Open Access Article
Open Access ArticleImage and data mining in reticular chemistry powered by GPT-4V†
Zhiling
Zheng
 abc,
Zhiguo
He
ab,
Omar
Khattab
d,
Nakul
Rampal
abc,
Matei A.
Zaharia
e,
Christian
Borgs
ce,
Jennifer T.
Chayes
cefgh and
Omar M.
Yaghi
abc,
Zhiguo
He
ab,
Omar
Khattab
d,
Nakul
Rampal
abc,
Matei A.
Zaharia
e,
Christian
Borgs
ce,
Jennifer T.
Chayes
cefgh and
Omar M.
Yaghi
 *abci
*abci
aDepartment of Chemistry, University of California, Berkeley, California 94720, USA. E-mail: yaghi@berkeley.edu
bKavli Energy Nanoscience Institute, University of California, Berkeley, California 94720, USA
cBakar Institute of Digital Materials for the Planet, University of California, Berkeley, California 94720, USA
dDepartment of Computer Science, Stanford University, Stanford, California 94305, USA
eDepartment of Electrical Engineering and Computer Sciences, University of California, Berkeley, California 94720, USA
fDepartment of Mathematics, University of California, Berkeley, California 94720, USA
gDepartment of Statistics, University of California, Berkeley, California 94720, USA
hSchool of Information, University of California, Berkeley, California 94720, USA
iKACST-UC Berkeley Center of Excellence for Nanomaterials for Clean Energy Applications, King Abdulaziz City for Science and Technology, Riyadh 11442, Saudi Arabia
First published on 2nd February 2024
Abstract
The integration of artificial intelligence into scientific research opens new avenues with the advent of GPT-4V, a large language model equipped with vision capabilities. In this study, we demonstrate that GPT-4V, accessible through the ChatGPT web user interface or an API, offers promising possibilities in navigating and mining complex data for metal–organic frameworks (MOFs) especially from graphical sources (e.g. sorption isotherms, powder X-ray diffraction patterns, thermogravimetric analysis graphs, etc.). Our approach involved an automated process of converting 346 scholarly articles into 6240 images, which represents a benchmark dataset in this task, followed by deploying GPT-4V to categorize and analyze these images using natural language prompts, which can be written by chemists or materials scientists with minimal prior coding knowledge. This methodology enabled GPT-4V to accurately identify and interpret key plots integral to MOF characterization, such as nitrogen isotherms, PXRD patterns, and TGA curves, among others, with accuracy and recall above 93%. The model's proficiency in extracting critical information from these plots not only underscores its capability in data mining but also highlights its potential to aid in the digitalization of experimental data and the creation of datasets for reticular chemistry. In addition, the trends and values of nitrogen isotherm data from the selected literature allowed for a comparison between theoretical and experimental porosity values for over 200 compounds, highlighting certain discrepancies and underscoring the importance of integrating computational and experimental data. This work highlights the potential of AI in accelerating scientific discovery by bridging the gap between computational tools and experimental research.
Introduction
The integration of artificial intelligence (AI) with the chemical sciences holds immense potential, and is accelerated by the rise of large language models (LLMs).1–7 These models have received substantial attention for the fact that they can be intuitively “programmed” or “taught” using daily conversational language, thereby assisting with diverse chemistry research tasks.8–20 It is envisioned that the evolution from text-only to more dynamic, multi-modal LLMs will result in even more powerful and convenient AI assistants across various applications.5,21–23The recent introduction of GPT-4V, with ‘V’ denoting its vision capability, stands as a testament to this progress.21,24 Trained on a vast and varied collection of multi-modal data, GPT-4V can process and respond to both textual and visual inputs.24–27 Its ability to interpret and analyze scientific literature, especially in identifying valuable data within graphical representations, makes it more attractive than traditional text-only models in natural language processing (NLP).10,28–30 These novel capabilities allow researchers from diverse backgrounds, including those with no specialized coding or computer vision expertise, to harness the power of GPT-4V through customized instructions applicable to various areas.19,31–33
Herein, we report the applications of GPT-4V in the field of reticular chemistry.34,35 We demonstrate how GPT-4V can seamlessly interpret and integrate complex data from both textual and graphical sources in the scientific literature (Fig. 1). This ability to extract and analyze critical information from various figures and plots dramatically increases the model's utility in data mining and information retrieval. In particular, distilling physical characterization results from graphical content reported in papers can be pivotal in establishing the collection of experimental data and guiding the discovery of new chemical compounds.34,36–39 Moreover, this methodology, while demonstrated in the context of reticular chemistry, has the potential to be generalized to other scientific disciplines for automated literature reading. This development underscores the expanding role of AI in fostering innovation and discovery, which will further bridge the gap between advanced computational tools and cutting-edge chemical research.40,41
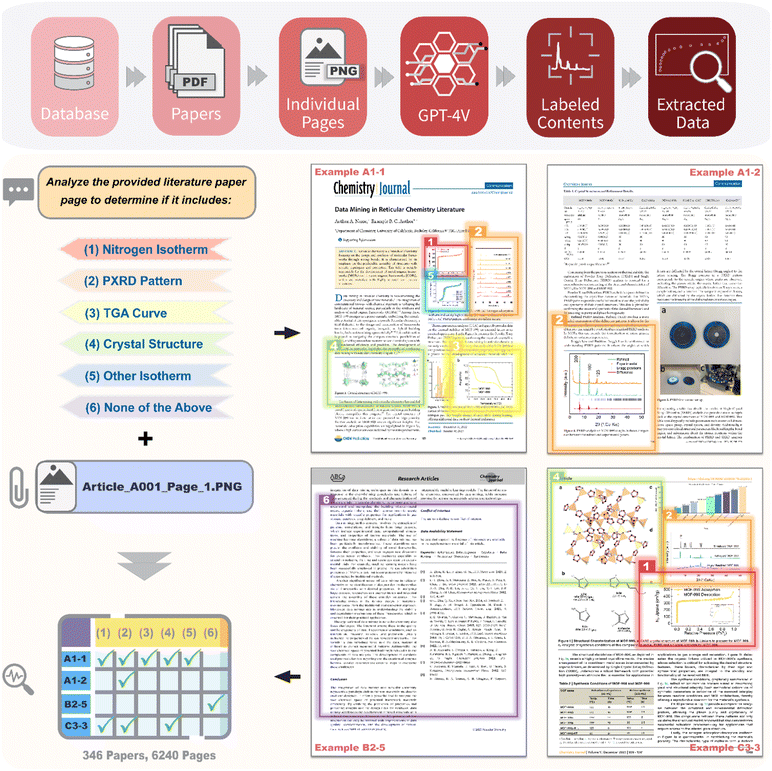 | ||
| Fig. 1 Schematic representation of a LLM-driven workflow (top) utilizing image input capabilities for the extraction and categorization of scientific data from 346 chemistry literature papers. Papers are converted into 6240 PNG images and paired with fixed textual prompts to guide GPT-4V in interpreting content within individual pages and responding with appropriate labels (left). The four example pages depicted are not actual published materials used in this study but are non-copyrighted images crafted for illustrative purposes (right), mimicking the layout of a standard scientific paper with diverse plots and figures that are similar to those found in the 346 articles and their ESI† files used for this study. The content of these figures is based on actual experimental data, extracted and replotted from earlier publications to accurately reflect the plot shapes for demonstration of the workflow. GPT-4V identifies features based on the described target plots, and the classification responses, which may include single or multiple labels, are compiled. These responses are associated with the DOI and corresponding page number of the paper, facilitating efficient data retrieval from the targeted pages. | ||
Methods
Data preparation
For this study, a comprehensive dataset comprising 6240 pages from 346 scholarly articles published in 22 peer-reviewed journals among 5 different publishers discussing metal–organic frameworks (MOFs) was meticulously curated from the CoRE MOF Database (2019)42 to ensure the diversity of writing styles, article formatting and page layout. The selection criteria focused on excluding papers reporting structures with zero accessible surface areas and those without publicly available DOI numbers. In this case, the criteria resulted in the selection of 346 downloadable articles which met the specified requirements on the accessible surface area from the 9147 papers listed in the CoRE MOF database. The selected PDFs were transformed into PNG format utilizing a Python script developed with the assistance of ChatGPT (ESI Fig. S16†). This conversion process included 4423 pages from the main text and 1817 pages from the ESI of those chosen papers (ESI Data Set).†Prompts and image mining considerations
