here. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction in another study of multidrug-resistant Pseudomonas aeruginosa and vancomycin-resistant Enterococcus faecium required UV doses of only 2 and 8 mJ cm−2, respectively.38 However, relative abundance of some ARB or ARGs increased after UV in WWTPs in some studies,16,32,63 indicating possible differential selective pressure by UV on ARB. The regrowth of ARB was also observed after a UV dose of 10 mJ cm−2.64,65 Recent studies have started exploring the impact of UV 222 nm on Gram-positive or Gram-negative bacteria, bacterial spores, and wastewater or drinking water samples61,66 but have not focused specifically on ARB. Due to limited research on the specific impacts of UV wavelengths on ARB and the generally improved disinfection efficacy of 222 nm over 254 nm for several bacteria, these findings address the importance to further explore mechanisms of different UV wavelengths on other ARB and ARGs.
log reduction in another study of multidrug-resistant Pseudomonas aeruginosa and vancomycin-resistant Enterococcus faecium required UV doses of only 2 and 8 mJ cm−2, respectively.38 However, relative abundance of some ARB or ARGs increased after UV in WWTPs in some studies,16,32,63 indicating possible differential selective pressure by UV on ARB. The regrowth of ARB was also observed after a UV dose of 10 mJ cm−2.64,65 Recent studies have started exploring the impact of UV 222 nm on Gram-positive or Gram-negative bacteria, bacterial spores, and wastewater or drinking water samples61,66 but have not focused specifically on ARB. Due to limited research on the specific impacts of UV wavelengths on ARB and the generally improved disinfection efficacy of 222 nm over 254 nm for several bacteria, these findings address the importance to further explore mechanisms of different UV wavelengths on other ARB and ARGs.
3.2 ARG damage
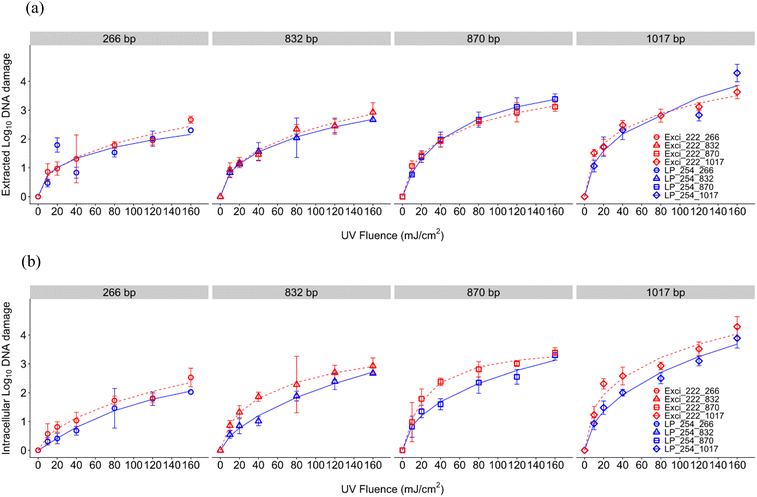
For DNA damage of the blt gene conferring multidrug resistance to B. subtilis, log10 DNA damage (calculated as the reduction of the ARG signal in qPCR) increased with UV dose for both UV wavelengths (Fig. 3). In general, shorter amplicons required a higher UV dose than longer amplicons to obtain the same reduction efficiency, which aligns with previous reports indicating the pattern of slower/lower ARG degradation using shorter qPCR amplicons43,67–71 but contrasts a study that found no relationship with amplicon length.62 This inverse relationship between amplicon length and loss of amplification is likely due to an increase in the number of nucleotides having more potential lesion targets. Compared to ARB inactivation, ARG damage was slower, which aligns with previous results.43,72 The role of cell structure could be implicated in the study where no relationship between genome damage and amplicon length was observed because the high abundance of protein in cells may contribute to the increased absorbance of UV at 222 nm, resulting in relatively less bacterial genome damage compared to viruses.62 A review identified ARGs in WWTPs and found that a significant reduction of ARB doesn't result in an efficient ARG removal, which could also explain the different rate constants between ARB and ARGs.73 Our results show greater damage of this ARG than a previous study in drinking water where 200 mJ cm−2 resulted in incomplete elimination of ARGs (1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction).64
log reduction).64
For the extracted DNA, ARG damage efficiencies were comparable between 254 nm and 222 nm (Fig. 3, Table S1†). Because the UV absorption spectrum of DNA shows absorbance increasing below 220 nm and peaking at 260 nm,74 the comparable ARG damage efficiencies of extracted DNA could be attributed to the fact that 254 nm and 222 nm have similar DNA absorbance when there are no other interfering or interacting compounds present. Due to limited interference of cellular materials or sample matrix components in solution, the impacts of which have been previously explored for 254 nm UV photoreactivity of nucleic acids,75 extracted DNA damage may be attributed solely to direct UV absorption. Extracted DNA degradation at 254 nm seen here is comparable to the previous research that informed our study design.43 Although He et al. used linear regression modeling including only UV doses up to 45 mJ cm−2 to exempt the tailing effect, we considered tailing by using the log logistic model. Due to the lack of sigmoidal shape, the linear rate constant in the log logistic model indicated by parameter b at the inflection point e doesn't adequately describe the gene damage kinetics alone, because the inflection point was