House dust mite extract forms a der p 2 corona on multi-walled carbon nanotubes: implications for allergic airway disease†
Judith
Dominguez
 a,
Samantha K.
Holmes
a,
Ryan D.
Bartone
b,
Logan J.
Tisch
b,
Robert M.
Tighe
a,
Samantha K.
Holmes
a,
Ryan D.
Bartone
b,
Logan J.
Tisch
b,
Robert M.
Tighe
 c,
James C.
Bonner
c,
James C.
Bonner
 b and
Christine K.
Payne
b and
Christine K.
Payne
 *a
*a
aThomas Lord Department of Mechanical Engineering and Materials Science, Duke University, Durham, North Carolina 27708, USA. E-mail: christine.payne@duke.edu
bToxicology Program, Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA
cDepartment of Medicine, Duke University School of Medicine, Durham, North Carolina 27710, USA
First published on 15th December 2023
Abstract
Multi-walled carbon nanotubes (MWCNTs) are used in materials for the construction, automotive, and aerospace industries. Workers and consumers are exposed to these materials via inhalation. Existing recommended exposure limits are based on MWCNT exposures that do not take into account more realistic co-exposures. Our goal was to understand how a common allergen, house dust mite extract, interacts with pristine MWCNTs and lung fluid proteins. We used gel electrophoresis, western blotting, and proteomics to characterize the composition of the allergen corona formed from house dust mite extract on the surface of MWCNTs. We found that the corona is dominated by der p 2, a protein associated with human allergic responses to house dust mites. Der p 2 remains adsorbed on the surface of the MWCNTs following subsequent exposures to lung fluid proteins. The high concentration of der p 2, localized on surface of MWCNTs, has important implications for house dust mite-induced allergies and asthma. This research provides a detailed characterization of the complex house dust mite-lung fluid protein coronas for future cellular and in vivo studies. These studies will help to address the molecular and biochemical mechanisms underlying the exacerbation of allergic lung disease by nanomaterials.
Environmental significanceMulti-walled carbon nanotubes (MWCNTs) are used in materials essential for the construction, automotive, and aerospace industries. Workers are exposed to these engineered nanomaterials during manufacturing. Consumers are exposed following environmental release of the materials. Inhalation is of specific concern as the major route of exposure in both the occupational and environmental setting. In 2013, the National Institute for Occupational Safety and Health (NIOSH) provided an overview of the occupational hazards associated with these materials and recommended exposure limits. Real-world human exposure will likely not be limited to pristine MWCNTs. A variety of organic and inorganic substances can adsorb on MWCNTs. For example, various types of air pollution and allergens will also be inhaled with the MWCNTs. Our research examined the complex interaction of MWCNTs, house dust mite allergens, and lung fluid proteins. By providing a detailed molecular characterization of this interaction, we hope to better understand observed increases in allergic airway disease caused by co-exposures to nanomaterials and allergens. |
Introduction
Multi-walled carbon nanotubes (MWCNTs) are essential components of electronics, plastics, sensors, and coatings with applications in the construction, automotive, and aerospace industries.1,2 Given this wide range of applications, it is important to understand the effects that MWCNTs may have on both workers in the manufacturing setting and consumers who are exposed to these materials following environmental release. Inhalation is of specific concern as this is the primary route of exposure.3–6 The pulmonary toxicity of MWCNTs in rodents is well-demonstrated and known to be dependent on the physicochemical characteristics of the MWCNTs.7–14 For example, rigid, rod-like MWCNTs are more toxic than flexible, tangled, MWCNTs.15 The National Institute for Occupational Safety and Health (NIOSH) provides an overview of the occupational hazards associated with these materials and recommended exposure limits.16 These recommended exposure limits are based solely on MWCNT exposure that do not take into account more realistic co-exposures. For example, allergic lung disease induced by common allergens, such as those from house dust mites (HDM; Dermatophagoides pteronyssinus), is exacerbated by MWCNT inhalation exposure in mice.17 These findings suggest that the current exposure limits to MWCNTs should take into account co-exposures. In addition, the mechanistic details underlying the observed toxicity of these nanomaterial–allergen co-exposures are lacking, especially regarding the initial steps of inhalation and the relationship of material properties to physiological and pathological outcomes.18,19Previous work has shown that blood serum proteins and cell lysates adsorb on the surface of MWCNTs forming a protein corona.20–23 The protein corona, rather than the bare nanomaterial, determines the subsequent biological events including cellular internalization and immune response.24–30 Our research focuses on the formation of an allergen corona as MWCNTs interact with allergens in the environment, followed by the interaction of these allergen–MWCNTs with lung fluid obtained from mice. The interaction of titanium dioxide and polystyrene nanoparticles with bronchoalveolar lavage fluid (BALF) has been characterized previously.31,32 For the titanium dioxide nanoparticles, a BALF corona led to increased expression of pro-inflammatory cytokines in macrophages.32 To the best of our knowledge, our work described below is the first to address a combined allergen and lung fluid corona.
We examine one common allergen, HDM extract. Allergies to HDM are estimated to affect 1–2% of the global population.33–35 Eighty-five percent of asthmatics are allergic to HDM.36 HDM extract is a multi-component allergen consisting of a mixture of proteins from mites, nymphs, fecal matter, and eggs.37–40 Of specific interest are two proteins, der p 1 and der p 2.38 The majority (∼80%) of people with HDM allergies are sensitive to der p 1 and der p 2.41 Sera samples from allergic subjects showed that 50% of IgE binding was directed at these two proteins.42 Der p 1 is a cysteine protease.43–46 Der p 2 is a myeloid differentiation factor-2-related lipid recognition domain lipid-binding family protein,47 which is found in the digestive track and feces of HDM.48 Previous work has suggested that a HDM corona on MWCNTs could be a factor in the exacerbated allergic airway disease observed in mice.49In vitro experiments examined gold nanoparticles (50 nm) conjugated to der p 1.50 Proteolytic assays showed that the der p 1-gold nanoparticle conjugates had increased proteolytic activity compared to der p 1,50 suggesting that allergen coronas may amplify allergic responses. This is supported by in vivo experiments showing that mice co-exposed to MWCNTs and HDM extract displayed increased allergic lung inflammation that was not observed with MWCNTs or low doses of HDM extract alone.17
In terms of human exposures, MWCNTs could encounter HDM in both manufacturing and consumer settings. HDM are ubiquitous in the indoor environment: they thrive in upholstered furniture, carpets, bedding, and dust. Previous work has found that MWCNTs were present in the office areas adjacent to production facilities.51 In these office areas, MWCNTs would certainly come into contact with HDM. This study did not measure HDM in the MWCNT production facility, but it is likely that such facilities also contain some HDM, especially in production areas that may contain dust. It is also possible that MWCNTs and HDM will come into contact during the life cycle of the nanomaterial, where there is release of MWCNTs from a matrix or during recycling.52 For example, the future use of MWCNTs in textiles would result in consumer exposure within homes that contain HDM.
