Open Access Article
Open Access ArticleMachine learning for hypothesis generation in biology and medicine: exploring the latent space of neuroscience and developmental bioelectricity
Thomas
O'Brien
a,
Joel
Stremmel
 a,
Léo
Pio-Lopez
b,
Patrick
McMillen
b,
Cody
Rasmussen-Ivey
b and
Michael
Levin
a,
Léo
Pio-Lopez
b,
Patrick
McMillen
b,
Cody
Rasmussen-Ivey
b and
Michael
Levin
 *bc
*bc
aStreams, Winona, MN 55987, USA
bAllen Discovery Center, Tufts University, 200 Boston Ave., Suite 4600, Medford, MA 02155, USA. E-mail: michael.levin@tufts.edu
cWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
First published on 4th January 2024
Abstract
Artificial intelligence is a powerful tool that could be deployed to accelerate the scientific enterprise. Here we address a major unmet need: use of existing scientific literature to generate novel hypotheses. We use a deep symmetry between the fields of neuroscience and developmental bioelectricity to evaluate a new tool, FieldSHIFT. FieldSHIFT is an in-context learning framework using a large language model to facilitate candidate scientific research from existing published studies, serving as a tool to generate hypotheses at scale. We release a new dataset for translating between the neuroscience and developmental bioelectricity domains and show how FieldSHIFT helps human scientists explore a latent space of papers that could exist, providing a rich field of suggested future research. We demonstrate the performance of FieldSHIFT for hypothesis generation relative to human-generated developmental biology research directions then test a key prediction of this model using bioinformatics, showing a surprising conservation of molecular mechanisms involved in cognitive behavior and developmental morphogenesis. By allowing scientists to rapidly explore symmetries and meta-parameters that exist in a corpus of scientific papers, we show how machine learning can potentiate human creativity and assist with one of the most interesting and crucial aspects of research: identifying insights from data and generating potential candidates for research agendas.
Introduction
The scientific enterprise depends critically not only on the skills needed to definitively test crisp hypotheses,1–7 but also on the much less well-understood step of acquiring insights and fruitful research directions.8,9 Many recent discussions10,11 have focused on the fact that while modern “big data” methods are generating a deluge of facts and measurements, it is increasingly more important to be “building the edifice of science, in addition to throwing bricks into the yard”.12 In other words, it is essential that we develop strategies for deriving novel insights and deep hypotheses that cross traditional (in many cases, artificial) boundaries between disciplines. We must improve, at a pace that keeps up with technological and data science advances, our ability to identify symmetries (invariances) between sets of observations and approaches – to unify and simplify where possible by identifying large-scale patterns in research literature and thus motivate and enhance new research programs (Fig. 1).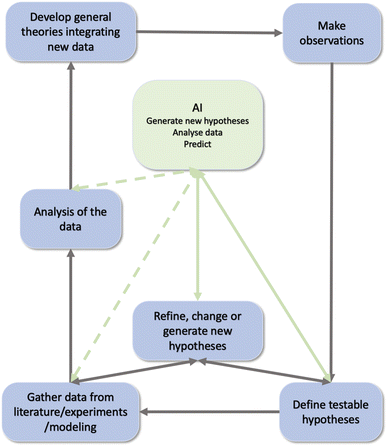 | ||
| Fig. 1 The scientific discovery cycle and the role of AI within it. Black arrows describe the scientific cycle, green dotted arrow the links with AI and green plain arrows the ones we are interested in this paper. Modified after ref. 111. | ||
One set of tools that is being rapidly developed and ubiquitously deployed originates in the advances of the field of artificial intelligence.13–15 Machine learning is clearly poised to help science.14,16–19 It is becoming widely recognized that “robot scientist” platforms can not only provide number crunching, automation, and high throughput, but could potentially also guide research by identifying what experiments to do next.20–22 Machine learning tools have been suggested to be essential for progress in the bioinformatics of shape23–26 (moving beyond molecular information to understand organ- and organism-level form and function), and developmental biology.27,28 Not surprisingly, much emphasis is currently placed on improving the factuality of the output of AI tools; this parallels the well-developed notions of strong inference and hypothesis-testing in science: the well-established algorithm of falsifying and removing incorrect ideas. What is much less well-understood, but ultimately as crucial, is the process that provides a pool of interesting hypotheses from which to winnow until provisional truths are found. Our contribution attempts to bolster this second component of the scientific process – ideation – with a new AI-based tool for human scientists that provides input into the canonical scientific method.
Recent work in this field includes the AI-based discovery of testable models of regenerative anatomical regulation,29 which were subsequently empirically validated,30 and similar efforts in genomics,22 chemistry31,32 and biomaterials.33 One of the most potentially important current gaps is the paucity of tools for assisting with the most creative aspect of the work: identifying deep commonalities between disparate functional datasets, to enable generalized insight from the ever-growing literature. Tools are needed that would take meta-hypotheses of human scientists and mine the published studies for information that can be used to turn those conceptual hunches into actionable research programs.
One interdisciplinary area in which computer-assisted discovery would be most welcome is real-time physiology that controls form and function in vivo. How large-scale anatomy and behavior arises from the operation of molecular processes is a canonical systems biology problem.34 It is currently handled by two distinct fields, with their own educational tracks, conferences, journals, funding bodies, and paradigms: neuroscience and developmental biology. Interestingly however, recent work has suggested that these may share an important underlying set of dynamics.35–37 Neuroscience has long understood that behavior and cognition are driven by the physiological information processing by neural networks, which signal via electrical events at their membranes.38–41 Likewise, a long history of classical work has suggested that bioelectric signaling of all kinds of cells is required as an instructive set of influences that help direct complex developmental and regenerative morphogenesis.42–44 Interestingly, recent advances in the molecular understanding of developmental bioelectricity45–47 has begun to blur the boundaries between those disciplines48,49 (Fig. 2).
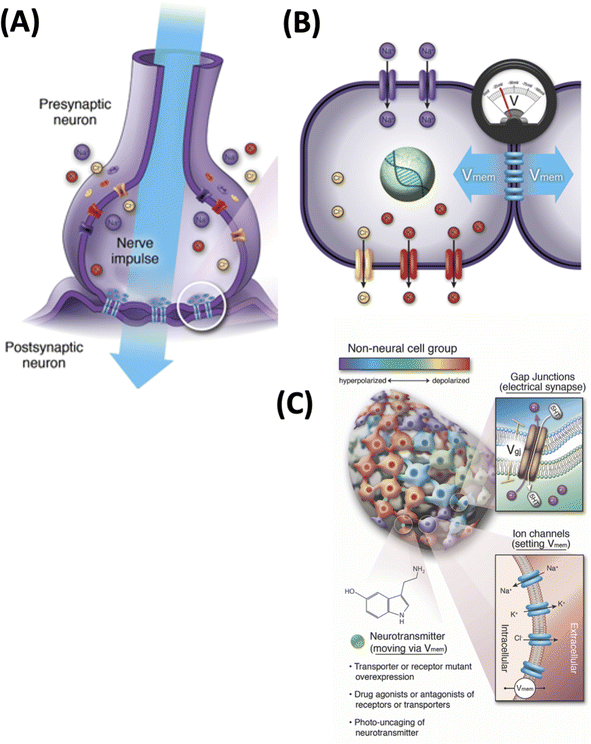 | ||
| Fig. 2 Bioelectricity of the brain and body. (A) Neuronal signaling that underlies cognition and behavior is implemented by ion channels and gap junctions, which together set the voltage potential of cells and the propagation of these voltage states through neural networks. (B) All cells in the body have ion channels, and most have gap junctional connections to their neighbors, establishing a bioelectrical network outside the brain. (C) Numerous studies45–47 have taken advantage of the isomorphism of function and mechanistic conservation between neurons and other cell types by exploiting classic tools of neuroscience in developmental biology outside the CNS. Specifically, analogs of synaptic plasticity (altering connectivity between cells by targeting gap junctions), intrinsic plasticity (directly modifying the voltage state of specific cells), and neurotransmitter signaling (which also takes place in non-neural tissues) have all been used. Optogenetics, ion channel mutations, and channel- and gap junction-modifying pharmacological agents have been used to induce specific changes in the processing of morphogenetic information. All images by Jeremy Guay of Peregrine Creative. (A) and (B) used with permission from ref. 36; (C) used with permission from ref. 57. | ||
Specifically, it has been conjectured that the immense functional capabilities of brains have their origin in the more ancient, and slower, bioelectric networks that began operation at the emergence of bacterial biofilms50–54 and were heavily exploited by evolution to orchestrate metazoan morphogenesis.55,56 The idea that the same algorithms for scaling cellular activity into larger competencies (such as regulative morphogenesis and intelligent behavior) are used in the brain and in the body (Fig. 3) has many deep implications, for example with respect to porting methods from neuroscience and behavioral science into biomedicine,35,57,58 to take advantage of neuroscience's successful paradigms for managing multi-scale causality and inferring effective interventions.
