Open Access Article
Open Access ArticleUnravelling the mechanism of apoptosis induced by copper(II) complexes of NN2-pincer ligands in lung cancer cells†
Athulya
Das
and
Muniyandi
Sankaralingam
 *
*
Bioinspired & Biomimetic Inorganic Chemistry Laboratory, Department of Chemistry, National Institute of Technology Calicut, Kozhikode-673601, Kerala, India. E-mail: msankaralingam@nitc.ac.in; sankarjan06@gmail.com
First published on 24th July 2024
Abstract
The invention of efficient chemotherapeutic drugs is essential for human health and development. Keeping this in mind, a series of copper(II) pincer complexes, 1–4, of ligands L1(H) = 2-morpholino-N-(quinolin-8-yl)acetamide, L2(H) = 2-di-n-propylamino-N-(quinolin-8-yl)acetamide, L3(H) = 2-di-n-butylamino-N-(quinolin-8-yl)acetamide and L4(H) = 2-di-n-benzylamino-N-(quinolin-8-yl)acetamide have been synthesized, characterized, and utilized for inhibiting cancer proliferation. Complexes 1–4 showed very efficient activity against lung (A549) and breast (MCF-7) cancer cells, which are the most frequently diagnosed cancers according to the WHO. Among them, 1 was highly active against lung cancer cells with an IC50 value of 8 μM, showing no toxicity towards common L929 fibroblast cell lines (IC50 > 1000 μM). Moreover, AO–EB staining inferred that this cellular demise was attributed to apoptosis, which was determined to be 25.91% of cells by flow cytometry at the IC50 concentration. Furthermore, carboxy-H2DCFDA staining revealed the involvement of ROS in the mechanism. Interestingly, JC-1 dye staining revealed a change in the potential of the mitochondrial membrane, which indicates the enhanced production of ROS in mitochondria. A deep search for the mechanism through in silico studies guided us to the fact that complexes 1–4 might perturb the function of complex I in mitochondria. Furthermore, the studies can be expanded towards clinical applications mainly with morpholine appended complex 1.
1. Introduction
According to the World Health Organization (WHO), cancer caused one among six deaths in 2018.1 Even though the survival rate is improving, there is a long way to go for a complete cure of the disease. Lung cancer and breast cancer have become very common but an ideal treatment method is yet to be established. Therefore, scientists are on a huge thrust to develop novel, efficient drugs against this malady. As a result, recent years have witnessed the rapid expansion of metallodrugs with a wide variety of metals including transition metals.2–7 The choice of a metal centre plays an important role because it can change the physicochemical properties of the complexes.8,9 Moreover, biomimetic first-row transition metal complexes are found to be very promising candidates as anticancer drugs.10 Among them, copper complexes always have a special place due to their high anticancer activity.11–14 One of the relevant components of metallodrugs is the heterocyclic ligands that are extremely biocompatible due to the structurally relevant natural molecules.15–17 For instance, aminoquinoline is one of the pharmacophores in medicinal chemistry involved in many applications including anticancer activities.18–20 Similarly, along with aminoquinoline, if bioactive molecules like morpholine are used, then the activity could be enhanced.21 Moreover, the solubility and lipophilicity can be tuned by propyl, butyl, and benzyl groups on the ligands.22In the past few years, several pincer complexes of different metals have been reported to have excellent anticancer activities that can be attributed to their unique structural orientation.23 For example, two of the aminoquinoline-based copper(II) pincer complexes were reported to have cytotoxic activity against A549 cells. Among them, one of the complexes had an IC50 value of 1 μM and the other was active only at a concentration as high as 1 mM.24 In another report, two more aminoquinoline based copper(II) pincer complexes were reported with IC50 values of around 0.46 and 0.43–0.90 μM, respectively, against MCF-7 and A549 cell lines.25 Another report described a series of terpyridine based copper(II) pincer complexes with good activity against A549 (IC50 = 4.2–100 μM) and MCF-7 (IC50 = 43–100 μM) cell lines.26 Similarly, cyclometalated copper(I) pincer complexes with phosphaadamantane derivatives were reported with very good anticancer activity against A549 and MCF-7 cells, for which the IC50 value ranges from 2.35 to 3.92 μM and 3.08 to 8.41 μM, respectively.27 Very recently, SNS donor copper(II) pincer complexes have been reported against A549 cells.28 Among them, the chloro and thiocyanate derivatives were more active with IC50 values of around 12.30 μM. However, the perchlorate derivative had an IC50 value of 14.08 μM. Thus, activity mainly changes depending on the ligand donors.
As previously mentioned, morpholine (1), dipropyl (2), dibutyl (3), and dibenzyl (4) derivatives of 8-aminoquinoline based copper(II) pincer complexes (Scheme 1) were synthesized, characterized, and tested for their anticancer efficacy against MCF-7 and A549 cell lines. It should be noted that complexes 1–3 have already been proved as better antimicrobial agents from our recent study.7 Here, with IC50 values ranging from 8 to 20 μM and 15 to 19.35 μM, all four complexes showed potential cytotoxicity against A549 and MCF-7 cells, respectively. The highly active complex 1 against A549 cells was found to display no toxicity towards the normal L929 fibroblast cell line (IC50 > 1000 μM). The dominant activity of 1 compared to its corresponding ligand L1(H) (IC50 = 77.65 μM) against A549 cells revealed the importance of the copper complexes. A detailed mechanistic investigations reveal that cell death through apoptosis, which was induced by the present copper complexes, produces excess ROS in mitochondria. Furthermore, we expect that clinical applications with morpholine appended complex 1 may be extended.29
2. Experimental
2.1. Materials
Chemicals purchased commercially were used without additional purification. Bromoacetyl bromide and 8-aminoquinoline were obtained from TCI Chemicals. Copper(II) bromide, triethylamine, morpholine, and diethyl ether were procured from Merck, India. Alfa Aesar supplied di-n-propylamine and di-n-butylamine. Dibenzylamine and DMSO (extra pure 99%) were procured from SRL, India. Acetone, methanol, dichloromethane, and ethyl acetate were procured from Qualigens, India. Using magnesium turnings and iodine, methanol was distilled. Calcium chloride and calcium hydride were used to distill acetone and DCM respectively. The cell lines A549 and MCF-7 were obtained from the National Centre for Cell Science (NCCS), Pune, India.2.2. Physical measurements
Agilent Cary 8454 UV-visible and Jasco 4700 ATR-FTIR spectrometers were utilized for recording the absorption spectra and infrared spectra of the compounds, respectively. NMR spectra were recorded using a Jeol 500 MHz NMR spectrometer. Mass data were measured using an Agilent 6545 Q-TOF LC/MS instrument and a Bruker Daltonics Esquire 6000. A PerkinElmer 2400 Series II analyser was used for CHN analysis. A Bruker BioSpin spectrometer, Germany (EMXmicro A200-9.5/12/S/W), was used to collect electron paramagnetic resonance (EPR) spectra at 100 K. Calibration of the g value was done using a manganese marker. Magnetic measurements viz. zero-field cooling (ZFC) and field cooling (FC) were performed using Bruker WIN EPR acquisition and processing software in the range of 100–300 K. A Bruker D8 Venture dual source diffractometer was employed to obtain single-crystal X-ray diffraction data using Mo-Kα radiation. An Olympus fluorescence microscope, CKX-53, Japan, was used to see the cells after anticancer mechanistic studies. All the incubations for anticancer studies were under a 5% CO2 atmosphere at 37 °C.2.3. Synthesis of ligands and complexes
The ligands L1(H), L2(H), and L3(H) and their copper(II) complexes 1, 2, and 3 were synthesized by following our very recent reports.6,7 The detailed synthetic steps are elaborated in the ESI.†![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 3265 cm−1 (N–H).
O) and 3265 cm−1 (N–H).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 3264 cm−1 (N–H).
O) and 3264 cm−1 (N–H).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 3245 cm−1 (N–H).
