Open Access Article
Open Access ArticleHigh-throughput DNA synthesis for data storage
Meng
Yu
abc,
Xiaohui
Tang
ac,
Zhenhua
Li
ac,
Weidong
Wang
c,
Shaopeng
Wang
d,
Min
Li
d,
Qiuliyang
Yu
e,
Sijia
Xie
 *abc,
Xiaolei
Zuo
*abc,
Xiaolei
Zuo
 *d and
Chang
Chen
*d and
Chang
Chen
 *abcf
*abcf
aInstitute of Medical Chips, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, 200025, Shanghai, China. E-mail: sijia.xie@shsmu.edu.cn; chang.chen@shsmu.edu.cn
bSchool of Microelectronics, Shanghai University, 201800, Shanghai, China
cShanghai Industrial μTechnology Research Institute, 201800, Shanghai, China
dInstitute of Molecular Medicine, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, 200127, Shanghai, China. E-mail: zuoxiaolei@sjtu.edu.cn
eShenzhen Key Laboratory for the Intelligent Microbial Manufacturing of Medicines, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, 518055, Shenzhen, China
fState Key Laboratory of Transducer Technology, Shanghai Institute of Microsystem and Information Technology, Chinese Academy of Sciences, 200050, Shanghai, China
First published on 18th March 2024
Abstract
With the explosion of digital world, the dramatically increasing data volume is expected to reach 175 ZB (1 ZB = 1012 GB) in 2025. Storing such huge global data would consume tons of resources. Fortunately, it has been found that the deoxyribonucleic acid (DNA) molecule is the most compact and durable information storage medium in the world so far. Its high coding density and long-term preservation properties make itself one of the best data storage carriers for the future. High-throughput DNA synthesis is a key technology for “DNA data storage”, which encodes binary data stream (0/1) into quaternary long DNA sequences consisting of four bases (A/G/C/T). In this review, the workflow of DNA data storage and the basic methods of artificial DNA synthesis technology are outlined first. Then, the technical characteristics of different synthesis methods and the state-of-the-art of representative commercial companies, with a primary focus on silicon chip microarray-based synthesis and novel enzymatic DNA synthesis are presented. Finally, the recent status of DNA storage and new opportunities for future development in the field of high-throughput, large-scale DNA synthesis technology are summarized.
1. Introduction
From ancient wall paintings to paper scrolls and books, it is widely recognized that storing and sharing information (or data) are essential requirements in the development of human civilization.1 Moreover, the invention of hard drives in the 1970s, denoting the beginning of information age, has brought explosive growth in data storage capabilities. According to the report from the International Data Corporation (IDC), the global data generated by 2025 are projected to reach a staggering 175 zettabytes (ZB), which is almost a three times increment over the data formed in 2021 (64 ZB).2,3 In such circumstances, a crisis in data storage may arise if the relevant technology fails to keep up with demand.Generally, data can be categorized into “hot data” (frequently accessed on a daily basis, for example, logs and emails) and “cold data” (characterized by long-term storage requirements, such as archives, surveillance videos, and backup files, etc., whose significance may only be obvious over time), based on their access frequency. Moreover, the statistics indicate that more than 60% of all data are going to become archival so that it is natural to observe that some “hot data” are transformed into “cold data” eventually. Before humans established the digital world, such cold data were stored in media such as paper, films and later on, tapes in specific warehouses under delicate environmental conditions (temperature, humidity, UV light exposure, etc.) to prevent damage or degradation of the physical form of these media. Although these cold data are rarely needed, they carry crucial archival and evidential information that can be highly valuable in certain circumstances where historical investigation is expected. Currently, in order to store (and to transfer, when necessary) these huge amounts of “cold data” with the growing trend, tapes and hard disk drives are commonly utilized thanks to their advantages of low cost, large scale, and environmental compatibility. However, these magnetic recording siblings can only serve for merely a few decades, while “cold data” are usually required to be stored for 50 and even more than 100 years.3 In order to preserve data integrity and mitigate the potential risk of data loss resulting from storage device failures, it is imperative to routinely transfer and back up data. The maintenance and periodic transference of “cold data” to new devices gradually become burdensome and risky in terms of data preservation. In recent years, the development of the Cloud technique has greatly eased the transportation as well as transfer of the big data among devices, even remotely, while its reliability and potential cost for long-term data storage are yet unclear. The extended lifespan of DNA largely mitigates this need for DNA data storage. Plus, storing massive amounts of data consumes huge quantities of energy. According to reports, data centers consumed 205 terawatt hours (TW h) of electricity in 2018 and this number is expected to be 210 TW h in 2023 and estimated to reach 1929 TW h in 2030.4–7 Therefore, investigating promising data storage technologies with higher capacity, lower cost, and long-term stability is still an urgent essential for today's information society. Fortunately, it is found that DNA may be a “game-breaker”.8
The most advanced transistor was realized by IBM in 2021, having a size down to 2 nm with the help of the 2-nm process.9 In a digital circuit, a bit is stored by a function unit (e.g., a flip-flop or a latch) which is made of several transistors. Thus, the denser the transistors are fabricated on a chip, the higher storage capacity the chip in principle will have. 3D memory chips were developed to further increase storage capacity by vertically stacking multiple layers. For example, Micron released a 232-layer 3D NAND with a chip size of about 70.1 mm2 which holds 1012 bit data.10 DNA is the storage medium for genetic information in biology. It is a macromolecular polymer composed of deoxyribonucleotides (hereafter referred to as nucleotides). Nucleotides are composed of phosphate, deoxyribose, and their attached bases. There are four types of bases in DNA: adenine (A), guanine (G), cytosine (C) and thymine (T). The sequence of these four bases on a single nucleotide strand of DNA holds the biological genetic information. A single nucleotide, which has a size of about 0.3 nm, is considered the most basic unit of biological genetic information. Currently, the digital coding density of DNA can reach max. 2 bits per base.11–15 According to the evaluation in the literature, the data density of flash can reach 1016 bits per cm3, and hard drive 1013 bits per cm3, while DNA up to 1022 bits per cm3 in principle.16–20 From this perspective, a nucleotide as the basic element for digital data storage is potentially a more effective candidate compared with the transistors, given that the size of the latter is not probable to be further reduced unless there will be sub-nanometer, large-scale semiconductor fabrication techniques. Of course, the overall capacity of DNA storage is limited by the length of a generated DNA strand, the number of copies required for each strand considering the practical aspects during synthesis, storing and sequencing, as well as the speed and throughput for DNA synthesis and sequencing, which will be discussed in detail later in this review. Additionally, if only the physical size is considered, DNA seems a much lighter-weighted medium than the conventional media such as tapes or hard disks. In this sense, the physical transfer of massive data in DNA would be much easier. Although it still requires intense development to facilitate an integrated and mature process flow of DNA data storage and transfer, in the foreseeable future, it is optimistic to predict that transferring data with DNA will be an easier and more resource-efficient way than using the current storage media. Besides, its high chemical stability, relatively low maintenance cost, and almost non-commutative nature of the format for billions of years also make DNA an excellent candidate for storing the “cold data” of human society.19,21 Given the rapid iteration in the field of information technology as well as biotechnology, it seems positive that DNA, the information carrier from ancient times, would be provided a way to be compatible with future technologies. Although the whole storage process is currently rather complex and costly, the technical difficulties are expected to be solved in the near future. Using DNA for data storage is considered to be the next-generation revolution in the storage of the “cold data”.
