The effect of nanoparticle surface charge on freshwater algae growth, reproduction, and lipid production†
Emma
McKeel
 a,
Hye-In
Kim
a,
Hye-In
Kim
 b,
Su-Ji
Jeon
b,
Su-Ji
Jeon
 b,
Juan Pablo
Giraldo
b,
Juan Pablo
Giraldo
 b and
Rebecca
Klaper
*a
b and
Rebecca
Klaper
*a
aSchool of Freshwater Sciences, University of Wisconsin-Milwaukee, 600 E Greenfield Ave Milwaukee, WI 53204, USA. E-mail: rklaper@uwm.edu
bDepartment of Botany and Plant Sciences, University of California Riverside, USA
First published on 16th December 2023
Abstract
Surface charge is a key characteristic of nanoparticles that has great potential to impact the interactions of nanoparticles and biological systems. Understanding the role charge plays in these interactions is key to determining the ecological risks of nanoparticle exposure and informing sustainable nanoparticle design. In this study, the model freshwater algae Raphidocelis subcapitata was exposed to carbon dots (CDs) functionalized with polymers to have positive, negative, or neutral surface charges to examine the impact of nanoparticle surface charge on nano–algae interactions. Traditional toxicological endpoints of survival and growth inhibition were measured. Additionally, morphological impacts on whole cells, individual organelles, and cellular components were quantified using high-content fluorescence microscopy, demonstrating one of the first uses of high-content imaging in microalgae. Results indicate that PEI functionalized, positively charged CDs are most toxic to green algae (EC50 42.306 μg L−1), but that CDs with negative charge induce sublethal impacts on algae. PEI-CD toxicity is hypothesized to be related to electrostatic interactions between CDs and the algal cell wall, which lead to significant cell aggregation. Interestingly, morphological data suggests that exposure to both positively and negatively charged CDs leads to increased neutral lipid droplet formation, a possible indicator of nutrient stress. Further investigation of the mechanisms underlying impacts of nanoparticle surface charge on algae biology can lead to more sustainable nanoparticle design and environmental protections.
Environmental significanceThe advancement of nanotechnology has also led to growing concerns regarding the potential environmental impacts of nanomaterials. This work elucidates the role that nanoparticle surface chemistry plays in interactions with freshwater microalga, key organisms in ecosystem health that are commonly used in ecotoxicological experiments. Our findings demonstrate that positively charged particles are most toxic to microalgae, but that sublethal morphological changes occur after exposure to non-toxic doses of nanoparticles. These findings can enable the design of nanoparticles that are safer for aquatic life and inform proper protections to prevent adverse ecological effects caused by algal nanoparticle exposure. |
Introduction
The rapid development of engineered nanomaterials in recent years has generated excitement surrounding their potential to revolutionize countless areas of society, from technology to agriculture and medicine. However, this development has also led to growing concerns surrounding the environmental impacts of nanoparticles (NPs) which have a high probability of making their way into the environment. Despite this, much is still unknown about how nanomaterial properties impact organism health in the environment. Understanding the risk nanomaterials pose to environmental health will require a better understanding of their adverse effects, and the mechanisms by which they occur. This task is made difficult by the diversity of NPs, which vary greatly in their composition and surface chemistry. Therefore, understanding what properties make a nanoparticle toxic can be a useful approach in the field of nanotoxicology.One such property with particular importance to nanoparticle toxicity is surface charge, which has been found to impact mechanisms of toxicity and rate of uptake.1,2 However, most cellular studies investigating nanoparticle charge have involved mammalian cell cultures. In mammalian cells, increased particle surface charge is associated with greater toxicity.3,4 Electrostatic interactions likely play a role in this increased toxicity, with positively charged particles interacting with negatively charged DNA and membranes.4,5 In environmental models, positively charged NPs have similarly been found to be more toxic than those with negative or neutral surface charge. In Daphnia magna, for example, gold NPs functionalized to have positive surface charge were orders of magnitude more toxic than those with negative surface charge, with exposure to 10 μg L−1 positively charged NPs resulting in 40% mortality at 48 hours.6 Less studied is the impact of NP charge on cellular interactions with microalgae, primary producers in aquatic ecosystems. Due to their cell walls, algae have different surface interactions with and barriers to nanoparticle uptake than other eukaryotic cells. Previous work has shown that nanoparticle charge can impact uptake and bioaccumulation of silver nanoparticles in the algae Chlorella vulgaris, with a higher uptake rate constant observed for positively charged NPs compared to negative.7 Furthermore, adsorption measurements have shown that positive and neutral particles show stronger adsorption to the cell wall of Raphidocelis subcapitata, exhibiting the importance of surface charge in nano–algae interactions.8 Such adsorption could contribute to a shading effect, reducing algal growth by inhibiting photosynthesis.9 However, many questions remain regarding the impact of charge on microalgal morphology and cellular components, and the mechanisms through which nanoparticle charge impacts microalgae.
Fortunately, as nanotechnology develops, so do methods for understanding the impacts of nanomaterials on biological organisms. One such method is high-content screening (HCS), also known as high-content imaging (HCI), simply defined as the usage of automated imaging and quantitative image analysis to investigate biological questions.10,11 These methods are gaining popularity in the realm of toxicology, particularly in pharmaceuticals research, and have been validated across diverse human-derived cell types.12 However, high-content imaging has also been applied in nanotoxicology—for example, in evaluating the impacts of transition metal oxide nanoparticles to zebrafish.13 In the field of algal toxicology, the analysis of morphological traits through fluorescence and bright-field imaging is a well-established method which has been used to evaluate cell size, lipid content, and cell division, among other features.14–16 However, traditional fluorescence imaging methods require time-consuming analysis and manual imaging, introducing human error and limiting the number of cells which can be observed. High-content imaging allows for the utilization of previously validated stains and fluorescent channels. Rather than imaging occurring manually, though, it is automated, reducing human bias and increasing throughput. This project is one of the first to utilize high-throughput fluorescence imaging to evaluate the impacts of nanoparticles on microalgae. In this work, specific attention will be paid to previously validated morphological endpoints, including impacts on cell size, nucleation state, and lipid droplet production.