O) and 3245 cm−1 (N–H).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (1) mixture to obtain the pure ligand with a yield of 60(2)% as a white powder. 1H NMR (500 MHz, CDCl3, 298 K) δ (ppm): 11.67 (s, 1H), 9.03 (s, 1H), 8.76–8.73 (m, 1H), 8.20–8.18 (dd, J = 8.20, 1.53 Hz, 1H), 7.67–7.65 (d, J = 6.87 Hz, 4H), 7.54–7.52 (m, 3H), 7.33–7.30 (t, J = 7.25 Hz, 4H), 7.25–7.22 (m, 2H), 3.76 (s, 4H), 3.38 (s, 2H). 13C NMR (125 MHz, CDCl3, 298 K) δ (ppm): 58.89, 59.09, 116.40, 121.59, 121.67, 127.34, 127.41, 128.07, 128.47, 129.22, 134.47, 136.21, 138.17, 138.79, 148.19, 169.99. ATR, cm−1 (Fig. S1†): 1676 cm−1 (C
ethyl acetate (1) mixture to obtain the pure ligand with a yield of 60(2)% as a white powder. 1H NMR (500 MHz, CDCl3, 298 K) δ (ppm): 11.67 (s, 1H), 9.03 (s, 1H), 8.76–8.73 (m, 1H), 8.20–8.18 (dd, J = 8.20, 1.53 Hz, 1H), 7.67–7.65 (d, J = 6.87 Hz, 4H), 7.54–7.52 (m, 3H), 7.33–7.30 (t, J = 7.25 Hz, 4H), 7.25–7.22 (m, 2H), 3.76 (s, 4H), 3.38 (s, 2H). 13C NMR (125 MHz, CDCl3, 298 K) δ (ppm): 58.89, 59.09, 116.40, 121.59, 121.67, 127.34, 127.41, 128.07, 128.47, 129.22, 134.47, 136.21, 138.17, 138.79, 148.19, 169.99. ATR, cm−1 (Fig. S1†): 1676 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and 3280 cm−1 (N–H).
O) and 3280 cm−1 (N–H).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), QTOF MS m/z (%) (Fig. S3†): 443.10 (calcd), 443.10 [Cu(L4)]+ (found). Elemental analysis calcd. for (C25H22N3OBrCu): C 57.31, H 4.23, N 8.02; found: C 57.22, H 4.21, N 7.99.
O), QTOF MS m/z (%) (Fig. S3†): 443.10 (calcd), 443.10 [Cu(L4)]+ (found). Elemental analysis calcd. for (C25H22N3OBrCu): C 57.31, H 4.23, N 8.02; found: C 57.22, H 4.21, N 7.99.
2.4. Single-crystal X-ray diffraction studies
By careful examination under an optical microscope, fine single crystals suitable for X-ray diffraction were chosen from the mother liquor of complex 1. Single crystals were mounted using a fibre loop and optically centered. The automatic cell determination routine was employed to collect reflections (24 frames at three different orientations of the detector) and the APEX3-SAINT program was used for determining the unit cell parameters.30The data were corrected for Lorentz-polarization effects. Semiempirical absorption correction (multi-scan) based on symmetry-equivalent reflections was applied using the SADABS program.31 The structures were solved by direct methods and refined by full-matrix least-squares, based on F2 using SHELX-2018 software package2 and the WinGX program.32 The crystallographic details of the data collected for 1 are given in Table 1.
| 1 | |
|---|---|
| a R 1 = [∑(||Fo| − |Fc||)/∑|Fo|]; wR2 = {[∑(w(Fo2 − Fc2)2)/∑(wFo4)]}1/2. | |
| Empirical formula | C15H18BrCuN3O3 |
| Formula weight, g mol−1 | 431.77 |
| Crystal habit, colour | Block, green |
| Crystal system | Monoclinic |
| Crystal size, mm | 0.042 × 0.030 × 0.025 |
| Space group | P21/n |
| a, Å | 7.9149(8) |
| b, Å | 18.466(2) |
| c, Å | 11.1514(11) |
| α, ° | 90 |
| β, ° | 94.607(7) |
| γ, ° | 90 |
| V (Å3) | 1624.6(3) |
| Z | 4 |
| ρ calcd/mg m−3 | 1.765 |
| F(000) | 868 |
| Temperature (K) | 298(2) |
| No. of reflections collected | 9012 |
| Independent reflections | 2055 [R(int) = 0.1366] |
| Radiation (Mo-Kα)/Å | 1.54178 |
| Goodness-of-fit on F2 | 1.203 |
| Number of refined parameters | 214 |
| R 1/wR2[I > 2σ(I)]a | R 1 = 0.1300, wR2 = 0.3196 |
| R 1/wR2 (all data) | R 1 = 0.2503, wR2 = 0.4030 |
2.5. MTT assay
The MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay can discern the metabolic activity using the colorimetric technique. In the presence of viable cells, MTT turns into purple formazan from yellow colour.33,34 Once the cell culture was trypsinized, a medium containing 10% fetal bovine serum (FBS) was used to adjust the cell count to 1 × 105 cells per mL. Then, they were filled in 96-well microliter plates in such a way that each well contained 50000 cells. A partial monolayer was formed after 24 h. This monolayer was rinsed with the medium and compounds dissolved in DMSO were added in serial dilutions from 20 μM to 0 μM. Furthermore, it was incubated for 24 h. Then, 100 μL of 5 mg per 10 mL MTT in PBS (phosphate buffered saline) was distributed in each well and incubation was continued for 4 h. Following the removal of the supernatant, 100 μL of DMSO was introduced to solubilize the formazan. The absorbance of this solution was determined using a microplate reader (Tecan plate reader) at 590 nm. The following formula was used to determine the extent of cell growth inhibition: | (1) |
Furthermore, the dose–response curve between different concentrations of compounds against the percentage inhibition was able to obtain the half-maximal inhibitory concentration (IC50). The IC50 was computed using GraphPad Prism 6 (GraphPad, San Diego, CA, USA). The IC50 values are calculated based on the mean of the triplicate absorptions. In addition, mechanistic studies were carried out for complex 1 against A549 cells due to its elevated activity compared to other complexes.
2.6. Acridine orange (AO)–ethidium bromide (EB) staining
Cells treated with 1 were examined for apoptosis using a dual AO–EB fluorescence staining method.35–37 In a nutshell, 4000 cells per well in 24 well plates were processed with 1 at the IC50 concentration and incubated for 24 h. The staining solution containing AO and EB at a concentration of 100 g mL−1 was added to each well at a volume of 500 μL after 24 h of incubation. The results were observed through a fluorescence microscope.2.7. Flow cytometry
Annexin V-FITC/PI staining is considered as a sophisticated technique that can distinguish cells of viable, apoptotic, and necrotic features.38–40 Once 105 cells per well were cultured under incubation for 24 h in a 6-well plate, they were treated with 1 at the IC50 concentration. The cells were trypsinized, and after 24 h of incubation, they were rinsed with PBS and dyed with Annexin V-FITC/PI in accordance with the instructions on the Annexin V-FITC apoptosis detection kit. The cells that have not undergone treatment with 1 were treated as the control. Furthermore, the extent of cell death was evaluated using a SYSMEX flow cytometer (Japan) and analyzed using FlowJo software.2.8. Determination of ROS
Carboxy-H2DCFDA (2′,7′-dichlorodihydrofluorescein diacetate) is normally non-fluorescent. Once ROS is present, it oxidises to show green fluorescence.41–43 A fresh stock solution of carboxy-H2DCFDA was prepared in sterile 100% ethanol right before use. After 24 hours, 1 at the IC50 concentration was treated with carboxy-H2DCFDA (1 μM) in a regular culture medium with reduced serum (2%) and incubated for 30 minutes. The medium containing carboxy-H2DCFDA was removed and washed with PBS twice. A fluorescence microscope was used to examine the cells.2.9. Determining the mitochondrial membrane potential (MMP) by JC-1 staining
Roughly 5 × 105 cells per mL were cultured on coverslips in a 6-well culture plate and incubated for 8 h.44–46 After 24 h of incubation with 1 at the IC50 concentration, 1–10 μM of JC-1 dye was added to the cell culture medium as a working stock solution and the plates were incubated (15–30 min). The dead cell percentage was immediately observed under a fluorescence microscope.2.10 Lipophilicity and Stability studies
The lipophilicity was determined by the shake-flask method using a pre-saturated 1-octanol-water solution which is detailed in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 47 The stability of the complexes was calculated according to their decay in DMSO and FBS using UV-vis spectroscopy. The absorption spectra of compounds dissolved in DMSO and 5% DMSO in FBS were recorded at 0 h and after 24 h. The percentage of decay was calculated from the differences in the absorption of both spectra.