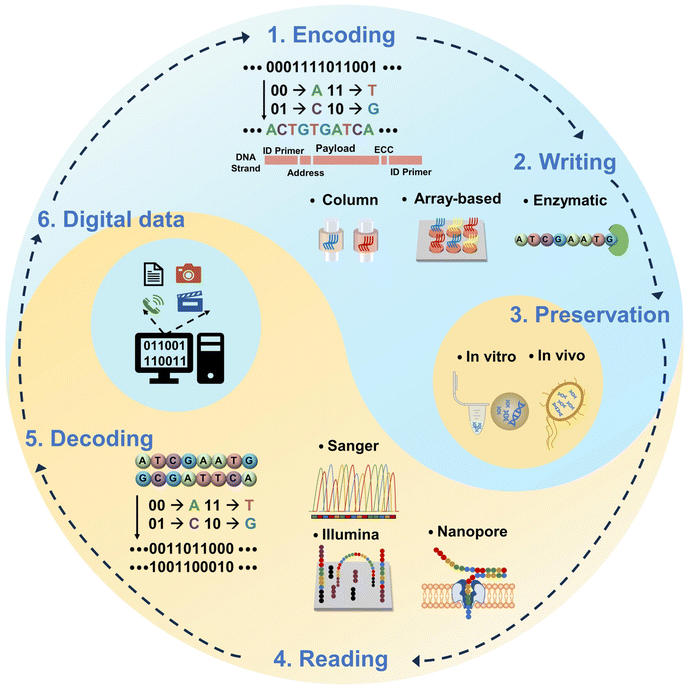
Furthermore, the rapid development of semiconductor technology has accelerated the iterative improvement of storage media. Memory chips based on integrated circuit technology have been developed for flash memory, memory cards, U-disks, hard disk devices, etc., whose capacity and processing speed have been greatly improved. Under this prerequisite, the combination of DNA storage with semiconductor technology is expected to create more superb possibilities for data storage. Similar to the data storage processes of semiconductor devices, DNA data storage also includes encoding, writing, preservation, reading, decoding, etc. Compared with the data storage mechanism of electronic devices, the digital data are “written” by DNA synthesis technology to couple nucleotides one by one according to designed sequences and then “read out” by DNA sequencing technology. The core of this pioneering massive data storage technology is to achieve high-throughput and high-speed data writing and reading. To realize this, a fast and massive data writing approach, namely DNA synthesis, is a critical aspect of this technique.
In the field of DNA data storage, there are already a large number of previous works on topics such as coding/decoding algorithms,14,17,22–24 error correction mechanisms,15,20,25 preservation methods,19,26,27 and overall reviews.12,18,28–35 Complementary to these articles and reviews, here, our discussion mainly focuses on the field of hardware manufacturing that is committed to the use of silicon-based chip technology to develop micro and nano scale integrated chips for high-throughput artificial DNA synthesis platforms: The history and current situation of DNA synthesis technology are introduced. Several high-throughput array-based DNA synthesis technologies are described in detail, and the state-of-the-art representative companies are listed. The advantages and limitations of each technology are comprehensively compared from the perspective of DNA data storage-oriented applications. Besides, we outline the evolution of next generation enzymatic DNA synthesis technology, as well as the new opportunities it brings to DNA data storage, together with the semiconductor chip technology. Finally, we analyze the performance gap between DNA data storage and current storage devices from the perspective of synthesis and sequencing, and propose future directions for the development of this technology.