We wrote instructions in natural language to guide GPT-4V for image classification and analysis. The primary objective was to enable GPT-4V to categorize each image with a label reflecting its content. These labels were designed to represent the most prevalent types of plots encountered in reticular chemistry, encompassing essential data and values. Key plots include the nitrogen adsorption–desorption isotherm (indicating porosity), the PXRD pattern (illustrating crystallinity), the TGA curve (demonstrating thermal stability), crystallographic structure rendering images (depicting crystal structure or topology), and other gas sorption isotherms (relevant for applications and gas uptake). Accordingly, these were established as label choices 1 through 6. It is noteworthy that a single page may contain multiple plots (e.g., PXRD and TGA concurrently), which requires GPT-4V's capability to perform an identification of multiple formats of plots. Therefore, the primary task assigned to GPT-4V involves identifying these specified elements within the image of the whole page, complemented by a structured response template. Furthermore, the crafting of prompts adhered to three guiding principles outlined in our previous publication:10 (i) minimizing hallucination, (ii) implementing detailed instructions, and (iii) ensuring structured output. The complete prompt is available in ESI Fig. S15.†Automation in paper reading
With the prompt finalized, GPT-4V's task was to “read” through all the pages from selected papers. This was accomplished by sequentially presenting each image of the whole page, along with the designated prompt, to the model and collecting its responses. This iterative process was automated using a for-loop structure via simple python code generated with the assistance of ChatGPT (ESI Fig. S17†). In particular, two options exist, and both were tested in this study: (i) interfacing with the web-based chatbot (ChatGPT powered by GPT-4V, version dated September 25, 2023) and (ii) utilizing an API to connect with the gpt-4-1106-vision-preview model. Both methods applied the same underlying base model and were facilitated through Python scripts capable of operating autonomously (Fig. S17 and S18†). Additionally, the process for parsing and extracting the labels from the responses given by GPT-4V was automated by Python script (Fig. S19 and S20†).Results and discussion
Initial assessment of GPT-4V's knowledge of reticular chemistry
At the outset of this study, we rigorously evaluated the proficiency of GPT-4V in recognizing and interpreting a range of figures typically found in reticular chemistry literature by asking GPT-4V to describe the respective figure sent along with the chat (Fig. S1–S14†). This knowledge assessment encompassed various physical characterization plots including nitrogen isotherms, PXRD patterns, TGA curves, NMR and IR spectra, as well as illustrative plots such as scatter plots, bar plots, and 2D or 3D molecular structures. In addition, synthesis schemes and real experimental images, like microscope and SEM images, were also analyzed. Each figure was presented to GPT-4V alongside a brief prompt requesting a description of the input figure in a conversational format.The responses from GPT-4V (ESI Fig. S1–S14†) demonstrated its remarkable capability to not only categorize these images accurately but also to elaborate on specific details, including notations, axis ranges, color coding and shape of symbols and lines, labels, legends, and to draw inferences from the provided information in the figure caption. This advanced level of contextual data interpretation and holistic analysis underscores the suitability of GPT-4V as a potent AI assistant for image and data mining in scientific literature.
Designing prompts for page content labelling
The core objective of this study was to investigate whether GPT-4V could automatically navigate through scientific papers, identifying specific information and aggregating it into a comprehensive dataset for further analysis. Our interest specifically centered on plots pivotal in the physical characterization of MOFs. These plots, namely nitrogen isotherms, PXRD patterns, TGA curves, crystal structure or topology illustrations, and other gas sorption isotherms, are instrumental in deducing key properties of chemical compounds, including permanent porosity, crystallinity, thermal stability, connectivity (topology), and sorbent selectivity towards gases. Successfully extracting and consolidating data from these plots amongst a vast volume of literature holds immense potential for advancing our understanding of structure–property relationships and in accelerating the discovery of novel compounds.8,34,37,38,43–48To this end, we designed a specific and extensive prompt for GPT-4V, targeting the aforementioned categories (Fig. 1 and ESI Fig. S15†). Unlike the brief prompts employed in the initial assessment of GPT-4V's knowledge in reticular chemistry (Fig. S1–S14†), the formal prompt used for image mining in this study was developed by employing the prompt engineering strategies that we previously reported,10,14 making it more extensive and comprehensive (see Methods section and ESI Fig. S15†). It is noteworthy that the prompt allowed for the possibility of multiple selections on a single page, as it is common for various plots to coexist in scientific literature. Additionally, GPT-4V was instructed to indicate the absence of these five categories when applicable. Therefore, in total six choices were provided for GPT-4V (Fig. 1). The development of these prompts was guided by general principles for prompt engineering in text mining.10
Performance evaluation of GPT-4V
In our workflow, each page of the selected literature was digitized into an image and then analyzed by GPT-4V (see Methods section). This process involved combining each image with a textual prompt, after which GPT-4V's responses were collected. Particularly, GPT-4V was “programmed” by instructions using human daily conservational language in the prompt to guide it to follow a specific response format (ESI Fig. S15†), allowing for the automatic labeling of each page based on its content. While copyright restrictions preclude the sharing of the actual images used in this study, we provide four representative examples that closely simulate the layout and content of the analyzed pages from actual published literature49–53 to illustrate this figure content identification process (Fig. 1). These examples demonstrate GPT-4V's ability to accurately recognize and label the desired plots on each page, regardless of the complexity of the information shown. It is important to note that in the actual evaluation, instead of using the four demo images shown in Fig. 1, we used 6240 single-page images, akin to “screenshots” that contain all the text and associated figures, if any, in that page as a whole image, from 346 articles. Consequently, the respective 6240 output answers from GPT-4V were collected (ESI Data Set†). In other words, during each interaction, each page was submitted separately in a new instance of conversation to yield the classification result of the given page. Essentially, GPT-4V could perform iterative classifications of each page, utilizing the same prompt but varying image content; this process was completed without human intervention (ESI Fig. S17 and S18†).The evaluation of GPT-4V's classification accuracy involved comparing its responses against a benchmarking ground truth dataset. This dataset was meticulously created by experts in reticular chemistry, who manually reviewed and labeled all 6240 images for the presence of specific content (choices 1 to 6). Performance metrics for each category were calculated individually, considering the multiple-choice nature of the task (ESI Data Set and Table S1†). For example, if the ground truth for an image was labeled as “1, 3, 4” and GPT-4V's response was “1, 4, 5,” the classifications for each category were assessed as follows: nitrogen isotherm (Choice 1) received a True Positive, PXRD pattern (Choice 2) a True Negative, TGA curve (Choice 3) a False Negative, crystal category (Choice 4) a True Positive, other gas sorption isotherm (Choice 5) a False Positive, and “none of the above” (Choice 6) a True Negative.