sometimes at a dose greater than 160 mJ cm−2 (Table S1†). For more direct comparison to studies with linear dose responses, we calculated D1 and D2 for each amplicon length using fitted log logistic models, and took the inverse to determine rate constants k1 and k2 (Table S1†). The k1 and k2 for 254 nm for extracted DNA damage were 0.050 and 0.011 cm2 mJ−1 (266 bp), 0.066 and 0.011 mJ cm2 (832 bp), 0.075 and 0.017 mJ cm−2 (870 bp), and 0.134 and 0.023 mJ cm−2 (1017 bp). The k1 and k2 for 254 nm for intracellular DNA damage were 0.019 and 0.066 cm2 mJ−1 (266 bp), 0.033 and 0.0109 mJ cm−2 (832 bp), 0.072 and 0.017 mJ cm−2 (870 bp), and 0.089 and 0.023 mJ cm−2 (1017 bp). As can be seen from these rate constants, we observed a faster gene damage rate with extracted DNA than intracellular DNA. He et al.'s rate constants for extracted DNA damage (0.02–0.088 mJ cm−2) for 254 nm fell between our observed k1 and k2 values, and rate constants for intracellular DNA damage were consistent with the extracted DNA (2019). Our rate constants also indicate faster gene damage with increasing amplicon length after UV disinfection, which was also illustrated by previous studies.43,69,76 In addition to different modeling, other slight differences from He et al. could be due to our differing experimental set up. We conducted collimated beam UV exposure experiments while they used a merry-go-round photoreactor, resulting in different reflection and divergence factors. The difference in reaction volume (3 ml Petri dish compared to 100 ml quartz tube) may have also affected kinetics due to different light penetration and therefore water factor.
Comparing between extracted and intracellular DNA damage, UV dose responses demonstrated slightly different patterns. In our study, intracellular DNA required higher doses to damage the ARG (D1) than extracted DNA as shown in Fig. 5(a) and Table S1.† However as can be seen when comparing dose responses, differences between modeled parameters were minimal with overlapping errors (Table S1†). Both extracted and intracellular DNA exhibited similar tailing (d parameter) for both wavelengths. Tailing has been observed after 254 nm previously,43 and has been hypothesized to be due to combined effects of formation and photoreversal of CPD but irreversible formation of 6–4 photoproducts.77 Since tailing parameters did not differ, wavelength impacts are likely minimal. These minimal impacts between extracted and intracellular ARG damage are supported by previous comparisons using two Gram-positive bacteria and two Gram-negative bacteria under 254 nm that indicated no significant differences between extracted DNA and intracellular gene damage.38 No significant difference was observed in 254 nm UV degradation of intracellular DNA or extracted DNA in another study.78 Another 254 nm study of Acinetobacter spp. showed similar deactivation rate constants of chromosomal methicillin-resistant mecA, but more tailing was observed in intracellular DNA,77 while the tailing effects for extracted and intracellular DNA were similar in our study as indicated by parameter d (Table S1†). Results are also comparable to another study that treated two ARGs (∼800 bps) at UV 254 nm; however intracellular ARGs had a slower rate of damage compared to extracted, with a key difference being that the ARG was encoded on a plasmid76 rather than on chromosomes as studied here. A study with a plasmid encoded amp gene in E. coli indicated that 37 mJ cm−2 at 254 nm UV was required for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 reduction of both extracted and intracellular ARGs (Yoon, Dodd, and Lee 2018),69 which was lower than the dose required in our study with a chromosomal ARG. Additionally, a recent investigation demonstrated that 254 nm UV treatment led to comparable rates of inactivation between extracellular and intracellular forms of chromosomal ARGs, but chromosomal ARGs degraded faster than plasmid ARGs.78 Taken together these results and the literature indicate that further study may be warranted to discern the impact of UV wavelengths on damaging plasmid encoded ARGs.
log10 reduction of both extracted and intracellular ARGs (Yoon, Dodd, and Lee 2018),69 which was lower than the dose required in our study with a chromosomal ARG. Additionally, a recent investigation demonstrated that 254 nm UV treatment led to comparable rates of inactivation between extracellular and intracellular forms of chromosomal ARGs, but chromosomal ARGs degraded faster than plasmid ARGs.78 Taken together these results and the literature indicate that further study may be warranted to discern the impact of UV wavelengths on damaging plasmid encoded ARGs.