2. Overview of DNA data storage
The idea of using molecules to store digital information was first proposed by American scientist Richard Feynman in a public lecture in 1959.36 In the mid-1960s, Mikhail Samiolvich Neiman and Norbert Wiener first proposed theoretically a miniature device for storing data with DNA molecules.37,38 However, the first validated work on DNA data storage was done by Davis in 1988, who encoded the “Microvenus” icon into 28-base-pair long double-stranded DNA (dsDNA) inserted into Escherichia coli (E. coli).39 The maximum storage capacity of the subsequent period was only a few tens of bytes.40–42 The major breakthroughs and real demonstrations occurred in the 2010s when Church et al. and Goldman et al. successfully stored several hundred kilobytes of data in DNA, taking advantage of array-based DNA synthesis technology.13,14 Recently, with the development of high-throughput DNA synthesis and sequencing technologies, the capacity of digital data (pictures, books, films, etc.) to be stored in DNA is breaking through to the MB level,23,43,44 and even large-scale storage of 200 MB has been achieved.45As mentioned above, data storage with DNA involves several steps (Fig. 1): encoding, writing, preservation, reading, and decoding.8,12,18,28–35
Specifically, “encoding” refers to the conversion of the binary digital data (0/1) of the initial data into DNA base sequences (A/G/C/T) arranged in specific order according to the corresponding coding algorithms, such as: Huffman-code,14,46 Fountain code,23 Reed Solomon (RS) code,15,45 and yin-yang code (YYC).47 Usually, the final base sequences don't only contain valid data fragments, but may also include assisting fragments such as (1) primers located at both ends of a strand. They are used for the polymerase chain reaction (PCR) when data amplification or random access is needed. To prevent a higher melting temperature for the PCR process and a larger loss of storage density, the length of this region is usually limited to less than 25 nt (nt represents the unit of oligonucleotide strand length), but cannot be much shorter without sacrificing diversity in addresses. When storing 1 GB of data with 200 nt long DNA molecules, the primer fraction ratio is estimated to be 6.5%.12 There are encoding schemes which skip the primers or only include them on one end of the synthesized sequences48–50 while some of them may potentially sacrifice the data storage capacity by occupying the valuable data payload space. (2) Address fragments. They help mark the locations of the data; (3) error correction code areas. They are for adding logical redundancy to deal with errors such as deletion, insertion, and substitutions that could happen during the process of synthesis and sequencing. They play an important role in retrieving lost sequences and correcting errors for reliable DNA-based data storage. Normally, the length of currently synthesized DNA is limited to 200 nt with high accuracy, so the total data have to be segmented and jigsawed into a whole piece later on. Therefore, the assisting regions in each segment bring a lot of redundancy as they occupy the space for data payload (about 20 nt at both the front and back ends), limiting the storage capacity and also decreasing the storage density (by about 15% per address region).23,49,51 In this sense, using a long nucleotide strand for DNA data storage would in principle ease the coding and decoding, and reduce the complexity of data generation and retrieval.
Following the sequence encoded in the previous step, “writing” refers to the process of coupling base monomers through chemical and biological reactions, resulting in the assembly of segments one by one. The obtained single-stranded DNA (ssDNA) fragments are known as an “oligo pool” equivalent to a database. This is a key step in actually turning digital data into molecular structures. The level of DNA synthesis technology determines the overall quality of data storage.
“Preservation” usually involves the assembly of synthesized ssDNA into more chemically stable dsDNA, which is subsequently purified and stored in vitro23,45 (in the forms of frozen powder or solution, or capsuled in micro/nanoparticles15,48) at a low temperature or preserved in vivo39,52–54 (in plasmids55 or artificial chromosomes56 in bacteria, or integrated in the genome of a living organism57–59) by genetic technology including transfection, clustered regularly interspaced short palindromic repeat-associated (CRISPR–Cas) or recombinases for long-term preservation.60,61 Recent research studies have mentioned the use of ssDNA attached to the dsDNA62 or in the storing sample49 for better data access. However, ssDNA is prone to coil conformation due to the lack of the double helix secondary structure, and it suffers from damage by environmental conditions (e.g., temperature, humidity, UV irradiation, oxidation, etc.) more easily than dsDNA. For in vitro preservation, although ssDNA can be stored as synthesized for up to 2 years63–66 under optimized conditions, dsDNA is still the “safer” option when longer preservation time is expected. Nevertheless, when using ssDNA as the data storage medium, the technical benefits of not first having to go through the complex process of preparing ssDNA into dsDNA may well outweigh its disadvantages in long term stability. Owing to the higher efficiency of DNA replication in living organisms, the information encoded into DNA stored in vivo can be replicated with greater accuracy and speed than through in vitro preservation.29,67–69 In addition, it has been shown that information preserved in vivo enables dynamic data access and editing of information with single-base resolution.52,70 However, further investigation is still needed for better understanding and optimizing the compatibility, stability, and functionality of the input DNA.70–72 In addition, specific laboratory environments are often necessary for ensuring the genetic stability as well as the viability of the living organisms, unless considering using the tenacious candidates.73 For DNA data storage applications, the most favorable method should be selected according to the frequency of access to data required in different scenarios. Generally, for DNA data storage, the amount of each sequence of the synthesized DNA is at a trace scale, because large-scale parallelization is needed to increase the speed and data density of the “writing step”. In order to ensure the effectiveness and reliability of the data, PCR amplification technology is necessary to increase the concentration of the synthesized products and backup the data as well.74–76
“Reading” means that the stored DNA molecules are extracted by biochemical methods, and the target base sequences are identified one by one to obtain the written coding data. DNA sequencing technology is used to read and splice the base sequences carried by DNA fragments in the oligonucleotide pool. In early studies, data stored in DNA required sequencing all of the molecules. Later on, PCR-based random-access techniques were developed,17,45 allowing random access to a portion of the data without sequencing all of the DNA in the oligonucleotide pool. As a new trend, array-based technologies for DNA data storage may ease the workload for the PCR because the synthesized DNA is confined or immobilized to the designed location on the array and can be addressed directly via the chip. Approaches such as spot-specified digital microfluidics,77 sequencing-by-synthesis,78 DNA microdisks,79 and SlipChip80 have contributed to a further step towards high manipulability and rapid access. The DNA is conjugated to the surface of the chip and is not damaged or lost during replication, which also allows for easy access and handling, reducing the need for PCR primer selection and large-scale PCR amplification.
Finally, “decoding” is the reconversion of base sequences into digital data and further restoration to the original format of the data. In the whole workflow of a storage cycle, biological and chemical reactions take on the function of writing/reading data.