In this study, the impact of NP charge on algae–nano interactions was investigated. Carbon dots (CDs) were used as a model particle for this work due to their intrinsic fluorescent properties, tunable surface chemistry, and emergent applications in nano-enabled agriculture.17,18 CDs were functionalized with polymers to vary their surface charge. Polyethylenimine (PEI) functionalized CDs were created with positive surface charge. Carboxylated polyethylenimine (CP) and polyvinylpyrrolidone (PVP) functionalized CDs were synthesized with negative surface charges. A versatile carbon-based nanoparticle, CDs are known for their photoluminescence, and have been studied for use in biomedicine and energy as well as for their optical properties.19 Functionalization of carbon dots extends these applications further. As an example, PEI functionalized CDs (with arginine–disulfide linkers) have proven effective in the delivery of CRISPR/Cas9 components.20 Carbon dots are also preferably used over other nanoparticles due to their reported low toxicity.21–23 Although CD toxicity to algae has been observed, it is typically at very high concentrations. An EC50 of 70 mg L−1 was observed after four days of CD exposure in the algae Chlorella vulgaris.24 Similarly, Yao et al. reported a 96 hour EC50 of 74.8 mg L−1 in the algae Scenedesmus obliquus.25 However, toxicity could vary depending on the diverse CD functional and structural properties of CDs, including charge.
The freshwater algae Raphidocelis subcapitata was the microalgal species chosen as a model for this study. R. subcapitata is popular in ecotoxicological bioassays due to its sensitivity to contaminants and high growth rate and is recommended for such assays by the Organization for Economic Cooperation and Development (OECD).26R. subcapitata has been used both to study the toxicity of nanoparticles to the algae itself and to assess the risk of trophic transfer of nanoparticles from R. subcapitata to zooplankton, such as Daphnia carinata.27
This study aims to understand the impact of nanoparticle charge on nano–algae interactions. The dose dependent toxicity of carbon dots and their impacts on algal morphology were investigated. To accomplish this, we used traditional measures of toxicity as well as high-content imaging, morphological profiling, and lipid production assays to investigate impacts of CD exposure on R. subcapitata.
Experimental
Synthesis of the core carbon dots
The carbon dots were prepared following previously reported protocols with modifications.28 Briefly, 1.92 g of citric acid was dissolved into 2 mL of DI water by stirring in a ceramic mortar. 0.675 mL of ammonium hydroxide was added drop wise into the citric acid mixture twice. 2.40 g of urea was added slowly into the reaction mixture and dissolved for 10 minutes. The reaction mixture was then transferred to a preheated oven and kept at 180 °C for 90 minutes. The dried mixture was allowed to return to room temperature, then gently crushed, and added to 40 mL of DI water with stirring for 150 rpm. The solution was then moved to a conical tube and bath-sonicated for 30 min by 37 Hz (60% power, Elmasonic P) to enhance the dispersion of nanoparticles. The crude solution was centrifuged at 4500 rpm (3999×g) for 30 min to remove the large aggregates. After decanting the supernatant, approximately 7.5 mL of precipitates remained. The supernatant was purified using a dialysis membrane (molecular weight cut off 3k, Spectrum) to remove any unreacted precursors or small molecular residues. During the dialysis, 3 L of DI water was replaced every 12 hours, totaling three replacements. After dialysis, the core CDs were transferred into a conical tube and then bath sonicated for 30 minutes by 37 Hz (60% power). The desired core CDs were obtained by filtration with a nanopore syringe filter (pore size 20 nm, Whatman) then stored in a refrigerator until further modifications.Functionalization of the core carbon dots
Functionalization of core CDs was also conducted as previously reported by Kim et al.28 To functionalize core CDs with PEI10k (PEI-CD), 16 mL of the core CD solution (5 mg mL−1) was suspended in a 20 mL glass vial and the pH adjusted to 12 with NaOH solution (6 M). This solution was then added to 3.2 mL of PEI10k (0.1 g mL−1) while vigorously stirring, and transferred to an oven (Isotherm, Fisher Scientific) set at 85 °C for 16 hours. To purify the desired PEI-CD by extraction method, the mixture was cooled down to room temperature, then transferred into a 500 mL of Erlenmeyer flask. While stirring the core CD, the same volume of ethanol and chloroform were subsequently added. The mixture was vigorously mixed with a Volex for 5 to 10 seconds and centrifuged at 4500 rpm (3999×g) for 5 minutes. The organic solvent upper layer was discarded, and the extraction step was repeated 5 times. After removing the organic solvent layer, any remaining organic solvent residue was removed by air-blowing overnight. The unreacted polymer was removed with 30k centrifugal filters 3 times. The PVP10k functionalized CD (PVP-CD) was synthesized with the same method of PEI-CD synthesis, with the replacement of ethanol with acetone. The carboxylic acid functionalized PEI-CD (CP-CD) was prepared by modification of the PEI-CD surface with succinic anhydride. 1.5 g of succinic anhydride was dissolved in 10 mL of DMF, and the succinic anhydride solution was rapidly added to 30 mL of PEI-CD solution (1 mg mL−1) in a 100 mL beaker. The reaction mixture was stirred for 16 hours. The resulting mixture was purified by extraction method with ethanol and chloroform. The extraction and filtration methods were the same as the methods used for PEI-CDs.Particle characterization
Carbon dots were characterized using dynamic light scattering (DLS) with a Zetasizer Nano device (Malvern PANalytical, United Kingdom). Size and zeta potential measurements were obtained in both 10 mM TES buffer with 0.1 mM NaCl and OECD algae media. Final particle concentration was 1 mg mL−1. For zeta potential, the Smoluchowski model was used. HCl was used to adjust pH to 7.0.Starter culture and strain
The freshwater algae Raphidocelis subcapitata (also known as Selenastrum capricornutum) was obtained from the UTEX Culture Collection of Algae (UTEX 1648). Algae was cultured in OECD media under continuous illumination in an incubator at 24 °C. 400 mL OECD media were inoculated weekly in a 1 L Erlenmeyer flask. These cultures were allowed to grow for three days prior to use for inoculating exposure cultures.Exposure setup