3.3 ARG transfer inhibition
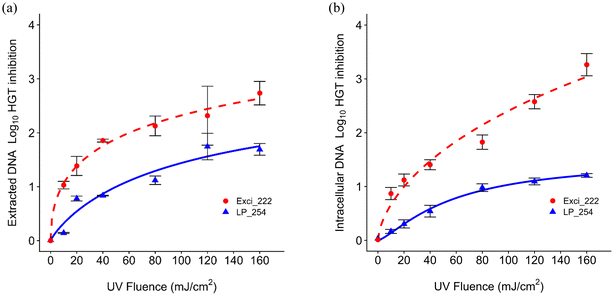
As shown in Fig. 4, the log10 inhibition of blt gene HGT generally increased with UV dose. The dose responses show that 222 nm was more effective than 254 nm on HGT inhibition as shown by D1 in Table S1† for both extracted and intracellular DNA inhibition (extracted: D1 = 8.57 mJ cm−2 at 222 nm and 50.23 mJ cm−2 at 254 nm; intracellular: D1 = 20.14 mJ cm−2 at 222 nm and 92.9 mJ cm−2 at 254 nm). The rate constants for HGT inhibition were lower than those for both cell inactivation, which aligns with previous results.79 Extracted DNA required lower doses than intracellular ARGs for HGT inhibition. The rate constants for HGT inhibition were similar to those for 266 bp and 832 bp ARG damage. He et al. also found that the HGT inhibition rate was similar to the gene damage for all amplicons, but was higher than the 260 bp gene damage.43Previous work demonstrated that the rate constant of transforming plasmid encoded ARGs was lower than that of CPD formation under 254 nm UV.69 Another study indicated that DNA integrity affecting the transformation and lower intracellular rate constant for transformation than that for gene damage may be due to gene repair.43 Plasmid encoded ARGs exposed to UV LEDs at 265 nm and 285 nm at 40 mJ cm−2 or 186 mJ cm−2 indicated that 265 nm was more efficient than 285 nm on inhibiting gene damage and gene transfer.70 No studies have explored the impacts of UV 222 on HGT. Our findings regarding more effective HGT inhibition at 222 nm could promote further exploration or application of 222 nm in water engineered systems.
3.4 Electrical energy comparisons
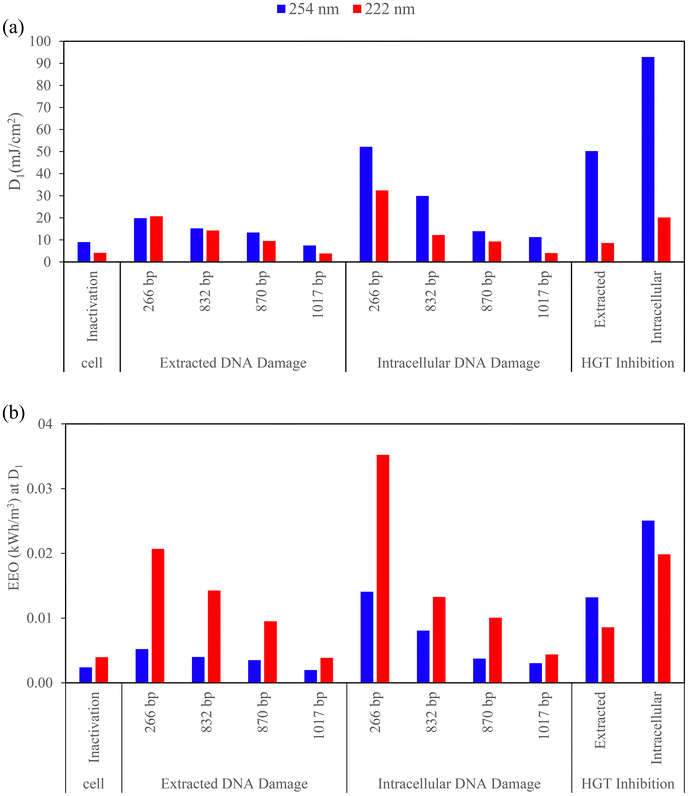
Comparisons of electrical energy per order, EEO, which is the electrical energy required for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 cell inactivation, ARG damage, and HGT inhibition, are summarized in Fig. 5 along with the summary of D1 used to calculate the EEO for each condition. A lower EEO value indicates greater energy efficiency.80 Most notably, EEO value for both extracted and intracellular HGT inhibition were higher for 254 nm LP than 222 nm KrCl, indicating that 222 nm is both more effective and more electrically efficient at preventing HGT of ARGs than 254 nm.
log10 cell inactivation, ARG damage, and HGT inhibition, are summarized in Fig. 5 along with the summary of D1 used to calculate the EEO for each condition. A lower EEO value indicates greater energy efficiency.80 Most notably, EEO value for both extracted and intracellular HGT inhibition were higher for 254 nm LP than 222 nm KrCl, indicating that 222 nm is both more effective and more electrically efficient at preventing HGT of ARGs than 254 nm.