Using DNA for data storage has several attractive advantages: (1) high storage density. Considering a coding density of 2 bits per base,13 DNA would have a theoretical data density of 6 bits per nm,33 given that a nucleotide is ∼0.3 nm long. If we only consider the nature of the DNA molecule and put aside the complexity of the practical aspects including data retrieval, 1 gram of DNA can store about 4.5 × 107 GB of data given that only a single copy of each unique DNA sequence presents the mixture, while the current technology only stores 10 terabyte (TB) on a 600 g HDD, which is 6 orders of magnitude difference.17,33,81 For the possibility of fully retrieving the data, to use as few as 10 copies of per sequence in the mixture would result in a storage density of 17 EB g−1,17 which is still a significant improvement compared with the current HDD. (2) Long preservation time and durability. Under suitable conditions (e.g., at room temperature in a dry atmosphere, or lyophilized powder), DNA can remain stable for thousands of years and withstand temperatures as low as −196 °C (liquid nitrogen) and as high as 250 °C (silica).82–85 As for magnetic, silicon-based storage devices, the requirements for humidity, temperature and magnetic fields in the environment are stringent and the lifetime usually does not exceed 50 years.15 However, long-term storage of DNA molecules also does face some risks.68 For example, the stored data may be contaminated by bacteria or human DNA.67 In addition, natural DNA is highly susceptible to degradation by microorganisms and nuclease enzymes in the natural environment, while environmental factors can cause strand breaks, hydrolytic damage and UV-induced cross-linking, all of which can lead to partial data loss. Mirror-image DNA has the same storage density as natural DNA, but also has a unique bio-orthogonality, which prevents it from being easily degraded by microorganisms and nucleases, and it is successfully utilized in orthogonal information storage.86–88 (3) Low maintenance cost and environmental friendliness. The inherent durability of DNA renders it highly amenable to preservation. Compared with the regular maintenance of conventional long-term storage equipment which consumes a lot of electricity, energy, and land resources, the energy to store the DNA is almost negligible.15,28,89 In addition, the data stored in DNA can be easily backed up by PCR technology. A helpful comparison of the main performance indicators for various storage media was given by Linda C. Meiser et al. in 2022 (Fig. 2).31 Although current DNA synthesis methods cannot completely avoid using toxic chemicals, even in the case of enzymatic synthesis, DNA is still a more friendly option for data storage media compared with its opponents, as DNA is biodegradable90 and requires less heavy metals and rare elements for synthesis.31
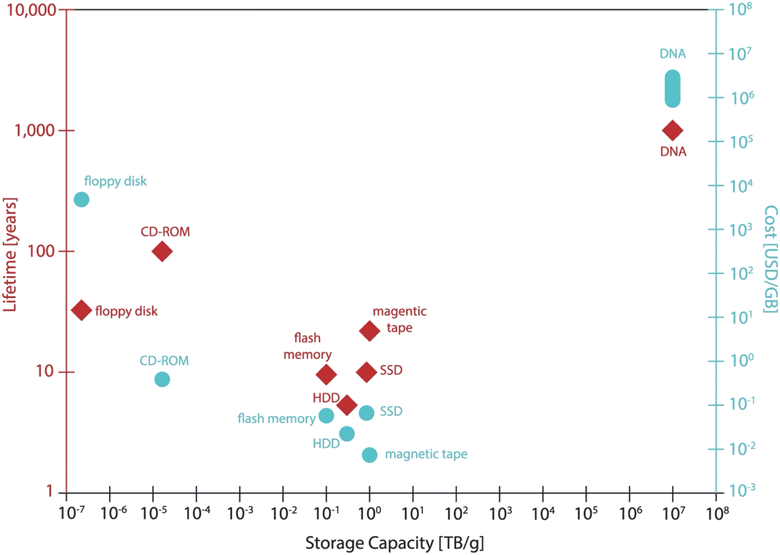 | ||
| Fig. 2 A comparison of the various storage methods in terms of lifetime, capacity and cost. The cost of mainstream media is derived from the average consumer market price. The data survey was carried out during the writing period of ref. 31. Reproduced from ref. 31 with permission from Copyright 2022 Springer Nature. | ||
In 2020, the world's leading enterprises including Microsoft Research, Illumina, Western Digital, Twist Bioscience, etc., founded the international organization “DNA Data Storage Alliance”. As the association is growing, the total number of members has now exceeded 40. It brings together the world's state-of-the-art information technology, DNA artificial synthesis, DNA sequencing, and integrated circuit manufacturing industries.
Their mission is to create and promote an interoperable storage ecosystem based on manufactured DNA as a data storage medium. The alliance launched its first version of white paper in 2021,3 outlining the background, strategy, and technical development of DNA data storage. It seems promising that the establishment of the consortium will accelerate the cross-fertilization and breakthrough progress of data encoding technology, high-throughput DNA synthesis technology, and sequencing technology, and will vigorously promote the process of DNA data storage technology.
However, current DNA data storage technology still faces several challenges: (1) low throughput and speed. At present, the throughput of synthesis technology and sequencing technology is far not high enough for data storage, particularly the synthesis technology. Enzymatic synthesis offers a higher speed for “data writing” compared with the chemical approach. It has been demonstrated that the coupling time of enzymatic synthesis can be minimized to 10–20 s, while that of the chemical phosphoramidite synthesis is usually in the range of 4–10 min.91,92 Lee et al. gave an estimate of 40 s per cycle for enzymatic synthesis, which is six times faster than phosphoramidite synthesis. However, this rate is still much slower than that of the state-of-the-art electronic devices.93,94 (2) Difficult data access. Unlike conventional storage devices, it is not yet feasible to access random parts of the data or modify them in DNA molecules on a single device. (3) Workload in large-scale data reproduction. Although the PCR is no doubt a powerful tool for nucleic acid amplification and is generally acknowledged to be a high-fidelity process, it introduces bias by e.g., the GC content in the strand, which may cause loss of the data strands containing a high GC content during amplification. This would lead to a significantly different proportion of the sequences when the PCR cycles are large,95,96 and affect both data storage capacity and retrieval efficiency. Also, it is still difficult to amplify highly repetitive sequences by the PCR.97 (4) High complexity and costs of integration. Most of today's DNA data storage strategies are realized on separate devices and locations for synthesis, preservation, replication, and sequencing sessions, making the process complex and time-consuming. Besides, although the average cost of sequencing genes per TB of data in 2021 was only $0.006 (calculated based on the production cost of sequencing one million bases, including equipment, reagents, administration, and overhead costs), significantly decreased from $5292.39 in 2001, according to the National Human Genome Research Institute (NHGRI),98 the cost of DNA synthesis is still orders of magnitude higher compared with the cost of sequencing. According to the estimation by Meiser et al., storing 1 MB encoded data into DNA would cost around $800 to $500023,28,31 in which the cost of DNA synthesis makes up the major proportion.12,99 Yet, tape storage costs just $16 per TB.33 Antkowiak et al. had given a detailed estimation on each step of the DNA data storage workflow in 2020.100 The high cost greatly prevents DNA storage from becoming a commercial product.13,28,100 Nevertheless, DNA data storage is still considered to be one of the most promising long-term storage solutions for the future, as the cost of synthesis and sequencing keeps falling dramatically and consistently over the years.
