 Open Access Article
Open Access ArticleIsotopes and otolith chemistry provide insight into the biogeochemical history of mercury in southern flounder across a salinity gradient†
Dana K.
Sackett
 *a,
Jared K.
Chrisp
b and
Troy M.
Farmer
b
*a,
Jared K.
Chrisp
b and
Troy M.
Farmer
b
aDepartment of Environmental Science and Technology, University of Maryland, 8127 Regents Dr, College Park, MD 20742, USA. E-mail: dsackett@umd.edu
bDepartment of Forestry and Environmental Conservation, Clemson University, 262 Lehotsky Hall, Clemson, SC 29634, USA
First published on 24th January 2024
Abstract
Methylmercury (MeHg) continues to pose a significant global health risk to wildlife and humans through fish consumption. Despite numerous advancements in understanding the mercury (Hg) cycle, questions remain about MeHg sources that accumulate in fish, particularly across transitional coastal areas, where harvest is prominent and Hg sources are numerous. Here we used a unique combination of Hg and nutrient isotopes, and otolith chemistry to trace the biogeochemical history of Hg and identify Hg sources that accumulated in an economically important fish species across Mobile Bay, Alabama (USA). Fish tissue Hg in our samples primarily originated from wet deposition within the watershed, and partly reflected legacy industrial Hg. Results also suggest that little Hg was lost through photochemical processes (<10% of fish tissue Hg underwent photochemical processes). Of the small amount that did occur, photodegradation of the organic form, MeHg, was not the dominant process. Biotic transformation processes were estimated to have been a primary driver of Hg fractionation (∼93%), with isotope results indicating methylation as the primary biotic fractionation process prior to Hg entering the foodweb. On a finer scale, individual lifetime estuarine habitat use influenced Hg sources that accumulated in fish and fish Hg concentrations, with runoff from terrestrial Hg sources having a larger influence on fish in freshwater regions of the estuary compared to estuarine regions. Overall, results suggest increases in Hg inputs to the Mobile Bay watershed from wet deposition, turnover of legacy sources, and runoff are likely to translate into increased uptake into the foodweb.
Environmental significanceDespite advancements in understanding the mercury cycle, questions remain about sources of methylmercury that accumulate in fish, particularly across a salinity gradient, where fishery harvest is prominent. Here, isotope and otolith data identified that Hg in an estuarine foodweb primarily originated from local and regional emissions that were deposited in the estuary watershed, and entered through freshwater runoff. These mercury inputs were largely methylated rather than being demethylated or photochemically degraded. Unique isotopic signatures also identified relative sources of Hg in fish collected from different regions across the estuary. In total, this research identified sources and drivers of Hg in a coastal food web and demonstrates the importance of understanding Hg biogeochemical pathways in a changing environment. |
Introduction
Mercury (Hg) poses significant global health risks to wildlife and humans through the consumption of fish contaminated with monomethylmercury (MeHg), a neurotoxic and bioavailable form of mercury that bioaccumulates and biomagnifies in foodwebs.1–3 Despite decades of research,4–6 questions remain about how Hg cycles through the environment and the sources of Hg that ultimately accumulates in fish tissue.7 This knowledge is particularly vital for estuarine and coastal species that make-up a large portion of fisheries for human consumption and thus human exposure to MeHg.8 Sources of MeHg in estuarine and coastal fish tissues are often uncertain and can come from more than one source, even in relatively small transitional coastal regions (e.g., freshwater run-off, estuarine sediments, marine sediments, anaerobic microcosms surrounding particulate organic matter in the water column, among others).9–12 Here we define source as meaning all possible sources of Hg in fish tissue, including but not limited to inorganic and organic Hg entering an ecosystem through deposition, runoff, and turnover of buried Hg, and areas prone to increased methylation and thus increased net MeHg accumulation in foodwebs. Further, many of these estuarine Hg sources are influenced by anthropogenic activities that provide opportunities for mitigation (e.g., limiting nutrient run-off, landscape restoration, reducing local emissions, remediation of legacy pollution).13 As such, filling these knowledge gaps, which largely stem from uncertainties in Hg sources and pathways (defined as Hg movement and transformations through the biogeochemical Hg cycle), is needed.11,14–16Specifically, the biogeochemical Hg cycle involves the release of three inorganic forms of Hg into the atmosphere, largely from anthropogenic sources (e.g., combustions of fossil fuels): elemental (Hg0), oxidized (Hg(II)), and particulate (PHg).7,17 While oxidized and particulate forms of inorganic Hg deposit relatively close to emission sites, the long atmospheric residence time of Hg0 allows it to travel to distant sites before transformation to Hg(II) and deposition.18,19 As such, Hg deposition in an estuarine ecosystem has the potential to include a mixture Hg emitted from distant, local, and regional sources. Direct releases of inorganic Hg to the terrestrial and aquatic environment from past industrial uses (e.g., industrial use of mercury catalysts) have also resulted in pools of legacy Hg in soils and sediments.12,13 Consequently, Hg(II) and PHg enter aquatic systems through rainfall (wet deposition), settlement (dry deposition), runoff, or turnover of previously deposited legacy Hg in sediment and soils, where it can be methylated by bacteria into MeHg.20,21 Net rates of methylation in aquatic systems can vary substantially due primarily to dissolved oxygen concentration, pH, local land practices, dissolved organic matter (DOM) concentrations and type, and microbial composition.22–25 These factors affect net methylation rates by influencing Hg transport, bioavailability, and whether Hg transformation processes will be dominated by MeHg and Hg(II) degradation and reduction (e.g., volatilization of Hg0 that can re-enter the atmosphere) or by methylation and bioaccumulation.25,26
Stable isotope applications developed over the last few decades provide tools to better understand the Hg biogeochemical cycle and identify sources of Hg that accumulate in fish tissue.11,27–29 Mercury, for instance, has seven naturally occurring stable isotopes that do not fractionate appreciably during trophic transfer.10,30 Mercury isotope signatures are therefore preserved when incorporated into the foodweb.10 Odd Hg isotopes undergo mass-independent fractionation (MIF) from photochemical processes (represented by Δ199Hg and Δ201Hg) and can be used to distinguish between the type of photochemical process that dominated Hg fractionation before entering the foodweb (e.g., photodegradation of MeHg to Hg(II) or photoreduction of Hg(II) to Hg0).31,32 Mass dependent fractionation is represented by δ202Hg and results from both abiotic and biotic processes. Because MDF occurs with both abiotic and biotic processes, and MIF of odd isotopes (e.g., Δ199Hg) is associated only with abiotic photochemical processes, the slope of Δ199Hg to δ202Hg in relation to previous well-established slopes of this relationship31 can provide insight into the extent of MDF that occurred as a result of photochemical processes; with the remaining portion of MDF attributable to microbially-driven biotic processes (i.e., methylation, and microbially-driven reductive and oxidative demethylation).25,31,32 Mass independent fractionation of even Hg isotope 200Hg (Δ200Hg) also provides insight into the relative importance of atmospherically derived Hg, wherein positive values reflect Hg sources from precipitation and negative values reflect gaseous elemental Hg exposed to upper-atmosphere higher-energy UV light during long-distance transport.21,33 Using these relationships, Hg isotopes have successfully linked fish tissue Hg to sources such as non-point urban runoff, atmospheric deposition and transport, terrestrial runoff, and industrial releases.9,11,12,21
Like Hg isotopes ratios, bulk C isotope ratios (δ13C) and source amino acid N isotope ratios (e.g., phenylalanine, δ15NPhe) remain largely unaltered during trophic transfer, preserving isotope signatures at the base of the foodweb. As such, C isotopes can distinguish whether energy supplying a foodweb originated from C3 versus C4 plants34 and whether sources originated from benthic or pelagic algae.35 Nitrogen isotope ratios for source amino acids (e.g., δ15NPhe) can determine the relative amount of nitrate-rich (e.g., benthic) versus nitrate-poor (e.g., atmospheric) sources at the base of the foodweb.36,37 Bulk N isotopes (δ15N), which average source and trophic amino acid N isotope ratios, have commonly been used in environmental toxicology and ecology to identify the trophic position of individual organisms, providing insight into contaminant bioaccumulation.35,37 Given that N, C, and Hg accumulate in individual fish through the same dietary items, together these data can refine our understanding of Hg sources that accumulate in an ecosystem foodweb.