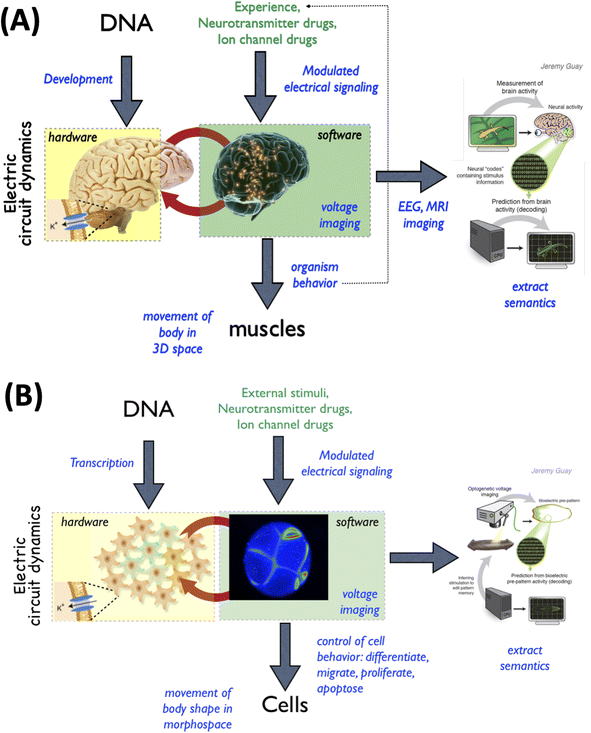 | ||
| Fig. 3 Functional invariants between neuroscience and developmental biology. The fields of neuroscience and developmental biology are addressed by separate communities, journals, funding bodies, and educational programs. While many details across these domains differ, it is increasingly seen that many tools and concepts used in neuroscience carry over and can be successfully used in developmental biology settings. This is because of a shared evolutionary history and a deep symmetry of both hardware and physiology across these domains. (A) In the nervous system, behavior is implemented in 3-dimensional space by electrical networks that process information so as to control muscle cells. The interplay between the cellular hardware and physiological software is the subject of a field of neuroscience known as neural decoding,112 where it is hoped that the cognitive content (memories, plans, preferences, etc.) of the 1st-person mind implemented by the electrophysiology can be read out by 3rd-person observers (scientists). (B) This same architecture was implemented by an evolutionarily more ancient system in which bioelectric networks in the body processed information so as to control the behavior of all cell types, moving the body configuration through morphospace (during development, regeneration, and remodeling).55,56 As with the brain, developmental biologists have been attempting to decode these patterns and re-write them,36,60 to be able to infer and modify the paths that cellular collective will take through morphospace. All images by Jeremy Guay of Peregrine Creative. | ||
The proposed symmetry between cellular swarms using collective dynamics to solve problems in anatomical morphospace and in 3D classical behavioral space has been empirically tested in specific contexts,34,47 and has significant implications for biomedicine and evolutionary biology.59,60 However, there has been no way to derive these hypotheses at scale from the plethora of functional literature of neuroscience and developmental biophysics. Specifically, it has been claimed that the deep similarity between the role of bioelectric networks in control of body shape (cellular collectives' behavior) and cognition (organism-level behavior) enables one to readily read neuroscience papers as if they were developmental biology papers, by only pivoting problem spaces (from 3D space to anatomical morphospace) and time scales (from milliseconds to minutes).49,56 However, there has never been an efficient way to implement this conjecture and actually explore the latent space of hypothetical papers that could provide candidate hypotheses and potentially fruitful research directions.
Here, we provide a first-generation tool, FieldSHIFT, that helps human scientists explore the mapping between developmental bioelectricity and neuroscience and begins the journey towards exploring the space of scientific studies far wider than what has already been written. FieldSHIFT is an in-context learning framework which uses a large language model to convert existing paper abstracts in neuroscience to the field of developmental biology, by appropriately replacing specific words and concepts. We show that this process generates useful, readable, insightful results that can be used to expand scientific intuition and identify testable hypotheses for novel work at the intersection of two fields. Furthermore, we test some of the resulting predictions using a bioinformatics approach, revealing a remarkable quantitative conservation of genes between developmental morphogenesis and cognitive behavior. The described system is a first step on the road to using AI tools as an imagination enhancing tool for research, deriving novel insights from expensive published experiments and letting scientists explore life-as-it-could be.61 We tested one of the predictions of its output, finding a quantitatively significant enrichment in overlap between specific genes implicated in both cognitive processes and morphogenesis.
There have been a handful of related research efforts using generative AI for hypothesis generation and cross-domain knowledge discovery. One such approach is MechGPT, a large language model for identifying and probing analogous concepts between scientific domains.62 Other approaches have focused on knowledge discovery across scientific domains, such as63 for translating between protein sequences and music note sequences with attention-based deep neural networks and64 for identifying new designs within the field of mechanical engineering via cross-domain patent mining using knowledge graphs and natural language processing. Our method focuses uniquely on generating scientific hypotheses in the form of abstracts via text translations.
Methods
We propose FieldSHIFT: a framework for domain translation with modular components in the form of training data and an AI language model for mapping text from one domain to another. We constructed a dataset of neuroscience and developmental biology sentences and associated concepts to test FieldSHIFT with two AI language models. We used trained biologists to both build the dataset and evaluate the model translations. While we focused on mapping neuroscience to developmental biology text, we believe a similar approach could be taken to apply FieldSHIFT to other domains.Data
As an exercise with students and other researchers, the Levin Lab has been translating neuroscience to development biology, by hand, for several decades. They'd do this by changing concepts like “brain” to “body”, “millisecond” to “minute”, and “neuron” to “cell”. We collected 70 such translation pairs. Using these concept pairs, we constructed translations varying from one sentence to abstract-length paragraphs by matching neuroscience passages with corresponding developmental biology passages. These passages include a variety of text from facts about these scientific domains to recently published abstracts from neuroscience and developmental biology articles. We collected 1437 translation samples in total. The average length of a sample input or output passage is 209 characters, and the standard deviation is 375. Below is an example pair pertaining to “brain” as the neuroscience concept and “body” as the developmental biology concept:Neuroscience: “The brain is a complex system of neurons that are interconnected by synapses.”
Developmental Biology: “The body is a complex system of cells that are interconnected by gap junctions.”
In addition to translations, in some cases we added the same developmental biology abstract as both input and output examples. The goal was to teach a model trained on these examples not to change anything in the case that development biology text is provided as input. The six abstracts (corresponding to papers65–70) we used in this way were selected by choosing developmental biology papers produced by the Levin Lab. We published the full dataset on Hugging Face for re-use by the community: https://huggingface.co/datasets/levinlab/neuroscience-to-dev-bio/tree/main.
To train and test models for domain translation, we randomly split the dataset according to the neuroscience concepts associated with each sample. An effective domain translator should be able to generalize beyond the concepts it sees during training, demonstrating general knowledge of both domains and an ability to both identify new concepts within the input domain and an ability to map those concepts to corresponding concepts in the output domain. As such, we split the concepts into 43 training, 6 validation, and 21 test concepts without overlap via random sampling. After splitting concepts, we split the passages associated with these concepts into 503 train, 50 validation, and 57 test passages by searching for each concept name within the passage (after normalizing the text by stripping punctuation, lowercasing, and ignoring extra whitespace characters). Because some concepts mapped to more passages, we repeated the process until we achieved close to this desired ratio of train, validation, and test samples (roughly 10× the number of train samples as validation and test) without looking at the resulting data until settling on a desirable ratio.