Prior to exposure, concentration of algae in the starter culture was estimated using optical density. A standard curve was prepared to find the absorption coefficient before beginning exposure replicates. To do so, six samples of algae at random concentrations were prepared. These were measured for absorbance at a wavelength of 680 nm in a 96-well plate, using an Agilent BioTek Synergy H4 Hybrid Microplate Reader (Agilent Technologies, California, USA). Absorbance was then graphed opposite cell per mL counts obtained using a hemacytometer. Using a linear line of best fit, the absorption constant was found to be 8 × 10−8. This constant was used to estimate concentration, in cells per mL, of starter culture in a 96-well plate, using Beer's law,| A = εcl | (1) |
After estimation of starter culture concentration, algae were diluted in OECD media to yield a final concentration of 5 × 104 cells per mL for exposure. Algae was then cultured at a total volume of 40 mL in 250 mL Erlenmeyer flasks, under the same conditions as the starter culture (24 °C, orbital shaker at 100 rpm, and continuous lighting) in the absence or presence of CDs. Each replicate of exposures consisted of one control and three concentrations of each CD (10, 50, and 100 μg L−1). Cell free controls of nanoparticles in OECD media were also prepared for each treatment. Six replicates of each treatment were performed.
Growth inhibition
Optical density of algal cells was measured every 24 hours by removing a 300 μL aliquot from each exposure flask and measuring its absorbance at 680 nm, along with the absorbance of a cell-free control for each treatment. Concentration of cells per milliliter was then estimated as described above in exposure setup. These estimates were utilized to calculate percent growth inhibition, as outlined by OECD guidelines.26 This involved first calculating the average specific growth rate for each flask over the 72 hour exposure period as seen in eqn (2): | (2) |
These calculations were then used to calculate percent growth inhibition at 72 hours, using the equation:
 | (3) |
Cell staining and high-content imaging
Algal cells exposed to PEI, CP, or PVP CDs and cells grown in the absence of CDs were sampled at 24 hour intervals for imaging. 500 μL samples were taken from each exposure flask. Each sample was then stained with BODIPY 505/515 (ref. 29) (final concentration 5 μM) and NucBlue Live Ready Probes Reagent (Invitrogen, Massachusetts, USA). Samples were then incubated in the dark for 15 min. After incubation, 300 μL samples were dispensed into a 96-well plate for imaging. Images were acquired using an ImageXpress Micro XLS Widefield High-Content Imaging System (Molecular Devices, California, USA) in three channels: Cy5, DAPI, and GFP. The Cy5 (excitation 628/40 nm, emission 692/40 nm) channel was used to visualize the natural fluorescence of the algal chlorophyll. DAPI (excitation 377/50 nm, emission 447/60 nm) allowed for visualization of the nuclei after staining with NucBlue. GFP (excitation 472/30 nm, emission 520/35 nm) captured the fluorescence of the BODIPY-stained neutral lipid droplets. Sixteen images were collected of each replicate, with the data compiled and treated as a single replicate.Image analysis
Utilizing MetaXpress high-content image acquisition and analysis software (Molecular Devices, California, USA) automated image analysis was performed to quantify morphological features. Each channel was analyzed for one of these three purposes: Cy5 for cell counts and cell size, GFP for lipid droplet counts and sizes, and DAPI for nuclei counts, as seen in Fig. 1. A custom module was created within the MetaXpress software to automate the process of analyzing these traits, the details of which can be found in ESI.† | ||
| Fig. 1 Example images from ImageXpress acquisition, showing each channel individually and as an overlay. Individual channels were utilized for analysis. Images processed using Fiji.50 | ||
Statistical analysis
All statistical analysis was completed in R.30,31 Prior to statistical analysis, a Shapiro–Wilk test was utilized to ensure normal distribution of data. All treatments were then compared to the control. Endpoints for which all data was normally distributed were analyzed using an unpaired t-test. A series of Wilcoxon rank sum test was used for data which was not normally distributed to compare treatments to the control. For nucleation data, MetaXpress software output included average counts of cells per nucleation state (1, 2, 4, or >4 nuclei) of each treatment. The percentage of cells in each state was then calculated, and a Kruskal–Wallis test followed by a Dunn test was used to compare these percentages across all treatments. For nanoparticle characterization, an ANOVA test and subsequent Tukey's test were used to compare zeta potentials, as this data followed a normal distribution. Shapiro–Wilk and a Dunn test were used for nanoparticle size data. A confidence level of 95% was used to determine statistical significance. For growth inhibition data, the drc package was used to calculate an EC50 value.32 Results were visualized using ggplot.33Results and discussion
Nanoparticle characterization
Carbon dot zeta potential, but not size, was significantly impacted by suspension in OECD media, as seen in Fig. 2. This result was observed for both PEI-CDs and CP-CDs. PEI-CDs exhibited a mean zeta potential of 20.89 ± 0.895 mV in TES buffer, compared to 11.438 ± 0.959 mV in OECD media. Similarly, CP-CDs exhibited a statistically significant shift towards 0 mV when suspended in OECD media, with a zeta potential of −30.487 ± 0.904 mV in TES buffer and −16.56 ± 0.489 mV in OECD media. No significant differences were observed between PVP-CDs in TEs buffer and PVP-CDs in OECD media. Particle size was not significantly impacted by media for any particles. | ||
| Fig. 2 (a) Carbon dot zeta potential measured in TES buffer and OECD algae media. Asteriks indicate significant media-dependent differences in size or zeta potential (p < 0.05). (b) Diameter of carbon dots measured in both TES buffer and OECD media. Figures created using ggplot.33 | ||
These results suggest that the properties of functionalized CDs are impacted by suspension in OECD media. Given the differences in composition between TES buffer and OECD 201 media as well as previous studies showing the behavior of nanoparticles in OECD algal media, this finding is unsurprising.34,35 However, these findings exhibit the importance of testing particle characteristics in test media when performing ecotoxicological experiments. Additional particle characterization data is available in ESI† Fig. S1.