3. DNA synthesis
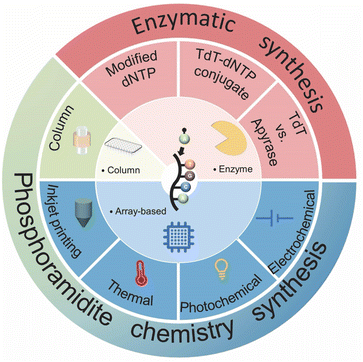
Fundamentally, two methods have been developed for artificial DNA synthesis: chemical and enzymatic. Based on these two methods, the technological route of artificial DNA synthesis can be divided into three generations (Fig. 3).91,94,101–103 The first generation is the traditional column-based solid-phase phosphoramidite chemistry synthesis. Owing to the development of microchip technology, high-throughput array-based synthesis technology based on phosphoramidite chemistry synthesis has blossomed in recent years, which is considered to be the second generation. Enzymatic synthesis, subsequently, could bring the synthetic biology industry into an exciting next stage.3.1. Chemical synthesis
The history of DNA synthesis began in the 1950s when Michelson and Todd published the first chemical synthesis of dinucleotides.104 Subsequently, phosphodiester105 and phosphotriester106 methods of oligo synthesis were developed. In 1981, Caruthers first described the solid-phase phosphoramidite method of oligo synthesis.107 In this method, nucleotides were covalently immobilized on a solid-phase carrier. Phosphoramidite monomers, each carrying a base group, are used as the synthesis unit. The monomers underwent a series of chemical reactions to extend the nucleotide strand in a controlled manner. So far, this is still the standard protocol for chemical synthesis of DNA. The conventional solid-phase phosphoramidite chemistry method consists of four cyclic steps, which are displayed in Fig. 4.94,101,108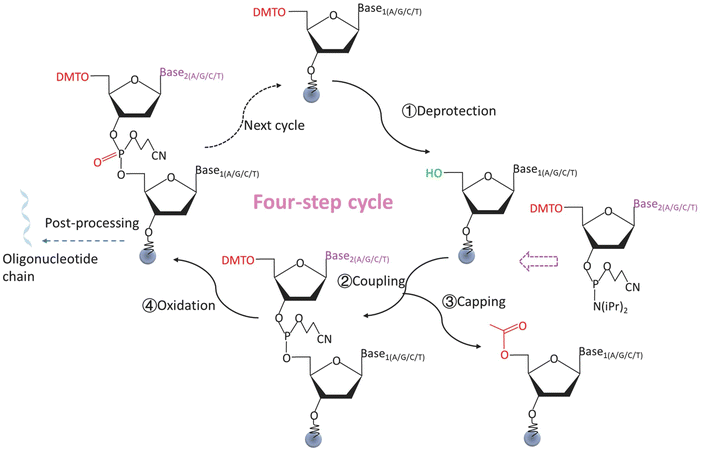 | ||
| Fig. 4 A four-step cycle for the synthesis of oligonucleotides by solid-phase phosphoramidite chemistry method. ① Deprotection. The DMT group at the 5′ end of an oligonucleotide monomer is removed, and the hydroxyl group is exposed to start the reaction. ② Coupling. The desired free nucleotide monomer is attached to the 5′ end hydroxyl group of the previous monomer. ③ Capping. The unreacted 5′ end hydroxyl groups of the oligonucleotide are sealed to prevent unwanted strand extention. ④ Oxidation. Oxidation reagent oxidizes the linkage bonds between the coupled monomers to a more stable state. The cycle is repeated until the target sequences are achieved. Reproduced from ref. 94 with permission from Copyright 2014 Springer Nature. | ||
Step 1: deprotection. The 5′ end of each phosphoramidite monomer has a protecting group (e.g., dimethoxytrityl (DMT)). This protecting group prevents a monomer from the chemical reaction which couples another monomer. In the deprotection step, the protecting group is removed under certain chemical conditions (e.g., an acid environment created by trichloroacetic acid (TCA)), exposing the 5′ end hydroxyl group that allows the subsequent reactions to occur.
Step 2: coupling. The 5′ end hydroxyl group of the monomer in step 1 couples with a newly added 5′ end protected monomer by forming a phosphite triester.
Step 3: capping. The uncoupled 5′ end hydroxyl group of the monomer in step 1 is acetylated in the presence of N-methylimidazole to prevent unwanted coupling (e.g., strand extension) in the next cycle.
Step 4: oxidation. The unstable phosphite triester bond between the successfully coupled monomers in step 2 is oxidized to the more stable phosphate by an oxidizing agent (e.g., a mixture of I2 in pyridine/H2O/THF). The strand is then ready for the next four-step nucleotide extension cycle.
Between each reaction step, there is a washing process commonly by acetonitrile which rinses off the excess reagents to start the next reaction. By cycling the above four steps, bases are added one per cycle in the designated order until the target oligonucleotide sequences are achieved.