Our approach, described below, was to characterize the protein corona formed on MWCNTs incubated with HDM extract, both alone and in combination with BALF. We focus on a single type of MWCNT (NC7000), which is the parent nanomaterial that can be further chemically or thermally purified and functionalized.14 We use gel electrophoresis, western blotting, and proteomics to characterize the composition of the protein corona. We found that HDM extract forms a corona on the surface of MWCNTs and that this corona is dominated by der p 2. Der p 2 remains adsorbed on the MWCNTs following subsequent exposures to BALF or albumin, the main protein present in BALF. The high concentration of der p 2, localized on surface of MWCNTs, has important implications for HDM-induced allergies and asthma. This research will help to address the molecular and biochemical mechanisms underlying the exacerbation of allergic lung disease by nanomaterials and provides a detailed characterization of these complex protein coronas for future cellular and in vivo studies.
Materials and methods
MWCNT characterization
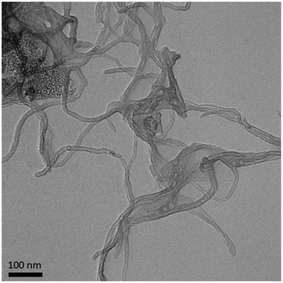
MWCNTs (NC7000, Nanocyl, Sambreville, Belgium) were used for all experiments. Stock solutions of MWCNTs were prepared by suspending dry MWCNTs (10 mg mL−1) in 1× phosphate buffered saline (PBS; 21300025, Thermo Fisher, Waltham, MA). Solutions were sonicated for five minutes with a cup horn sonicator (40% amplitude; Q500 Sonicator, Q Sonica, Newtown, CT) to help suspend the MWCNTs.Transmission electron microscopy (TEM; Talos F200X, Thermo Fisher) was used to quantify the diameter of the MWCNTs. A MWCNT solution (100 μg mL−1 in PBS) was drop-cast onto carbon coated copper grids (FCF200-Cu, Electron Microscopy Sciences, Hatfield, PA). Images were collected with 50k× magnification at 200 kV. ImageJ was used for image analysis.53 X-ray photoelectron spectroscopy (XPS; Kratos Analytical Axis Ultra, Shimadzu Corporation, Kyoto, Japan) at the Shared Materials Instrumentation Facility at Duke University was used to measure the elemental composition of the MWCNTs.
Rodent bronchoalveolar lavage fluid (BALF)
Our procedure for obtaining BALF has been described previously.32 All procedures were approved by the Duke University Institutional Animal Care and Use Committee (IACUC) and were performed under an IACUC approved animal protocol (A053-21-03). All animal experiments were conducted in accordance with the American Association for the Accreditation of Laboratory Animal Care guidelines. In brief, C57BL/6 male mice (8–10 weeks) were purchased from Jackson Laboratories (Bar Harbor, ME). Bronchoalveolar lavages were performed following a published protocol.45 Prior to lavage, mice were deeply anesthetized with an intraperitoneal injection of ketamine (100 mg kg−1), xylazine (100 mg kg−1), and saline (0.9%), dosed by weight (350–500 μL). The chest and trachea were dissected to expose the lungs and trachea. Tubing (PE-60, #9565S30, Thomas Scientific, Swedesboro, NJ) was inserted into the trachea and attached to a 12-inch infusion set (#SV-25BLK, Terumo, Tokyo, Japan) connected to a 10 mL syringe held on a ring stand. Lungs were passively filled to 20 cm H2O with PBS to reach total lung capacity. The BALF was then passively drained and placed on ice for further processing. BALF used for protein corona formation was pooled from 10–20 mice to reduce mouse-to-mouse variation.Protein corona formation and concentration
Protein coronas were formed by incubating MWCNTs with proteins for 30 minutes at room temperature (RT) on an orbital shaker (700 rpm, 88882006, Digital Microplate Shaker, Thermo Fisher). Proteins used included HDM (Dermatophagoides pteronyssinus, XPB91D3A2.5, Lot 414145, Stallergenes Greer, Lenoir, NC), bovine serum albumin (BSA; A2153-50G, Sigma-Aldrich, St. Louis, MO), and BALF. To remove unbound protein, samples were washed three times by centrifugation (15–30 minutes, 4 °C, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) and resuspension in PBS. Removal of unbound and weakly bound proteins results in a “hard” corona. To create sequential coronas, the corona formation procedure was repeated using a second protein. Protein concentrations were measured with a colorimetric assay (Pierce 660 nm Protein Assay, referred to as a 660 nm assay, 2260, Thermo Fisher Scientific) and quantified with a plate reader (SpectraMax iD3, Molecular Devices, San Jose, CA).
000 rpm) and resuspension in PBS. Removal of unbound and weakly bound proteins results in a “hard” corona. To create sequential coronas, the corona formation procedure was repeated using a second protein. Protein concentrations were measured with a colorimetric assay (Pierce 660 nm Protein Assay, referred to as a 660 nm assay, 2260, Thermo Fisher Scientific) and quantified with a plate reader (SpectraMax iD3, Molecular Devices, San Jose, CA).
Gel electrophoresis
The compositions of the protein coronas were visualized using gel electrophoresis. Protein–MWCNT complexes were resuspended in Laemmli SDS-sample buffer (4×, reducing, BP-110R, Boston BioProducts, Milford, MA). Proteins were denatured by boiling samples at 95 °C for 5 minutes before loading into a tris-glycine sodium dodecyl sulfate (SDS) precast polyacrylamide gel (4561096, Bio-Rad Laboratories, Hercules, California). A 10–250 kDa molecular weight marker (Precision Plus Protein Dual Color Standards, 1610374, Bio-Rad) was included in the gel. A voltage of 230 V was applied for 30 minutes. The gel was rinsed by microwaving in deionized water three times (1 minute heat, 1 minute rocking at RT, water replaced). The gel was then stained (SimplyBlue Safe Stain, LC6060, Thermo Fisher) by microwaving for 45 seconds and rocking for 5 minutes at RT. Deionized water was used to destain the gel (rocking overnight, RT) before imaging (PhotoDoc-It; Analytik Jena, Jena, Germany).Western blotting
Gels for western blotting were transferred to a 0.2 mm PVDF membrane (1704156, Bio-Rad Laboratories) using the Trans-Blot Turbo Transfer System (1704150, Bio-Rad Laboratories) and run at 2.5 A for 7 min. Membranes were blocked (5% dry milk in tris-buffered saline with 0.1% Tween buffer) for 1 hour and then incubated overnight in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of a polyclonal anti-der p 2 (AA 18-146) antibody (ABIN7141165, Antibodies-online Inc., Limerick, PA). Following primary antibody incubation, the membranes were washed and incubated in 1
1000 dilution of a polyclonal anti-der p 2 (AA 18-146) antibody (ABIN7141165, Antibodies-online Inc., Limerick, PA). Following primary antibody incubation, the membranes were washed and incubated in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 dilution with horseradish peroxidase-conjugated secondary anti-rabbit antibody (#7074, Cell Signaling Technology, Danvers, MA). Enhanced chemiluminescence (ECL) Prime Western Blotting Detection Reagent (Cytiva, Marlborough, MA) was used to for HRP-induced chemiluminescence, and the resulting signals were imaged using an Amersham Imager 680 (GE Life Sciences, Marlborough, MA).