Domain translation with machine learning
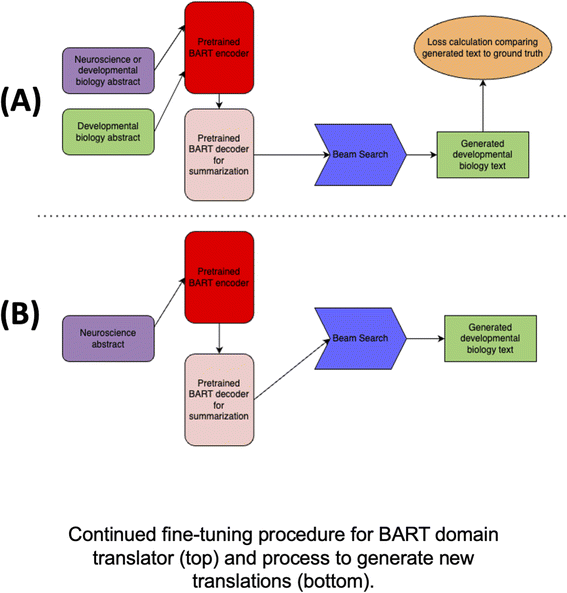
We implemented and tested two domain translation approaches for research paper abstracts using pretrained language models to identify a suitable backend model for FieldSHIFT. The first is inspired by neural machine translation (NMT) and abstractive summarization and uses the open-source BART71 pretrained Transformer language model (LM).72 The pretrained BART model is designed for summarizing text. Rather than have the model generate a summary of an input sample, we fine-tuned it to map neuroscience text to developmental biology text using the domain translation dataset. The second model we implemented uses in-context learning73 to prompt the GPT-4 (ref. 74) large language model (LLM) to complete the task of translating neuroscience text into developmental biology text. We used human evaluation from trained biologists to test the effectiveness of these approaches by having each model generate translations and counting successful ones according to expert judgement. For more details on the testing procedure, see the model validation section.Transformer LMs, and recently, LLMs, which demonstrate emergent capabilities when scaled to billions of parameters,75 are highly expressive deep neural networks used to represent sequences, especially text data, and have achieved state-of-the-art performance on a variety of natural language tasks.76–78 LMs are typically pretrained to learn general representations of text from large, unannotated corpora, then fine-tuned for specific, downstream natural language tasks. Both NMT and summarization involve the task of text generation, conditioning on an input text sequence to generate an output text sequence. We exploit this symmetry in our first domain translation approach using BART by continuing to fine-tune an encoder–decoder LM72 already trained for summarization. This neural architecture embeds discrete textual characters from an input sequence in a vector space. It then generates an encoded representation of the text in a hidden, latent space in a manner that captures word order (encoder layer). Next, it decodes the latent space, conditioning on previously generated words and the encoder hidden state (decoder layer). Finally, it predicts the next word in the output sequence using the decoder output. Greedily predicting the most likely word at each step is unlikely to generate the best overall output sentence, while selecting the most likely output sentence in its entirety is computationally infeasible. As such, we used Beam Search79 to select the top-k most likely output words given an input sentence and encoder–decoder model parameters.
We continued fine-tuning the pretrained BART LM71 first fine-tuned as an abstractive summarizer (and available via Hugging Face transformers80) for domain translation by feeding the summarizer text pairs specific to our translation task. We selected the BART LM for its pretraining procedure designed for text generation, its performance on summarization benchmarks, and its public availability. A key advantage of the BART architecture is the way it combines ideas from other popular and effective Transformer neural networks, specifically bidirectional encoding via corruption of input text as in 77 and autoregressive decoding as in the GPT family of models.81
Recent decoder-only GPT models such as GPT-3,81 InstructGPT,82 and GPT-4 have proven even more powerful than encoder–decoder models when scaled to billions of parameters and trained on sufficiently large corpora.83 Furthermore, Instruction Fine-Tuning and Reinforcement Learning with Human Feedback82 have led to human-like, and in some cases, beyond human-level performance on text generation tasks with decoder-only LLMs. The key idea of these models is to frame all downstream tasks as text generation with no intermediate encoding step and align the outputs from the model to human expectations using Reinforcement Learning to reward the model for generating desirable text. Once such a model has been pretrained and instruction fine-tuned, given some starting input, such as a request or a series of patterns, the LLM can generate text which fulfills the request or completes the pattern in a stunningly human-like fashion. We selected and used the GPT-4 LLM based on its superior performance to other LLMs.74
Model training
Here we describe the core ideas and training procedures for the two domain translation models we used within FieldSHIFT: fine-tuned BART and GPT-4.The core idea of the BART-based domain translator is to learn a mapping from one text domain to another. This learning process is illustrated in the top section of Fig. 4. The process for generating new developmental biology text using the trained model is illustrated in the bottom section. During continued fine-tuning of the BART model, we fed two types of text pairs which we call positive and negative samples. Positive samples are neuroscience abstracts/sentences we want to translate into developmental biology abstracts/sentences, and negative samples are developmental biology abstracts/sentences which should remain untouched. This teaches the model to leave existing developmental biology texts as they are. An additional benefit of this strategy is that the model sees more examples of developmental biology text during continued fine-tuning, which comes “for free” as no human-time is required to generate these negative pairs. We sourced both from the domain translation dataset described above.
We used Beam Search with k = 5 beams to generate the next output token from the predicted probabilities of each token given by the decoder at each decoding step. We used the final predicted text sequence and the encoded ground truth text consisting of either the same developmental biology abstract as was provided as input text (for negative samples) or the corresponding output developmental biology text from a human-generated pair (for positive samples) to compute cross-entropy loss, then updated the weights of the BART encoder–decoder over many epochs of training using the AdamW optimizer84 with a learning rate of 0.00005, weight decay of 0.01, batch size per device of 1, and 128 gradient accumulation steps. We selected the final model according to cross-entropy loss on the validation set after 8 epochs of no improvement. The 503 training, 50 validation, and 57 test examples generated via our neuroscience concept split were used to train, validate, and test the model.
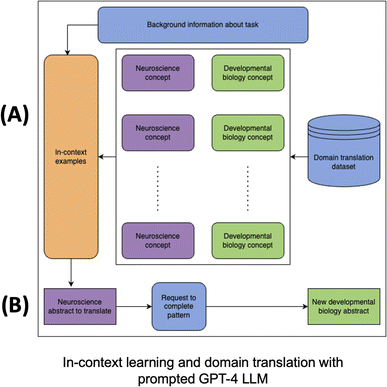
The core idea of the GPT-4-based domain translator is also to learn mappings from neuroscience to developmental biology text. It does this via text generation and in-context learning as shown in Fig. 5. When prompting the model, we provided translation example concept mappings such as those listed below. The full set of concepts was sourced from our domain translation dataset.
• Neuron > Cell
• Neural > Cellular
• Behavior > Morphogenesis
To use the GPT-4 LLM as a domain-translator, we provided in-context examples and asked the LLM to complete the pattern. We did not update the weights of GPT-4, rather we simply ran inference using training examples within the prompt. We sampled these in-context examples from the train concepts of our domain translation dataset and validated the pattern completions on abstracts corresponding to test concepts. In this way, we measured whether GPT-4 can generalize and translate new concepts not seen among the in-context examples. We hypothesized that the very large pretraining set (much of the text on the internet) used to build GPT-4 contains many neuroscience and developmental biology concepts, and thus, the model is able to use information stored within its layers pertaining to these concepts in order to both identify new concepts from the same domain, i.e., a neuroscience concept in the test set but not in the train set, and map these concepts to developmental biology concepts not provided as sample outputs in the in-context examples.
The following illustrates how we tested GPT-4. It includes a very simple way to prompt GPT-4 to translate from neuroscience to developmental biology. It consists of our prompts and GPT responses.
Prompt
You are a translator—you translate neuroscience to developmental biology. There are deep symmetries between these fields and we want to use them to create new hypotheses in developmental biology.The output should read as developmental biology, meaning that any neuroscience concept, such as “hippocampus”, should be translated into the most relevant/related developmental biology term.