Since the 1980s, based on the aforementioned synthetic principles, the carrier linkers, reagents, and deprotection methods used in synthesis have been continuously optimized. Established industrial DNA solid-phase chemical synthesis uses controlled pore glass (CPG) or polystyrene (PS) beads filled in a synthesis column as the solid carrier for nucleotide strands.109,110 Automated synthesizers drive the synthesis reagents in a unidirectional flow into the synthesis columns by controlled air pressure or a peristaltic pump. Solenoid valves can precisely eject milliliter (mL) and even microliter (μL) levels of reagents. In the meantime, multiple synthesis columns are utilized simultaneously, allowing for parallel and crossover synthesis. After synthesis, the target products are separated from the carrier, and the terminal protecting group is removed under alkaline conditions. The treatment is generally concentrated ammonia, while liquids such as methylamine are also used. Finally, depending on the needs of the application, direct elution and purification steps are performed for subsequent use. Typically, purification techniques such as polyacrylamide gel electrophoresis (PAGE) or high-performance liquid chromatography (HPLC) can effectively remove the wrong strands from the initial products. The products within a single column correspond to the same target sequence, namely the yield of a single column is between 2 nmol and 1000 nmol. In the case of large-scale column-based synthesis, the throughput of common automated parallel synthesis ranges from 96–1536 oligonucleotide sequences.101,111,112
Chemical synthesis methods are already well-developed and have been industrialized for many years. However, a number of errors may occur during synthesis: (1) deletions (0.1%). It is caused by a failure in coupling a nucleotide when the deprotection is insufficient or the coupling efficiency is low, leading to a missing base in the designed sequence; (2) insertions (0.1%). This means that additional, unwanted nucleotides are added. When the terminal-protected phosphoramidites undergo unintended deprotection due to cross-contamination of the reagent or a wrong treatment, it results in unwanted active sites on the strand to-be-extended, leading to an insertion error; (3) substitutions (0.5%). It occurs when another base is coupled instead of the intended base, mostly induced by reagent contamination or incomplete washing between the synthesis steps. Eventually, because of the limitation of chemical reactivity, the usual stepwise efficiency of the chemical synthesis is only 95%–99.5%.100,113 In addition, depurination may occur during deprotection, meaning that excess acid causes loss of purine bases, leading to hydrolysis of the DNA strands and ultimately to the decrease of yield and purity of the targeted products.114–116 As the depurination damage may occur in each synthesis cycle, it may accumulate as the sequence extension continues. Thus, the longer the synthesis sequences, the greater the probability of depurination would be. This greatly limits the yield of long products by chemical synthesis as well.109 Totally, a high error rate leads to low yield (yield = coupling efficiency(length−1)), where the coupling efficiency indicates the proportion of correct monomers added at each synthesis cycle. Clearly, the longer the strand, the lower the yield obtained due to the accumulation of errors. A small numerical increase of the coupling efficiency would lead to a great increase in the yield of the full-length product. For example, when the desired strand length is 200 nt, the full-length yield obtained by 99.3% coupling efficiency is only 24.7%, while 99.9% coupling efficiency corresponds to 81.9%. The length of the nucleotide strands synthesized based on phosphoramidite chemistry is usually limited to 200 nt and no commercial product has been yet announced to exceed 300 nt in practice without DNA assembly.92,117 Therefore, to synthesize a long target nucleotide sequence chemically, the sequence has to be split into short fragments of 50–100 bases, which are synthesized respectively, and then assembled. At present, in addition to producing short fragments such as primers and probes, the synthetic biology industry has begun to explore synthetic genomes and DNA data storage, placing higher demands on the length and quality of synthetic DNA. There are several concerns about the practical aspects that may hinder further industrialization of utilizing chemical DNA synthesis for data storage. For example, phosphoramidite chemistry requires demanding laboratory environment maintenance (e.g., strict humidity and inert atmosphere control), expensive reagents (e.g., phosphoramidites, whose price is up to 81 US$ g−1), long synthesis cycles, and generates large amounts of hazardous waste (e.g., acetonitrile which contaminates water and soil, pyridine and furan that are harmful mammal nervous system) during the synthesis process.118 In large-scale chemical synthesis, the purchase of raw materials, the waste stream, as well as the post-processing (e.g., purification and assembly) all contribute to tremendous costs that strongly limit the industrialization of DNA data storage. Although similar cost and environmental concerns also exist in the manufacturing industry of semiconductor memory chips, the technical limitations of chemical synthesis are becoming more and more critical.
3.2. Enzymatic synthesis
In biological organisms, polymerases are able to synthesize DNA naturally, but they are template-dependent and cannot synthesize arbitrary target sequences from a single base. Terminal deoxynucleotidyl transferase (TdT) is the only known polymerase whose primary activity is to indiscriminately add deoxynucleotide triphosphates (dNTPs) to the 3′ end of ssDNA in the absence of a template strand, making it a natural candidate for enzymatic oligonucleotide synthesis.34,119–123 In 1960, Bollum first described that TdT could act as a ssDNA polymerase.120 It was subsequently demonstrated that TdT could be used for template-free de novo DNA synthesis. Later on, Bollum proposed in 1962 a theoretical model for TdT-based DNA synthesis: TdT adds a nucleotide monomer with an acetyl protection group at the 3′ end by coupling its 5′ end to the 3′-OH of the previous nucleotide. The acetyl group plays a similar role to the DMT in the solid-phase phosphoramidite chemistry method, which can prevent elongation of the strand and thus realizes controlled strand extension.123,124A main advantage of enzymatic DNA synthesis is that it is carried out under mild aqueous conditions which effectively reduces DNA damage such as depurination and results in fewer by-products, making the achievement of longer target nucleotide strands possible. TdT-based enzymatic DNA synthesis has been extensively pursued by numerous companies (a detailed description is provided in Section 5). As reported, the coupling efficiency of enzymatic DNA synthesis can exceed 99%, which is comparable to phosphoramidite chemistry,125 and a coupling efficiency of about 99.9% calculated from the overall yield of 85% for 300 nt full-length sequences has been advertised.126 In terms of synthesis speed, the time of coupling a single base by phosphoramidite chemistry is about 4–10 min, while enzymatic synthesis by the TdT–dNTP conjugate can take only 10–20 s.91,92,127 Notably, the synthesis speed of different enzymatic synthesis pathways varies greatly.91 It's clear that enzymatic synthesis has the potential to produce longer strands with high accuracy at faster cycle times compared with chemical synthesis.92,122 Currently, the TdT-based enzymatic synthesis routes are being enthusiastically investigated and the mechanism of the enzyme is well studied. However, there are several issues that are necessary to be considered carefully: how to add nucleotides in a controlled and precise manner? What moieties are used to modify the monomers? How much do the unreacted initiators contribute to the deletion error rate? What is the probability of side reactions occurring in the synthesis process? What level of scale can be achieved for target products?122 How to improve the enzyme activity of the native TdT on 3′-end blocked dNTPs? In addition, further exploratory improvements in enzyme engineering and optimization of enzyme cycle reactions, etc., are still needed for large-scale industrialization. For example, Lu et al. demonstrated a two-step cyclic synthetic route using an engineered Zonotrichia albicollis (ZaTdT) enzyme with an average stepwise coupling efficiency of 98.7% for extending single nucleotides, which has some potential applications. The catalytic activity of this engineered enzyme was 3-fold higher than that of the normal TdT enzyme.128 Verardo et al.129 from DNA Script recently reported their approach to large-scale industrialization of TdT-based enzymatic DNA synthesis, which will be discussed in detail later in this review. This is a significant step towards the industrialization and parallelization of enzyme synthesis.