47 The stability of the complexes was calculated according to their decay in DMSO and FBS using UV-vis spectroscopy. The absorption spectra of compounds dissolved in DMSO and 5% DMSO in FBS were recorded at 0 h and after 24 h. The percentage of decay was calculated from the differences in the absorption of both spectra.
2.11. Molecular docking
The interaction of complexes 1–4 with complex I of mitochondria was performed using AutoDock Vina.48 The protein structure, 5XTD, was obtained from the Protein Data Bank. The active site was included completely by creating grid box dimensions of 46 Å × 66 Å × 74 Å and the same was used for all the complexes. The Gaussian 09 software package at the B3LYP level of theory and the basis sets 6-31G for C, H, O, and N and LANL2DZ for the metal centre were used to optimize the geometry of the complexes.49 The output was visualized and analysed using Autodock tools 1.5.7 and Discovery Studio.503. Results and discussion
3.1. Synthesis and characterization of ligands and copper(II) complexes
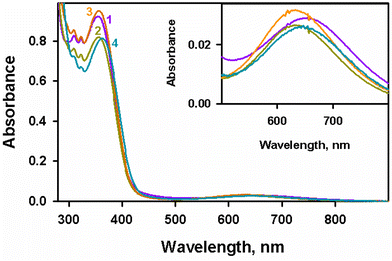
As per the literature method, ligands L1(H), L2(H), L3(H) and L4(H) were obtained by reacting 2-bromo-N-(quinoline-8-yl)acetamide with morpholine, diisopropylamine, dibutylamine, and dibenzylamine, respectively.6,7 Then, by stirring a methanolic solution of an equivalent amount of ligand, CuBr2, and triethylamine, copper(II) complexes 1–4 were synthesized as reported.7 The isolated compounds were characterized using advanced analytical techniques. For instance, the removal of a N–H peak during complex formation was clearly visible from the disappearance of the peak around 3245–3280 cm−1 in the ATR-IR spectra of 4 compared to the corresponding ligand L4(H). This observation was similar for 1–3 and L1(H)–L3(H).7 Mass spectrometry also revealed the formation of 4. A square planar coordination geometry of the copper(II) centre of 1–4 in solution was indicated by a highly intense peak around 356–364 nm and a peak corresponding to a d–d transition around 631–655 nm in the UV-visible spectra. Complex 4 was observed with g-tensors (g|| = 2.293 and g⊥ = 2.039) and coupling constants (A|| = 194 × 10−4 cm−1) gained from the axial symmetry EPR spectra that match very well with the recently reported EPR spectra of 1–3.7 Furthermore, structural characterization of complex 1 by single-crystal X-ray crystallography revealed a distorted square pyramidal geometry that can be formulated as [Cu(L1)Br(H2O)].3.2. Infrared spectra of 1–4 with L1(H)–L4(H)
Ligands L1(H)–L4(H) exhibited two major peaks around 3245–3280 and 1676–1694 cm−1 in the functional group region, which are assigned to N–H (L1(H), 3265 cm−1; L2(H), 3264 cm−1; L3(H), 3245 cm−1; and L4(H), 3280 cm−1) and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (L1(H), 1694 cm−1; L2(H), 1676 cm−1; L3(H), 1680 cm−1; and L4(H), 1676 cm−1) groups. Upon complexation with CuBr2 in the presence of TEA, the N–H peak completely disappeared in 1–4, revealing N− (amido) coordination to the copper centre (Fig. S1 and S2†). Furthermore, the decrease in C
O (L1(H), 1694 cm−1; L2(H), 1676 cm−1; L3(H), 1680 cm−1; and L4(H), 1676 cm−1) groups. Upon complexation with CuBr2 in the presence of TEA, the N–H peak completely disappeared in 1–4, revealing N− (amido) coordination to the copper centre (Fig. S1 and S2†). Furthermore, the decrease in C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching frequencies (1587–1597 cm−1) was also noticed after complexation, indicating that the electron density localized more towards the copper centre. All these results were in concordance with the data recently reported by us.7
O stretching frequencies (1587–1597 cm−1) was also noticed after complexation, indicating that the electron density localized more towards the copper centre. All these results were in concordance with the data recently reported by us.7
3.3. Electronic properties of complexes
Complexes 1–4 showed two peaks in the UV-visible spectra in methanol at room temperature. The observed highly intense peaks at around 356–364 nm (1, 356 nm (ε, 4610 M−1 cm−1); 2, 357 nm (ε, 4095 M−1 cm−1); 3, 357 nm (ε, 4755 M−1 cm−1); and 4, 364 nm (ε, 4070 M−1 cm−1)) are assigned to the charge transfer transition from the ligand to the copper(II) centre (LMCT) (Fig. 1). The other broad peaks at around 631–655 nm (1, 655 nm (ε, 145 M−1 cm−1); 2, 631 nm (ε, 130 M−1 cm−1); 3, 633 nm (ε, 160 M−1 cm−1); and 4, 647 nm (ε, 135 M−1 cm−1)) with a low intensity are assigned to the d–d transition (2B1g → 2Eg in square planar copper(II) complexes).7,51The frozen EPR spectra of complex 4 exhibit a well-refined axial symmetry (Fig. 2), with g-tensors g|| = 2.293 and g⊥ = 2.039 and coupling constants (A|| = 194 × 10−4 cm−1), which were comparable to those reported for complexes 1–3 (g|| = 1, 2.285; 2, 2.283; and 3, 2.274 and g⊥ = 1, 2.042; 2, 2.039; and 3, 2.033) and coupling constants (A|| in × 10−4 cm−1 = 1, 197; 2, 197; and 3, 197).7 In addition to low g|| and high A|| values, it is observed that the f quotient (118 cm−1) falls within 105–135 cm−1, which suggests that the planar nature of complex 4 is similar to the other complexes 1–3.7,52 Moreover, the square-based geometry of the complexes was perceptible from the g|| > g⊥ values, which indicates that dx2−y2 is the ground state.53
 | ||
Fig. 2 A comparison of the frozen EPR spectrum of 4 with that of 1–3 in DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (4 methanol (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) measured at 100 K.7 1 v/v) measured at 100 K.7 | ||
3.4. Molecular geometry of 1
The molecular structure of complex 1 is shown in Fig. 3 with the atom numbering scheme, principal bond lengths and bond angles provided in Table 2. It was observed that 1 exhibits a distorted square pyramidal geometry with a monoclinic crystal system and the P21/n space group. The tridentate chelating ligand is coordinated in a facial fashion towards the copper(II) centre with the nitrogen atoms of quinolyl, amido and morpholine, along with a bromine atom (Br) and a water molecule (O(3)). This water molecule (O(3)) might be present in the solvent of crystallization, revealed by the lack of a peak corresponding to OH in the IR spectra of 1 (Fig. S2† (1)). Moreover, the coordination around the copper(II) of 1 with bond angles ranging from 91.7° to 110.4° that deviate from the ideal square pyramidal angles of 90° and 180° was observed. Here, the coordination around copper gives a trigonality index (τ5) of 0.2 (τ5 = (β – α)/60, where α = N(2)–Cu(1)–Br = 152° and β = N(1)–Cu(1)–N(3) = 164°).54 As expected, the bond length of Cu(II)–Nquinoline (N(1)) is shorter than the bond distance of Cu(II)–Nmorpholine (N(3)) due to the sp2 hybridized quinoline ring. | ||
| Fig. 3 ORTEP diagram of 1 showing 30% probability of thermal ellipsoids, and the labelling scheme of the atoms. Hydrogen atoms in the chelating ligands are omitted for clarity. | ||
| Bond lengths/Å | |
|---|---|
| Cu–N(1) | 2.004(16) |
| Cu–N(2) | 1.909(17) |
| Cu–N(3) | 2.070(16) |
| Cu–Br | 2.456(5) |
