Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The O2-stable [FeFe]-hydrogenase CbA5H reveals high resilience against organic solvents†
Martin
Gerbaulet
 ,
Anja
Hemschemeier
,
Anja
Hemschemeier
 and
Thomas
Happe
and
Thomas
Happe
 *
*
Faculty of Biology and Biotechnology, Photobiotechnology, Ruhr University Bochum, Universitätsstr. 150, 44801 Bochum, Germany. E-mail: thomas.happe@ruhr-uni-bochum.de
First published on 7th November 2024
Abstract
[FeFe]-Hydrogenases are highly efficient hydrogen-(H2) converting enzymes which play pivotal roles for H2 cycling in natural habitats, but which are also of interest for sustainable approaches to generate or employ H2 gas. [FeFe]-Hydrogenases harbor a unique active site metal cofactor, the H-cluster, whose di-iron site by itself is nearly inactive but, as part of the protein, allows high turnover rates. Understanding this essential interplay of protein and co-factor might help to install [FeFe]-hydrogenases in biotechnological applications. The catalytic unit of the H-Cluster can be synthesized chemically and incorporated into [FeFe]-hydrogenase precursors, which allows to introduce non-natural metals or ligands and study their impact on catalytic activity. However, these compounds are often not water-soluble and have to be added to the proteins in solvents known to destabilize polypeptides. The resilience of [FeFe]-hydrogenases against organic solvents has hardly been investigated. To address this knowledge gap, we characterized the stability of the [FeFe]-hydrogenase CbA5H from Clostridium beijerinckii in several organic solvents (dimethylsulfoxide (DMSO), acetone, acetonitrile as well as water-miscible short-chain alcohols). These solvents are required to dissolve co-factor analogues and are also employed in chemical syntheses that might be combined with biocatalysts such as hydrogenases for more sustainable industrial processes. In the medium time-frame, CbA5H is remarkably stable in high concentrations of acetone and acetonitrile and also withstands intermediate concentrations of DMSO, ethanol and methanol. Combined with the unusual O2 stability and high temperature and pressure tolerance, this makes CbA5H a candidate for its use in non-aqueous reaction environments.
Introduction
Hydrogenases are redox enzymes that reversibly interconvert protons, electrons and molecular hydrogen (H2). They are grouped into [Fe]-, [NiFe]-, and [FeFe]-hydrogenases depending on the metal ions they employ in their catalytic center.1 [FeFe]-hydrogenases usually show the highest turn-over rates.2,3 These enzymes employ a biologically unique iron sulfur (FeS-) cluster combination as cofactor termed the H-cluster. This cluster comprises a cubane [4Fe–4S] cluster ([4Fe]H) coupled through a coordinating cysteine residue to a special diiron cluster ([2Fe]H). The two Fe-ions of [2Fe]H, termed proximal and distal with respect to [4Fe]H, are bridged by an azodithiolate moiety and are further coordinated by three CO and two CN− ligands.4,5 Catalysis takes place at the open coordination site at the distal Fe ion, which can also be targeted by inhibitors. Several known inhibitors are reversible, such as gaseous carbon monoxide (CO)6 and formaldehyde,7–9 whereas molecular oxygen (O2) destroys most [FeFe]-hydrogenases irreversibly.10,11 In living organisms, the [4Fe]H, sub-cluster of the H-cluster is believed to be assembled by the house-keeping Fe–S cluster biosynthetic machinery, whereas [2Fe]H is built and integrated into a hydrogenase precursor already containing [4Fe]H by the dedicated maturases HydE, HydF and HydG.12 However, the [2Fe]H sub-cluster can also be synthesized chemically, and this chemical mimic can be incorporated into a [4Fe]H-containing [FeFe]-hydrogenase.13,14 This procedure allows detailed studies on the interaction of cofactor and hydrogenase polypeptide.15,16 In addition, many variants of [2Fe]H have been synthesized and integrated into [FeFe]-hydrogenase precursors with the aim of studying, for example, the effects of different Fe ion ligands or even different metals.17–19Although limited by their oxygen lability, [FeFe]-hydrogenases possess high biotechnological potential. Their high efficiency, combined with reliance on abundant and readily available resources (Fe and S) are a possible alternative to platinum for industrial hydrogen production.20 To date, these enzymes have not been implemented into industrial applications of an economic scale, but much research is being done to allow this to happen. One approach is to develop miniature models of [FeFe]-hydrogenases that might be easier to provide on a large scale.21 However, so far, no purely chemical compound could be synthesized that shows sufficient activity to be of economic interest.22–25 Therefore, understanding the roles of the coordination spheres provided by the [FeFe]-hydrogenase polypeptide is pivotal, and pursued by introducing changes to the protein, but also, as mentioned above, by employing chemically different cofactors. Several semi-artificial hydrogenases have been created based on water-soluble cofactor variants such as a selenium substituted cofactor.26 Many chemically synthesized cofactor mimics, however, are insoluble in water and have to be added to hydrogenase precursors in highly concentrated solvents.27–29 If and how these solvents affect [FeFe]-hydrogenases has hardly been investigated, and since the addition of even small quantities of organic solvents can cause protein inactivation,30,31 their concentration is often kept low in maturation experiments.
Biocatalysis in the presence of organic solvents has been an active research field for more than 40 years as low-water content environments can minimize water dependent side reactions, tune substrate specificity and impact enzyme regio- and enantioselectivity.30 As natural soluble enzymes have evolved to fold and function in water, their outer amino acid residues commonly interact with water molecules. This first hydration shell is crucial for enzyme activity and stability and alteration of water content in this first hydration shell can cause destabilization of the protein structure.32 Organic solvents applied in biocatalysis are usually divided into two classes by hydrophobicity, described by their partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). Polar solvents are defined by a log
P). Polar solvents are defined by a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P smaller than 2, non-polar solvents by a log
P smaller than 2, non-polar solvents by a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P greater than 4. Both classes differ in their interaction with enzymes: Polar solvents are often detrimental to enzyme integrity as they are capable of stripping away enzyme-bound water from the first hydration shell. This is usually accompanied by further penetration of the solvent into the enzyme, causing structural reorganization of the protein.33,34 Non-polar solvents are not capable of stripping away water molecules from the enzyme and are generally the more hospitable solvation environment for biocatalysis. However, molecules of these solvents are often located at hydrophobic regions of enzymes where they can alter orientations of hydrophobic amino acid residues. This can lead to different substrate specificities and activities.35 Therefore, the presence of organic solvents in reaction or storage media can introduce additional challenges to the stability of enzymes, which is why enzymes applied in industrial processes must be highly resilient.
P greater than 4. Both classes differ in their interaction with enzymes: Polar solvents are often detrimental to enzyme integrity as they are capable of stripping away enzyme-bound water from the first hydration shell. This is usually accompanied by further penetration of the solvent into the enzyme, causing structural reorganization of the protein.33,34 Non-polar solvents are not capable of stripping away water molecules from the enzyme and are generally the more hospitable solvation environment for biocatalysis. However, molecules of these solvents are often located at hydrophobic regions of enzymes where they can alter orientations of hydrophobic amino acid residues. This can lead to different substrate specificities and activities.35 Therefore, the presence of organic solvents in reaction or storage media can introduce additional challenges to the stability of enzymes, which is why enzymes applied in industrial processes must be highly resilient.
The identification or generation of more robust [FeFe]-hydrogenases that withstand harsh conditions is likely essential for these enzymes to reach biotechnological applicability. Resistance against O2 is one important aspect, but so far, no [FeFe]-hydrogenase has been identified or engineered that shows high H2 turnover activity in the presence of air. However, the [FeFe]-hydrogenase CbA5H from Clostridium beijerinckii can assume a conformational state in which the H-cluster is shielded against O2 by a cysteine residue that binds to the open coordination site of [2Fe]H.36–39 Although the enzyme is catalytically inactive in this state it can be easily reactivated under reducing conditions, which therefore allows a much easier handling of CbA5H.36 Indeed, this robustness was recently exploited to employ the air-purified enzyme in the regeneration of flavin mononucleotide (FMN) and nicotinamide adenine dinucleotide (phosphate) (NAD(P)+), which then served as co-substrates for additional enzymes that converted substrates of industrial interest.40,41 In addition to air, industrial processes might include harsh environmental conditions, detergents, or organic co-solvents.42 Several hydrogenases are resilient against high temperatures, non-physiological pH values and denaturating agents such as sodium dodecyl sulfate (SDS), urea and guanidine.43,44 Investigations of hydrogenase stability in organic solvents have been rather scarce45–47 although, in addition to purely technical issues as noted above, solvents might even stimulate hydrogenase activity as shown for the [NiFe]-hydrogenase HynSL from the purple sulfur bacterium Thiocapsa roseopersicina.47 Not least, stability against organic solvents could define novel solvation environments capable of dissolving alternative electron donors and acceptors, such as photosensitizers on Ru(II)-tris-bipyridine basis.48
Here, we analyzed the stability of the C. beijerinckii [FeFe]-hydrogenase CbA5H towards diverse solvents. We aim to describe novel solvation environments for [FeFe]-hydrogenases to increase their applicability as biocatalysts. The O2 stability of CbA5H already provides an important step towards this goal.36,37 We examined its stability in different organic solvents commonly used for cofactor solvation and synthesis.14,49 Furthermore, we investigated its response to environments with increasing hydrophobic properties as these might prove beneficial for biocatalysis.