A similar EEO was required between wavelengths for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log extracted DNA HGT inhibition (Fig. S3†), but UV 222 nm required a lower dose (60.14 mJ cm−2 for 222 nm and 236.81 mJ cm−2 for 254 nm). EEO is greatly impacted by having high wall plug efficiency (WPE) to use less energy for disinfection,81 and the maximum WPE for 222 nm was lower than 254 nm. A competitive EEO was required between wavelengths for ARB inactivation as shown in Fig. 5b and S3b† (0.004 (222 nm) and 0.002 (254 nm) kW h m−3 for D1, 0.013 (222 nm) and 0.008 (254 nm) kW h m−3 for D2). Although UV 222 nm had a lower WPE and water factor, UV 222 nm showed competitive energy efficiency on ARG damage. Our results align with another study about virus disinfection that indicated LP and excimer lamps to be competitive in terms of disinfection energy requirements.53
log extracted DNA HGT inhibition (Fig. S3†), but UV 222 nm required a lower dose (60.14 mJ cm−2 for 222 nm and 236.81 mJ cm−2 for 254 nm). EEO is greatly impacted by having high wall plug efficiency (WPE) to use less energy for disinfection,81 and the maximum WPE for 222 nm was lower than 254 nm. A competitive EEO was required between wavelengths for ARB inactivation as shown in Fig. 5b and S3b† (0.004 (222 nm) and 0.002 (254 nm) kW h m−3 for D1, 0.013 (222 nm) and 0.008 (254 nm) kW h m−3 for D2). Although UV 222 nm had a lower WPE and water factor, UV 222 nm showed competitive energy efficiency on ARG damage. Our results align with another study about virus disinfection that indicated LP and excimer lamps to be competitive in terms of disinfection energy requirements.53
HGT inhibition had a similar EEO to ARG damage of shorter amplicons, and ARB disinfection had a similar EEO to ARG damage of longer amplicons. For ARG damage, shorter amplicons required higher energy than longer amplicons for both extracted and intracellular DNA at both UV wavelengths, which aligns with the lowered rate constants of damaging shorter amplicons. In general, for ARG damage, the EEO was higher for 222 nm and for intracellular DNA, but the variation was almost overlapped with 254 nm. Due to the nonlinearity of dose responses, differences were examined when comparing D1 and corresponding EEO (Fig. 5b) with D2 and corresponding EEO (Fig. S3b†). UV doses required for extracted DNA damage with shorter amplicons were similar between 254 nm and 222 nm for D1, but 222 nm was more effective than 254 nm when comparing D2. The EEO values here for disinfection at 254 nm are within the range of those reported for Escherichia coli, MS2 coliphage, human adenovirus type 2, and Bacillus pumilus spores (0.006–0.297 kW h m−3 for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction at 254 nm).52 In our study, 0.008 kW h m−3 was required for 2
log reduction at 254 nm).52 In our study, 0.008 kW h m−3 was required for 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction of B. subtilis at 254 nm while 222 nm required 0.013 kW h m−3. In their study, the EEO to obtain a 2
log reduction of B. subtilis at 254 nm while 222 nm required 0.013 kW h m−3. In their study, the EEO to obtain a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log reduction for B. pumilus was 0.297 kW h m−3 using LP 254 nm and 0.373 kW h m−3 using MP which were higher compared with our study for B. subtilis, possibly due to resistance differences among Bacillus species. In their study, LP and MP UV lamps were more electrically efficient than UV LEDs. EEO also varied among different species in their study, where LP was the most efficient for inactivating E. coli and MS2 and MP and LP were equally efficient for inactivating HAdV2 and B. pumilus. More recently, the reported WPE and energy efficiency of LEDs are still low, WPEs of 0.003 (255 nm), 0.007 (265 nm) and 0.016 (285 nm) were applied to analyze the energy consumption of UV LEDs on P22 virus and E. coli and 255 nm had the lowest energy efficiency.82 Another UV LED study with 265, 280 and 300 wavelength disinfected Pseudomonas aeruginosa, Legionella pneumophila and surrogate microorganisms and compared the energy efficiency at 3
log reduction for B. pumilus was 0.297 kW h m−3 using LP 254 nm and 0.373 kW h m−3 using MP which were higher compared with our study for B. subtilis, possibly due to resistance differences among Bacillus species. In their study, LP and MP UV lamps were more electrically efficient than UV LEDs. EEO also varied among different species in their study, where LP was the most efficient for inactivating E. coli and MS2 and MP and LP were equally efficient for inactivating HAdV2 and B. pumilus. More recently, the reported WPE and energy efficiency of LEDs are still low, WPEs of 0.003 (255 nm), 0.007 (265 nm) and 0.016 (285 nm) were applied to analyze the energy consumption of UV LEDs on P22 virus and E. coli and 255 nm had the lowest energy efficiency.82 Another