An additional limitation to understanding Hg sources to fish tissue across a salinity gradient is knowledge about fish habitat use, which can influence the sources of Hg that an individual fish is exposed to. Otoliths, bony structures in fish ears, accrue calcium carbonate throughout the life of an individual with annual marks (annuli) separating regions deposited in specific years of life. Within each annual region in the otolith, trace elements (e.g., strontium [Sr], barium [Ba]) from the surrounding environment (i.e., water) are incorporated (after passing through multiple membranes and biological barriers in the gills, blood, and endolymph) in approximate proportion to their concentration in the environment by randomly substituting for calcium (Ca) in the calcium carbonate matrix of the otoliths.38,39 Once incorporated into the otolith, the concentration of these elements is permanent as otoliths are inert and are not resorbed during periods of stress, providing a lifetime record of salinity exposure for an individual fish.38–40 As a result, otolith chemistry (defined here as the ratio of Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca measured across otolith annuli) has been a useful tool in providing inferences about habitat use and residency patterns of fishes across estuarine salinity gradients.41,42 In sum, using otolith chemistry in combination with tissue Hg isotopes, total tissue Hg, and nutrient isotopes (defined here as C and N isotope data) can help to refine the spatial scale at which we distinguish sources of Hg in fish tissues across transitional ecosystems, such as an estuarine salinity gradient; where numerous and variable Hg sources may contribute to mercury concentrations in economically important coastal fisheries.8,43
Ca measured across otolith annuli) has been a useful tool in providing inferences about habitat use and residency patterns of fishes across estuarine salinity gradients.41,42 In sum, using otolith chemistry in combination with tissue Hg isotopes, total tissue Hg, and nutrient isotopes (defined here as C and N isotope data) can help to refine the spatial scale at which we distinguish sources of Hg in fish tissues across transitional ecosystems, such as an estuarine salinity gradient; where numerous and variable Hg sources may contribute to mercury concentrations in economically important coastal fisheries.8,43
In this study, we targeted southern flounder (Paralichthys lethostigma); a species that inhabits a wide variety of estuarine salinities (i.e., freshwater to hypersaline)41,44,45 in the northern Gulf of Mexico, where they support economically important recreational and commercial fisheries. Juvenile southern flounder inhabit freshwater and estuarine environments to adult sizes before moving offshore to overwinter and spawn.46 Because southern flounder inhabit freshwater, estuarine, and marine environments,47,48 understanding sources of Hg to this species provides insight on how Hg is transferred through foodwebs across these various ecosystems and to humans. As such, our objectives were to quantify the sources and pathways of Hg bioaccumulation for this economically important, and frequently consumed species in the Mobile Bay, Alabama (USA) estuary and to identify finer scale sources of Hg in individual fish across the salinity gradient using Hg isotopes, bulk N isotopes, amino acid compound-specific N isotopes, bulk C isotopes, and otolith chemistry.
Materials and methods
Sample collection
The Mobile-Tensaw River is the primary source of freshwater to Mobile Bay and is considered the fourth largest river system in the continental United States. Three additional smaller sources of freshwater also contribute to the bay: Dog River (237 km2 watershed), Fowl River (184 km2 watershed), and Week's Bay (521 km2).50,51 Samples were collected from ten sites located along a 60+ km seasonal salinity gradient of saltmarshes, bays, tidal creeks, and freshwater ecosystems. Sites were sampled one to two times monthly during May–July of 2018 and March, May–July of 2019. Within the Mobile Bay estuary, three salinity sampling regions (freshwater–oligohaline [<5‰], mesohaline [5–18‰], polyhaline [18–30‰]) were targeted. Sample sites within each region were randomly stratified by available suitable southern flounder habitat types (e.g., mud flats, oyster beds, tidal marsh).Southern flounder between 270 mm and 547 mm TL (average age in years = 0.9 ± 0.4) were collected using gill nets, electrofishing, and hook-and-line. Gillnets (30 m by 2.4 m with 127 mm stretch mesh) targeted large juveniles and adults (≥100 mm TL) at meso- to polyhaline sites by soaking nets for four hours (two 2 hour sets). Nets were set parallel to shore with a hook towards shore at the downstream end. At freshwater sites, pulsed DC boat electrofishing (Midwest Lake Electrofishing Systems Infinity Box) was used along shorelines. Six 15 minute boom mounted electrofishing transects were conducted during each site visit. Hook-and-line sampling was conducted after all standardized sampling was completed or in areas not accessible by the previous methods. At each site, date, time, GPS coordinates, and water depth were recorded at the beginning and end of each sampling transect or gillnet set. All southern flounder collections were conducted according to use guidelines outlined in IACUC protocol #AUP2018-001 at Clemson University.
Chemical analyses of fish tissue and otoliths collected from Mobile Bay
Dorsal axial tissue samples (N = 23) were processed using metals-cleaned stainless-steel instruments and freeze-dried following EPA standard protocols for measuring total Hg (USEPA method 7473).52 Previous research analyzed southern flounder muscle tissue collected at four sites across the freshwater–oligohaline portions of the Mobile-Tensaw River Delta for both total Hg and MeHg and found that MeHg comprised 87% (±20% SD) of total Hg and did not differ across the Mobile-Tensaw River Delta.50 We therefore assumed that muscle tissue from the same species in this study were consistent with these results. However, it should be noted that southern flounder collected in this study were on average larger and from additional sites (i.e., Lower Mobile Bay, Weeks Bay; Fig. 1) not sampled by Farmer et al.,50 and thus could have had higher proportions of MeHg to total Hg in muscle tissue.44 Dried muscle tissue samples were ground to a homogenous powder and analyzed for total dry weight Hg concentrations using a Prodigy-7 dual-view ICP optical emission spectrometer. Ti was used as an internal standard for mass bias correction. The procedural blank was 0.03 ppb, equivalent to ∼0.5% of the analytical signal. Seven of 23 samples were run in duplicate for total Hg. The mean relative standard deviation of duplicates was 9.98% ± 5.5% SD. The mean relative difference for DORM-2 Hg standard reference material (certified value as 4640 ± 260 ng g−1 dry weight) was 1% ± 1% SD (N = 2; values = 4543 and 4640.2 ng g−1 dry weight). Total dry weight Hg concentrations were converted to total wet weight Hg concentrations using the percent moisture measured for each sample. Total wet weight Hg are reported as μg g−1 ww in this study.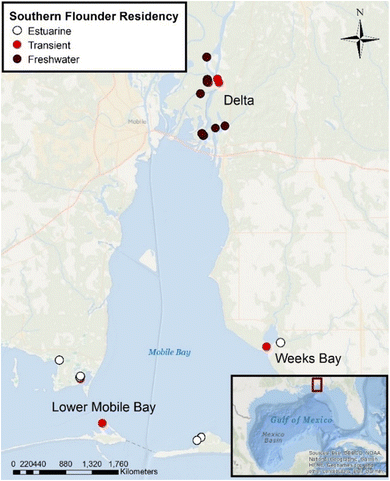 | ||
| Fig. 1 Southern flounder collection sites across the Mobile-Tensaw River Delta and Mobile Bay, Alabama (USA) for isotope analyses. Sites were sampled in three locations: the Delta (freshwater riverine areas of the Mobile-Tensaw River Delta), Week's Bay (small estuary that drains into lower Mobile Bay and experiences freshwater to polyhaline conditions), and Lower Mobile Bay (sites located within the lower Mobile Bay with salinities ranging from mesohaline–polyhaline). Lifetime salinity exposure classifications are indicated by color on the map and were classified based on otolith chemistry and water chemistry analyses.42 | ||
All N and C isotope analyses were conducted by the Biogeochemical Stable Isotope Laboratory in the School of Ocean and Earth Science and Technology at the University of Hawaii, with methods described in Dale et al.,53 Sackett et al.,37 and Wall et al.54 Briefly, bulk N and C isotopes (δ15N and δ13C) were analyzed using a continuous flow isotope ratio mass spectrometer (Costech ECS 4010 Elemental Combustion System using a Zero Blank Autosampler) coupled with a Thermo Scientific Delta V Advantage through a Thermo Scientific Conflo IV. Isotopic values are reported in δ-notation relative to atmospheric N2 and V-PDB (Vienna Peedee Belemnite), for N and C respectively. A glycine and an in-house white muscle tissue tuna standard, characterized for δ15N and δ13C using international reference materials (δ15N: USGS 32, USGS 34, USGS 35, NIST 3; and δ13C: NBS 18, NBS 19, NIST 1547) and verified by an independent laboratory, were used to quantify and correct isotopic results. Known standard values were 11.4 δ15N and −36.5 δ13C for glycine and 14.2 δ15N and −16.2 δ13C for tuna. Standards were run in triplicate before samples were analyzed and then with every 7 samples thereafter. All isotope data accuracy was ≤±0.2‰. Procedural blanks were 0.06 ppb or 1.3% of the analytical signal.