Results
Time-dependent stability of CbA5H in acetone, DMSO or acetonitrile
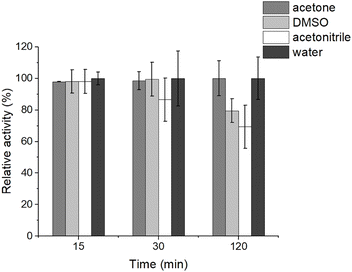
We first tested the stability of CbA5H that was incubated in 30% (v/v in water) solutions of solvent for up to two hours to gain an indicator for suitable incubation periods for subsequent analyses. Here, we focused on solvents employed for the chemical synthesis of [2Fe]H or its analogues (acetonitrile) or its solvation (dimethyl sulfoxide (DMSO) and acetone), which fall into the category of polar solvents (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values: DMSO = −1.35; acetonitrile = −0.34; acetone = −0.24; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). We judged enzyme stability from the H2 production activity of solvent-incubated enzyme compared to the control in an artificial electron delivery system containing excess methylviologen and sodium dithionite (details are described in the Experimental section). The activity of CbA5H remained unchanged after 2 h of incubation in 30% (v/v) acetone (Fig. 1). Incubation in 30% (v/v) DMSO resulted in a moderately decreased hydrogenase activity of 80 ± 7% after 2 h, whereas no effect on enzymatic activity was observed after 30 min (Fig. 1). In contrast, incubation in 30% (v/v) acetonitrile resulted in decreased activity of 87 ± 14% already after 30 min, and further decreased H2 production rates (69 ± 14%) after 2 h of incubation (Fig. 1). Note that we tested whether the solvents we used here would affect the in vitro hydrogenase activity assay in the highest concentrations employed (see below), but this was not significantly the case (Table 1).
P values: DMSO = −1.35; acetonitrile = −0.34; acetone = −0.24; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). We judged enzyme stability from the H2 production activity of solvent-incubated enzyme compared to the control in an artificial electron delivery system containing excess methylviologen and sodium dithionite (details are described in the Experimental section). The activity of CbA5H remained unchanged after 2 h of incubation in 30% (v/v) acetone (Fig. 1). Incubation in 30% (v/v) DMSO resulted in a moderately decreased hydrogenase activity of 80 ± 7% after 2 h, whereas no effect on enzymatic activity was observed after 30 min (Fig. 1). In contrast, incubation in 30% (v/v) acetonitrile resulted in decreased activity of 87 ± 14% already after 30 min, and further decreased H2 production rates (69 ± 14%) after 2 h of incubation (Fig. 1). Note that we tested whether the solvents we used here would affect the in vitro hydrogenase activity assay in the highest concentrations employed (see below), but this was not significantly the case (Table 1).
| Solvent | Activity (%) |
|---|---|
| Acetone | 107.4 ± 7.7 |
| Acetonitrile | 93.1 ± 7.4 |
| DMSO | 107.4 ± 7.7 |
| Methanol | 96.5 ± 11.3 |
| Ethanol | 110.0 ± 5.9 |
| 1-Propanol | 100.3 ± 6.7 |
| 2-Propanol | 103.3 ± 8.7 |
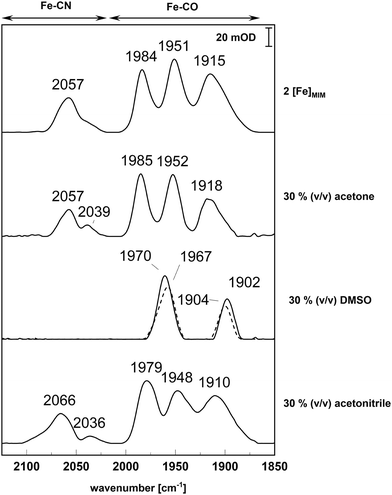
To distinguish whether any effects we observed on hydrogenase activity might simply be due to susceptibility of the cofactor alone, we employed Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) spectroscopy to test whether the used solvents would affect the chemical [2Fe]H analogue, which we term ‘[2Fe]MIM’ (‘MIM’ referring to ‘mimic’) here. ATR-FTIR is sensitive towards the vibrations of the CO- and CN− ligands between about 2200 and 1700 cm−1 and can be used to assess the integrity of [2Fe]H within [FeFe]-hydrogenases or of free [2Fe]MIM compounds.36,50 The spectra of [2Fe]MIM did hardly change after a 120 min incubation in 30% (v/v) acetone or acetonitrile (Fig. 2) save for minor peaks appearing in the region sensitive for the CN− ligands (2036 cm−1). Incubation for 120 min in 30% (v/v) DMSO, however, led to a loss of the signals of the CN− ligands as well as of the typically observed three Fe-CO signals. Instead, two prominent peaks at 1902 cm−1 and 1970 cm−1 arose (Fig. 2). Because of this strong deviation of the typical [2Fe]MIM spectrum, we performed a control experiment during which we dried a 30% (v/v) DMSO solution for a longer period of time. Indeed, the signals at 1902 cm−1 and 1970 cm−1 appeared again and thus probably stem from DMSO accumulating on the crystal (Fig. 2).
Whereas maturating [FeFe]-hydrogenase precursors with chemical mimics is usually conducted in the timeframe of minutes up to an hour,13,51 industrial biocatalytic processes would probably take place for several hours. We were curious about how long CbA5H would withstand incubation in 30% (v/v) solutions of the above-mentioned solvents. The experiments revealed that the H2 production activity of CbA5H decreased more strongly from the four-hour timepoint on (Fig. S1†), particularly in acetone and acetonitrile. Its remaining activity appeared to stabilize in acetone (22 ± 1% of activity left after 24 h), but hardly any activity remained after incubating CbA5H in acetonitrile for 24 h (1 ± 0.4%; Fig. S1†). Its activity in 30% (v/v) of DMSO remained at about 80% for 8 h but decreased to 37 ± 4% at the 24 h timepoint (Fig. S1†).
We also investigated the long-term effects of the same solvent solutions on two additional [FeFe]-hydrogenases that are routinely investigated in literature, namely HydA1 from Chlamydomonas reinhardtii and CpI from Clostridium pasteurianum. HydA1 lost about 80% of its activity after only 4 h of incubation and displayed residual activities of 21 ± 7% in acetone, 22 ± 9% in DMSO and 14 ± 5% in acetonitrile at this time-point (Fig. S2†) so that it was not investigated for longer periods of time. In contrast, CpI appeared to be more resilient and retained residual activities of 46 ± 5% and 20 ± 7% after incubation for 24 h in 30% (v/v) solutions of acetone and acetonitrile, respectively, and even 80 ± 6% in DMSO (Fig. S3†).
Concentration-dependent stability of CbA5H in acetone, DMSO and acetonitrile
We then tested for effects of different concentrations of the same solvents on CbA5H activity and decided on a timeframe of 30 min. As before, the enzyme was pre-incubated in the indicated solvent solutions, and its hydrogenase activity was determined afterwards. Due to this experimental setup, some solvent was always transferred into the reaction mixture of the hydrogenase activity assay. Although the final concentrations of the solvents in this reaction mixture reached maximally 0.45% (v/v), we tested whether this might affect the assay itself. To this end, the activity of CbA5H was compared in activity assays with or without 0.45% (v/v) of the employed solvents, but its activities were not significantly different, indicating that the solvents did not have a strong effect on the assay (Table 1).The incubation of CbA5H in different concentrations of solvents revealed that incubating CbA5H for 30 min in acetone solutions of up to 70% (v/v) had hardly an effect on subsequent H2 generation activities, and after incubation in 90% (v/v) acetone, CbA5H still showed an activity of 86 ± 16% compared to the enzyme incubated in water (Fig. 3). The activity of the [FeFe]-hydrogenase was also quite stable in up to 50% (v/v) DMSO, whereas the H2 production rate decreased in 70% (v/v) DMSO (53 ± 8%). Incubation in 90% (v/v) DMSO resulted in an almost complete loss of hydrogenase activity (Fig. 3).
 | ||
| Fig. 3 H2 production activity of the [FeFe]-hydrogenase CbA5H after incubation in the indicated concentrations (all in (v/v) in water) of acetone (dark grey), DMSO (light grey) or acetonitrile (white). The experimental procedure was the same as described in the caption of Fig. 1, except that the time of incubation in the solvents was 30 min in all cases. The columns show the averages of two independent experiments employing biological duplicates. The standard deviation is indicated by the error bars. The activities were always calculated to those of the CbA5H enzyme incubated in water, which were set to 100% and equalled 2200 U × mg−1. | ||
Acetonitrile had a noticeably different effect on CbA5H in that it did not result in a continuously decreasing activity (Fig. 3). Instead, while the percentual H2 production rates decreased stepwise from 90 ± 12% in 10% (v/v) acetonitrile to only 10 ± 4% in 50% (v/v) acetonitrile, they recovered in 70 and 90% (v/v) acetonitrile, reaching 66 ± 10% and 70 ± 10% of the activity determined of the control (Fig. 3). Again, we investigated the effect of these solvents on HydA1 and CpI and documented similar effects on their residual H2 production capabilities (Fig. S4 and S5†).