| Cu–O(3) | 2.251(16) |
| Bond angles [°] | |
| N(1)–Cu–N(2) | 80.2(9) |
| N(1)–Cu–N(3) | 164.0(8) |
| N(2)–Cu–N(3) | 83.8(8) |
| N(1)–Cu–Br | 95.7(6) |
| N(2)–Cu–Br | 152.0(6) |
| N(3)–Cu–Br | 98.1(6) |
| N(1)–Cu–O(3) | 91.7(6) |
| N(2)–Cu–O(3) | 110.4(7) |
| N(3)–Cu–O(3) | 94.5(6) |
| O(3)–Cu–Br | 97.3(5) |
Very recently, complexes 2 and 3 have been reported with a square planar geometry that is slightly distorted and crystallize with the space groups C2/c and P21/n, respectively, in a monoclinic system.7 Upon examining the bond parameters of 2 and 3, it is found that the bond angles and bond lengths are comparable to those of complex 1. It should be noted that in all complexes (1–3), the Cu–Namine (2.070(16)–2.0901(16) Å) bond is lengthier than the Cu–Nquinoline (2.004(16)–2.0457(16) Å) bond, which is consistent with the sp3 and sp2 hybridizations, respectively. Moreover, these complexes deviate from their ideal structures with trigonality indexes of τ5 = 0.2 for 1, τ4 = 0.16 for 2, and 0.24 for 3. Also, the distorted square pyramidal coordination of 1 was found to be similar to that of the recently reported zinc(II) complex [Zn(L1)Cl(H2O)] with a τ5 value of 0.257.6
3.5. Activity of 1–4 against lung (A549) and breast (MCF-7) cancer cell lines
 | ||
| Fig. 4 (A) Viability of A549 under complex 1 and (B) IC50 values of ligand L1(H) and complexes 1–4 effective against A549 and MCF-7 cancer cell lines. | ||
Similarly, complexes [Cu(κ3-L)(Cl)2]·3H2O, [Cu(κ3-L)(SCN)2], and [Cu(κ3-L)(ClO4)2], where L = 2,6-bis[[(4-bromophenyl)thio]methyl]pyridine, show results (IC50 = 12.38–14.98 μM) comparable to the present complexes against A549 cells under incubation for 48 h.29 Other complexes [Cu(CMQA)(H2O)] where CMQA = N-carboxymethyl-L-methionine-N′-8-quinolylamide, [CuCl2(1-methyl-1H-pyrrol-2-yl-terpy)], and [CuCl2(R-dtpy)] where R = furan-2-yl, thiophen-2-yl, 1-methyl-1H-pyrrol-2-yl and dtpy = 2,6-di(thiazol-2-yl)pyridine show less activity against A549 cells (IC50 = 64.3–100 μM) compared to the present complexes 1–4.25,27
In a similar way, comparing the reported activity of copper pincer complexes against MCF-7 revealed that [Cu(Me)(N-TP)(PF6)], [Cu(Et)(N-TP)(PF6)], [Cu(iPr)(N-TP)(PF6)], [Cu(L)Cl](ClO4), and [Cu(L)Br2] have IC50 values in between 0.46 and 8.41 μM, at an incubation time of 48 h. At the same time, it is observed that complexes [CuCl2(R-terpy)] and [CuCl2(R-dtpy)] where R = furan-2-yl, thiophen-2-yl, and 1-methyl-1H-pyrrol-2-yl are less active than 1–4 with IC50 values ranging between 43 and 100 μM against MCF-7 at 48 h of incubation.27 Very recently, we have reported the anticancer activity of zinc complexes of ligands L1(H)–L3(H) against A549 and MCF-7 cells and found that the IC50 values range from 16.35 to 17.95 and 33.35 to 40 μM, respectively.6 This diminished activity of zinc analogues indicates the supremacy of copper complexes in cancer therapy. Moreover, among the copper(II) complexes, 1 of L1(H) has the highest activity but in the case of zinc(II) complexes, 3 of L3(H) has the best activity. From this, it is understood that the ligand system also affects the activity along with the choice of metal ions.
Furthermore, the quantification of apoptosis in A549 cells induced by 1 was marked using flow cytometry. A549 cells treated with 1, at the IC50 concentration, showed a different extent of cell death compared with the control, as shown in Fig. 7. This resulted in 2.61% (Q3) of cells under apoptosis, 23.3% (Q2) of cells in the late apoptotic stage, and 5.47% (Q1) of necrotic cells (Fig. 7). The impact of 1 is clearly visible by comparing the results with that of the control (Q2 = 0.076%, Q3 = 0.41%, and Q1 = 3.1%). These findings altogether revealed that apoptosis is the primary mechanism by which the complex caused cell death. Similarly, the reported terpyridine based copper(II) pincer complexes were active against A549 and MCF-7 cell lines and were able to induce apoptosis in A549 cells.27 Similarly, the reported zinc(II) complex of L3(H) had a slightly low percentage of cells under apoptotic (1.37%), late apoptotic (21.4%), and necrotic (4.94%) cells compared to that of the present copper(II) complexes in the flow cytometry experiment.6
 | ||
| Fig. 8 H2DCFDA stained A549 cells: (A) control and (B) after being treated with 1 at the IC50 concentration. | ||
ROS-induced damage is more prominent in mitochondrial DNA than nuclear DNA.57 The JC-1 assay is a very effective way to find depolarization of the mitochondrial membrane. Once the JC-1 dye enters the mitochondria of a healthy cell, it emits red fluorescence (∼590 nm) of J-aggregates in healthy cells and green fluorescence (∼525 nm) of their monomeric form in unhealthy cells. Fig. 9 explains the change in the MMP. Moreover, this change is due to the increased production of ROS which causes membrane permeabilization and results in the release of cytochrome c.58 This change in the MMP is an indication of the initial phase of apoptosis.59 Moreover, it is observed that the recently reported zinc(II) complex of L3(H) promotes the production of excess ROS and changes in the MMP.6 However, the higher activity of the present copper complexes 1–3 than their zinc analogues might be due to the inherent redox-active nature of copper.
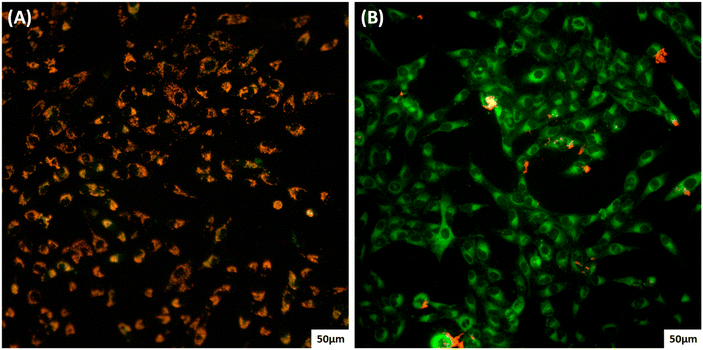 | ||
| Fig. 9 JC-1 staining assay of A549 cells: (A) control and (B) after being treated with 1 at the IC50 concentration. | ||
As alluded to above, the ROS formation and the change in the MMP were confirmed experimentally, and thus we propose the mechanism of anticancer activity of 1–4 with the generation of ROS in mitochondria by perturbing the function of complex I as reported earlier.60–62 Being the first enzyme of the respiratory chain, complex I has a vital role in cellular energy production.63 This is possible only if 1–4 are capable of anchoring near complex I. To support this postulate, we examined molecular docking with complex I of PDB ID: 5XTD with our copper complexes.64
Interestingly, we found that 1 binds very near to the flavin mononucleotide (FMN) site of complex I (Fig. 10A); conversely, 2–4 bind away from this site with docking scores of −7.3, −6.6, −8.2, and −8.4 kcal mol−1, respectively (Fig. S12–S14†) under the same grid box sizes.65 Complex 1 binds to Gly350 (2.32 Å) and Leu349 (2.86 Å) amino acids through a strong hydrogen bonding. In complex 2, the donor–donor bond between His376 and the amide nitrogen is very strong with a distance of 2.90 Å. Complex 3 interacted with His74 and Ala76 much more strongly with bond distances of 3.58 Å (carbon) and 3.86 Å (alkyl), respectively. A π–π stacking interaction (3.80 Å) was observed with 4 and His736. In a similar way, the interaction of 4 with Val174 was observed with two strong π–sigma interactions with distances of 3.88 and 3.95 Å. This indicates that 1–4 can interact with complex I of mitochondria and possibly inhibit the activity. Specifically, the strong binding of 1 near the FMN site compared to the other complexes 2–4 might be the reason for its enhanced anticancer activity.