2000 dilution with horseradish peroxidase-conjugated secondary anti-rabbit antibody (#7074, Cell Signaling Technology, Danvers, MA). Enhanced chemiluminescence (ECL) Prime Western Blotting Detection Reagent (Cytiva, Marlborough, MA) was used to for HRP-induced chemiluminescence, and the resulting signals were imaged using an Amersham Imager 680 (GE Life Sciences, Marlborough, MA).
Proteomic analysis
Proteins were removed from the MWCNT surface by incubation with 5% SDS buffer for 5 minutes at 95 °C. A 660 nm assay was used to determine protein concentrations prior to submission for proteomic analysis. Individual bands for proteomic analysis were excised from the polyacrylamide gels and submitted without further processing.The compositions of the protein coronas on MWCNTs were determined using liquid chromatography with tandem mass spectrometry (LC-MS/MS). Proteomic analysis was conducted by the Proteomics and Metabolomics Core Facility, part of the Duke Center for Genomics and Computational Biology. A UPLC column (75 μm × 250 mm, nanoAcquity, Waters Corporation; 400 nL min−1) with a 60 minute elution time was used for NanoFlow liquid chromatography with an acetonitrile gradient (5–40%) and 0.1% formic acid. In-line tandem mass spectrometry was used to analyze peptide fragments (Orbitrap Fusion Lumos, Thermo Fisher). We analyzed the LC-MS/MS data using MaxQuant (v2.1.0, Max Planck Institute, Munich, Germany).54,55 The raw LC-MS/MS spectra were searched using the integrated Andromeda search engine against the Swiss-Prot Dermatophagoides pteronyssinus (European house dust mite; #6956) and Bos taurus (bovine; #9913) canonical protein databases from UniProt, accessed on June 5th, 2023. A custom contaminants file was used while searching, which contained a relevant subset of the Common Repository of Adventitious Proteins (cRAP) database.56 For protein and peptide quantification and identification, default MaxQuant parameters were used including a 0.01 false discovery rate, a minimum peptide length of 7 amino acids, a maximum peptide length of 25 amino acids, oxidation and acetyl groups as variable modifications, and carbamidomethyl as a fixed modification.
Perseus (v2.0.3.1, Max Planck Institute) was used to analyze and filter proteomic data.57 Proteins were excluded if they were contaminants, quality control standards, or were not observed in at least 2 samples. A quantitative internal standard was not used for these experiments. The intensities of all proteins identified within a sample were summed to obtain the relative abundance of each protein within each sample triplicate. Averages and standard deviations were calculated from the abundances of the sample triplicates. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository with the dataset identifier PXD047039.58
Results and discussion
MWCNT characterization
The diameter of the MWCNTs was measured from TEM images (Fig. 1). Dynamic light scattering is not suitable for these filamentous, non-spherical samples. The MWCNTs have an average diameter of 11.9 ± 0.6 nm (n = 6), in good agreement with previous TEM experiments showing a diameter of 12 nm.14 XPS analysis of MWCNTs revealed a composition of 98.33% carbon, 1.49% oxygen, and 0.18% aluminum (Table 1). A previous analysis of these MWCNTs with XPS found similar levels of carbon (97.8%) and oxygen (1.4%).14 Trace amounts of aluminum are attributed to the synthesis process, in which metal oxides are present during catalytic chemical vapor deposition.3,59–62| Sample | Elemental analysis (%) | ||||
|---|---|---|---|---|---|
| C | O | Al | N | Na | |
| MWCNT | 98.33 | 1.49 | 0.18 | — | — |
| HDM–MWCNT | 72.01 | 22.08 | 0.20 | 2.93 | 2.31 |
HDM extract forms a corona on MWCNTs
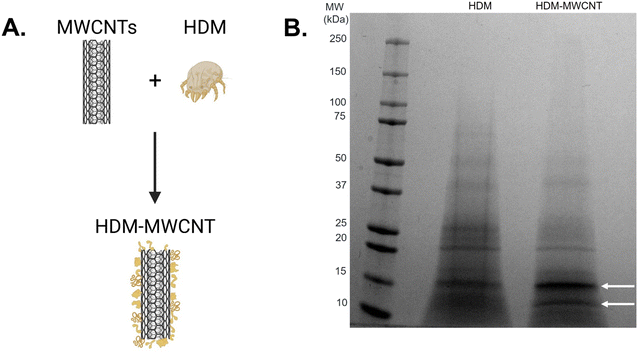
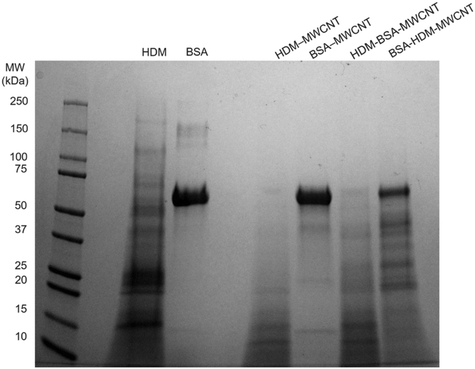
We first characterized the corona that forms on MWCNTs following incubation with HDM extract using gel electrophoresis (Fig. 2). MWCNTs (1 mg mL−1) were incubated with HDM (1 mg mL−1) in PBS (30 minutes, RT) then washed to remove unbound protein, as described in Materials and methods (Fig. 2A and S1†). XPS shows that MWCNTs with a HDM corona (HDM–MWCNT) are similar in composition to the pristine MWCNTs, but with an increase in the percentage of oxygen (22.08%) and the addition of nitrogen (2.93%) and sodium (2.31%) (Table 1), likely reflecting the presence of protein and PBS used to form the protein corona. Gel electrophoresis was used to separate and identify the specific proteins that comprise the HDM–MWCNT corona (Fig. 2B). In comparison to HDM alone, the corona formed on MWCNTs (HDM–MWCNT) shows an increase in the band at ∼15 kDa and the appearance of a ∼12 kDa band. These bands were excised and submitted for proteomics. Based on spectral counts, they were identified to be der p 2 (14 kDa) and der p 30 (12 kDa) (Tables S1 and S2†). In addition, western blotting confirms the ∼15 kDa band is der p 2 (Fig. S2†).Concentration of the HDM corona
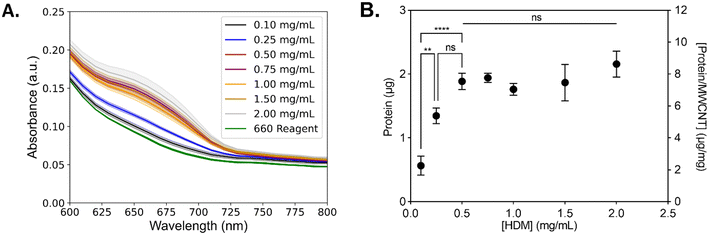