Make sure the translation makes sense as developmental biology text and is using real terms that exist in developmental biology literature.
Make sure the translation doesn't include any neuroscience words, such as a part of the brain. Do the best you can to find the most relevant translation.
For example, here are some terms and their translations:
Neuron > Cell
Neural > Cellular
Behavior > Morphogenesis
Millisecond > Minute
Msec > Min
… (link to full prompt below dialogue)
And here's some training data. The “prompt” is the neuroscience and “completion” is the developmental biology.
idx,prompt,completion
156,Live 3D Brain Function Mapping,Live 3D Body Function Mapping
397,How many neuroscientists are there in the world that specialize in human behavior?,How many developmental biologists are there in the world that specialize in human morphogenesis?
… (link to full prompt below dialogue)
Are you ready to translate neuroscience into developmental biology?
Typical GPT response
Yes, I am ready to translate neuroscience into developmental biology. Please provide the text or concepts you would like to translate, and I will do my best to provide a relevant and accurate translation in the field of developmental biology.Prompt
(Developmental biology text here)GPT response
(Translation)Link to full prompt: https://github.com/listenaddress/domain-translator/blob/main/prompt.txt/.
Link to full dataset from which the examples in the prompt were selected: https://huggingface.co/datasets/levinlab/neuroscience-to-dev-bio/tree/main.
We also tested a version of the GPT-4-based domain translator with chain-of-thought prompting and self-critique.85 This approach takes advantage of the chatbot interface available for GPT-4 and asks questions via multiple prompts to arrive at a final translation. We employ self-critique, asking the model to revise its initial translation, to arrive at a final translation from neuroscience to developmental biology text. In the results section, we refer to this variation of the GPT-4 domain translator as GPT-4 with self-critique.
We used the ChatGPT interface for GPT-4 provided by OpenAI with the March 23, 2023, API version. We used the default temperature of 0.7 when generating translations. Note that we used language like “are you ready to translate…” because of OpenAI and Microsoft documentation suggesting that the GPT-4 model is more effective when used in a role-playing fashion, encouraging and preparing the model like one might a human before a test.
Model validation
For each test concept in our domain translation dataset, we searched the term and chose the first abstract that included the term. Specifically, we collected one paper per test concept and selected samples using the following structure in a Google Scholar query: “〈research domain〉” “〈specific topic〉”. For example, to find a paper on neurodegeneration to test, this was our search: “neuroscience” “neurodegeneration”. We then selected the first result. There were five test phrases, such as “Control of anatomy by bioelectric signaling”, “Change of ion channel expression” and “Axonal-dendritic chemical synapses” that didn't yield any exact matches. For those we did a follow up search for the closest matches with a query like this: “neuroscience” “Axonal-dendritic chemical synapses”. We then selected the first result.To judge the quality of the translations we had two professional cell/developmental biologists grade the text. Both annotators graded real developmental biology abstracts and GPT-4 generated abstracts using the self-critique version of the domain translator. Only one annotator graded BART-generated abstracts following initial results suggesting superiority of the GPT-4 approach. When comparing abstracts, the annotators were given the following instructions:
For each abstract you'll consider the following: does this abstract seem interesting, offering a way to think about an aspect of developmental biology or new ideas that could be investigated? The grade can either be a 1 (yes) or a 0 (no).
To sample developmental biology abstracts, we used the same concept-based search approach described above, e.g., searching for “developmental biology” “cellular”. The goal was to provide a baseline understanding of how often developmental biology abstracts are seen as interesting/valuable directions of study. All abstracts were provided without specifying their origin. That is, the annotators did not know which methods generated each abstract they were grading. We report the count of 1 s and 0 s for each method in the results section and P-values for these counts using two-sample T-tests to compare sample proportions.
Bioinformatics
We extracted the whole set of the unique genes related to gene ontologies ‘Developmental process’ (GO:0032502) and ‘Behavior’ (GO:0007610) in humans, Drosophila, the zebrafish (Danio rerio), and the mice (Mus musculus) using Ensembl database and computed the percentage of genes of ‘Behavior’ included in the set of genes annotated as ‘Developmental process’.86,87 As a control, we also computed the overlap between all the pairs of gene ontologies that are the children at level one of the gene ontology ‘Biological process’ (GO:0008150) as a control for each organism, or in other words all the gene ontologies at the same level of ‘Developmental process’. We removed from the set of combinations the gene ontologies that should not be used for annotation of genes (as defined on the Gene ontology website); the removal includes: ‘Regulation of biological processes’ (GO:0050789), ‘Negative regulation of biological process’ (GO:0048519), ‘Positive regulation of biological process’ (GO:0048518), ‘Biological regulation’ (GO:0065007), ‘Response to stimulus’ (GO:0050896), ‘Cellular process’ (GO:0009987), ‘Multicellular organismal process’ (GO:0032501), ‘Metabolic process’ (GO:0008152), ‘Growth’ (GO:0040007), ‘Biological process involved in intraspecies interaction between organisms’ (GO:0051703), and ‘Biological process involved in intraspecies interaction between organisms’ (GO:0044419). We applied a one-sample T-test to compare the distribution of genetic overlap between all pairs of gene ontologies we extracted and the genetic overlap between ‘Developmental process’ and ‘Behavior’ sets of genes. The code for retrieving and analyzing the data can be found at: https://github.com/LPioL/DevCog_GeneticOverlap.Results
Comparing domain translators
A quantitative evaluation of domain translation methods via human annotation suggests the effectiveness of ML-based domain translation. According to both annotators (annotator 1 and 2), the GPT-4-based domain translator with self-critique generated good translations according to the annotation criteria for over half of the neuroscience abstracts provided. The GPT-4 approach performed significantly better than the BART approach (P = 0.017) according to annotator 1 who compared ML methods. Both annotators graded more real abstracts as good than ML-generated abstracts, however, this difference was significant only for annotator 1 (Tables 1 and 2).| Model | Count of good translations (out of 36) | Proportion of good translations | Comparison to real abstracts: P-value | Comparison to BART: P-value | Comparison to GPT-4 with self-critique: P-value |
|---|---|---|---|---|---|
| BART | 11 | 0.306 | <0.001 | 1.000 | 0.017 |
| GPT-4 with self-critique | 21 | 0.583 | 0.001 | 0.017 | 1.000 |
| Real | 33 | 0.917 | 1.000 | <0.001 | 0.001 |
| Model | Count of good translations (out of 36) | Proportion of good translations | Comparison to real abstracts: P-value | Comparison to GPT-4 with self-critique: P-value |
|---|---|---|---|---|
| GPT-4 with self-critique | 19 | 0.528 | 0.641 | 1.000 |
| Real | 21 | 0.583 | 1.000 | 0.641 |
While there were more false positives when generating abstracts with ML, the approach is far more scalable than humans writing abstracts. The GPT-4-based domain translator performed best of the two ML approaches tested and can output a new abstract in about five seconds. Compared to human-written abstracts, generalizing from our results, the chance of producing a good abstract is reduced by about 39%. We compute this as 1 − 20/33 where we average the 21 and 19 good translations from GPT-4 from each annotator. However, assuming these abstracts can be quickly verified and that they are sometimes very interesting and very different from humans in their content, we believe this is an incredibly useful system, enabling the generation of thousands of scientific hypotheses within a single day.
Compute budget and limited human oversight (involving less work than writing abstracts) become the main constraints on hypothesis generation using this ML-based approach to domain translation. Additional constraints may include the diversity of input papers and an upper bound on the total number of symmetries between domains of which GPT-4 is aware. While these claims warrant further research and our results represent initial findings, the value of ML-based domain translation is promising given the inherent ability to scale.
The inter-annotator agreement for the combined set of GPT-4 with self-critique and real abstracts was 0.639 with Cohen's Kappa 0.235. The low Cohen's Kappa indicates the subjectivity of this exercise, even when grading real developmental biology abstracts. Future work may look to increase the number of annotators or annotations for comparing domain translation methods (Table 3).
| Agent | Count of good translations (out of 36) | Proportion of good translations |
|---|---|---|
| GPT-4 | 21 | 0.583 |
| GPT-4 with self-critique | 21 | 0.583 |
We found no difference between GPT-4 and GPT-4 with self-critique according to an evaluation from the first annotator. It's possible future work on the series of prompts used for self-critique could improve model performance. We leave this as future work.