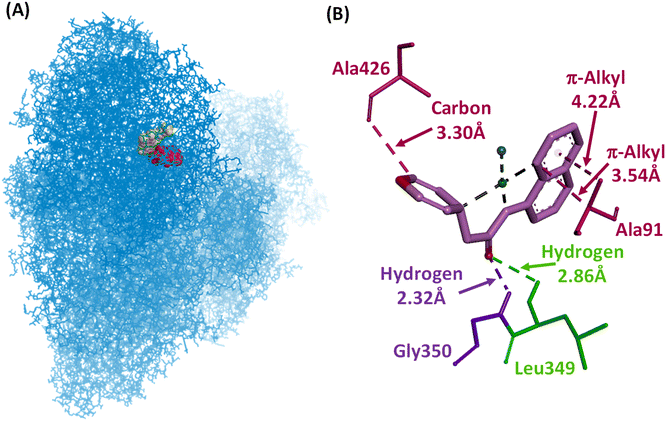 | ||
| Fig. 10 (A) Molecular docked conformation and (B) interactions of 1 at the active site of 5XTD. | ||
Overall, complexes 1–4 exhibit promising activity against A549 and MCF-7 cell lines. Mechanistic investigation reveals that the cell death of A549 is induced by the most active complex 1 because of the excess production of ROS in the mitochondria and thereby apoptosis. The observation is well supported by experimental and in silico studies.
3.6. Lipophilicity analysis
Complex 4 and the corresponding ligand L4(H) were found to have a partition coefficient, P (or log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow), with values of 3.14 and 10.04, respectively, determined by the shake flask method.47 Compounds 4 and L4(H) were found to be highly lipophilic compared to the other complexes 1, −0.51; 2, 1.07; and 3, 1.83 and ligands L1(H), 2.71; L2(H), 0.67; and L3(H), 0.55.6,7 It should be noted that 1 has a slightly hydrophilic nature and shows the best activity. At the same time, 4 has higher lipophilicity which might not be good for a drug (Fig. S15†). This indicates how the lipo–hydro balance matters for drug molecules. In the present compounds, it is seen that the comparatively highly lipophilic ligands are less active than the copper complexes. The lipophilicity–anticancer activity correlation of the present complexes is useful in the future to design better metal complexes in this field. Moreover, the formation constant (Kf) of complexes 1–4 was calculated in methanol (Fig. S16†). The Kf value of 4 (Kf = (5.14 ± 0.97) × 105 M−1) compared to those of the other complexes 1–3 is in the order 4 < 1 [(6.21 ± 4.68) × 105] < 2 [(7.19 ± 2.42) × 107] < 3 [(5.50 ± 3.86) × 109].7 The order of formation constant in methanol is concordant with the lipophilicity values, where the most hydrophobic 4 has the lowest Kf value.
Kow), with values of 3.14 and 10.04, respectively, determined by the shake flask method.47 Compounds 4 and L4(H) were found to be highly lipophilic compared to the other complexes 1, −0.51; 2, 1.07; and 3, 1.83 and ligands L1(H), 2.71; L2(H), 0.67; and L3(H), 0.55.6,7 It should be noted that 1 has a slightly hydrophilic nature and shows the best activity. At the same time, 4 has higher lipophilicity which might not be good for a drug (Fig. S15†). This indicates how the lipo–hydro balance matters for drug molecules. In the present compounds, it is seen that the comparatively highly lipophilic ligands are less active than the copper complexes. The lipophilicity–anticancer activity correlation of the present complexes is useful in the future to design better metal complexes in this field. Moreover, the formation constant (Kf) of complexes 1–4 was calculated in methanol (Fig. S16†). The Kf value of 4 (Kf = (5.14 ± 0.97) × 105 M−1) compared to those of the other complexes 1–3 is in the order 4 < 1 [(6.21 ± 4.68) × 105] < 2 [(7.19 ± 2.42) × 107] < 3 [(5.50 ± 3.86) × 109].7 The order of formation constant in methanol is concordant with the lipophilicity values, where the most hydrophobic 4 has the lowest Kf value.
4. Conclusion
Currently, growing interest in biological applications of pincer-type metal complexes proves that they can act as drugs with enhanced anticancer activity. To attain such activity, a series of 8-aminoquinoline based pincer ligands and their copper(II) complexes 1–4 were synthesized, characterized and studied for their anticancer activity. The complexes were very efficient against A549 lung and MCF-7 breast carcinoma cells. Interestingly, 1 was very selective against lung cancer cells with an IC50 value of 8 μM and it is noteworthy that it showed no toxicity against the normal fibroblast cell line L929 (IC50 > 1000 μM). Furthermore, the involvement of ROS in the mechanism of anticancer activity of 1 was determined by carboxy-H2DCFDA staining. Moreover, the mechanism of cell death was found to be apoptosis using the AO–EB staining assay, and through flow cytometry, it is calculated to be 25.91%. The depolarization of the MMP observed by JC-1 marking indicated elevated ROS production in the mitochondria. Furthermore, the most probable mechanism was found to be the perturbation in the function of complex I of mitochondria by in silico studies. Overall, the studies give promising results to extend complex 1 to clinical applications.Author contributions
Athulya Das: data curation, formal analysis, investigation, resources, software, visualization, writing – original draft, and writing – review & editing. Muniyandi Sankaralingam: conceptualization, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing – original draft, and writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Crystallographic data for 1 have been deposited at the repository namely CCDC under 2311348 and can be obtained from https://www.ccdc.cam.ac.uk/data_request/cif or by emailing E-mail: data_request@ccdc.cam.ac.uk.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
MS sincerely acknowledges the Science and Engineering Research Board (SERB) for the CRG grant (CRG/2023/002850). MS and AD sincerely thank the Department of Science and Technology, New Delhi, for awarding the DST-Inspire Faculty research grant (IFA-17-CH286) and the DST-Inspire Fellowship (IF-170136), respectively. The authors thank DST SAIF-IITM for helping with single crystal XRD analysis. We also thank the Department of Chemistry IITM for recording mass spectra. We would like to thank Prof. M. Sankar and his student Mr. Rajesh Kumar for allowing us to record the EPR spectra. We also thank NIT Calicut for the NMR (CMC) and HPC (CCMS) facilities and Thasnim P Mohammed, a PhD Scholar under the guidance of Dr MS in BBICL, for optimizing the geometries. We are pleased to thank the Biome Live Analytical Centre, Karaikudi, for anticancer studies.References
- WHO , Cancer. https://www.who.int/health-topics/cancer#tab=tab_1, accessed 17.03.2023.
- S. Sen, M. Won, M. S. Levine, Y. Noh, A. C. Sedgwick, J. S. Kim, J. L. Sessler and J. F. Arambula, Metal-Based Anticancer Agents as Immunogenic Cell Death Inducers: The Past, Present, and Future, Chem. Soc. Rev., 2022, 51, 1212–1233 RSC.
- D. Hernández-Romero, S. Rosete-Luna, A. López-Monteon, A. Chávez-Piña, N. Pérez-Hernández, J. Marroquín-Flores, A. Cruz-Navarro, G. Pesado-Gómez, D. Morales-Morales and R. Colorado-Peralta, First-Row Transition Metal Compounds Containing Benzimidazole Ligands: An Overview of Their Anticancer and Antitumor Activity, Coord. Chem. Rev., 2021, 439, 213930 CrossRef.
- A. Das, A. Rajeev, S. Bhunia, M. Arunkumar, N. Chari and M. Sankaralingam, Synthesis, Characterization and Antimicrobial Activity of Nickel(II) Complexes of Tridentate N3 Ligands, Inorg. Chim. Acta, 2021, 526, 120515 CrossRef.