UV LED study with 265, 280 and 300 wavelength disinfected Pseudomonas aeruginosa, Legionella pneumophila and surrogate microorganisms and compared the energy efficiency at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 inactivation with 254 nm LP UV, a WPE of 0.333 was applied for 254 nm LP while UV LEDs had lower values (0.006 for 265 nm, 0.019 for 280 nm, and 0.026 for 300 nm UV-LEDs, respectively). The results indicated higher energy efficiency using LP 254 nm which, was one to two orders of magnitude larger than those of UV LEDs, and 280 nm was the most effective among UV LEDs.83 The higher electrical energy consumption of UV LEDs over LP due to the lower WPE was also indicated in a study disinfecting waterborne fungal spores.84 For field applications of UV, the energy requirement for an optimized wastewater treatment process ranges from 0.057–0.172 kW h m−3 and depends on many factors including the disinfection process, while the energy requirement for UV disinfection alone was 0.03 kW h m−3,85 which was at the same magnitude with our lab-scale study. The EEO requirement for treating chemicals in WWTPs was three orders of magnitude higher than the requirement for treating microorganisms.86,87 The different energy requirements for field applications and chemical removal compared to lab scale cell inactivation and gene damage indicate areas of potential further exploration on the effects of energy consumption on factors including real water matrices such as dissolved organic matter, nitrate, and potential toxic by-products.
log10 inactivation with 254 nm LP UV, a WPE of 0.333 was applied for 254 nm LP while UV LEDs had lower values (0.006 for 265 nm, 0.019 for 280 nm, and 0.026 for 300 nm UV-LEDs, respectively). The results indicated higher energy efficiency using LP 254 nm which, was one to two orders of magnitude larger than those of UV LEDs, and 280 nm was the most effective among UV LEDs.83 The higher electrical energy consumption of UV LEDs over LP due to the lower WPE was also indicated in a study disinfecting waterborne fungal spores.84 For field applications of UV, the energy requirement for an optimized wastewater treatment process ranges from 0.057–0.172 kW h m−3 and depends on many factors including the disinfection process, while the energy requirement for UV disinfection alone was 0.03 kW h m−3,85 which was at the same magnitude with our lab-scale study. The EEO requirement for treating chemicals in WWTPs was three orders of magnitude higher than the requirement for treating microorganisms.86,87 The different energy requirements for field applications and chemical removal compared to lab scale cell inactivation and gene damage indicate areas of potential further exploration on the effects of energy consumption on factors including real water matrices such as dissolved organic matter, nitrate, and potential toxic by-products.
4. Conclusions
Our results indicated that 222 nm UV light exhibits higher efficacy over 254 nm in inactivating ARB, damaging intracellular ARG, and inhibiting HGT of ARB, and greater electrical efficiency of inhibiting HGT and competitive electrical efficiency of disinfecting ARB, suggesting 222 nm as a promising alternative or complement to existing disinfection for enhancing water treatment processes. Further explorations on other ARB and both plasmid and chromosomally encoded ARGs with multiple UV sources in various simulated and realistic water and wastewater treatment relevant matrices should be performed to optimize the UV efficiency or design protocols in engineered water systems.Data availability
The research data associated with the ARB UV disinfection study are available on the Figshare website (https://figshare.com/account/home#/projects/210526).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by startup support to Dr. Natalie Hull from the Department of Civil, Environmental, and Geodetic Engineering and the Sustainability Institute at The Ohio State University. I would also like to acknowledge Judith Straathof and Zanna Leciejewski for UV experiment training and Daniel Ma for help with troubleshooting.References
- D. G. J. Larsson and C. F. Flach, Nat. Rev. Microbiol., 2022, 20, 257–269 CrossRef CAS.
- A. Pruden, R. Pei, H. Storteboom and K. H. Carlson, Environ. Sci. Technol., 2006, 40, 7445–7450 CrossRef CAS PubMed.
- P. J. Vikesland, A. Pruden, P. J. J. Alvarez, D. Aga, H. Bürgmann, X. Li, C. M. Manaia, I. Nambi, K. Wigginton, T. Zhang and Y.-G. Zhu, Environ. Sci. Technol., 2017, 51, 13061–13069 CrossRef CAS PubMed.
- C. U.S. Department of Health and Human Services, Antibiotic resistance threats in the United States, 2019, Atlanta, Georgia, 2019 Search PubMed.
- I. C. Stanton, A. Bethel, A. F. C. Leonard, W. H. Gaze and R. Garside, Environ. Evid., 2022, 11, 8 CrossRef PubMed.
- N. G. Godijk, M. C. J. Bootsma and M. J. M. Bonten, BMC Infect. Dis., 2022, 22, 482 Search PubMed.
