Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Antibiotic resistance response of activated sludge to sulfamethoxazole: insights from the intracellular and extracellular DNA fractions†
M.
Martínez-Quintela
 *ab,
D.
Calderón-Franco
*ab,
D.
Calderón-Franco
 b,
M. C. M.
van Loosdrecht
b,
M. C. M.
van Loosdrecht
 b,
S.
Suárez
a,
F.
Omil
a and
D. G.
Weissbrodt
b,
S.
Suárez
a,
F.
Omil
a and
D. G.
Weissbrodt
 bc
bc
aCRETUS, Department of Chemical Engineering, Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Galicia, Spain. E-mail: miguel.martinez1b@gmail.com
bDepartment of Biotechnology, Delft University of Technology, Delft, The Netherlands
cDepartment of Biotechnology and Food Science, Norwegian University of Science and Technology, Trondheim, Norway
First published on 1st May 2024
Abstract
In activated sludge, the antibiotic resistance genes (ARGs) can be present either in the intracellular (iDNA) or extracellular DNA fraction (exDNA). Recent advances in the exDNA extraction methodology allow a better profiling of the pool of ARGs. However, little is known about how stress conditions modify the distribution of ARGs between both DNA fractions. Here, we performed two batch tests for analyzing the effects of two different stress conditions, namely nutrient starvation and high concentrations of sulfamethoxazole (1, 10 and 150 mg L−1) in activated sludge. We tracked by qPCR the resulting relative abundances of four target genes, namely the universal 16S rRNA gene, the class 1 integron-integrase gene intI1, and the sulfonamide resistance genes sul1 and sul2 in both the iDNA and exDNA fractions. In the exDNA pool, unlike starvation, which provoked a decrease of 1–2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA in the concentration of sul1 and intI1, the presence of sulfamethoxazole did not influence the abundances of sul1 and sul2. However, high concentrations of sulfamethoxazole (150 mg L−1) selected for microorganisms harboring sul1 and, more remarkably, sul2 genes in their iDNA during their exponential growth phase. The abundances of intI1 and sul1 were positively correlated in the exDNA fraction (r > 0.7), whereas no significant correlation (p < 0.05) between the abundance of these two genes was found in the iDNA fraction of the sludge. High SMX concentrations influenced the abundance of ARGs in the iDNA; their abundance in the exDNA was influenced by nutrient limitations. Further studies should consider the profiling of exDNA fractions because of the relationship between ARGs and mobile genetic elements. Besides, the surveillance of antimicrobial resistance is encouraged in wastewater treatment plants facing high antibiotic concentrations.
log10 [copies] per ng DNA in the concentration of sul1 and intI1, the presence of sulfamethoxazole did not influence the abundances of sul1 and sul2. However, high concentrations of sulfamethoxazole (150 mg L−1) selected for microorganisms harboring sul1 and, more remarkably, sul2 genes in their iDNA during their exponential growth phase. The abundances of intI1 and sul1 were positively correlated in the exDNA fraction (r > 0.7), whereas no significant correlation (p < 0.05) between the abundance of these two genes was found in the iDNA fraction of the sludge. High SMX concentrations influenced the abundance of ARGs in the iDNA; their abundance in the exDNA was influenced by nutrient limitations. Further studies should consider the profiling of exDNA fractions because of the relationship between ARGs and mobile genetic elements. Besides, the surveillance of antimicrobial resistance is encouraged in wastewater treatment plants facing high antibiotic concentrations.
Water impactThe extracellular DNA is commonly overlooked when assessing antibiotic resistance in wastewater treatments. This study assesses the distribution of the antibiotic resistance genes in both the intracellular and extracellular DNA fractions when the microbial population is exposed to high concentrations of antibiotics. Results will help to establish surveillance campaigns for better assessing the antibiotic resistance in WWTPs. |
1. Introduction
Antibiotic consumption has greatly increased since 2000 worldwide, especially for human health purposes.1,2 Among them, sulfamethoxazole (SMX) is one of the most consumed antibiotics both by humans and animals.3 SMX inhibits the folic acid biosynthesis, hindering the bacterial growth.4 Its presence in wastewater is very well documented,5 and it has been included in the 2020 update of the EU Watch List as a concerning emerging contaminant. The overuse of antibiotics is considered the main driver of antibiotic resistance (AR), one of the major threats to human health in the current century.The problem is not restricted to clinical settings since bacteria can move freely between human, animal, and natural environments. These compartments must be comprehensively taken into account when evaluating AR, following the One-Health approach.6 Wastewater treatment plants (WWTPs) should act as barriers to limit the spread of both, antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs), to the environment.7–9 WWTP effluents, and wastewater catchment areas, are currently considered as one of the major release points of ARB and ARGs to the environment.10 Microorganisms present in WWTPs are considered as reservoirs of ARB since the mixture of animal and environmental bacteria coupled with the presence of trace concentrations of chemical stressors like antibiotics or heavy metals, creates a perfect environment for AR development and horizontal gene transfer (HGT).11–13 Although some studies have confirmed the capability of WWTPs to decrease the concentration of ARB and ARGs between influent and effluent,7,14–16 others have reported a higher number of copies of some ARGs in WWTP effluents.17,18 Hultman et al. (2018)19 have detected some bacterial families in the WWTP effluent harboring one ARG (tetM) that was absent from the influent, suggesting possible HGT events in the activated sludge. The factors influencing the removal efficiency of ARB and ARGs, and HGT events are still unclear.
ARGs can be present either in the intracellular or extracellular DNA fractions. Extracellular DNA (exDNA) is commonly released from the intracellular DNA (iDNA) during cell lysis (passive release) or excreted from live cells (active release).20,21 Such exDNA can be present as a structural component in microbial aggregates, as part of the matrix of extracellular polymeric substances (EPS)22 or floating freely in the water body.23 Recent advances in methods for the extraction of exDNA allow for a deeper analysis of the composition and distribution of both DNA fractions in WWTPs and natural environments.24 The amount of exDNA detected in nutrient-rich wastewater environments is commonly of 1–2 orders of magnitude below iDNA.24,25 In contrast, a higher concentration (around 20%) of exDNA than iDNA has been detected in sediments in natural environments with nutrient scarcity.26 An increase (up to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 gene copies per mL) of some ARGs (ermB and sul1) in the exDNA pool has been detected across a WWTP process, eventually discharged in the effluent.8 The exDNA pool can therefore not be underestimated in AR surveillance in wastewater systems.
log10 gene copies per mL) of some ARGs (ermB and sul1) in the exDNA pool has been detected across a WWTP process, eventually discharged in the effluent.8 The exDNA pool can therefore not be underestimated in AR surveillance in wastewater systems.
Once DNA is released from the bacterial cell, it is susceptible to be degraded abiotically or due to the action of extracellular nucleases.27 Several studies have stated that exDNA could persist without losing its integrity in natural environments for hours or days if it is free-floating, or even years if it is bounded to soils or surfaces.27–30 Therefore, exDNA can also be considered a reservoir of ARGs and MGEs because of its high prevalence and stability.
Whether free or bound, exDNA can contribute to the spreading of ARGs via natural transformation in both pure and mixed cultures.31,32 Natural transformation is a mechanism of HGT in which bacterial cells in a competent state can take up, integrate, and functionally express genetic materials from foreign DNA.32,33 In the presence of stressors, this mechanism may be enhanced since some antibiotics and heavy metals can damage the cell membrane or DNA, creating the need to incorporate external DNA for reparation purposes.20 Antibiotics and heavy metals can also influence the ARG content in the iDNA fraction in high-density populations continuously exposed to them, such as those developing during biological treatments in WWTPs.34 ARG transfer via HGT processes between colonies of Escherichia coli has been detected after applying 10 μg L−1 of tetracycline.35 Moreover, a list of antibiotics that may promote ARG selection processes in WWTPs in the range of 0.008 to 64 μg L−1 has been reported.36 The relationship between the distribution of the sul1 and sul2 genes, two of the most targeted ARGs related with sulfonamide resistance in WWTPs,7,8,16,17 and the presence of SMX has not been previously assessed. The concentration of certain ARGs in both the iDNA and exDNA fractions can increase during the wastewater treatment process, suggesting HGT events through the WWTP.8,19 It is important to identify which WWTP conditions induce a shift in the ARB hosts and possible ARB reservoirs in order to delineate measures to reduce the dissemination of AR in the environment.