- (a) A. Das, T. P. Mohammed, R. Kumar, S. Bhunia and M. Sankaralingam, Carbazole Appended Trans-Dicationic Pyridinium Porphyrin Finds Supremacy in DNA Binding/Photocleavage over a Non-Carbazolyl Analogue, Dalton Trans., 2022, 51, 12453–12466 RSC; (b) A. Das, T. P. Mohammed and M. Sankaralingam, Biological Activity of Copper Porphyrins, Coord. Chem. Rev., 2024, 506, 215661 CrossRef CAS.
- A. Das and M. Sankaralingam, Are Zn(II) pincer complexes efficient apoptosis inducers? A deep insight into their activity against A549 lung cancer cells, Dalton Trans., 2023, 52, 14465–14476 RSC.
- A. Das, R. Sangavi, S. Gowrishankar, R. Kumar and M. Sankaralingam, Deciphering the Mechanism of MRSA Targeting Copper(II) Complexes of NN2 Pincer Type Ligands, Inorg. Chem., 2023, 62, 18926–18939 CrossRef CAS PubMed.
- (a) M. Sankaralingam, Y.-M. Lee, W. Nam and S. Fukuzumi, Amphoteric reactivity of metal–oxygen complexes in oxidation reactions, Coord. Chem. Rev., 2018, 365, 41–59 CrossRef CAS; (b) M. Sankaralingam, Y.-M. Lee, D. G. Karmalkar, W. Nam and S. Fukuzumi, A mononuclear non-heme manganese(III)-aqua complex as a new active oxidant in hydrogen atom transfer reactions, J. Am. Chem. Soc., 2018, 140, 12695–12699 CrossRef CAS PubMed; (c) M. Sankaralingam, Y.-M. Lee, Y. Pineda-Galvan, D. G. Karmalkar, M. S. Seo, S. H. Jeon, Y. Pushkar, S. Fukuzumi and W. Nam, Redox reactivity of a mononuclear manganese-oxo complex binding calcium ion and other redox-inactive metal ions, J. Am. Chem. Soc., 2019, 141, 1324–1336 CrossRef CAS; (d) M. Sankaralingam, M. Balamurugan and M. Palaniandavar, Alkane and alkene oxidation reactions catalyzed by nickel(II) complexes: effect of ligand factors, Coord. Chem. Rev., 2020, 403, 213085 CrossRef; (e) W. P. Sohtun, S. Muthuramalingam, M. Sankaralingam, M. Velusamy and R. Mayilmurugan, Copper(II) Complexes of Tripodal Ligand Scaffold (N3O) as Functional Models for Phenoxazinone Synthase, J. Inorg. Biochem., 2021, 216, 111313 CrossRef CAS; (f) T. P. Mohammed and M. Sankaralingam, Reactivities of high valent manganese-oxo porphyrins in aqueous medium, Tetrahedron, 2022, 103, 132483 CrossRef CAS; (g) A. Rajeev, M. Balamurugan and M. Sankaralingam, Rational design of first-row transition metal complexes as the catalysts for oxidation of arenes: A homogeneous approach, ACS Catal., 2022, 12, 9953–9982 CrossRef CAS; (h) A. Rajeev and M. Sankaralingam, Highlights of Oxygen Atom Transfer Reactions Catalysed by Nickel Complexes, Oxygen Atom Transfer Reactions, 2023, vol. 1, pp. 62–90 Search PubMed; (i) S. Patra, A. Das, S. Bhunia, A. Rajeev and M. Sankaralingam, Electrocatalytic production of hydrogen using nickel complexes with tridentate N3 ligands, Catal. Today, 2023, 423, 113972 CrossRef CAS; (j) Y. Wang, H. Huang, Q. Zhang and P. Zhang, Chirality in Metal-Based Anticancer Agents, Dalton Trans., 2018, 47, 4017–4026 RSC.
- (a) M. Sankaralingam, M. Balamurugan, M. Palaniandavar, P. Vadivelu and C. H. Suresh, Nickel(II) Complexes of Pentadentate N5 Ligands as Catalysts for Alkane Hydroxylation by Using m-CPBA as Oxidant: A Combined Experimental and Computational Study, Chem. – Eur. J., 2014, 20, 11346–11361 CrossRef PubMed; (b) M. Sankaralingam, P. Vadivelu and M. Palaniandavar, Novel nickel(II) complexes of sterically modified linear N4 ligands: effect of ligand stereoelectronic factors and solvent of coordination on nickel(II) spin-state and catalytic alkane hydroxylation, Dalton Trans., 2017, 46, 7181–7193 RSC; (c) M. Sankaralingam, Y.-M. Lee, W. Nam and S. Fukuzumi, Selective Oxygenation of Cyclohexene by Dioxygen via an Iron(V)-Oxo Complex-Autocatalyzed Reaction, Inorg. Chem., 2017, 56, 5096–5104 CrossRef PubMed; (d) C. Saracini, D. D. Malik, M. Sankaralingam, Y.-M. Lee, W. Nam and S. Fukuzumi, Enhanced Electron-Transfer Reactivity of a Long-Lived Photoexcited State of a Cobalt–Oxygen Complex, Inorg. Chem., 2018, 57, 10945–10952 CrossRef PubMed; (e) S. Muthuramalingam, M. Sankaralingam, M. Velusamy and R. Mayilmurugan, Catalytic Conversion of Atmospheric CO2 into Organic Carbonates by Nickel(II) Complexes of Diazepane-Based N4 Ligands, Inorg. Chem., 2019, 58, 12975–12985 CrossRef PubMed; (f) X. Lu, Y.-M. Lee, M. Sankaralingam, S. Fukuzumi and W. Nam, Catalytic Four-Electron Reduction of Dioxygen by Ferrocene Derivatives with a Nonheme Iron(III) TAML Complex, Inorg. Chem., 2020, 59, 18010–18017 CrossRef PubMed.
- (a) H. Lim, C. Oh, M.-S. Park, H.-B. Park, C. Ahn, W. K. Bae, K. H. Yoo and S. Hong, Hint from an Enzymatic Reaction: Superoxide Dismutase Models Efficiently Suppress Colorectal Cancer Cell Proliferation, J. Am. Chem. Soc., 2023, 145, 16058–16068 CrossRef PubMed; (b) Y. Lee, C. Oh, J. Kim, M.-S. Park, W. K. Bae, K. H. Yoo and S. Hong, Bioinspired Nonheme Iron Complex That Triggers Mitochondrial Apoptotic Signalling Pathway Specifically for Colorectal Cancer Cells, Chem. Sci., 2022, 13, 737–747 RSC.
- (a) E. J. Ge, A. I. Bush, A. Casini, P. A. Cobine, J. R. Cross, G. M. DeNicola, Q. P. Dou, K. J. Franz, V. M. Gohil, S. Gupta, S. G. Kaler, S. Lutsenko, V. Mittal, M. J. Petris, R. Polishchuk, M. Ralle, M. L. Schilsky, N. K. Tonks, L. T. Vahdat, L. Van Aelst, D. Xi, P. Yuan, D. C. Brady and C. J. Chang, Connecting Copper and Cancer: From Transition Metal Signalling to Metalloplasia, Nat. Rev. Cancer, 2022, 22, 102–113 CrossRef CAS PubMed; (b) C. Rajarajeswari, R. Loganathan, M. Palaniandavar, E. Suresh, A. Riyasdeen and M. A. Akbarsha, Copper(II) Complexes with 2NO and 3N Donor Ligands: Synthesis, Structures and Chemical Nuclease and Anticancer Activities, Dalton Trans., 2013, 42, 8347–8363 RSC; (c) A. Sîrbu, O. Palamarciuc, M. V. Babak, J. M. Lim, K. Ohui, E. A. Enyedy, S. Shova, D. Darvasiová, P. Rapta, W. H. Ang and V. B. Arion, Copper(II) Thiosemicarbazone Complexes Induce Marked ROS Accumulation and Promote Nrf2-Mediated Antioxidant Response in Highly Resistant Breast Cancer Cells, Dalton Trans., 2017, 46, 3833–3847 RSC.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Advances in Copper Complexes as Anticancer Agents, Chem. Rev., 2013, 114, 815–862 CrossRef.
- X. Mingjin, Z. Qihua, L. Fangfang, Y. Junru, Z. Jihong, X. Jingyuan and S. Xueru, Schiff Bases Copper(II) Anticancer Complex and its Preparation Method and Application, Chinese Patent, 107759491B, 2017 Search PubMed.
- K. M. K. Al-Zaydi, A. I. D. Al-Sulami and M. T. J. Basha, Heterocyclic Diazenyl Pyridinone Copper(II) Complexes as Pharmacological Antitumor Agents, US Patent, US11104692B1, 2021 Search PubMed.
- O. Afzal, S. Kumar, M. R. Haider, M. R. Ali, R. Kumar, M. Jaggi and S. A. Bawa, Review on Anticancer Potential of Bioactive Heterocycle Quinoline, Eur. J. Med. Chem., 2015, 97, 871–910 CrossRef CAS PubMed.
- O. O. Ajani, K. T. Iyaye and O. T. Ademosun, Recent Advances in Chemistry and Therapeutic Potential of Functionalized Quinoline Motifs – a Review, RSC Adv., 2022, 12, 18594–18614 RSC.
- N. T. A. Dawoud, E. M. El-Fakharany, A. E. Abdallah, H. El-Gendi and D. R. Lotfy, Synthesis, and Docking Studies of Novel Heterocycles Incorporating the Indazolylthiazole Moiety as Antimicrobial and Anticancer Agents, Sci. Rep., 2022, 12, 3424 CrossRef CAS PubMed.
- V. R. Solomon, S. Pundir and H. Lee, Examination of Novel 4-Aminoquinoline Derivatives Designed and Synthesized by a Hybrid Pharmacophore Approach to Enhance Their Anticancer Activities, Sci. Rep., 2019, 9, 6315 CrossRef.
- B. Kundu, S. K. Das, S. P. Chowdhuri, S. Pal, D. Sarkar, A. Ghosh, A. Mukherjee, D. Bhattacharya, B. B. Das and A. Talukdar, Discovery and Mechanistic Study of Tailor-Made Quinoline Derivatives as Topoisomerase 1 Poison with Potent Anticancer Activity, J. Med. Chem., 2019, 62, 3428–3446 CrossRef PubMed.
- G. Gasser, I. Ott and N. Metzler-Nolte, Organometallic Anticancer Compounds, J. Med. Chem., 2011, 54, 3–25 CrossRef.
- A. P. Kourounakis, D. Xanthopoulos and A. Tzara, Morpholine as a Privileged Structure: A Review on the Medicinal Chemistry and Pharmacological Activity of Morpholine Containing Bioactive Molecules, Med. Res. Rev., 2020, 40, 709–752 CrossRef.
- (a) N. Takahashi, T. Honda and T. Ohba, Anticancer and Superoxide Scavenging Activities of P-Alkylaminophenols Having Various Length Alkyl Chains, Bioorg. Med. Chem., 2006, 14, 409–417 CrossRef PubMed; (b) F. Hackenberg and M. Tacke, Benzyl-Substituted Metallocarbene Antibiotics and Anticancer Drugs, Dalton Trans., 2014, 43, 8144–8153 RSC.
- S. Wu, Z. Wu, Q. Ge, X. Zheng and Z. Yang, Antitumor Activity of Tridentate Pincer and Related Metal Complexes, Org. Biomol. Chem., 2021, 19, 5254 RSC.
- S. Zhang, C. Tu, X. Wang, Z. Yang, J. Zhang, L. Lin, J. Ding and Z. Guo, Novel Cytotoxic Copper(II) Complexes of 8-Aminoquinoline Derivatives: Crystal Structure and Different Reactivity towards Glutathione, Eur. J. Inorg. Chem., 2004, 2004, 4028–4035 CrossRef.
- J. Lu, Q. Sun, J.-L. Li, L. Jiang, W. Gu, X. Liu, J.-L. Tian and S.-P. Yan, Two Water-Soluble Copper(II) Complexes: Synthesis, Characterization, DNA Cleavage, Protein Binding Activities and in Vitro Anticancer Activity Studies, J. Inorg. Biochem., 2014, 137, 46–56 CrossRef CAS.
- K. Czerwińska, B. Machura, S. Kula, S. Krompiec, K. Erfurt, C. Roma-Rodrigues, A. R. Fernandes, L. S. Shul'pina, N. S. Ikonnikov and G. B. Shul'pin, Copper(II) Complexes of Functionalized 2,2′:6′,2′′-terpyridines and 2,6-di(Thiazol-2-yl)pyridine: Structure, Spectroscopy, Cytotoxicity and Catalytic Activity, Dalton Trans., 2017, 46, 9591–9604 RSC.
- L. Tabrizi and H. Chiniforoshan, Synthesis and C–H Activation Reactions of Cyclometalated Copper(I) Complexes with NCN Pincer and 1,3,5-Triaza-7-Phosphaadamantane Derivatives: In Vitro Antimicrobial and Cytotoxic Activity, New J. Chem., 2017, 41, 10972–10984 RSC.
- H. G. Sogukomerogullari and S. Akkoc, Copper(II) Complexes with Thioether based SNS Pincer Ligand: Synthesis, Characterization and Antiproliferative Activity, J. Struct. Chem., 2023, 64, 157–167 CrossRef CAS.
- A. Das and M. Sankaralingam, Method for preparing a potent copper(II) complex as an apoptosis inducer in lung cancer, Indian Patent, 518352, 2024 Search PubMed.
- SADABS, Ver. 2.05, SMART, Ver. 5.631, and SAINT, v 6.45, Bruker AXS Inc., Madison, WI, 2003 Search PubMed.
- G. M. Sheldrick, Crystal structure refinement with SHELXL, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- L. J. Farrugia, WinGX and ORTEP for Windows: an update, J. Appl. Crystallogr., 2012, 45, 849–854 CrossRef CAS.
- M. Alley, D. A. Scudiero, A. Monks, M. L. Hursey, M. Czerwinski, D. L. Fine, B. J. Abbott, J. A. Mayo, R. H. Shoemaker and M. R. Boyd, Feasibility of Drug Screening with Panels of Human Tumor Cell Lines Using a Microculture Tetrazolium Assay, Cancer Res., 1988, 48, 589–601 CAS.
- Z. Mao, Z. Liu, L. Chen, J. Yang, B. Zhao, Y. M. Jung, X. Wang and C. Zhao, Predictive Value of the Surface-Enhanced Resonance Raman Scattering-Based MTT Assay: A Rapid and Ultrasensitive Method for Cell Viability in Situ, Anal. Chem., 2013, 85, 7361–7368 CrossRef CAS PubMed.
- G. M. Asong, F. Amissah, C. Voshavar, A. T. Nkembo, E. Ntantie, N. S. Lamango and S. Y. Ablordeppey, A Mechanistic Investigation on the Anticancer Properties of SYA013, a Homopiperazine Analogue of Haloperidol with Activity against Triple Negative Breast Cancer Cells, ACS Omega, 2020, 5, 32907–32918 CrossRef CAS PubMed.
- D. Ribble, N. B. Goldstein, D. A. Norris and Y. G. Shellman, A simple technique for quantifying apoptosis in 96-well plates, BMC Biotechnol., 2005, 5, 32907–32918 CrossRef.
- K. Liu, P.-C. Liu, R. Liu and X. Wu, Dual AO/EB Staining to Detect Apoptosis in Osteosarcoma Cells Compared with Flow Cytometry, Med. Sci. Monit. Basic Res., 2015, 21, 15–20 CrossRef PubMed.
- I. Vermes, C. Haanen and C. Reutelingsperger, Flow Cytometry of Apoptotic Cell Death, J. Immunol. Methods, 2000, 243, 167–190 CrossRef CAS PubMed.
- V. V. Shynkar, A. S. Klymchenko, C. Kunzelmann, G. Duportail, C. D. Muller, A. P. Demchenko, J.-M. Freyssinet and Y. Mely, Fluorescent Biomembrane Probe for Ratiometric Detection of Apoptosis, J. Am. Chem. Soc., 2007, 129, 2187–2193 CrossRef CAS.
- D. Wlodkowic, J. Skommer and Z. Darzynkiewicz, Flow Cytometry-Based Apoptosis Detection, Methods Mol. Biol., 2009, 559, 19–32 CrossRef CAS.