The concentration of HDM proteins that adsorb on the surface of MWCNTs was quantified using a 660 nm protein assay (Fig. 3). MWCNTs (1 mg mL−1) were incubated with increasing concentrations of HDM (0.1–2.0 mg mL−1). Samples at lower incubation concentrations (0.1 mg mL−1 and 0.25 mg mL−1 HDM) were pooled (×3) for measurement to provide sufficient protein for the assay. The resulting concentration of protein adsorbed on the MWCNTs increases from 0.1 mg mL−1 to 0.5 mg mL−1 and then remains constant with increasing HDM. The maximum concentration of protein in the corona is 7.69 ± 0.80 μg mg−1 MWCNT, calculated as the average of the samples formed with 0.5–2.0 mg mL−1 of HDM. This increasing corona concentration as a function of increasing incubation protein concentration is in agreement with other corona studies. Similar assays with polystyrene and silica nanoparticles have shown that increasing the incubation protein concentrations leads to increasing corona concentrations,63–66 with rare exceptions.63 However, the maximum corona concentration is low compared to previous studies. For example, titanium nanoparticles incubated with fetal bovine serum form a corona of 50 μg protein/mg of titanium dioxide nanoparticles.32 It is possible that HDM, which is a mixture of mites, nymphs, fecal matter, and eggs,37 results in maximum corona formation at relatively low corona concentrations values. In comparison, much of the previous work has focused on human plasma or bovine serum relevant to cell culture and biomedical applications.Composition of the HDM corona
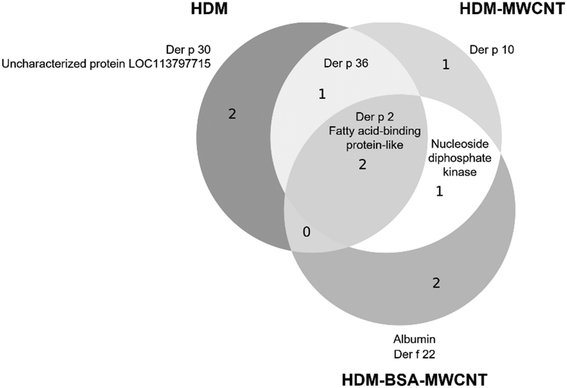
Although gel electrophoresis provides a visualization of the corona proteins (Fig. 2B), proteomics provides a more detailed description of the composition of the protein corona (Table 2). Proteomic analysis is shown as the normalized abundance for HDM alone and the HDM corona (HDM–MWCNT). 79 proteins were identified in HDM and 46 proteins were identified in the HDM–MWCNT coronas. The top-10 of each sample (n = 3) are listed. Previous proteomics studies of HDM alone have identified a complex mixture of proteins.39,40 Der p 30, der p 2, der p 36, der p 1, and der p 10 have all previously been identified as highly abundant proteins in mite bodies or extracts.39,40 Der f 27, observed in D. farinae,40 was not observed at high abundance in D. pteronyssinus previously.| Protein | MW (kDa) | HDM (%) | HDM–MWCNT (%) |
|---|---|---|---|
| Der p 30 (ferritin) | 12 | 29.3 ± 13.3 | 1.00 ± 1.51 |
| Uncharacterized protein LOC113797715 | 191 | 18.1 ± 6.71 | 0.39 ± 0.29 |
| Der p 2 (NPC2 family) | 14 | 12.7 ± 9.01 | 49.6 ± 15.2 |
| Fatty acid-binding protein-like | 15 | 5.97 ± 4.61 | 7.49 ± 9.79 |
| Der p 36 (C2 domain containing protein) | 25 | 3.73 ± 3.14 | 14.3 ± 0.98 |
| Der f 27 (serpin) | 47 | 2.03 ± 0.85 | 1.16 ± 1.34 |
| Der p 1 (cysteine protease) | 24 | 1.79 ± 0.54 | 0.17 ± 0.17 |
| Der p 10 (tropomyosin) | 33 | 1.69 ± 1.53 | 3.90 ± 3.89 |
| Peptidase 1-like | 39 | 1.62 ± 0.31 | — |
| Fructose-bisphosphate aldolase | 39 | 1.44 ± 2.35 | — |
| Nucleoside diphosphate kinase | 18 | 0.91 ± 0.68 | 4.03 ± 0.66 |
| Der p 7 (bactericidal permeability increasing-like) | 24 | 0.12 ± 0.067 | 3.09 ± 1.09 |
| Der f 22 (group 2-like) | 18 | — | 2.46 ± 3.04 |
| Der p 40 (thioredoxin-like) | 12 | — | 1.10 ± 0.76 |
We compare the protein abundance in HDM to the corona formed on MWCNTs. For example, der p 30 is the most highly abundant protein (29.3 ± 13.3%) in the HDM extract, but it was observed at very low levels in the corona (1.00 ± 1.51%). Der p 2 is much less abundant in HDM (12.7 ± 9.01%), but the most abundant allergen in the HDM corona (49.6 ± 15.2%). Der p 36 was the second most highly abundant protein in the corona (14.3 ± 0.98%), although not highly abundant in HDM extract (3.73 ± 3.14%), showing that it is enriched in the corona. Der p 1, which, along with der p 2, is a major human allergen,38,42 was observed at low abundance in this lot of HDM (1.79 ± 0.54%) and the corona (0.17 ± 0.17%). The same analysis, using a different lot of HDM, also showed der p 2 was the most abundant protein in the corona (51.6 ± 15.6%), although there were lot-to-lot variations in the HDM and resulting corona (Table S3†). This lot-to-lot variation in HDM is expected based on previous work.67
A direct comparison of gel electrophoresis and proteomics is complicated by overlapping molecular weights of proteins and different levels of protein staining. For example, based on proteomics data, der p 30 is highly abundant in HDM extract (29.3 ± 13.3%), but difficult to resolve in the gel (Fig. 2B). Gel electrophoresis of the corona results in fewer fragments and a cleaner gel resulting in a visible band at 12 kDa, although the abundance of der p 30 is relatively low (1.00 ± 1.51%) by proteomics. Proteomics of the excised bands at 12 kDa and 15 kDa confirms the presence of der p 30 in the corona and the enrichment of der p 2 (Fig. 2B and Tables S1 and S2†).