Examples of abstracts generated
Examining some specific cases, BART and GPT performed the best when translating words in or near the space of words they'd seen before. Specific neuroscience terms that often don't have clear translations such as “trigeminal autonomic cephalalgias” would often be erroneously translated or not translated at all. One of the most interesting translations was the following:This is a very interesting translation, because it suggests the use of a predictive coding model (a predictive gradient) to understand progressive morphogenesis.88,89 This is testable, as biophysical or biochemical representations of cell expectations (homeostatic setpoints, in the sense of priors90–96) can be looked for experimentally.
To illustrate the abilities and struggles of the two domain translation models, below are three translation examples, from easy to difficult. Both approaches correctly translated the first, only GPT-4 correctly translated the second, and neither correctly translated the third.
“Neurogeneration” test – easy
“Nervous system” test – medium
“Autonomic reflexes” test – hard
The examples above show BART and GPT-4 have an easier time translating words they've seen in the training data than esoteric neuroscience words they haven't seen. They also show the kinds of translations that set GPT-4 apart in our experiments. For example, in the medium case, GPT-4 translated “cortex” into “tissue organization”, while our BART model didn't recognize “cortex” as a neuroscience word and attempt to translate it.
Hypothesis testing: molecular conservation between cognition and morphogenesis
One clear hypothesis that can be generated by FieldSHIFT is that the molecular mechanisms behind developmental bioelectricity and that of cognitive/behavioral phenomena should be conserved. That is, the same genes should be found as implicated in both roles; this is novel because only the idea of “morphogenesis as behavior in anatomical space”37,56 predicts any association between genes across what are normally thought of as two very distinct fields of biology. Interestingly, GPT (August 3rd version of GPT-4) itself generated this hypothesis after doing all the translations and being asked to come up with a new idea as follows:Input: (Base input as described above) You are a translator—you translate neuroscience to developmental biology…
Input: Let's start by generating a hypothesis based on everything you know about developmental biology and genetics; instead of making a translation, generate a novel and testable hypothesis about the genes involved in learning and morphogenesis.
Reply: There exists a set of evolutionarily conserved genes that are activated both during early morphogenetic processes and later in the organism's life during learning and memory formation. These genes mediate cellular plasticity mechanisms, which are vital for both the formation of complex body structures during embryonic development and the flexibility and adaptability of mature organisms to new environmental stimuli.
Thus, we next decided to test this prediction using bioinformatics, focusing for the purposes of this study on the genes thought to underlie phenomena in behavioral science and developmental biology.
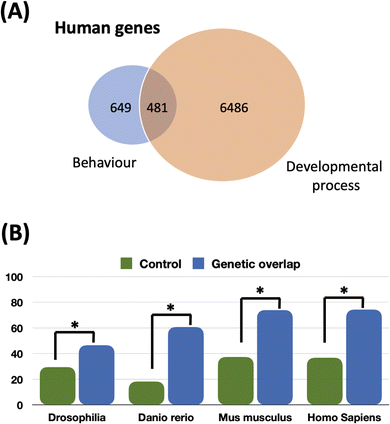
We computed the genetic overlap between the set of genes annotated as ‘Behaviour’ and the set of genes annotated as ‘Developmental process’ in different species widely dispersed within the phylogenetic tree including Drosophila, zebrafish, mice and homo sapiens. We found that 74.1% of the ‘Behavior’ genes (649 genes) are included in the ‘Developmental process’ set of genes (6486 genes). From Drosophila to humans, this genetic overlap is increasing: 46.5% for Drosophila, 60.6% for the zebrafish, 73.8% for the mouse and 74.1% for humans (see Fig. 6A and B). We applied a one-sample T-test to compare the distribution of genetic overlap between all pairs of gene ontologies we extracted and the genetic overlap between ‘Developmental process’ and ‘Behavior’ sets of genes for each organism. Statistical analysis of overlap among all pairs of gene ontologies (at the same level of ‘Developmental process’) for these four organisms is significantly smaller (p < 0.05) than the overlap we observed among the predictive categories of development and behavior. Thus, we conclude that during evolution there is an increase of the genetic overlap between ‘Behavior’ and ‘Developmental process’. In other words, genes used for development and morphogenesis have been co-opted for cognition. The mapping between behavior and development/morphogenesis can also be seen at the genetic level.
Discussion
Machine learning for domain translation
We proposed FieldSHIFT, a framework for domain translation. To implement FieldSHIFT, we built a domain translation dataset and compared two ML approaches for domain translation using this data and expert evaluation of the translations. Expert evaluation identified that the GPT-4-based approach produced good translations, leading to a useful hypothesis or insight about developmental biology, in about 20 out of every 33 tries. This suggests that new hypotheses in developmental biology can be produced at scale in an automated fashion, using humans to review and extend the initial ideas. We found that curating a high-quality dataset and designing prompts was the bulk of the work for making the system effectively translate between domains.While the GPT-4-based approach which used in-context learning performed better than the BART-based approach, the recent public release of open-source LLMs such as Llama 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 97 and Falcon98 present opportunities to perform domain translation by fine-tuning on arbitrarily many examples of translated text. BART is an early LM for text generation, though its size makes it a cost-effective choice compared to fine-tuning a large, open-source LLM. Still, because the exact implementation of the GPT-4 LLM is not publicly disclosed, and because fine-tuning can provide greater control and performance on specific tasks than in-context learning, future efforts should consider the use of open LLM architectures for domain translation. We note also that multimodal strategies for training and applying LLMs to a mix of image and text data could be valuable for domain translation. In addition to example abstracts or articles, researchers could include images or videos pertaining to experiments or experimental findings as input to a system like FieldSHIFT, or a domain translator could output images or video clips in addition to text as literal visions of scientific hypothesis and their potential outcomes.
97 and Falcon98 present opportunities to perform domain translation by fine-tuning on arbitrarily many examples of translated text. BART is an early LM for text generation, though its size makes it a cost-effective choice compared to fine-tuning a large, open-source LLM. Still, because the exact implementation of the GPT-4 LLM is not publicly disclosed, and because fine-tuning can provide greater control and performance on specific tasks than in-context learning, future efforts should consider the use of open LLM architectures for domain translation. We note also that multimodal strategies for training and applying LLMs to a mix of image and text data could be valuable for domain translation. In addition to example abstracts or articles, researchers could include images or videos pertaining to experiments or experimental findings as input to a system like FieldSHIFT, or a domain translator could output images or video clips in addition to text as literal visions of scientific hypothesis and their potential outcomes.