- P. De Voogt, Reviews of Environmental Contamination and Toxicology, 2015, vol. 236 Search PubMed.
- S. Bombaywala, A. Mandpe, S. Paliya and S. Kumar, Environ. Sci. Pollut. Res., 2021, 28, 24889–24916 Search PubMed.
- K. Liguori, I. Keenum, B. C. Davis, J. Calarco, E. Milligan, V. J. Harwood and A. Pruden, Environ. Sci. Technol., 2022, 56, 9149–9160 CrossRef CAS PubMed.
- J. Wang, L. Chu, L. Wojnárovits and E. Takács, Sci. Total Environ., 2020, 744, 140997 CrossRef CAS PubMed.
- S. Perveen, C. Pablos, K. Reynolds, S. Stanley and J. Marugán, Sci. Total Environ., 2023, 856, 159024 CrossRef CAS PubMed.
- E. Sanganyado and W. Gwenzi, Sci. Total Environ., 2019, 669, 785–797 CrossRef CAS PubMed.
- M. Kalli, C. Noutsopoulos and D. Mamais, Water, 2023, 15(11), 2084 CrossRef CAS.
- M. Kang, J. Yang, S. Kim, J. Park, M. Kim and W. Park, Sci. Total Environ., 2022, 811, 152331 CrossRef CAS PubMed.
- T. Zhang, Y. Hu, L. Jiang, S. Yao, K. Lin, Y. Zhou and C. Cui, Chem. Eng. J., 2019, 358, 589–597 CrossRef CAS.
- H. Wang, J. Wang, S. Li, G. Ding, K. Wang, T. Zhuang, X. Huang and X. Wang, Water Res., 2020, 185, 116290 Search PubMed.
- A. K. Mittal, R. Bhardwaj, P. Mishra and S. K. Rajput, Open Biotechnol. J., 2020, 14, 107–112 CrossRef CAS.
- C. Bouki, D. Venieri and E. Diamadopoulos, Ecotoxicol. Environ. Saf., 2013, 91, 1–9 CrossRef CAS PubMed.
- J. Zheng, C. Su, J. Zhou, L. Xu, Y. Qian and H. Chen, Chem. Eng. J., 2017, 317, 309–316 CrossRef CAS.
- C. Li, F. Ling, M. Zhang, W. T. Liu, Y. Li and W. Liu, J. Environ. Sci., 2017, 51, 21–30 Search PubMed.
- O. M. Lee, H. Y. Kim, W. Park, T. H. Kim and S. Yu, J. Hazard. Mater., 2015, 295, 201–208 Search PubMed.
- P. Shi, S. Jia, X. X. Zhang, T. Zhang, S. Cheng and A. Li, Water Res., 2013, 47, 111–120 CrossRef CAS.
- J. Zhang, W. Li, J. Chen, W. Qi, F. Wang and Y. Zhou, Chemosphere, 2018, 203, 368–380 Search PubMed.
- C. Xi, Y. Zhang, C. F. Marrs, W. Ye, C. Simon, B. Foxman and J. Nriagu, Appl. Environ. Microbiol., 2009, 75, 5714–5718 CrossRef CAS PubMed.
- J. Oh, D. E. Salcedo, C. A. Medriano and S. Kim, J. Environ. Sci., 2014, 26, 1238–1242 Search PubMed.
- S. Li, B. S. Ondon, S. H. Ho, Q. Zhou and F. Li, J. Water Process Eng., 2023, 53, 103907 Search PubMed.
- Y. Shen, J. Luo, A. Di Cesare, N. Guo, S. Zou and Y. Yang, Environ. Pollut., 2024, 345, 123427 CrossRef CAS PubMed.
- K. Gopal, S. S. Tripathy, J. L. Bersillon and S. P. Dubey, J. Hazard. Mater., 2007, 140, 1–6 Search PubMed.
- T. Furukawa, A. Jikumaru, T. Ueno and K. Sei, Water, 2017, 9, 1–9 Search PubMed.
- L. Shen, T. M. Griffith, P. O. Nyangaresi, Y. Qin, X. Pang, G. Chen, M. Li, Y. Lu and B. Zhang, J. Hazard. Mater., 2020, 386, 121968 Search PubMed.
- W. Lin, S. Li, S. Zhang and X. Yu, Water Res., 2016, 91, 331–338 CrossRef CAS.
- M. T. Guo, Q. Bin Yuan and J. Yang, Chemosphere, 2013, 93, 2864–2868 Search PubMed.
- R. P. Sinha and D. P. Häder, Photochem. Photobiol. Sci., 2002, 1, 225–236 Search PubMed.
- M. Oliveira, P. Truchado, R. Cordero-García, M. I. Gil, M. A. Soler, A. Rancaño, F. García, A. Álvarez-Ordóñez and A. Allende, Antibiotics, 2023, 12(2), 400 CrossRef CAS.