Overall, the comparison between HDM and HDM–MWCNTs shows that der p 2 is highly concentrated on the surface of MWCNTs, which may be important for allergic airway disease. Der p 1, which is the other major human allergen, was identified in the protein corona at very low abundance.38,41,42
Interaction of HDM–MWCNTs with lung fluid proteins
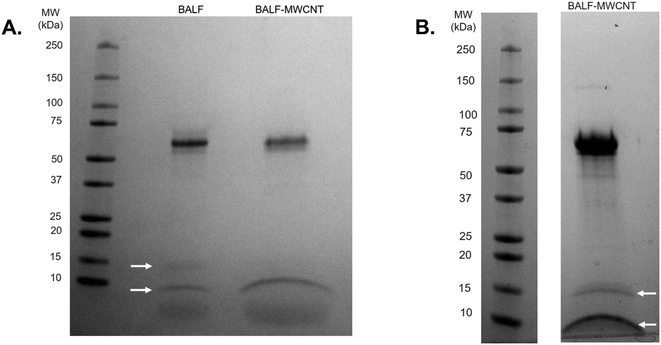
The results above describe the protein corona formed when HDM proteins adsorb on the surface of MWCNTs. The most relevant human exposure to these HDM–MWCNT complexes is through inhalation. To probe this exposure mechanism, we investigated the interaction of BALF with MWCNTs and HDM–MWCNTs. As a first step, we confirmed that a BALF corona formed on pristine MWCNTs. Gel electrophoresis and a protein concentration assay were used to analyze the composition and concentration of the BALF protein corona (Fig. 4). A BALF corona concentration of 35.3 ± 0.85 μg protein per mg MWCNT was measured. Albumin (69 kDa) was the most abundant protein in BALF and in the BALF corona (BALF–MWCNT) in agreement with previous work using proteomics to characterize the protein corona formed on MWCNTs incubated with BALF (24 h, 37 °C, 0.1% Pluronic).68 At high BALF concentrations, two other bands are visible in both BALF and the BALF corona at ∼15 kDa and ∼8 kDa, along with a low molecular weight smear.We then investigated the interaction of BALF with HDM–MWCNTs. For these experiments, we formed sequential protein coronas by first preparing HDM–MWCNTs and then incubating the HDM–MWCNTs with BALF to form a HDM–BALF–MWCNT corona (Fig. 5). Although the reverse corona (BALF–HDM–MWCNT) is less likely to be formed in a realistic setting, we also generated these coronas for comparison. Gel electrophoresis was used to visualize these sequential coronas (Fig. 6).
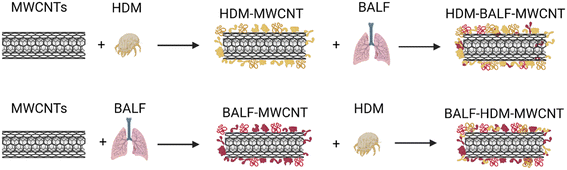 | ||
| Fig. 5 Schematic showing the formation of sequential coronas formed from HDM and BALF incubated with MWCNTs. The protein listed first is the initial protein used to form the corona. | ||
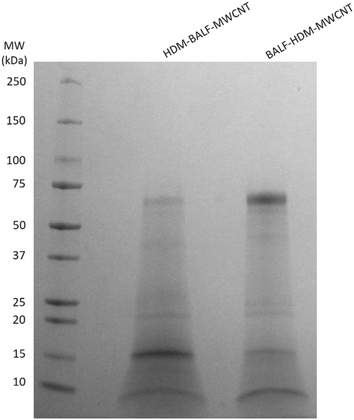
Gel electrophoresis shows that an initial HDM corona on MWCNTs will result in a HDM–BALF–MWCNT corona with a band at ∼70 kDa (likely albumin), as well as ∼15 kDa and ∼8 kDa (Fig. 6). A 12 kDa, possibly der p 30, is very faint. An initial BALF corona results in a BALF–HDM–MWCNT corona with similar bands to HDM–BALF–MWCNT, but different relative intensities (Fig. 6). The ∼15 kDa band dominates HDM–BALF–MWCNT and the ∼70 kDa band dominates BALF–HDM–MWCNT.
Interaction of HDM–MWCNTs with albumin
Gel electrophoresis following the incubation of HDM–MWCNTs with BALF suggests that der p 2 is present in the HDM–BALF–MWCNT coronas and is not displaced from the surface of MWCNTs by the proteins present in BALF (Fig. 6). To probe this observation, we used albumin, specifically bovine serum albumin (BSA, 66 kDa), as a representative BALF protein. The use of this single representative protein, instead of the BALF mixture, makes proteomics tractable. Repeating the formation of sequential coronas, now using BSA in place of BALF, shows a similar result. An initial HDM corona leads to a HDM-dominated corona (bands at ∼15 kDa and ∼12 kDa) with a faint ∼70 kDa band. An initial BSA corona leads to a complex corona dominated by a ∼70 kDa band and previously unobserved proteins (Fig. 7). Der p 2 (14 kDa) is not clearly visible in the BSA–HDM–MWCNT sample.The proteomics data shows that the HDM–BSA–MWCNT corona contains a high concentration of albumin (39.7 ± 4.38%) and der p 2 (36.0 ± 2.91%) (Table 3, Fig. 8). Der p 30 is present at low levels (0.781 ± 0.81%). These results with BSA are in good agreement with the BALF gel electrophoresis experiments (HDM–BALF–MWCNT; Fig. 6), which showed a strong band at ∼15 kDa, similar to the molecular weight of der p 2 (14 kDa) and no clear der p 30 band (12 kDa).
| Protein | MW (kDa) | HDM–BSA–MWCNT (%) | BSA–HDM–MWCNT (%) |
|---|---|---|---|
| Albumin | 69 | 39.7 ± 4.38 | 98.1 ± 0.72 |
| Der p 2 (NPC2 family) | 14 | 36.0 ± 2.91 | 0.129 ± 0.10 |
| Der f 22 (group 2-like) | 18 | 4.37 ± 6.44 | — |
| Fatty acid-binding protein-like | 15 | 3.52 ± 1.75 | — |
| Nucleoside diphosphate kinase | 18 | 2.82 ± 1.76 | — |
| Der p 30 (ferritin) | 12 | 0.781 ± 0.81 | 0.293 ± 0.50 |
| Probable methylmalonate-semialdehyde dehydrogenase | 59 | — | 0.293 ± 0.32 |
| Der p 11 (paramyosin) | 102 | 1.15 ± 0.61 | 0.129 ± 0.02 |
| Probable serine/threonine-protein kinase | 191 | — | 0.130 ± 0.084 |
The BSA–HDM–MWCNT corona is less physiologically relevant as we expect the MWCNTs to be exposed to HDM and then lung fluid. The initial exposure to BSA leads to an albumin-dominated corona (98.1 ± 0.72%) with a unique collection of HDM corona proteins (Table 3). Der p 2 was not detected in the BSA–HDM–MWCNT coronas, suggesting albumin inhibits the binding of der p 2 binding to the MWCNT surface. This points towards potential surface treatments for MWCNTs that could mitigate any enhanced allergic response. The BALF–HDM–MWCNT gel electrophoresis did show a band at ∼15 kDa, but it is likely that this is a BALF protein (Fig. 4).