Bioelectricity: conserved mechanisms and algorithms between control of body and behavior
It has been previously hypothesized that the algorithms of behavior (active inference, perceptual control, collective decision-making, memory and learning, navigation of problem spaces, preferences, pursuit of goal states with degrees of flexible response, etc.) are used to direct morphogenesis and may even derive from it evolutionarily.55,56,99 Here, we produce a tool that can be used to explore the symmetries and commonalities between these two fields, and in fact any other two fields. By translating abstracts of papers from one set of vocabulary into another, we generate new ways to think about data and potential new hypotheses. We show here that this works in the case of the parallels between behavioral science and the science of morphogenesis, and generated a specific new hypothesis: not only the algorithms, but the molecular mechanisms (genes) are also conserved. While empirical (functional) testing will be needed to conclusively test this idea, we performed bioinformatics experiments that generated new data supporting the hypothesis: implications of the same genes between the topics of morphogenesis and cognition occurs at a frequency much greater than between other random categories of genes. This finding thus suggests a number of genes and their products to be tested in novel morphogenetic assays using genetic and pharmacological tools. This work is currently under way in our lab.Latent space of scientific papers
We believe that this work is just the beginning of the AI-guided exploration of the latent space around scientific papers, in the sense of the “adjacent possible”.100,101 Each real scientific paper in the literature provides access to an associated set of possible papers in which one or more aspects are changed – in effect, exploring various symmetries of concepts in specific problem spaces. These papers are not meant to be taken literally, since they do not provide real-world data, but instead as tools to spur creativity, provide testable novel hypotheses, suggest studies to be carried out, and perhaps most importantly, by reflecting approaches from specific studies into different disciplines, dissolve barriers between fields and knowledge silos. These “virtual” papers are not published in scientific journals (which should be reserved for studies that have actually been carried out), but are accessible via machine learning and represent another useful repository (whose goal is to advance creativity and novel thinking – a second-order function complementing first order traditional papers containing specific knowledge and data). While it is essential for the scientific literature to be clearly demarcated as to whether a study was actually done or is only hypothetical, we believe that maintaining artificial barriers between the creative ideation and hypothesis testing phases of science is counterproductive. An early example of this realization came from the James V. McConnell (discoverer of the ability of memories to move through tissues102,103), who edited a journal called The Journal of Biological Psychology which contained peer-reviewed primary papers in one half of each issue, while the second half (printed up-side-down) contained perspectives, drawings, poetry, etc.Limitations of the study and next steps
One limitation of the study is the number of papers we were able to manually curate; the machine learning will improve as more papers are added in the future. Especially with the rapid development in large language models, we can expect significant improvements in the quality of output in the future. While a number of ways exist in which machine learning may disrupt current scientific structures,104 the platform described herein can be safely utilized to generate candidate research programs, as long as it is clearly kept in mind that the system we describe is not claimed to generate truths by analysis of existing data; it is a system for generating novel hypotheses that must be empirically tested.While the current evaluation is based on human assessments of the quality of translations between the fields of neuroscience and developmental biology, we acknowledge that experimental validation of generated hypotheses would be necessary to fully realize the power of our method. That is, a complete evaluation of FieldSHIFT would require conducting experiments to address hypotheses from our domain translator and assessing the value of the findings relative to human-generated experimental directions. Not only the findings of the experiments but also the methods, complexity, and feasibility of testing FieldSHIFT-generated hypotheses should be compared to human-directed research. Work of this kind is already underway at the Levin Lab, and we look forward to sharing these results in future work.
Conclusion
The increased overlap we observed between genes implicated in morphogenesis and behavior has a number of implications for experimental work on the evolution and biomedical targeting of many of those genes. More broadly, our approach has identified a method for generating testable hypotheses in the space of developmental bioelectricity, which has implications for birth defects, traumatic injury, and cancer.57,105–107 Future work will incorporate this kind of workflow into machine science pipelines, and empirically test the creative products of exploring the structures of concepts among diverse fields of inquiry. It is clear however that this approach in exploring the latent space around existing scientific literature extends far beyond bioelectricity and can be applied to numerous domains. Machine learning and AI are poised to significantly potentiate every aspect of research.20,21,108–110 One of the critical roles will be not as mere number crunching, but as an essential aid to the first key step: hypothesis generation. We foresee extremely fruitful collaborations between naturally evolved (human scientists) and engineered (AI) scientists.Data availability
Data and processing scripts for this paper are available at Hugging Face, at https://huggingface.co/datasets/levinlab/neuroscience-to-dev-bio/tree/main.Author contributions
T. O'B.: conceptualization, data curation, formal analysis, investigation, software, writing. J. S.: conceptualization, data curation, formal analysis, investigation, software, writing. L. P.-L.: conceptualization, formal analysis, investigation, software. P. M.: data curation, validation. C. R.-I.: data curation, validation. M. L.: conceptualization, formal analysis, investigation, writing, project administration, supervision, funding acquisition.Conflicts of interest
M. L. is a scientific co-founder of two companies (Morphoceuticals, and Astonishing Labs) which explore the symmetries between neuroscience and developmental biology. J. S. is an employee of Optum, a healthcare technology company.Acknowledgements
We thank Giovanni Pezzulo for helpful discussions, and Juanita Mathews for manually rating AI-derived text. We also thank Julia Poirier for assistance with the manuscript. M. L. gratefully acknowledges support via Grant 62212 from the John Templeton Foundation. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation.References
- J. R. Platt, Strong Inference: Certain systematic methods of scientific thinking may produce much more rapid progress than others, Science, 1964, 146(3642), 347–353 CrossRef CAS PubMed.
- D. L. Jewett, What's wrong with single hypotheses?: Why it is time for Strong-Inference-PLUS, Scientist, 2005, 19(21), 10 Search PubMed.
- R. H. Davis, Strong Inference: rationale or inspiration?, Perspect. Biol. Med., 2006, 49(2), 238–250 CrossRef PubMed.
- V. T. DeVita Jr, Strong inference, Nat. Clin. Pract. Oncol., 2008, 5(4), 177 CrossRef PubMed.
- V. T. DeVita Jr, More on strong inference, Nat. Clin. Pract. Oncol., 2008, 5(5), 239 CrossRef PubMed.
- D. A. Beard and M. J. Kushmerick, Strong inference for systems biology, PLoS Comput. Biol., 2009, 5(8), e1000459 CrossRef PubMed.
- D. S. Fudge, Fifty years of J. R. Platt's strong inference, J. Exp. Biol., 2014, 217(Pt 8), 1202–1204 CrossRef PubMed.
- D. Bohm and L. Nichol, On creativity, Routledge, London, New York, 1998, vol. xxiv, p. 125 Search PubMed.
- G. Pólya, How to solve it; a new aspect of mathematical method, Doubleday, Garden City, N.Y., 2nd edn Doubleday anchor books, 1957, p. 253 Search PubMed.
- C. D. Stern, Reflections on the past, present and future of developmental biology, Dev. Biol., 2022, 488, 30–34 CrossRef CAS PubMed.
- M. Bizzari, et al., A call for a better understanding of causation in cell biology, Nat. Rev. Mol. Cell Biol., 2019, 20(5), 261–262 CrossRef CAS PubMed.
- B. K. Forscher, Chaos in the Brickyard, Science, 1963, 142(3590), 339 CrossRef CAS PubMed.
- Y. Gil, et al., Amplify scientific discovery with artificial intelligence, Science, 2014, 346, 171–172 CrossRef CAS PubMed.
- A. M. Deiana, et al., Applications and Techniques for Fast Machine Learning in Science, Front. Big Data, 2022, 5, 787421 CrossRef PubMed.
- I. Chattopadhyay and H. Lipson, Data smashing: uncovering lurking order in data, J. R. Soc., Interface, 2014, 11(101), 20140826 CrossRef PubMed.
- A. D. J. van Dijk, et al., Machine learning in plant science and plant breeding, iScience, 2021, 24(1), 101890 CrossRef PubMed.
- J. Westermayr, et al., Perspective on integrating machine learning into computational chemistry and materials science, J. Chem. Phys., 2021, 154(23), 230903 CrossRef CAS PubMed.
- S. Zhong, et al., Machine Learning: New Ideas and Tools in Environmental Science and Engineering, Environ. Sci. Technol., 2021, 55(19), 12741–12754 CAS.
- E. Gianti and S. Percec, Machine Learning at the Interface of Polymer Science and Biology: How Far Can We Go?, Biomacromolecules, 2022, 23(3), 576–591 CrossRef CAS PubMed.
- A. Sparkes, et al., Towards Robot Scientists for autonomous scientific discovery, Autom. Exp., 2010, 2, 1 CrossRef PubMed.
- L. N. Soldatova, et al., An ontology for a Robot Scientist, Bioinformatics, 2006, 22(14), e464–e471 CrossRef CAS PubMed.
- R. D. King, et al., Functional genomic hypothesis generation and experimentation by a robot scientist, Nature, 2004, 427(6971), 247–252 CrossRef CAS PubMed.
- D. Lobo, et al., Limbform: a functional ontology-based database of limb regeneration experiments, Bioinformatics, 2014, 30(24), 3598–3600 CrossRef CAS PubMed.
- D. Lobo, et al., A bioinformatics expert system linking functional data to anatomical outcomes in limb regeneration, Regeneration, 2014, 1(2), 37–56 CrossRef PubMed.
- D. Lobo, T. J. Malone and M. Levin, Planform: an application and database of graph-encoded planarian regenerative experiments, Bioinformatics, 2013, 29(8), 1098–1100 CrossRef CAS PubMed.