- M. Munir, K. Wong and I. Xagoraraki, Water Res., 2011, 45, 681–693 Search PubMed.
- E. A. Auerbach, E. E. Seyfried and K. D. McMahon, Water Res., 2007, 41, 1143–1151 Search PubMed.
- M. T. Guo and C. Kong, Chemosphere, 2019, 224, 827–832 Search PubMed.
- C. W. McKinney and A. Pruden, Environ. Sci. Technol., 2012, 46, 13393–13400 CrossRef CAS PubMed.
- W. Ben, L. Liu, Y. Gao, Z. Sun, J. Li and Z. Qiang, ACS ES&T Water, 2024, 4(4), 1680–1687 Search PubMed.
- E. R. Blatchley, D. J. Brenner, H. Claus, T. E. Cowan, K. G. Linden, Y. Liu, T. Mao, S.-J. Park, P. J. Piper, R. M. Simons and D. H. Sliney, Crit. Rev. Environ. Sci. Technol., 2022, 1–21 Search PubMed.
- J. Xu and C. Huang, Environ. Sci. Technol. Lett., 2023, 10(6), 543–548 CrossRef CAS PubMed.
- D. Wang, T. Oppenländer, M. G. El-Din and J. R. Bolton, Photochem. Photobiol., 2010, 86, 176–181 Search PubMed.
- H. He, P. Zhou, K. K. Shimabuku, X. Fang, S. Li, Y. Lee and M. C. Dodd, Environ. Sci. Technol., 2019, 53, 2013–2026 CrossRef CAS PubMed.
- H. Ul Haq, W. Huang, Y. Li, T. Zhang, S. Ma, Y. Zhang, Y. Song, D. Lin and B. Tian, Biologia, 2022, 77, 1151–1160 CrossRef CAS.
- A. M. Earl, R. Losick and R. Kolter, Trends Microbiol., 2008, 16, 269–275 CrossRef CAS PubMed.
- N. Fluman and E. Bibi, Biochim. Biophys. Acta, Proteins Proteomics, 2009, 1794, 738–747 Search PubMed.
- N. M. Hull, A. L. Reens, C. E. Robertson, L. F. Stanish, J. K. Harris, M. J. Stevens, D. N. Frank, C. Kotter and N. R. Pace, Water Res., 2015, 69, 318–327 CrossRef CAS.
- J. R. Bolton and K. G. Linden, J. Environ. Eng., 2003, 129, 209–215 CrossRef CAS.
- K. G. Linden and J. L. Darby, J. Environ. Eng., 1997, 123, 1142–1149 CrossRef CAS.
- D. Ma and N. Hull, Polychromatic UV Fluence (Dose) Response Determination V.4, protocols.io, 2024, DOI:10.17504/protocols.io.n92ldzqoov5b/v4.
- K. Sholtes and K. G. Linden, Water Res., 2019, 165, 114965 CrossRef CAS PubMed.
- S. E. Beck, H. Ryu, L. A. Boczek, J. L. Cashdollar, K. M. Jeanis, J. S. Rosenblum, O. R. Lawal and K. G. Linden, Water Res., 2017, 109, 207–216 CrossRef CAS PubMed.
- N. M. Hull and K. G. Linden, Water Res., 2018, 143, 292–300 CrossRef CAS PubMed.
- EPA, Environmental Protection, 2006, 2, 1–436 Search PubMed.
- W. Taylor, E. Camilleri, D. L. Craft, G. Korza, M. R. Granados, J. Peterson, R. Szczpaniak, S. K. Weller, R. Moeller, T. Douki, W. W. K. Mok and P. Setlow, Appl. Environ. Microbiol., 2020, 86, e03039-19 CrossRef PubMed.
- K. Narita, K. Asano, K. Naito, H. Ohashi, M. Sasaki, Y. Morimoto, T. Igarashi and A. Nakane, J. Hosp. Infect., 2020, 105, 459–467 CrossRef PubMed.
- T. Li, Y. Zhang, J. Gan, X. Yu and L. Wang, J. Hazard. Mater., 2023, 452, 131292 CrossRef CAS PubMed.
- A. C. Eischeid and K. G. Linden, Appl. Environ. Microbiol., 2011, 77, 1145–1147 CrossRef CAS PubMed.
- S. E. Beck, R. A. Rodriguez, K. G. Linden, T. M. Hargy, T. C. Larason and H. B. Wright, Environ. Sci. Technol., 2014, 48, 591–598 CrossRef CAS PubMed.
- R. S. N. Tavares, D. Adamoski, A. Girasole, E. N. Lima, A. da Silva Justo-Junior, R. Domingues, A. C. C. Silveira, R. E. Marques, M. de Carvalho, A. L. B. Ambrosio, A. F. P. Leme and S. M. G. Dias, J. Photochem. Photobiol., B, 2023, 243, 112713 CrossRef CAS PubMed.