The summarized metrics, including accuracy, precision, recall and F1 score, are presented in Table 1. Notably, it displays promising accuracy rates above 94% for all categories. This indicates that GPT-4V can correctly classify a high percentage of figure content across all 6 categories, which encompass 5 different plots types and 1 NOTA category. Precision ranges from 88% to 97% for five of the plot types; however, it is notably lower at 61% for the “other gas sorption isotherm” category. This reduced precision is attributed to the category's broad scope and occasional mislabeling of IR and NMR spectra as such. This highlights an opportunity for further refinement in the content of prompts, aiming to enhance GPT-4V's understanding of the definition of gas sorption isotherm and ensuring it does not mistakenly identify other spectra as isotherm plots. The recall values, between 94% and 99% for all six classes, suggest the model is effective at capturing a high percentage of figure content relevant to each specific class. The F1 scores range from 93% to 95%, demonstrating a well-balanced and robust performance in both precision and recall. However, the “other gas sorption isotherm” is an exception with a 76% F1 score, which is foreseen due to its low recall performance, indicating a certain limitation of the model in this specific class, possibly due to insufficiently detailed instruction in the prompt.
| Plot type | Accuracy | Precision | Recall | F1 score |
|---|---|---|---|---|
| Nitrogen isotherm | 99.5% | 90% | 96.7% | 93.5% |
| Power X-ray diffraction | 99.2% | 94.3% | 98.4% | 96.3% |
| Thermogravimetric analysis | 99.2% | 87.8% | 99.3% | 93.2% |
| Crystal structure or topology | 98.1% | 93.2% | 97.1% | 95.1% |
| Other gas sorption isotherm | 95.0% | 61.4% | 99.5% | 76.0% |
| None of the above | 94.3% | 96.7% | 93.7% | 95.1% |
Additionally, similar accuracy rates were observed in GPT-4V's performance via both the web user interface (WUI) and API (Table S2†), a testament to the uniformity of the underlying base model (gpt-4-1106-vision-preview). Here we acknowledge that the base model is still in preview version, yet the obtained results are very promising. The automated process for reading papers and collecting output classifications through both the WUI and API offers diverse operational choices with consistently high performance. We note that in this study, no fine-tuning strategies were applied, and the performance primarily resulted from carefully crafted text-based instructions after prompt engineering. This approach is becoming increasingly important in the realm of scientific research involving LLMs.32,54–57 Impressively, we found that this also holds true for multimodal models handling both text and graphical inputs such as GPT-4V in this study.
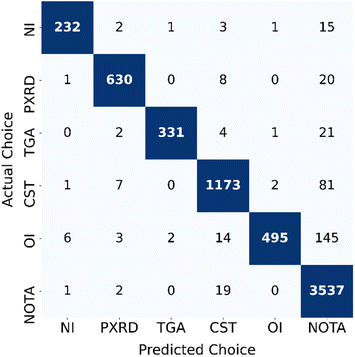
Furthermore, the relation between the ground truth and GPT-4V's classification output was visualized in a confusion matrix (Fig. 2). Among the 6240 pages analyzed, GPT-4V identified approximately 232 plots that were with nitrogen isotherms, 630 that were PXRD patterns, and 331 that were TGA traces. It suggests a substantial volume of data within the analyzed literature. Moreover, 3537 pages were classified as lacking the plots of interest, which can be a strategy to be applied in the future to help human researchers streamline the review process for a specific type of literature plot by excluding less important pages and focusing only on the rest of the content-rich pages. We also estimated the time of this automated literature processing approach (ESI Tables S3 and S4†). On average, processing a 20-page paper took approximately 2 minutes at a rate of 5.5 seconds per image via API and this process can be parallelized (ESI Table S3†).