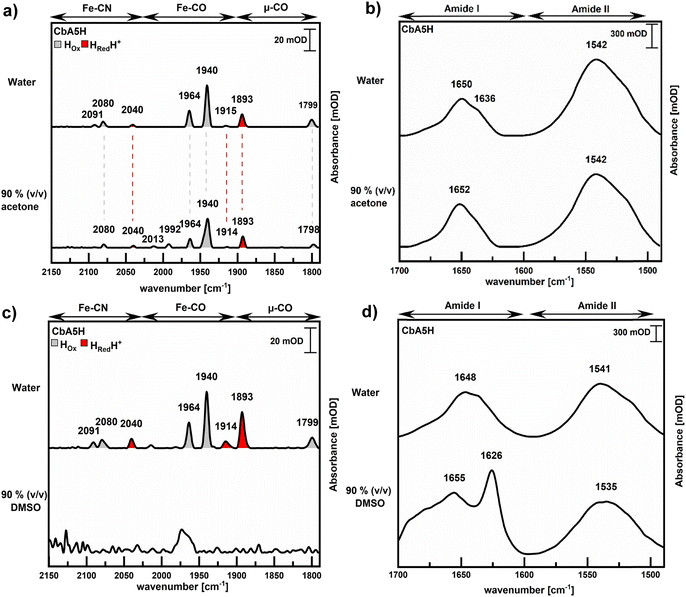
ATR-FTIR spectroscopy on solvent-incubated enzymes was conducted to determine whether changes in activity of solvent-incubated CbA5H could be ascribed to different states or to degradation of the H-cluster, or to strong changes in secondary structure elements (Fig. 4). During catalysis, the H-cluster assumes different redox states, which can be observed in the infrared by specific absorption signals of the CO- and CN− ligands between about 2100 cm−1 and 1800 cm−1. When CbA5H was purified under anoxic conditions, the most prominent states observed are the so-termed state Hox (bridging CO (μ-CO): 1799 cm−1; Fe–CO: 1940 cm−1 and 1964 cm−1; Fe–CN: 2080 cm−1 and 2091 cm−1) and HredH+ (μ-CO: 1810 cm−1; Fe–CO: 1893 cm−1, 1916 cm−1; Fe–CN: 2041 cm−1, 2075 cm−1), which can appear side-by-side in the infrared spectrum.37 Upon exposure to oxygen or oxidants, CbA5H enters the catalytically inactive Hinact state (μ-CO: 1840 cm−1; Fe–CO: 1992 cm−1 and 2011 cm−1; Fe–CN: 2080 cm−1 and 2107 cm−1), in which the open coordination site at the H-cluster is coordinated to the cysteine residue at position 367.36–39
Secondary structures of polypeptides are often analysed by FTIR through observing the amide I and II bands. The amide I band is mainly composed of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the peptide backbone (∼1650 cm−1) whereas the amide II band originates from N–H bending vibrations (1550 cm−1). Both bands are strongly affected by the secondary structure of the backbone and can be used for predictions of protein secondary structures.52,53 We used solvent concentrations equal to the concentrations in the corresponding protein sample for blank measurements.
O stretching vibrations of the peptide backbone (∼1650 cm−1) whereas the amide II band originates from N–H bending vibrations (1550 cm−1). Both bands are strongly affected by the secondary structure of the backbone and can be used for predictions of protein secondary structures.52,53 We used solvent concentrations equal to the concentrations in the corresponding protein sample for blank measurements.
When the spectra of CbA5H before and after incubation in 90% (v/v) acetone were compared, both showed the typical mixture of H-cluster redox states as listed above and were hardly distinguishable, except signals at 2013 cm−1 und 1992 cm−1 that were only present in the acetone-treated enzyme (Fig. 4a). We further investigated the shape of the amide I and II bands before and after incubation of CbA5H in solvent in order to document the effect of the solvents on protein secondary structure. Comparing the signals in the amide regions revealed no strong differences in the amide I and II bands of CbA5H incubated in water or in 90% (v/v) acetone, save for a minor decrease of the intensity of a shoulder at 1636 cm−1 (Fig. 4b).
In contrast, the spectra of CbA5H after incubation in 90% (v/v) DMSO displayed loss of specific H-cluster signals (Fig. 4c) as well as changes in the shape of the amide I band with a new peak at 1626 cm−1 and broadening of the shape between 1650 cm−1 and 1700 cm−1. In addition, the amide II band shifted from 1541 cm−1 to 1535 cm−1 (Fig. 4d).
ATR-FTIR spectroscopy of CbA5H incubated in acetonitrile reflected the activity measurements in that the signals of the H-cluster of the enzyme incubated in 50% (v/v) acetonitrile were lower than those recorded of CbA5H incubated in 30 or 70% (v/v) acetonitrile (Fig. 5a). While the amide II band of the protein incubated in the acetonitrile solutions shifted only slightly and lay between 1537–1542 cm−1, the amide I band decreased in intensity at 1653 cm−1 after the enzyme had been incubated in 50% (v/v) acetonitrile and a new peak was distinguishable at 1620 cm−1 (Fig. 5b).
 | ||
| Fig. 5 H-Cluster signals (a) and amide bands I and II (b) of CbA5H monitored by ATR-FTIR. The measurements were done as described for Fig. 4, except that the indicated concentrations of acetonitrile were used for pre-incubation of CbA5H. For each measurement, a blank was conducted with the same solvent concentrations that were employed for incubating the protein sample. | ||
Stability of CbA5H in short chain alcohols
We also investigated the response of CbA5H to solvents with gradually increasing hydrophobic properties as defined by their partition coefficient log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (log
P (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values: methanol = −0.74; ethanol = −0.31; 2-propanol = +0.05; 1-propanol = +0.25; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). As described for the solvents employed above, the [FeFe]-hydrogenase was incubated for 30 min in different concentrations of the solvents, and then its H2 production activity was determined compared to the control (Fig. 6). As before, we first tested whether these solvents would affect the hydrogenase activity assay in the highest concentrations that were transferred to the reaction mixture with the pre-incubated enzyme (0.45% (v/v)). However, the activity assays without and with these alcoholic solvents resulted in very similar values (Table 1).
P values: methanol = −0.74; ethanol = −0.31; 2-propanol = +0.05; 1-propanol = +0.25; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). As described for the solvents employed above, the [FeFe]-hydrogenase was incubated for 30 min in different concentrations of the solvents, and then its H2 production activity was determined compared to the control (Fig. 6). As before, we first tested whether these solvents would affect the hydrogenase activity assay in the highest concentrations that were transferred to the reaction mixture with the pre-incubated enzyme (0.45% (v/v)). However, the activity assays without and with these alcoholic solvents resulted in very similar values (Table 1).
When the CbA5H enzyme was incubated for 30 min in 10 or 30% (v/v) methanol or ethanol, its activity remained at nearly 100% of the control activity. However, when the hydrogenase was exposed to the same solvents at higher concentrations, it showed a stepwise decrease of activity, particularly at 70 and 90% (v/v) (Fig. 6). CbA5H activity was stronger impaired after incubation in 2-propanol, and particularly 1-propanol, which resulted in lower residual activities already in solutions of 30% (v/v) and 10% (v/v), respectively (Fig. 6). Incubation in 1-propanol resulted in almost complete loss of hydrogenase activity already in 30% (v/v) solutions. CbA5H hardly tolerated incubation in any of the alcohols when they were present as 70 and 90% (v/v) solutions and only displayed residual activities of 10 to 15% compared to the control activity (Fig. 6).
As before, we analyzed whether the alcoholic solvents affected the integrity of the H-cluster or introduced changes to the amide bands by ATR-FTIR. Similar to what was observed in the activity assays, incubation of CbA5H in 30% (v/v) of methanol (Fig. S6a†) or ethanol (Fig. S7a†) had only little effect on the vibrational patterns of the H-cluster and introduced only minor changes to the amide signals. In contrast, no H-cluster signals were left after incubation in 70% (v/v) of either alcohol, and the amide bands were changed regarding their shape and the position of their maxima (Fig. S6 and S7†). The amide I signal of CbA5H incubated for 30 min in 70% (v/v) methanol decreased strongly in intensity and split into two peaks with maxima at 1655 cm−1 and 1621 cm−1, whereas the amide II band showed a broader profile with its maximum shifted to 1529 cm−1 (Fig. S6†). Incubation in 70% (v/v) ethanol resulted in CbA5H displaying a shifted absorption maximum of the amide I band (from 1648 cm−1 to 1653 cm−1) and the appearance of a shoulder between 1610 cm−1 and 1625 cm−1 (Fig. S7b†). The shape of the amide II band of this CbA5H protein had broadened, and the peak centre had shifted from 1542 cm−1 to 1539 cm−1 (Fig. S7b†).