For Hg isotope analyses, aliquots of homogenous dried muscle tissue samples were digested using 3 mL of aqua regia in a 40 mL amber glass vessel by heating on a hot plate at 120 °C for 4 hours. After digestion, the supernatant was extracted and made-up to 20 mL with 0.002 M BrCl. Digested samples were analyzed for Hg isotopes using a Nu Plasma II MC-ICP-MS in standard wet plasma cones. An ESI ICP Hydride generation system was used to generate Hg0 vapor by quantitative reduction of Hg(II) in solution using stannous chloride (3% w/v in 1 M HCl). The Hg0 vapor was introduced directly into the ICP. The Tl internal standard was nebulized using a 50 μL min−1 self-aspirating micro-concentric PFA nebulizer and a cyclonic/Scott dual spray chamber. Instrument tuning and gain calibration of the detectors was performed daily. An NIST cinnabar standard (RM 8610) was used to verify data accuracy and agreed with values reported in the literature (N = 5; mean, 1 SD; δHg202 = −0.50 ± 0.03, δHg201 = −0.44 ± 0.04, δHg200 = −0.27 ± 0.05, δHg199 = −0.16 ± 0.04, ΔHg199 = −0.04 ± 0.03, ΔHg201 = −0.06 ± 0.04).55 Mercury isotope analyses were conducted at the Trent University Water Quality Centre, Peterborough ON Canada. Hg isotope ratios were reported in per mil (‰), referenced to the NIST SRM 3133 mercury standard, where:
| δxxxHg = {[(xxxHg/198Hg)sample/(xxxHg/198Hg)NIST3133] − 1} × 1000 | (1) |
| Δ199Hg = δ199Hg − (δ202Hg × 0.2520) | (2) |
| Δ200Hg = δ200Hg − (δ202Hg × 0.5024) | (3) |
| Δ201Hg = δ201Hg − (δ202Hg × 0.7520) | (4) |
Odd Hg isotopes undergo MIF from photochemical processes (represented by Δ199Hg and Δ201Hg) and can be used to distinguish between the type of photochemical transformation that dominated Hg fractionation.
Amino acid compound-specific stable isotope (AA-CSIA) analyses were conducted at the Biogeochemical Facility at the University of Hawaii following the same protocols reported in Dale et al.,53 Sackett et al.,37 and Wall et al.54 Briefly, dried tissue samples were subjected to acid hydrolysis, esterification of the carboxyl terminus and trifluoracetylation of the amine group prior to being introduced into a Delta V or MAT 253 mass spectrometer interfaced with a Trace GC gas chromatograph through a GC-C III combustion furnace (980 °C), reduction furnace (650 °C) and liquid N2 cold trap. Internal reference compounds, L-2-aminoadipic acid (AAA, δ15N = −6.2‰) and L-(+)-norleucine (Nor, δ15N = 19.06‰) of known nitrogen isotope compositions, were co-injected with samples and used to normalize the measured isotope values of unknown amino acids and determine accuracy and precision. Amino acids targeted here included glycine (Gly), phenylalanine (Phe), lysine (Lys), leucine (Leu), and glutamic acid (Glu). All amino acids were analyzed in triplicate and isotope values are reported in δ-notation relative to atmospheric N2. The mean relative standard deviations (RSD) for internal reference values L-(+)-norleucine and L-2-aminoadipic acid were 0.99% and 7.4%, respectively. The mean measured values for internal reference compounds Nor and AAA were 19.3‰ ± 0.19‰ SD (difference of −0.2‰) and −4.8‰ ± 0.35‰ SD (difference of −1.4‰), respectively. Mean RSD for triplicate amino acid results used in this study were low, indicating high precision in analytical methods and study results (glycine = 3.1%, leucine = 1.4%, phenylalanine = 8.6%, glutamic acid = 0.85%).
The elemental composition of fish ear bones (otoliths) and water samples were used to determine the lifetime salinity exposure of each southern flounder collected across the salinity gradient in Mobile Bay as part of a separate study.42 As such, detailed otolith chemistry and residency pattern classification methods for these samples are described in Chrisp et al.42 Briefly, following methods from Farmer et al.,41 otoliths were ablated from the core to the distal edge of the otolith along a straight transect parallel to the sulcal groove. Otolith chemistry analysis targeted Ca and Sr concentrations every 0.6 seconds using an Agilent 7700x quadrupole inductively coupled plasma mass spectrometer (ICPMS) coupled to a 213 nm Nd:YAG NWR laser. Using Ca (37.69%) as an internal standard, elemental concentrations were converted to molar ratios. Water samples were processed for elemental concentrations of Ca and Sr with the same ICPMS system in solution mode coupled with an Agilent autosampler. Concentrations of each element measured in water were converted to molar ratios using Ca as a standard to compare with otolith chemistry.42,56,57 Using these data, southern flounder were classified into one of three lifetime residency patterns (i.e., freshwater, estuarine or transient) based on lifetime salinity exposure (as inferred from Sr to Ca molar ratios: Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca) across the otolith transect. To accomplish this, nonlinear models were fit to the data to quantify relationships between water Sr
Ca) across the otolith transect. To accomplish this, nonlinear models were fit to the data to quantify relationships between water Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca versus ambient sample site salinity. Predicted values from these nonlinear models were then multiplied by a partition coefficient to develop a link between expected otolith Sr
Ca versus ambient sample site salinity. Predicted values from these nonlinear models were then multiplied by a partition coefficient to develop a link between expected otolith Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca and ambient salinity.42 Finally, a regime shift algorithm was used to detect any significant shifts in Sr
Ca and ambient salinity.42 Finally, a regime shift algorithm was used to detect any significant shifts in Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca and thus, salinity, across the lifetime of the individual.58,59 Otolith Sr
Ca and thus, salinity, across the lifetime of the individual.58,59 Otolith Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca equivalent to ≤1 psu salinity was classified as a freshwater resident for a given time step, while otolith Sr
Ca equivalent to ≤1 psu salinity was classified as a freshwater resident for a given time step, while otolith Sr![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ca equivalent to >1 psu salinity was classified as an estuarine resident for a given time step. If more than 90% of the time steps across the lifetime of an individual fell into one classification (i.e., freshwater or estuarine), residency patterns were assigned to that classification. If neither classification comprised 90% of the transect, then a ‘transient’ classification was assigned to indicate a fish that either moved between freshwater and estuarine habitats or a fish that resided in an area that experienced seasonal changes in salinity.
Ca equivalent to >1 psu salinity was classified as an estuarine resident for a given time step. If more than 90% of the time steps across the lifetime of an individual fell into one classification (i.e., freshwater or estuarine), residency patterns were assigned to that classification. If neither classification comprised 90% of the transect, then a ‘transient’ classification was assigned to indicate a fish that either moved between freshwater and estuarine habitats or a fish that resided in an area that experienced seasonal changes in salinity.