GPT-4V's interpretation of nitrogen isotherm data
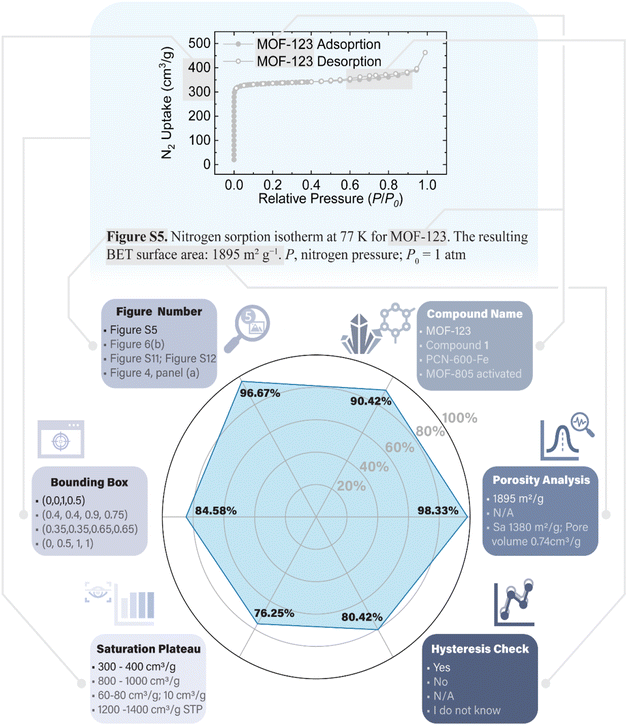
Building upon the successful labeling of page contents, we next directed our focus towards utilizing GPT-4V for the detailed interpretation and analysis of pages featuring nitrogen isotherm plots. To achieve this, we refined our prompt strategy, incorporating additional and specific verbal instructions (ESI Fig. S15†). These enhanced prompts guided GPT-4V to not only recognize nitrogen isotherms but also to extract and report key descriptors from each plot. These descriptors included the figure number, compound name, surface area or pore volume value reported by the author, the presence of hysteresis in the adsorption–desorption curve, the saturation plateaus of the isotherm, and an estimation of a bounding box encompassing the figure (Fig. 3). A critical aspect of this approach, aiming to minimize hallucinations, was the instruction for GPT-4V to strictly utilize the information available on the page image, defaulting to answer “N/A” for any unobtainable or ambiguous data. It turned out that GPT-4V demonstrated a remarkable ability to extract these details by analyzing the isotherm, its axes, legends, and accompanying text content.To validate the accuracy of GPT-4V's nitrogen isotherm analysis, we manually reviewed over 200 responses from pages containing nitrogen isotherms in our selected papers (ESI† data set). This evaluation was conducted for each of the six descriptors independently. The results indicated high accuracy levels for the figure number (96.67%), compound name (90.42%), and porosity analysis (98.33%). We hypothesize that GPT-4V's image processing capabilities, potentially incorporating optical character recognition (OCR) tools, played a pivotal role in these tasks. Given GPT-4V's proficiency with text, it likely excelled in tasks where textual information was directly “readable” from the image. Conversely, the other three descriptors, namely hysteresis presence, saturation plateaus, and bounding box estimation, showed generally satisfactory performance, ranging between 76.25% and 84.58%. These tasks, inherently more challenging and nuanced, required a comprehensive analysis of all image elements.58 Nonetheless, the overall performance was impressive, especially considering the simplicity with which researchers could instruct GPT-4V using natural language.
Accelerating the creation of digital datasets in reticular chemistry
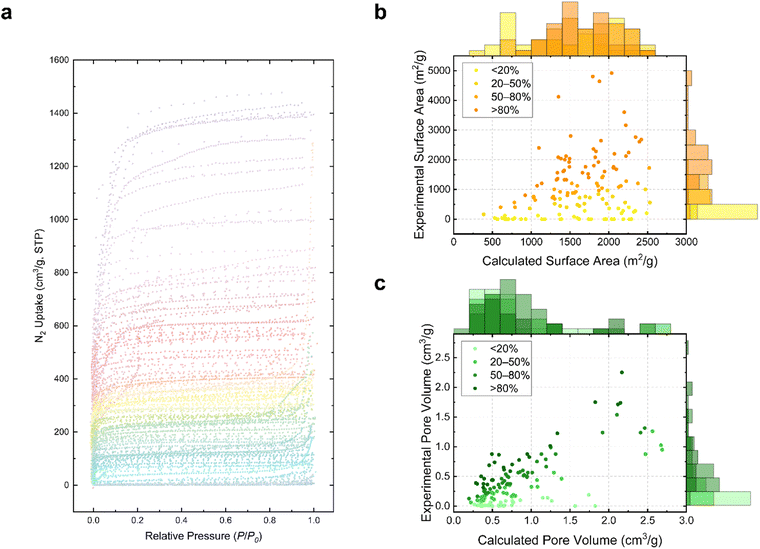
Our study further demonstrates the potential of GPT-4V in accelerating the digitalization of experimental data for reticular compounds. This is particularly evident from the sorption measurement outcomes gleaned from community-published literature. Once pages featuring nitrogen isotherm plots are identified, each corresponding curve, typically presented in a non-digital format (e.g., scanned or plotted images), can be meticulously extracted using data extraction tools like WebPlotDigitizer.59 The extracted data points are then systematically compiled and stored (ESI Data Set†). While we currently make all mined sorption data available in a spreadsheet dataset format, it is feasible that digitalized data can be converted into AIF files as a first step toward building the MOF sorption data library.60Fig. 4a presents a collection of nitrogen isotherm data points manually extracted from the qualified pages in our study as a proof-of-concept, showcasing a diverse array of isotherm types and porosity characteristics. Leveraging the CoRE MOF Database,36,42 which provides computational results for compounds discussed in these papers, we matched each adsorption–desorption curve's corresponding compound with their CCDC number, accessible surface area, and pore volume. This enabled us to visualize and compare the experimental porosity (indicated by the nitrogen isotherm curves) with the calculated values (derived from the CoRE MOF Database). The results, illustrated in scatter plots (Fig. 4b and c), represent each compound as a data point, where the x-axis denotes the theoretical values calculated from CCDC CIF structures,36,42 and the y-axis represents the experimentally reported surface area or pore volume inferred from the extracted isotherm obtained from the graphs identified by GPT-4V. The distributions of x and y values of the data points are shown in the respective bar charts. It is crucial to note that our focus was more on the general trends across MOF compounds selected for this study rather than on pinpointing the exact surface area and porosity for each compound, as variations may arise due to differences in calculation models and assumptions, and the re-digitalized data points at very low relative pressures may decrease the accuracy of the calculation.60–65Interestingly, despite the use of experimentally determined crystal structures in the CoRE MOF database,36,42 discrepancies between theoretical predictions and actual experimental outcomes were observed. For instance, some compounds exhibited a high theoretical porosity based on their refined CCDC structures but failed to demonstrate such porosity experimentally, possibly due to factors like structural collapse during activation, inaccessible pore environments, or suboptimal synthesis conditions. While variations of surface area, porosity or measured isotherms may exist among different research groups and preparations,66 such a phenomenon does not necessarily indicate a reproducibility issue, but rather a failure to reach the full potential and optimal conditions of the MOF in those studies. As a demonstration, Table 2 presents eight representative examples illustrating these variances, with some compounds showing excellent agreement between theoretical and experimental values (e.g., TAKCAM, OTIHOQ), while others displayed certain deviations (e.g., BOHXED, TAKTAD, and TOCJAY).