- Y. Qiu, E. Rojas, R. A. Murray, J. Irigoyen, D. Gregurec, P. Castro-Hartmann, J. Fledderman, I. Estrela-Lopis, E. Donath and S. E. Moya, Cell Uptake, Intracellular Distribution, Fate and Reactive Oxygen Species Generation of Polymer Brush Engineered CeO2−XNPs, Nanoscale, 2015, 7, 6588–6598 RSC.
- C. Xu, R. Song, P. Lu, J. Chen, Y. Zhou, G. Shen, M. Jiang and W. Zhang, A pH-Responsive Charge-Reversal Drug Delivery System with Tumor-Specific Drug Release and ROS Generation for Cancer Therapy, Int. J. Nanomedicine., 2020, 15, 65–80 CrossRef CAS PubMed.
- H. Zhao, J. Huang, Y. Li, X. Lv, H. Zhou, H. Wang, Y. Xu, C. Wang, J. Wang and Z. Liu, ROS-Scavenging Hydrogel to Promote Healing of Bacteria Infected Diabetic Wounds, Biomaterials, 2020, 258, 120286 CrossRef CAS.
- (a) M. Reers, S. T. Smiley, C. Mottola-Hartshorn, A. Chen, M. Lin and L. B. Chen, Mitochondrial Membrane Potential Monitored by JC-1 Dye, Methods Enzymol., 1995, 260, 406–417 CAS.
- P. C. Saha, T. Chatterjee, R. Pattanayak, R. S. Das, A. Mukherjee, M. Bhattacharyya and S. Guha, Targeting and Imaging of Mitochondria Using Near-Infrared Cyanine Dye and Its Application to Multicolor Imaging, ACS Omega, 2019, 4, 14579–14588 CrossRef CAS.
- F. Sivandzade, A. Bhalerao and L. Cucullo, Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe, Bio-Protoc., 2019, 9, e3128 Search PubMed.
- M. F. Harris and J. L. Logan, Determination of Log Kow Values for Four Drugs, J. Chem. Educ., 2014, 91, 915–918 CrossRef CAS.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09 Revision A.02, Gaussian Inc., Wallingford CT, 2009 Search PubMed.
- Accelrys Software Inc, Discovery Studio Modeling Environment, Release 4.0, Accelrys Software Inc., San Diego, 2013 Search PubMed.
- (a) A. S. Thennarasu, T. P. Mohammed and M. Sankaralingam, Mononuclear Copper(II) Schiff Base Complexes as Effective Models for Phenoxazinone Synthase, New J. Chem., 2022, 46, 21684–21694 RSC; (b) T. P. Mohammed, A. George, M. P. Sivaramakrishnan, P. Vadivelu, S. Balasubramanian and M. Sankaralingam, Deciphering the Effect of Amine versus Imine Ligands of Copper(II) Complexes in 2-Aminophenol Oxidation, J. Inorg. Biochem., 2023, 247, 112309 CrossRef CAS PubMed.
- (a) B. J. Hathaway and D. E. Billing, The Electronic Properties and Stereochemistry of Mono-Nuclear Complexes of the Copper(II) Ion, Coord. Chem. Rev., 1970, 5, 143–207 CrossRef CAS; (b) E. Garribba and G. Micera, The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment, J. Chem. Educ., 2006, 83, 1229 CrossRef CAS.
- (a) E. Billig, R. Williams, I. Bernal, J. H. Waters and H. B. Gray, The Electronic Structures of Square-Planar Metal Complexes. II. The Complexes of Maleonitriledithiolate with Copper(II), Nickel(II), Palladium(II), and Platinum(II), Inorg. Chem., 1964, 3, 663–666 CrossRef CAS; (b) M. M. Bhadbhade and D. Srinivas, Effects on Molecular Association, Chelate Conformation, and Reactivity toward Substitution in Copper Cu(5-X-Salen) Complexes, Salen2− = N,N′-Ethylenebis(Salicylidenaminato), X = H, CH3O, and Cl: Synthesis, X-Ray Structures, and EPR Investigations, Inorg. Chem., 1993, 32, 5458–5466 CrossRef CAS; (c) G. Verquin, G. Fontaine, E. Abi-Aad, E. Zhilinskaya, A. Aboukaïs and J.-L. Bernier, EPR Study of Copper(II) Complexes of Hydroxysalen Derivatives in Order to be Used in the DNA Cleavage, J. Photochem. Photobiol., B., 2007, 86, 272–278 CrossRef CAS PubMed.
- A. G. Blackman, E. B. Schenk, R. E. Jelley, E. H. Krenske and L. R. Gahan, Five-Coordinate Transition Metal Complexes and the Value of τ5: Observations and Caveats, Dalton Trans., 2020, 49, 14798–14806 RSC.
- G. Abbas, G. Pandey, K. B. Singh and N. Gautam, One-Pot Surface Modification of β-Cu2O NPs for Biocatalytic Performance against A-549 Lung Carcinoma Cell Lines through Docking Analysis, ACS Omega, 2021, 6, 29380–29393 CrossRef CAS PubMed.
- S. E. Salamifar and R. Y. Lai, Use of Combined Scanning Electrochemical and Fluorescence Microscopy for Detection of Reactive Oxygen Species in Prostate Cancer Cells, Anal. Chem., 2013, 85, 9417–9421 CrossRef CAS PubMed.
- M. d. P. S. Idelchik, U. Begley, T. J. Begley and J. A. Melendez, Mitochondrial ROS Control of Cancer, Semin. Cancer Biol., 2017, 47, 57–66 CrossRef CAS.
- E. Gottlieb, S. M. Armour, M. H. Harris and C. B. Thompson, Mitochondrial Membrane Potential Regulates Matrix Configuration and Cytochrome c Release during Apoptosis, Cell Death Differ., 2003, 10, 709–717 CrossRef CAS PubMed.
- S. N. Krishnan, J. L. Rosales and K.-Y. Lee, ROS-Mediated Cancer Cell Killing through Dietary Phytochemicals, Oxid. Med. Cell. Longevity, 2019, 2019, 1–16 Search PubMed.
- S. S. Sabharwal and P. T. Schumacker, Mitochondrial ROS in Cancer: Initiators, Amplifiers or an Achilles’ Heel?, Nat. Rev. Cancer, 2014, 14, 709–721 CrossRef CAS.
- S. Mohapatra, G. Das, V. Gupta, P. Mondal, M. Nitani, Y. Ie, S. Chatterjee, Y. Aso and S. Ghosh, Power of an Organic Electron Acceptor in Modulation of Intracellular Mitochondrial Reactive Oxygen Species: Inducing JNK- and Caspase-Dependent Apoptosis of Cancer Cells, ACS Omega, 2021, 6, 7815–7828 CrossRef CAS.
- B. Perillo, M. Di Donato, A. Pezone, E. Di Zazzo, P. Giovannelli, G. Galasso, G. Castoria and A. Migliaccio, ROS in Cancer Therapy: The Bright Side of the Moon, Exp. Mol. Med., 2020, 52, 192–203 CrossRef CAS.
- R. G. Efremov, R. Baradaran and L. A. Sazanov, The Architecture of Respiratory Complex I, Nature, 2010, 465, 441–445 CrossRef CAS PubMed.
- R. Guo, S. Zong, M. Wu, J. Gu and M. Yang, Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2, Cell, 2017, 170, 1247–1257 CrossRef CAS PubMed.
- P. Saura and V. R. I. Kaila, Energetics and Dynamics of Proton-Coupled Electron Transfer in the NADH/FMN Site of Respiratory Complex I, J. Am. Chem. Soc., 2019, 141, 5710–5719 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed procedure for synthesis and lipophilicity determination. Table S1 and Fig. S1–S16 include IR spectra of L1(H)–L4(H) and 1–4, the mass spectrum of 4, phase images, and percentage viability diagrams of A549 and MCF-7 treated with complexes and ligand L1(H), phase images and percentage viability diagrams of L929 treated with 1, UV-visible spectra of L4(H) and 4 for lipophilicity calculation, UV-visible spectra for complex 4 formation constant calculation, and molecular docking interactions of 2–4 with 5XTD. CCDC 2311348. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4dt01075b |
| This journal is © The Royal Society of Chemistry 2024 |