Effect of CDs on algae growth
Samples treated with PEI-CDs showed a dose-dependent increase in growth inhibition (Fig. 3). A Wilcoxon rank sum test was used to compare each treatment to the control, and statistically significant differences from the control were observed for all three PEI-CD treated samples (p < 0.05). Mean percent growth inhibitions were 11.69 ± 4.50%, 65.95 ± 9.24%, and 93.79 ± 10.16% for treatments of 10, 50, and 100 μg L−1, respectively. An EC50 of approximately 42.306 μg L−1 was calculated. No concentrations of CP or PVP-CDs resulted in significant growth inhibition, indicating that concentrations between 0 and 100 μg L−1 of these particles are not high enough to induce toxicity in R. subcapitata. | ||
| Fig. 3 Percent growth inhibition at 72 hours. Treatments are abbreviated as dose (μg L−1) followed by CD functionalization. Control treatment was abbreviated as CTRL. Asterisks represent a significant difference from control (p < 0.05).33 | ||
In addition to decreased growth, cells exposed to PEI-CDs exhibited significant clumping (see S2 in ESI†). Colocalization between PEI-CDs and R. subcapitata chloroplasts was further supported by confocal imaging (S3). This observation suggests electrostatic interactions between positively charged PEI-CDs and the negatively charged R. subcapitata cell, which has a zeta potential of approximately −40 mV in deionized water.8 Furthermore, such clumping and interactions with the algal cell wall can also provide insight into the mechanism by which PEI-CDs induce toxicity. It has previously been suggested that nanoparticle interactions with algal cell walls, and the aggregation caused by this interaction, can induce changes in growth, cell division, and photosynthetic activity.36,37 Given the significant aggregation observed in R. subcapitata cultures exposed to PEI-CDs, it is possible that such an interaction is also responsible for growth inhibition in this case. Regardless, the nanoparticle-induced aggregation of algal cells can have significant environmental effects. Such aggregates settle rapidly and could reduce available food sources for zooplankton, impacting food webs.38 Further work is required to determine the precise mechanism of growth inhibition and cell-particle interaction.
Impact of CDs on algae development
Changes in cell size have been associated with cell cycle disruption in microalgae,39 and have been observed after exposure to pollutants including nickel oxide nanoparticles.16 These changes can be further examined by tracking the nucleation state or DNA content of algal cells dividing by multiple fission.15,40R. subcapitata divides by multiple fission, with cells typically undergoing an increase in size, followed by a series of nuclear divisions. Commonly, this results in the formation of four autospores, but up to eight and as few as two autospores have also been observed to result from these divisions.41 An outline of a typical R. subcapitata cell cycle is shown in Fig. 4a. | ||
| Fig. 4 (a) A typical R. subcapitata cell cycle, wherein cells undergo growth, two nuclear divisions, and finally release daughter cells. (b) Nucleation state graphed by treatment, alongside (c) example images taken using an ImageXpress imaging system. (d) Average cell area in μm2. Asterisks indicate significant difference from the control (p < 0.05). (e) Example images of cells with varying sizes, taken with ImageXpress system. Magenta color represents the chloroplast (imaged in the Cy5 channel), green neutral lipid droplets stained with BODIPY 505-515 (imaged in the GFP channel), and blue Hoechst-stained nuclei (imaged in the DAPI channel). All example images processed with Fiji.50 | ||
The percentage of algae cells per nucleation state by CD treatment is shown in Fig. 4b, accompanied by example images in Fig. 4c. No significant differences were observed for any nucleation state (p = 0.8964 (1 nucleus), 0.7315 (2 nuclei), 0.9482 (4 nuclei), and 0.2821 (>4 nuclei)). This data suggests that CD exposure at the tested concentrations does not significantly impact the R. subcapitata cell cycle.
Changes in algae cell size were approximated by changes in cell area. This method allows for high-throughput screening for possible cell cycle changes or physiological impacts to algae after nanoparticle exposure. Limitations to this method do exist, as the area measured is based on the fluorescence of the chlorophyll, rather than a cytoplasm stain or brightfield imaging. However, similar approximations (such as using nuclear size to approximate cell area)42 are useful in developing high-content imaging assays for less studied cell types, and the use of high-content fluorescent imaging to document cell or organelle size is well documented.42–44 As seen in Fig. 4d, cell area changed significantly when compared to the control (Wilcoxon rank sum tests), particularly in the case of 50 μg L−1 PEI (p = 0.0260), 100 μg L−1 PEI (p = 0.00433), and 50 μg L−1 PVP (p = 0.0411) treatments. In the case of PEI-CD exposed cells, area decreases as CD concentration increases. This result can likely be explained by the dose-dependent increase in growth inhibition associated with PEI-CD exposure. The increase in R. subcapitata cell area associated with PVP-CDs can be associated with cell cycle interruption, particularly after cells undergo a growth phase. In this case, it would be expected that nucleation state data would reflect such changes. It is possible that this increased cell size is the result of another cellular impact, which requires further investigation to determine. Cells exposed to CP-CDs exhibited average areas very similar to the control and were not significantly different from it. Therefore, there is no evidence of cell cycle interruption in CP-CD exposed samples.
Lipid droplet accumulation
Algae are noted for their production of lipid droplets, particularly triacylglycerols (TAGs), which are of interest in biofuels production. The synthesis of lipids in algae occurs to the highest extent under stress conditions, posing a challenge to production of biofuels.45 Nitrogen deficiency, for example, is one of the most cited conditions which induce lipid droplet accumulation across many strains of algae.14,46 Oxidative stress has also been observed to increase lipid accumulation, as exhibited in the case of a halophilic strain of Dunaliella salina.47 Yet another mechanism by which algal lipid metabolism can be impacted is through an immune response. Overproduction of highly saturated TAGs and increased BODIPY fluorescence has been observed in the marine algae Emiliania huxleyi following viral infection.48 Therefore, although nutrient stress is by far the most studied mechanism by which environmental factors can influence algal accumulation of neutral lipids, there are potentially many other variables which can lead to lipid droplet accumulation.Significant changes in lipid content were observed in numerous samples with varying CD functionalization (Fig. 5). Wilcoxon rank sum tests were utilized in statistical analysis of lipid droplet count data, while unpaired t-tests were used for lipid droplet size data. After exposure to 10 μg L−1 of PEI-CDs, a significant increase in lipid droplet count (p = 0.0411) and size (p = 0.0307) was observed. Similarly, significant changes to lipid droplet count occurred after exposure to 10 μg L−1 CP-CDs (p = 0.00216), 50 μg L−1 CP-CDs (p = 0.00866), and 50 μg L−1 PVP-CDs (p = 0.0411). Lipid droplet size increased relative to the control in 100 μg L−1 CP-CDs (p = 0.0226) and 50 μg L−1 PVP-CDs (p = 0.0443). Increased accumulation of lipid droplets is expected for PEI-CD exposure, as PEI-CDs significantly increase growth inhibition at low concentrations. However, at 50 and 100 μg L−1 concentrations of PEI-CDs, no increase in lipid content is observed. This could be explained by the very high growth inhibition in these samples, leaving few living cells available for imaging. Interestingly CP and PVP-CDs at lower concentration exposures also resulted in increased lipid droplet accumulation. This indicates that despite a lack of growth inhibition resulting from exposure to PVP and CP-CDs, these nanoparticles still impact microalgae.
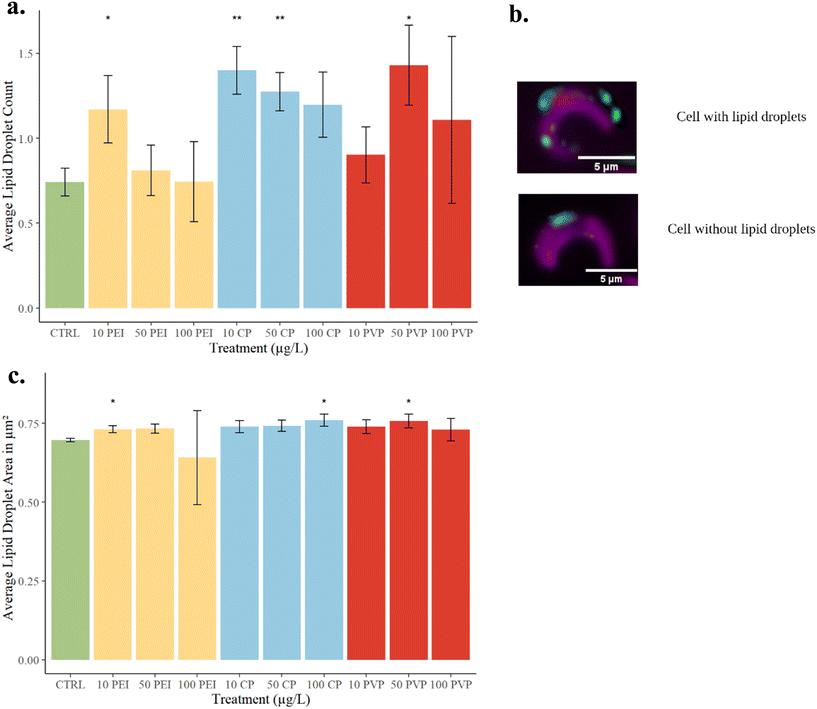 | ||
| Fig. 5 (a) Average lipid droplet count per cell. (b) Example images of cells with and without lipid droplets, taken using ImageXpress imaging system. Magenta color represents the chloroplast (imaged in the Cy5 channel), green neutral lipid droplets stained with BODIPY 505-515 (imaged in the GFP channel), and blue Hoechst-stained nuclei (imaged in the DAPI channel). (c) Average lipid droplet size in μm2. Asterisks indicate statistically significant difference from the control (p < 0.05). Images further processed using Fiji.50 | ||
The presence of lipid droplet accumulation in CD exposed algae also could provide insight into the mechanism by which CDs interact with R. subcapitata. Given the strong link between nutrient deficits, particularly nitrogen deficiency, and lipid droplet formation, it is possible that CD exposure is inducing nutrient stress. Nutrient adsorption onto carbon-based materials, including graphene oxide and nano biochar, has been observed in previous algal exposures.9,49 Nitrogen was among those nutrients which were found to be impacted by adsorption to carbon-based materials.43 Oxidative stress or another mechanism, such as an immune response cannot, however, be ruled out as potential mechanisms.
Conclusions
We tested the impact of charge on nanoparticle interactions with the microalgae R. subcapitata using functionalized carbon dots. Positively charged PEI-carbon dots were more toxic than CDs with negative zeta potentials, with PEI-CDs exhibiting an EC50 of approximately 42.306 μg L−1. Although carbon dots are generally considered to be a low toxicity nanoparticle, functionalization with positively charged polymers could significantly increase toxicity. Tuning surface charge density of positively charged nanoparticles can be a pathway to control their binding to algae cells and toxicity. In the case of R. subcapitata, we also observed significant cell aggregation after exposure to PEI-CDs. We hypothesize that this clumping is due to electrostatic interactions between the negative algal cells and positive PEI CDs. Furthermore, this clumping may be contributing to PEI-CD toxicity by limiting cell mobility, division, and photosynthesis. Analysis of morphological traits was also carried out using high-content imaging, demonstrating a novel use of this technique to quantify morphological changes to microalgae. Cell size, nucleation state, and lipid droplet production were the key developmental and biochemical traits studied with this technique. While evidence of significant cell cycle interruption was not observed, increased cell size occurred in cells treated with 50 μg L−1. Cells treated with 50 or 100 μg L−1 PEI-CDs exhibited decreased cell size. Interestingly, enhanced lipid droplet accumulation occurred in PEI 10 μg L−1, CP 10 and 50 μg L−1, and PVP 50 μg L−1 treatments. Often associated with nutrient or other stress, accumulation of neutral lipid droplets is also of interest in biofuels research. Carbon-based materials have previously been shown to adsorb nutrients from algal media, reducing nutrient availability for cells.9,49 Further research could determine whether this is the mechanism by which lipid droplet accumulation was induced, or if another mechanism is responsible.Understanding the impact of nanoparticle charge on nano–algae interactions will enable the design of sustainable nanoparticles and a better understanding of the ecological impacts of nanoparticles entering waterways. As primary producers in aquatic ecosystems, algae are not just model species, but also key species required to maintain the health of our waterways. Future studies should investigate the mechanisms by which functionalized CDs interact with and impact microalgae, as well as test the impact of nanoparticle characteristics such as size, shape, and charge density on interactions with algae. Furthermore, this work demonstrated the potential for high-content imaging to contribute to the field of nanotoxicology, identifying potential modes of action by which nanoparticles impact biological systems. The further development of this tool for use in test organisms of environmental significance should be a focus of future research.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant No. CHE-2001611, the NSF Center for Sustainable Nanotechnology. The CSN is part of the Centers for Chemical Innovation Program. Figures created with Biorender.References
- E. Fröhlich, The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles, Int. J. Nanomed., 2012, 7, 5577–5591 CrossRef PubMed.
- N. Schaeublin, A. Schrand, J. J. Schlager and S. M. Hussain, Surface charge of nanoparticles mediates mechanism of toxicity, Nanoscale, 2011, 3, 410–420 RSC.
- M. Havrdova, K. Hola, J. Skopalik, K. Tomankova, M. Petr, K. Cepe, K. Polakova, J. Tucek, A. B. Bourlinos and R. Zboril, Toxicity of carbon dots-Effect of surface functionalization on the cell viability, reactive oxygen species generation and cell cycle, Carbon, 2016, 99, 238–248 CrossRef CAS.
- Y.-W. Huang, M. Cambre and H.-J. Lee, The Toxicity of Nanoparticles Depends on Multiple Molecular and Physicochemical Mechanisms, Int. J. Mol. Sci., 2017, 18, 2702 CrossRef PubMed.
- A. Sukhanova, S. Bozrova, P. Sokolov, M. Berestovoy, A. Karaulov and I. Nabiev, Dependence of Nanoparticle Toxicity on Their Physical and Chemical Properties, Nanoscale Res. Lett., 2018, 13, 44 CrossRef PubMed.
- T. A. Qiu, J. S. Bozich, S. E. Lohse, A. M. Vartanian, L. M. Jacob, B. M. Meyer, I. L. Gunsolus, N. J. Niemuth, C. J. Murphy, C. L. Haynes and R. D. Klaper, Gene expression as an indicator of the molecular response and toxicity in the bacterium Shewanella oneidensis and the water flea Daphnia magna exposed to functionalized gold nanoparticles, Environ. Sci.: Nano, 2015, 2, 615–629 RSC.
- J. Zhang, Q. Xiang, L. Shen, J. Ling and C. Zhou, Surface charge-dependent bioaccumulation dynamics of silver nanoparticles in freshwater algae, Chemosphere, 2020, 247, 125936 CrossRef CAS PubMed.
- T. M. Nolte, N. B. Hartmann, J. M. Kleijn, J. Garnæs, D. van de Meent, A. Jan Hendriks and A. Baun, The toxicity of plastic nanoparticles to green algae as influenced by surface modification, medium hardness and cellular adsorption, Aquat. Toxicol., 2017, 183, 11–20 CrossRef CAS PubMed.
- J. Zhao, X. Cao, Z. Wang, Y. Dai and B. Xing, Mechanistic understanding toward the toxicity of graphene-family materials to freshwater algae, Water Res., 2017, 111, 18–27 CrossRef CAS PubMed.