ATR-FTIR spectra of CbA5H incubated in 2-propanol (30 and 50% (v/v)) showed a gradual decrease in H-cluster signal intensities (Fig. S8a†), yet little changes to the amide signals (Fig. S8b†). After incubation in 70% (v/v) 2-propanol, however, the H-cluster signals were hardly detectable and the amide I band had split into two signals at 1655 cm−1 and 1624 cm−1. Additionally, the shape of the amide II band had broadened similar to the shape change after incubation in 70% (v/v) methanol and ethanol, respectively (Fig. S8b†). Incubation of CbA5H in 10% (v/v) 1-propanol resulted in an enzyme with lower intensities of the vibrational pattern of the H-cluster compared to incubation in water, whereas incubation in 30% (v/v) 1-propanol resulted in protein samples that showed no specific signals for the H-cluster anymore (Fig. S9a†). Incubation of the [FeFe]-hydrogenase in 10% (v/v) 1-propanol had little effect on the amide I and II bands, but after incubation in 30% (v/v) 1-propanol, several separate peaks could be detected in the amide I (1648 cm−1, 1676 cm−1, 1685 cm−1) and amide II band (1535 cm−1, 1546 cm−1, 1560 cm−1).
Discussion
Biocatalysts that can tolerate solvents may be integrated into industrial processes that couple chemical and biological catalysts,54 and in some instances, as mentioned in the introduction, organic solvents even improve biocatalytic performance.30 This latter effect has indeed also been observed for a hydrogenase of the [NiFe]-type.47 However, stability of enzymes in solvents is not only required for several applied biocatalytic fields, but may also assist basic research, as solvents can be used as probes for understanding aspects of enzymatic mechanisms.55 The stability of enzymes in the given solvents is a prerequisite for these approaches. Here, we tested the stability of the enzyme CbA5H, one of the few O2-stable [FeFe]-hydrogenases, in a variety of organic solvents that are of relevance in chemical syntheses and catalyses.[FeFe]-Hydrogenases owe their high specific activity to their specialized active site metal-cluster, the H-cluster. Therefore, as is the case for many enzymes with inorganic cofactors, organic solvents may influence their activity on both the level of the polypeptide and the cofactor. As has been shown for a multitude of enzymes34,56,57 organic solvents may influence the conformation or conformational dynamics of [FeFe]-hydrogenases or even result in their denaturation. They might also attack, or coordinate to, the cofactor, similar to several (ir)reversible inhibitors of [FeFe]-hydrogenases that often target the open coordination site at the distal Fe ion10,58,59 or the amine bridgehead.9 On the other hand, the protein environment might shield the cofactor.
The latter effect can indeed be well studied in the case of [FeFe]-hydrogenases, because, as explained in the introduction, natural and modified versions of the diiron-subsite of the H-cluster can be chemically synthesized and integrated into the protein post-translationally.13,14,19 Thereby, solvent effects on the free analogues (or ‘mimics’, as we term them here) and on the assembled enzymes can be compared. We first tested the medium-term (two hours) effects of solvents common for chemical mimic syntheses or solubilization, namely acetone, DMSO and acetonitrile on both holo-[FeFe]-hydrogenase and free 2[Fe]MIM. We could observe that while 30% (v/v) acetone and acetonitrile had hardly any effects on the ATR-FTIR spectra of the free mimic after 120 min of incubation (Fig. 1 and 2), 30% (v/v) DMSO resulted in a drastically changed spectrum, which, according to a comparison to dried 30% (v/v) DMSO solutions, very likely represented signals from DMSO (Fig. 2). Because none of the other signals typical for 2[Fe]MIM were visible, for example in the region where the CN− ligands usually absorb, it is likely that most of the mimic had degraded. Similar effects of aqueous DMSO solutions were reported for a mono-cyanide derivative of 2[Fe]MIM, which degraded in 50% (v/v) DMSO in water over the course of two hours.27 In contrast, treatment of holo-CbA5H with 30% (v/v) DMSO for two hours resulted in an activity loss of only about 20%, suggesting that the protein shielded the cofactor, either by excluding the solvent or by protecting it from solvent attack through its own interactions with the metal site. In contrast, incubation of the assembled holo-enzyme in 30% (v/v) of acetonitrile for two hours resulted in a notable decrease of hydrogenase activity (Fig. 1), while the same incubation of the free 2[Fe]MIM revealed only minor signs of degradation in the ATR-FTIR spectrum (Fig. 2), indicating that the negative effect of this acetonitrile concentration on the enzyme was due to a simultaneous degradation of cofactor and enzyme structure.
We then tested concentration-dependent effects of various solvents on the stability of CbA5H, and again employed its remaining activity as well as ATR-FTIR spectroscopy to assess the enzyme's state after solvent treatment. In addition to the well-understood H-cluster signals, we recorded the amide I and II bands to gain information on effects on the polypeptide. Viewing all of these features side-by-side, a general conclusion we can make is that the solvents we tested here always showed similar extents of effects on protein secondary structures and H-cluster integrity, paralleled by equivalent changes in hydrogenase activity. The solvents we tested can all be categorized as polar according to their log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (DMSO: −1.35; acetonitrile: −0.34; acetone: −0.24; methanol: −0.74; ethanol: −0.32; 2-propanol: + 0.05; 1-propanol: + 0.25; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). However, their chemical characteristics and their corresponding effects on enzymes differ, because both structure and reactive group of solvents modulate their interaction with proteins.35,57,60 The –OH group of alcohols can form H-bonds with enzymes, which can lead to competition for enzyme-bound water molecules or affect the structure directly by enhancing enzymatic flexibility and thereby promoting unfolding. Acetone and acetonitrile can have this effect, too, and they have additionally been shown to be capable of penetrating into the active center of proteins and changing its structural organization.56 DMSO, on the other hand, appears to mostly introduce structural changes to enzymes by targeting and solvating exposed hydrophobic amino acid residues.35,61
P values (DMSO: −1.35; acetonitrile: −0.34; acetone: −0.24; methanol: −0.74; ethanol: −0.32; 2-propanol: + 0.05; 1-propanol: + 0.25; values were obtained from the National Center for Biotechnology Information's PubChem database; https://pubchem.ncbi.nlm.nih.gov/). However, their chemical characteristics and their corresponding effects on enzymes differ, because both structure and reactive group of solvents modulate their interaction with proteins.35,57,60 The –OH group of alcohols can form H-bonds with enzymes, which can lead to competition for enzyme-bound water molecules or affect the structure directly by enhancing enzymatic flexibility and thereby promoting unfolding. Acetone and acetonitrile can have this effect, too, and they have additionally been shown to be capable of penetrating into the active center of proteins and changing its structural organization.56 DMSO, on the other hand, appears to mostly introduce structural changes to enzymes by targeting and solvating exposed hydrophobic amino acid residues.35,61
For the effect of the here analysed solvents on CbA5H, the shapes of the amide I bands, and the signals recorded in the signal range sensitive for vibrations of the H-cluster help to explain the response of the enzyme towards the respective solvent. Perhaps most notable in the context of protein conformation are observed changes of the amide I band, which is an important indicator for a protein's secondary structure elements.53 In all solvent solutions that caused a loss of subsequently measured hydrogenase activity of CbA5H, the amide I band split into two peaks (90% DMSO (Fig. 4), 50% acetonitrile (Fig. 5), 70% methanol (Fig. S6†), 70% 2-propanol (Fig. S8†); all percent values in (v/v)) or revealed a prominent shoulder at lower wavenumbers (70% (v/v) ethanol (Fig. S7†)). Whereas α-helices usually absorb the strongest at 1655 cm−1, β-sheets result in strong absorption at around 1630 cm−1 and 1690 cm−1,62 although the number of strands and whether they are antiparallel or parallel affects the position of the main band.53 Our observation of signals arising in the range between 1620 cm−1 and 1630 cm−1 in CbA5H proteins after incubation in detrimental solvent solutions therefore suggests the formation of highly ordered intermolecular β-sheets in the naturally mostly α-helical enzyme.38 Indeed, similar observations have been made, and similar conclusions drawn, in the context of pressure- and temperature-dependent degradation of CbA5H.63 In contrast to the latter study, however, we saw no clear increase in the range of 1690 cm−1, suggesting that the β-sheets induced by solvents differ from those that arise in high temperature.63 In addition to these prominent changes of the amide I bands, in most of the same cases we observed a broadening of the amide II band. This indicates that N–H bending was affected by the deleterious solvent solutions, which likely altered the local H-bonding network.62
In contrast, the amide I and II bands of CbA5H solutions that retained high activity after incubation in different solvents and/or different concentrations of solvents showed little or no changes. This suggests that these protein populations maintained the native structure and indicates that the respective (concentration of) solvent(s) were tolerated by the enzyme. Whether, on a molecular level, this stability towards certain solvent environments is related to the high temperature and pressure tolerance of CbA5H63 requires additional studies, such as Molecular Dynamics simulations that have, for example, often been implemented to analyse effects of solvents on protein flexibility.15,35
It is noteworthy that all solvent solutions that resulted in loss of hydrogenase activity as well as changes to the amide I and II bands of the preincubated CbA5H populations affected the H-cluster signals of the ATR-FTIR spectra correspondingly, in that the signals lost intensity or disappeared altogether. At this point, we cannot distinguish whether the H-cluster – or parts thereof – was expelled from the respective proteins upon their conformational changes or whether it was degraded, or both. Note that the free 2[Fe]MIM results in much less intense ATR-FTIR signals than the H-cluster within the hydrogenase polypeptide, which can be seen when comparing the signals and concentrations of free mimic (5 mM; Fig. 2) and hydrogenase (roughly 4 μM) (e.g.Fig. 4). However, as the H-cluster signals of the less affected CbA5H solutions did hardly show changed patterns, save for two peaks after incubation in 90% (v/v) of acetone, we can assume that none of the solvent molecules changed the redox state of the H-cluster or formed a covalent bond to it. This supports our hypothesis that the solvents analysed here mostly or at least initially affected the protein structure, as opposed to the active site cofactor.
Comparing the effects of the different organic solvents we tested here, the general trend appeared that they affected the [FeFe]-hydrogenase CbA5H the more the higher their concentrations, although some solvents, such as 2-propanol, were detrimental at lower concentrations than others. However, acetone and acetonitrile were exceptions. CbA5H was remarkably resistant against acetone, showing hardly impaired activity, H-cluster signals or amide bands even after incubation in 90% (v/v) acetone (Fig. 3 and 4). It should be noted that 90% (v/v) acetone solutions are commonly used for protein precipitation, although the incubation is usually done at sub-zero temperatures and for longer periods of time. For the time being we cannot distinguish whether CbA5H stays soluble in this acetone concentration or whether it precipitates in a way that allows an almost complete reactivation of the enzyme upon its transfer to the hydrogenase activity reaction mixture. However, both scenarios would imply a high resilience of the native CbA5H structure to this high concentration of acetone. Acetonitrile, on the other hand, resulted in inversed bell-shaped effects on all levels tested – activity, H-cluster, amide bands – in that all parameters showed a progressively impaired enzyme status from 10 to 50% (v/v) but rather healthy proteins in 70 and 90% (v/v) of acetonitrile (Fig. 3 and Fig. 5). The ATR-FTIR spectrum of the enzyme incubated in 50% (v/v) acetonitrile revealed lower intensities of the H-cluster signals in combination with a split amide I band, suggesting that this solvent mixture changed the polypeptide structure, which also led to a loss of H-cluster or to its degradation. As these changes were not observed in lower or higher acetonitrile concentrations, it seems that the most hostile environment for the enzyme is a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture of acetonitrile and water. Indeed this inverse bell-shaped stability pattern in dependence of solvent concentration in water has often been observed, for example in the case of lysozyme and subtilisin,64 and is believed to be based on the interplay of conformational mobility and stability of the protein.35,65 At a certain water content, many enzymes behave as in aqueous solutions, whereas above a certain threshold of the water concentration, the enzyme becomes too flexible and starts to unfold.55,66 Here, we assume that at acetonitrile concentrations below 50% (v/v) the conformational stability of CbA5H was sufficiently resilient to counteract disrupting effects of solvent molecules whereas at concentrations higher than 50% (v/v) its conformational mobility would be reduced, effectively counteracting conformational instability.
50 mixture of acetonitrile and water. Indeed this inverse bell-shaped stability pattern in dependence of solvent concentration in water has often been observed, for example in the case of lysozyme and subtilisin,64 and is believed to be based on the interplay of conformational mobility and stability of the protein.35,65 At a certain water content, many enzymes behave as in aqueous solutions, whereas above a certain threshold of the water concentration, the enzyme becomes too flexible and starts to unfold.55,66 Here, we assume that at acetonitrile concentrations below 50% (v/v) the conformational stability of CbA5H was sufficiently resilient to counteract disrupting effects of solvent molecules whereas at concentrations higher than 50% (v/v) its conformational mobility would be reduced, effectively counteracting conformational instability.
Domain arrangements and oligomeric structures of [FeFe]-hydrogenase vary greatly.67 We could find only few studies addressing the solvent tolerance of these H2-converting enzymes so that we cannot predict whether [FeFe]-hydrogenases would generally be suited for biocatalysis in non-aqueous environments. When comparing our findings to previous studies on different hydrogenases it appears that CbA5H is somewhat more resilient than these hydrogenases in the short-term: A hydrogenase from Chromatium has been reported to withstand incubation in 50% DMSO, but to be completely inactivated after incubation in 80% solutions,44 and a similar behavior was reported for a hydrogenase isolated from Megasphaera elsdenii.46 Our own study shows that the model hydrogenases HydA1 from C. reinhardtii and CpI from C. pasteurianum, which both have very different structures than CbA5H, retained quite similar levels of activity after short-term incubation in different concentrations of acetone, DMSO and acetonitrile as CbA5H (Fig. S6 and S7†). This high stability during short term exposure could be of particular interest for maturation experiments of [FeFe]-hydrogenases with modified versions of 2[Fe]MIM that are only soluble in high concentrations of organic solvents. However, although the enzymes analyzed here tolerate high concentrations of organic solvents when in their maturated holo-form, the solvents might affect the maturation process itself. This would have to be tested before conclusions on the activity of a certain cofactor mimic could be drawn.
While most scientific approaches that require the presence of organic solvents would be completed after a few hours at most, integrating [FeFe]-hydrogenases into industrial approaches would probably require their long-term stability in said solvents. Exposure of CbA5H for up to 24 hours to acetone, DMSO and acetonitrile (all 30% (v/v)) revealed that CbA5H lost about 50% of its activity after being incubated in DMSO for 16 hours, in acetone for 8 hours, and in acetonitrile for 2 hours (Fig. S1†). In these experiments, C. pasteurianum CpI proved to be more resilient (Fig. S3†), whereas the small algal [FeFe]-hydrogenase HydA1 was much more susceptible and lost about 50% of its activity already after 2 hours (Fig. S2†). In perspective, a [NiFe]-hydrogenase from Thiocapsa roseopersicina revealed resistance against highly concentrated acetone and acetonitrile solutions as well as against intermediate concentrations of methanol, ethanol and DMSO over time frames of 20 hours.47 [FeFe]-Hydrogenases are usually much more active than other hydrogenase types, and CbA5H has the additional advantage of being stable in air. However, for long-term applications the enzyme would require further stabilizing modifications, which could include protein engineering or immobilization on carrier matrices.68,69
Conclusion
The [FeFe]-hydrogenase CbA5H displays stability towards several of the organic solvents analyzed here, particularly towards high concentrations of acetone and acetonitrile (up to 90% (v/v)), medium to high DMSO concentrations (50 to 70% (v/v)) and low to medium concentrations of ethanol and methanol (30 to 50% (v/v)). Although the enzyme lost most of its activity after long-term incubation in acetone, DMSO and acetonitrile, CbA5H has the additional advantage of being O2-stable, which would make any handling of the enzyme on an industrial scale much easier. Its stability in several solvents could facilitate its application in semi-natural approaches, such as in combination with photosensitizers or artificial light harvesting systems.70–72 For example, very recently, the [FeFe]-hydrogenase CpI was combined with a photo-absorbant cyanamide-modified graphitic carbon nitride matrix that resulted in light-dependent hydrogen production coupled to the selective oxidation of alcohols.42 Catalysts systems like these could greatly benefit from organic solvent environments as this could increase the range of mediators and substrates.Experimental
Enzyme production and purification
The [FeFe]-hydrogenases analysed here (Clostridium beijerinckii CbA5H, Chlamydomonas reinhardtii HydA1 and Clostridium pasteurianum CpI) were heterologously produced in Escherichia coli as reported before.73,74E. coli BL21(DE3) ΔiscR cells74 were transformed by electroporation (2.5 kV; 25 μF; 600 Ω; 13.8 ms) with pET21b expression vectors into which the hydrogenase-encoding sequences had been introduced. E. coli cells were incubated in 500 μL LB medium for 1 h and then plated on LB agar–agar plates containing 40 μg × mL−1 kanamycin and 100 μg × mL−1 ampicillin for selection. The plates were incubated overnight at 37 °C. From these plates, colonies were picked and transferred to 100 mL precultures (LB-3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.4, 40 μg × mL−1 kanamycin and 100 μg × mL−1 ampicillin) and incubated at 37 °C overnight, shaking at 130 rpm.Expression cultures contained 0.5% (w/v) glucose, 2 mM ammonium iron III citrate, 40 μg × mL−1 kanamycin and 100 μg × mL−1 ampicillin in a total volume of 1 L. Cultures were inoculated with the precultures to an OD600 of 0.05 and incubated at 37 °C for 2 h, shaking at 130 rpm. After reaching an OD600 of 0.5, the cultures were transferred into 1 L Schott flasks and placed into an anoxic chamber (Coy Laboratory Products Inc., https://coylab.com/) containing an atmosphere of N2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2 98
H2 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). Sodium fumarate (4 g × L−1) and cysteine (0.3 g × L−1) were added, and the E. coli cultures were incubated for at least 30 min in the anoxic chamber at room temperature (RT). Then, expression of the constructs was induced by adding 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and the expression cultures were incubated at RT overnight upon stirring with a magnetic stir bar.
2). Sodium fumarate (4 g × L−1) and cysteine (0.3 g × L−1) were added, and the E. coli cultures were incubated for at least 30 min in the anoxic chamber at room temperature (RT). Then, expression of the constructs was induced by adding 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and the expression cultures were incubated at RT overnight upon stirring with a magnetic stir bar.
Harvesting of the E. coli cells and purification of the Strep-tagged [FeFe]-hydrogenase were carried out under anoxic conditions (c(O2) < 35 ppm) in the anoxic chamber, except for centrifugation steps. Cells were harvested in 1 L centrifugation flasks for 20 min at 4700 × g and 4 °C. The pellets were resuspended in 100 mM Tris-HCl, pH 6.8, with 2 mM sodium dithionite (NaDT), 10 mg lysozyme and 1% (v/v) Triton X-100, and incubated on ice for 20 min. Cell lysis was done via sonication (Branson Sonifier 250, 5 cycles à 45 seconds, output level 3, 2 min breaks between cycles, solutions were kept on ice during the process). Lysate separation was performed by ultracentrifugation (120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 4 °C, 60 min), and afterwards, the supernatant was cleared by passing it through 0.2 μm pore size sterile filters.
000 × g, 4 °C, 60 min), and afterwards, the supernatant was cleared by passing it through 0.2 μm pore size sterile filters.
Proteins were purified using the Strep-Tactin system. 5 mL gravity flow columns containing Strep-Tactin® Superflow® high capacity resin (IBA Lifesciences GmbH, https://www.iba-lifesciences.com) were loaded with the cleared lysate and washed with 20 mL of 100 mM Tris-HCl, pH 8, 2 mM NaDT. Target proteins were eluted by the same buffer, but supplemented with 2.5 mM desthiobiotin. Protein concentrations were determined employing Quick Start Bradford Protein Assay reagent (BioRad, https://www.bio-rad.com). Enzyme solutions were stored anoxically (in 100 mM Tris-HCl, pH 8, 2 mM NaDT, 2.5 mM desthiobiotin) at −80 °C for further use.
[FeFe]-Hydrogenase maturation
By employing the heterologous production protocol described above, [FeFe]-hydrogenases are equipped with the [4Fe4S]-subcluster of the H-cluster only. For subsequent completion of the H-cluster, purified enzymes were maturated in vitro as described before.13 The proteins were incubated with a 10-fold molar excess of the chemically synthesized [2Fe]H sub-cluster termed ‘[2Fe]MIM’ here ([2Fe2[μ-(SCH2)2NH](CN)2(CO)4]2−)75 for 1 h at 4 °C. Maturated enzymes were separated from excess mimic by size exclusion chromatography using NAP5 columns (NAP 5 column, GE healthcare, https://www.gehealthcare.de). Enzymes were eluted in 100 mM potassium phosphate buffer, pH 6.8, containing 2 mM of NaDT. Enzyme solutions were concentrated using 30 kDa spin filters (Amicon Ultra 0.5 mL, Merck Millipore, https://www.merckmillipore.com). Final protein concentrations were determined as described above.Incubation of [FeFe]-hydrogenases in solvents
The recombinant purified and maturated [FeFe]-hydrogenases (960 ng in the case of subsequent in vitro activity assays, and 3 μg for subsequent spectroscopy; see below) were incubated in closed 1.5-mL reaction tubes in a volume of 20 μL of water with different volumetric percentages (v/v) of solvent (0, 10, 30, 50, 70 or 90%) at 23 °C in the anoxic glove-box containing a N2/H2 atmosphere (98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). Incubation times differed and are indicated in the text. Prior to employing the solvents for the incubation of the enzymes, oxygen was removed from the liquids by purging 1 mL of solvent in 8 mL sealed headspace flasks with Argon gas for 8 minutes.
2). Incubation times differed and are indicated in the text. Prior to employing the solvents for the incubation of the enzymes, oxygen was removed from the liquids by purging 1 mL of solvent in 8 mL sealed headspace flasks with Argon gas for 8 minutes.
Hydrogenase activity assay
Hydrogenase activity was assayed employing an artificial electron delivery system. Ten μL of the [FeFe]-hydrogenase solutions prepared and incubated as described above were added to 100 mM potassium phosphate buffer, pH 6.8, 100 mM NaDT and 10 mM methyl viologen in a total volume of 2 mL in 8 mL headspace flasks. To test for any effects of the solvents on the assay itself, the maximum concentration of the given solvents that were transferred to the assay (0.45% (v/v)) were added to the same reaction mixtures. Maturated and not solvent-exposed CbA5H hydrogenase was added to these mixtures as well as to assays without solvents. In all cases, the flasks were sealed with red-rubber Suba-Seal Septa (Sigma-Aldrich/Merck; https://www.sigmaaldrich.com), purged with pure argon for 3 min and then incubated in a shaking water bath at 37 °C for 30 min. Afterwards, 400 μL of the headspace gas were injected into a gas chromatograph equipped for H2 detection (model GC-2010-Plus (Shimadzu; https://www.shimadzu.de) equipped with a purged packed inlet, Molesieve 5A column (30 m, ID 0.53 mm, film 25 mm) and thermal conductivity detector, using argon as carrier gas). Specific activity was calculated in units (U) per mg enzyme, U being defined as the production of 1 μmol H2 per minute. All measurements were done in technical duplicates employing two independent protein batches.Attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy
ATR-FTIR measurements were performed as described before.76 In brief, a Bruker Tensor II spectrometer (Bruker Optik) equipped with a BioATR cell II (Harrick, https://harricksci.com/) carrying a double-reflection ZnSe/Si crystal was employed. The scan resolution was set to 2 cm−1. Both sample incubation and measurement were carried out under a N2/H2 atmosphere (98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). Samples were dried for 5 to 10 min until H-cluster signals were clearly distinguishable from background absorption. Before analysing the protein samples, the system was blanked against the same solvent concentration employed for incubating the protein sample or the free cofactor mimic [2Fe]MIM. Data analysis and peak determination was performed using Origin Pro 2023, and H-cluster signal heights were normalized to the amide II band.
2). Samples were dried for 5 to 10 min until H-cluster signals were clearly distinguishable from background absorption. Before analysing the protein samples, the system was blanked against the same solvent concentration employed for incubating the protein sample or the free cofactor mimic [2Fe]MIM. Data analysis and peak determination was performed using Origin Pro 2023, and H-cluster signal heights were normalized to the amide II band.
Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
MG: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. AH: conceptualization, supervision, writing – review & editing. TH: conceptualization, funding acquisition, project administration, resources, supervision, writing – review & editing.Conflicts of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.Acknowledgements
M. G. and T. H. acknowledge the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy - EXC 2033 - 390677874 - RESOLV. T. H. thanks the DFG (HA 2555/10-1) and the VolkswagenStiftung (Az 98621) for financial support. We thank Shanika Yadav and Ulf-Peter Apfel (Faculty for Chemistry and Biochemistry, Ruhr University Bochum) for synthesizing and providing the [2Fe]H mimic for in vitro maturation.References
- P. M. Vignais and B. Billoud, Occurrence, classification, and biological function of hydrogenases: an overview, Chem. Rev., 2007, 107, 4206–4272, DOI:10.1021/cr050196r.
- S. V. Hexter, F. Grey and T. Happe, et al., Electrocatalytic mechanism of reversible hydrogen cycling by enzymes and distinctions between the major classes of hydrogenases, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11516–11521, DOI:10.1073/pnas.1204770109.
- J. T. Kleinhaus, F. Wittkamp and S. Yadav, et al., [FeFe]-Hydrogenases: maturation and reactivity of enzymatic systems and overview of biomimetic models, Chem. Soc. Rev., 2021, 50, 1668–1784, 10.1039/d0cs01089h.
- J. W. Peters, W. N. Lanzilotta and B. J. Lemon, et al., X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution, Science, 1998, 282, 1853–1858, DOI:10.1126/science.282.5395.1853.
- Y. Nicolet, C. Piras and P. Legrand, et al., Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center, Structure, 1999, 7, 13–23, DOI:10.1016/s0969-2126(99)80005-7.
- C. Greco, M. Bruschi and J. Heimdal, et al., Structural insights into the active-ready form of [FeFe]-hydrogenase and mechanistic details of its inhibition by carbon monoxide, Inorg. Chem., 2007, 46, 7256–7258, DOI:10.1021/ic701051h.
- A. F. Wait, C. Brandmayr and S. T. Stripp, et al., Formaldehyde—a rapid and reversible inhibitor of hydrogen production by [FeFe]-hydrogenases, J. Am. Chem. Soc., 2011, 133, 1282–1285, DOI:10.1021/ja110103p.
- C. E. Foster, T. Krämer and A. F. Wait, et al., Inhibition of [FeFe]-Hydrogenases by Formaldehyde and Wider Mechanistic Implications for Biohydrogen Activation, J. Am. Chem. Soc., 2012, 134, 7553–7557, DOI:10.1021/ja302096r.
- J. Duan, A. Veliju and O. Lampret, et al., Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases, J. Am. Chem. Soc., 2023, 145, 26068–26074, DOI:10.1021/jacs.3c07800.
- A. Kubas, C. Orain and D. Sancho, et al., Mechanism of O2 diffusion and reduction in FeFe hydrogenases, Nat. Chem., 2017, 9, 88–95, DOI:10.1038/nchem.2592.
- S. Yadav, R. Haas and E. B. Boydas, et al., Oxygen Sensitivity of [FeFe]-Hydrogenase: A Comparative Study of Active Site Mimics Inside vs. Outside the Enzyme, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2021-d91v1-v2.
- R. D. Britt, G. Rao and L. Tao, Biosynthesis of the catalytic H-cluster of [FeFe] hydrogenase: the roles of the Fe-S maturase proteins HydE, HydF, and HydG, Chem. Sci., 2020, 11, 10313–10323, 10.1039/d0sc04216a.
- J. Esselborn, C. Lambertz and A. Adamska-Venkates, et al., Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic, Nat. Chem. Biol., 2013, 9, 607–609, DOI:10.1038/nchembio.1311.
- G. Berggren, A. Adamska and C. Lambertz, et al., Biomimetic assembly and activation of [FeFe]-hydrogenases, Nature, 2013, 499, 66–69, DOI:10.1038/nature12239.
- A. Adamska-Venkatesh, S. Roy and J. F. Siebel, et al., Spectroscopic Characterization of the Bridging Amine in the Active Site of [FeFe] Hydrogenase Using Isotopologues of the H-Cluster, J. Am. Chem. Soc., 2015, 137, 12744–12747, DOI:10.1021/jacs.5b06240.
- R. Gilbert-Wilson, J. F. Siebel and A. Adamska-Venkatesh, et al., Spectroscopic Investigations of [FeFe] Hydrogenase Maturated with [57Fe2(adt)(CN)2(CO)4]2−, J. Am. Chem. Soc., 2015, 137, 8998–9005, DOI:10.1021/jacs.5b03270.
- M. L. K. Sanchez, S. Wiley and E. Reijerse, et al., Time-Resolved Infrared Spectroscopy Reveals the pH-Independence of the First Electron Transfer Step in the [FeFe] Hydrogenase Catalytic Cycle, J. Phys. Chem. Lett., 2022, 13, 5986–5990, DOI:10.1021/acs.jpclett.2c01467.
- C. Sommer, C. P. Richers and W. Lubitz, et al., A [RuRu] Analogue of an [FeFe]-Hydrogenase Traps the Key Hydride Intermediate of the Catalytic Cycle, Angew. Chem., Int. Ed., 2018, 57, 5429–5432, DOI:10.1002/anie.201801914.
- J. F. Siebel, A. Adamska-Venkatesh and K. Weber, et al., Hybrid [FeFe]-hydrogenases with modified active sites show remarkable residual enzymatic activity, Biochemistry, 2015, 54, 1474–1483, DOI:10.1021/bi501391d.
- A. K. Jones, E. Sillery and S. P. J. Albracht, et al., Direct comparison of the electrocatalytic oxidation of hydrogen by an enzyme and a platinum catalyst, Chem. Commun., 2002, 866–867, 10.1039/b201337a.
- C. Esmieu, P. Raleiras and G. Berggren, From protein engineering to artificial enzymes - biological and biomimetic approaches towards sustainable hydrogen production, Sustainable Energy Fuels, 2018, 2, 724–750, 10.1039/c7se00582b.
- J. Ekström, M. Abrahamsson and C. Olson, et al., Bio-inspired, side-on attachment of a ruthenium photosensitizer to an iron hydrogenase active site model, Dalton Trans., 2006, 4599–4606, 10.1039/b606659c.
- R. Goy, U.-P. Apfel and C. Elleouet, et al., A Silicon-Heteroaromatic System as Photosensitizer for Light-Driven Hydrogen Production by Hydrogenase Mimics, Eur. J. Inorg. Chem., 2013, 2013, 4466–4472, DOI:10.1002/ejic.201300537.
- W.-G. Wang, F. Wang and H.-Y. Wang, et al., Photocatalytic Hydrogen Evolution by [FeFe] Hydrogenase Mimics in Homogeneous Solution, Chem. – Asian J., 2010, 5, 1796–1803, DOI:10.1002/asia.201000087.
- J. Liu and W. Jiang, Photoinduced hydrogen evolution in supramolecular devices with a rhenium photosensitizer linked to FeFe-hydrogenase model complexes, Dalton Trans., 2012, 41, 9700–9707, 10.1039/c2dt30468f.
- L. Kertess, F. Wittkamp and C. Sommer, et al., Chalcogenide substitution in the 2Fe cluster of FeFe-hydrogenases conserves high enzymatic activity, Dalton Trans., 2017, 46, 16947–16958, 10.1039/C7DT03785F.
- C. Esmieu and G. Berggren, Characterization of a monocyanide model of FeFe hydrogenases - highlighting the importance of the bridgehead nitrogen for catalysis, Dalton Trans., 2016, 45, 19242–19248, 10.1039/c6dt02061e.
- M. Wang, L. Chen and L. Sun, Recent progress in electrochemical hydrogen production with earth-abundant metal complexes as catalysts, Energy Environ. Sci., 2012, 5, 6763–6778, 10.1039/c2ee03309g.
- S. Gao, Y. Liu and Y. Shao, et al., Iron carbonyl compounds with aromatic dithiolate bridges as organometallic mimics of [FeFe] hydrogenases, Coord. Chem. Rev., 2020, 402, 213081, DOI:10.1016/j.ccr.2019.213081.
- A. M. Klibanov, Improving enzymes by using them in organic solvents, Nature, 2001, 409, 241–246, DOI:10.1038/35051719.
- A. Tjernberg, N. Markova and W. J. Griffiths, et al., DMSO-related effects in protein characterization, J. Biomol. Screening, 2006, 11, 131–137, DOI:10.1177/1087057105284218.
- K. Rezaei, E. Jenab and F. Temelli, Effects of water on enzyme performance with an emphasis on the reactions in supercritical fluids, Crit Rev Biotechnol, 2007, 27, 183–195, DOI:10.1080/07388550701775901.
- P. Trodler and J. Pleiss, Modeling structure and flexibility of Candida antarctica lipase B in organic solvents, BMC Struct. Biol., 2008, 8, 9, DOI:10.1186/1472-6807-8-9.
- F. Secundo, S. Fialà and M. W. Fraaije, et al., Effects of water miscible organic solvents on the activity and conformation of the Baeyer-Villiger monooxygenases from Thermobifida fusca and Acinetobacter calcoaceticus: a comparative study, Biotechnol. Bioeng., 2011, 108, 491–499, DOI:10.1002/bit.22963.
- S. Wang, X. Meng and H. Zhou, et al., Enzyme Stability and Activity in Non-Aqueous Reaction Systems: A Mini Review, Catalysts, 2016, 6, 32, DOI:10.3390/catal6020032.
- S. Morra, M. Arizzi and F. Valetti, et al., Oxygen Stability in the New [FeFe]-Hydrogenase from Clostridium beijerinckii SM10 (CbA5H), Biochemistry, 2016, 55, 5897–5900, DOI:10.1021/acs.biochem.6b00780.
- P. S. Corrigan, J. L. Tirsch and A. Silakov, Investigation of the Unusual Ability of the [FeFe] Hydrogenase from Clostridium beijerinckii to Access an O2-Protected State, J. Am. Chem. Soc., 2020, 142, 12409–12419, DOI:10.1021/jacs.0c04964.
- M. Winkler, J. Duan and A. Rutz, et al., A Safety cap protects hydrogenase from oxygen attack, Nat. Commun., 2021, 12, 756, DOI:10.1038/s41467-020-20861-2.
- A. Rutz, C. K. Das and A. Fasano, et al., Increasing the O2 Resistance of the [FeFe]-Hydrogenase CbA5H through Enhanced Protein Flexibility, ACS Catal., 2023, 13, 856–865, DOI:10.1021/acscatal.2c04031.
- F. Gasteazoro, G. Catucci and L. Barbieri, et al., Cascade reactions with two non-physiological partners for NAD(P) H regeneration via renewable hydrogen, Biotechnol. J., 2024, 19, e2300567, DOI:10.1002/biot.202300567.
- S. E. Cleary, S. Hall and R. Galan-Bataller, et al., Scalable Bioreactor Production of an O2-Protected [FeFe]-Hydrogenase Enables Simple Aerobic Handling for Clean Chemical Synthesis, ChemCatChem, 2024, 16, e202400193, DOI:10.1002/cctc.202400193.
- Y. Liu, C. Pulignani and S. Webb, et al., Electrostatic [FeFe]-hydrogenase–carbon nitride assemblies for efficient solar hydrogen production, Chem. Sci., 2024, 6088–6094, 10.1039/D4SC00640B.
- T. Happe and J. D. Naber, Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii, Eur. J. Biochem., 1993, 475–481 CrossRef CAS PubMed.
- T. Strekas, B. C. Antanaitis and A. I. Krasna, Characterization and stability of hydrogenase from Chromatium, Biochim. Biophys. Acta, 1980, 1–9 CAS.
- G. R. F. Orton, S. Belazregue and J. K. Cockcroft, et al., Biomimics of [FeFe]-hydrogenases with a pendant amine: Diphosphine complexes [Fe2(CO)4{μ-S(CH2)nS}{κ2-(Ph2PCH2)2NR}] (n = 2, 3; R = Me, Bn) towards H2 oxidation catalysts, J. Organomet. Chem., 2023, 991, 122673, DOI:10.1016/j.jorganchem.2023.122673.
- C. van Dijk, H. J. Grande and S. G. Mayhew, et al., Properties of the hydrogenase of Megasphaera elsdenii, Eur. J. Biochem., 1980, 107, 251–261, DOI:10.1111/j.1432-1033.1980.tb04645.x.
- L. T. Serebryakova, N. A. Zorin and A. A. Karyakin, Improvement of hydrogenase enzyme activity by water-miscible organic solvents, Enzyme Microb. Technol., 2009, 44, 329–333, DOI:10.1016/j.enzmictec.2008.12.004.
- G.-F. Luo, Y. Biniuri and W.-H. Chen, et al., Artificial Photosynthesis with Electron Acceptor/Photosensitizer-Aptamer Conjugates, Nano Lett., 2019, 19, 6621–6628, DOI:10.1021/acs.nanolett.9b02880.
- S. Ezzaher, J.-F. Capon and F. Gloaguen, et al., Influence of a pendant amine in the second coordination sphere on proton transfer at a dissymmetrically disubstituted diiron system related to the [2Fe]H subsite of [FeFe]H2ase, Inorg. Chem., 2009, 48, 2–4, DOI:10.1021/ic801369u.
- W. Roseboom, A. L. Lacey and V. M. Fernandez, et al., The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy, J. Biol. Inorg. Chem., 2006, 11, 102–118, DOI:10.1007/s00775-005-0040-2.
- G. Caserta, A. Adamska-Venkatesh and L. Pecqueur, et al., Chemical assembly of multiple metal cofactors: The heterologously expressed multidomain [FeFe]-hydrogenase from Megasphaera elsdenii, Biochim. Biophys. Acta, 2016, 1857, 1734–1740, DOI:10.1016/j.bbabio.2016.07.002.
- J. Bandekar, Amide modes and protein conformation, Biochim. Biophys. Acta, 1992, 1120, 123–143, DOI:10.1016/0167-4838(92)90261-b.
- A. Barth, Infrared spectroscopy of proteins, Biochim. Biophys. Acta, 2007, 1767, 1073–1101, DOI:10.1016/j.bbabio.2007.06.004.
- E. L. Bell, W. Finnigan and S. P. France, et al., Biocatalysis, Nat. Rev. Methods Primers, 2021, 1, 46, DOI:10.1038/s43586-021-00044-z.
- M. Silberstein, S. Dennis and L. Brown, et al., Identification of substrate binding sites in enzymes by computational solvent mapping, J. Mol. Biol., 2003, 332, 1095–1113, DOI:10.1016/j.jmb.2003.08.019.
- M. Z. Kamal, P. Yedavalli and M. V. Deshmukh, et al., Lipase in aqueous-polar organic solvents: Activity, structure, and stability, Protein Sci., 2013, 22, 904–915, DOI:10.1002/pro.2271.
- Y. Liu, X. Zhang and H. Tan, et al., Effect of pretreatment by different organic solvents on esterification activity and conformation of immobilized Pseudomonas cepacia lipase, Process Biochem., 2010, 45, 1176–1180, DOI:10.1016/j.procbio.2010.03.023.
- B. J. Lemon and J. W. Peters, Binding of exogenously added carbon monoxide at the active site of the iron-only hydrogenase (CpI) from Clostridium pasteurianum, Biochemistry, 1999, 38, 12969–12973, DOI:10.1021/bi9913193.
- J. Duan, A. Hemschemeier and D. J. Burr, et al., Cyanide Binding to [FeFe]-Hydrogenase Stabilizes the Alternative Configuration of the Proton Transfer Pathway, Angew. Chem., Int. Ed., 2023, 62, e202216903, DOI:10.1002/anie.202216903.
- S. Bucciarelli, E. S. Sayedi and S. Osella, et al., Disentangling the role of solvent polarity and protein solvation in folding and self-assembly of α-lactalbumin, J. Colloid Interface Sci., 2020, 561, 749–761, DOI:10.1016/j.jcis.2019.11.051.
- T. Arakawa, Y. Kita and S. N. Timasheff, Protein precipitation and denaturation by dimethyl sulfoxide, Biophys. Chem., 2007, 131, 62–70, DOI:10.1016/j.bpc.2007.09.004.
- J. Kong and S. Yu, Fourier Transformed Infrared Spectroscopic Analysis of Protein Secondary Structures, Acta Biochim. Biophys. Sin., 2007, 39, 549–559, DOI:10.1111/j.1745-7270.2007.00320.x.
- K. Edenharter, M. W. Jaworek and V. Engelbrecht, et al., H2 production under stress: [FeFe]-hydrogenases reveal strong stability in high pressure environments, Biophys. Chem., 2024, 308, 107217, DOI:10.1016/j.bpc.2024.107217.
- K. Griebenow and A. M. Klibanov, On Protein Denaturation in Aqueous−Organic Mixtures but Not in Pure Organic Solvents, J. Am. Chem. Soc., 1996, 118, 11695–11700, DOI:10.1021/ja961869d.
- D. Lousa, A. M. Baptista and C. M. Soares, A molecular perspective on nonaqueous biocatalysis: contributions from simulation studies, Phys. Chem. Chem. Phys., 2013, 15, 13723–13736, 10.1039/c3cp51761f.
- C. M. Soares, V. H. Teixeira and A. M. Baptista, Protein Structure and Dynamics in Nonaqueous Solvents: Insights from Molecular Dynamics Simulation Studies, Biophys. J., 2003, 84, 1628–1641, DOI:10.1016/S0006-3495(03)74972-8.
- H. Land, M. Senger and G. Berggren, et al., Current State of [FeFe]-Hydrogenase Research: Biodiversity and Spectroscopic Investigations, ACS Catal., 2020, 10, 7069–7086, DOI:10.1021/acscatal.0c01614.
- V. Stepankova, S. Bidmanova and T. Koudelakova, et al., Strategies for Stabilization of Enzymes in Organic Solvents, ACS Catal., 2013, 3, 2823–2836, DOI:10.1021/cs400684x.
- A. S. Bommarius and M. F. Paye, Stabilizing biocatalysts, Chem. Soc. Rev., 2013, 42, 6534–6565, 10.1039/c3cs60137d.
- W. Yang, G. E. Vansuch and Y. Liu, et al., Surface-Ligand “Liquid” to “Crystalline” Phase Transition Modulates the Solar H2 Production Quantum Efficiency of CdS Nanorod/Mediator/Hydrogenase Assemblies, ACS Appl. Mater. Interfaces, 2020, 12, 35614–35625, DOI:10.1021/acsami.0c07820.
- M. V. Pavliuk, M. Lorenzi and D. R. Morado, et al., Polymer Dots as Photoactive Membrane Vesicles for [FeFe]-Hydrogenase Self-Assembly and Solar-Driven Hydrogen Evolution, J. Am. Chem. Soc., 2022, 144, 13600–13611, DOI:10.1021/jacs.2c03882.
- D. Adam, L. Bösche and L. Castañeda-Losada, et al., Sunlight-Dependent Hydrogen Production by Photosensitizer/Hydrogenase Systems, ChemSusChem, 2017, 10, 894–902, DOI:10.1002/cssc.201601523.
- J. M. Kuchenreuther, C. S. Grady-Smith and A. S. Bingham, et al., High-Yield Expression of Heterologous [FeFe] Hydrogenases in Escherichia coli, PLoS One, 2010, 5, e15491, DOI:10.1371/journal.pone.0015491.
- C. J. Schwartz, J. L. Giel and T. Patschkowski, et al., IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 14895–14900, DOI:10.1073/pnas.251550898.
- H. Li and T. B. Rauchfuss, Iron Carbonyl Sulfides, Formaldehyde, and Amines Condense To Give the Proposed Azadithiolate Cofactor of the Fe-Only Hydrogenases, J. Am. Chem. Soc., 2002, 124, 726–727, DOI:10.1021/ja016964n.
- V. Engelbrecht, K. Liedtke and A. Rutz, et al., One isoform for one task? The second hydrogenase of Chlamydomonas reinhardtii prefers hydrogen uptake, Int. J. Hydrogen Energy, 2021, 46, 7165–7175, DOI:10.1016/j.ijhydene.2020.11.231.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cy01018c |
| This journal is © The Royal Society of Chemistry 2024 |