- D. Lobo, T. J. Malone and M. Levin, Towards a bioinformatics of patterning: a computational approach to understanding regulative morphogenesis, Biol. Open, 2013, 2(2), 156–169 CrossRef PubMed.
- P. Villoutreix, What machine learning can do for developmental biology, Development, 2021, 148(1), dev188474 CrossRef CAS PubMed.
- B. C. Feltes, et al., Perspectives and applications of machine learning for evolutionary developmental biology, Mol. Omics, 2018, 14(5), 289–306 RSC.
- D. Lobo and M. Levin, Inferring Regulatory Networks from Experimental Morphological Phenotypes: A Computational Method Reverse-Engineers Planarian Regeneration, PLoS Comput. Biol., 2015, 11(6), e1004295 CrossRef PubMed.
- D. Lobo, J. Morokuma and M. Levin, Computational discovery and in vivo validation of hnf4 as a regulatory gene in planarian regeneration, Bioinformatics, 2016, 32(17), 2681–2685 CrossRef CAS PubMed.
- S. Asche, et al., A robotic prebiotic chemist probes long term reactions of complexifying mixtures, Nat. Commun., 2021, 12(1), 3547 CrossRef CAS PubMed.
- B. Burger, et al., A mobile robotic chemist, Nature, 2020, 583(7815), 237–241 CrossRef CAS PubMed.
- A. Vasilevich and J. de Boer, Robot-scientists will lead tomorrow's biomaterials discovery, Curr. Opin. Biomed. Eng., 2018, 6, 74–80 CrossRef.
- G. Pezzulo and M. Levin, Top-down models in biology: explanation and control of complex living systems above the molecular level, J. R. Soc., Interface, 2016, 13(124) CrossRef PubMed.
- G. Pezzulo and M. Levin, Re-membering the body: applications of computational neuroscience to the top-down control of regeneration of limbs and other complex organs, Integr. Biol., 2015, 7(12), 1487–1517 CrossRef CAS PubMed.
- M. Levin and C. J. Martyniuk, The bioelectric code: An ancient computational medium for dynamic control of growth and form, Biosystems, 2018, 164, 76–93 CrossRef PubMed.
- M. Levin, Bioelectric networks: the cognitive glue enabling evolutionary scaling from physiology to mind, Anim. Cognit., 2023 Search PubMed.
- M. P. van den Heuvel, L. H. Scholtens and R. S. Kahn, Multiscale Neuroscience of Psychiatric Disorders, Biol. Psychiatry, 2019, 86(7), 512–522 CrossRef PubMed.
- D. Marr, Vision: a computational investigation into the human representation and processing of visual information, W.H. Freeman, San Francisco, 1982. vol. xvii, p. 397 Search PubMed.
- D. Marr and E. Hildreth, Theory of edge detection, Proc. R. Soc. London, Ser. B, 1980, 207(1167), 187–217 CAS.
- B. Sengupta, et al., Towards a Neuronal Gauge Theory, PLoS Biol., 2016, 14(3), e1002400 CrossRef PubMed.
- H. S. Burr and F. S. C. Northrop, The electro-dynamic theory of life, Q. Rev. Biol., 1935, 10(3), 322–333 CrossRef CAS.
- E. J. Lund, Bioelectric fields and growth, Univ. of Texas Press, Austin, 1947, vol. xii, p. 391 Search PubMed.
- R. Nuccitelli, K. Robinson and L. Jaffe, On electrical currents in development, Bioessays, 1986, 5(6), 292–294 CrossRef CAS PubMed.
- E. Bates, Ion Channels in Development and Cancer, Annu. Rev. Cell Dev. Biol., 2015, 31, 231–247 CrossRef CAS PubMed.
- M. P. Harris, Bioelectric signaling as a unique regulator of development and regeneration, Development, 2021, 148(10), dev180794 CrossRef CAS PubMed.
- M. Levin, Bioelectric signaling: Reprogrammable circuits underlying embryogenesis, regeneration, and cancer, Cell, 2021, 184(4), 1971–1989 CrossRef CAS PubMed.
- M. Levin, The Computational Boundary of a “Self”: Developmental Bioelectricity Drives Multicellularity and Scale-Free Cognition, Front. Psychol., 2019, 10(2688), 2688 CrossRef PubMed.
- M. Levin, Technological Approach to Mind Everywhere: An Experimentally-Grounded Framework for Understanding Diverse Bodies and Minds, Front. Syst. Neurosci., 2022, 16, 768201 CrossRef PubMed.
- C. Y. Yang, et al., Encoding Membrane-Potential-Based Memory within a Microbial Community, Cell Syst., 2020, 10(5), 417–423 CrossRef CAS PubMed.
- R. Martinez-Corral, et al., Metabolic basis of brain-like electrical signalling in bacterial communities, Philos. Trans. R. Soc., B, 2019, 374(1774), 20180382 CrossRef CAS PubMed.
- A. Prindle, et al., Ion channels enable electrical communication in bacterial communities, Nature, 2015, 527(7576), 59–63 CrossRef CAS PubMed.
- J. W. Larkin, et al., Signal Percolation within a Bacterial Community, Cell Syst., 2018, 7(2), 137–145 CrossRef CAS PubMed.
- J. Liu, et al., Coupling between distant biofilms and emergence of nutrient time-sharing, Science, 2017, 356(6338), 638–642 CrossRef CAS PubMed.
- C. Fields, J. Bischof and M. Levin, Morphological Coordination: A Common Ancestral Function Unifying Neural and Non-Neural Signaling, Physiology, 2020, 35(1), 16–30 CrossRef PubMed.
- C. Fields and M. Levin, Competency in Navigating Arbitrary Spaces as an Invariant for Analyzing Cognition in Diverse Embodiments, Entropy, 2022, 24(6), e2406089 CrossRef PubMed.
- J. Mathews and M. Levin, The body electric 2.0: recent advances in developmental bioelectricity for regenerative and synthetic bioengineering, Curr. Opin. Biotechnol., 2018, 52, 134–144 CrossRef CAS PubMed.
- S. Biswas, et al., Gene Regulatory Networks Exhibit Several Kinds of Memory: Quantification of Memory in Biological and Random Transcriptional Networks, iScience, 2021, 24(3), 102131 CrossRef CAS PubMed.
- K. G. Sullivan, M. Emmons-Bell and M. Levin, Physiological inputs regulate species-specific anatomy during embryogenesis and regeneration, Commun. Integr. Biol., 2016, 9(4), e1192733 CrossRef PubMed.
- M. Levin, Endogenous bioelectrical networks store non-genetic patterning information during development and regeneration, J. Physiol., 2014, 592(11), 2295–2305 CrossRef CAS PubMed.
- C. G. Langton, Artificial life: an overview. Complex adaptive systems, MIT Press, Cambridge, Mass, 1995, vol. xi, p. 340, [6] of plates Search PubMed.
- M. Buehler, MechGPT, a Language-Based Strategy for Mechanics and Materials Modeling that Connects Knowledge Across Scales, Disciplines, and Modalities, Appl. Mech. Rev., 2023, 1–82 CAS.
- M. J. Buehler, Unsupervised cross-domain translation via deep learning and adversarial attention neural networks and application to music-inspired protein designs, Patterns, 2023, 4(3), 100692 CrossRef CAS PubMed.
- F. Ye, et al., Cross-domain Knowledge Discovery based on Knowledge Graph and Patent Mining, J. Phys.: Conf. Ser., 2021, 1744(4), 042155 CrossRef.
- D. S. Adams and M. Levin, Endogenous voltage gradients as mediators of cell-cell communication: strategies for investigating bioelectrical signals during pattern formation, Cell Tissue Res., 2013, 352(1), 95–122 CrossRef CAS PubMed.
- M. Levin, et al., Left/right patterning signals and the independent regulation of different aspects of Situs in the chick embryo, Dev. Biol., 1997, 189(1), 57–67 CrossRef CAS PubMed.
- M. Levin, et al., A molecular pathway determining left-right asymmetry in chick embryogenesis, Cell, 1995, 82(5), 803–814 CrossRef CAS PubMed.
- S. Sundelacruz, M. Levin and D. L. Kaplan, Role of membrane potential in the regulation of cell proliferation and differentiation, Stem Cell Rev. Rep., 2009, 5(3), 231–246 CrossRef PubMed.