- S. Li and M. Xia, Review of high-content screening applications in toxicology, Arch. Toxicol., 2019, 93, 3387–3396 CrossRef CAS PubMed.
- G. P. Way, H. Sailem, S. Shave, R. Kasprowicz and N. O. Carragher, Evolution and impact of high content imaging, SLAS Discovery, 2023, 28, 292–305 CrossRef CAS.
- C. Willis, J. Nyffeler and J. Harrill, Phenotypic Profiling of Reference Chemicals across Biologically Diverse Cell Types Using the Cell Painting Assay, SLAS Discovery, 2020, 25, 755–769 CrossRef CAS PubMed.
- S. Lin, Y. Zhao, T. Xia, H. Meng, Z. Ji, R. Liu, S. George, S. Xiong, X. Wang, H. Zhang, S. Pokhrel, L. Mädler, R. Damoiseaux, S. Lin and A. E. Nel, High Content Screening in Zebrafish Speeds up Hazard Ranking of Transition Metal Oxide Nanoparticles, ACS Nano, 2011, 5, 7284–7295 CrossRef CAS PubMed.
- S. Harchouni, B. Field and B. Menand, AC-202, a highly effective fluorophore for the visualization of lipid droplets in green algae and diatoms, Biotechnol. Biofuels, 2018, 11, 120 CrossRef PubMed.
- M. D. Machado and E. V. Soares, Reproductive cycle progression arrest and modification of cell morphology (shape and biovolume) in the alga Pseudokirchneriella subcapitata exposed to metolachlor, Aquat. Toxicol., 2020, 222, 105449 CrossRef CAS PubMed.
- C. A. Sousa, H. M. V. M. Soares and E. V. Soares, Toxic effects of nickel oxide (NiO) nanoparticles on the freshwater alga Pseudokirchneriella subcapitata, Aquat. Toxicol., 2018, 204, 80–90 CrossRef CAS PubMed.
- I. Santana, S.-J. Jeon, H.-I. Kim, M. R. Islam, C. Castillo, G. F. H. Garcia, G. M. Newkirk and J. P. Giraldo, Targeted Carbon Nanostructures for Chemical and Gene Delivery to Plant Chloroplasts, ACS Nano, 2022, 16, 12156–12173 CrossRef CAS PubMed.
- Y. Yao, L. Yue, X. Cao, F. Chen, J. Li, B. Cheng, C. Wang and Z. Wang, Carbon Dots Embedded in Nanoporous SiO 2 Nanoparticles for Enhancing Photosynthesis in Agricultural Crops, ACS Appl. Nano Mater., 2023, 6, 110–118 CrossRef CAS.
- J. Liu, R. Li and B. Yang, Carbon Dots: A New Type of Carbon-Based Nanomaterial with Wide Applications, ACS Cent. Sci., 2020, 6, 2179–2195 CrossRef CAS PubMed.
- I. Hashemzadeh, A. Hasanzadeh, F. Radmanesh, B. Khodadadi Chegeni, E. S. Hosseini, J. Kiani, A. Shahbazi, M. Naseri, Y. Fatahi, H. Nourizadeh, B. K. Y. Azar, A. R. Aref, Y. Liu, M. R. Hamblin and M. Karimi, Polyethylenimine-Functionalized Carbon Dots for Delivery of CRISPR/Cas9 Complexes, ACS Appl. Bio Mater., 2021, 4, 7979–7992 CrossRef CAS PubMed.
- G. Ge, L. Li, D. Wang, M. Chen, Z. Zeng, W. Xiong, X. Wu and C. Guo, Carbon dots: synthesis, properties and biomedical applications, J. Mater. Chem. B, 2021, 9, 6553–6575 RSC.
- S. C. Ray, A. Saha, N. R. Jana and R. Sarkar, Fluorescent Carbon Nanoparticles: Synthesis, Characterization, and Bioimaging Application, J. Phys. Chem. C, 2009, 113, 18546–18551 CrossRef CAS.
- J. Xu, Y. Zhou, G. Cheng, M. Dong, S. Liu and C. Huang, Carbon dots as a luminescence sensor for ultrasensitive detection of phosphate and their bioimaging properties: Carbon dots as a luminescence sensors, Luminescence, 2015, 30, 411–415 CrossRef CAS PubMed.
- M. Zhang, H. Wang, P. Liu, Y. Song, H. Huang, M. Shao, Y. Liu, H. Li and Z. Kang, Biotoxicity of degradable carbon dots towards microalgae Chlorella vulgaris, Environ. Sci.: Nano, 2019, 3316–3323 RSC.
- K. Yao, X. Lv, G. Zheng, Z. Chen, Y. Jiang, X. Zhu, Z. Wang and Z. Cai, Effects of Carbon Quantum Dots on Aquatic Environments: Comparison of Toxicity to Organisms at Different Trophic Levels, Environ. Sci. Technol., 2018, 52, 14445–14451 CrossRef CAS PubMed.
- OECD, Test No. 201: Freshwater Alga and Cyanobacteria, Growth Inhibition Test. OECD Guidelines for the Testing of Chemicals, Section 2, OECD Publishing, Paris, 2011 Search PubMed.
- S. Lekamge, A. F. Miranda, A. S. Ball, R. Shukla and D. Nugegoda, The toxicity of coated silver nanoparticles to Daphnia carinata and trophic transfer from alga Raphidocelis subcapitata, PLoS One, 2019, 14, e0214398 CrossRef CAS PubMed.
- K. Kim, S.-J. Jeon, P. Hu, C. M. Anastasia, W. F. Beimers, J. P. Giraldo and J. A. Pedersen, Sulfolipid density dictates the extent of carbon nanodot interaction with chloroplast membranes, Environ. Sci.: Nano, 2022, 9, 2691–2703 RSC.
- J. Rumin, H. Bonnefond, B. Saint-Jean, C. Rouxel, A. Sciandra, O. Bernard, J. P. Cadoret and G. Bougaran, The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae, Biotechnol. Biofuels, 2015, 8, 1–16 CrossRef CAS.
- R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2021 Search PubMed.
- RStudio Team, RStudio: Integrated Development for R. RStudio, PBC, Boston, MA, 2020 Search PubMed.
- C. Ritz, F. Baty, J. C. Streibig and D. Gerhard, Dose-Response Analysis Using R, PLoS One, 2015, 10, e0146021 CrossRef.
- H. Wickham, ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag New York, 2016, ISBN 978-3-319-24277-4, https://ggplot2.tidyverse.org Search PubMed.
- N. Manier, M. Garaud, P. Delalain, O. Aguerre-Chariol and P. Pandard, Behaviour of ceria nanoparticles in standardized test media – influence on the results of ecotoxicological tests, J. Phys.: Conf. Ser., 2011, 304, 012058 CrossRef.
- Y. Nur, J. R. Lead and M. Baalousha, Evaluation of charge and agglomeration behavior of TiO2 nanoparticles in ecotoxicological media, Sci. Total Environ., 2015, 535, 45–53 CrossRef CAS.
- S. Mizukami-Murata, Y. Suzuki, K. Sakurai and H. Yamashita, Freshwater alga Raphidocelis subcapitata undergoes metabolomic changes in response to electrostatic adhesion by micrometer-sized nylon 6 particles, Environ. Sci. Pollut. Res., 2021, 28, 66901–66913 CrossRef CAS.
- F. Perreault, N. Bogdan, M. Morin, J. Claverie and R. Popovic, Interaction of gold nanoglycodendrimers with algal cells (Chlamydomonas reinhardtii) and their effect on physiological processes, Nanotoxicology, 2012, 6, 109–120 CrossRef CAS PubMed.
- B. Campos, C. Rivetti, P. Rosenkranz, J. M. Navas and C. Barata, Effects of nanoparticles of TiO2 on food depletion and life-history responses of Daphnia magna, Aquat. Toxicol., 2013, 130–131, 174–183 CrossRef CAS PubMed.
- M. D. Machado and E. V. Soares, Modification of cell volume and proliferative capacity of Pseudokirchneriella subcapitata cells exposed to metal stress, Aquat. Toxicol., 2014, 147, 1–6 CrossRef CAS PubMed.
- A. C. Almeida, T. Gomes, J. A. B. Lomba and A. Lillicrap, Specific toxicity of azithromycin to the freshwater microalga Raphidocelis subcapitata, Ecotoxicol. Environ. Saf., 2021, 222, 112553 CrossRef CAS PubMed.
- T. Yamagishi, H. Yamaguchi, S. Suzuki, Y. Horie and N. Tatarazako, Cell reproductive patterns in the green alga Pseudokirchneriella subcapitata (=Selenastrum capricornutum) and their variations under exposure to the typical toxicants potassium dichromate and 3, 5-DCP, PLoS One, 2017, 1–12 Search PubMed.
- B. Knapp, I. Rebhan, A. Kumar, P. Matula, N. A. Kiani, M. Binder, H. Erfle, K. Rohr, R. Eils, R. Bartenschlager and L. Kaderali, Normalizing for individual cell population context in the analysis of high-content cellular screens, BMC Bioinf., 2011, 12, 485 CrossRef CAS PubMed.
- A. Capus, M. Monnerat, L. C. Ribeiro, W. De Souza, J. L. Martins and C. Sant'Anna, Application of high-content image analysis for quantitatively estimating lipid accumulation in oleaginous yeasts with potential for use in biodiesel production, Bioresour. Technol., 2016, 203, 309–317 CrossRef CAS.
- G. K. Y. Chan, T. L. Kleinheinz, D. Peterson and J. G. Moffat, A Simple High-Content Cell Cycle Assay Reveals Frequent Discrepancies between Cell Number and ATP and MTS Proliferation Assays, PLoS One, 2013, 8, e63583 CrossRef CAS PubMed.
- Y. Li-Beisson, J. J. Thelen, E. Fedosejevs and J. L. Harwood, The lipid biochemistry of eukaryotic algae, Prog. Lipid Res., 2019, 74, 31–68 CrossRef CAS.
- M. J. Griffiths and S. T. L. Harrison, Lipid productivity as a key characteristic for choosing algal species for biodiesel production, J. Appl. Phycol., 2009, 21, 493–507 CrossRef CAS.
- K. Yilancioglu, M. Cokol, I. Pastirmaci, B. Erman and S. Cetiner, Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain, PLoS One, 2014, 9(3), e91957 CrossRef PubMed.
- S. Malitsky, C. Ziv, S. Rosenwasser, S. Zheng, D. Schatz, Z. Porat, S. Ben-Dor, A. Aharoni and A. Vardi, Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol, New Phytol., 2016, 210, 88–96 CrossRef CAS PubMed.
- X. Huang, S. Zhu, H. Zhang, Y. Huang, X. Wang, Y. Wang and D. Chen, Biochar Nanoparticles Induced Distinct Biological Effects on Freshwater Algae via Oxidative Stress, Membrane Damage, and Nutrient Depletion, ACS Sustainable Chem. Eng., 2021, 9, 10761–10770 CrossRef CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00353a |
| This journal is © The Royal Society of Chemistry 2024 |