Conclusions
This work aimed to model real-world exposures to MWCNTs by considering the interaction of HDM proteins with MWCNTs. A combination of XPS (Table 1), gel electrophoresis (Fig. 2), protein concentration assay (Fig. 3), western blotting (Fig. S2†), and proteomics (Table 2 and Tables S1 and S2†) shows that HDM proteins adsorb on the surface of MWCNTs forming a protein corona, similar to the coronas observed for nanomaterials used in biomedical applications.29,69–71 Much previous work has focused on serum proteins. The protein corona dictates the subsequent biological response to these nanomaterials. It is likely that the HDM corona will similarly determine the cellular and in vivo response to HDM–MWCNTs. We find that der p 2, a major HDM allergen,42 becomes highly concentrated on the surface of MWCNTs, which may be important for allergic airway disease. When the HDM–MWCNTs are then exposed to BALF or BSA (Fig. 5), which serves as a representative lung fluid protein, we find that der p 2 remains highly abundant (36.0 ± 2.91% with BSA) (Fig. 6 and 7, Table 3). This shows that der p 2 is not displaced from the surface of MWCNTs by lung fluid proteins and suggests that any physiological response due to the highly concentrated der p 2 will not be mitigated by lung fluid, although future functional assays will be necessary to probe the allergic potential of der p 2 adsorbed on the surface of MWCNTs. Previous in vivo experiments showed that mice co-exposed to MWCNTs and HDM extract displayed increased eosinophilic lung inflammation that was not observed with MWCNTs or HDM alone,72 suggesting der p 2 is functional. The results described in this manuscript suggest that the high local concentration of der p 2 on the MWCNTs may deliver the allergen to immune cells exacerbating allergic lung inflammation. Future work will extend these studies to MWCNTs following common purification methods and chemical functionalizations to determine if these results are general. In addition, a variety of carbon-based nanomaterials present in the environment (ultrafine particulate matter, wildfire smoke) exacerbate asthma.73,74 Our findings may have broad implications for understanding the underlying mechanism.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This study received funding from the NIH-NIEHS (R01ES032443). R. D. B. and L. J. T. were partially funded by NIEHS Training Grant T32ES007046. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under DGE 2139754 to J. D. We thank the Duke University School of Medicine for the use of the Proteomics and Metabolomics Shared Resource, which provided proteomics service, with special thanks to Erik Soderblom and Greg Waitt for technical advice. TEM was performed at the Chapel Hill Analytical and Nanofabrication Laboratory, CHANL, a member of the North Carolina Research Triangle Nanotechnology Network (RTNN), which is supported by the National Science Foundation, Grant ECCS-1542015, as part of the National Nanotechnology Coordinated Infrastructure (NNCI).References
- V. T. Rathod, J. S. Kumar and A. Jain, Polymer and ceramic nanocomposites for aerospace applications, Appl. Nanosci., 2017, 7, 519–548 CrossRef CAS.
- Carbon nanotubes: market size, share & trends analysis report by product (SWNT, MWNT), by application (polymers, energy, electrical & electronics), and segment forecasts, 2015–2022, San Francisco, CA, 2022.
- K. Donaldson, R. Aitken, L. Tran, V. Stone, R. Duffin, G. Forrest and A. Alexander, Carbon nanotubes: a review of their properties in relation to pulmonary toxicology and workplace safety, Toxicol. Sci., 2006, 92, 5–22 CrossRef CAS PubMed.
- J. P. Ryman-Rasmussen, M. F. Cesta, A. R. Brody, J. K. Shipley-Phillips, J. I. Everitt, E. W. Tewksbury, O. R. Moss, B. A. Wong, D. E. Dodd and M. E. Andersen, Inhaled carbon nanotubes reach the subpleural tissue in mice, Nat. Nanotechnol., 2009, 4, 747–751 CrossRef CAS PubMed.
- I. G. Canu, K. Batsungnoen, A. Maynard and N. Hopf, State of knowledge on the occupational exposure to carbon nanotubes, Int. J. Hyg. Environ. Health, 2020, 225, 113472 CrossRef PubMed.
- M. D. Wright, A. J. Buckley and R. Smith, Estimates of carbon nanotube deposition in the lung: improving quality and robustness, Inhalation Toxicol., 2020, 32, 282–298 CrossRef CAS PubMed.
- J. Muller, F. Huaux, N. Moreau, P. Misson, J.-F. Heilier, M. Delos, M. Arras, A. Fonseca, J. B. Nagy and D. Lison, Respiratory toxicity of multi-wall carbon nanotubes, Toxicol. Appl. Pharmacol., 2005, 207, 221–231 CrossRef CAS PubMed.
- D. W. Porter, A. F. Hubbs, R. R. Mercer, N. Wu, M. G. Wolfarth, K. Sriram, S. Leonard, L. Battelli, D. Schwegler-Berry and S. Friend, Mouse pulmonary dose-and time course-responses induced by exposure to multi-walled carbon nanotubes, Toxicology, 2010, 269, 136–147 CrossRef CAS PubMed.
- M. F. Cesta, J. P. Ryman-Rasmussen, D. G. Wallace, T. Masinde, G. Hurlburt, A. J. Taylor and J. C. Bonner, Bacterial lipopolysaccharide enhances PDGF signaling and pulmonary fibrosis in rats exposed to carbon nanotubes, Am. J. Respir. Cell Mol. Biol., 2010, 43, 142–151 CrossRef CAS PubMed.
- J. P. Ryman-Rasmussen, E. W. Tewksbury, O. R. Moss, M. F. Cesta, B. A. Wong and J. C. Bonner, Inhaled multiwalled carbon nanotubes potentiate airway fibrosis in murine allergic asthma, Am. J. Respir. Cell Mol. Biol., 2009, 40, 349–358 CrossRef CAS PubMed.
- K.-i. Inoue, E. Koike, R. Yanagisawa, S. Hirano, M. Nishikawa and H. Takano, Effects of multi-walled carbon nanotubes on a murine allergic airway inflammation model, Toxicol. Appl. Pharmacol., 2009, 237, 306–316 CrossRef CAS PubMed.
- E. M. Rydman, M. Ilves, A. J. Koivisto, P. A. Kinaret, V. Fortino, T. S. Savinko, M. T. Lehto, V. Pulkkinen, M. Vippola and K. J. Hämeri, Inhalation of rod-like carbon nanotubes causes unconventional allergic airway inflammation, Part. Fibre Toxicol., 2014, 11, 1–17 CrossRef PubMed.
- S. G. Han, R. Andrews and C. G. Gairola, Acute pulmonary response of mice to multi-wall carbon nanotubes, Inhalation Toxicol., 2010, 22, 340–347 CrossRef CAS PubMed.
- A. J. Taylor-Just, M. D. Ihrie, K. S. Duke, H. Y. Lee, D. J. You, S. Hussain, V. K. Kodali, C. Ziemann, O. Creutzenberg and A. Vulpoi, The pulmonary toxicity of carboxylated or aminated multi-walled carbon nanotubes in mice is determined by the prior purification method, Part. Fibre Toxicol., 2020, 17, 1–18 CrossRef PubMed.
- K. S. Duke, A. J. Taylor-Just, M. D. Ihrie, K. A. Shipkowski, E. A. Thompson, E. C. Dandley, G. N. Parsons and J. C. Bonner, STAT1-dependent and-independent pulmonary allergic and fibrogenic responses in mice after exposure to tangled versus rod-like multi-walled carbon nanotubes, Part. Fibre Toxicol., 2017, 14, 1–15 CrossRef PubMed.
- OSHA Fact Sheet: Working safely with nanomaterials, U.S. Department of Labor, Washington D.C.
- H. Y. Lee, D. J. You, A. J. Taylor-Just, L. Tisch, R. D. Bartone, H. M. Atkins, L. M. Ralph, S. Antoniak and J. C. Bonner, Role of the protease-activated receptor-2 (PAR2) in the exacerbation of house dust mite-induced murine allergic lung disease by multi-walled carbon nanotubes, Part. Fibre Toxicol., 2023, 20, 32 CrossRef CAS PubMed.
- E. D. Kuempel, M.-C. Jaurand, P. Møller, Y. Morimoto, N. Kobayashi, K. E. Pinkerton, L. M. Sargent, R. C. Vermeulen, B. Fubini and A. B. Kane, Evaluating the mechanistic evidence and key data gaps in assessing the potential carcinogenicity of carbon nanotubes and nanofibers in humans, Crit. Rev. Toxicol., 2017, 47, 1–58 CrossRef CAS PubMed.
- A. V. Singh, P. Laux, A. Luch, C. Sudrik, S. Wiehr, A.-M. Wild, G. Santomauro, J. Bill and M. Sitti, Review of emerging concepts in nanotoxicology: Opportunities and challenges for safer nanomaterial design, Toxicol. Mech. Methods, 2019, 29, 378–387 CrossRef CAS PubMed.
- M. Nicoletti, C. Capodanno, C. Gambarotti and E. Fasoli, Proteomic investigation on bio-corona of functionalized multiwalled carbon nanotubes, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 2293–2303 CrossRef CAS PubMed.
- L. García-Hevia, M. Saramiforoshani, J. Monge, N. Iturrioz-Rodríguez, E. Padín-González, F. González, L. González-Legarreta, J. González and M. L. Fanarraga, The unpredictable carbon nanotube biocorona and a functionalization method to prevent protein biofouling, J. Nanobiotechnol., 2021, 19, 1–13 CrossRef PubMed.
- X. Cai, R. Ramalingam, H. San Wong, J. Cheng, P. Ajuh, S. H. Cheng and Y. W. Lam, Characterization of carbon nanotube protein corona by using quantitative proteomics, Nanomedicine, 2013, 9, 583–593 CrossRef CAS PubMed.
- K. Bhattacharya, S. P. Mukherjee, A. Gallud, S. C. Burkert, S. Bistarelli, S. Bellucci, M. Bottini, A. Star and B. Fadeel, Biological interactions of carbon-based nanomaterials: From coronation to degradation, Nanomedicine, 2016, 12, 333–351 CrossRef CAS PubMed.
- C. C. Fleischer and C. K. Payne, Nanoparticle-cell interactions: Molecular structure of the protein corona and cellular outcomes, Acc. Chem. Res., 2014, 47, 2651–2659 CrossRef CAS PubMed.
- C. D. Walkey and W. C. W. Chan, Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment, Chem. Soc. Rev., 2012, 41, 2780–2799 RSC.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- K. Nienhaus and G. U. Nienhaus, Towards a molecular-level understanding of the protein corona around nanoparticles–recent advances and persisting challenges, Curr. Opin. Biomed. Eng., 2019, 10, 11–22 CrossRef.
- L. Kobos and J. Shannahan, Biocorona-induced modifications in engineered nanomaterial–cellular interactions impacting biomedical applications, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, e1608 Search PubMed.
- C. K. Payne, A protein corona primer for physical chemists, J. Chem. Phys., 2019, 151, 130901 CrossRef PubMed.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, A decade of the protein corona, ACS Nano, 2017, 11, 11773–11776 CrossRef CAS PubMed.
- H. Whitwell, R.-M. Mackay, C. Elgy, C. Morgan, M. Griffiths, H. Clark, P. Skipp and J. Madsen, Nanoparticles in the lung and their protein corona: the few proteins that count, Nanotoxicology, 2016, 10, 1385–1394 CrossRef CAS PubMed.
- K. M. Poulsen, M. C. Albright, N. J. Niemuth, R. M. Tighe and C. K. Payne, Interaction of TiO2 nanoparticles with lung fluid proteins and the resulting macrophage inflammatory response, Environ. Sci.: Nano, 2023, 10, 2427–2436 RSC.
- W. R. Thomas, B. J. Hales and W.-A. Smith, House dust mite allergens in asthma and allergy, Trends Mol. Med., 2010, 16, 321–328 CrossRef CAS PubMed.
- A. Jacquet, The role of innate immunity activation in house dust mite allergy, Trends Mol. Med., 2011, 17, 604–611 CrossRef CAS PubMed.
- M. A. Calderón, A. Linneberg, J. Kleine-Tebbe, F. De Blay, D. H. F. de Rojas, J. C. Virchow and P. Demoly, Respiratory allergy caused by house dust mites: what do we really know?, J. Allergy Clin. Immunol., 2015, 136, 38–48 CrossRef PubMed.
- T. A. Platts-Mills, A. L. de Weck, R. Aalberse, J. Bessot, B. Bjorksten, E. Bischoff, J. Bousquet, J. Van Bronswijk, G. ChannaBasavanna and M. Chapman, Dust mite allergens and asthma—a worldwide problem, J. Allergy Clin. Immunol., 1989, 83, 416–427 CrossRef PubMed.
- W. R. Thomas, W.-A. Smith, B. J. Hales, K. L. Mills and R. M. O'Brien, Characterization and immunobiology of house dust mite allergens, Int. Arch. Allergy Immunol., 2002, 129, 1–18 CrossRef CAS PubMed.
- M. D. Chapman and T. Platts-Mills, Purification and characterization of the major allergen from Dermatophagoides pteronyssinus-antigen P1, J. Immunol., 1980, 125, 587–592 CrossRef CAS.
- R. Waldron, J. McGowan, N. Gordon, C. McCarthy, E. B. Mitchell and D. A. Fitzpatrick, Proteome and allergenome of the European house dust mite Dermatophagoides pteronyssinus, PLoS One, 2019, 14, e0216171 CrossRef PubMed.
- V. Bordas-Le Floch, M. Le Mignon, L. Bussières, K. Jain, A. Martelet, V. Baron-Bodo, E. Nony, L. Mascarell and P. Moingeon, A combined transcriptome and proteome analysis extends the allergome of house dust mite Dermatophagoides species, PLoS One, 2017, 12, e0185830 CrossRef PubMed.
- M. Reithofer and B. Jahn-Schmid, Allergens with protease activity from house dust mites, Int. J. Mol. Sci., 2017, 18, 1368 CrossRef PubMed.
- M. Weghofer, W. Thomas, G. Pittner, F. Horak, R. Valenta and S. Vrtala, Comparison of purified Dermatophagoides pteronyssinus allergens and extract by two-dimensional immunoblotting and quantitative immunoglobulin E inhibitions, Clin. Exp. Allergy, 2005, 35, 1384–1391 CrossRef CAS PubMed.
- W. R. Thomas, T. K. Heinrich, W.-A. Smith and B. J. Hales, Pyroglyphid house dust mite allergens, Protein Pept. Lett., 2007, 14, 943–953 CrossRef CAS PubMed.
- K. Meno, P. B. Thorsted, H. Ipsen, O. Kristensen, J. N. Larsen, M. D. Spangfort, M. Gajhede and K. Lund, The crystal structure of recombinant proDer p 1, a major house dust mite proteolytic allergen, J. Immunol., 2005, 175, 3835–3845 CrossRef CAS PubMed.
- S. De Halleux, E. Stura, L. VanderElst, V. Carlier, M. Jacquemin and J.-M. Saint-Remy, Three-dimensional structure and IgE-binding properties of mature fully active Der p 1, a clinically relevant major allergen, J. Allergy Clin. Immunol., 2006, 117, 571–576 CrossRef CAS PubMed.
- M. Chruszcz, M. D. Chapman, L. D. Vailes, E. A. Stura, J.-M. Saint-Remy, W. Minor and A. Pomés, Crystal structures of mite allergens Der f 1 and Der p 1 reveal differences in surface-exposed residues that may influence antibody binding, J. Mol. Biol., 2009, 386, 520–530 CrossRef CAS PubMed.
- U. Derewenda, J. Li, Z. Derewenda, Z. Dauter, G. Mueller, G. Rule and D. Benjamin, The crystal structure of a major dust mite allergen Der p 2, and its biological implications, J. Mol. Biol., 2002, 318, 189–197 CrossRef CAS PubMed.
- G. M. Park, S. M. Lee, I. Y. Lee, H. I. Ree, K. S. Kim, C. S. Hong and T. S. Yong, Localization of a major allergen, Der p 2, in the gut and faecal pellets of Dermatophagoides pteronyssinus, Clin. Exp. Allergy, 2000, 30, 1293–1297 CrossRef CAS PubMed.
- M. D. Ihrie, K. S. Duke, K. A. Shipkowski, D. J. You, H. Y. Lee, A. J. Taylor-Just and J. C. Bonner, STAT6-dependent exacerbation of house dust mite-induced allergic airway disease in mice by multi-walled carbon nanotubes, NanoImpact, 2021, 22, 100309 CrossRef CAS PubMed.
- I. Radauer-Preiml, A. Andosch, T. Hawranek, U. Luetz-Meindl, M. Wiederstein, J. Horejs-Hoeck, M. Himly, M. Boyles and A. Duschl, Nanoparticle-allergen interactions mediate human allergic responses: protein corona characterization and cellular responses, Part. Fibre Toxicol., 2015, 13, 1–15 CrossRef PubMed.
- E. Kuijpers, C. Bekker, W. Fransman, D. Brouwer, P. Tromp, J. Vlaanderen, L. Godderis, P. Hoet, Q. Lan and D. Silverman, Occupational exposure to multi-walled carbon nanotubes during commercial production synthesis and handling, Ann. Occup. Hyg., 2016, 60, 305–317 CrossRef CAS PubMed.
- B. Nowack, R. M. David, H. Fissan, H. Morris, J. A. Shatkin, M. Stintz, R. Zepp and D. Brouwer, Potential release scenarios for carbon nanotubes used in composites, Environ. Int., 2013, 59, 1–11 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- J. Cox and M. Mann, MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol., 2008, 26, 1367–1372 CrossRef CAS PubMed.
- S. Tyanova, T. Temu and J. Cox, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics, Nat. Protoc., 2016, 11, 2301–2319 CrossRef CAS PubMed.
- cRAP protein sequences: The Global Proteome Machine, 2012.
- S. Tyanova, T. Temu, P. Sinitcyn, A. Carlson, M. Y. Hein, T. Geiger, M. Mann and J. Cox, The Perseus computational platform for comprehensive analysis of (prote) omics data, Nat. Methods, 2016, 13, 731–740 CrossRef CAS PubMed.
- Y. Perez-Riverol, A. Csordas, J. Bai, M. Bernal-Llinares, S. Hewapathirana, D. J. Kundu, A. Inuganti, J. Griss, G. Mayer and M. Eisenacher, The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res., 2019, 47, D442–D450 CrossRef CAS PubMed.
- R. P. Silvy, C. Pirlot and B. Culot, Catalyst system for a multi-walled carbon nanotube production process, US Pat., 7923615, 2011 Search PubMed.
- R. P. Silvy, F. Liegeois, B. Culot and S. Lambert, Method of synthesising a support catalyst for the production of carbon nanotubes, US Pat., 7754181, 2010 Search PubMed.
- A.-C. Dupuis, The catalyst in the CCVD of carbon nanotubes—a review, Prog. Mater. Sci., 2005, 50, 929–961 CrossRef CAS.
- C. M. White, R. Banks, I. Hamerton and J. F. Watts, Characterisation of commercially CVD grown multi-walled carbon nanotubes for paint applications, Prog. Org. Coat., 2016, 90, 44–53 CrossRef CAS.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. B. Bombelli and K. A. Dawson, Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles, J. Am. Chem. Soc., 2011, 133, 2525–2534 CrossRef CAS PubMed.
- K. Partikel, R. Korte, N. C. Stein, D. Mulac, F. C. Herrmann, H.-U. Humpf and K. Langer, Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles, Eur. J. Pharm. Biopharm., 2019, 141, 70–80 CrossRef CAS PubMed.
- C. Gräfe, A. Weidner, M. vd Lühe, C. Bergemann, F. H. Schacher, J. H. Clement and S. Dutz, Intentional formation of a protein corona on nanoparticles: Serum concentration affects protein corona mass, surface charge, and nanoparticle–cell interaction, Int. J. Biochem. Cell Biol., 2016, 75, 196–202 CrossRef PubMed.
- K. M. Poulsen and C. K. Payne, Concentration and composition of the protein corona as a function of incubation time and serum concentration: an automated approach to the protein corona, Anal. Bioanal. Chem., 2022, 414, 7265–7275 CrossRef CAS PubMed.
- J. M. Cyphert-Daly, Z. Yang, J. L. Ingram, R. M. Tighe and L. G. Que, Physiologic response to chronic house dust mite exposure in mice is dependent on lot characteristics, J. Allergy Clin. Immunol., 2019, 144, 1428–1432 CrossRef PubMed.
- G. M. Hilton, PhD, North Carolina State University, 2017 Search PubMed.
- S. Runa, M. Hussey and C. K. Payne, Nanoparticle–cell interactions: Relevance for public health, J. Phys. Chem. B, 2018, 122, 1009–1016 CrossRef CAS PubMed.
- J. Shannahan, The biocorona: a challenge for the biomedical application of nanoparticles, Nanotechnol. Rev., 2017, 6, 345–353 CAS.
- D. Docter, S. Strieth, D. Westmeier, O. Hayden, M. Gao, S. K. Knauer and R. H. Stauber, No king without a crown–impact of the nanomaterial-protein corona on nanobiomedicine, Nanomedicine, 2015, 10, 503–519 CrossRef CAS PubMed.
- M. D. Ihrie, A. J. Taylor-Just, N. J. Walker, M. D. Stout, A. Gupta, J. S. Richey, B. K. Hayden, G. L. Baker, B. R. Sparrow and K. S. Duke, Inhalation exposure to multi-walled carbon nanotubes alters the pulmonary allergic response of mice to house dust mite allergen, Inhalation Toxicol., 2019, 31, 192–202 CrossRef CAS PubMed.
- O. K. Kurt, J. Zhang and K. E. Pinkerton, Pulmonary health effects of air pollution, Curr. Opin. Pulm. Med., 2016, 22, 138 CrossRef CAS PubMed.
- C. E. Reid, M. Brauer, F. H. Johnston, M. Jerrett, J. R. Balmes and C. T. Elliott, Critical review of health impacts of wildfire smoke exposure, Environ. Health Perspect., 2016, 124, 1334–1343 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00666b |
| This journal is © The Royal Society of Chemistry 2024 |