- D. J. Blackiston, K. A. McLaughlin and M. Levin, Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle, Cell Cycle, 2009, 8(21), 3519–3528 CrossRef PubMed.
- M. Levin and M. Mercola, Gap junctions are involved in the early generation of left-right asymmetry, Dev. Biol., 1998, 203(1), 90–105 CrossRef CAS PubMed.
- M. Lewis, et al., BART: Denoising Sequence-to-Sequence Pre-training for Natural Language Generation, Translation, and Comprehension, in 58th Annual Meeting of the Association for Computational Linguistics, 2020 Search PubMed.
- A. Vaswani, et al., Attention Is All You Need, in 31st Conference on Neural Information Processing Systems (NIPS 2017), Curran Associates Inc, Long Beach, CA, 2017 Search PubMed.
- Q. Dong, et al., A Survey on In-Context Learning, arXiv, 2023, preprint, arXiv:2301.00234v3, DOI:10.48550/arXiv.2301.00234.
- OpenAI, GPT-4 Technical Report, arXiv, 2023, preprint, arXiv.2303.08774, DOI:10.48550/arXiv.2303.08774.
- T. B. Brown, et al., Language Models are Few-Shot Learners, in 34th Conference on Neural Information Processing Systems (NeurIPS 2020), 2020 Search PubMed.
- A. Wang, et al., SuperGLUE: A Stickier Benchmark for General-Purpose Language Understanding Systems, in 33rd International Conference on Neural Information Processing Systems, Vancouver, Canada, 2019 Search PubMed.
- J. Devlin, et al., BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding, in 17th Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Minneapolis, MN, 2019 Search PubMed.
- Z. Tan, et al., Neural machine translation: A review of methods, resources, and tools, AI Open, 2020, 1, 5–21 CrossRef.
- M. Freitag and Y. Al-Onaizan, Beam Search Strategies for Neural Machine Translation, in First Workshop on Neural Machine Translation, Association for Computational Linguistics, Vancouver, Canada, 2017 Search PubMed.
- T. Wolf, et al., Transformers: State-of-the-Art Natural Language Processing, in Conference on Empirical Methods in Natural Language Processing: System Demonstrations, 2020 Search PubMed.
- A. Radford, et al., Language Models are Unsupervised Multitask Learners, OpenAI, 2019 Search PubMed.
- L. Ouyang, et al., Training Language Models to Follow Instructions with Human Feedback, in 36th Conference on Neural Information Processing Systems (NeurIPS 2022), 2022, New Orleans, LA Search PubMed.
- J. Kaplan, et al., Scaling Laws for Neural Language Models, arXiv, 2020, preprint, arXiv.2001.08361, DOI:10.48550/arXiv.2001.08361.
- I. Loshchilov and F. Hutter, Decoupled Weight Decay Regularization, in 5th International Conference on Learning Representations, International Conference on Learning Representations (ICLR), Toulon, France, 2017 Search PubMed.
- J. Wei, et al., Chain-of-Thought Prompting Elicits Reasoning in Large Language Models, arXiv, 2023, preprint, arXiv.2201.11903, DOI:10.48550/arXiv.2201.11903.
- The Gene Ontology Consortium, The Gene Ontology resource: enriching a GOld mine, Nucleic Acids Res., 2021, 49(D1), D325–D334 CrossRef PubMed.
- M. Ashburner, et al., Gene ontology: tool for the unification of biology. The Gene Ontology Consortium, Nat. Genet., 2000, 25(1), 25–29 CrossRef CAS PubMed.
- G. Dodig-Crnkovic, Cognition as Morphological/Morphogenetic Embodied Computation In Vivo, Entropy, 2022, 24(11) CrossRef PubMed.
- K. Friston, et al., Knowing one’s place: a free-energy approach to pattern regulation, J. R. Soc., Interface, 2015, 12(105), 20141383 CrossRef PubMed.
- L. Pio-Lopez, et al., The scaling of goals from cellular to anatomical homeostasis: an evolutionary simulation, experiment and analysis, Interface Focus, 2023, 13(3), 20220072 CrossRef PubMed.
- L. Pio-Lopez, et al., Active inference, morphogenesis, and computational psychiatry, Front. Comput. Neurosci., 2022, 16, 988977 CrossRef PubMed.
- D. A. Friedman, et al., Active Inferants: An Active Inference Framework for Ant Colony Behavior, Front. Behav. Neurosci., 2021, 15, 647732 CrossRef PubMed.
- G. Pezzulo, F. Rigoli and K. J. Friston, Hierarchical Active Inference: A Theory of Motivated Control, Trends Cognit. Sci., 2018, 22(4), 294–306 CrossRef PubMed.
- M. Kirchhoff, et al., The Markov blankets of life: autonomy, active inference and the free energy principle, J. R. Soc., Interface, 2018, 15(138), 20171792 CrossRef PubMed.
- A. Constant, et al., A variational approach to niche construction, J. R. Soc., Interface, 2018, 15(141), 20170685 CrossRef PubMed.
- M. Allen and K. J. Friston, From cognitivism to autopoiesis: towards a computational framework for the embodied mind, Synthese, 2018, 195(6), 2459–2482 CrossRef PubMed.
- H. Touvron, et al., Llama 2: Open Foundation and Fine-Tuned Chat Models, arXiv, 2023, preprint, arXiv:2307.09288, DOI:10.48550/arXiv.2307.09288.
- G. Penedo, et al., The RefinedWeb Dataset for Falcon LLM: Outperforming Curated Corpora with Web Data, and Web Data Only, arXiv, 2023, preprint, arXiv:2306.01116, DOI:10.48550/arXiv.2306.01116.
- S. Grossberg, Communication, Memory, and Development, in Progress in Theoretical Biology, ed. R. Rosen and F. Snell, 1978 Search PubMed.
- O. Witkowski and T. Ikegami, How to Make Swarms Open-Ended? Evolving Collective Intelligence Through a Constricted Exploration of Adjacent Possibles, Artif. Life, 2019, 25(2), 178–197 CrossRef PubMed.
- S. A. Kauffman, The origins of order : self organization and selection in evolution, Oxford University Press, New York, 1993, vol. xviii, p. 709 Search PubMed.
- J. V. McConnell, A. L. Jacobson and D. P. Kimble, The effects of regeneration upon retention of a conditioned response in the planarian, J. Comp. Physiol. Psychol., 1959, 52, 1–5 CAS.
- J. V. McConnell and J. M. Shelby, Memory transfer experiments in invertebrates, in Molecular mechanisms in memory and learning, ed. G. Ungar, Plenum Press, New York, 1970, pp. 71–101 Search PubMed.
- E. Gibney, Could machine learning fuel a reproducibility crisis in science?, Nature, 2022, 608(7922), 250–251 CrossRef CAS PubMed.
- L. Pio-Lopez and M. Levin, Morphoceuticals: Perspectives for discovery of drugs targeting anatomical control mechanisms in regenerative medicine, cancer and aging, Drug Discovery Today, 2023, 28(6), 103585 CrossRef CAS PubMed.
- J. Mathews, et al., Cellular signaling pathways as plastic, proto-cognitive systems: Implications for biomedicine, Patterns, 2023, 4(5), 100737 CrossRef PubMed.
- E. Lagasse and M. Levin, Future medicine: from molecular pathways to the collective intelligence of the body, Trends Mol. Med., 2023 Search PubMed.
- D. Qi, et al., An ontology for description of drug discovery investigations, J. Integr. Bioinform., 2010, 7(3), 126 Search PubMed.
- R. D. King, et al., The automation of science, Science, 2009, 324(5923), 85–89 CrossRef CAS PubMed.
- J. Grizou, et al., A curious formulation robot enables the discovery of a novel protocell behavior, Sci. Adv., 2020, 6(5), eaay4237 CrossRef CAS PubMed.
- D. Lianghao and B. Margarete, How much sharing is enough? cognitive patterns in building interdisciplinary collaborations, in Social Network Analysis, CRC Press, 2017 Search PubMed.
- H. Y. Lu, et al., Multi-scale neural decoding and analysis, J. Neural. Eng., 2021, 18(4), 045013 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |