Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A closer look at amyloid ligands, and what they tell us about protein aggregates†
Timothy S.
Chisholm
 and
Christopher A.
Hunter
and
Christopher A.
Hunter
 *
*
Yusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge, CB2 1 EW, UK. E-mail: herchelsmith.orgchem@ch.cam.ac.uk
First published on 20th December 2023
Abstract
The accumulation of amyloid fibrils is characteristic of neurodegenerative diseases such as Alzheimer's disease (AD) and Parkinson's disease. Detecting these fibrils with fluorescent or radiolabelled ligands is one strategy for diagnosing and better understanding these diseases. A vast number of amyloid-binding ligands have been reported in the literature as a result. To obtain a better understanding of how amyloid ligands bind, we have compiled a database of 3457 experimental dissociation constants for 2076 unique amyloid-binding ligands. These ligands target Aβ, tau, or αSyn fibrils, as well as relevant biological samples including AD brain homogenates. From this database significant variation in the reported dissociation constants of ligands was found, possibly due to differences in the morphology of the fibrils being studied. Ligands were also found to bind to Aβ(1–40) and Aβ(1–42) fibrils with similar affinities, whereas a greater difference was found for binding to Aβ and tau or αSyn fibrils. Next, the binding of ligands to fibrils was shown to be largely limited by the hydrophobic effect. Some Aβ ligands do not fit into this hydrophobicity-limited model, suggesting that polar interactions can play an important role when binding to this target. Finally several binding site models were outlined for amyloid fibrils that describe what ligands target what binding sites. These models provide a foundation for interpreting and designing site-specific binding assays.
Introduction
Amyloidogenic proteins are an expansive class of polypeptides that self-assemble into fibrillar, highly structured, β-sheet rich aggregates.1–5 The modern biophysical definition of an amyloid fibril encompasses an unbranched protein fibre with a repeating substructure composed of β-strands aligned perpendicular to the fibre axis, forming an extended cross-β sheet structure. The accumulation and deposition of amyloid fibrils are characteristic hallmarks of a range of neurodegenerative diseases.6–15 The most common fibrils implicated in these diseases are composed of amyloid-β (Aβ) peptide and tau in Alzheimer's disease (AD), and α-synuclein (αSyn) in Parkinson's disease (PD).Detailed structural information of amyloid fibrils has been historically challenging to obtain due to the difficulty in obtaining crystalline samples for X-ray studies.16 More recent applications of cryo-electron microscopy techniques have negated the need for crystalline samples and revealed high-resolution structures for several amyloid fibrils.17–21 These structures show that the fibrils are typically composed of two protofilaments, each containing an extended β-sheet structure.1,16,21 Structural investigations of amyloid fibrils have consistently demonstrated a significant degree of polymorphism, where a single amyloid protein can aggregate to form a range of fibril structures.1,20–26 This polymorphism can arise due to differences in the structures of individual protofilaments, the number of protofilaments, and the arrangement of protofilaments relative to one another. Amyloid structure is therefore not strictly dictated by the primary sequence of the polypeptide, but is instead a complex property that is dependent on environmental conditions during aggregation.24,25,27–36
A range of amyloid-binding ligands have been reported for studying and imaging amyloid fibrils.3,37–46 Amyloid-binding ligands are typically either fluorescent or radiolabelled to allow for binding to be detected and measured. Many fluorescent ligands, such as Thioflavin T (ThT, 1) and Congo Red (CR, 2) (Fig. 1), are solvatochromic and exhibit a change in fluorescence properties upon binding to amyloid fibrils. Fluorescent ligands have a range of applications in microscopy, and in identifying or characterising amyloid fibrils. These changes in fluorescence properties can also be used to measure the proportion of bound ligand in binding assays.
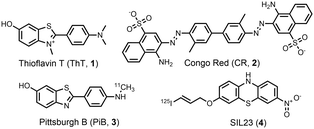 | ||
| Fig. 1 Examples of amyloid-binding ligands: fluorescent ligands thioflavin T (ThT, 1) and Congo Red (CR, 2), and radiolabelled ligands Pittsburgh B (PiB, 3) and SIL23 (4). | ||
Radiolabelled ligands are important for the detection of amyloids in vivo. These ligands are designed as radiotracers for positron emission tomography (PET) by incorporating 3H, 11C or 18F (e.g.3), or for single-photon emission computerized tomography (SPECT) by incorporating 125I (e.g.4). Binding assays of radiotracers are typically performed by isolating the radioligand–fibril complex by filtration and determining the concentration of this species by measuring radioactivity.47–49
The field has focused on developing high-affinity ligands that selectively bind to aggregates comprised of a specific protein. High binding affinities are a necessary feature for PET ligands to provide strong signal-to-noise in imaging, and for detecting dilute aggregates in biofluids.3,41 Selective binding is important as many diseases, such as Alzheimer's disease and Parkinson's disease, are characterised by aggregates comprised of different proteins that are useful to differentiate. Lower-affinity and less-selective ligands, such as ThT, can also be useful as universal markers of amyloids for imaging and biophysical studies.
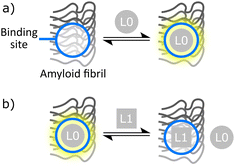
Fluorescence or radiometric binding assays are frequently employed to measure the dissociation constant (Kd) of amyloid ligands. Two common experiments are saturation binding assays and competition binding assays (Fig. 2). Saturation binding assays involve titrating a ligand directly into a sample of amyloid fibril and measuring the proportion of bound ligand. Competition binding assays involve first binding a reporter ligand (L0 in Fig. 2) that is either fluorescent or radioactive to an amyloid fibril. Then, a second competing ligand (L1 in Fig. 2) is added. The change in signal due to displaced L0 is used to indirectly monitor binding of L1, which means that this experiment can be used to study L1 ligands that are not fluorescent or radioactive.
Although a large number of ligand–amyloid fibril interactions have been reported in the literature, a compilation and review of these data has yet to be performed. Here we present an analysis of literature data on amyloid-binding ligands and use this information to provide some insights into the properties required for ligands to bind to amyloid fibrils and the nature of the ligand binding sites that are present on different types of amyloid fibril.
Database of amyloid ligands
We compiled a database of 3457 measurements of ligand–amyloid fibril interactions from the scientific literature.50–385 The requirements to be included in this database were (1) a known chemical structure, (2) a structure that does not contain metals (metalloligands have been thoroughly reviewed elsewhere in the literature43,386–391), (3) a Kd or IC50 measurement, and (4) a fibril target related to Aβ, tau, or αSyn. Nearly half of the database reports on binding to Aβ fibrils formed in vitro, although recent years has seen a shift in focus toward tau and αSyn fibrils formed in vitro (Table 1). There is also a significant amount of data on binding to AD brain homogenates. Most of the reported Kd values in the database are in the nanomolar range, with smaller numbers of picomolar or micromolar values (Fig. 3). The database contains 2,076 unique ligands, which are structurally diverse. The most common structural cores are illustrated in Fig. 4.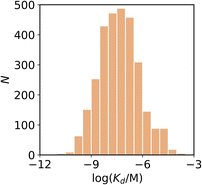 | ||
| Fig. 3 Frequency distribution of log(Kd/M) values in the database plotted as the number of ligands (N). | ||
We will use this database to investigate four questions. First, how reproducible are the Kd measurements reported in the literature? Second, how selective are ligands for different types of fibril? Third, what properties of ligands lead to high affinity binding? And fourth, what can we learn about the nature of the binding sites present on amyloid fibrils?
Variation between reported measurements
Reproducibility underpins the scientific method yet for many amyloid ligands, such as ThT 1, a wide range of Kd values have been reported. The variability in Kd values in the database was investigated by comparing Kd values for ligands binding to a specific target and measured using the same method: fluorescence saturation assays, radioligand saturation assays, or radioligand competition assays. Only binding measurements corresponding to a one-site model were used, and Kd values measured using competition binding assays were only compared for reporting ligands from the same structural class (Fig. 4). The results for all of the ligand–fibril combinations with three or more Kd measurements are illustrated in Fig. 5, and the structures of the ligands are shown in Fig. 6. | ||
| Fig. 6 Structures of ligands listed in Fig. 5 with three or more Kd measurements reported in the literature. | ||
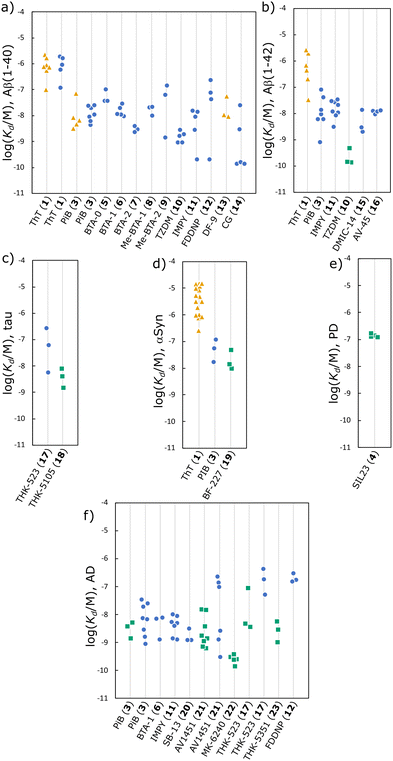
For some ligands, the measurements of dissociation constant are consistent between different publications. For example, the results for BTA ligands 3, 5, 6, 7, 8, 10 (see Fig. 6 for structures) show excellent reproducibility for binding to Aβ(1–40) (Fig. 5(a)), and similarly for 16 binding to αSyn (Fig. 5(d)) and 22 binding to AD brain homogenates (Fig. 5(f)). However, the measured dissociations constants for ligand–fibril combinations can vary by more than an order of magnitude. For example, the dissociation constants measured for 3 binding to Aβ(1–42) range from 0.8–77 nM (Fig. 5(b)), and similar variation is found for 3 and 21 binding to AD brain homogenates (Fig. 5(f)), and for 1 binding to αSyn (Fig. 5(d)).
Where multiple measurements of Kd are reported in a single publication, such as for 22 binding AD brain homogenates, the results are usually relatively consistent. The variability seen in Fig. 5 may therefore come primarily from differences in sample preparation or assay methodology between publications. A main source of variability for amyloid binding is that fibrils composed of the same protein can have different morphologies. Different publications often use different fibril aggregation procedures, or do not describe the aggregation procedure in detail. For measurements using biological fibrils, the method of isolating fibrils from brain homogenates varies and may also influence fibril structure. Different studies may therefore be screening different morphologies. Ideally, fibrils should be characterised using techniques such as transmission electron microscopy (TEM), circular dichroism (CD), or limited proteolysis, which can reveal differences and similarities in morphology.
Differences in assay conditions can also influence the measurement of ligand dissociation constants. For radioligand assays, washing steps are used to separate bound and free ligand. Poorly optimised washing steps may remove bound ligand, leading to incorrect quantification of binding.47,48 For fluorescence competition assays, 1 is commonly used as a reporting ligand. However, 1 is typically sold as an impure mixture with dye content as low as 65%, so purification by recrystallisation is required for quantitative measurements.392 Additionally 1 typically shows only micromolar affinities for amyloid fibrils, but at concentrations above 4–5 μM in aqueous solution 1 forms micelles and self-fluoresces.392–394 Accurately measuring the micromolar binding of 1 is therefore challenging, possibly explaining the large variability in Kd values for 1 binding to αSyn. An additional source of error may be the methods used to analyse titration data. Many publications do not describe the procedure used to fit titration data to a binding isotherm, and some use a two-site binding model to fit data without a clear justification or comparison to a one-site model.
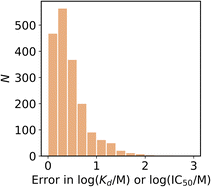
The variation of measurements reported within individual studies, i.e. the reported error, is illustrated in Fig. 7. The median error in the reported Kd was 15% or 0.35 log units, and only 9% of the reported Kd measurements had an error greater than 1 log unit. In contrast, 42% of the ligands listed in Fig. 5 have Kd measurements with a range of more than 1 log unit. The variations reported within individual studies is therefore less than the variation between studies. To accurately represent this variation it is important that truly independent replicates are performed, and that the meaning of provided error values are stated. It should be clear whether a reported error is, for example, a confidence interval or a standard error.
Steps can therefore be taken to reduce, or better understand, the variability in reported Kd values: the method for fibril aggregation and isolation should be reported in detail; fibrils should be characterised using techniques such as TEM, CD, and limited proteolysis; the method used for the binding assay should be reported in detail; the method used to fit titration data should be reported, as well as justification for the use of isotherms that are more complex than a one-site model. In other words, the variation in Kd values highlighted in Fig. 5 is most likely due to differences in what was actually measured in different reports of the same ligand–fibril interaction.
Ligand selectivity
Amyloid ligands that selectively bind to a specific type of fibril are useful for imaging and diagnostic applications. To evaluate binding selectivity between different fibril targets, ligands in the database with Kd values for two or more types of fibril were identified. As above, only Kd values measured using the same assay method and a one-site binding model were compared, and Kd values measured using competition binding assays were only compared for reporting ligands from the same structural class (see Fig. 4). When multiple Kd values were reported for the same ligand–fibril combination with the same assay method, the average value was used.If the ligand binding profiles for different types of amyloid fibril are similar, we can assume that the nature of the binding sites that are present are likely to be similar. The root mean square difference (RMSD) in ligand binding affinity as defined by eqn (1) was therefore used for pairwise comparison of different amyloid fibrils.
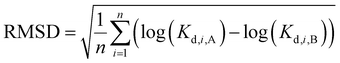 | (1) |
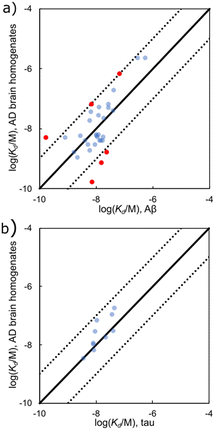
Fig. 8 shows that there is an excellent correlation between the results obtained for ligand binding to Aβ(1–40) and Aβ(1–42) fibrils. The RMSD between the log(Kd/M) for Aβ(1–40) and the log(Kd/M) for Aβ(1–42) is 0.46, and most of the datapoints fall very close to the y = x line. This result suggests that the binding sites on Aβ(1–40) and Aβ(1–42) fibrils are structurally similar, so the data for ligands binding to both Aβ fibril isoforms will be combined and collectively referred to as Aβ fibrils. There are two outliers highlighted in red in Fig. 8, and the structures of these ligands are shown in Fig. 9. Thiazine red (ThR, 24) and curcumin (25) show selectivity between the two Aβ isoforms with an affinity for Aβ(1–40) that is more than an order of magnitude higher than for Aβ(1–42).
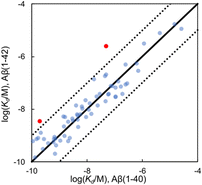 | ||
| Fig. 8 Comparison of values of Kd measured for ligand binding to Aβ(1–40) and Aβ(1–42) fibrils (65 ligands, RMSD = 0.46 log units). The solid line is y = x, and the dotted lines correspond to ± 1 log unit. Red datapoints are outliers that bind to one fibril target at least one order-of-magnitude stronger than the other fibril target. The chemical structures of these outliers are shown in Fig. 9. | ||
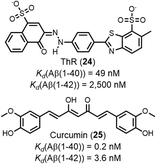 | ||
| Fig. 9 Structures of ligands that show selectively between Aβ(1–40) fibrils and Aβ(1–42) fibrils, highlighted as red datapoints in in Fig. 8. | ||
Fig. 10 compares dissociation constants measured for the same ligand binding to Aβ, αSyn, and tau. The ligand affinities for Aβ and tau fibrils are broadly similar with most datapoints clustered around the y = x line and an overall RMSD of 0.55 log units (Fig. 10(a)). However, when binding of a ligand to either Aβ or tau is compared with the affinity for αSyn fibrils, the correlation is much weaker (RMSD = 0.66 log units for Fig. 10(b), and 0.79 log units for Fig. 10(c)). These results suggest that the ligand binding sites found on Aβ and tau have a high degree of similarity, but αSyn fibrils are quite different, which would make αSyn selective ligands an attractive target.
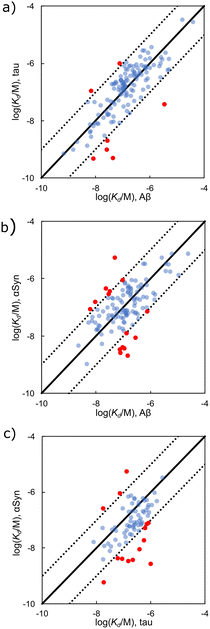 | ||
| Fig. 10 Comparison of Kd values measured for ligand binding to (a) Aβ and tau fibrils (139 ligands, RMSD = 0.55 log units), (b) Aβ and αSyn fibrils (119 ligands, RMSD = 0.66 log units), and (c) tau and αSyn fibrils (76 ligands, RMSD = 0.79 log units). The solid line is y = x, and the dotted lines correspond to ± 1 log unit. Red datapoints bind to one fibril type at least one order-of-magnitude stronger than the other (structures in Fig. 11 and 12). | ||
The structures of the ligands that are outliers in Fig. 10 (highlighted in red) are shown in Fig. 10–12. The ligands in Fig. 11 show selectivity for Aβ over αSyn. Ligands 29 and 30 also show selectivity for Aβ over tau.54,55 The ligands in Fig. 12 show selectivity for tau over Aβ, although 36 only bound to soluble rather than insoluble tau aggregates.56 Ligands 33, 34, and 35 also show selectivity for tau over αSyn. Fig. 13 shows the ligands that selectivity bind to αSyn fibrils.54,55,57–61
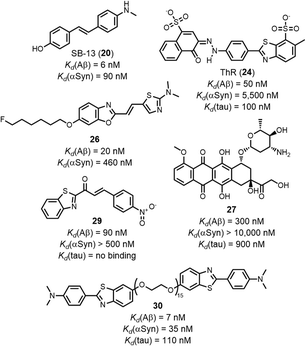 | ||
| Fig. 11 Structures of ligands that show selective binding for Aβ fibrils, highlighted as red datapoints in in Fig. 10(a) and(b). | ||
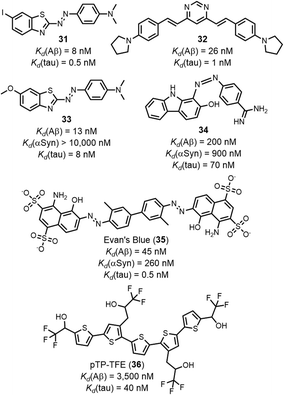 | ||
| Fig. 12 Structures of ligands that show selective binding for tau fibrils, highlighted as red datapoints in in Fig. 10(a) and (c). | ||
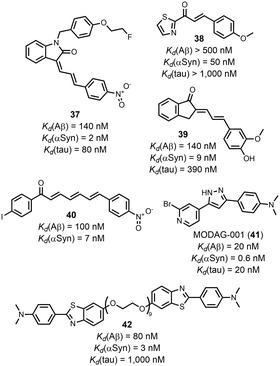 | ||
| Fig. 13 Structures of ligands that show selective binding for αSyn fibrils, highlighted as red datapoints in in Fig. 10(b) and (c). | ||
Fig. 14 compares the Kd values measured for ligands binding to AD brain homogenates with the Kd values for binding to Aβ and tau fibrils. Relatively small datasets are available, although several selective ligands were identified. The benzoheterocycles flutemetamol (43), AZD2184 (44), and 45 are all selective for AD brain homogenates. IMSB (46) has a greater affinity for Aβ fibrils, and a screen of several indanones by Nan et al. demonstrated that all had a higher affinity for Aβ fibrils (e.g.47).62 Amyloid ligands are often screened against fibrils formed in vitro (e.g. Aβ or tau fibrils) as a substitute for fibrils formed in vivo (e.g. AD brain homogenates). Fig. 14 suggests that binding affinities against in vitro Aβ and tau fibrils are similar to binding affinities against AD brain homogenates, although a larger dataset would help define the limitations of this comparison. For example, different types of ligands target different binding sites, and some binding sites on in vitro fibrils may more closely resemble biological binding sites than others. Additionally, different publications appear to be screening ligands against different fibril morphologies. Some in vitro preparations will be more representative of biological fibrils than others. Recently in vitro aggregation procedures have been reported that produce fibrils with biological morphologies, which would lead to more biologically relevant binding assays.28
 | ||
| Fig. 14 Comparison of values of Kd measured for ligand binding to (a) Aβ fibrils and AD brain homogenates (32 ligands RMSD = 0.70 log units), and (b) tau fibrils and AD brain homogenates (7 ligands, RMSD = 0.40 log units). The solid line is y = x, and the dotted lines correspond to ± 1 log unit. Red datapoints are outliers that bind to one fibril target at least one order-of-magnitude stronger than the other fibril target. The chemical structures of these outliers are shown in Fig. 15. | ||
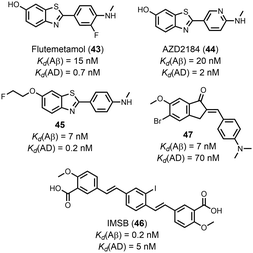 | ||
| Fig. 15 Structures of ligands that show selectively between Aβ fibrils and AD brain homogenates, highlighted as red datapoints in Fig. 14. | ||
Selectivity was also achieved by using different types of binding assays. Honson et al. demonstrated that 24 selectively targeted tau (Kd = 200 nM) over Aβ(1–42) (Kd > 2000 nM) fibrils when measured by a fluorescence displacement assay with thioflavin S (ThS, 54). However, 24 exhibited a similar affinity to both fibrils (Kd = 1000–2500 nM) when measured by a fluorescence saturation assay.63 This difference in affinity may be due to different assays targeting different binding sites.
Ligand hydrophobicity
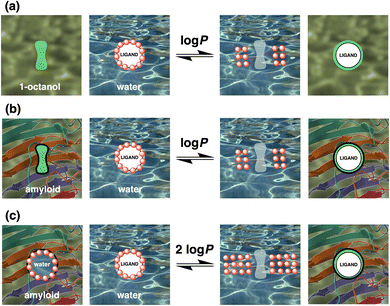
The relationship between Kd and various molecular descriptors of the ligands (e.g. molecular weight, topological polar surface area, the number of rotatable bonds) was explored. The only descriptor found to have any relationship with Kd was log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (see ESI†). Amyloid ligands are thought to bind in hydrophobic grooves on the surface of fibrils,392 and if the hydrophobic effect is largely responsible for binding, there may be some relationship between the log(Kd/M) and the n-octanol–water partition coefficient (log
P (see ESI†). Amyloid ligands are thought to bind in hydrophobic grooves on the surface of fibrils,392 and if the hydrophobic effect is largely responsible for binding, there may be some relationship between the log(Kd/M) and the n-octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of the ligands. The value of log
P) of the ligands. The value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P provides a measure of the free energy change associated with replacing the solvation shell of a ligand dissolved in water with a non-polar environment (Fig. 16(a)), and simple calculations based on chemical structure provide reasonably reliable values of log
P provides a measure of the free energy change associated with replacing the solvation shell of a ligand dissolved in water with a non-polar environment (Fig. 16(a)), and simple calculations based on chemical structure provide reasonably reliable values of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P.396 In general non-polar groups, such as hydrocarbons, increase the value of log
P.396 In general non-polar groups, such as hydrocarbons, increase the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, while polar functional groups, such as hydroxyls, are well-solvated by water and decrease the value of log
P, while polar functional groups, such as hydroxyls, are well-solvated by water and decrease the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P.
P.
Fig. 17 compares the values of Kd measured for ligands binding to AD brain homogenates, Aβ fibrils, tau fibrils, and αSyn fibrils with the corresponding values of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. Although there is no direct correlation between the two parameters, high affinity ligands tend to have a log
P. Although there is no direct correlation between the two parameters, high affinity ligands tend to have a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 3 or more. There appears to be a sharp cut-off in Fig. 17(a), indicated by the red line, suggesting that there is a lower limit on the value of Kd that can be achieved for a given value of log
P value of 3 or more. There appears to be a sharp cut-off in Fig. 17(a), indicated by the red line, suggesting that there is a lower limit on the value of Kd that can be achieved for a given value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P. Many hydrophobic ligands lie to the right of this line and exhibit lower binding affinities, but there are some polar ligands that lie to the left of the line and exhibit a higher binding affinity. The slope of this red line is −2, corresponding to where the log(Kd/M) of a ligand is proportional to twice the log
P. Many hydrophobic ligands lie to the right of this line and exhibit lower binding affinities, but there are some polar ligands that lie to the left of the line and exhibit a higher binding affinity. The slope of this red line is −2, corresponding to where the log(Kd/M) of a ligand is proportional to twice the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of the ligand.
P of the ligand.
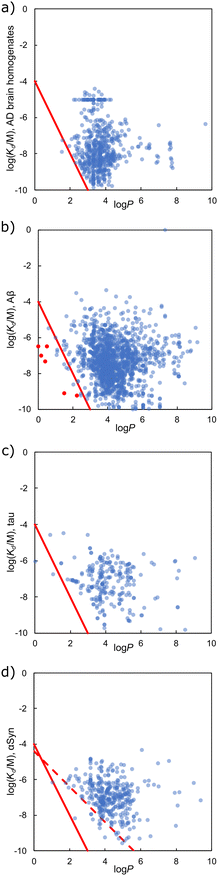 | ||
Fig. 17 Comparison of ligand log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values395 with values of log(Kd/M) measured for binding to (a) AD brain homogenates, (b) Aβ fibrils, (c) tau fibrils, and (d) αSyn fibrils. The slope of the solid red lines is −2, and the slope of the dotted red line is −1. Examples of outliers with a binding affinity greater than that predicted by the solid red line are highlighted in red. The chemical structures of these outliers are shown in Fig. 18. P values395 with values of log(Kd/M) measured for binding to (a) AD brain homogenates, (b) Aβ fibrils, (c) tau fibrils, and (d) αSyn fibrils. The slope of the solid red lines is −2, and the slope of the dotted red line is −1. Examples of outliers with a binding affinity greater than that predicted by the solid red line are highlighted in red. The chemical structures of these outliers are shown in Fig. 18. | ||
A slope of −2 would be expected for idealised ligand binding that is based entirely on the hydrophobic effect. If a ligand was completely desolvated on binding into a completely non-polar pocket on the surface of a fibril, then the binding affinity would be directly related to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, if the binding pocket on the fibril did not require any desolvation (Fig. 16(b)). However, ligand binding involves not only desolvation of the ligand, it also requires desolvation of the binding pocket. If the binding pocket was perfectly complementary to the ligand, then the molecular surface that would be desolvated on the fibril would be the same as the surface of the ligand. The contribution due to desolvation of both the ligand and the binding pocket would therefore be twice the value of log
P, if the binding pocket on the fibril did not require any desolvation (Fig. 16(b)). However, ligand binding involves not only desolvation of the ligand, it also requires desolvation of the binding pocket. If the binding pocket was perfectly complementary to the ligand, then the molecular surface that would be desolvated on the fibril would be the same as the surface of the ligand. The contribution due to desolvation of both the ligand and the binding pocket would therefore be twice the value of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for the ligand (Fig. 16(c)).
P for the ligand (Fig. 16(c)).
Ligands that are not perfectly complementary to the binding site or are not fully encapsulated inside the pocket on the surface of the fibril will have lower affinities because they fail to maximise the hydrophobic interactions that are possible. Fig. 17(a) shows that increasing the hydrophobicity of the ligand does not necessarily improve affinity, because hydrophobicity can be added in regions that do not contact the binding site. For example, there are ligands with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values ranging from 2 to 8 that all achieve the same log
P values ranging from 2 to 8 that all achieve the same log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (Kd/M) of −8. However, there are no ligands with a log
(Kd/M) of −8. However, there are no ligands with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value less than 2 that achieve a log
P value less than 2 that achieve a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (Kd/M) of −8.
(Kd/M) of −8.
Although there is more scatter in the Aβ data shown in Fig. 17(b), a similar cut-off appears to apply to these measurements. There are two differences between Fig. 17(a) and (b). Firstly, the cut-off for the majority of the datapoints in Fig. 17(b) appears to be displaced to the right by about one log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P unit compared with the red cut-off line. Notably, when binding of the same ligand to AD brain homogenates and Aβ fibrils was compared, significant differences were not observed (Fig. 14(a)). However, Fig. 14 considered only a subset of ligands that were screened against both targets, whereas the data shown in Fig. 17 is for all ligands in the database. The other notable difference between Fig. 17(a) and (b) is that a small number of ligands lie to the left of the cut-off line and have higher affinities for Aβ fibrils than the hydrophobic effect alone would allow (red datapoints in Fig. 17(b)). Polar interactions with some of the functional groups on these more polar ligands (Fig. 18) may play an important role in binding. Ligands that make binding site-specific polar interactions are particularly attractive lead compounds for the development of ligands with increased fibril selectivity. Alternatively, polar groups like the sugars in 27, 51 and 52 may sit outside the binding pocket and have a minimal impact on binding, whilst significantly decreasing log
P unit compared with the red cut-off line. Notably, when binding of the same ligand to AD brain homogenates and Aβ fibrils was compared, significant differences were not observed (Fig. 14(a)). However, Fig. 14 considered only a subset of ligands that were screened against both targets, whereas the data shown in Fig. 17 is for all ligands in the database. The other notable difference between Fig. 17(a) and (b) is that a small number of ligands lie to the left of the cut-off line and have higher affinities for Aβ fibrils than the hydrophobic effect alone would allow (red datapoints in Fig. 17(b)). Polar interactions with some of the functional groups on these more polar ligands (Fig. 18) may play an important role in binding. Ligands that make binding site-specific polar interactions are particularly attractive lead compounds for the development of ligands with increased fibril selectivity. Alternatively, polar groups like the sugars in 27, 51 and 52 may sit outside the binding pocket and have a minimal impact on binding, whilst significantly decreasing log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of the ligand.
P of the ligand.
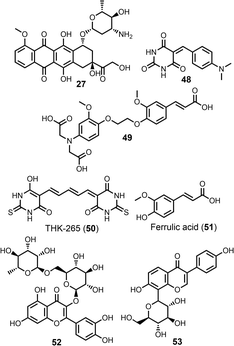 | ||
| Fig. 18 Structures of polar Aβ ligands with high binding affinities, highlighted as red datapoints in Fig. 17(b). | ||
Fewer datapoints are available for ligand binding to tau and αSyn fibrils. The relationship between Kd and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P for tau fibrils is similar to that observed for AD brain homogenates and Aβ fibrils. For αSyn fibrils, the datapoints are significantly displaced to the right of the cut-off line (Fig. 17(d)). For ligands with similar hydrophobicities, binding affinities for αSyn fibrils were generally lower than observed for the other fibrils. This result suggests that there may be some fundamental difference in the structure of the binding sites present on αSyn fibrils, which is consistent with the differences in ligand binding affinities highlighted in Fig. 10. For example, if the αSyn binding pocket were partially collapsed in the absence of ligand, then the hydrophobic contribution to binding would be substantially reduced. In the limit where no desolvation of the binding pocket was required, because it was fully collapsed and opened only to accommodate a ligand, then the maximum log(Kd/M) of a ligand would become proportional to twice the log
P for tau fibrils is similar to that observed for AD brain homogenates and Aβ fibrils. For αSyn fibrils, the datapoints are significantly displaced to the right of the cut-off line (Fig. 17(d)). For ligands with similar hydrophobicities, binding affinities for αSyn fibrils were generally lower than observed for the other fibrils. This result suggests that there may be some fundamental difference in the structure of the binding sites present on αSyn fibrils, which is consistent with the differences in ligand binding affinities highlighted in Fig. 10. For example, if the αSyn binding pocket were partially collapsed in the absence of ligand, then the hydrophobic contribution to binding would be substantially reduced. In the limit where no desolvation of the binding pocket was required, because it was fully collapsed and opened only to accommodate a ligand, then the maximum log(Kd/M) of a ligand would become proportional to twice the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of the ligand. The red dotted line in Fig. 17(b) shows this situation and provides a rather good description of an alternative cut-off line for the αSyn data. Of course, it is also possible that only relatively weak αSyn ligands have been discovered to date.
P of the ligand. The red dotted line in Fig. 17(b) shows this situation and provides a rather good description of an alternative cut-off line for the αSyn data. Of course, it is also possible that only relatively weak αSyn ligands have been discovered to date.
Binding site heterogeneity
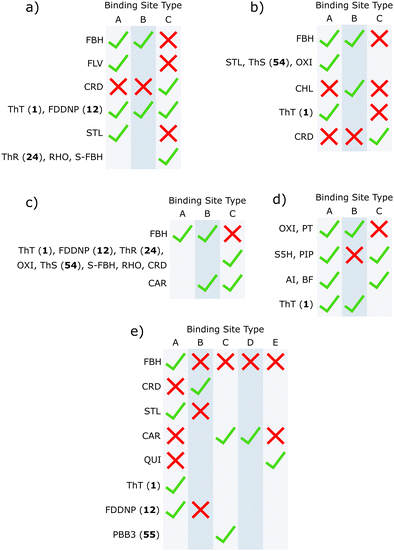
Competition binding assays are a useful tool for identifying different binding sites on amyloid fibrils. Typically, a competing ligand is titrated into a solution of fibril bound to a reporting ligand that produces either a fluorescent or radioactive signal (Fig. 2). Binding of the competing ligand (L1) is observed by monitoring changes in the signal produced by the reporting ligand (L0). Using this assay format, ligands that target the same or different binding sites can be identified. Several publications have used this approach and show that some ligands independently bind to amyloid fibrils but do not compete with one another, indicating the presence of different types of binding sites.64–67,397 Similarly, if the dissociation constant measured for a competing ligand changes as a function of the reporter ligand, then the presence of multiple binding sites with different ligand affinities can be inferred.A small number of binding site models have previously been proposed for amyloid fibrils based on competition binding assays and a range of analytical and computational techniques.65–67,397–406 Lockhart et al., Ye et al., and LeVine all reported Aβ(1–40) binding site models and include multiple ThT 1 binding sites, a separate high-affinity FBH site, and a site accessible to both ThT 1 and CRD.66,67,401 Ye et al. also proposed that CRD and FBH share a binding site, which other studies do not show.67 Hsieh et al. proposed a binding site model for αSyn fibrils with three sites, denoted site 2, site 9, and site 3/13.65 S5H were shown to bind to site 2, with a weaker affinity for site 9, while PIP were shown to bind to site 9, with a weaker affinity for site 2. OXI and PHT targeted site 3/13. Cai et al. identified that FBH, QUI, and CAR all have separate high-affinity binding sites in AD brain homogenates.397 However, all of these binding sites consider a relatively small subset of amyloid binding ligands and do not consider other data from the literature.
Here, data from different competition binding assays is collated to propose binding site models for each of these fibril types. The ligands in the database were first grouped by structural class (Fig. 4), as ligands with a shared structural core are assumed to target the same subset of fibril binding sites. Structural class BSB was found to consistently target the same binding sites as CRD, and so these ligands are treated as a single CRD class in the analysis that follows. Key reporter ligands commonly used in competition assays are shown in Fig. 19.
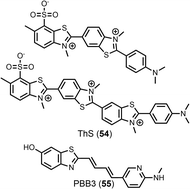 | ||
| Fig. 19 Structures of additional ligands commonly used in competition assays. ThS 54 is used as a mixture of compounds, with the two most abundant components shown. | ||
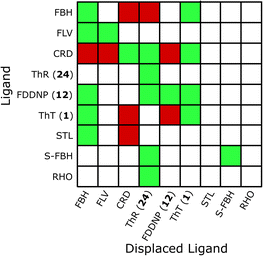
Heatmaps were used to summarise competition binding experiments for each fibril type. Fig. 20 shows the heatmap for Aβ(1–40) fibrils (see ESI† for other fibrils). Green tiles are used to show that the ligand class on the y-axis displaces the ligand class on the x-axis. Red tiles are used to show that no displacement was observed, and white tiles indicate experiments that have not been carried out. The data in the heatmap can be used to construct the simplest binding site model that satisfies the competition binding assay data for each fibril. Analysis of the data in Fig. 20 requires three different binding sites. For example, FBH bind to Aβ(1–40) fibrils with a high affinity at a site we define as site A. Ligands FLV, STL, ThT and FDDNP displace FBH and therefore also bind to site A. Several reports suggest that FBH targets a second site on Aβ(1–40) fibrils that is also shared with ThT, which we define as site B. Ligand CRD is not displaced by either FBH or ThT, which means that there is a third site, which we define as site C. Some reports suggest that ThT and CRD share a binding site,66,260 so ThT may also bind to site C. This assignment is supported by reports that show that ThT completely displaces FBH, but FBH partially displaces ThT.66 However, it is also worth noting that other reports find no competition between ThT and CRD.166,325 The three binding site model shown in Fig. 21(a) accounts for all of the competition experiments shown in the heatmap in Fig. 20. A similar analysis was carried out for Aβ(1–42), tau, αSyn, and AD brain homogenates to give the binding site models summarised in Fig. 21 (see ESI† for details).
Using the results from reported competition binding assays we have constructed models of the binding sites present on different types of amyloid fibrils. Each site bound a different combination of ligands, and many ligands were shown to bind to multiple sites. While different binding studies may employ morphologically distinct fibrils, these models were supported by data from multiple sources. Some inconsistencies were observed between publications, such as the orthogonal binding of ThT 1 and CRD to Aβ(1–40) fibrils. These inconsistencies may suggest that certain binding sites are particularly sensitive to fibril morphology and could therefore inform the development of morphology-selective ligands. These binding site models also allow specific binding sites to be targeted by designing competition assays using specific combinations of ligands.
Conclusions
In summary we have compiled a comprehensive database of amyloid-binding ligands, comprising a total of 3457 experimental dissociation constants for 2076 unique compounds. This represents the largest reported database of ligands that bind amyloid fibrils involved in neurodegenerative diseases. Our hope is that this database will be used to guide ligand development and to improve our understanding of how ligands interact with amyloid fibrils.The reproducibility of binding measurements in this database was then investigated. Many examples of ligands with binding constants spanning more than an order of magnitude were found. This variability may be a result of ligands being assayed against different fibril morphologies, which must be considered when comparing binding studies.
Ligands were then shown to bind to Aβ(1–40) and Aβ(1–42) fibrils with similar affinities, whereas a greater difference was found for binding to Aβ or tau and αSyn fibrils. This result may be due to αSyn fibrils possessing the most unique binding sites, or due to the larger number of studies developing αSyn-selective ligands. Ligands were then found to bind AD brain homogenates similarly to Aβ and tau fibrils that were formed in vitro, although more work is needed to confidently determine how representative these in vitro fibrils are of biological fibrils for binding studies.
The binding of ligands to fibril binding sites was then found to be limited by the hydrophobic effect. Near complete desolvation of both the ligand and binding site is observed for high-affinity ligands binding to AD brain homogenates. For Aβ fibrils, some ligands did not fit into this hydrophobicity-limited model suggesting that polar interactions can contribute to binding this target. For αSyn fibrils, relatively little desolvation of the binding site was required for binding to occur.
Finally, binding site models were constructed for Aβ(1–40), Aβ(1–42), tau, and αSyn fibrils, and AD brain homogenates. Three to five binding sites were found for each fibril, and the ligands that target each site were identified. This result provides a framework for interpreting the results of binding assays, and for designing new site-specific assays.
Author contributions
The manuscript was written through contributions of all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Cambridge Trust Prince of Wales Scholarship for funding T. S. C.Notes and references
- J. Greenwald and R. Riek, Structure, 2010, 18, 1244–1260 CrossRef CAS PubMed.
- Z. L. Almeida and R. M. M. Brito, Molecules, 2020, 25, 1195 CrossRef CAS PubMed.
- C. A. Mathis, B. J. Lopresti, M. D. Ikonomovic and W. E. Klunk, Semin. Nucl. Med., 2017, 47, 553–575 CrossRef PubMed.
- J. Vaquer-Alicea and M. I. Diamond, Annu. Rev. Biochem., 2019, 88, 785–810 CrossRef CAS PubMed.
- P. C. Ke, R. Zhou, L. C. Serpell, R. Riek, T. P. J. Knowles, H. A. Lashuel, E. Gazit, I. W. Hamley, T. P. Davis, M. Fändrich, D. E. Otzen, M. R. Chapman, C. M. Dobson, D. S. Eisenberg and R. Mezzenga, Chem. Soc. Rev., 2020, 49, 5473–5509 RSC.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2017, 86, 27–68 CrossRef CAS PubMed.
- F. U. Hartl, Annu. Rev. Biochem., 2017, 86, 21–26 CrossRef CAS PubMed.
- P. Sweeney, H. Park, M. Baumann, J. Dunlop, J. Frydman, R. Kopito, A. McCampbell, G. Leblanc, A. Venkateswaran, A. Nurmi and R. Hodgson, Transl. Neurodegener., 2017, 6, 6 CrossRef PubMed.
- A. Alzheimer, Neurol. Cent., 1906, 23, 1129–1136 Search PubMed.
- A. Alzheimer, Allg. Z. Psychiatr. Psych.-Gerichtl. Med., 1907, 64, 146–148 Search PubMed.
- G. R. Lafora, Trab. Lab. Invest. Biol. Univers. Madr., 1913, 11, 29–42 Search PubMed.
- G. R. Lafora, Trab. Lab. Invest. Biol. Univers. Madr., 1913, 11, 43–54 Search PubMed.
- F. H. Lewy, in Handbuch der Neurologie, Dritter Band, Spezielle Neurologie I, ed. M. Lewandowsky, Julius Springer, Berlin, 1912, pp.920–933 Search PubMed.
- C. Trétiakoff, Contribution a l’etude l’anatomie pathologique du locus Niger de soemmering: avec quelques déductions relatives à la pathogénie des troubles du tonus musculaire et de la maladie de Parkinson, Jouve, 1919 Search PubMed.
- M. G. Spillantini, M. L. Schmidt, V. M.-Y. Lee, J. Q. Trojanowski, R. Jakes and M. Goedert, Nature, 1997, 388, 839–840 CrossRef CAS PubMed.
- R. N. Rambaran and L. C. Serpell, Prion, 2008, 2, 112–117 CrossRef PubMed.
- M. Sunde, L. C. Serpell, M. Bartlam, P. E. Fraser, M. B. Pepys and C. C. F. Blake, J. Mol. Biol., 1997, 273, 729–739 CrossRef CAS PubMed.
- B. C. Creekmore, Y. W. Chang and E. B. Lee, J. Neuropathol. Exp. Neurol., 2021, 80, 514–529 CrossRef CAS PubMed.
- M. G. Iadanza, M. P. Jackson, E. W. Hewitt, N. A. Ranson and S. E. Radford, Nat. Rev. Mol. Cell Biol., 2018, 19, 755–773 CrossRef CAS PubMed.
- D. Li and C. Liu, Nat. Rev. Neurosci., 2022, 23, 523–534 CrossRef CAS PubMed.
- M. Fändrich, M. Schmidt and N. Grigorieff, Trends Biochem. Sci., 2011, 36, 338–345 CrossRef PubMed.
- B. H. Toyama and J. S. Weissman, Annu. Rev. Biochem., 2011, 80, 557–585 CrossRef CAS PubMed.
- A. K. Paravastu, R. D. Leapman, W. M. Yau and R. Tycko, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18349–18354 CrossRef CAS PubMed.
- M. Y. Suvorina, O. M. Selivanova, E. I. Grigorashvili, A. D. Nikulin, V. V. Marchenkov, A. K. Surin and O. V. Galzitskaya, J. Alzheimer's Dis., 2015, 47, 583–593 CAS.
- M. Fändrich, J. Meinhardt and N. Grigorieff, Prion, 2009, 3, 89–93 CrossRef PubMed.
- J. P. Colletier, A. Laganowsky, M. Landau, M. Zhao, A. B. Soriaga, L. Goldschmidt, D. Flot, D. Cascio, M. R. Sawaya and D. Eisenberg, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16938–16943 CrossRef CAS PubMed.
- W. Hoyer, T. Antony, D. Cherny, G. Heim, T. M. Jovin and V. Subramaniam, J. Mol. Biol., 2002, 322, 383–393 CrossRef CAS PubMed.
- S. Lövestam, F. A. Koh, B. van Knippenberg, A. Kotecha, A. G. Murzin, M. Goedert and S. H. W. Scheres, eLife, 2022, 11, e76494 CrossRef PubMed.
- V. H. Finder, I. Vodopivec, R. M. Nitsch and R. Glockshuber, J. Mol. Biol., 2010, 396, 9–18 CrossRef CAS PubMed.
- T. Härd, J. Phys. Chem. Lett., 2014, 5, 607–614 CrossRef PubMed.
- P. I. Kitov, J. M. Sadowska, G. Mulvey, G. D. Armstrong, H. Ling, N. S. Pannu, R. J. Read and D. R. Bundle, Nature, 2000, 403, 669–672 CrossRef CAS PubMed.
- G. Bhak, J. Lee, T. H. Kim, S. Lee, D. Lee and S. R. Paik, Biochem. J., 2014, 464, 259–269 CrossRef CAS PubMed.
- A. T. Petkova, R. D. Leapman, Z. Guo, W. M. Yau, M. P. Mattson and R. Tycko, Science, 2005, 307, 262–265 CrossRef CAS PubMed.
- L. Bousset, A. Alik, A. Arteni, A. Böckmann, B. H. Meier and R. Melki, Methods Mol. Biol., 2023, 2551, 345–355 CrossRef PubMed.
- R. Guerrero-Ferreira, N. M. I. Taylor, A. A. Arteni, P. Kumari, D. Mona, P. Ringler, M. Britschgi, M. E. Lauer, A. Makky, J. Verasdock, R. Riek, R. Melki, B. H. Meier, A. Böckmann, L. Bousset and H. Stahlberg, eLife, 2019, 8, e48907 CrossRef CAS PubMed.
- P. Ragonis-Bachar and M. Landau, Curr. Opin. Struct. Biol., 2021, 68, 184–193 CrossRef CAS PubMed.
- C. A. Mathis, N. S. Mason, B. J. Lopresti and W. E. Klunk, Semin. Nucl. Med., 2012, 42, 423–432 CrossRef PubMed.
- N. S. Mason, C. A. Mathis and W. E. Klunk, J. Labelled Compd. Radiopharm., 2013, 56, 89–95 CrossRef CAS PubMed.
- A. Leuzy, K. Chiotis, L. Lemoine, P. G. Gillberg, O. Almkvist, E. Rodriguez-Vieitez and A. Nordberg, Mol. Psychiatry, 2019, 24, 1112–1134 CrossRef CAS PubMed.
- L. Saint-Aubert, L. Lemoine, K. Chiotis, A. Leuzy, E. Rodriguez-Vieitez and A. Nordberg, Mol. Neurodegener., 2017, 12, 19 CrossRef PubMed.
- A. Kallinen and M. Kassiou, Nucl. Med. Biol., 2022, 114–115, 115–127 CrossRef CAS PubMed.
- D. Willbold, B. Strodel, G. F. Schröder, W. Hoyer and H. Heise, Chem. Rev., 2021, 121, 8285–8307 CrossRef CAS PubMed.
- A. Aliyan, N. P. Cook and A. A. Martí, Chem. Rev., 2019, 119, 11819–11856 CrossRef CAS PubMed.
- A. Kaur, E. J. New and M. Sunde, ACS Sens., 2020, 5, 2268–2282 CrossRef CAS PubMed.
- F. Tang, K. Wang, X. Liu, X. Zhang, W. Zhou, Z. Mu, T. Zhang, W. Shu, Y. Liu and H. Xiao, Chem. – Eur. J., 2023, 29, e202300592 CrossRef CAS PubMed.
- D. Su, W. Diao, J. Li, L. Pan, X. Zhang, X. Wu and W. Mao, ACS Chem. Neurosci., 2022, 13, 540–551 CrossRef CAS PubMed.
- J. J. Maguire, R. E. Kuc and A. P. Davenport, Methods Mol. Biol., 2012, 897, 31–77 CrossRef CAS PubMed.
- A. P. Davenport and F. D. Russell, Current Directions in Radiopharmaceutical Research and Development, Springer, Netherlands, 1996, pp.169–179 Search PubMed.
- D. B. Bylund and M. L. Toews, Am. J. Physiol. Lung Cell. Mol. Physiol., 1993, 265, L421–L519 CrossRef CAS PubMed.
- Y. Wang, W. E. Klunk, M. L. Debnath, G. F. Huang, D. P. Holt, L. Shao and C. A. Mathis, J. Mol. Neurosci., 2004, 24, 55–62 CrossRef CAS PubMed.
- J. Kang and K. Han, Bull Korean Chem. Soc., 2001, 22, 1065 CAS.
- L. Lemoine, P.-G. Gillberg, M. Svedberg, V. Stepanov, Z. Jia, J. Huang, S. Nag, H. Tian, B. Ghetti, N. Okamura, M. Higuchi, C. Halldin and A. Nordberg, Alzheimer's Res. Ther., 2017, 9, 96 CrossRef PubMed.
- A. Manook, B. H. Yousefi, A. Willuweit, S. Platzer, S. Reder, A. Voss, M. Huisman, M. Settles, F. Neff, J. Velden, M. Schoor, H. von der Kammer, H. J. Wester, M. Schwaiger, G. Henriksen and A. Drzezga, PLoS One, 2012, 7, e31310 CrossRef CAS PubMed.
- C. J. Hsieh, K. Xu, I. Lee, T. J. A. Graham, Z. Tu, D. Dhavale, P. Kotzbauer and R. H. Mach, ACS Omega, 2018, 3, 4486–4493 CrossRef CAS PubMed.
- E. Sanna, M. Rodrigues, S. G. Fagan, T. S. Chisholm, K. Kulenkampff, D. Klenerman, M. G. Spillantini, F. I. Aigbirhio and C. A. Hunter, Chem. Sci., 2021, 12, 8892–8899 RSC.
- Y. Zhao, O. Tietz, W.-L. L. Kuan, A. K. Haji-Dheere, S. Thompson, B. Vallin, E. Ronchi, G. Tóth, D. Klenerman and F. I. Aigbirhio, Chem. Sci., 2020, 11, 4773–4778 RSC.
- W. Chu, D. Zhou, V. Gaba, J. Liu, S. Li, X. Peng, J. Xu, D. Dhavale, D. P. Bagchi, A. D’Avignon, N. B. Shakerdge, B. J. Bacskai, Z. Tu, P. T. Kotzbauer and R. H. Mach, J. Med. Chem., 2015, 58, 6002–6017 CrossRef CAS PubMed.
- X. Sun, P. Admane, Z. A. Starosolski, J. L. Eriksen, A. V. Annapragada and E. A. Tanifum, ChemMedChem, 2022, 17, e202100611 CrossRef CAS PubMed.
- S. Kaide, H. Watanabe, S. Iikuni, M. Hasegawa, K. Itoh and M. Ono, ACS Chem. Neurosci., 2021, 13, 16–26 CrossRef PubMed.
- S. Kaide, H. Watanabe, S. Iikuni, M. Hasegawa and M. Ono, ACS Chem. Neurosci., 2022, 13, 2982–2990 CrossRef CAS PubMed.
- L. Kuebler, S. Buss, A. Leonov, S. Ryazanov, F. Schmidt, A. Maurer, D. Weckbecker, A. M. Landau, T. P. Lillethorup, D. Bleher, R. S. Saw, B. J. Pichler, C. Griesinger, A. Giese and K. Herfert, Eur. J. Nucl. Med. Mol. Imaging, 2021, 48, 1759–1772 CrossRef CAS PubMed.
- D. D. Nan, C. S. Gan, C. W. Wang, J. P. Qiao, X. M. Wang and J. N. Zhou, Eur. J. Med. Chem., 2016, 124, 117–128 CrossRef CAS PubMed.
- N. S. Honson, R. L. Johnson, W. Huang, J. Inglese, C. P. Austin and J. Kuret, Neurobiol. Dis., 2007, 28, 251–260 CrossRef CAS PubMed.
- Z. P. Zhuang, M. P. Kung, C. Hou, D. M. Skovronsky, T. L. Gur, K. Plössl, J. Q. Trojanowski, V. M. Y. Lee and H. F. Kung, J. Med. Chem., 2001, 44, 1905–1914 CrossRef CAS PubMed.
- C. J. Hsieh, J. J. Ferrie, K. Xu, I. Lee, T. J. A. Graham, Z. Tu, J. Yu, D. Dhavale, P. Kotzbauer, E. J. Petersson and R. H. Mach, ACS Chem. Neurosci., 2018, 9, 2521–2527 CrossRef CAS PubMed.
- A. Lockhart, L. Ye, D. B. Judd, A. T. Merrittu, P. N. Lowe, J. L. Morgenstern, G. Hong, A. D. Gee and J. Brown, J. Biol. Chem., 2005, 280, 7677–7684 CrossRef CAS PubMed.
- L. Ye, J. L. Morgenstern, A. D. Gee, G. Hong, J. Brown and A. Lockhart, J. Biol. Chem., 2005, 280, 23599–23604 CrossRef CAS PubMed.
- E. D. Agdeppa, V. Kepe, J. Liu, G. W. Small, S. C. Huang, A. Petrič, N. Satyamurthy and J. R. Barrio, Mol. Imaging Biol., 2003, 5, 404–417 CrossRef PubMed.
- T. Akasaka, H. Watanabe, S. Kaide, S. Iikuni, M. Hasegawa and M. Ono, Bioorg. Med. Chem. Lett., 2022, 64, 128679 CrossRef CAS PubMed.
- D. Alagille, H. Dacosta, R. M. Baldwin and G. D. Tamagnan, Bioorg. Med. Chem. Lett., 2011, 21, 2966–2968 CrossRef CAS PubMed.
- J. An, P. Verwilst, H. Aziz, J. Shin, S. Lim, I. Kim, Y. K. Kim and J. S. Kim, Bioact. Mater., 2022, 13, 239–248 CAS.
- U. R. Anumala, J. Gu, F. Lo Monte, T. Kramer, R. Heyny-Von Haußen, J. Hölzer, V. Goetschy-Meyer, C. Schön, G. Mall, I. Hilger, C. Czech, J. Herms and B. Schmidt, Bioorg. Med. Chem., 2013, 21, 5139–5144 CrossRef CAS PubMed.
- D. P. Bagchi, L. Yu, J. S. Perlmutter, J. Xu, R. H. Mach, Z. Tu and P. T. Kotzbauer, PLoS One, 2013, 8, e55031 CrossRef CAS PubMed.
- H. Benzeid, E. Mothes, E. M. Essassi, P. Faller and G. Pratviel, C. R. Chim., 2012, 15, 79–85 CrossRef CAS.
- J. Bian, Y.-Q. Liu, J. He, X. Lin, C.-Y. Qiu, W.-B. Yu, Y. Shen, Z.-Y. Zhu, D.-Y. Ye, J. Wang and Y. Chu, Eur. J. Med. Chem., 2021, 226, 113887 CrossRef CAS PubMed.
- B. Blass, ACS Med. Chem. Lett., 2014, 5, 619–620 CrossRef CAS PubMed.
- S. Sanabria Bohórquez, J. Marik, A. Ogasawara, J. N. Tinianow, H. S. Gill, O. Barret, G. Tamagnan, D. Alagille, G. Ayalon, P. Manser, T. Bengtsson, M. Ward, S.-P. Williams, G. A. Kerchner, J. P. Seibyl, K. Marek and R. M. Weimer, Eur. J. Nucl. Med. Mol. Imaging, 2019, 46, 2077–2089 CrossRef PubMed.
- A. Boländer, D. Kieser, C. Voss, S. Bauer, C. Schön, S. Burgold, T. Bittner, J. Hölzer, R. Heyny-Von Haußen, G. Mall, V. Goetschy, C. Czech, H. Knust, R. Berger, J. Herms, I. Hilger and B. Schmidt, J. Med. Chem., 2012, 55, 9170–9180 CrossRef PubMed.
- E. Boukherrouba, C. Larosa, K.-A. Nguyen, J. Caburet, L. Lunven, H. Bonnet, A. Fortuné, A. Boumendjel, B. Boucherle, S. Chierici and M. Peuchmaur, Eur. J. Med. Chem., 2022, 231, 114139 CrossRef CAS PubMed.
- S. R. Byeon, Y. J. Jin, S. J. Lim, J. H. Lee, K. H. Yoo, K. J. Shin, S. J. Oh and D. J. Kim, Bioorg. Med. Chem. Lett., 2007, 17, 4022–4025 CrossRef CAS PubMed.
- L. Cai, F. T. Chin, V. W. Pike, H. Toyama, J. S. Liow, S. S. Zoghbi, K. Modell, E. Briard, H. U. Shetty, K. Sinclair, S. Donohue, D. Tipre, M. P. Kung, C. Dagostin, D. A. Widdowson, M. Green, W. Gao, M. M. Herman, M. Ichise and R. B. Innis, J. Med. Chem., 2004, 47, 2208–2218 CrossRef CAS PubMed.
- L. Cai, J. Cuevas, S. Temme, M. M. Herman, C. Dagostin, D. A. Widdowson, R. B. Innis and V. W. Pike, J. Med. Chem., 2007, 50, 4746–4758 CrossRef CAS PubMed.
- L. Cai, B. Qu, B. T. Hurtle, S. Dadiboyena, R. Diaz-Arrastia and V. W. Pike, ACS Chem. Neurosci., 2016, 7, 897–911 CrossRef CAS PubMed.
- M. S. Celej, E. A. Jares-Erijman and T. M. Jovin, Biophys. J., 2008, 94, 4867–4879 CrossRef CAS PubMed.
- R. Chandra, M. P. Kung and H. F. Kung, Bioorg. Med. Chem. Lett., 2006, 16, 1350–1352 CrossRef CAS PubMed.
- R. Chandra, S. Oya, M. P. Kung, C. Hou, L. W. Jin and H. F. Kung, J. Med. Chem., 2007, 50, 2415–2423 CrossRef CAS PubMed.
- Y. S. Chang, J. M. Jeong, Y. S. Lee, H. W. Kim, R. B. Ganesha, Y. J. Kim, D. S. Lee, J. K. Chung and M. C. Lee, Nucl. Med. Biol., 2006, 33, 811–820 CrossRef CAS PubMed.
- W. M. Chang, M. Dakanali, C. C. Capule, C. J. Sigurdson, J. Yang and E. A. Theodorakis, ACS Chem. Neurosci., 2011, 2, 249–255 CrossRef CAS PubMed.
- C. J. Chen, K. Bando, H. Ashino, K. Taguchi, H. Shiraishi, O. Fujimoto, C. Kitamura, S. Matsushima, M. Fujinaga, M. R. Zhang, H. Kasahara, T. Minamizawa, C. Jiang, M. Ono, M. Higuchi, T. Suhara, K. Yamada and B. Ji, Bioorg. Med. Chem., 2014, 22, 4189–4197 CrossRef CAS PubMed.
- C. J. Chen, K. Bando, H. Ashino, K. Taguchi, H. Shiraishi, K. Shima, O. Fujimoto, C. Kitamura, S. Matsushima, K. Uchida, Y. Nakahara, H. Kasahara, T. Minamizawa, C. Jiang, M. R. Zhang, M. Ono, M. Tokunaga, T. Suhara, M. Higuchi, K. Yamada and B. Ji, J. Nucl. Med., 2015, 56, 120–126 CrossRef CAS PubMed.
- Y. F. Chen, J. Bian, P. Zhang, L. L. Bu, Y. Shen, W. B. Yu, X. H. Lu, X. Lin, D. Y. Ye, J. Wang and Y. Chu, Bioorg. Med. Chem., 2020, 28, 115358 CrossRef CAS PubMed.
- Y. Chen, Q. Ouyang, Y. Li, Q. Zeng, B. Dai, Y. Liang, B. Chen, H. Tan and M. Cui, Eur. J. Med. Chem., 2022, 227, 113968 CrossRef CAS PubMed.
- X. Chen, Y. Li, J. Kang, T. Ye, Z. Yang, Z. Liu, Q. Liu, Y. Zhao, G. Liu and J. Pan, J. Lumin., 2023, 256, 119661 CrossRef CAS.
- Y. Cheng, M. Ono, H. Kimura, S. Kagawa, R. Nishii, H. Kawashima and H. Saji, ACS Med. Chem. Lett., 2010, 1, 321–325 CrossRef CAS PubMed.
- Y. Cheng, M. Ono, H. Kimura, S. Kagawa, R. Nishii and H. Saji, Bioorg. Med. Chem. Lett., 2010, 20, 6141–6144 CrossRef CAS PubMed.
- Y. Cheng, B. Zhu, Y. Deng and Z. Zhang, Anal. Chem., 2015, 87, 4781–4787 CrossRef CAS PubMed.
- Y. Cheng, B. Y. Zhu, X. Li, G. B. Li, S. Y. Yang and Z. R. Zhang, Bioorg. Med. Chem. Lett., 2015, 25, 4472–4476 CrossRef CAS PubMed.
- S. Chia, Z. Faidon Brotzakis, R. I. Horne, A. Possenti, B. Mannini, R. Cataldi, M. Nowinska, R. Staats, S. Linse, T. P. J. Knowles, J. Habchi and M. Vendruscolo, Mol. Pharmaceutics, 2023, 20, 183–193 CrossRef CAS PubMed.
- D. T. Chien, S. Bahri, A. K. Szardenings, J. C. Walsh, F. Mu, M.-Y. Su, W. R. Shankle, A. Elizarov and H. C. Kolb, J. Alzheimer's Dis., 2013, 34, 457–468 CAS.
- S. R. Choi, G. Golding, Z. Zhuang, W. Zhang, N. Lim, F. Hefti, T. E. Benedum, M. R. Kilbourn, D. Skovronskyhank and H. F. Kung, J. Nucl. Med., 2009, 50, 1887–1894 CrossRef CAS PubMed.
- J. W. Choi, Y. H. Ju, Y. H. Ju, Y. Choi, Y. Choi, S. J. Hyeon, C. G. Gadhe, J. H. Park, M. S. Kim, S. Baek, Y. Kim, K. D. Park, K. D. Park, A. N. Pae, A. N. Pae, H. Ryu, H. Ryu, C. J. Lee, C. J. Lee, B. R. Cho, B. R. Cho and B. R. Cho, ACS Chem. Neurosci., 2020, 11, 1801–1810 CrossRef CAS PubMed.
- R. Chongzhao, X. Xiaoyin, S. B. Raymond, B. J. Ferrara, K. Neal, B. J. Bacskai, Z. Medarova and A. Moore, J. Am. Chem. Soc., 2009, 131, 15257–15261 CrossRef PubMed.
- S. C. Young, J. L. Soo, J. O. Seung, H. M. Dae, J. K. Dong, C. G. Cho and H. Y. Kyung, Bull. Korean Chem. Soc., 2008, 29, 1765–1768 CrossRef.
- A. Cornejo, J. Caballero, M. Simirgiotis, V. Torres, L. Sánchez, N. Díaz, M. Guimaraes, M. Hernández, C. Areche, S. Alfaro, L. Caballero and F. Melo, J. Enzyme Inhib. Med. Chem., 2021, 36, 154–162 CrossRef CAS PubMed.
- M. C. Cui, Z. J. Li, R. K. Tang and B. L. Liu, Bioorg. Med. Chem., 2010, 18, 2777–2784 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, B. Liu and H. Saji, Bioorg. Med. Chem., 2011, 19, 4148–4153 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, B. Liu and H. Saji, J. Med. Chem., 2011, 54, 2225–2240 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, B. L. Liu and H. Saji, Bioorg. Med. Chem. Lett., 2011, 21, 980–982 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, H. Kawashima, B. L. Liu and H. Saji, Nucl. Med. Biol., 2011, 38, 313–320 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, B. Liu and H. Saji, Bioorg. Med. Chem. Lett., 2011, 21, 4193–4196 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Kimura, M. Ueda, Y. Nakamoto, K. Togashi, Y. Okamoto, M. Ihara, R. Takahashi, B. Liu and H. Saji, J. Med. Chem., 2012, 55, 9136–9145 CrossRef CAS PubMed.
- M. Cui, X. Wang, P. Yu, J. Zhang, Z. Li, X. Zhang, Y. Yang, M. Ono, H. Jia, H. Saji and B. Liu, J. Med. Chem., 2012, 55, 9283–9296 CrossRef CAS PubMed.
- M. Cui, M. Ono, H. Watanabe, H. Kimura, B. Liu and H. Saji, J. Am. Chem. Soc., 2014, 136, 3388–3394 CrossRef CAS PubMed.
- Y. Dai, T. Fang, Y. Xu, T. Jiang and J. Qiao, Chem. Biol. Drug Des., 2022, 1–12 Search PubMed.
- P. Dao, F. Ye, Y. Liu, Z. Y. Du, K. Zhang, C. Z. Dong, B. Meunier and H. Chen, ACS Chem. Neurosci., 2017, 8, 798–806 CrossRef CAS PubMed.
- L. Declercq, S. Celen, J. Lecina, M. Ahamed, T. Tousseyn, D. Moechars, J. Alcazar, M. Ariza, K. Fierens, A. Bottelbergs, J. Mariën, R. Vandenberghe, I. J. Andres, K. Van Laere, A. Verbruggen and G. Bormans, Mol. Imaging, 2016, 15, 1–15 CrossRef CAS PubMed.
- L. Declercq, F. Rombouts, M. Koole, K. Fierens, J. Mariën, X. Langlois, J. I. Andrés, M. Schmidt, G. MacDonald, D. Moechars, W. Vanduffel, T. Tousseyn, R. Vandenberghe, K. Van Laere, A. Verbruggen and G. Bormans, J. Nucl. Med., 2017, 58, 975–981 CrossRef CAS PubMed.
- A. A. Deeg, A. M. Reiner, F. Schmidt, F. Schueder, S. Ryazanov, V. C. Ruf, K. Giller, S. Becker, A. Leonov, C. Griesinger, A. Giese and W. Zinth, Biochim. Biophys. Acta, Gen. Subj., 2015, 1850, 1884–1890 CrossRef CAS PubMed.
- X. H. Duan, J. P. Qiao, Y. Yang, M. C. Cui, J. N. Zhou and B. L. Liu, Bioorg. Med. Chem., 2010, 18, 1337–1343 CrossRef CAS PubMed.
- M. V. Fawaz, A. F. Brooks, M. E. Rodnick, G. M. Carpenter, X. Shao, T. J. Desmond, P. Sherman, C. A. Quesada, B. G. Hockley, M. R. Kilbourn, R. L. Albin, K. A. Frey and P. J. H. Scott, ACS Chem. Neurosci., 2014, 5, 718–730 CrossRef CAS PubMed.
- J. J. Ferrie, Z. Lengyel-Zhand, B. Janssen, M. G. Lougee, S. Giannakoulias, C.-J. Hsieh, V. V. Pagar, C.-C. Weng, H. Xu, T. J. A. Graham, V. M.-Y. Lee, R. H. Mach and E. J. Petersson, Chem. Sci., 2020, 11, 12746–12754 RSC.
- D. P. Flaherty, S. M. Walsh, T. Kiyota, Y. Dong, T. Ikezu and J. L. Vennerstrom, J. Med. Chem., 2007, 50, 4986–4992 CrossRef CAS PubMed.
- D. P. Flaherty, T. Kiyota, Y. Dong, T. Ikezu and J. L. Vennerstrom, J. Med. Chem., 2010, 53, 7992–7999 CrossRef CAS PubMed.
- M. T. Fodero-Tavoletti, D. P. Smith, C. A. Mclean, P. A. Adlard, K. J. Barnham, L. E. Foster, L. Leone, K. Perez, M. Cortés, J. G. Culvenor, Q.-X. Li, K. M. Laughton, C. C. Rowe, C. L. Masters, R. Cappai and V. L. Villemagne, J. Neurosci., 2007, 27, 10365–10371 CrossRef CAS PubMed.
- M. T. Fodero-Tavoletti, R. S. Mulligan, N. Okamura, S. Furumoto, C. C. Rowe, Y. Kudo, C. L. Masters, R. Cappai, K. Yanai and V. L. Villemagne, Eur. J. Pharmacol., 2009, 617, 54–58 CrossRef CAS PubMed.
- M. T. Fodero-Tavoletti, N. Okamura, S. Furumoto, R. S. Mulligan, A. R. Connor, C. A. Mclean, D. Cao, A. Rigopoulos, G. A. Cartwright, G. O’keefe, S. Gong, P. A. Adlard, K. J. Barnham, C. C. Rowe, C. L. Masters, Y. Kudo, R. Cappai, K. Yanai and V. L. Villemagne, Brain, 2011, 134, 1089–1100 CrossRef PubMed.
- H. Fu, L. Yu, M. Cui, J. Zhang, X. Zhang, Z. Li, X. Wang, J. Jia, Y. Yang, P. Yu, H. Jia and B. Liu, Bioorg. Med. Chem., 2013, 21, 3708–3714 CrossRef CAS PubMed.
- H. Fu, M. Cui, P. Tu, Z. Pan and B. Liu, Chem. Commun., 2014, 50, 11875–11878 RSC.
- H. Fu, M. Cui, L. Zhao, P. Tu, K. Zhou, J. Dai and B. Liu, J. Med. Chem., 2015, 58, 6972–6983 CrossRef CAS PubMed.
- H. Fu, P. Tu, L. Zhao, J. Dai, B. Liu and M. Cui, Anal. Chem., 2016, 88, 1944–1950 CrossRef CAS PubMed.
- W. Fu, C. Yan, Z. Guo, J. Zhang, H. Zhang, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2019, 141, 3171–3177 CrossRef CAS PubMed.
- T. Fuchigami, N. Kobashi, M. Haratake, M. Kawasaki and M. Nakayama, Eur. J. Med. Chem., 2013, 60, 469–478 CrossRef CAS PubMed.
- T. Fuchigami, Y. Yamashita, M. Haratake, M. Ono, S. Yoshida and M. Nakayama, Bioorg. Med. Chem., 2014, 22, 2622–2628 CrossRef CAS PubMed.
- T. Fuchigami, A. Ogawa, Y. Yamashita, M. Haratake, H. Watanabe, M. Ono, M. Kawasaki, S. Yoshida and M. Nakayama, Bioorg. Med. Chem. Lett., 2015, 25, 3363–3367 CrossRef CAS PubMed.
- S. Furumoto, N. Okamura, K. Furukawa, M. Tashiro, Y. Ishikawa, K. Sugi, N. Tomita, M. Waragai, R. Harada, T. Tago, R. Iwata, K. Yanai, H. Arai and Y. Kudo, Mol. Imaging Biol., 2013, 15, 497–506 CrossRef PubMed.
- C. Gan, L. Zhou, Z. Zhao and H. Wang, Med. Chem. Res., 2013, 22, 4069–4074 CrossRef CAS.
- C. Gan, Z. Zhao, D. D. Nan, B. Yin and J. Hu, Eur. J. Med. Chem., 2014, 76, 125–131 CrossRef CAS PubMed.
- C. Gan, J. Hu, D. D. Nan, S. Wang and H. Li, Future Med. Chem., 2017, 9, 1587–1596 CrossRef CAS PubMed.
- J. Gu, U. R. Anumala, F. Lo Monte, T. Kramer, R. Heyny Von Haußen, J. Hölzer, V. Goetschy-Meyer, G. Mall, I. Hilger, C. Czech and B. Schmidt, Bioorg. Med. Chem. Lett., 2012, 22, 7667–7671 CrossRef CAS PubMed.
- J. Gu, U. R. Anumala, R. Heyny-von Haußen, J. Hölzer, V. Goetschy-Meyer, G. Mall, I. Hilger, C. Czech and B. Schmidt, ChemMedChem, 2013, 8, 891–897 CrossRef CAS PubMed.
- H. Han, C. G. Cho and P. T. Lansbury, J. Am. Chem. Soc., 1996, 118, 4506 CrossRef CAS.
- R. Harada, N. Okamura, S. Furumoto, T. Tago, M. Maruyama, M. Higuchi, T. Yoshikawa, H. Arai, R. Iwata, Y. Kudo and K. Yanai, Eur. J. Nucl. Med. Mol. Imaging, 2013, 40, 125–132 CrossRef CAS PubMed.
- R. Harada, N. Okamura, S. Furumoto, T. Yoshikawa, H. Arai, K. Yanai and Y. Kudo, Mol. Imaging Biol., 2014, 16, 19–27 CrossRef PubMed.
- S. T. Harrison, J. Mulhearn, S. E. Wolkenberg, P. J. Miller, S. S. O’Malley, Z. Zeng, D. L. Williams, E. D. Hostetler, S. Sanabria-Bohórquez, L. Gammage, H. Fan, C. Sur, J. C. Culberson, R. J. Hargreaves, J. J. Cook, G. D. Hartman and J. C. Barrow, ACS Med. Chem. Lett., 2011, 2, 498–502 CrossRef CAS PubMed.
- S. H. Hausner, D. Alagille, A. O. Koren, L. Amici, J. K. Staley, K. P. Cosgrove, R. M. Baldwin and G. D. Tamagnan, Bioorg. Med. Chem. Lett., 2009, 19, 543–545 CrossRef CAS PubMed.
- M. Hintersteiner, A. Enz, P. Frey, A. L. Jaton, W. Kinzy, R. Kneuer, U. Neumann, M. Rudin, M. Staufenbiel, M. Stoeckli, K. H. Wiederhold and H. U. Gremlich, Nat. Biotechnol., 2005, 49, 2725–2730 Search PubMed.
- N. Okamura, R. Harada, A. Ishiki, A. Kikuchi, T. Nakamura and Y. Kudo, Clin. Transl. Imaging, 2018, 6, 305–316 CrossRef PubMed.
- N. S. Honson, R. L. Johnson, W. Huang, J. Inglese, C. P. Austin and J. Kuret, Neurobiol. Dis., 2007, 28, 251–260 CrossRef CAS PubMed.
- E. D. Hostetler, A. M. Walji, Z. Zeng, P. Miller, I. Bennacef, C. Salinas, B. Connolly, L. Gantert, H. Haley, M. Holahan, M. Purcell, K. Riffel, T. G. Lohith, P. Coleman, A. Soriano, A. Ogawa, S. Xu, X. Zhang, E. Joshi, J. Della Rocca, D. Hesk, D. J. Schenk and J. L. Evelhoch, J. Nucl. Med., 2016, 57, 1599–1606 CrossRef CAS PubMed.
- S. S. Hou, J. Yang, J. H. Lee, Y. Kwon, M. Calvo-Rodriguez, K. Bao, S. Ahn, S. Kashiwagi, A. T. N. Kumar, B. J. Bacskai and H. S. Choi, Nat. Biomed. Eng., 2023, 7, 270–280 CrossRef CAS PubMed.
- L. Ji, X. Zhou, R. Liu, L. Wang, Y. Hu, J. Gu, Z. Li, C. Li, T. Huang and Y. Yu, Sens. Actuators, B, 2023, 380, 133415 CrossRef CAS.
- J. Jia, J. Song, J. Dai, B. Liu and M. Cui, ACS Chem. Neurosci., 2016, 7, 1275–1282 CrossRef CAS PubMed.
- A. E. Johnson, F. Jeppsson, J. Sandell, D. Wensbo, J. A. M. Neelissen, A. Juréus, P. Ström, H. Norman, L. Farde and S. P. S. Svensson, J. Neurochem., 2009, 108, 1177–1186 CrossRef CAS PubMed.
- L. Josephson, N. Stratman, Y. Liu, F. Qian, S. H. Liang, N. Vasdev and S. Patel, Mol. Imaging, 2018, 17, 1–6 CrossRef CAS PubMed.
- A. Juréus, B. M. Swahn, J. Sandell, F. Jeppsson, A. E. Johnson, P. Johnström, J. A. M. Neelissen, D. Sunnemark, L. Farde and S. P. S. Svensson, J. Neurochem., 2010, 114, 784–794 CrossRef PubMed.
- S. Kaide, M. Ono, H. Watanabe, Y. Shimizu, Y. Nakamoto, K. Togashi, A. Yamaguchi, H. Hanaoka and H. Saji, Bioorg. Med. Chem., 2018, 26, 3352–3358 CrossRef CAS PubMed.
- S. Kaide, H. Watanabe, Y. Shimizu, S. Iikuni, Y. Nakamoto, M. Hasegawa, K. Itoh and M. Ono, ACS Chem. Neurosci., 2020, 11, 4254–4261 CrossRef CAS PubMed.
- D. Kim, H. Moon, S. H. Baik, S. Singha, Y. W. Jun, T. Wang, K. H. Kim, B. S. Park, J. Jung, I. Mook-Jung and K. H. Ahn, J. Am. Chem. Soc., 2015, 137, 6781–6789 CrossRef CAS PubMed.
- W. E. Klunk, M. L. Debnath and J. W. Pettegrew, Neurobiol. Aging, 1994, 15, 691–698 CrossRef CAS PubMed.
- W. E. Klunk, M. L. Debnath and J. W. Pettegrew, Neurobiol. Aging, 1995, 16, 541–548 CrossRef CAS PubMed.
- W. E. Klunk, Y. Wang, G. Feng Huang, M. L. Debnath, D. P. Holt and C. A. Mathis, Life Sci., 2001, 69, 1471–1484 CrossRef CAS PubMed.
- W. E. Klunk, B. J. Bacskai, C. A. Mathis, S. T. Kajdasz, M. E. McLellan, M. P. Frosch, M. L. Debnath, D. P. Holt, Y. Wang and B. T. Hyman, J. Neuropathol. Exp. Neurol., 2002, 61, 797–805 CrossRef CAS PubMed.
- W. E. Klunk, Y. Wang, G. Feng Huang, M. L. Debnath, D. P. Holt, L. Shao, R. L. Hamilton, M. D. Ikonomovic, S. T. DeKosky and C. A. Mathis, J. Neurosci., 2003, 23, 2086–2092 CrossRef CAS PubMed.
- H. Kroth, F. Oden, J. Molette, H. Schieferstein, F. Capotosti, A. Mueller, M. Berndt, H. Schmitt-Willich, V. Darmency, E. Gabellieri, C. Boudou, T. Juergens, Y. Varisco, E. Vokali, D. T. Hickman, G. Tamagnan, A. Pfeifer, L. Dinkelborg, A. Muhs and A. Stephens, Eur. J. Nucl. Med. Mol. Imaging, 2019, 46, 2178–2189 CrossRef CAS PubMed.
- Y. Kudo, N. Okamura, S. Furumoto, M. Tashiro, K. Furukawa, M. Maruyama, M. Itoh, R. Iwata, K. Yanai and H. Arai, J. Nucl. Med., 2007, 48, 553–561 CrossRef CAS PubMed.
- H. F. Kung, C. W. Lee, Z. P. Zhuang, M. P. Kung, C. Hou and K. Plössl, J. Am. Chem. Soc., 2001, 123, 12740–12741 CrossRef CAS PubMed.
- M. P. Kung, C. Hou, Z. P. Zhuang, D. M. Skovronsky, B. Zhang, T. L. Gur, J. Q. Trojanowski, V. M. Y. Lee and H. F. Kung, J. Mol. Neurosci., 2002, 19, 7–10 CrossRef CAS PubMed.
- M. P. Kung, C. Hou, Z. P. Zhuang, B. Zhang, D. Skovronsky, J. Q. Trojanowski, V. M. Y. Lee and H. F. Kung, Brain Res., 2002, 956, 202–210 CrossRef CAS PubMed.
- M. P. Kung, Z. P. Zhuang, C. Hou, L. W. Jin and H. F. Kung, J. Mol. Neurosci., 2003, 20, 249–253 CrossRef CAS PubMed.
- M. P. Kung, C. Hou, Z. P. Zhuang, D. Skovronsky and H. F. Kung, Brain Res., 2004, 1025, 98–105 CrossRef CAS PubMed.
- C. W. Lee, Z. P. Zhuang, M. P. Kung, K. Plössl, D. Skovronsky, T. Gur, C. Hou, J. Q. Trojanowski, V. M. Y. Lee and H. F. Kung, J. Med. Chem., 2001, 44, 2270–2275 CrossRef CAS PubMed.
- C. W. Lee, M. P. Kung, C. Hou and H. F. Kung, Nucl. Med. Biol., 2003, 30, 573–580 CrossRef CAS PubMed.
- J. H. Lee, S. R. Byeon, Y. S. Kim, S. J. Lim, S. J. Oh, D. H. Moon, K. H. Yoo, B. Y. Chung and D. J. Kim, Bioorg. Med. Chem. Lett., 2008, 18, 5701–5704 CrossRef CAS PubMed.
- B. C. Lee, J. S. Kim, B. S. Kim, J. Y. Son, S. K. Hong, H. S. Park, B. S. Moon, J. H. Jung, J. M. Jeong and S. E. Kim, Bioorg. Med. Chem., 2011, 19, 2980–2990 CrossRef CAS PubMed.
- I. Lee, J. Yang, J. H. Lee and Y. S. Choe, Bioorg. Med. Chem. Lett., 2011, 21, 5765–5769 CrossRef CAS PubMed.
- L. Lemoine, L. Saint-Aubert, A. Marutle, G. Antoni, J. P. Eriksson, B. Ghetti, N. Okamura, I. Nennesmo, P. G. Gillberg, A. Nordberg, G. Gillberg and A. Nordberg, Acta Neuropathol. Commun., 2015, 3, 40 CrossRef PubMed.
- Z. Lengyel-Zhand, J. J. Ferrie, B. Janssen, C. J. Hsieh, T. Graham, K. Y. Xu, C. M. Haney, V. M. Y. Lee, J. Q. Trojanowski, E. J. Petersson and R. H. Mach, Chem. Commun., 2020, 56, 3567–3570 RSC.
- C. Wai, T. Leung, F. Guo, Y. Hong, E. Zhao, R. Tsz, K. Kwok, N. Lik, C. Leung, S. Chen, N. N. Vaikath, O. Mukhtar El-Agnaf, Y. Tang, W.-P. Gai and B. Z. Tang, Chem. Commun., 2015, 51, 1869 Search PubMed.
- Z. Li, M. Cui, J. Zhang, J. Dai, X. Zhang, P. Chen, H. Jia and B. Liu, Eur. J. Med. Chem., 2014, 84, 628–638 CrossRef CAS PubMed.
- Y. Li, D. Xu, S.-L. Ho, H.-W. Li, R. Yang and M. S. Wong, Biomaterials, 2016, 94, 84–92 CrossRef CAS PubMed.
- Z. Li, X. Zhang, X. Zhang, M. Cui, J. Lu, X. Pan and X. Zhang, J. Med. Chem., 2016, 59, 10577–10585 CrossRef CAS PubMed.
- Y. Li, J. Yang, H. Liu, J. Yang, L. Du, H. Feng, Y. Tian, J. Cao and C. Ran, Chem. Sci., 2017, 8, 7710–7717 RSC.
- Y. Li, D. Xu, A. Sun, S. L. Ho, C. Y. Poon, H. N. Chan, O. T. W. Ng, K. K. L. Yung, H. Yan, H. W. Li and M. S. Wong, Chem. Sci., 2017, 8, 8279–8284 RSC.
- Y. Li, C. Chen, D. Xu, C. Y. Poon, S. L. Ho, R. Zheng, Q. Liu, G. Song, H. W. Li and M. S. Wong, ACS Omega, 2018, 3, 6812–6819 CrossRef CAS PubMed.
- K. Liu, T. L. Guo, J. Chojnacki, H. G. Lee, X. Wang, S. L. Siedlak, W. Rao, X. Zhu and S. Zhang, ACS Chem. Neurosci., 2012, 3, 141–146 CrossRef CAS PubMed.
- X.-Y. Liu, X.-J. Wang, L. Shi, Y.-H. Liu, L. Wang, K. Li, Q. Bu, X.-B. Cen and X.-Q. Yu, Anal. Chem., 2022, 94, 7665–7673 CrossRef CAS PubMed.
- T. Liu, Y. Li, Y. Wang, X.-X. Yan, J. Dai and M. Cui, Eur. J. Med. Chem., 2023, 246, 114991 CrossRef CAS PubMed.
- C. Lois, I. Gonzalez, K. A. Johnson and J. C. Price, Brain Imaging Behav., 2019, 13, 333–344 CrossRef PubMed.
- G. Lv, B. Cui, H. Lan, Y. Wen, A. Sun and T. Yi, Chem. Commun., 2015, 51, 125–128 RSC.
- P. Lv, C. L. Xia, N. Wang, Z. Q. Liu, Z. S. Huang and S. L. Huang, Bioorg. Med. Chem., 2018, 26, 4693–4705 CrossRef CAS PubMed.
- R. Mallesh, J. Khan, K. Pradhan, R. Roy, N. R. Jana, P. Jaisankar and S. Ghosh, ACS Chem. Neurosci., 2022, 13, 2503–2516 CrossRef CAS PubMed.
- M. Maruyama, H. Shimada, T. Suhara, H. Shinotoh, B. Ji, J. Maeda, M. R. Zhang, J. Q. Trojanowski, V. M. Y. Lee, M. Ono, K. Masamoto, H. Takano, N. Sahara, N. Iwata, N. Okamura, S. Furumoto, Y. Kudo, Q. Chang, T. C. Saido, A. Takashima, J. Lewis, M. K. Jang, I. Aoki, H. Ito and M. Higuchi, Neuron, 2013, 79, 1094–1108 CrossRef CAS PubMed.
- C. A. Mathis, B. J. Bacskai, S. T. Kajdasz, M. E. McLellan, M. P. Frosch, B. T. Hyman, D. P. Holt, Y. Wang, G. F. Huang, M. L. Debnath and W. E. Klunk, Bioorg. Med. Chem. Lett., 2002, 12, 295–298 CrossRef CAS PubMed.
- C. A. Mathis, Y. Wang, D. P. Holt, G. F. Huang, M. L. Debnath and W. E. Klunk, J. Med. Chem., 2003, 46, 2740 CrossRef CAS PubMed.
- K. Matsumura, M. Ono, S. Hayashi, H. Kimura, Y. Okamoto, M. Ihara, R. Takahashi, H. Mori and H. Saji, MedChemComm, 2011, 2, 596–600 RSC.
- K. Matsumura, M. Ono, H. Kimura, M. Ueda, Y. Nakamoto, K. Togashi, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, ACS Med. Chem. Lett., 2012, 3, 58–62 CrossRef CAS PubMed.
- K. Matsumura, M. Ono, M. Yoshimura, H. Kimura, H. Watanabe, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, Bioorg. Med. Chem., 2013, 21, 3356–3362 CrossRef CAS PubMed.
- A. Maurer, A. Leonov, S. Ryazanov, K. Herfert, L. Kuebler, S. Buss, F. Schmidt, D. Weckbecker, R. Linder, D. Bender, A. Giese, B. J. Pichler and C. Griesinger, ChemMedChem, 2020, 15, 411–415 CrossRef CAS PubMed.
- Y. Maya, M. Ono, H. Watanabe, M. Haratake, H. Saji and M. Nakayama, Bioconjugate Chem., 2009, 20, 95–101 CrossRef CAS PubMed.
- J. Miao, M. Miao, Y. Jiang, M. Zhao, Q. Li, Y. Zhang, Y. An, K. Pu and Q. Miao, Angew. Chem., Int. Ed., 2023, 62, e202216351 CrossRef CAS PubMed.
- P. Miranda-Azpiazu, M. Svedberg, M. Higuchi, M. Ono, Z. Jia, D. Sunnemark, C. S. Elmore, M. Schou and A. Varrone, Brain Res., 2020, 1749, 147131 CrossRef CAS PubMed.
- G. Ribeiro Morais, H. Vicente Miranda, I. C. Santos, I. Santos, T. F. Outeiro and A. Paulo, Bioorg. Med. Chem., 2011, 19, 7698–7710 CrossRef PubMed.
- R. J. A. Nabuurs, V. V. Kapoerchan, A. Metaxas, S. De Jongh, M. De Backer, M. M. Welling, W. Jiskoot, A. D. Windhorst, H. S. Overkleeft, M. A. Van Buchem, M. Overhand and L. Van Der Weerd, Bioorg. Med. Chem., 2014, 22, 2469–2481 CrossRef CAS PubMed.
- K. Nakagawa, H. Watanabe, S. Kaide and M. Ono, ACS Med. Chem. Lett., 2022, 13, 1598–1605 CrossRef CAS PubMed.
- R. Narlawar, M. Pickhardt, S. Leuchtenberger, K. Baumann, S. Krause, T. Dyrks, S. Weggen, E. Mandelkow and B. Schmidt, ChemMedChem, 2008, 3, 165–172 CrossRef CAS PubMed.
- L.-M. Needham, J. Weber, J. A. Varela, J. W. B. Fyfe, D. T. Do, C. K. Xu, L. Tutton, R. Cliffe, B. Keenlyside, D. Klenerman, C. M. Dobson, C. A. Hunter, K. H. Müller, K. O’Holleran, S. E. Bohndiek, T. N. Snaddon and S. F. Lee, Chem. Sci., 2020, 11, 4578–4583 RSC.
- E. E. Nesterov, J. Skoch, B. T. Hyman, W. E. Klunk, B. J. Bacskai and T. M. Swager, Angew. Chem., Int. Ed., 2005, 44, 5452–5456 CrossRef CAS PubMed.
- B. Neumaier, S. Deisenhofer, C. Sommer, C. Solbach, S. N. Reske and F. Mottaghy, Appl. Radiat. Isot., 2010, 68, 1066–1072 CrossRef CAS PubMed.
- N. Okamura, T. Suemoto, H. Shimadzu, M. Suzuki, T. Shiomitsu, H. Akatsu, T. Yamamoto, M. Staufenbiel, K. Yanai, H. Arai, H. Sasaki, Y. Kudo and T. Sawada, J. Neurosci., 2004, 24, 2535–2541 CrossRef CAS PubMed.
- N. Okamura, T. Suemoto, S. Furumoto, M. Suzuki, H. Shimadzu, H. Akatsu, T. Yamamoto, H. Fujiwara, M. Nemoto, M. Maruyama, H. Arai, K. Yanai, T. Sawada and Y. Kudo, J. Neurosci., 2005, 25, 10857–10862 CrossRef CAS PubMed.
- N. Okamura, M. Mori, S. Furumoto, T. Yoshikawa, R. Harada, S. Ito, Y. Fujikawa, H. Arai, K. Yanai and Y. Kudo, J. Alzheimer's Dis., 2011, 23, 37–48 CAS.
- N. Okamura, S. Furumoto, R. Harada, T. Tago, T. Yoshikawa, M. Fodero-Tavoletti, R. S. Mulligan, V. L. Villemagne, H. Akatsu, T. Yamamoto, H. Arai, R. Iwata, K. Yanai and Y. Kudo, J. Nucl. Med., 2013, 54, 1420–1427 CrossRef CAS PubMed.
- M. Ono, M.-P. Kung, C. Hou and H. F. Kung, Nucl. Med. Biol., 2002, 29, 633–642 CrossRef CAS PubMed.
- M. Ono, A. Wilson, J. Nobrega, D. Westaway, P. Verhoeff, Z. P. Zhuang, M. P. Kung and H. F. Kung, Nucl. Med. Biol., 2003, 30, 565–571 CrossRef CAS PubMed.
- M. Ono, M. Haratake, M. Nakayama, Y. Kaneko, K. Kawabata, H. Mori, M. P. Kung and H. F. Kung, Nucl. Med. Biol., 2005, 32, 329–335 CrossRef CAS PubMed.
- M. Ono, N. Yoshida, K. Ishibashi, M. Haratake, Y. Arano, H. Mori and M. Nakayama, J. Med. Chem., 2005, 48, 7253–7260 CrossRef CAS PubMed.
- M. Ono, H. Kawashima, A. Nonaka, T. Kawai, M. Haratake, H. Mori, M. P. Kung, H. F. Kung, H. Saji and M. Nakayama, J. Med. Chem., 2006, 49, 2725–2730 CrossRef CAS PubMed.
- M. Ono, Y. Maya, M. Haratake, K. Ito, H. Mori and M. Nakayama, Biochem. Biophys. Res. Commun., 2007, 361, 116–121 CrossRef CAS PubMed.
- M. Ono, M. Hori, M. Haratake, T. Tomiyama, H. Mori and M. Nakayama, Bioorg. Med. Chem., 2007, 15, 6388–6396 CrossRef CAS PubMed.
- M. Ono, Y. Maya, M. Haratake and M. Nakayama, Bioorg. Med. Chem., 2007, 15, 444–450 CrossRef CAS PubMed.
- M. Ono, M. Haratake, H. Mori and M. Nakayama, Bioorg. Med. Chem., 2007, 15, 6802–6809 CrossRef CAS PubMed.
- M. Ono, M. Haratake, H. Saji and M. Nakayama, Bioorg. Med. Chem., 2008, 16, 6867–6872 CrossRef CAS PubMed.
- M. Ono, R. Watanabe, H. Kawashima, Y. Cheng, H. Kimura, H. Watanabe, M. Haratake, H. Saji and M. Nakayama, J. Med. Chem., 2009, 52, 6394–6401 CrossRef CAS PubMed.
- M. Ono, R. Watanabe, H. Kawashima, T. Kawai, H. Watanabe, M. Haratake, H. Saji and M. Nakayama, Bioorg. Med. Chem., 2009, 17, 2069–2076 CrossRef CAS PubMed.
- M. Ono, S. Hayashi, H. Kimura, H. Kawashima, M. Nakayama and H. Saji, Bioorg. Med. Chem., 2009, 17, 7002–7007 CrossRef CAS PubMed.
- M. Ono, M. Ishikawa, H. Kimura, S. Hayashi, K. Matsumura, H. Watanabe, Y. Shimizu, Y. Cheng, M. Cui, H. Kawashima and H. Saji, Bioorg. Med. Chem. Lett., 2010, 20, 3885–3888 CrossRef CAS PubMed.
- M. Ono, S. Hayashi, K. Matsumura, H. Kimura, Y. Okamoto, M. Ihara, R. Takahashi, H. Mori and H. Saji, ACS Chem. Neurosci., 2011, 2, 269–275 CrossRef CAS PubMed.
- M. Ono, Y. Cheng, H. Kimura, M. Cui, S. Kagawa, R. Nishii and H. Saji, J. Med. Chem., 2011, 54, 2971–2979 CrossRef CAS PubMed.
- M. Ono, H. Watanabe, H. Kimura and H. Saji, ACS Chem. Neurosci., 2012, 3, 319–324 CrossRef CAS PubMed.
- M. Ono, Y. Cheng, H. Kimura, H. Watanabe, K. Matsumura, M. Yoshimura, S. Iikuni, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, PLoS One, 2013, 8, e74104 CrossRef CAS PubMed.
- M. Ono, Y. Doi, H. Watanabe, M. Ihara, A. Ozaki and H. Saji, RSC Adv., 2016, 6, 44305–44312 RSC.
- M. Ono, N. Sahara, K. Kumata, B. Ji, R. Ni, S. Koga, D. W. Dickson, J. Q. Trojanowski, V. M.-Y. Lee, M. Yoshida, I. Hozumi, Y. Yoshiyama, J. C. Van Swieten, A. Nordberg, T. Suhara, M.-R. Zhang and M. Higuchi, Brain J. Neurol., 2017, 140, 764–780 Search PubMed.
- Y. D. Park, J. H. Park, M. G. Hur, S. W. Kim, J. J. Min, S. H. Park, Y. J. Yoo, Y. J. Yoon and S. D. Yang, Bioorg. Med. Chem. Lett., 2012, 22, 4106–4110 CrossRef CAS PubMed.
- K.-S. Park, Y. Seo, M. K. Kim, K. Kim, Y. K. Kim, H. Choo and Y. Chong, Org. Biomol. Chem., 2015, 13, 11194 RSC.
- Y. Dae Park, J.-J. Kim, S. Lee, C.-H. Park, H.-W. Bai and S. S. Lee, J. Anal. Methods Chem., 2018, 2018, 1651989 Search PubMed.
- A. Petrič, S. A. Johnson, H. V. Pham, Y. Li, S. Cěh, A. Golobič, E. D. Agdeppa, G. Timbol, J. Liu, G. Keum, N. Satyamurthy, V. Kepe, K. N. Houk and J. R. Barrio, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16492–16497 CrossRef PubMed.
- J. P. Qiao, C. S. Gan, C. W. Wang, J. F. Ge, D. D. Nan, J. Pan and J. N. Zhou, ChemBioChem, 2012, 13, 1652–1662 CrossRef CAS PubMed.
- L. Qin, J. Vastl and J. Gao, Mol. BioSyst., 2010, 6, 1791–1795 RSC.
- W. Qu, M. P. Kung, C. Hou, T. E. Benedum and H. F. Kung, J. Med. Chem., 2007, 50, 2157–2165 CrossRef CAS PubMed.
- W. Qu, M. P. Kung, C. Hou, S. Oya and H. F. Kung, J. Med. Chem., 2007, 50, 3380–3387 CrossRef CAS PubMed.
- W. Qu, M. P. Kung, C. Hou, L. W. Jin and H. F. Kung, Bioorg. Med. Chem. Lett., 2007, 17, 3581–3584 CrossRef CAS PubMed.
- W. Qu, S. R. Choi, C. Hou, Z. Zhuang, S. Oya, W. Zhang, M. P. Kung, R. Manchandra, D. M. Skovronsky and H. F. Kung, Bioorg. Med. Chem. Lett., 2008, 18, 4823–4827 CrossRef CAS PubMed.
- K. Rajasekhar, N. Narayanaswamy, N. A. Murugan, G. Kuang, H. Ågren and T. Govindaraju, Sci. Rep., 2016, 6, 23668 CrossRef CAS PubMed.
- K. Rajasekhar, N. Narayanaswamy, N. A. Murugan, K. Viccaro, H. G. Lee, K. Shah and T. Govindaraju, Biosens. Bioelectron., 2017, 98, 54–61 CrossRef CAS PubMed.
- L. Rejc, L. Šmid, V. Kepe, Č. Podlipnik, A. Golobič, M. Bresjanac, J. R. Barrio, A. Petrič and J. Košmrlj, J. Med. Chem., 2017, 60, 8741–8757 CrossRef CAS PubMed.
- W. Ren, J. Zhang, C. Peng, H. Xiang, J. Chen, C. Peng, W. Zhu, R. Huang, H. Zhang and Y. Hu, Bioconjugate Chem., 2018, 29, 3459–3466 CrossRef CAS PubMed.
- L. E. Rojo, J. Alzate-Morales, I. N. Saavedra, P. Davies and R. B. MacCioni, J. Alzheimer's Dis., 2010, 19, 573–589 CAS.
- E. K. Ryu, Y. S. Choe, K. H. Lee, Y. Choi and B. T. Kim, J. Med. Chem., 2006, 49, 6111–6119 CrossRef CAS PubMed.
- P. M. Seidler, D. R. Boyer, M. R. Sawaya, P. Ge, W. S. Shin, M. A. DeTure, D. W. Dickson, L. Jiang and D. S. Eisenberg, Nat. Commun., 2022, 13, 5451 CrossRef CAS PubMed.
- N. Seneca, L. Cai, J.-S. Liow, S. Zoghbi, R. Gladding, J. Little, P. Aisen, J. Hong, V. Pike and R. Innis, Curr. Radiopharm., 2010, 2, 129–136 CrossRef.
- Y. Seo, K. S. Park, T. Ha, M. K. Kim, Y. J. Hwang, J. Lee, H. Ryu, H. Choo and Y. Chong, ACS Chem. Neurosci., 2016, 7, 1474–1481 CrossRef CAS PubMed.
- K. Serdons, C. Terwinghe, P. Vermaelen, K. Van Laere, H. Kung, L. Mortelmans, G. Bormans and A. Verbruggen, J. Med. Chem., 2009, 52, 1428–1437 CrossRef CAS PubMed.
- K. Serdons, T. Verduyckt, D. Vanderghinste, J. Cleynhens, P. Borghgraef, P. Vermaelen, C. Terwinghe, F. Van Leuven, K. Van Laere, H. Kung, G. Bormans and A. Verbruggen, Bioorg. Med. Chem. Lett., 2009, 19, 602–605 CrossRef CAS PubMed.
- K. Serdons, K. Van Laere, P. Janssen, H. F. Kung, G. Bormans and A. Verbruggen, J. Med. Chem., 2009, 52, 7090–7102 CrossRef CAS PubMed.
- K. Serdons, T. Verduyckt, D. Vanderghinste, P. Borghgraef, J. Cleynhens, F. Van Leuven, H. Kung, G. Bormans and A. Verbruggen, Eur. J. Med. Chem., 2009, 44, 1415–1426 CrossRef CAS PubMed.
- X. Shao, G. M. Carpenter, T. J. Desmond, P. Sherman, C. A. Quesada, M. Fawaz, A. F. Brooks, M. R. Kilbourn, R. L. Albin, K. A. Frey and P. J. H. Scott, ACS Med. Chem. Lett., 2012, 3, 936–941 CrossRef CAS PubMed.
- S. K. Sonawane, H. Chidambaram, D. Boral, N. V. Gorantla, A. A. Balmik, A. Dangi, S. Ramasamy, U. K. Marelli and S. Chinnathambi, Sci. Rep., 2020, 10, 12579 CrossRef CAS PubMed.
- F. Sozmen, S. Kolemen, H.-O. Kumada, M. Ono, H. Saji and E. U. Akkaya, RSC Adv., 2014, 4, 51032–51037 RSC.
- K. A. Stephenson, R. Chandra, Z.-P. Zhuang, C. Hou, S. Oya, M.-P. Kung and H. F. Kung, Bioconjugate Chem., 2007, 18, 238–246 CrossRef CAS PubMed.
- T. Suemoto, N. Okamura, T. Shiomitsu, M. Suzuki, H. Shimadzu, H. Akatsu, T. Yamamoto, Y. Kudo and T. Sawada, Neurosci. Res., 2004, 48, 65–74 CrossRef CAS PubMed.
- G. S. M. Sundaram, D. D. Dhavale, J. L. Prior, P. Yan, J. Cirrito, N. P. Rath, R. Laforest, N. J. Cairns, J. M. Lee, P. T. Kotzbauer and V. Sharma, Sci. Rep., 2016, 6, 35636 CrossRef CAS PubMed.
- J. Sutharsan, M. Dakanali, C. Capule, M. Haidekker, J. Yang and E. Theodorakis, ChemMedChem, 2010, 5, 56–60 CrossRef CAS PubMed.
- B. M. Swahn, D. Wensbo, J. Sandell, D. Sohn, C. Slivo, D. Pyring, J. Malmström, E. Arzel, M. Vallin, M. Bergh, F. Jeppsson, A. E. Johnson, A. Juréus, J. Neelissen and S. Svensson, Bioorg. Med. Chem. Lett., 2010, 20, 1976–1980 CrossRef CAS PubMed.
- B. M. Swahn, J. Sandell, D. Pyring, M. Bergh, F. Jeppsson, A. Juréus, J. Neelissen, P. Johnström, M. Schou and S. Svensson, Bioorg. Med. Chem. Lett., 2012, 22, 4332–4337 CrossRef CAS PubMed.
- T. Tago, S. Furumoto, N. Okamura, R. Harada, H. Adachi, Y. Ishikawa, K. Yanai, R. Iwata and Y. Kudo, J. Nucl. Med., 2016, 57, 608–614 CrossRef CAS PubMed.
- R. Tao, N. Wang, T. Shen, Y. Tan, Y. Ren, W. Wei, M. Liao, D. Tan, C. Tang, N. Xu, H. Wang, X. Liu and X. Li, Theranostics, 2022, 12, 2549–2559 CrossRef CAS PubMed.
- P. W. Thompson, L. Ye, J. L. Morgenstern, L. Sue, T. G. Beach, D. J. Judd, N. J. Shipley, V. Libri and A. Lockhart, J. Neurochem., 2009, 109, 623–630 CrossRef CAS PubMed.
- Y. Tu, K. Chai, J. Wu, Y. Hu, S. Shi, D. Yang and T. Yao, Sens. Actuators, B, 2023, 380, 133406 CrossRef CAS.
- N. Vasdev, P. Cao, E. M. Van Oosten, A. A. Wilson, S. Houle, G. Hao, X. Sun, N. Slavine, M. Alhasan, P. P. Antich, F. J. Bonte and P. Kulkarni, MedChemComm, 2012, 3, 1228–1230 RSC.
- M. Verdurand, E. Levigoureux, W. Zeinyeh, L. Berthier, M. Mendjel-Herda, F. Cadarossanesaib, C. Bouillot, T. Iecker, R. Terreux, S. Lancelot, F. Chauveau, T. Billard and L. Zimmer, Mol. Pharmaceutics, 2018, 15, 3153–3166 CrossRef CAS PubMed.
- C. Vermeiren, P. Motte, D. Viot, G. Mairet-Coello, J.-P. Courade, M. Citron, J. Mercier, J. Hannestad and M. Gillard, Mov. Disord, 2018, 33, 273–281 CrossRef CAS PubMed.
- K. D. Volkova, V. B. Kovalska, A. O. Balanda, M. Y. Losytskyy, A. G. Golub, R. J. Vermeij, V. Subramaniam, O. I. Tolmachev and S. M. Yarmoluk, Bioorg. Med. Chem., 2008, 16, 1452–1459 CrossRef CAS PubMed.
- Y. Wang, C. A. Mathis, G. F. Huang, D. P. Holt, M. L. Debnath and W. E. Klunk, J. Labelled Compd. Radiopharm., 2002, 45, 647–664 CrossRef CAS.
- Y. Wang, W. E. Klunk, G. F. Huang, M. L. Debnath, D. P. Holt and C. A. Mathis, J. Mol. Neurosci., 2002, 19, 11–16 CrossRef PubMed.
- H. Watanabe, M. Ono, R. Ikeoka, M. Haratake, H. Saji and M. Nakayama, Bioorg. Med. Chem., 2009, 17, 6402–6406 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, M. Haratake, N. Kobashi, H. Saji and M. Nakayama, Bioorg. Med. Chem., 2010, 18, 4740–4746 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, H. Kimura, S. Kagawa, R. Nishii, T. Fuchigami, M. Haratake, M. Nakayama and H. Saji, Bioorg. Med. Chem. Lett., 2011, 21, 6519–6522 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, H. Kimura, K. Matsumura, M. Yoshimura, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, Bioorg. Med. Chem. Lett., 2012, 22, 5700–5703 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, K. Matsumura, M. Yoshimura, H. Kimura and H. Saji, Mol. Imaging, 2013, 12, 338–347 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, H. Kimura, K. Matsumura, M. Yoshimura, S. Iikuni, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, MedChemComm, 2014, 5, 82–85 RSC.
- H. Watanabe, M. Ono and H. Saji, Chem. Commun., 2015, 51, 17124–17127 RSC.
- H. Watanabe, M. Ono, S. Iikuni, H. Kimura, Y. Okamoto, M. Ihara and H. Saji, RSC Adv., 2015, 5, 1009–1015 RSC.
- H. Watanabe, T. Ariyoshi, A. Ozaki, M. Ihara, M. Ono and H. Saji, Bioorg. Med. Chem., 2017, 25, 6398–6403 CrossRef CAS PubMed.
- H. Watanabe, M. Ono, T. Ariyoshi, R. Katayanagi and H. Saji, ACS Chem. Neurosci., 2017, 8, 1656–1662 CrossRef CAS PubMed.
- H. Wongso, M. Ono, T. Yamasaki, K. Kumata, M. Higuchi, M.-R. Zhang, M. J. Fulham, A. Katsifis and P. A. Keller, RSC Med. Chem., 2023, 14, 858 RSC.
- M. C. Wren, T. Lashley, E. Årstad and K. Sander, Acta Neuropathol. Commun., 2018, 6, 34 CrossRef PubMed.
- C. Wu, J. Wei, K. Gao and Y. Wang, Bioorg. Med. Chem., 2007, 15, 2789–2796 CrossRef CAS PubMed.
- C.-F. Xia, J. Arteaga, G. Chen, U. Gangadharmath, L. F. Gomez, D. Kasi, C. Lam, Q. Liang, C. Liu, V. P. Mocharla, F. Mu, A. Sinha, H. Su, A. K. Szardenings, J. C. Walsh, E. Wang, C. Yu, W. Zhang, T. Zhao and H. C. Kolb, Alzheimers Dement., 2013, 9, 666–676 CrossRef PubMed.
- T. Xie, Y. Li, C. Tian, C. Yuan, B. Dai, S. Wang, K. Zhou, J. Liu, H. Tan, Y. Liang, J. Dai, B. Chen and M. Cui, J. Med. Chem., 2022, 65, 14527–14538 CrossRef CAS PubMed.
- J. W. Yan, J. Y. Zhu, K. X. Zhou, J. S. Wang, H. Y. Tan, Z. Y. Xu, S. B. Chen, Y. T. Lu, M. C. Cui and L. Zhang, Chem. Commun., 2017, 53, 9910–9913 RSC.
- Y. Yang, L. Zhu, X. Chen and H. Zhang, J. Mol. Graphics Modell., 2010, 29, 538–545 CrossRef CAS PubMed.
- Y. Yang, X. H. Duan, J. Y. Deng, B. Jin, H. M. Jia and B. L. Liu, Bioorg. Med. Chem. Lett., 2011, 21, 5594–5597 CrossRef CAS PubMed.
- W. Yang, Y. Wong, O. T. W. Ng, L.-P. Bai, D. W. J. Kwong, Y. Ke, Z.-H. Jiang, H.-W. Li, K. K. L. Yung and M. S. Wong, Angew. Chem., Int. Ed., 2012, 51, 1804–1810 CrossRef PubMed.
- Y. Yang, H.-M. Jia and B.-L. Liu, Molecules, 2012, 17, 4252–4265 CrossRef CAS PubMed.
- Y. Yang, M. Cui, X. Zhang, J. Dai, Z. Zhang, C. Lin, Y. Guo and B. Liu, J. Med. Chem., 2014, 57, 6030–6042 CrossRef CAS PubMed.
- Y. Yang, H. Fu, M. Cui, C. Peng, Z. Liang, J. Dai, Z. Zhang, C. Lin and B. Liu, Eur. J. Med. Chem., 2015, 104, 86–96 CrossRef CAS PubMed.
- Y. Yang, X. Zhang, M. Cui, J. Zhang, Z. Guo, Y. Li, X. Zhang, J. Dai and B. Liu, Sci. Rep., 2015, 5, 12084 CrossRef CAS PubMed.
- J. Yang, B. Zhu, W. Yin, Z. Han, C. Zheng, P. Wang and C. Ran, Chem. Sci., 2020, 11, 5238–5245 RSC.
- L. Ye, J. L. Morgenstern, J. R. Lamb and A. Lockhart, Biochem. Biophys. Res. Commun., 2006, 347, 669–677 CrossRef CAS PubMed.
- L. Ye, A. Velasco, G. Fraser, T. G. Beach, L. Sue, T. Osredkar, V. Libri, M. G. Spillantini, M. Goedert and A. Lockhart, J. Neurochem., 2008, 105, 1428–1437 CrossRef CAS PubMed.
- M. Yoshimura, M. Ono, K. Matsumura, H. Watanabe, H. Kimura, M. Cui, Y. Nakamoto, K. Togashi, Y. Okamoto, M. Ihara, R. Takahashi and H. Saji, ACS Med. Chem. Lett., 2013, 4, 596–600 CrossRef CAS PubMed.
- B. H. Yousefi, A. Manook, A. Drzezga, B. V. Reutern, M. Schwaiger, H. J. Wester and G. Henriksen, J. Med. Chem., 2011, 54, 949–956 CrossRef CAS PubMed.
- L. Yu, J. Cui, P. K. Padakanti, L. Engel, D. P. Bagchi, P. T. Kotzbauer and Z. Tu, Bioorg. Med. Chem., 2012, 20, 4625–4634 CrossRef CAS PubMed.
- P. Yu, M. Cui, X. Wang, X. Zhang, Z. Li, Y. Yang, J. Jia, J. Zhang, M. Ono, H. Saji, H. Jia and B. Liu, Eur. J. Med. Chem., 2012, 57, 51–58 CrossRef CAS PubMed.
- X. Yue, D. D. Dhavale, J. Li, Z. Luo, J. Liu, H. Yang, R. H. Mach, P. T. Kotzbauer and Z. Tu, Bioorg. Med. Chem. Lett., 2018, 28, 1011–1019 CrossRef CAS PubMed.
- N. Yue, H. Fu, Y. Chen, X. Gao, J. Dai and M. Cui, Eur. J. Med. Chem., 2022, 243, 114715 CrossRef CAS PubMed.
- F. Zeng, J. A. Southerland, R. J. Voll, J. R. Votaw, L. Williams, B. J. Ciliax, A. I. Levey and M. M. Goodman, Bioorg. Med. Chem. Lett., 2006, 16, 3015–3018 CrossRef CAS PubMed.
- Q. Zeng, Y. Chen, Y. Yan, R. Wan, Y. Li, H. Fu, Y. Liu, S. Liu, X.-X. Yan and M. Cui, Anal. Chem., 2022, 94, 15261–15269 CrossRef CAS PubMed.
- Z. Zha, S. R. Choi, K. Ploessl, B. P. Lieberman, W. Qu, F. Hefti, M. Mintun, D. Skovronsky and H. F. Kung, J. Med. Chem., 2011, 54, 8085–8098 CrossRef CAS PubMed.
- W. Zhang, S. Oya, M. P. Kung, C. Hou, D. L. Maier and H. F. Kung, J. Med. Chem., 2005, 48, 5980–5988 CrossRef CAS PubMed.
- W. Zhang, S. Oya, M. P. Kung, C. Hou, D. L. Maier and H. F. Kung, Nucl. Med. Biol., 2005, 32, 799–809 CrossRef CAS PubMed.
- W. Zhang, M. P. Kung, S. Oya, C. Hou and H. F. Kung, Nucl. Med. Biol., 2007, 34, 89–97 CrossRef CAS PubMed.
- X. Zhang, Y. Tian, C. Zhang, X. Tian, A. W. Ross, R. D. Moir, H. Sun, R. E. Tanzi, A. Moore and C. Ran, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9734–9739 CrossRef CAS PubMed.
- J. Zhang, A. Sandberg, X. Wu, S. Nyströ, M. Lindgren, P. Konradsson and P. Hammarströ, ACS Omega, 2017, 2, 4693–4704 CrossRef CAS PubMed.
- J. Zhang, J. Wang, A. Sandberg, X. Wu, S. Nyström, H. LeVine, P. Konradsson, P. Hammarström, B. Durbeej and M. Lindgren, ChemPhysChem, 2018, 19, 3001–3009 CrossRef CAS PubMed.
- J. Zhang, A. Sandberg, A. Konsmo, X. Wu, S. Nyström, K. P. R. Nilsson, P. Konradsson, H. LeVine, M. Lindgren and P. Hammarström, Chem. – Eur. J., 2018, 24, 7210–7216 CrossRef CAS PubMed.
- J. Zhang, A. Konsmo, A. Sandberg, X. Wu, S. Nyströ, U. Obermü, B. M. Wegenast-Braun, P. Konradsson, M. Lindgren and P. Hammarströ, J. Med. Chem., 2019, 62, 2038–2048 CrossRef CAS PubMed.
- W. Zhang, J. Arteaga, D. K. Cashion, G. Chen, U. Gangadharmath, L. F. Gomez, D. Kasi, C. Lam, Q. Liang, C. Liu, V. P. Mocharla, F. Mu, A. Sinha, A. Katrin Szardenings, E. Wang, J. C. Walsh, C. Xia, C. Yu, T. Zhao and H. C. Kolb, J. Alzheimer's Dis., 2012, 31, 601–612 CAS.
- W. Zhen, H. Han, M. Anguiano, C. A. Lemere, C. G. Cho and P. T. Lansbury, J. Med. Chem., 1999, 42, 2805–2815 CrossRef CAS PubMed.
- M. Q. Zheng, D. Z. Yin, J. P. Qiao, L. Zhang and Y. X. Wang, J. Fluorine Chem., 2008, 129, 210–216 CrossRef CAS.
- K. Zhou, H. Fu, L. Feng, M. Cui, J. Dai and B. Liu, Chem. Commun., 2015, 51, 11665–11668 RSC.
- K. Zhou, H. Bai, L. Feng, J. Dai and M. Cui, Anal. Chem., 2017, 89, 9432–9437 CrossRef CAS PubMed.
- K. Zhou, Y. Li, Y. Peng, X. Cui, J. Dai and M. Cui, Anal. Chem., 2018, 90, 8576–8582 CrossRef CAS PubMed.
- Z. P. Zhuang, M. P. Kung, C. Hou, K. Plössl, D. Skovronsky, T. L. Gur, J. Q. Trojanowski, V. M. Y. Lee and H. F. Kung, Nucl. Med. Biol., 2001, 28, 887–894 CrossRef CAS PubMed.
- Z.-P. Zhuang, M.-P. Kung, C. Hou, C.-W. Lee, J. Q. Trojanowski, V. M.-Y. Lee and H. F. Kung, J. Labelled Compd. Radiopharm., 2001, 44, S29–S32 Search PubMed.
- Z. P. Zhuang, M. P. Kung, A. Wilson, C. W. Lee, K. Plössl, C. Hou, D. M. Holtzman and H. F. Kung, J. Med. Chem., 2003, 46, 237–243 CrossRef CAS PubMed.
- Z. P. Zhuang, M. P. Kung and H. F. Kung, J. Med. Chem., 2006, 49, 2841–2844 CrossRef CAS PubMed.
- T. S. Chisholm, M. Mackey and C. A. Hunter, J. Am. Chem. Soc., 2023, 145, 15936–15950 CrossRef CAS PubMed.
- S.-P. Wey, C.-C. Weng, K.-J. Lin, C.-H. Yao, T.-C. Yen, H. F. Kung, D. Skovronsky and M.-P. Kung, Nucl. Med. Biol., 2009, 36, 411–417 CrossRef CAS PubMed.
- Y. Wang, C. A. Mathis, G.-F. Huang, M. L. Debnath, D. P. Holt, L. Shao and W. E. Klunk, J. Mol. Neurosci., 2003, 20, 255–260 CrossRef PubMed.
- N. Okamura, T. Suemoto, T. Shiomitsu, M. Suzuki, H. Shimadzu, H. Akatsu, T. Yamamoto, H. Arai, H. Sasaki, K. Yanai, M. Staufenbiel, Y. Kudo and T. Sawada, J. Mol. Neurosci., 2004, 24, 247–255 CrossRef CAS PubMed.
- Y. Wang, T. T. Huynh, N. Bandara, H.-J. Cho, B. E. Rogers and L. M. Mirica, Dalton Trans., 2022, 51, 1216–1224 RSC.
- L. Sun, A. K. Sharma, B.-H. Han and L. M. Mirica, ACS Chem. Neurosci., 2020, 11, 2741–2752 CrossRef CAS PubMed.
- L. Sun, H.-J. Cho, S. Sen, A. S. Arango, T. T. Huynh, Y. Huang, N. Bandara, B. E. Rogers, E. Tajkhorshid and L. M. Mirica, J. Am. Chem. Soc., 2021, 143, 10462–10476 CrossRef CAS PubMed.
- Z. Yu, W. Guo, S. Patel, H.-J. Cho, L. Sun and L. M. Mirica, Chem. Sci., 2022, 13, 12818–12830 RSC.
- M. Rana, A. Pareek, S. Bhardwaj, G. Arya, S. Nimesh, H. Arya, T. K. Bhatt, S. Yaragorla and A. K. Sharma, RSC Adv., 2020, 10, 28827–28837 RSC.
- K. Terpstra, Y. Wang, T. T. Huynh, N. Bandara, H.-J. Cho, B. E. Rogers and L. M. Mirica, Inorg. Chem., 2022, 61, 20326–20336 CrossRef CAS PubMed.
- H. F. Kung, S. R. Choi, W. Qu, W. Zhang and D. Skovronsky, J. Med. Chem., 2010, 53, 933–941 CrossRef CAS PubMed.
- N. Bandara, A. K. Sharma, S. Krieger, J. W. Schultz, B. H. Han, B. E. Rogers and L. M. Mirica, J. Am. Chem. Soc., 2017, 139, 12550–12558 CrossRef CAS PubMed.
- Y. Huang, H.-J. Cho, N. Bandara, L. Sun, D. Tran, B. E. Rogers and L. M. Mirica, Chem. Sci., 2020, 11, 7789–7799 RSC.
- A. K. Sharma, J. Kim, J. T. Prior, N. J. Hawco, N. P. Rath, J. Kim and L. M. Mirica, Inorg. Chem., 2014, 53, 11367–11376 CrossRef CAS PubMed.
- R. Leuma Yona, S. Mazères, P. Faller and E. Gras, ChemMedChem, 2008, 3, 63–66 CrossRef PubMed.
- C. Rodríguez-Rodríguez, M. A. Telpoukhovskaia, J. Alí-Torres, L. Rodríguez-Santiago, Y. Manso, G. A. Bailey, J. Hidalgo, M. Sodupe and C. Orvig, Metallomics, 2015, 7, 83–92 CrossRef PubMed.
- N. Pravin, R. Kumar, S. Tripathi, P. Kumar, G. M. Mohite, A. Navalkar, R. Panigrahi, N. Singh, L. G. Gadhe, S. Manchanda, M. Shimozawa, P. Nilsson, J. Johansson, A. Kumar, S. K. Maji and M. Shanmugam, J. Neurochem., 2021, 156, 1003–1019 CrossRef CAS PubMed.
- A. S. DeToma, J. Krishnamoorthy, Y. Nam, H. J. Lee, J. R. Brender, A. Kochi, D. Lee, V. Onnis, C. Congiu, S. Manfredini, S. Vertuani, G. Balboni, A. Ramamoorthy and M. H. Lim, Chem. Sci., 2014, 5, 4851–4862 RSC.
- K. Cisek, J. R. Jensen, N. S. Honson, K. N. Schafer, G. L. Cooper and J. Kuret, Biophys. Chem., 2012, 170, 25–33 CrossRef CAS PubMed.
- Q. Li, J. Min, Y.-H. Ahn, J. Namm, E. M. Kim, R. Lui, H. Y. Kim, Y. Ji, H. Wu, T. Wisniewski and Y.-T. Chang, ChemBioChem, 2007, 8, 1679–1687 CrossRef CAS PubMed.
- S. J. Jung, Y. D. Park, J. H. Park, S. D. Yang, M. G. Hur and K. H. Yu, Med. Chem. Res., 2013, 22, 4263–4268 CrossRef CAS.
- M. A. Telpoukhovskaia, C. Rodríguez-Rodríguez, J. F. Cawthray, L. E. Scott, B. D. G. Page, J. Alí-Torres, M. Sodupe, G. A. Bailey, B. O. Patrick and C. Orvig, Metallomics, 2014, 6, 249–262 CrossRef CAS PubMed.
- H. Fu, C. Peng, Z. Liang, J. Dai, B. Liu and M. Cui, Chem. Commun., 2016, 52, 12745–12748 RSC.
- H. Fu, P. Tu, L. Zhao, J. Dai, B. Liu and M. Cui, Anal. Chem., 2016, 88, 1944–1950 CrossRef CAS PubMed.
- M.-L. Pan, M. T. Mukherjee, H. H. Patel, B. Patel, C. C. Constantinescu, M. R. Mirbolooki, C. Liang and J. Mukherjee, Synapse, 2016, 70, 163–176 CrossRef CAS PubMed.
- E. Hellström-Lindahl, P. Westermark, G. Antoni and S. Estrada, Amyloid, 2014, 21, 21–27 CrossRef PubMed.
- E. Fisher, Y. Zhao, R. Richardson, M. Janik, A. K. Buell, F. I. Aigbirhio and G. Tóth, ACS Chem. Neurosci., 2017, 8, 2088–2095 CrossRef CAS PubMed.
- N. Gour, V. Kshtriya, S. Gupta, B. Koshti, R. Singh, D. Patel and K. B. Joshi, ACS Appl. Bio Mater., 2019, 2, 4442–4455 CrossRef CAS PubMed.
- S. Rivera-Marrero, L. Fernández-Maza, S. León-Chaviano, M. Sablón-Carrazana, A. Bencomo-Martínez, A. Perera-Pintado, A. Prats-Capote, F. Zoppolo, I. Kreimerman, T. Pardo, L. Reyes, M. Balcerzyk, G. Dubed-Bandomo, D. Mercerón-Martínez, L. A. Espinosa-Rodríguez, H. Engler, E. Savio and C. Rodríguez-Tanty, Curr. Radiopharm., 2019, 12, 58–71 CrossRef CAS PubMed.
- M. Xu, R. Li, X. Li, G. Lv, S. Li, A. Sun, Y. Zhou and T. Yi, J. Mater. Chem. B, 2019, 7, 5535–5540 RSC.
- H.-L. Yang, S.-Q. Fang, Y.-W. Tang, C. Wang, H. Luo, L.-L. Qu, J.-H. Zhao, C.-J. Shi, F.-C. Yin, X.-B. Wang and L.-Y. Kong, Eur. J. Med. Chem., 2019, 179, 736–743 CrossRef CAS PubMed.
- J. Zhu, L. Zhou, Y. Li, S. Chen, J. Yan and L. Zhang, Anal. Chim. Acta, 2017, 961, 112–118 CrossRef CAS PubMed.
- G. S. M. Sundaram, K. Garai, N. P. Rath, P. Yan, J. R. Cirrito, N. J. Cairns, J.-M. Lee and V. Sharma, Org. Lett., 2014, 16, 3640–3643 CrossRef CAS PubMed.
- P. Dao, F. Ye, Z. Y. Du, Q. Chen, K. Zhang, C. Z. Dong, B. Meunier and H. Chen, Dyes Pigm., 2017, 147, 130–140 CrossRef CAS.
- G. Lv, Y. Xu, J. Yang, W. Li, C. Li and A. Sun, Analyst, 2020, 145, 6579–6585 RSC.
- N. R. Marzano, K. M. Wray, C. L. Johnston, B. P. Paudel, Y. Hong, A. van Oijen and H. Ecroyd, ACS Chem. Neurosci., 2020, 11, 4191–4202 CrossRef CAS PubMed.
- H. Sun, H. Leng, J. Liu, G. Roy, J. Yan and L. Zhang, Sens. Actuators, B, 2020, 305, 127509 CrossRef CAS.
- H. Tan, Y. Qiu, H. Sun, J. Yan and L. Zhang, Chem. Commun., 2019, 55, 2688–2691 RSC.
- J. Yan, J. Zhu, K. Zhou, J. Wang, H. Tan, Z. Xu, S. Chen, Y. Lu, M. Cui and L. Zhang, Chem. Commun., 2017, 53, 9910–9913 RSC.
- Y. Wang, Y. Qiu, A. Sun, Y. Xiong, H. Tan, Y. Shi, P. Yu, G. Roy, L. Zhang and J. Yan, Anal. Chim. Acta, 2020, 1133, 109–118 CrossRef CAS PubMed.
- S. Rivera-Marrero, A. Bencomo-Martínez, E. Orta Salazar, M. Sablón-Carrazana, L. García-Pupo, F. Zoppolo, F. Arredondo, R. Dapueto, M. Daniela Santi, I. Kreimerman, T. Pardo, L. Reyes, L. Galán, S. León-Chaviano, L. A. Espinosa-Rodríguez, R. Menéndez-Soto del Valle, E. Savio, S. Díaz Cintra and C. Rodríguez-Tanty, Bioorg. Med. Chem., 2020, 28, 115700 CrossRef CAS PubMed.
- P. Gaur, M. Galkin, A. Kurochka, S. Ghosh, D. A. Yushchenko and V. V. Shvadchak, ACS Chem. Neurosci., 2021, 12, 1293–1298 CrossRef CAS PubMed.
- H. Kaur, M. R. Felix, C. Liang and J. Mukherjee, Bioorg. Med. Chem. Lett., 2021, 46, 128164 CrossRef CAS PubMed.
- J. Wu, C. Shao, X. Ye, X. Di, D. Li, H. Zhao, B. Zhang, G. Chen, H.-K. Liu and Y. Qian, ACS Sens., 2021, 6, 863–870 CrossRef CAS PubMed.
- J. An, P. Verwilst, H. Aziz, J. Shin, S. Lim, I. Kim, Y. K. Kim and J. S. Kim, Bioact. Mater., 2022, 13, 239–248 CAS.
- D. Fang, X. Wen, Y. Wang, Y. Sun, R. An, Y. Zhou, D. Ye and H. Liu, Theranostics, 2022, 12, 3178–3195 CrossRef CAS PubMed.
- Y. Guo, H. Leng, Q. Chen, J. Su, W. Shi, C. Xia, L. Zhang and J. Yan, Sens. Actuators, B, 2022, 372, 132648 CrossRef CAS.
- R. Mallesh, J. Khan, K. Pradhan, R. Roy, N. R. Jana, P. Jaisankar and S. Ghosh, ACS Chem. Neurosci., 2022, 13, 2503–2516 CrossRef CAS PubMed.
- Y. Wang, D. Mei, X. Zhang, D.-H. Qu and J. Mei, Sci. China: Chem., 2022, 65, 339–352 CrossRef CAS.
- H. Tan, K. Zhou, J. Yan, H. Sun, M. Pistolozzi, M. Cui and L. Zhang, Sens. Actuators, B, 2019, 298, 126903 CrossRef CAS.
- I. Kocsis, E. Sanna and C. A. Hunter, Org. Lett., 2021, 23, 647–650 CrossRef CAS PubMed.
- L. Esposito, M. C. Yadon, J. Cummings, F. M. Hudson, T. Lake, F. F. Hefti, G. Golding, S.-R. Choi, X. Li, A. D. Snow, H. Qubai and C. Judy, WO2012118935A1, 2012.
- W. E. Klunk, C. A. Mathis and Y. Wang, US Pat., US10137210B2, 2018 Search PubMed.
- Y. Kudo, S. Furumoto and N. Okamura, WO2012057312A1, 2011.
- I. Tooyama, H. Taguchi, S. Morikawa, M. Urushitani, D. Yanagisawa, T. Nagae, N. Shirai, K. Hirao, M. Kato, H.Kimura and T. Okada, WO2010098502A1, 2010.
- W. Abbas, E. Hostetler, T. Greshock, J. Li, K. P. Moore, I. Bennacef, J. Mulheam, H. Selnick, K. Yang and J. Fu, WO2015188368A1, 2015.
- B. W. Caprathe, J. L. Gilmore, S. J. Hays, J. C. Jaen and H. Levine III, US Pat., US6001331A, 1999.
- H. F. Kung, M.-P. Kung and Z.-P. Zhuang, US Pat., US7807135B2, 2010.
- H. F. Kung, personal communication.
- D. J. Hayne, S. Lim and P. S. Donnelly, Chem. Soc. Rev., 2014, 43, 6701–6715 RSC.
- K. Chen and M. Cui, MedChemComm, 2017, 8, 1393–1407 RSC.
- J.-F. Morfin, S. Lacerda, C. F. G. C. Geraldes and É. Tóth, Sens. Diagn., 2022, 1, 627–647 RSC.
- S. Lacerda, J.-F. Morfin, C. F. G. C. Geraldes and É. Tóth, Dalton Trans., 2017, 46, 14461–14474 RSC.
- O. Krasnovskaya, D. Spector, A. Zlobin, K. Pavlov, P. Gorelkin, A. Erofeev, E. Beloglazkina and A. Majouga, Int. J. Mol. Sci., 2020, 21, 9190 CrossRef CAS PubMed.
- M. Groenning, J. Chem. Biol., 2010, 3, 1–18 CrossRef PubMed.
- R. Khurana, C. Coleman, C. Ionescu-Zanetti, S. A. Carter, V. Krishna, R. K. Grover, R. Roy and S. Singh, J. Struct. Biol., 2005, 151, 229–238 CrossRef CAS PubMed.
- C. Xue, T. Y. Lin, D. Chang and Z. Guo, R. Soc. Open Sci., 2017, 4, 160696 CrossRef PubMed.
- E. D. Agdeppa, V. Kepe, J. Liu, S. Flores-Torres, N. Satyamurthy, A. Petric, G. M. Cole, G. W. Small, S. C. Huang and J. R. Barrio, J. Neurosci., 2001, 21, RC189–RC189 CrossRef CAS PubMed.
- S. A. Wildman and G. M. Crippen, J. Chem. Inf. Comput. Sci., 1999, 39, 868–873 CrossRef CAS.
- L. Cai, B. Qu, B. T. Hurtle, S. Dadiboyena, R. Diaz-Arrastia and V. W. Pike, ACS Chem. Neurosci., 2016, 7, 897–911 CrossRef CAS PubMed.
- N. A. Murugan, A. Nordberg and H. Ågren, ACS Chem. Neurosci., 2021, 12, 2437–2447 CrossRef CAS PubMed.
- N. A. Murugan, A. Nordberg and H. Ågren, ACS Chem. Neurosci., 2018, 9, 1757–1767 CrossRef CAS PubMed.
- N. A. Murugan, C. Halldin, A. Nordberg, B. Långström and H. Ågren, J. Phys. Chem. Lett., 2016, 7, 3313–3321 CrossRef CAS PubMed.
- H. Levine, Amyloid, 2005, 12, 5–14 CrossRef CAS PubMed.
- S. K. Mishra, Y. Yamaguchi, M. Higuchi and N. Sahara, Int. J. Mol. Sci., 2020, 22, 349 CrossRef PubMed.
- R. Ni, P. G. Gillberg, A. Bergfors, A. Marutle and A. Nordberg, Brain, 2013, 136, 2217–2227 CrossRef PubMed.
- A. Sidhu, J. Vaneyck, C. Blum, I. Segers-Nolten and V. Subramaniam, Amyloid, 2018, 25, 189–196 CrossRef CAS PubMed.
- E. Smith and I. Collins, Future Med. Chem., 2015, 7, 159–183 CrossRef CAS PubMed.
- P. Duan, K. J. Chen, G. Wijegunawardena, A. J. Dregni, H. K. Wang, H. Wu and M. Hong, J. Am. Chem. Soc., 2022, 144, 1416–1430 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details on database preparation, heatmaps summarising competition binding experiments, and spreadsheet containing the ligand database. See DOI: https://doi.org/10.1039/d3cs00518f |
| This journal is © The Royal Society of Chemistry 2024 |