In this work, we evaluated the distribution of sulfonamide resistance genes (sul1 and sul2), widely reported in wastewater, and the class 1 integron-integrase (intI1) genetic marker in both iDNA and exDNA fractions of activated sludge after a short-term exposure to sulfamethoxazole. Additionally, we aimed to elucidate resistance selection processes using the same target genes in activated sludge under different sulfamethoxazole concentrations. To support the experimental investigations, we used mathematical models to examine the effect and biodegradation of SMX in the activated sludge.
2. Materials and methods
2.1. Experimental setup
The inoculum for both experiments was obtained from the activated sludge of a WWTP located in Amsterdam West, The Netherlands. Each experiment was performed in 1 L Erlenmeyer flasks with 0.4 L of working volume at room temperature (~20 °C). Before inoculation, the biomass was filtered and washed to remove small particles. To ensure proper air transfer, flasks were placed in an incubator (Edmund Bühler) with a 120–130 rpm stirring rate. In each experiment, pH was manually controlled daily between 6.8 (initial) and 8.5 by addition of HCl at 1 mol L−1, to maintain the bacterial activity. A schematic representation of the experimental design is depicted in Fig. 1.Incubations and sampling points. Two different incubations were set up: one control (no antibiotic) and one supplied with SMX at an initial concentration of 1 mg L−1 in the mixed liquor. For each incubation, 3 sampling points were considered (1, 3, and 7 days) in duplicate, which implied the preparation of six flasks per incubation (total of 12 flasks). At each time point, the mixed liquor content of the whole flask (0.4 L) was used for performing the DNA extractions. Two additional flasks were prepared to measure the initial conditions (day 0) shared in both incubations.
Growth conditions and substrates pulses. In each flask, 0.4 L of activated sludge was inoculated. The initial concentration of volatile suspended solids (VSS) was about 4.40 g VSS L−1. Flasks were continuously stirred through orbital shaking in an Edmund Bühler GmbH (K5-15) incubator to ensure a proper air transfer to maintain aerobic conditions. To maintain the biomass activity, sodium acetate (initial concentration in flask at 1.2 g COD L−1) and ammonium chloride (130 mg N-NH4+ L−1) were pulsed to each flask daily until day 3 (included). Then, the supply of substrates to the microbial cultures was stopped to analyze the effect of nutrient starvation and cell lysis over the last 4 days of the experimental periods.
Antibiotic spikes. A stock solution of 7.8 g L−1 of SMX was prepared in methanol. 51 μL of this stock solution were spiked daily to obtain a SMX concentration of 1 mg L−1 until day 3 (included). A high concentration of antibiotic was used in comparison with those found in wastewater to enhance the antibiotic effect in the biomass. After day 3, antibiotic pulses were stopped to maintain starvation conditions.
Chemical and biomolecular analyses. Conventional parameters (chemical oxygen demand – COD, ammonium, nitrite) were analyzed at four different time points (0, 4, 8, and 24 h) during the first day, to check if the biomass was active. The concentration of total suspended solids (TSS) and VSS were analyzed on days 0 and 7 to measure biomass growth. The concentration of SMX was analyzed at the beginning of the experiment and before the addition of a new antibiotic pulse. The ARG content of both iDNA and exDNA fractions and the antibiotic concentration were analyzed at each time point (0, 1, 3 and 7 d) by qPCR and liquid chromatography, respectively, following the analytical methods described below.
Biomass acclimation. Initially, flasks were inoculated with 1.4 g VSS L−1 of activated sludge biomass, which was about 4-times lower concentration than in the first experiment in order to monitor the effect of the antibiotic spiked during the exponential growth phase of the biomass. During the first week, the biomass was only fed with nutrients without antibiotic to acclimate it to a synthetic medium composed of easily biodegradable carbon, nitrogen, and phosphorus sources. To stimulate biomass growth, nutrients were provided in each flask in a single pulse containing a high concentration of sodium acetate (3 g COD L−1) and non-limiting concentrations of ammonium chloride (400 mg N-NH4+ L−1) and dihydrogen potassium phosphate (40 mg P-PO43− L−1). This resulted in a better nutrient balance (100 g COD
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 g N-NH4+
13 g N-NH4+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.3 g P-PO43−) when compared to the conventional stoichiometric COD
1.3 g P-PO43−) when compared to the conventional stoichiometric COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N
N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio of nutrients required for activated sludge (100 g COD
P ratio of nutrients required for activated sludge (100 g COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5–10 g N
5–10 g N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 g P).37 The ammonium nitrogen was therefore not limiting the growth of ordinary heterotrophic organisms (OHOs) in this case.
1 g P).37 The ammonium nitrogen was therefore not limiting the growth of ordinary heterotrophic organisms (OHOs) in this case.
Antibiotic supply. Once the nutrients were depleted, biomass was washed three times with tap water to renew the medium, and the antibiotic exposure was applied. For this purpose, 0.5 g VSS L−1 of the pre-acclimatized biomass was used and fed with the same nutrient pulse as for the biomass cultivation period. A control (no antibiotic supply) and two different levels of antibiotic exposure (high enough to provoke any effect on the microbial community) were considered: 10 mg L−1 and 150 mg L−1. The spike was done only at the start of the experiment.
Chemical and biomolecular analyses. Conventional parameters (COD, ammonium and nitrite) and pH were followed at different times points (0, 24, 48, 72 h) during both the biomass cultivation and the antibiotic exposure periods. Biomass growth and SMX concentration were also checked daily during the first three days. Mixed liquor samples were collected each day to monitor the evolution of the targeted ARGs in the iDNA fraction.
2.2. iDNA and exDNA extraction
iDNA was extracted from 1–2 mL of mixed liquor from each flask. After centrifugation, a dry pellet of 0.10–0.25 g wet weight was obtained. The extraction of iDNA was performed using the NucleoSpin® PowerSoil kit (Macherey-Nagel, USA) according to the manufacturer's instructions. Each extraction of iDNA was normalized using 0.25 g of wet weight from each biomass pellet.To retrieve exDNA, a sufficient volume (0.4 L) of mixed liquor was centrifugated and filtered through a PVDF 0.2 μm membrane filter. The sample volume for exDNA was first optimized to obtain a sufficient yield for performing qPCR analyses. The optimization results are described in section 3.1. The filtrate was loaded on a positively charged 1 mL diethylaminoethyl cellulose (DEAE-C) anion-exchange chromatographic column (BIA Separations, Slovenia). A liquid chromatography pump (Shimadzu, LC-8A) was used to load the sample into the column at a flow rate of 3 mL min−1, after equilibration of the column (procedure described in Table 1).
| Buffers and solutions | ||||
|---|---|---|---|---|
| Equilibrium buffer | Elution buffer | Regenerating buffer | Cleaning solution | Storage solution |
| - Tris 50 mmol L−1 | - Tris 50 mmol L−1 | - Tris 50 mmol L−1 | - NaOH 1 mol L−1 | - 20% EtOH |
| - EDTA 10 mmol L−1 | - EDTA 10 mmol L−1 | - EDTA 10 mmol L−1 | - NaCl 2 mol L−1 | |
| - pH: 7.2 | - NaCl 1.5 mol L−1 | - NaCl 2 mol L−1 | ||
| - pH: 7.2 | - pH: 7.2 | |||
| Procedures | ||||
|---|---|---|---|---|
| Equilibration | Regeneration | Cleaning in place | Sanitization | Storage |
| - 10 CV of equilibrium buffer | - 20 CV of regenerating buffer | - 10 CV of MilliQ water | - 10 CV of cleaning solution | - 10 CV of equilibrium buffer |
| - 10 CV of elution buffer | - 20 CV of equilibrium buffer | - 10 CV of cleaning solution | - Maintain for 2–12 hours | - 10 CV of MilliQ water |
| - 10 CV equilibrium buffer | - 10 CV of MilliQ water | - 5 CV of cleaning solution | - 20 CV of storage solution | |
| - 10 CV of equilibrium buffer | - 10 CV of MilliQ water | |||
| - 10 CV of MilliQ water | −10 CV of equilibrium buffer | |||
| −10 CV of MilliQ water | ||||
After all the sample was loaded, the exDNA retained in the column was eluted during 20–30 min with an elution buffer (Table 1). Then, a volume of absolute ice-cold ethanol of 2–2.5 times the eluted volume was added to precipitate the DNA pellet. The supernatant was removed, and the pellet was washed two times with 70% ethanol, and the pellet was finally left to air dry. To improve the purity of the exDNA extract, the pellet was incubated with proteinase K for 2 h to remove residual proteins. The protein-digested pellet was finally purified using a GeneJET NGS Cleanup Kit (Thermo Scientific, USA). After applying the Cleanup Kit, 0.1 mL of volume with the purified exDNA was obtained.
Buffers and procedures needed for column equilibration, regeneration, cleaning, and storage are described in Table 1. The equilibration procedure is required to prepare the column before the sample load. The regeneration procedure is performed after the elution of the sample. Cleaning and sanitization are washing procedures. The storage procedure is used to preserve the column integrity after the workout. This method has been previously described in Calderón-Franco et al. (2021).24
2.3. Analytical methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60, v/v) at pH 2.2 was used as the mobile phase. The injection volume was 100 μL, the column operated at 30 °C, and the detector worked at 275 nm to detect SMX. The limit of quantification of the method was 50 μg SMX L−1.
60, v/v) at pH 2.2 was used as the mobile phase. The injection volume was 100 μL, the column operated at 30 °C, and the detector worked at 275 nm to detect SMX. The limit of quantification of the method was 50 μg SMX L−1.
All qPCRs were performed in a qTOWER3 Real-time PCR machine (Westburg, DE). qPCRs were conducted in 20 μL of reaction mixtures comprising: 10 μL of IQTM SYBR Green Supermix (Bio-Rad), 2 μL of DNA sample, 0.2 μL of forward primer, 0.2 μL of reverse primer (both at 10 μmol L−1) and 7.6 μL of molecular grade water (Sigma Aldrich, USA). Three qPCR technical replicates were prepared for each sample. PCR conditions consisted of an initial denaturation at 95 °C for 10 min followed by 40 cycles of DNA template denaturation for 15 s at 95 °C, primer annealing for 30 s at 60 °C for intI1 and sul1, 55 °C for 16S rRNA and 61 °C for sul2 genes, and primer extension at 72 °C for 10 s. A technical duplicate of at least 6 dilution series of synthetic DNA fragments (IDT, USA) (Table S1†) containing the studied genes was included in each qPCR analysis to create the standard curve. Forward and reverse primers are described in Table S2.†
2.4. Statistical analysis
To determine differences in the presence and abundance of ARGs in the microbiological samples, statistical tests were performed using the R software 4.1.0 and RStudio (https://www.rstudio.com/) at 95% confidence level (p < 0.05). One-way ANOVA analyses were performed first to analyze significant differences among the samples. A post hoc Holm correction analysis was performed to adjust the p-value and determine which specific samples were significantly different. A Kruskal–Wallis test was applied when the normality assumption for applying the one-way ANOVA analysis was not satisfied.2.5. Mathematical modelling simulations of SMX biodegradation and growth inhibition
| ρgrowth,OHO = μmax,OHO·SAc/(KAc + SAc)·KMIC,SMX/(KMIC,SMX + SSMX)·XOHO | (1) |
| ρdecay,OHO = bOHO·XOHO | (2) |
| ρbiodegradation,SMX = kbiol,SMX·SSMXn·XOHO (with n = 1) | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 m/m), COD-limiting (100
7 m/m), COD-limiting (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 m/m) and nitrogen-limiting (100
20 m/m) and nitrogen-limiting (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 m/m) COD
1 m/m) COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratios, exposing low-concentrated biomasses (0.250 g VSS L−1) to an antibiotic concentration of 150 mg SMX L−1. It was assumed that SMX exerted the same bacteriostatic effect on the different microbial guilds.
N ratios, exposing low-concentrated biomasses (0.250 g VSS L−1) to an antibiotic concentration of 150 mg SMX L−1. It was assumed that SMX exerted the same bacteriostatic effect on the different microbial guilds.
3. Results and discussion
3.1. Validation of the exDNA extraction method at different volumes and concentrations of mixed liquor
The exDNA extraction method was first tested to evaluate its sensibility to different sample volumes or biomass concentrations. Calderón-Franco et al. (2021)24 have shown that the anion-exchange chromatographic method using a diethylaminoethyl cellulose (DEAE-C) anion-exchange column can help recover more than 4 μg of exDNA from WWTP influent, effluent, and activated sludge using at least 1 L of sample. Here, a lower amount of sample volume was tested (0.25 and 0.5 L) as well as two different raw (∼3.5 gVSS L−1) and diluted (∼1 gVSS L−1) concentrations of activated sludge. Results on the retrieved exDNA concentrations and yields are depicted in Table 2.| Initial volume of activated sludge | Raw sludge | Diluted sludge | |
|---|---|---|---|
| exDNA concentration after extraction (ng μL−1) | 0.25 L | 100.00 | n.m. |
| 0.5 L | 104.00 | 45.20 | |
| 1 L | 92.10 | 104.00 | |
| exDNA yield (μg) | 0.25 L | 5.00 | n.m. |
| 0.5 L | 10.40 | 2.26 | |
| 1 L | 18.42 | 5.20 |
As expected, the amount of exDNA obtained in each extraction decreased depending on the initial sample volume and biomass concentration. The amount of exDNA extracted was proportional to the amount of biomass in the sample (R2: 0.99): the yield (5.2 μg of DNA) obtained for 1 L of diluted sludge (i.e., from ∼1 gVSS of biomass) was similar to the one obtained for 0.25 L of raw sludge (5.0 μg of DNA) (i.e., from ∼0.88 gVSS of biomass). In all volumes and concentrations tested, a sufficient amount of exDNA was extracted. Such amounts (>1 μg DNA) allowed further downstream analysis of the pool of exDNA by qPCR, and can also be used for metagenomics analysis of the resistome (not conducted here). With these results, we decided to perform the following experiments with 0.4 L of activated sludge.24
3.2. ARGs content was influenced by SMX in the iDNA and by starvation in the exDNA
The first batch experiment was run in flasks containing 4.4 gVSS L−1 of activated sludge with pulse addition of acetate-based COD (at 1.2 g COD L−1) and ammonium (at 130 mg N-NH4+ L−1) as primary substrates.The consumptions of COD and NH4+ were followed on the first day of the experiment. Ammonium was consumed entirely after 1 d, but the soluble COD concentration remained at 200 mg L−1. Maybe the lack of phosphate or other trace mineral component limited the complete COD consumption. A volumetric maximum rate of COD consumption of 130 mg COD L−1 h−1 was measured in the first 8 h of the batch. Considering a theoretical COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N
N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio of 100
P ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (m/m/m) for a balanced growth of ordinary heterotrophic organisms (OHOs),37 around 50 mg N-NH4+ would have been consumed per 1 g of CODS of acetate. The conversion of ammonium could be due to both assimilation into OHO biomass and nitrification. The biomass concentration slightly increased from 4.4 gVSS L−1 in the inoculum to ∼4.9 gVSS L−1 on day 7 in all experiments.
1 (m/m/m) for a balanced growth of ordinary heterotrophic organisms (OHOs),37 around 50 mg N-NH4+ would have been consumed per 1 g of CODS of acetate. The conversion of ammonium could be due to both assimilation into OHO biomass and nitrification. The biomass concentration slightly increased from 4.4 gVSS L−1 in the inoculum to ∼4.9 gVSS L−1 on day 7 in all experiments.
SMX was spiked at an initial concentration of 1 mg L−1 in the mixed liquor every day until day 3. Before the addition of a new pulse, the concentrations of SMX were always below the limit of quantification of the method (<50 μg L−1). According to the consumption of the conventional parameters (COD and N), which followed the same consumption rate between the control and the SMX exposed flasks, the bacterial activity was not inhibited due to the antibiotic presence. The antibiotic measurements indicated a daily consumption of SMX, suggesting the biotransformation of the compound. Biodegradation rather than sorption of SMX has been previously reported.42 Aquasim simulations were performed with the conditions of the experiment and the parameters explained in section 2.1.1. A total consumption of SMX was predicted after 20 h of batch.
The amount of exDNA extracted varied significantly (p < 0.05) throughout the experiment. Less exDNA was recovered on day 7 (3.95 ± 0.82 ng DNA mL−1) than on day 0 (13.53 ± 0.86 ng DNA mL−1). We hypothesize that the prolonged starvation conditions during the batch (where the water matrix was intentionally not renewed over the 7 days) led to the degradation of the residual exDNA or the starved biomass took up the exDNA as carbon source. DNA degradation can be caused by nucleases present in the sludge or released during cell lysis.27 In addition, bacterial inactivity could have resulted in lesser release of exDNA.43 Substantial analytical efforts will be needed in future for addressing the different mechanisms of exDNA release, uptake and degradation in microbial cultures.
Distributions of 16S rRNA, sul1, sul2, and intI1 genes in both iDNA and exDNA fractions of the first set of batch experiments under control and SMX treatment conditions are depicted in Fig. 2. Attending to the general trends in both incubations (control and SMX exposed flasks), very few differences in the abundance of the 16S rRNA gene were detected between days 0 and 7. This matched with the relatively low variation in biomass measured over the batch, which was inoculated at a relatively high concentration like an activated sludge tank. However, the rest of the targeted genes experienced a slight increase (∼0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [gene copies] per ng DNA) in the number of copies in the iDNA fraction between these days.
log10 [gene copies] per ng DNA) in the number of copies in the iDNA fraction between these days.
Focusing on the differences between the SMX exposed flasks and the control flasks, the number of copies of sul1 and sul2 in the iDNA fraction in the flasks exposed to SMX after 7 days was significantly higher (p < 0.005) than on the starting day. This fact suggests a possible selection of microorganisms carrying those ARGs in their iDNA in the presence of the antibiotic. No significant differences (p < 0.05) were found in the case of intI1 between the different incubations.
In the exDNA fraction, the 16S rRNA gene barely varied from 5.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [gene copies] per ng DNA during these 7 days. There was a non-significant increase from 5.5 to 6
log10 [gene copies] per ng DNA during these 7 days. There was a non-significant increase from 5.5 to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [gene copies] per ng DNA on day 1 in the control flask, but the number of copies progressively dropped during the following days back to the initial value on day 7. The ANOVA did not show any significant difference (p < 0.05) in the 16S rRNA gene copies in neither the control nor the SMX-exposed flasks.
log10 [gene copies] per ng DNA on day 1 in the control flask, but the number of copies progressively dropped during the following days back to the initial value on day 7. The ANOVA did not show any significant difference (p < 0.05) in the 16S rRNA gene copies in neither the control nor the SMX-exposed flasks.
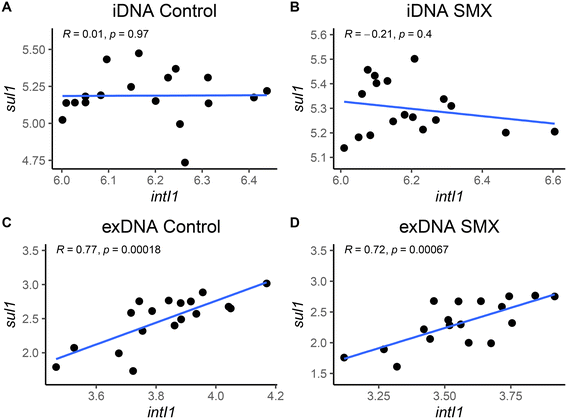
The intI1 and sul1 genes exhibited similar profiles in the exDNA fraction of both the control and SMX treatment incubations: a progressive drop in their number of copies per ng of DNA was detected throughout the experimental period. A correlation between the number of copies per ng of DNA of each gene (intI1 and sul1) were performed in both DNA fractions (Fig. 3). The gene concentrations correlated better in the exDNA fraction (r > 0.7) than in the iDNA fraction (r < 0.01), independent of the stress condition applied (either starvation or antibiotic exposure). It has been previously reported that class 1 integrons contain sul1 in their gene cassettes.39 The correlation between the abundances of intI1 and sul1 genes on exDNA suggest that most of sul1 genes present in this fraction were embedded in class 1 integrons. The experimental profiles highlighted significant differences among all days (p < 0.005) for the intI1 gene and sul1 gene in the SMX-exposed flasks, especially between days 3 and 7. sul1 was below the limit of detection on the last day. The number of copies of sul2 in the exDNA fraction increased (>1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 ng−1 DNA) significantly (p < 0.005) from day 1 to day 7 to in the control flasks. In the SMX-exposed flasks, the significant (p < 0.05) increase in sul2 occurred between day 0 and 1, but in the following days the concentration was similar to the concentration of day 1 (∼1.75
log10 ng−1 DNA) significantly (p < 0.005) from day 1 to day 7 to in the control flasks. In the SMX-exposed flasks, the significant (p < 0.05) increase in sul2 occurred between day 0 and 1, but in the following days the concentration was similar to the concentration of day 1 (∼1.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 gene copies per ng DNA).
log10 gene copies per ng DNA).
The DNA release was not affected by the presence of the antibiotic but likely by the loss of bacterial activity. The exposure to 1 mg L−1 of sulfamethoxazole did neither significantly impact the bacterial activity nor the presence of sul1 and sul2 genes in exDNA. However, the presence of SMX selected for microorganisms carrying sul1 and sul2 in their iDNA over the non-resistant ones. As follow-up, we performed a second experiment to examine the ARGs concentration during the exponential phase of the biomass growth and focusing only on the iDNA fraction, since no significant differences were detected in the exDNA for the SMX resistance genes. The results are reported in the section 3.3 hereafter.
3.3. High SMX concentrations can select for antibiotic-resistant bacteria in activated sludge
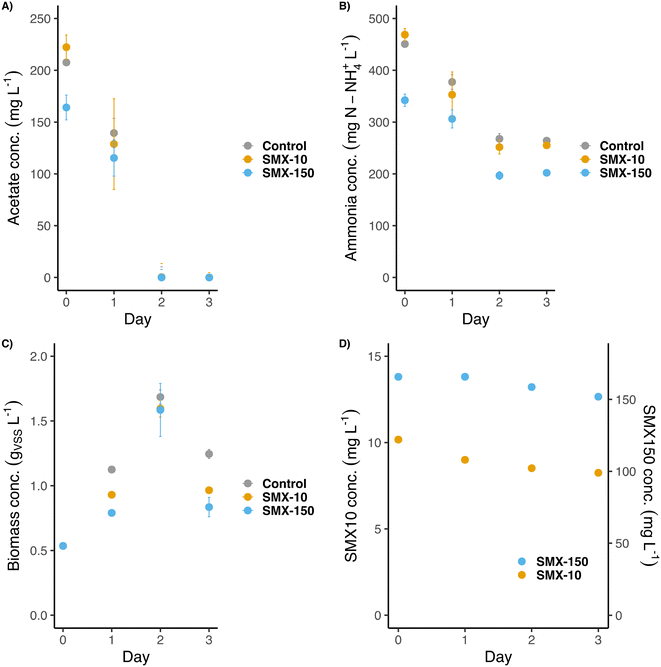
The second batch experiment was inoculated at a 9-times lower concentration of previously acclimated biomass (0.5 gVSS L−1) for promoting growth and a selective enrichment during the experiment. Nutrients were supplied as one single initial pulse at a high concentration of acetate (3 g COD L−1) and non-limiting concentrations of ammonium (400 mg N-NH4+ L−1) and phosphate (40 mg P-PO43− L−1). The aim was to provide enough nutrients to promote the biomass growth over several days. Phosphate was also added here to avoid growth limitations. Nutrients were consumed progressively during the first two days. Acetate was set as the limiting compound (Fig. 4A). The high concentrations of SMX applied to the flasks (10 and 150 mg L−1), i.e., 10 to 150 times higher than in the previous experiment (1 mg L−1), did not affect the microbial activity. After 2 days, acetate was fully consumed, indicating that maybe the lack of phosphate was a limiting compound for the COD removal in the previous experiment. On day 2 growth and ammonium consumption also stopped (around 150 mg N-NH4+ L−1 were consumed in each flask). Likely, nitrifiers were more sensitive to the antibiotic than OHOs, as it was previously reported,44,45 since only traces of nitrite were detected (<1 mg N-NO2− L−1) in the SMX exposed flasks during the experiment. A slight increase in the ammonium concentration was detected between 48 h and 72 h (Fig. 4B), suggesting cell decay due to the lack of COD. Whereas Aquasim simulations (see section 3.4 hereafter) predicted a full biodegradation of SMX over 30–50 h under 1 and 10 mg L−1 of concentration (also considering an inhibition of growth by 50% under acute SMX treatment according to Katipoglu-Yazan et al. (2021)46), the measured concentrations of SMX only slightly decreased during the first 3 days, namely 10–20% of the initial concentrations. In the flasks exposed to 10 mg L−1 of SMX, its concentration decreased down to 8 mg L−1 on day 3. In the case of flasks exposed to 150 mg L−1 of SMX, its final concentration after 3 days was around 140 mg L−1. During these experiments, the SMX concentrations remained high over the experimental growth periods of 2 days (Fig. 4D).The biomass grew from its starting concentration (∼0.5 gVSS L−1) to a maximum of ∼1.65 gVSS L−1 on day 2 (Fig. 4C). Afterward, cell decay provoked a decrease in the biomass concentration to ∼0.9 gVSS L−1 on day 3. Although all flasks reached the same final biomass concentration, the growth rate patterns differed: flasks exposed to SMX grew slightly slower (0.011–0.016 gVSS L−1 h−1) than the control one (0.025 gVSS L−1 h−1). This suggested a partial inhibition of bacterial growth under antibiotic treatment. Still, the biomasses could quickly adapt to these antibiotic conditions and reach similar final concentrations as the non-exposed flasks.
According to the biomass dynamics, the qPCR analysis of the targeted genes was focused on the first 3 days (Fig. 5). The abundance of the 16S rRNA gene followed in each flask a similar trend as the biomass concentration (Fig. 5A). Since the biomass of the control flask grew initially faster, the concentration of this gene on day 1 was higher than in the two SMX treatment flasks. However, on day 2, the 16S rRNA gene copies of the three flasks were similar (∼7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA), matching with the almost equal biomass concentrations obtained in the control and SMX exposed batches.
log10 [copies] per ng DNA), matching with the almost equal biomass concentrations obtained in the control and SMX exposed batches.
The abundances of the intI1 and sul1 genes were very similar among the flasks (4.0–4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10[copies] per ng DNA) (Fig. 5B and C). However, they differed in their distribution over time. A slight significant (p < 0.05) increment was detected on day 2 in intI1 between the control and exposed flasks (from 4.0 to 4.5
log10[copies] per ng DNA) (Fig. 5B and C). However, they differed in their distribution over time. A slight significant (p < 0.05) increment was detected on day 2 in intI1 between the control and exposed flasks (from 4.0 to 4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA), but it was compensated on day 3. For sul1, the same significant (p < 0.05) difference between the control and exposed flasks was detected on day 2. On day 3 the concentration of sul1 in the control flasks decreased down to ∼4.25
log10 [copies] per ng DNA), but it was compensated on day 3. For sul1, the same significant (p < 0.05) difference between the control and exposed flasks was detected on day 2. On day 3 the concentration of sul1 in the control flasks decreased down to ∼4.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA. Unlike for the control and the SMX-10 flasks, the abundance of sul1 significantly differed (p < 0.005) between day 0 and day 3 in the SMX-150 flasks. The SMX treatments selected for microorganisms carrying sul1 and intI1 genes on their iDNA, i.e., potentially able to develop resistance and grow. Thus, the exposition to SMX resulted in an increased abundance of both sul1 and intI1 genes in the bacterial community.
log10 [copies] per ng DNA. Unlike for the control and the SMX-10 flasks, the abundance of sul1 significantly differed (p < 0.005) between day 0 and day 3 in the SMX-150 flasks. The SMX treatments selected for microorganisms carrying sul1 and intI1 genes on their iDNA, i.e., potentially able to develop resistance and grow. Thus, the exposition to SMX resulted in an increased abundance of both sul1 and intI1 genes in the bacterial community.
The sul2 concentration also differed depending on the presence of the antibiotic. In the control, the concentration barely varied from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA (initial concentration at day 0) (Fig. 5D). In both flasks exposed to SMX, the abundance of sul2 increased up to a value of 6.5
log10 [copies] per ng DNA (initial concentration at day 0) (Fig. 5D). In both flasks exposed to SMX, the abundance of sul2 increased up to a value of 6.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 [copies] per ng DNA on day 2 (p < 0.005). Such sul2 concentration was maintained even along the cell decay that occurred between days 2 and 3. The high concentrations of SMX selected for bacteria that significantly carried sul2. The selection of microorganisms harboring sul2 compared with those harboring sul1 is remarkable. Since their resistance mechanism is the same (target protection) (Rizzo et al., 2013),10 a possible explanation for the large selection of sul2 over sul1 could be related to their different location. The sul1 gene is usually embedded in class 1 integrons (also displayed by the aforementioned correlation in abundance profiles of sul1 and intI1 genes), whereas the sul2 gene is usually located in small plasmids.47 Thus, sul1 and sul2 are not necessary assembled in the same genetic cluster and can be present on different organisms independently, explaining the differences found between them. Besides, integrons are not considered mobile: they need to be inserted in other MGEs (like transposons or plasmids) to spread and replicate.39,48 Plasmids usually contain genes for their self-replication, but they are also present in several copies in the bacterial cells (multi-copy plasmids).49 Therefore, it is more probable that genes embedded in plasmids will proliferate faster than those embedded in integrons in a nutrient-rich bacterial community. Further experiments would be needed to confirm this hypothesis.
log10 [copies] per ng DNA on day 2 (p < 0.005). Such sul2 concentration was maintained even along the cell decay that occurred between days 2 and 3. The high concentrations of SMX selected for bacteria that significantly carried sul2. The selection of microorganisms harboring sul2 compared with those harboring sul1 is remarkable. Since their resistance mechanism is the same (target protection) (Rizzo et al., 2013),10 a possible explanation for the large selection of sul2 over sul1 could be related to their different location. The sul1 gene is usually embedded in class 1 integrons (also displayed by the aforementioned correlation in abundance profiles of sul1 and intI1 genes), whereas the sul2 gene is usually located in small plasmids.47 Thus, sul1 and sul2 are not necessary assembled in the same genetic cluster and can be present on different organisms independently, explaining the differences found between them. Besides, integrons are not considered mobile: they need to be inserted in other MGEs (like transposons or plasmids) to spread and replicate.39,48 Plasmids usually contain genes for their self-replication, but they are also present in several copies in the bacterial cells (multi-copy plasmids).49 Therefore, it is more probable that genes embedded in plasmids will proliferate faster than those embedded in integrons in a nutrient-rich bacterial community. Further experiments would be needed to confirm this hypothesis.
3.4. Mathematical modelling of SMX effect and biodegradation in activated sludge
Mathematical modelling was used to support the batch experiments and analyze the effect and biodegradation of SMX in the activated sludge.A simple first model was developed and simulated to predict the biomass inhibition period length during the two sets of batch experiments conducted by exposing at either high (4.4 g VSS L−1) or low (0.5 g VSS L−1) initial biomass concentrations to low and high initial concentrations of SMX (0, 1, 10, 150 mg SMX L−1) (Fig. S1 and S2 in ESI†). Simulations highlighted that an antibiotic concentrations of 1 mg SMX L−1 does not exert an important inhibition of the heterotrophic biomass when compared to the control simulation without antibiotic, during which biomass grew steadily over the first day. Under the simulation conditions, decay of biomass is driven by substrate limitation as soon as acetate is depleted. A concentration of 10 mg SMX L−1 results in an about twice longer lag time. A high concentration of 150 mg SMX L−1 induces a prolonged bacteriostatic inhibition (100 days under the simulation conditions) after which it starts to biodegrade along with the progressive slow development of the heterotrophic biomass.
We used a second model to assess the influence of nitrogen loading (which was a main difference between the first and second sets of batches) on SMX biodegradation and effect on growth, inhibition, and substrates conversions by activated sludge microorganisms (Fig. 6). We had previously developed this extended model to analyze microbial selection processes in sludge, involving ordinary heterotrophic organisms (OHOs), nitrifiers as aerobic ammonium-oxidizing organisms (AOOs) and nitrite-oxidizing organisms (NOOs), as well as denitrifying heterotrophic organisms (DHOs) (Weissbrodt et al. 2023).44 While the first model used was implemented with observed parameter values (10–20 °C) retrieved from activated sludge models,41 this second model has been implemented with stoichiometric and biokinetic parameters calculated based on thermodynamics of microbial growth under biochemical reference conditions (25 °C, pH 7.0, 1 atm). Thermodynamics sets the best boundary conditions for growth and therefore differences in model outputs can be foreseen by maximizing conversion rates, resulting in shorter simulated conversion periods. Despite this difference, this expanded model helped examine the trends in biomass growth and SMX-inhibition under different ammonium loading along the sequential microbial selections of heterotrophs, nitrifiers and denitrifiers.
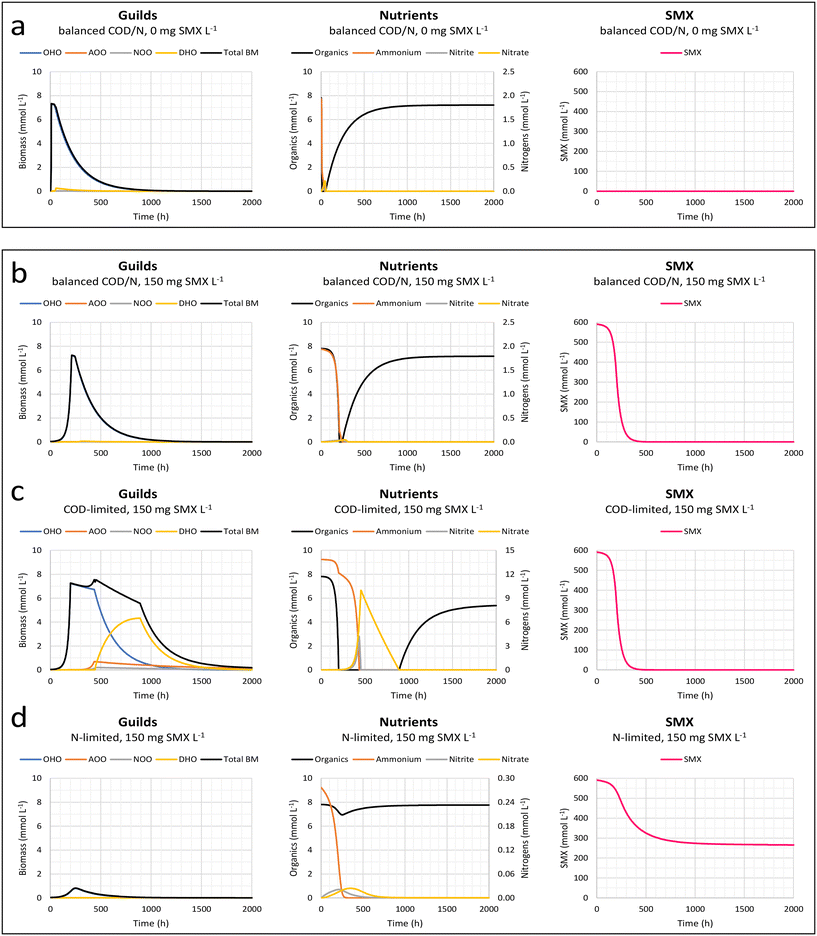 | ||
Fig. 6 Effect of nitrogen loading on SMX biodegradation and effect on the growth of the guilds of ordinary heterotrophic organisms (OHOs), nitrifiers (AOOs and NOOs), and denitrifiers (DHOs) and their substrates conversions. The second model developed based on microbial growth thermodynamics (Weissbrodt et al. 2023)44 was used. Simulations were performed without antibiotic (a) and with antibiotic (b–d), under balanced COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N conditions (100 N conditions (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 m/m) (a and b) or COD limitation (100 7 m/m) (a and b) or COD limitation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 m/m) (c) and nitrogen limitation (100 50 m/m) (c) and nitrogen limitation (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 m/m) (d). Simulations exposed an initially low-concentrated biomass (0.250 mg VSS L−1) to a high concentration of sulfamethoxazole (150 mg SMX L−1). COD 1 m/m) (d). Simulations exposed an initially low-concentrated biomass (0.250 mg VSS L−1) to a high concentration of sulfamethoxazole (150 mg SMX L−1). COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N ratios (100 N ratios (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 7, 50 m/m) were varied by changing the initial concentration of ammonium (5, 35, 250 mg N-NH4+ L−1) while maintaining the initial concentration of acetate to 500 mg COD L−1. Simulations were performed using mol-based and hour time units, and run over a long time period of 2000 h to examine microbial growth, SMX-inhibition, substrate-limitation, and decay processes. 1, 7, 50 m/m) were varied by changing the initial concentration of ammonium (5, 35, 250 mg N-NH4+ L−1) while maintaining the initial concentration of acetate to 500 mg COD L−1. Simulations were performed using mol-based and hour time units, and run over a long time period of 2000 h to examine microbial growth, SMX-inhibition, substrate-limitation, and decay processes. | ||
Under balanced COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) N conditions (100
N conditions (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 m/m) and in absence of antibiotic (Fig. 6a), the biomass develops rapidly over the first 10 h. OHOs get selected in the batch because of their fast growth rate. When acetate and ammonium are depleted, OHOs stop to grow. Residuals of ammonium and nitrogen oxides (nitrite, nitrate) are marginal, and nitrifiers and denitrifiers cannot develop.
7 m/m) and in absence of antibiotic (Fig. 6a), the biomass develops rapidly over the first 10 h. OHOs get selected in the batch because of their fast growth rate. When acetate and ammonium are depleted, OHOs stop to grow. Residuals of ammonium and nitrogen oxides (nitrite, nitrate) are marginal, and nitrifiers and denitrifiers cannot develop.
Under antibiotic supply while maintaining nutrient balance (Fig. 6b), the imposed high concentration of 150 mg SMX L−1 inhibits over the first 100 h all the microbial guilds forming the total biomass. The same bacteriostatic effect was implemented for each microorganism in the simulations. Progressively, OHOs grow by dissimilating acetate and assimilating ammonium. This biomass growth results in SMX biodegradation. Under the simulation conditions, OHOs stop to grow after 250 h and decay as soon as acetate and ammonium are simultaneously depleted. Biomass decay results in the release of additional organics; however, since hardly any nitrogen oxides could be built up by nitrifiers, the denitrifiers cannot develop. SMX gets fully abated after 500 h.
Under COD limitation (i.e., low COD/N ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 m/m) (Fig. 6c), after OHOs have depleted acetate, a substantial residual concentration of ammonium is available for nitrifiers. Nitrifiers produce nitrogen oxides that denitrifiers can use together with the organics released by the decay of OHOs. Under the simulation conditions, DHO make a substantial fraction of the total biomass. When ammonium and nitrogen oxides are depleted, nitrifiers and then denitrifiers decay. SMX biodegradation follows the same pattern as under balanced growth conditions: since the same initial concentration of acetate was used (while increasing the initial ammonium concentration), SMX gets primarily degraded during the OHO growth period.
50 m/m) (Fig. 6c), after OHOs have depleted acetate, a substantial residual concentration of ammonium is available for nitrifiers. Nitrifiers produce nitrogen oxides that denitrifiers can use together with the organics released by the decay of OHOs. Under the simulation conditions, DHO make a substantial fraction of the total biomass. When ammonium and nitrogen oxides are depleted, nitrifiers and then denitrifiers decay. SMX biodegradation follows the same pattern as under balanced growth conditions: since the same initial concentration of acetate was used (while increasing the initial ammonium concentration), SMX gets primarily degraded during the OHO growth period.
Under nitrogen limitation (i.e., high COD/N ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 m/m) (Fig. 6d), OHOs only marginally grow until their ammonium nitrogen source is depleted. Since OHOs scavenge this low amount of ammonium, nitrifiers and further denitrifiers cannot establish. An only low total biomass concentration develops, which starts to decay after 250 h. Therefore, the volumetric rate of SMX biodegradation is rapidly limited by the low biomass concentration, and SMX biodegradation is incomplete (55% abatement only).
1 m/m) (Fig. 6d), OHOs only marginally grow until their ammonium nitrogen source is depleted. Since OHOs scavenge this low amount of ammonium, nitrifiers and further denitrifiers cannot establish. An only low total biomass concentration develops, which starts to decay after 250 h. Therefore, the volumetric rate of SMX biodegradation is rapidly limited by the low biomass concentration, and SMX biodegradation is incomplete (55% abatement only).
3.5. Integration of experimental and modelling results to study the effect and biodegradation of SMX and the development of AMR in activated sludge depending on growth conditions
These modelling results on the SMX effect and biodegradation in the activated sludge can be further related to AMR processes and the detection of resistance determinants in either the intracellular or extracellular DNA fractions depending on the growth and decay processes occurring in the sludge. SMX exerts a bacteriostatic effect but gets biodegraded simultaneously to biomass growth. The lowest concentrations of 1–10 mg SMX L−1 tested and simulated in this study only lead to a relatively short inhibition of biomass growth. These concentrations are still high when compared to the actual μg L−1 level of SMX in municipal wastewater.5,50In the second set of experiments, SMX was hardly biodegraded over 3 days (Fig. 4D) but acetate was already depleted after 2 days through biomass growth (Fig. 4A–C). Modelling curves show that SMX biodegradation is coupled to biomass growth but with a slight delay. However, this delay is not sufficient to describe the relatively smooth growth of biomass measured under the 3 days of antibiotic treatment. The model computed under the best growth thermodynamic boundaries indicated full acetate depletion after 10 days. The discrepancy between experimental and modeling results can originate from different causes. Either the activated sludge microorganisms harbor higher minimum inhibitory concentrations (MIC50 values) than the reference median of 8 mg SMX L−1 used in the model, or they carried or early developed antibiotic resistance by vertical and/or horizontal transfer of sul genes in the experiments.
In addition, the model is useful to predict the effects of nutrient imbalances on patterns of microbial growth, inhibition by SMX, and biomass decay induced by substrate limitations. In the activated sludge, the microbial decay is a continuous process occurring parallel to biomass growth. However, decay is exacerbated when growth is hampered by substrate depletion over a prolonged period. Decay can release a substantial fraction of extracellular DNA materials carrying AMR determinants in the mixed liquor and WWTP effluent.8 Modelling predictions can help better manage the length of wastewater treatment process phases to reduce biomass decay by switching to another process stage when substrates are depleted. According to simulation results, not only the nutrient imbalances but their respective concentration levels are importantly governing biomass growth and decay. Since hampering biomass growth, nitrogen limitation resulted in low biomass production and decay, but also in an only partial biodegradation of SMX. In a previous study, nutrient limitations have been shown to reduce conjugative transfers related to AMR development.51 AMR development in mixed cultures like activated sludge is complex and depends on a range of conditions and processes occurring in the biomass beyond the sole antibiotic level. Future research should further combine experimental and modelling approaches to better investigate and capture AMR phenomena in relation to the dynamic environmental conditions prevailing in the activated sludge.
4. Conclusions
Here, we showed the impact of antibiotic treatments with SMX on the abundances of marker genes coding for sulfonamide resistance (sul1, sul2) and class 1 integron-integrase (intI1) in an activated sludge, with an analytical insight from both the iDNA and exDNA pools. Collectively our results highlighted that:• A previously developed anion-exchange chromatography method for the isolation of exDNA from wastewater environments was validated for activated sludge fractions at different biomass concentrations and volumes. Enough exDNA template (>2 μg DNA) for downstream molecular analysis was obtained with a 2-fold lower volume (0.5 L) and biomass concentration (1 gVSS L−1), showing the robustness of the method.
• Exposure to SMX (at 1 mg L−1) did not impact the amount of exDNA present in the sludge. However, the exDNA concentration dropped by 70% during biomass starvation under nutrient limitations.
• The abundances of intI1 and sul1 genes in the exDNA fraction of the first batch exposed to 1 mg L−1 of SMX were positively correlated (r > 0.7): most of the extracellular sul1 genes were suggested to be embedded in the class 1 integrons.
• Starvation under nutrient limitation resulted in a decrease (of 1–2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log gene copies per ng DNA) of the intI1 and sul1 genes in the exDNA fraction, while sul2 in the control flasks conversely increased (of 1
log gene copies per ng DNA) of the intI1 and sul1 genes in the exDNA fraction, while sul2 in the control flasks conversely increased (of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log gene copies per ng DNA).
log gene copies per ng DNA).
• Under nutrient-rich conditions, the exposure to high SMX concentrations (>10 mg L−1) selected for the growth of microorganisms that carried the sulfonamide resistance genes sul1 (usually located on class 1 integrons) and especially sul2 (usually located on small plasmids) on their iDNA pool.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Spanish Government (Agencia Estatal de Investigación) through ANTARES (PID2019-110346RB-C21) project. M. Martinez-Quintela also expresses his gratitude to the same agency for awarding a research scholarship (BES-2017-080503) and for a research visit at the TU Delft for conducting this work. The TU Delft team was financially supported by the Biotechnology and Safety Program of the Ministry of Infrastructure and Water Management and the project TARGETBIO of the NWO-TTW program Biotechnology & Safety (grant no. 15812) of the Applied and Engineering Sciences (TTW) Division of the Dutch Research Council (NWO).References
- E. Y. Klein, T. P. Van Boeckel, E. M. Martinez, S. Pant, S. Gandra and S. A. Levin, et al., Global increase and geographic convergence in antibiotic consumption between 2000 and 2015, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(15), E3463–E3470 CrossRef CAS PubMed.
- A. J. Browne, M. G. Chipeta, G. Haines-Woodhouse, E. P. A. Kumaran, B. H. K. Hamadani and S. Zaraa, et al., Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study, Lancet Planet. Health, 2021, 5(12), e893–e904 CrossRef PubMed.
- J. Wang, L. Chu, L. Wojnárovits and E. Takács, Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview, Sci. Total Environ., 2020, 744, 140997 CrossRef CAS PubMed.
- Y. Jia, S. K. Khanal, H. Zhang, G. H. Chen and H. Lu, Sulfamethoxazole degradation in anaerobic sulfate-reducing bacteria sludge system, Water Res., 2017, 119, 12–20 CrossRef CAS PubMed.
- N. H. Tran, M. Reinhard and K. Y. H. Gin, Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions-a review, Water Res., 2018, 133, 182–207 CrossRef CAS PubMed.
- A. Miłobedzka, C. Ferreira, I. Vaz-Moreira, D. Calderón-Franco, A. Gorecki and S. Purkrtova, et al., Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health cycle, J. Hazard. Mater., 2022, 424 Search PubMed.
- R. Pallares-Vega, H. Blaak, R. van der Plaats, Husman A. M. de Roda, Leal L. Hernandez and M. C. M. van Loosdrecht, et al., Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: A cross-sectional study, Water Res., 2019, 161, 319–328 CrossRef CAS PubMed.
- D. Calderón-Franco, R. Sarelse, S. Christou, M. Pronk, D. Calder and M. C. M. Van Loosdrecht, et al., Metagenomic profiling and transfer dynamics of antibiotic resistance determinants in a full-scale granular sludge wastewater treatment plant, Water Res., 2022, 219, 118571 CrossRef PubMed.
- H. Bürgmann, D. Frigon, W. H. Gaze, C. M. Manaia, A. Pruden and A. C. Singer, et al., Water and sanitation: An essential battlefront in the war on antimicrobial resistance, FEMS Microbiol. Ecol., 2018, 94(9), 1–14 CrossRef PubMed.
- L. Rizzo, C. Manaia, C. Merlin, T. Schwartz, C. Dagot and M. C. Ploy, et al., Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review, Sci. Total Environ., 2013, 447, 345–360 CrossRef CAS PubMed.
- A. Karkman, T. T. Do, F. Walsh and M. P. J. Virta, Antibiotic-Resistance Genes in Waste Water, Trends Microbiol., 2018, 26(3), 220–228 CrossRef CAS PubMed.
- J. Bengtsson-Palme, R. Hammarén, C. Pal, M. Östman, B. Björlenius and C. F. Flach, et al., Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics, Sci. Total Environ., 2016, 572, 697–712 CrossRef CAS PubMed.
- C. M. Manaia, J. Rocha, N. Scaccia, R. Marano, E. Radu and F. Biancullo, et al., Antibiotic resistance in wastewater treatment plants: Tackling the black box, Environ. Int., 2018, 115, 312–324 CrossRef CAS PubMed.
- F. Lira, I. Vaz-Moreira, J. Tamames, C. M. Manaia and J. L. Martínez, Metagenomic analysis of an urban resistome before and after wastewater treatment, Sci. Rep., 2020, 10(1), 1–9 CrossRef PubMed.
- M. Quintela-Baluja, M. Abouelnaga, J. Romalde, J. Q. Su, Y. Yu and M. Gomez-Lopez, et al., Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters, Water Res., 2019, 162, 347–357 CrossRef CAS PubMed.
- R. Pallares-Vega, L. Hernandez Leal, B. N. Fletcher, E. Vias-Torres, M. C. M. van Loosdrecht and D. G. Weissbrodt, et al., Annual dynamics of antimicrobials and resistance determinants in flocculent and aerobic granular sludge treatment systems, Water Res., 2021, 190, 116752 CrossRef CAS PubMed.
- A. Di Cesare, E. M. Eckert, S. D'Urso, R. Bertoni, D. C. Gillan and R. Wattiez, et al., Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants, Water Res., 2016, 94, 208–214 CrossRef CAS PubMed.
- S. Rodriguez-Mozaz, S. Chamorro, E. Marti, B. Huerta, M. Gros and A. Sànchez-Melsió, et al., Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river, Water Res., 2015, 69, 234–242 CrossRef CAS PubMed.
- J. Hultman, M. Tamminen, K. Pärnänen, J. Cairns, A. Karkman and M. Virta, Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent, FEMS Microbiol. Ecol., 2018, 94(4), 1–10 CrossRef PubMed.
- A. Zarei-Baygi and A. L. Smith, Intracellular versus extracellular antibiotic resistance genes in the environment: Prevalence, horizontal transfer, and mitigation strategies, Bioresour. Technol., 2021, 319(2020), 124181 CrossRef CAS PubMed.
- D. Calderón-Franco, Q. Lin, M. C. M. van Loosdrecht, B. Abbas and D. G. Weissbrodt, Anticipating Xenogenic Pollution at the Source: Impact of Sterilizations on DNA Release From Microbial Cultures, Front. Bioeng. Biotechnol., 2020, 8, 1–13 Search PubMed.
- P. Rusanowska, A. Cydzik-Kwiatkowska and I. Wojnowska-Baryła, Microbial Origin of Excreted DNA in Particular Fractions of Extracellular Polymers (EPS) in Aerobic Granules, Water, Air, Soil Pollut., 2019, 230(8), 203 CrossRef.
- Y. Zhang, A. Li, T. Dai, F. Li, H. Xie and L. Chen, et al., Cell-free DNA: A Neglected Source for Antibiotic Resistance Genes Spreading from WWTPs, Environ. Sci. Technol., 2018, 52(1), 248–257 CrossRef CAS PubMed.
- D. Calderón-Franco, M. C. M. van Loosdrecht, T. Abeel and D. G. Weissbrodt, Free-floating extracellular DNA: Systematic profiling of mobile genetic elements and antibiotic resistance from wastewater, Water Res., 2021, 189, 116592 CrossRef PubMed.
- Q. B. Yuan, Y. M. Huang, W. B. Wu, P. Zuo, N. Hu and Y. Z. Zhou, et al., Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads, Environ. Int., 2019, 131, 104986 CrossRef CAS PubMed.
- D. Mao, Y. Luo, J. Mathieu, Q. Wang, L. Feng and Q. Mu, et al., Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation, Environ. Sci. Technol., 2014, 48(1), 71–78 CrossRef CAS PubMed.
- A. Torti, M. A. Lever and B. B. Jørgensen, Origin, dynamics, and implications of extracellular DNA pools in marine sediments, Mar. Genom., 2015, 24, 185–196 CrossRef PubMed.
- K. M. Nielsen, P. J. Johnsen, D. Bensasson and D. Daffonchio, Release and persistence of extracellular DNA in the environment, Environ. Biosaf. Res., 2007, 6(1–2), 37–53 CrossRef CAS PubMed.
- C. M. Thomas and K. M. Nielsen, Mechanisms of, and barriers to, horizontal gene transfer between bacteria, Nat. Rev. Microbiol., 2005, 3(9), 711–721 CrossRef CAS PubMed.
- M. Nagler, H. Insam, G. Pietramellara and J. Ascher-Jenull, Extracellular DNA in natural environments: features, relevance and applications, Appl. Microbiol. Biotechnol., 2018, 102(15), 6343–6356 CrossRef CAS PubMed.
- C. J. H. Von Wintersdorff, J. Penders, J. M. Van Niekerk, N. D. Mills, S. Majumder and L. B. Van Alphen, et al., Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer, Front. Microbiol., 2016, 7, 1–10 Search PubMed.
- M. Winter, A. Buckling, K. Harms, P. J. Johnsen and M. Vos, Antimicrobial resistance acquisition via natural transformation: context is everything, Curr. Opin. Microbiol., 2021, 64, 133–138 CrossRef CAS PubMed.
- J. M. A. Blair, M. A. Webber, A. J. Baylay, D. O. Ogbolu and L. J. V. Piddock, Molecular mechanisms of antibiotic resistance, Nat. Rev. Microbiol., 2015, 13(1), 42–51 CrossRef CAS PubMed.
- J. Bengtsson-Palme, E. Kristiansson and D. G. J. Larsson, Environmental factors influencing the development and spread of antibiotic resistance, FEMS Microbiol. Rev., 2018, 42(1), 68–80 CrossRef CAS PubMed.
- J. Jutkina, C. Rutgersson, C. F. Flach and D. G. Joakim Larsson, An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance, Sci. Total Environ., 2016, 548–549, 131–138 CrossRef CAS PubMed.
- J. Bengtsson-Palme and D. G. J. Larsson, Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation, Environ. Int., 2016, 86, 140–149 CrossRef CAS PubMed.
- Metcalf & Eddy I., Wastewater Engineering: Treatment and Reuse, McGraw-Hill, 5th edn, 2014 Search PubMed.
- E. W. Rice, L. Bridgewater, A. P. H. Association, A. W. W. Association and W. E. Federation, Standard methods for the examination of water and wastewater, ed. E. W. Rice, American Water Works Association, Washington, D.C, 22nd edn, 2012 Search PubMed.
- T. M. Ghaly, M. R. Gillings, A. Penesyan, Q. Qi, V. Rajabal and S. G. Tetu, The natural history of integrons, Microorganisms, 2021, 9(11), 1–12 CrossRef PubMed.
- P. Reichert, Aquasim - A Tool for Simulation and Data Analysis of Aquatic Systems, Water Sci. Technol., 1994, 30(2), 21–30 CrossRef CAS.
- M. Henze, W. Gujer, T. Mino, T. Matsuo, M. C. Wentzel and G. V. R. Marais, et al., Activated Sludge Model No. 2D, ASM2D, Water Sci. Technol., 1999, 39(1), 165–182 CrossRef CAS.
- D. M. Kennes-Veiga, L. Gónzalez-Gil, M. Carballa and J. M. Lema, The organic loading rate affects organic micropollutants' cometabolic biotransformation kinetics under heterotrophic conditions in activated sludge, Water Res., 2021, 189, 116587 CrossRef CAS PubMed.
- M. Nagler, S. M. Podmirseg, G. W. Griffith, H. Insam and J. Ascher-Jenull, The use of extracellular DNA as a proxy for specific microbial activity, Appl. Microbiol. Biotechnol., 2018, 102(6), 2885–2898 CrossRef CAS PubMed.
- D. G. Weissbrodt, M. C. M. van Loosdrecht and Y. Comeau, Basic microbiology and metabolism. Biological Wastewater Treatment, IWA Publishing, 2nd edn, 2023, pp. 11–76 Search PubMed.
- S. Schmidt, J. Winter and C. Gallert, Long-term effects of antibiotics on the elimination of chemical oxygen demand, nitrification, and viable bacteria in laboratory-scale wastewater treatment plants, Arch. Environ. Contam. Toxicol., 2012, 63(3), 354–364 CrossRef CAS PubMed.
- T. Katipoglu-Yazan, E. Ubay-Cokgor and D. Orhon, Acute inhibitory impact of sulfamethoxazole on mixed microbial culture: Kinetic analysis of substrate utilization biopolymer storage nitrification and endogenous respiration, Biochem. Eng. J., 2021, 167, 107911 CrossRef CAS.
- O. Sköld, Sulfonamide resistance: Mechanisms and trends, Drug Resistance Updates, 2000, 3(3), 155–160 CrossRef PubMed.
- S. Domingues, G. J. Da Silva and K. M. Nielsen, Global dissemination patterns of common gene cassette arrays in class 1 integrons, Microbiology, 2015, 161(7), 1313–1337 CrossRef CAS PubMed.
- M. Shintani, Z. K. Sanchez and K. Kimbara, Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy, Front. Microbiol., 2015, 6, 1–16 CrossRef PubMed.
- M. Carballa, F. Omil, J. M. Lema, M. Llompart, C. García-Jares and I. Rodríguez, et al., Behavior of pharmaceuticals, cosmetics and hormones in a sewage treatment plant, Water Res., 2004, 38(12), 2918–2926 CrossRef CAS PubMed.
- R. Pallares-Vega, G. Macedo, M. S. M. Brouwer, L. Hernandez Leal, P. van der Maas and M. C. M. van Loosdrecht, et al., Temperature and Nutrient Limitations Decrease Transfer of Conjugative IncP-1 Plasmid pKJK5 to Wild Escherichia coli Strains, Front. Microbiol., 2021, 12, 656250 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ew00591g |
| This journal is © The Royal Society of Chemistry 2024 |