| Representative compound | Metal | CCDC code | Surface area (m2 g−1) | Reference | ||
|---|---|---|---|---|---|---|
| CoRE MOF databasea | Re-digitized experimental sorption curvesb | Literaturec | ||||
| a Based on the accessible surface area calculated for each compound's crystal structure as reported in the CoRE MOF 2019 database42 using nitrogen as the probe. b Based on data extracted from the experimental nitrogen isotherm reported in the respective literature, results are rounded to the nearest ten. c Based on the surface area reported by the author in the literature. | ||||||
| ZJU-70a | Cu | DORNAB | 2520 | 1790 | 1791 | 67 |
| MOF-802 | Zr | BOHXED | 2070 | 0 | <20 | 68 |
| Hf-L7 | Hf | TAKCAM | 2070 | 2200 | 2270 | 69 |
| MFM-300(Ga2) | Ga | TAKTAD | 1600 | 480 | 400 | 70 |
| MIL-100 (Al) | Al | BUSPIP | 1350 | 2090 | 2152 | 71 |
| [Cd(L4)]·1.5DMF | Cd | OTIHOQ | 1330 | 1320 | 1376 | 72 |
| Bio-MOF-101 | Zn | TOCJAY | 1100 | 2400 | 4410 | 73 |
| {[Ni6(N3)12L6]·(H2O)13}∞ | Ni | NUZCER | 790 | 400 | 309 | 74 |
Our findings reveal that reliance solely on computational results for material selection can be sometimes misleading, as many compounds exhibit experimental performances that deviate substantially from theoretical predictions, even when based on experimentally determined structures (Fig. 4). It should be noted that these experimentally-determined non-porous compounds are not necessarily useless, they in fact serve as valuable negative data points in the mined nitrogen isotherm dataset. By acknowledging these limitations and combining computational methods with experimental data, researchers can gain more comprehensive insights into reticular chemistry, discern trends, and make informed predictions.75–77 We envision that leveraging GPT-4V's capabilities to search for more experimental data—not limited to nitrogen isotherm but extending to other isotherms like water, CO2 capture, methane sorption—and other critical plots like TGA curves and PXRD from literature, in tandem with theoretical insights and computational science, can significantly advance the discovery and development of high-performance reticular compounds with desirable properties and functionalities.
To achieve this, it should be acknowledged that one limitation of our workflow is that the efficiency of using GPT-4V hinges significantly on the skillful crafting of prompts.10,58 It has always been a challenge to write good instructions to guide LLMs.78 Precision in prompt design is essential to achieve specific and accurate results in desirable format, underscoring the need for clear, detailed, and well-articulated instructions. In addition, idiosyncratic and subtle prompt cues can influence the LLMs without changing the underlying meaning, and consequently sometimes equivalent-looking prompts having very different quality.79,80 In this context, the emerging tool DSPy (Declarative Self-Improving Python)81 represents a significant advancement as compared with human written prompts and a future direction for this study. DSPy streamlines the process of prompt creation, combining techniques for both prompting and fine-tuning language models. Applying DSPy for prompt optimization necessitates the existence of a relatively small development set and some data annotation that DSPy will use to conduct the optimization automatically. Conceptually, this innovative system could enable researchers to commence their inquiries with a basic scaffolding of the high-level steps of the design and use DSPy to automate the instruction of LLMs, which can be considered a solution to further strengthen our workflow and improve the performance of LLMs. In the near future, we envision this combined approach can further increase the accuracy of the tasks demonstrated in this study, expand scope of the type of data to be mined from the literature, and hold the promise of making advanced natural language processing tools more accessible and adaptable to diverse research scenarios.
Concluding remarks
This study demonstrated the application of GPT-4V in text, image, and data mining within the realm of reticular chemistry. Our findings reveal that GPT-4V, when paired with carefully constructed prompts, is capable of processing page images and accurately identifying content in a fast and accurate manner. This capability can further extend to isolating pages with specific plots for in-depth analysis and building up datasets for experimental measurements from the literature. Importantly, the key features of LLMs like GPT-4V—being “programmable” through everyday natural language and possessing pre-trained domain knowledge—reduce traditional barriers associated with coding expertise and model training for specific plot or figure recognition. This adaptability is highlighted by the ease of transitioning from analysing one type of data, such as TGA curves, to another, like water isotherms, simply by modifying the instructional prompt. We envision that this workflow can be transferable and move beyond reticular chemistry to a broader spectrum of scientific disciplines. Nevertheless, there are certain limitations to consider. For instance, the model exhibits reduced accuracy in more complex tasks like the interpretation of graphs, indicating a need for further refinement in prompt engineering strategies. The lack of benchmarking datasets may increase the difficulty in coming up with effective prompts to guide LLMs. On the other hand, we envision that to make the instruction on GPT-4V more effective, the integration of advanced platform like DSPy into our workflow in the future will open new avenues in scientific data mining and points to a future where AI becomes an integral, user-friendly tool in the advancement of scientific knowledge.Data availability
The code, analysis scripts, and datasets supporting this article have been uploaded as part of the ESI.†Author contributions
Z. Z. and O. M. Y conceived and designed the study. Z. Z. and Z. H. performed the data curation. Z. Z. conducted the investigation and developed the methodology. Z. Z. and O. M. Y. carried out the formal analysis and visualization. O. M. Y. supervised the project. Z. Z. wrote the initial draft of the paper. All authors contributed significantly and collaboratively to the data analysis, and the review and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the Defense Advanced Research Projects Agency (DARPA) under contract HR0011-21-C-0020. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of DARPA. Z. Z. expresses gratitude to Mr Kefan Dong (Stanford University), Mr Jiayi Weng (OpenAI), Ms. Oufan Zhang (UC Berkeley), Mr Zichao Rong (UC Berkeley) and Ms. Zeqi Gu (Cornell University) for their valuable discussions. Z. Z. also acknowledges financial support from a Kavli ENSI Graduate Student Fellowship.Notes and references
- A. Birhane, A. Kasirzadeh, D. Leslie and S. Wachter, Nat. Rev. Phys., 2023, 5, 277–280 CrossRef.
- A. D. White, Nat. Rev. Chem, 2023, 7, 457–458 CrossRef CAS PubMed.
- S. Bubeck, V. Chandrasekaran, R. Eldan, J. Gehrke, E. Horvitz, E. Kamar, P. Lee, Y. T. Lee, Y. Li and S. Lundberg, Sparks of artificial general intelligence: Early experiments with gpt-4, arXiv, 2023, preprint, arXiv:2303.12712, DOI:10.48550/arXiv:2303.12712.
- Microsoft Research AI4Science, Microsoft Azure Quantum, The Impact of Large Language Models on Scientific Discovery: a Preliminary Study using GPT-4, arXiv, 2023, preprint, arXiv.2311.07361, DOI:10.48550/arXiv.2311.07361.
- OpenAI, GPT-4 technical report, arXiv, 2023, preprint, arXiv:2303.08774v3, DOI:10.48550/arXiv:2303.08774v3.
- R. Anil, A. M. Dai, O. Firat, M. Johnson, D. Lepikhin, A. Passos, S. Shakeri, E. Taropa, P. Bailey and Z. Chen, Palm 2 technical report, arXiv, 2023, preprint, arXiv:2305.10403, DOI:10.48550/arXiv.2305.10403.
- H. Touvron, L. Martin, K. Stone, P. Albert, A. Almahairi, Y. Babaei, N. Bashlykov, S. Batra, P. Bhargava and S. Bhosale, Llama 2: Open foundation and fine-tuned chat models, arXiv, 2023, preprint, arXiv:2307.09288, DOI:10.48550/arXiv.2307.09288.
- Z. Zheng, O. Zhang, H. L. Nguyen, N. Rampal, A. H. Alawadhi, Z. Rong, T. Head-Gordon, C. Borgs, J. T. Chayes and O. M. Yaghi, ACS Cent. Sci., 2023, 9, 2161–2170 CrossRef CAS PubMed.
- K. M. Jablonka, Q. Ai, A. Al-Feghali, S. Badhwar, J. D. Bocarsly, A. M. Bran, S. Bringuier, L. C. Brinson, K. Choudhary and D. Circi, Digital Discovery, 2023, 2, 1233–1250 RSC.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- Y. Kang and J. Kim, ChatMOF: An Autonomous AI System for Predicting and Generating Metal-Organic Frameworks, arXiv, 2023, preprint, arXiv:2308.01423, DOI:10.48550/arXiv:2308.01423.
- S. Liu, J. Wang, Y. Yang, C. Wang, L. Liu, H. Guo and C. Xiao, ChatGPT-powered Conversational Drug Editing Using Retrieval and Domain Feedback, arXiv, 2023, preprint, arXiv:2305.18090, DOI:10.48550/arXiv:2305.18090.
- A. M. Bran, S. Cox, A. D. White and P. Schwaller, ChemCrow: Augmenting large-language models with chemistry tools, arXiv, 2023, preprint, arXiv:2304.05376, DOI:10.48550/arXiv:2304.05376.
- Z. Zheng, Z. Rong, N. Rampal, C. Borgs, J. T. Chayes and O. M. Yaghi, Angew. Chem., Int. Ed., 2023, 62, e202311983 CrossRef CAS PubMed.
- M. Thway, A. Low, S. Khetan, H. Dai, J. Recatala-Gomez, A. P. Chen and K. Hippalgaonkar, Digital Discovery, 2024 10.1039/D3DD00202K.
- Z. Zheng, A. H. Alawadhi, S. Chheda, S. E. Neumann, N. Rampal, S. Liu, H. L. Nguyen, Y.-h. Lin, Z. Rong, J. I. Siepmann, L. Gagliardi, A. Anandkumar, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 28284–28295 CrossRef CAS PubMed.
- G. M. Hocky and A. D. White, Digital Discovery, 2022, 1, 79–83 RSC.
- Z. Xie, X. Evangelopoulos, Ö. H. Omar, A. Troisi, A. I. Cooper and L. Chen, Chem. Sci., 2024, 15, 500–510 RSC.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Nat. Commun., 2023, 14, 7964 CrossRef CAS PubMed.
- K. Cruse, V. Baibakova, M. Abdelsamie, K. Hong, C. J. Bartel, A. Trewartha, A. Jain, C. M. Sutter-Fella and G. Ceder, Chem. Mater., 2024, 36(2), 772–785 CrossRef CAS PubMed.
- Z. Yang, L. Li, K. Lin, J. Wang, C.-C. Lin, Z. Liu and L. Wang, The dawn of lmms: Preliminary explorations with gpt-4v (ision), arXiv, 2023, preprint, arXiv:2309.17421, DOI:10.48550/arXiv.2309.17421.
- Z. Yan, K. Zhang, R. Zhou, L. He, X. Li and L. Sun, Multimodal ChatGPT for Medical Applications: an Experimental Study of GPT-4V, arXiv, 2023, preprint, arXiv:2310.19061, DOI:10.48550/arXiv.2310.19061.
- C. Wu, S. Yin, W. Qi, X. Wang, Z. Tang and N. Duan, Visual chatgpt: Talking, drawing and editing with visual foundation models, arXiv, 2023, preprint, arXiv:2303.04671, DOI:10.48550/arXiv.2303.04671.
- OpenAI, GPT-4V(ision) System Card, 2023, accessed 2023-09-25 Search PubMed.
- C. Wu, J. Lei, Q. Zheng, W. Zhao, W. Lin, X. Zhang, X. Zhou, Z. Zhao, Y. Zhang and Y. Wang, Can gpt-4v (ision) serve medical applications? case studies on gpt-4v for multimodal medical diagnosis, arXiv, 2023, preprint, arXiv:2310.09909, DOI:10.48550/arXiv:2310.09909.
- N. Wake, A. Kanehira, K. Sasabuchi, J. Takamatsu and K. Ikeuchi, GPT-4V (ision) for Robotics: Multimodal Task Planning from Human Demonstration, arXiv, 2023, preprint, arXiv:2311.12015, DOI:10.48550/arXiv:2311.12015.
- Y. Shi, D. Peng, W. Liao, Z. Lin, X. Chen, C. Liu, Y. Zhang and L. Jin, Exploring ocr capabilities of gpt-4v (ision): A quantitative and in-depth evaluation, arXiv, 2023, preprint, arXiv:2310.16809, DOI:10.48550/arXiv:2310.16809.
- S. Park, B. Kim, S. Choi, P. G. Boyd, B. Smit and J. Kim, J. Chem. Inf. Model., 2018, 58, 244–251 CrossRef CAS PubMed.
- H. Park, Y. Kang, W. Choe and J. Kim, J. Chem. Inf. Model., 2022, 62, 1190–1198 CrossRef CAS PubMed.
- Y. Luo, S. Bag, O. Zaremba, A. Cierpka, J. Andreo, S. Wuttke, P. Friederich and M. Tsotsalas, Angew. Chem., Int. Ed., 2022, 61, e202200242 CrossRef CAS PubMed.
- Z. Wu, J. Chen, Y. Li, Y. Deng, H. Zhao, C.-Y. Hsieh and T. Hou, J. Chem. Inf. Model., 2023, 63(24), 7617–7627 CrossRef CAS PubMed.
- D. Vidhani,The Art of Asking Question: Mastering Human-AI (HAI) Duet in Chemistry Through Prompt Engineering, 2024, DOI:10.21203/rs.3.rs-3825267/v1.
- M. Ansari and S. M. Moosavi, Agent-based Learning of Materials Datasets from Scientific Literature, arXiv, 2023, preprint, arXiv:2312.11690, DOI:10.48550/arXiv:2312.11690.
- H. Lyu, Z. Ji, S. Wuttke and O. M. Yaghi, Chem, 2020, 6, 2219–2241 CAS.
- O. M. Yaghi and Z. Zheng, Reticular Chemistry and New Materials, World Scientific, 2024 Search PubMed.
- Y. G. Chung, J. Camp, M. Haranczyk, B. J. Sikora, W. Bury, V. Krungleviciute, T. Yildirim, O. K. Farha, D. S. Sholl and R. Q. Snurr, Chem. Mater., 2014, 26, 6185–6192 CrossRef CAS.
- A. S. Rosen, V. Fung, P. Huck, C. T. O'Donnell, M. K. Horton, D. G. Truhlar, K. A. Persson, J. M. Notestein and R. Q. Snurr, npj Comput. Mater., 2022, 8, 112 CrossRef CAS.
- S. Kancharlapalli and R. Q. Snurr, ACS Appl. Mater. Interfaces, 2023, 15(23), 28084–28092 CrossRef CAS PubMed.
- P. Z. Moghadam, Y. G. Chung and R. Q. Snurr, Nat. Energy, 2024, 1–13 Search PubMed.
- D. A. Boiko, R. MacKnight, B. Kline and G. Gomes, Nature, 2023, 624, 570–578 CrossRef CAS PubMed.
- B. A. Koscher, R. B. Canty, M. A. McDonald, K. P. Greenman, C. J. McGill, C. L. Bilodeau, W. Jin, H. Wu, F. H. Vermeire, B. Jin, T. Hart, T. Kulesza, S.-C. Li, T. S. Jaakkola, R. Barzilay, R. Gómez-Bombarelli, W. H. Green and K. F. Jensen, Science, 2023, 382, eadi1407 CrossRef CAS PubMed.
- Y. G. Chung, E. Haldoupis, B. J. Bucior, M. Haranczyk, S. Lee, H. Zhang, K. D. Vogiatzis, M. Milisavljevic, S. Ling, J. S. Camp, B. Slater, J. I. Siepmann, D. S. Sholl and R. Q. Snur, J. Chem. Eng. Data, 2019, 64, 5985–5998 CrossRef CAS.
- A. Nandy, G. Terrones, N. Arunachalam, C. Duan, D. W. Kastner and H. J. Kulik, Sci. Data, 2022, 9, 74 CrossRef CAS PubMed.
- S. M. Moosavi, A. Nandy, K. M. Jablonka, D. Ongari, J. P. Janet, P. G. Boyd, Y. Lee, B. Smit and H. J. Kulik, Nat. Commun., 2020, 11, 1–10 CrossRef PubMed.
- A. Nandy, S. Yue, C. Oh, C. Duan, G. G. Terrones, Y. G. Chung and H. J. Kulik, Matter, 2023, 6, 1585–1603 CrossRef CAS.
- A. Nandy, C. Duan and H. J. Kulik, J. Am. Chem. Soc., 2021, 143, 17535–17547 CrossRef CAS PubMed.
- J. C. Tan, T. D. Bennett and A. K. Cheetham, Proc. Natl. Acad. Sci. U.S.A., 2010, 107, 9938–9943 CrossRef CAS PubMed.
- R. Batra, C. Chen, T. G. Evans, K. S. Walton and R. Ramprasad, Nat. Mach., 2020, 2, 704–710 Search PubMed.
- B. Wang, X.-L. Lv, D. Feng, L.-H. Xie, J. Zhang, M. Li, Y. Xie, J.-R. Li and H.-C. Zhou, J. Am. Chem. Soc., 2016, 138, 6204–6216 CrossRef CAS PubMed.
- W. Song, Z. Zheng, A. H. Alawadhi and O. M. Yaghi, Nat. Water, 2023, 1, 626–634 CrossRef.
- B. F. Abrahams, A. D. Dharma, M. J. Grannas, T. A. Hudson, H. E. Maynard-Casely, G. R. Oliver, R. Robson and K. F. White, Inorg. Chem., 2014, 53, 4956–4969 CrossRef CAS PubMed.
- Z. Zheng, H. L. Nguyen, N. Hanikel, K. K.-Y. Li, Z. Zhou, T. Ma and O. M. Yaghi, Nat. Protoc., 2023, 18, 136–156 CrossRef CAS PubMed.
- Z. Zheng, N. Hanikel, H. Lyu and O. M. Yaghi, J. Am. Chem. Soc., 2022, 144, 22669–22675 CrossRef CAS PubMed.
- K. Hatakeyama-Sato, N. Yamane, Y. Igarashi, Y. Nabae, T. Hayakawa, Prompt engineering of GPT-4 for chemical research: what can/cannot be done?, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-s1x5p.
- A. G. Parameswaran, S. Shankar, P. Asawa, N. Jain and Y. Wang, Revisiting Prompt Engineering via Declarative Crowdsourcing, arXiv, 2023, preprint, arXiv:2308.03854, DOI:10.48550/arXiv:2308.03854.
- L. Reynolds and K. McDonell, presented in part at the Extended Abstracts of the 2021 CHI Conference on Human Factors in Computing Systems, 2021 Search PubMed.
- Y. Zhou, A. I. Muresanu, Z. Han, K. Paster, S. Pitis, H. Chan and J. Ba, Large language models are human-level prompt engineers, arXiv, 2022, preprint, arXiv:2211.01910, DOI:10.48550/arXiv:2211.01910.
- M. Mitchell, A. B. Palmarini and A. Moskvichev, Comparing Humans, GPT-4, and GPT-4V On Abstraction and Reasoning Tasks, arXiv, 2023, preprint, arXiv.2311.09247, DOI:10.48550/arXiv.2311.09247.
- A. Rohatgi, WebPlotDigitizer, https://automeris.io/WebPlotDigitizer, accessed 2022-09 Search PubMed.
- J. D. Evans, V. Bon, I. Senkovska and S. Kaskel, Langmuir, 2021, 37, 4222–4226 CrossRef CAS PubMed.
- T. Düren, F. Millange, G. Férey, K. S. Walton and R. Q. Snurr, J. Phys. Chem. C, 2007, 111, 15350–15356 CrossRef.
- G. Hai and H. Wang, Coord. Chem. Rev., 2022, 469, 214670 CrossRef CAS.
- K. S. Walton and R. Q. Snurr, J. Am. Chem. Soc., 2007, 129, 8552–8556 CrossRef CAS PubMed.
- P. Sinha, A. Datar, C. Jeong, X. Deng, Y. G. Chung and L.-C. Lin, J. Phys. Chem. C, 2019, 123, 20195–20209 CrossRef CAS.
- F. Ambroz, T. J. Macdonald, V. Martis and I. P. Parkin, Small Methods, 2018, 2, 1800173 CrossRef.
- J. W. M. Osterrieth, J. Rampersad, D. Madden, N. Rampal, L. Skoric, B. Connolly, M. D. Allendorf, V. Stavila, J. L. Snider, R. Ameloot, J. Marreiros, C. Ania, D. Azevedo, E. Vilarrasa-Garcia, B. F. Santos, X.-H. Bu, Z. Chang, H. Bunzen, N. R. Champness, S. L. Griffin, B. Chen, R.-B. Lin, B. Coasne, S. Cohen, J. C. Moreton, Y. J. Colón, L. Chen, R. Clowes, F.-X. Coudert, Y. Cui, B. Hou, D. M. D'Alessandro, P. W. Doheny, M. Dincă, C. Sun, C. Doonan, M. T. Huxley, J. D. Evans, P. Falcaro, R. Ricco, O. Farha, K. B. Idrees, T. Islamoglu, P. Feng, H. Yang, R. S. Forgan, D. Bara, S. Furukawa, E. Sanchez, J. Gascon, S. Telalović, S. K. Ghosh, S. Mukherjee, M. R. Hill, M. M. Sadiq, P. Horcajada, P. Salcedo-Abraira, K. Kaneko, R. Kukobat, J. Kenvin, S. Keskin, S. Kitagawa, K.-i. Otake, R. P. Lively, S. J. A. DeWitt, P. Llewellyn, B. V. Lotsch, S. T. Emmerling, A. M. Pütz, C. Martí-Gastaldo, N. M. Padial, J. García-Martínez, N. Linares, D. Maspoch, J. A. Suárez del Pino, P. Moghadam, R. Oktavian, R. E. Morris, P. S. Wheatley, J. Navarro, C. Petit, D. Danaci, M. J. Rosseinsky, A. P. Katsoulidis, M. Schröder, X. Han, S. Yang, C. Serre, G. Mouchaham, D. S. Sholl, R. Thyagarajan, D. Siderius, R. Q. Snurr, R. B. Goncalves, S. Telfer, S. J. Lee, V. P. Ting, J. L. Rowlandson, T. Uemura, T. Iiyuka, M. A. van der Veen, D. Rega, V. Van Speybroeck, S. M. J. Rogge, A. Lamaire, K. S. Walton, L. W. Bingel, S. Wuttke, J. Andreo, O. Yaghi, B. Zhang, C. T. Yavuz, T. S. Nguyen, F. Zamora, C. Montoro, H. Zhou, A. Kirchon and D. Fairen-Jimenez, Adv. Mater., 2022, 34, 2201502 CrossRef CAS PubMed.
- X. Duan, C. Wu, S. Xiang, W. Zhou, T. Yildirim, Y. Cui, Y. Yang, B. Chen and G. Qian, Inorg. Chem., 2015, 54, 4377–4381 CrossRef CAS PubMed.
- H. Furukawa, F. Gandara, Y.-B. Zhang, J. Jiang, W. L. Queen, M. R. Hudson and O. M. Yaghi, J. Am. Chem. Soc., 2014, 136, 4369–4381 CrossRef CAS PubMed.
- R. J. Marshall, C. L. Hobday, C. F. Murphie, S. L. Griffin, C. A. Morrison, S. A. Moggach and R. S. Forgan, J. Mater. Chem. A, 2016, 4, 6955–6963 RSC.
- C. P. Krap, R. Newby, A. Dhakshinamoorthy, H. García, I. Cebula, T. L. Easun, M. Savage, J. E. Eyley, S. Gao, A. J. Blake, W. Lewis, P. H. Beton, M. R. Warren, D. R. Allan, M. D. Frogley, C. C. Tang, G. Cinque, S. Yang and M. Schröder, Inorg. Chem., 2016, 55, 1076–1088 CrossRef CAS PubMed.
- C. Volkringer, D. Popov, T. Loiseau, G. Férey, M. Burghammer, C. Riekel, M. Haouas and F. Taulelle, Chem. Mater., 2009, 21, 5695–5697 CrossRef CAS.
- B. Liu, H.-F. Zhou, L. Hou, J.-P. Wang, Y.-Y. Wang and Z. Zhu, Inorg. Chem., 2016, 55, 8871–8880 CrossRef CAS PubMed.
- T. Li, M. T. Kozlowski, E. A. Doud, M. N. Blakely and N. L. Rosi, J. Am. Chem. Soc., 2013, 135, 11688–11691 CrossRef CAS PubMed.
- Q. Yu, Y.-F. Zeng, J.-P. Zhao, Q. Yang, B.-W. Hu, Z. Chang and X.-H. Bu, Inorg. Chem., 2010, 49, 4301–4306 CrossRef CAS PubMed.
- L. Gagliardi and O. M. Yaghi, Chem. Mater., 2023, 35, 5711–5712 CrossRef CAS.
- A. A. AlGhamdi, Mol. Front. J., 2023, 1–4 Search PubMed.
- A. S. Rosen, J. M. Notestein and R. Q. Snurr, Curr. Opin. Chem. Eng., 2022, 35, 100760 CrossRef.
- H. Nori, Y. T. Lee, S. Zhang, D. Carignan, R. Edgar, N. Fusi, N. King, J. Larson, Y. Li and W. Liu, Can Generalist Foundation Models Outcompete Special-Purpose Tuning? Case Study in Medicine, arxiv, 2023, preprint, arxiv:2311.16452, DOI:10.48550/arXiv.2311.16452.
- S. Huang, S. Mamidanna, S. Jangam, Y. Zhou and L. H. Gilpin, Can large language models explain themselves? a study of llm-generated self-explanations, arXiv, 2023, preprint, arXiv:2310.11207, DOI:10.48550/arXiv:2310.11207.
- G. Yona, R. Aharoni and M. Geva, Narrowing the Knowledge Evaluation Gap: Open-Domain Question Answering with Multi-Granularity Answers, arXiv, 2024, preprint, arXiv:2401.04695, DOI:10.48550/arXiv:2401.04695.
- O. Khattab, A. Singhvi, P. Maheshwari, Z. Zhang, K. Santhanam, S. Vardhamanan, S. Haq, A. Sharma, T. T. Joshi and H. Moazam, Dspy: Compiling declarative language model calls into self-improving pipelines, arXiv, 2023, preprint, arXiv.2310.03714, DOI:10.48550/arXiv.2310.03714.
Footnote |
| † Electronic supplementary information (ESI) available: Full prompts designed to guide GPT-4V; additional examples showcasing GPT-4V's performance in reading various figure inputs and its corresponding responses; Python code used to automate the data mining and analysis processes; detailed information on the selected papers in this study, including the ground truth and the classification output for each page in a spread-sheet format; extracted nitrogen isotherms in this study. See DOI: https://doi.org/10.1039/d3dd00239j |
| This journal is © The Royal Society of Chemistry 2024 |