Statistical analyses
To examine sources of nitrogen to southern flounder across Mobile Bay, and the relative trophic position of individual fish, we calculated the weighted mean δ15N value for source amino acids using glycine, lysine, and phenylalanine measured from AA-CSIA and bulk δ15N data following the same methods as Sackett et al.37 Briefly, we calculated the weighted mean δ15N value for source amino acids glycine, lysine, and phenylalanine using AA-CSIA: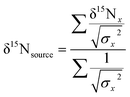 | (5) |
| TP = ((δ15NGlu − δ15NPhe − β)/TEF) + 1 | (6) |
Southern flounder tissue Hg isotopes, δ15Nsource, δ13C, total Hg concentrations, and relative trophic position were used to evaluate differences in Hg sources, dietary sources of N and C, and relative trophic position across sampling regions and lifetime residency classifications of fish across Mobile Bay. The Δ199Hg/Δ201Hg ratio resulting from the regression relationship between Δ199Hg to Δ201Hg was used to determine whether MIF was primarily driven by photodegradation of MeHg (Δ199Hg/Δ201Hg ratios of 1.2 to 1.36) or the photoreduction of Hg(II) to Hg0 (Δ199Hg/Δ201Hg ratios of ∼1.0) prior to Hg being ultimately methylated and incorporated into the foodweb.10,31,32 The Δ199Hg/δ202Hg ratio was also evaluated in relation to experimentally derived values from Bergquist and Blum31 to distinguish between the relative contribution of photochemical versus biotic fractionation processes that occurred prior to Hg entering the foodweb.10 Previous research has also used these Hg isotope ratios (i.e. Δ199Hg/Δ201Hg ratio and Δ199Hg/δ202Hg ratio) to provide an understanding of these Hg biogeochemical processes.9–11,62 Also, like previous work,11 we included results from several of these published studies for comparison to the results presented here in Fig. 2. Based on laboratory trials by Bergquist and Blum,31 a Δ199Hg/δ202Hg ratio of 2.43 is representative of Hg undergoing only MIF from photochemical processes, while a ratio of 0 is representative of little to no MIF or MDF from photochemical processes, and instead suggests microbial Hg transformations dominated MDF prior to Hg entering the foodweb. As such, dividing Δ199Hg/δ202Hg ratio from southern flounder tissue by the slope representative of 100% MIF from photochemical processes (i.e., 2.43; eqn (7)) gives insight into the relative proportion of photochemical processes that fractionated Hg prior to its incorporation into the foodweb. Subtracting this proportion by 1 in-turn provides the remaining proportion of Hg fractionation that was driven by microbial processes (eqn (7)) rather than photochemical processes. It should be noted that while there is evidence that dark abiotic demethylation can also occur under controlled laboratory settings and contribute to MDF, the strong reductants and oxidants used in these experiments are typically absent from the natural environment.25 It is therefore unlikely that dark abiotic demethylation contributed to demethylation processes in this study or Hg MDF values. Additionally, the foundational study on Hg isotopes by Bergquist and Blum31 showed no abiotic dark demethylation in experiments, even when Hg concentrations were high. Therefore, using the Δ199Hg/δ202Hg ratio representative of 100% MIF from photochemical processes (2.43)31 in comparison to the measured Δ199Hg/δ202Hg ratio as described above allows for the calculation of:
| % of biotically driven fractionation = (1 − ([Δ199Hg/δ202Hg ratio]/2.43)) × 100 | (7) |
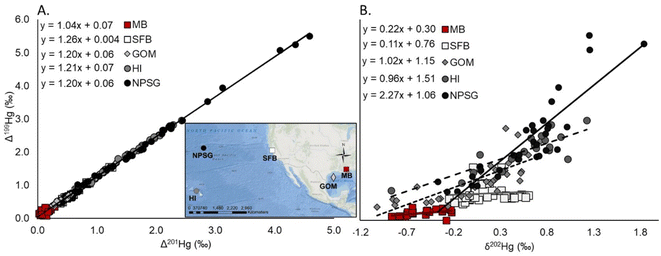 | ||
| Fig. 2 Regressions of fish tissue Hg isotope values as indicators of (A) the type of photochemical processes that occurred (P < 0.01 for all relationships) and (B) the relative amount of photochemical versus microbial processes that occurred to fish tissue Hg prior to its incorporation into the foodweb (P < 0.01 for all relationships except MB and SFB).31 These data were collected from Mobile Bay, Alabama (MB, N = 23, (A) R2 = 0.64, (B) P = 0.09), San Francisco Bay (SFB, N = 35 (A) R2 > 0.99, (B) P = 0.57),9 the Gulf of Mexico (GOM, N = 32, (A) R2 > 0.99, (B) R2 = 0.61),62 the waters surrounding Hawaii (HI, N = 19, (A) R2 > 0.99, (B) R2 = 0.72),11 and the North Pacific Subtropical Gyre (NPSG, N = 33, (A) R2 > 0.99, (B) R2 = 0.78).10 | ||
Using this equation, we estimated the relative percent of microbial versus photochemical processes that fractionated Hg in fish tissue prior to entering the foodweb.
Total Hg, fish total length, δ13C, δ15NSource, relative TP, δ202Hg, Δ199Hg, Δ200Hg, and Δ201Hg were compared for significant differences among southern flounder lifetime residency classifications to understand how different habitat uses across the estuary may have influenced Hg sources to fish tissues (Table 1).42 To better understand sources of Hg among southern flounder collected across the salinity gradient in Mobile Bay, Δ199Hg with Δ201Hg, Δ199Hg with δ202Hg, and Δ199Hg with δ13C, were also analyzed with k-means cluster analysis (α = 0.05; k = 3 clusters). Additionally, a multiple linear regression model (N = 23) was used to predict how lifetime salinity exposure as a proxy for lifetime estuarine habitat use, total length, and trophic position impacted the concentration of total Hg in fish tissue (α = 0.05). However, given that fish total length and trophic position can be interactive (e.g., the effect of total length can often depend on trophic position) and are well known primary drivers of total Hg in fish tissue,22,37 we included an interaction between total length and relative trophic position in this model to ensure these measures did not confound results (total Hg values that accounted for this interaction are summarized as least squares means of total Hg in Table 1). This model was also re-run using sample collection sites (Delta = freshwater habitats within Mobile-Tensaw River Delta that drains the 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 km2 watershed, Week's Bay = shallow estuary with 521 km2 watershed that drains into lower Mobile Bay with freshwater–polyhaline conditions, and Lower Mobile Bay = lower portion of Mobile Bay with mesohaline–polyhaline salinities; Fig. 1) in place of lifetime residency classifications, to determine if fish capture location was more important in explaining tissue Hg concentrations than lifetime estuarine habitat use based on salinity. Models were compared using Akaike's Information Criterion for small sample size (AICc), where the model with the lowest AICc value is the model that best explains the variation in the data.63 Total Hg data were normal and homoscedastic.
000 km2 watershed, Week's Bay = shallow estuary with 521 km2 watershed that drains into lower Mobile Bay with freshwater–polyhaline conditions, and Lower Mobile Bay = lower portion of Mobile Bay with mesohaline–polyhaline salinities; Fig. 1) in place of lifetime residency classifications, to determine if fish capture location was more important in explaining tissue Hg concentrations than lifetime estuarine habitat use based on salinity. Models were compared using Akaike's Information Criterion for small sample size (AICc), where the model with the lowest AICc value is the model that best explains the variation in the data.63 Total Hg data were normal and homoscedastic.
| Salinity exposure classifications | N | Total Hg (μg g−1 ww) | LSM total Hga (μg g−1 ww) | Total lengtha (mm) | Bulk δ13Ca (‰) | δ15Nsourcea (‰) | Relative TP | δ202Hga (‰) | Δ199Hg (‰) | Δ201Hg (‰) | Δ200Hg (‰) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a ANOVA significantly different among salinity exposure classifications, P < 0.05. b Because the δ15Nsource sample size (N) here is two the parenthetical value represents the range. | |||||||||||
| Freshwater | 8 | 0.08 (0.01) | 0.09 (0.01) | 315.3 (10.9) | −28.38 (0.54) | 0.89 (0.04) | 12.74 (0.26) | −0.62 (0.04) | 0.17 (0.02) | 0.07 (0.01) | 0.03 (0.01) |
| Transient | 8b | 0.06 (0.004) | 0.06 (0.01) | 387.4 (17.8) | −22.73 (1.34) | 0.82 (0.05b) | 11.79 (0.34) | −0.55 (0.08) | 0.20 (0.05) | 0.14 (0.03) | 0.01 (0.01) |
| Estuarine | 7 | 0.07 (0.02) | 0.06 (0.01) | 397.6 (35.2) | −20.93 (0.50) | 0.71 (0.06) | 11.93 (0.43) | −0.37 (0.05) | 0.20 (0.06) | 0.13 (0.05) | 0.05 (0.01) |
| All fish | 23 | 0.07 (0.005) | 0.07 (0.005) | 365.39 (14.6) | −24.14 (0.84) | 0.81 (0.04) | 12.16 (0.21) | −0.52 (0.04) | 0.19 (0.02) | 0.11 (0.02) | 0.03 (0.01) |
Results and discussion
Biogeochemical history of Hg across Mobile Bay
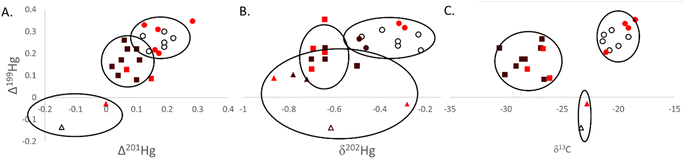 | ||
| Fig. 3 Isotopic data ((A) Δ199Hg to Δ201Hg; (B) Δ199Hg to δ202Hg; (C) Δ199Hg to δ13C) measured in southern flounder muscle tissue across the salinity gradient in Mobile Bay, Alabama (USA). Lifetime salinity exposure classifications were assigned to an individual based on otolith chemistry and water chemistry analyses and are indicated by color (white = estuarine, bright red = transient, and black = freshwater; colors are consistent with those in Fig. 1).42k-means cluster analyses were conducted for each pair of isotope relationships depicted. Hollow ellipses represent the 95% confidence area for each grouping. Data points assigned to distinct clusters are based on shape, with circles representing a cluster, squares representing a second cluster, and triangles a third cluster. | ||
Cluster analysis using Δ199Hg and δ13C resulted in very distinctive groupings of tissue data (Fig. 3C). Sources of carbon among lifetime salinity exposure classifications were significantly different (P < 0.01; Table 1) and drove much of the separation in these data (Fig. 3C). Mean δ13C was lowest for freshwater residents at −28.4‰ (SE = 0.54‰). δ13C values between −22‰ and −30‰ are indicative of C3 plant carbon sources from terrestrial freshwater runoff.34,35 Consequently, these results show that individuals classified as freshwater residents and those classified as transient individuals that were captured in the same region as freshwater residents (Fig. 3C) relied heavily on foodwebs based on freshwater terrestrial C3 plant carbon sources (e.g., agricultural runoff from prominent Alabama crops such as cotton, peanuts, soybeans, and fruit are C3 plants).77 Those fish classified as estuarine residents occupied more saline habitats throughout their life history and were therefore furthest from freshwater outflows in Mobile Bay. Estuarine residents also had the highest mean δ13C at −20.9‰ (SE = 0.50‰) among lifetime residency classifications (P < 0.01); signifying a lesser influence of terrestrial carbon inputs (−22‰ to −30‰) and the mixing of terrestrial carbon sources with marine benthic sources (−17‰).34,35 Marine benthic sources of carbon are more enriched in the heavier isotope (C13) with δ13C values of approximately −17‰ compared to marine phytoplankton with δ13C values of approximately −22‰.34 This distinction occurs because of the larger boundary layer that surrounds benthic algae (>1 mm) compared to pelagic algae (∼10 μm), which limits access to the preferred lighter 12C isotope for photosynthesis. Southern flounder are a benthic species, further supporting that the higher δ13C values for estuarine residents were likely from the mixing of freshwater terrestrial (δ13C ∼ −28‰) and marine benthic algae (δ13C ∼ −17‰) carbon sources. The low level of photochemical degradation indicated by odd Hg isotopes across the entire Mobile Bay estuary supports the notion that freshwater terrestrial inputs high in DOC likely caused more turbid waters and less sunlight penetration for photochemical processes, even to those habitats occupied by estuarine residents in the lower reaches of Mobile Bay.78 δ15Nsource data further supported these conclusions with significant differences in source N among lifetime salinity exposure classifications (P = 0.047; Table 1). The highest values were found for freshwater residents and lowest for estuarine residents (Table 1). High (or less negative) δ15Nsource data suggest a more nitrate-rich source of nitrogen (e.g., agricultural runoff) than lower (or more negative) values, which suggest a more nitrate-poor source (e.g., atmospheric nitrogen), to a foodweb.36,37 As such, the high positive δ15Nsource values in Mobile Bay would further suggest a terrestrial and thus more nitrate-rich base to the foodwebs across the entire estuary that was more pronounced in freshwater residents that occupied habitats closer to the source of those inputs. Together these data demonstrate that terrestrial freshwater run-off was a primary source of N, C, and Hg to Mobile Bay foodwebs, and that those individuals that resided in the freshwater portion of the estuary through their life history were more influenced by these terrestrial inputs. Those individuals with lifetime residency furthest from freshwater terrestrial inputs were also influenced by terrestrial N, C, and Hg inputs but had mixed terrestrial sources with marine benthic N, C, and Hg sources.
For all cluster analyses (Fig. 3), transient individuals (those that experienced both the freshwater and more saline estuarine conditions throughout their life history) were largely grouped with estuarine and freshwater residents based on their location of capture (Fig. 1). These results suggest that fish captured together likely occupied the same habitat and relied on the same food sources for much of their life history, but that those individuals classified as transient ventured into other habitats more so than their freshwater residency and estuarine residency counterparts. As with most groupings, each could be broken-down to a finer scale, particularly for southern flounder residency, which likely exists on more of a spectrum. For instance, in all clusters, two individuals were consistently and significantly grouped together despite different lifetime salinity exposure classifications (Fig. 3). Both individuals were captured in Week's Bay (Fig. 1), suggesting isotope signatures identifying sources of carbon and tissue Hg were specific to this bay. Previous research has suggested that nutrient-rich, acidic, low salinity riverine inputs to Weeks Bay forms a wedge that floats on top of denser, high salinity, high pH water that flows into the upper portion of this bay.79 The mixing zone of these two layers of water has warm, low dissolved oxygen conditions with low oxidation–reduction potential, all of which are favorable for Hg methylation.22,23,79 This study also suggested that wet deposition was the primary source of inorganic Hg to the watershed of Weeks Bay, a notion supported by positive tissue Δ200Hg values measured in our study (Δ200Hg = 0.06 for both individuals).79 As such these results suggest that Hg in fish from Week's Bay had been deposited in the Week's Bay watershed where it underwent very little to no photochemical processes through terrestrial freshwater run-off with carbon that was depleted in C13 (δ13C ∼ −28‰),35 and efficiently methylated in Week's Bay near the benthos, where benthic carbon sources were more enriched in C13 (δ13C ∼ −17‰). The mixing of these different sources of carbon and Hg to the Weeks Bay foodweb likely resulted in unique and intermediate δ13C values (mean δ13C = 23.1‰) and unique Hg isotope signatures in fish tissue (Fig. 3).
Differences in the biogeochemical histories and sources of tissue Hg among lifetime residency groups also resulted in differences in total tissue Hg concentrations. Results from a multiple linear regression demonstrated that when fish length and relative trophic position were accounted for, southern flounder lifetime residency classifications had significantly different concentrations of tissue Hg (P < 0.01, N = 23, R2 = 0.58 Table 1), with freshwater residents having significantly higher concentrations than estuarine or transient fish (Table 1). Previous studies have also observed negative relationships between the salinity where fish were captured and tissue Hg concentrations.50,80 These results were also significant (P = 0.01, N = 23, R2 = 0.47) when three general sample locations (Delta, Week's Bay, and Lower Mobile Bay; Fig. 1) were used in place of lifetime residency exposure classifications based on salinity; however, lifetime residency classifications had a lower AICc value (ΔAICc using sampling locations = 5.5) and thus were better at explaining the variation in southern flounder tissue Hg concentrations. Results showed that the more direct biogeochemical path of methylated Hg being incorporated into the foodweb from terrestrial freshwater runoff in the Mobile-Tensaw River Delta (Fig. 1) resulted in higher Hg accumulation in the foodweb in the freshwater portion of the estuary and, specifically, in southern flounder with lifetime freshwater residency patterns.
Conclusions
Southern flounder results across Mobile Bay have important implications for consumptive health risks. Primary sources of Hg in fish tissue in Mobile Bay serve as a proxy for Hg entering and accumulating in Mobile Bay foodwebs. Here results found sources of fish tissue Hg largely stemmed from local and regional atmospheric Hg that entered the estuary through wet deposition within the watershed, and runoff from terrestrial environments, with turn-over and run-off of legacy Hg sources from direct industrial releases at least partly contributing to fish Hg body burdens (for a simplified depiction of these results see Fig. 4). Our results also indicate Mobile Bay conditions are conducive to Hg methylation. As such, additional Hg loading to Mobile Bay or its watershed from wet deposition and runoff, or storm systems that turnover and release legacy Hg from sediment is likely to result in increased net MeHg and uptake into foodwebs.13 Of further concern, large storm systems (e.g., hurricane) are increasing in frequency and intensity because of climate change81–84 and can increase each of the fish tissue Hg sources identified in Mobile Bay (local and regional wet deposition and terrestrial runoff, legacy Hg turn-over). Because southern flounder also move offshore to spawn during the fall and winter, freshwater residents and at least some transient and estuarine residents, are expected biological vectors transporting Hg from terrestrial watersheds to marine ecosystems in the Gulf of Mexico. Moreover, southern flounder are fundamental to Alabama's commercial and recreational fishery with the majority of those harvested having transient salinity exposure classifications.42 Because transient individuals in this study often had Hg sources similar to estuarine and freshwater residents collected in similar locations, it is likely that harvested fish contain Hg sources from across the estuarine ecosystem. As such, increases in Hg inputs to this aquatic ecosystem are likely to translate into higher concentrations in Mobile Bay foodwebs, predators that consume prey from this ecosystem, and in human consumers of those fish. Lastly, this study demonstrates the utility of combining N, C, and Hg isotopes with otolith chemistry to better understand the biogeochemical cycle that ultimately accumulates Hg in economically important coastal fisheries.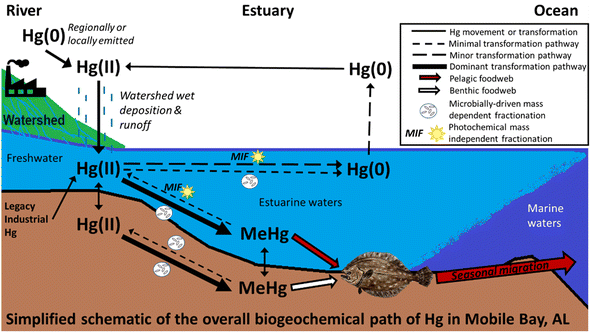 | ||
| Fig. 4 Mercury (Hg), C, and N isotopes, and otolith chemistry identified methylmercury (MeHg) sources and pathways before accumulating in an estuarine foodweb. | ||
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Alabama Marine Resources Division for providing the funds for field collections during this project (Federal Aid in Sport Fish Restoration Project F18AF00231 to M. Catalano and T. Farmer). Additional support was provided by the Clemson University Creative Inquiry and Undergraduate Research Program. We would also like to thank Hayla Evans of the Water Quality Centre at Trent University for assistance with Hg isotope data and analyses. Further thanks goes to Dr Brain Popp and Natalie Wallsgrove for the invaluable assistance with amino acid compound specific and bulk nutrient isotope data. We also thank the anonymous reviewers who helped to improve the final version of this paper. Last but certainly not least, a very special thanks goes to Dr D. Derek Aday for his outstanding editorial prowess and skill at incorporating humor into every edit.References
- L. Trasande, P. J. Landrigan and C. Schechter, Public health and economic consequences of methyl mercury toxicity to the developing brain, Environ. Health Perspect., 2005, 113, 590–596, DOI:10.1289/ehp.7743.
- D. Mergler, H. A. Anderson, L. Hing Man Chan, K. R. Mahaffey, M. Murray, M. Sakamoto and A. H. Stern, Methylmercury exposure and health effects in humans: a worldwide concern, Ambio, 2007, 36, 3–11 CrossRef.
- D. C. Evers and E. Sunderland, Technical Information Report on Mercury Monitoring in Biota, United Nations Environment Programme, Geneva, Switzerland, 2019 Search PubMed.
- S. Normura, Epidemiology of Minamata Disease. Minamata Disease. Study Group of Minamata Disease, Kumamoto University, Japan, 1968 Search PubMed.
- G. C. Compeau and R. Bartha, Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediments, Appl. Environ. Microbiol., 1985, 50, 498–502 CrossRef.
- D. R. Engstrom, Fish respond when the mercury rises, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 16394–16395 CrossRef PubMed.
- M. S. Gustin, D. C. Evers, M. S. Bank, C. R. Hammerschmidt, A. Pierce, N. Basu, J. Blum, P. Bustamante, C. Chen, C. T. Driscoll, M. Horvat, D. Jaffe, J. Pacyna, N. Pirrone and N. Selin, Importance of integration and implementation of emerging and future mercury research into the Minamata Convention, Environ. Sci. Technol., 2016, 50, 2767–2770, DOI:10.1021/acs.est.6b00573.
- E. M. Sunderland, M. Li and K. Bullard, Decadal Changes in the Edible Supply of Seafood and Methylmercury Exposure in the United States, Environ. Health Perspect., 2018, 126, 017006, DOI:10.1289/EHP2644.
- G. E. Gehrke, J. D. Blum, D. G. Slotton and B. K. Greenfield, Mercury isotopes link mercury in San Francisco Bay forage fish to surface sediments, Environ. Sci. Technol., 2011, 45, 1264–1270 CrossRef PubMed.
- J. D. Blum, B. N. Popp, J. C. Drazen, C. A. Choy and M. W. Johnson, Methylmercury production below the mixed layer in the North Pacific Ocean, Nat. Geosci., 2013, 6, 879–884 CrossRef.
- D. K. Sackett, J. C. Drazen, B. N. Popp, C. A. Choy, J. D. Blum and M. W. Johnson, Carbon, nitrogen, and mercury isotope evidence for the biogeochemical history of mercury in Hawaiian marine bottomfish, Environ. Sci. Technol., 2017, 51, 13976–13984, DOI:10.1021/acs.est.7b04893.
- S. E. Janssen, J. C. Hoffman, R. F. Lepak, D. P. Krabbenhoft, C. A. Eagles-Smith, G. Peterson, J. M. Ogorek, J. F. DeWild, A. Cotter, M. Pearson, M. T. Tate, R. B. Yeardley Jr and M. A. Mills, Examining historical mercury sources in the Saint Louis River estuary: how legacy contamination influences biological mercury levels in Great Lakes coastal regions, Sci. Total Environ., 2021, 779, 146284, DOI:10.1016/j.scitotenv.2021.146284.
- C. S. Eckley, C. C. Gilmour, S. Janssen, T. P. Luxton, P. M. Randall, L. Whalin and C. Austin, The assessment and remediation of mercury contaminated sites: a review of current approaches, Sci. Total Environ., 2020, 707, 136031, DOI:10.1016/j.scitotenv.2019.136031.
- N. Gandhi, R. W. K. Tang, S. P. Bhavsar and G. B. Arhonditsis, Fish mercury levels appear to be increasing lately: a report from 40 years of monitoring in the Province of Ontario, Canada, Environ. Sci. Technol., 2014, 48, 5404–5414 CrossRef PubMed.
- K. Sundseth, J. M. Pacyna, A. Banel, E. G. Pacyna and A. Rautio, Climate change impacts on the environment and human exposure to mercury in the Arctic, Int. J. Environ. Res. Public Health, 2015, 12, 3579–3599, DOI:10.3390/ijerph120403579.
- A. T. Schartup, C. P. Thackray, A. Qureshi, C. Dassuncao, K. Gillespie, A. Hanke and E. M. Sunderland, Climate change and overfishing increase neurotoxicant in marine predators, Nature, 2019, 572, 648–650 CrossRef CAS PubMed.
- X. Ren, W. T. Luke, P. Kelley, M. D. Cohen, M. L. Olson, J. Walker, R. Cole, M. Archer, R. Artz and A. A. Stein, Long-Term Observations of Atmospheric Speciated Mercury at a Coastal Site in the Northern Gulf of Mexico during 2007–2018, Atmosphere, 2020, 11, 268 CrossRef CAS.
- C. R. Hammerschmidt and W. F. Fitzgerald, Methylmercury in freshwater fish linked to atmospheric mercury deposition, Environ. Sci. Technol., 2006, 40, 4562–4573 Search PubMed.
- M. A. Engle, M. T. Tate, D. P. Krabbenhoft, A. Kolker, M. L. Olson, E. S. Edgerton, J. F. DeWild and A. K. McPherson, Characterization and cycling of atmospheric mercury along the central US Gulf Coast, Appl. Geochem., 2008, 23, 419–437 CrossRef.
- C. C. Gilmour, E. A. Henry and R. Mitchell, Sulfate stimulation of mercury methylation in fresh- water sediments, Environ. Sci. Technol., 1992, 26, 2281–2287 CrossRef.
- J. D. Blum, L. S. Sherman and M. W. Johnson, Mercury isotopes in earth and environmental sciences, Annu. Rev. Earth Planet. Sci., 2014, 42, 249–269 CrossRef.
- D. K. Sackett, D. D. Aday, J. A. Rice and W. G. Cope, A statewide assessment of mercury dynamics in North Carolina water bodies and fish, Trans. Am. Fish. Soc., 2009, 138, 1328–1341 CrossRef.
- S. Y. Kwon, J. D. Blum, C. Y. Chen, D. E. Meattey and R. P. Mason, Mercury isotope study of sources and exposure pathway of methylmercury in estuarine foodwebs in the Northeastern U.S, Environ. Sci. Technol., 2014, 48, 10089–10097 CrossRef.
- M. Podar, C. C. Gilmour, C. C. Brandt, A. Soren, S. D. Brown, B. R. Crable, A. V. Palumbo, A. C. Somenahally and D. A. Elias, Global prevalence and distribution of genes and microorganisms involved in mercury methylation, Sci. Adv., 2014, 1, e1500675, DOI:10.1126/sciadv.1500675.
- T. Barkay and B. Gu, Demethylation-the other side of the mercury methylation coin: A critical review, ACS Environ. Au, 2022, 2, 77–97, DOI:10.1021/acsenvironau.1c00022.
- M. Amyot, G. Mierle, D. Lean and D. J. McQueen, Effect of solar radiation on the formation of dissolved gaseous mercury in temperate lakes, Geochim. Cosmochim. Acta, 1997, 61, 975–987 CrossRef.
- J. D. Blum, J. C. Drazen, M. W. Johnson and A. L. Jamieson, Mercury isotopes identify near-surface marine mercury in deep-sea trench biota, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29292–29298, DOI:10.1073/pnas.2012773117.
- B. L. Lee, S. Y. Kwon, R. Yin, M. Li, S. Jung, S. H. Lim, J. H. Lee, K. W. Kim, K. D. Kim and J. W. Jang, Internal dynamics of inorganic and methylmercury in a marine fish: Insights from mercury stable isotopes, Environ. Pollut., 2020, 267, 115588, DOI:10.1016/j.envpol.2020.115588.
- Y. H. Yang, S. Y. Kwon, M. Tsz-Ki, L. C. Motta, S. J. Washburn, J. Park, M.-S. Kim and K.-H. Shin, Ecological traits of fish for mercury biomonitoring: insights from compound-specific nitrogen and stable mercury isotopes, Environ. Sci. Technol., 2022, 56, 10808–10817, DOI:10.1021/acs.est.2c02532.
- S. Y. Kwon, J. D. Blum, M. J. Carvan, N. Basu, J. A. Head, C. P. Madenjian and S. R. David, Absence of fractionation of mercury isotopes during trophic transfer of methylmercury to freshwater fish in captivity, Environ. Sci. Technol., 2012, 46, 7527–7534 CrossRef PubMed.
- B. A. Bergquist and J. D. Blum, Mass-dependent and -independent fractionation of Hg isotopes by photoreduction in aquatic systems, Science, 2007, 318, 417, DOI:10.1126/science.1148050.
- B. A. Bergquist and J. D. Blum, The odds and evens of mercury isotopes: applications of mass-dependent and mass-independent isotope fractionation, Elements, 2009, 5, 353–357, DOI:10.2113/gselements.5.6.353.
- R. F. Lepak, J. C. Hoffman, S. E. Janssen, D. P. Krabbenhoft, J. M. Ogorek, J. F. DeWild, M. T. Tate, C. L. Babiarz, R. Yin, E. W. Murphy, D. R. Engstrom and J. P. Hurley, Mercury source changes and food web shifts alter contamination signatures of predatory fish from Lake Michigan, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23600–23608 CrossRef CAS.
- R. L. France, Carbon-13 enrichment in benthic compared to planktonic algae: foodweb implications, Mar. Ecol.: Prog. Ser., 1995, 124, 307–312 CrossRef.
- T. E. Cerling, J. M. Harris, B. J. MacFadden, M. G. Leakey, J. Quade, V. Eisenmann and J. R. Ehleringer, Global vegetation change through the Miocene-Pliocene boundary, Nature, 1997, 389, 153–158 CrossRef CAS.
- O. A. Sherwood, M. F. Lehmann, C. J. Schubert, D. B. Scott and M. D. McCarthy, Nutrient regime shift in the western North Atlantic indicated by compound-specific δ15N of deep-sea gorgonian corals, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1011–1015 CrossRef.
- D. K. Sackett, J. C. Drazen, C. A. Choy, B. Popp and G. L. Pitz, Mercury sources and trophic ecology for Hawaiian bottomfish, Environ. Sci. Technol., 2015, 49, 6909–6918, DOI:10.1021/acs.est.5b01009.
- T. S. Elsdon and B. M. Gillanders, Consistency of patterns between laboratory experiments and field collected fish in otolith chemistry: an example and applications for salinity reconstructions, Mar. Freshwater Res., 2005, 56, 609–617 CrossRef.
- T. S. Elsdon, B. K. Wells, S. E. Campana, B. M. Gillanders, C. M. Jones, K. E. Limburg, D. H. Secor, S. R. Thorrold and B. D. Walther, Otolith chemistry to describe movements and life-history parameters of fishes: hypotheses, assumptions, limitations and inferences, in Oceanography and Marine Biology, CRC Press, 2008, pp. 303–336 Search PubMed.
- T. R. Nelson and S. P. Powers, Elemental concentrations of water and otoliths as salinity proxies in a northern Gulf of Mexico estuary, Estuaries Coasts, 2020, 43, 843–864 CrossRef.
- T. M. Farmer, D. R. DeVries and J. E. Gagnon, Using seasonal variation in otolith microchemical composition to indicate largemouth bass and southern flounder residency patterns across an estuarine salinity gradient, Trans. Am. Fish. Soc., 2013, 142, 1415–1429 CrossRef.
- J. K. Chrisp, T. R. Nelson, D. K. Sackett and T. M. Farmer, Southern flounder life history diversity and contributions to fisheries from differing estuarine salinity zones, Mar. Coast. Fish: Dynam. Manag. Ecosys. Sci., 2023, 15, e10243, DOI:10.1002/mcf2.10243.
- S. M. Garcia and IdL. Moreno, Global Overview of Marine Fisheries, in Responsible Fisheries in the Marine Ecosystem, ed. Sinclair M. and Valdimarsson G., Food and Agriculture Organization of the United Nations (FAO), CABI Publishing, Cambridge MA, 2003 Search PubMed.
- S. G. Rogers, T. E. Targett and S. B. Van Sant, Fish-nursery use in Georgia salt-marsh estuaries: the influence of springtime freshwater conditions, Trans. Am. Fish. Soc., 1984, 113, 595–606, DOI:10.1577/1548-8659(1984)113<595:FUIGSE>2.0.CO;2.
- R. L. Allen and D. M. Baltz, Distribution and microhabitat use by flatfishes in a Louisiana estuary, Environ. Biol. Fishes, 1997, 50, 85–103 CrossRef.
- J. K. Craig, W. E. Smith, F. S. Scharf and J. P. Monaghan, Estuarine residency and migration of southern flounder inferred from conventional tag returns at multiple spatial scales, Mar. Coast. Fish: Dynam. Manag. Ecosys. Sci., 2015, 7, 450–463 CrossRef.
- B. F. Froeschke, B. Sterba-Boatwright and G. W. Stunz, Assessing southern flounder (Paralichthys lethostigma) long-term population trends in the northern Gulf of Mexico using time series analyses, Fish. Res., 2011, 108, 291–298 CrossRef.
- A. M. Flowers, S. D. Allen and L. M. Mark AL Lee, Stock Assessment of Southern Flounder (Paralichthys lethostigma) in the South Atlantic, 1989–2017, Report number: NCDMF SAP-SAR-2019-01, 2019, pp. 1–228, DOI:10.13140/RG.2.2.24489.44641.
- L. Besnard, G. L. Croizier, F. Galvan-Magana, D. Point, E. Kraffe, J. Ketchum, R. O. M. Rincon and G. Schaal, Foraging depth depicts resource partitioning and contamination level in a pelagic shark assemblage: Insights from mercury stable isotopes, Environ. Pollut., 2021, 283, 117066, DOI:10.1016/j.envpol.2021.117066.
- T. M. Farmer, R. A. Wright and D. R. DeVries, Mercury concentration in two estuarine fish populations across a seasonal salinity gradient, Trans. Am. Fish. Soc., 2010, 139, 1896–1912 CrossRef.
- G. L. Lescord, T. A. Johnston, B. A. Branfireun and J. M. Gunn, Percentage of methylmercury in the muscle tissue of freshwater fish varies with body size and age among species, Environ. Toxicol. Chem., 2018, 37, 2682–2691, DOI:10.1002/etc.4233.
- USEPA (United States Environmental Protection Agency), Mercury in Solids and Solutions by Thermal Decomposition, Amalgamation, and Atomic Absorption Spectrophotometry, 2007, Method 7473 Search PubMed.
- J. J. Dale, N. J. Wallsgrove, B. N. Popp and K. N. Holland, Nursery habitat use and foraging ecology of the brown stingray Dasyatis lata determined from stomach contents, bulk and amino acid stable isotopes, Mar. Ecol.: Prog. Ser., 2011, 433, 221–236 CrossRef.
- C. B. Wall, N. J. Wallsgrove, R. D. Gates and B. N. Popp, Amino acid 13C and 15N reveal distinct species-specific patterns of trophic plasticity in a marine symbiosis, Limnol. Oceanogr., 2021, 66, 2033–2050, DOI:10.1002/lno.11742.
- C. A. Gonzalez and S. J. Choquette, Report of Investigation, Reference Material 8610, Mercury Isotopes in UM-Almaden Mono-Elemental Secondary Standard, NIST (National Institute of Standards and Technology), Gaithersburg, MD, 2017 Search PubMed.
- H. P. Longerich, D. Gunther and S. E. Jackson, Elemental fractionation in laser ablation inductively coupled plasma mass spectrometry, Fresenius. J. Anal. Chem., 1996, 355, 538–542, DOI:10.1007/s0021663550538.
- T. R. Nelson and S. P. Powers, Elemental concentrations of water and otoliths as salinity proxies in a northern Gulf of Mexico estuary, Estuaries Coasts, 2020, 43, 843–864, DOI:10.1007/s12237-019-00686-z.
- S. N. Rodionov, A sequential algorithm for testing climate regime shifts, Geophys. Res. Lett., 2004, 31, 1–4 CrossRef.
- M. E. Seeley and B. D. Walther, Facultative oligohaline habitat use in a mobile fish inferred from scale chemistry, Mar. Ecol.: Prog. Ser., 2018, 598, 233–245 CrossRef.
- Y. Chikaraishi, N. O. Ogawa, Y. Kashiyama, Y. Takano, H. Suga, A. Tomitani, H. Miyashita, H. Kitazato and N. Ohkouchi, Determination of aquatic food-web structure based on compound specific nitrogen isotopic composition of amino acids, Limnol. Oceanogr.: Methods, 2009, 7, 740–750 CrossRef.
- C. J. Bradley, N. J. Wallsgrove, C. A. Choy, J. C. Drazen, E. D. Hetherington, D. K. Hoen and B. N. Popp, Trophic position estimates of marine teleosts using amino acid compound specific isotopic analysis, Limnol. Oceanogr.: Methods, 2015, 13, 476–493 CrossRef.
- D. B. Senn, E. J. Chesney, J. D. Blum, M. S. Bank, A. Maage and J. P. Shine, Stable isotope (N, C, Hg) study of methylmercury sources and trophic transfer in the northern Gulf of Mexico, Environ. Sci. Technol., 2010, 44, 1630–1637 CrossRef PubMed.
- K. P. Burnham and D. R. Anderson, Model Selection and Inference—A Practical Information-Theoretic Approach, Springer-Verlag, New York, 1998 Search PubMed.
- M. Li, A. T. Schartup, A. P. Valberg, J. D. Ewald, D. P. Krabbenhoft, R. Yin, P. H. Balcom and E. M. Sunderland, Environmental origins of methylmercury accumulated in subarctic estuarine fish indicated by mercury stable isotopes, Environ. Sci. Technol., 2016, 50, 11559–11568, DOI:10.1021/acs.est.6b03206.
- R. Ft Lepak, S. E. Janssen, R. Yin, D. P. Krabbenhoft, J. M. Ogorek, J. F. DeWild, M. T. Tate, T. M. Holsen and J. P. Hurley, Factors affecting mercury stable isotopic distribution in piscivorous fish of the Laurentian Great Lakes, Environ. Sci. Technol., 2018, 52, 2768–2776, DOI:10.1021/acs.est.7b06120.
- L. C. Motta, J. D. Blum, B. N. Popp, J. C. Drazen and H. G. Close, Mercury stable isotopes in flying fish as a monitor of photochemical degradation of methylmercury in the Atlantic and Pacific Oceans, Mar. Chem., 2020, 223, 103790, DOI:10.1016/j.marchem.2020.103790.
- P. Rodriguez-Gonzalez, V. N. Epov, R. Bridou, E. Tessier, R. Guyoneaud, M. Monperrus and D. Amouroux, Species-specific stable isotope fractionation of mercury during Hg(II) methylation by an anaerobic bacteria (Desulfobulbus propionicus) under dark conditions, Environ. Sci. Technol., 2009, 43, 9183–9188 CrossRef PubMed.
- M. Jimenez-Moreno, V. Perrot, V. N. Epov, M. Monperrus and D. Amouroux, Chemical kinetic isotope fractionation of mercury during abiotic methylation of Hg(II) by methylcobalamin in aqueous chloride media, Chem. Geol., 2013, 336, 26–36 CrossRef.
- MBNEP (Mobile Bay National Estuary Program), Legacy Contaminants: Legacy Contaminants in and near the Mobile Tensaw Delta. A Division of The Dauphin Island Sea Lab, https://www.mobilebaynep.com/the_issues/legacy-contaminants, accessed 9/10/2022 Search PubMed.
- R. Harris, C. Pollman, W. Landing, D. Evans, D. Axelrad, D. Hutchinson, S. L. Morey, D. Rumbold, D. Dukhovskoy, D. H. Adams, K. Vijayaraghavan, C. Holmes, R. D. Atkinson, T. Myers and E. Sunderland, Mercury in the Gulf of Mexico: Sources to receptors, Environ. Res., 2012, 119, 42–52 CrossRef.
- NADP (National Atmospheric Deposition Program), Annual mercury deposition network maps by year, Deposition: 2018, 2019, 2020, https://nadp.slh.wisc.edu/maps-data/mdn-gradient-maps/, accessed 9/19/2022 Search PubMed.
- T. J. Butler, M. D. Cohen, F. M. Vermeylen, G. E. Likens, D. Schmeltz and R. S. Artz, Regional precipitation mercury trends in the eastern USA, 1998–2005: declines in the Northeast and Midwest, no trend in the Southeast, Atmos. Environ., 2008, 42, 1582–1592 CrossRef.
- S. Coburn, B. Dix, E. Edgerton, C. D. Holmes, D. Kinnison, Q. Liang, A. ter Schure, S. Y. Wang and R. Volkamer, Mercury oxidation from bromine chemistry in the free troposphere over the southeastern US, Atmos. Chem. Phys., 2016, 16, 3743–3760 CrossRef.
- S. J. Klapstein and N. J. O'Driscoll, Methylmercury biogeochemistry in freshwater ecosystems: a review focusing on DOM and photodemethylation, Bull. Environ. Contam. Toxicol., 2018, 100, 14–25 CrossRef PubMed.
- X. Y. Zhang, Y. B. Li, G. Feng, C. Tai, Y. G. Yin, Y. Cai and J. F. Liu, Probing the DOM-mediated photodegradation of methylmercury by using organic ligands with different molecular structures as the DOM model, Water Res., 2018, 138, 264–271 CrossRef.
- J. L. W. Cowan, J. R. Pennock and W. R. Boynton, Seasonal and interannual patterns of sediment-water nutrient and oxygen fluxes in Mobile Bay, Alabama (USA): regulating factors and ecological significance, Mar. Ecol.: Prog. Ser., 1996, 141, 229–245 CrossRef.
- USDA (United States Department of Agriculture), 2022 State Agriculture Overview: Alabama, https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=ALABAMA, 2022, Accessed May 1, 2023 Search PubMed.
- B. P. DiMento and R. P. Mason, Factors controlling the photochemical degradation of methylmercury in coastal and oceanic waters, Mar. Chem., 2017, 196, 116–125, DOI:10.1016/j.marchem.2017.08.006.
- R. H. Monrreal, Hydrology and water chemistry in Weeks Bay, Alabama: implications for mercury bioaccumulation, MS thesis, Auburn University, Auburn, Alabama, 2007.
- D. G. Rumbold, T. R. Lange, D. Richard, G. DelPizzo and N. Hass, Mercury biomagnification through foodwebs along a salinity gradient down-estuary from a biological hotspot, Estuarine, Coastal Shelf Sci., 2018, 200, 116–125 CrossRef.
- P. J. Webster, G. T. Holland, J. A. Curry and H. R. Chang, Changes in tropical cyclone number, duration, and intensity in a warming environment, Science, 2005, 309, 1844–1846 CrossRef PubMed.
- D. Schiedek, B. Sundelin, J. W. Readman and R. W. Macdonald, Interactions between climate change and contaminants, Mar. Pollut. Bull., 2007, 54, 1845–1856 CrossRef.
- P. D. Noyes, M. K. McElwee, H. D. Miller, B. W. Clark, L. A. Van Tiem, K. C. Walcott, K. N. Erwin and E. D. Levin, The toxicology of climate change: environmental contaminants in a warming world, Environ. Int., 2009, 35, 971–986 CrossRef PubMed.
- T. R. Knutson, J. L. McBride, J. Chan, K. Emanuel, G. Holland, C. Landsea, I. Held, J. P. Kossin, A. K. Srivastava and M. Sugi, Tropical cyclones and climate change, Nat. Geosci., 2010, 3, 157–163 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3em00482a |
| This journal is © The Royal Society of Chemistry 2024 |