3.3. Technological development for DNA data storage
DNA synthesis technology is a key step in the process of DNA data storage. The speed, throughput, accuracy, and cost of synthesis all contribute to determining the availability of DNA data storage.There are two strategies for improving the throughput of DNA synthesis: one is to simply increase the number of channels for the above column-based synthesis and expand the scale of parallel synthesis; the other is to increase the synthesis density and miniaturize the system as a whole. Miniaturized array-based synthesis allows more sequences to be synthesized in parallel in a limited space while reducing the amount of consumed liquids. The scale of the products at a single site in the array is much lower than that in the column. Furthermore, array-based synthesis costs only $0.00001–$0.0001 per base, while column-based synthesis costs $0.05–$0.10 per base which is 2–4 orders of magnitude higher.94,101 Array-based DNA synthesis is oriented to the synthetic biology field of gene splicing, library building, and other applications that require trace level (e.g., fmol) as well as multiple sequences. The automation and continuous miniaturization of the instrument further enhance the throughput of array-based synthesis, which precisely provides a more suitable platform for DNA artificial synthesis whose application is data storage.
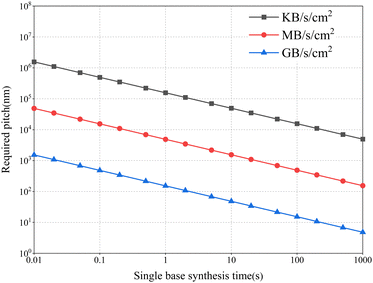
Fig. 5 shows the density of synthetic arrays required to achieve high-speed writing of large amounts of data. The total amount of data written per unit area can be calculated using the following equation:
| C = Eυριt |
Assuming that the amount of data shown in the figure needs to be achieved over an area of 1 square centimeter (cm2), and if a base can be encoded as 2 bits, a single synthesis site effectively encodes a nucleotide length of 100 nt, and 1 base could be synthesized at a rate of 1 base per second, then, to achieve TB (1 TB = 240 B) level data writing in one day, the scale of the array sites needs to be below the submicron level. However, current coding density is only able to reach 2 bits per base pair.11–15 What's more, the “encoding” and “decoding” steps also lead to errors. To restore the original data, in addition to the information-containing fragments, a certain length of data redundancy sequence needs to be added to the synthesized DNA strand. This requires the length of the synthetic sequence to be longer than the effective coding sequence.35 In sum, the above description implies that a much higher array density is required to achieve the TB level of data per day. To achieve such high-density arrays, micro and nanochips based on integrated circuit fabrication are the most optimal strategy.
Here, we aim to list and evaluate the diverse technological routes of utilizing integrated micro and nanoscale chips for DNA artificial synthesis. By weighing the pros and cons of each unique route, we hope that this review could provide a basic perspective on the trends in high-throughput DNA synthesis.
4. Array-based DNA synthesis chip
In the early 1990s, Affymetrix (acquired by Thermo Fisher) utilized integrated circuit technology to achieve high-density, 25 nt oligonucleotide synthesis on a single chip, opening up the route to DNA array-based synthesis technology.130,131 After decades of technological innovation, a variety of DNA array-based synthesis technologies have been developed and commercialized. These technologies are based on the four-step phosphoramidite chemistry but with different deprotection mechanisms. These are realized by involving terminal protecting groups of different natures in the synthesis process, such as pH-sensitive protecting groups,80,116,132–135 temperature-sensitive protecting groups32,136,137 or photolabile groups.138–140 Several array-based DNA synthesis methods are briefly illustrated in Fig. 6. Each of these has its advantages and disadvantages in terms of synthesis density, coupling efficiency, length, fidelity, time, and cost. These parameters are also evaluation indicators of DNA synthesis technologies. The following sections will further describe several mainstream technologies and the state of technological development of representative companies employing these technologies.4.1. Inkjet printing synthesis
Piezoelectric inkjet DNA synthesis was first proposed by Blanchard and Hood.141 This approach loads the monomer reagents into tiny nozzles as the “ink”, and uses an inkjet printer, namely a microdroplet generator, to precisely deposit reagents to the surface of a functionalized substrate to achieve large-scale parallel synthesis of oligonucleotide sequences.108,142 In brief, program-controlled nozzles move rapidly above the chip and spray chemical reagents to specified synthesis sites one by one according to a designed sequence as displayed in Fig. 7.143 The inkjet system is able to deposit the required monomer type to each site all over the entire chip rapidly in one round of injection in the coupling step, while the steps of deprotection, capping, oxidation and cleaning are carried out in the tiling mode through multiple channels. Generally, a piezoelectric printhead can control the liquid volume to the picoliter (pl) level. The droplets are spread on the array substrate with a diameter of tens to hundreds of microns in tens of microseconds. Owing to the small volume of the printed liquids, the reagent addition time is only hundreds of milliseconds. High-speed motors control the rapid movement of the microarray in the front and rear directions, and tens of thousands of dots can be printed in minutes. Additionally, 1,4-dicyanobutane (a more viscous, non-volatile solvent than acetonitrile) is used to dissolve the phosphoramidite monomers and the catalyst. This slows down the solvent evaporation, thus prolonging the reaction time between the reagent and substrate, ensuring coupling efficiency. However, as the nucleotide strand length extends, the surface properties of the array substrate may change, altering the size and location of the fallen droplets which might result in cross-contamination between adjacent sites. Therefore, a flat chip structure can't realize extremely high-density load. Moreover, a large number of deletions begin to accumulate once the strand length exceeds 50 nt.108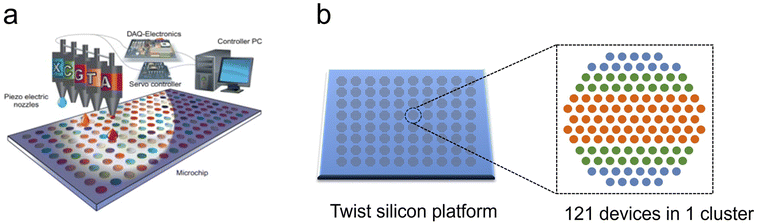 | ||