- Z. Jing, Z. Lu, D. Santoro, Z. Zhao, Y. Huang, Y. Ke, X. Wang and W. Sun, Chem. Eng. J., 2022, 447, 137584 CrossRef CAS.
- B. Ma, K. Bright, L. Ikner, C. Ley, S. Seyedi, C. P. Gerba, M. D. Sobsey, P. Piper and K. G. Linden, Photochem. Photobiol., 2022, 99, 975–982 CrossRef PubMed.
- Z. Yang, P. Liu, H. Wei, H. Li, J. Li, X. Qiu, R. Ding and X. Guo, Sci. Total Environ., 2022, 825, 153918 CrossRef CAS PubMed.
- R. Destiani and M. R. Templeton, AIMS Environ. Sci., 2019, 6, 222–241 CAS.
- M. F. Dias, G. da Rocha Fernandes, M. Cristina de Paiva, A. Christina de Matos Salim, A. B. Santos and A. M. Amaral Nascimento, Water Res., 2020, 174, 115630 CrossRef CAS PubMed.
- A. C. Lorenzo-Leal, W. Tam, A. Kheyrandish, M. Mohseni and H. Bach, BioMed Res. Int., 2023, 2023, 1–8 Search PubMed.
- M. Nihemaiti, Y. Yoon, H. He, M. C. Dodd, J. P. Croué and Y. Lee, Water Res., 2020, 182, 1–11 CrossRef PubMed.
- Y. Yoon, H. He, M. C. Dodd and Y. Lee, Water Res., 2021, 202, 117408 CrossRef CAS PubMed.
- Y. Yoon, M. C. Dodd and Y. Lee, Environ. Sci.: Water Res. Technol., 2018, 4, 1239–1251 RSC.
- M. Umar, Int. J. Environ. Res. Public Health, 2022, 19(3), 1636 CrossRef CAS PubMed.
- X. Meng, F. Li, L. Yi, M. Y. Dieketseng, X. Wang, L. Zhou and G. Zheng, J. Hazard. Mater., 2022, 430, 128502 CrossRef CAS PubMed.
- R. T. Robinson, N. Mahfooz, O. Rosas-Mejia, Y. Liu and N. M. Hull, Sci. Rep., 2022, 12, 1–10 CrossRef PubMed.
- M. Pazda, J. Kumirska, P. Stepnowski and E. Mulkiewicz, Sci. Total Environ., 2019, 697, 134023 CrossRef CAS PubMed.
- N. F. Gray, Ultraviolet Disinfection, Elsevier, 2nd edn, 2013 Search PubMed.
- Z. Qiao, Y. Ye, P. H. Chang, D. Thirunarayanan and K. R. Wigginton, Environ. Sci. Technol., 2018, 52, 10408–10415 CrossRef CAS PubMed.
- Y. Yoon, H. J. Chung, D. Y. Wen Di, M. C. Dodd, H. G. Hur and Y. Lee, Water Res., 2017, 123, 783–793 CrossRef CAS PubMed.
- Y. Choi, H. He, M. C. Dodd and Y. Lee, Environ. Sci. Technol., 2021, 55, 2541–2552 CrossRef CAS PubMed.
- D. Das, A. Bordoloi, M. P. Achary, D. J. Caldwell and R. P. S. Suri, Sci. Total Environ., 2022, 833, 155205 CrossRef CAS PubMed.
- P. H. Chang, B. Juhrend, T. M. Olson, C. F. Marrs and K. R. Wigginton, Environ. Sci. Technol., 2017, 51, 6185–6192 CrossRef CAS PubMed.
- D. Li, Z. Feng, B. Zhou, H. Chen and R. Yuan, Sci. Total Environ., 2022, 844, 157162 CrossRef CAS PubMed.
- M. Kneissl, III-Nitride Ultraviolet Emitters: Technology and Applications, 2016, vol. 227 Search PubMed.
- V. Martino, K. Ochsner, P. Peters, D. H. Zitomer and B. K. Mayer, Environ. Eng. Sci., 2021, 38, 458–468 CrossRef CAS.
- S. Rattanakul and K. Oguma, Water Res., 2018, 130, 31–37 CrossRef PubMed.
- Q. Wan, G. Wen, R. Cao, X. Xu, H. Zhao, K. Li, J. Wang and T. Huang, Water Res., 2020, 173, 115553 CrossRef CAS PubMed.
- P. Gikas, J. Environ. Manage., 2017, 203, 621–629 CrossRef CAS PubMed.
- K. M. S. Hansen and H. R. Andersen, Int. J. Photoenergy, 2012, 2012, 1–9 CrossRef.
- A. C. Mecha, M. S. Onyango, A. Ochieng and M. N. B. Momba, Chemosphere, 2017, 186, 669–676 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ew00530a |
| This journal is © The Royal Society of Chemistry 2025 |