| Fig. 7 Schematic diagram of inkjet printing synthesis platform. (a) A program controls the motion of the inkjet print heads and prints trace amounts of phosphoramidite reagents on the slide surface.143 The slides are packed with tens of thousands of reaction chambers. Each of them can carry out a conventional four-step synthesis of phosphoramidite chemistry. Reproduced from ref. 143 with permission from Copyright 2013 Elsevier. (b) Twist's silicon-based DNA Synthesis platform. There are thousands of clusters on the chip, each consisting of 121 surface sites, performing different sequence synthesis.146 | ||
Agilent is the forerunner in commercializing inkjet printing technology and has been a leader in the synthesis of long oligonucleotides in the past decade. In 2001, Timothy R. Hughes et al. first used inkjet printing technology to synthesize 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 oligonucleotide strands with a length of 25 nt on a single 25 mm × 75 mm chip (glass wafer) with a coupling efficiency of 94–98%.144 Currently, Agilent's advanced SurePrint platform uses a proprietary, non-contact industrial inkjet printing process in which oligo monomers are deposited uniformly onto specially prepared glass slides, enabling high-fidelity, high-throughput parallel DNA synthesis of up to 244
000 oligonucleotide strands with a length of 25 nt on a single 25 mm × 75 mm chip (glass wafer) with a coupling efficiency of 94–98%.144 Currently, Agilent's advanced SurePrint platform uses a proprietary, non-contact industrial inkjet printing process in which oligo monomers are deposited uniformly onto specially prepared glass slides, enabling high-fidelity, high-throughput parallel DNA synthesis of up to 244![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 oligonucleotide strands, simultaneously. It has industry-leading fidelity of up to 1
000 oligonucleotide strands, simultaneously. It has industry-leading fidelity of up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2400 and allows synthesis of long oligos of about 230 nt.142
2400 and allows synthesis of long oligos of about 230 nt.142
Emily Leproust et al. (Co-Founder of Twist Bioscience) have developed “a proprietary semiconductor-based synthetic DNA manufacturing process” that uses a high-throughput silicon-based platform to miniaturize the chemical reaction conditions required for DNA synthesis. This miniaturization platform can reduce the reaction volume by a factor of one millionth, while increasing the throughput by a factor of 1000 and even up to 696![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The chip synthesizes oligonucleotides on specially treated micron-sized through-holes and uses high-speed inkjet print heads to deliver trace amounts of reagents. The chemical reaction size is dramatically reduced from 15 μL in a 96-well plate to 10 pl on the silica-based platform.145
000. The chip synthesizes oligonucleotides on specially treated micron-sized through-holes and uses high-speed inkjet print heads to deliver trace amounts of reagents. The chemical reaction size is dramatically reduced from 15 μL in a 96-well plate to 10 pl on the silica-based platform.145
Since inkjet printers consume a low amount of reagent and produce relatively little waste when running, they make the synthesis method more environmentally friendly. This chip is stated to have the ability to synthesize 9600 genes on a single in silico chip, meanwhile, traditional synthesis methods using 96-well plates can only produce one gene with the same length (up to 300 nt per oligo) of physical space. Each chip contains thousands of discrete clusters and each of the clusters contains 121146 individually addressable surfaces that are capable of synthesizing one type of unique oligonucleotide sequence, enabling high-throughput synthesis of millions of oligonucleotide sequences in the length range of 120–300 nt with yields exceeding 0.2 fmol. The average error rate is up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 nt. Twist has announced a milestone technical achievement in the successful synthesis of 200 nt oligonucleotide strands on a chip for DNA data storage. In 2021, twist announced its ability to synthesize DNA on a silicon chip with sites spaced 1 micron apart. This is so far the highest synthesis site density for inkjet printing synthesis.147
3000 nt. Twist has announced a milestone technical achievement in the successful synthesis of 200 nt oligonucleotide strands on a chip for DNA data storage. In 2021, twist announced its ability to synthesize DNA on a silicon chip with sites spaced 1 micron apart. This is so far the highest synthesis site density for inkjet printing synthesis.147
Recently, Verardo et al. integrated an inkjet printing system with enzymatic DNA synthesis and have achieved a synthesis length of 21 nt with an estimated cycle efficiency of 98.9%, while allowing for parallelization of more than 2000 sites.129 Thanks to inkjet printing, the reagent consumption per cycle was reduced to low quantities at the micromolar level. They also optimized the nozzle setup to improve the printing performance. Additionally, they developed a low-viscosity ink that managed to avoid damaging the enzyme activity, and optimized additives in the ink to prevent evaporation and minimize secondary structure formation of the ssDNA.
At present, the synthesis of DNA by inkjet printing is the mainstream choice for commercial products. Further increasing the synthesis throughput requires reducing the synthesis site size as well as the nozzle size. However, to ensure precise delivery of low-volume reagents and avoid splashing during injection, the nozzle size cannot be continuously minimized, which limits the increase of synthesis density. Generally, in piezoelectric printing, the droplet volume can be precisely controlled by adjusting the drive voltage and pulse waveform to achieve the pl level. The distribution and spacing of the nozzles affect the uniformity and actual volume of the droplets. When the distance is close, satellite droplets will be produced to interfere with the neighboring sites. Surface tension influences reagent dispersion, and the viscosity of the reagents affects the average size of droplets or even prevents droplet generation. In addition to this, problems such as low droplet orientation, nozzle plugging, wettability of the nozzle inner, and the nozzle-to-substrate height are present. Developing and maintaining smaller and more complex printing equipment may be a challenge both technologically and financially.
4.2. Thermal synthesis
Thermally controlled oligonucleotide synthesis is based on temperature-sensitive protecting groups. Temperature is a key factor in controlling the start or end of the reaction, and it also influences the quality of products.32,137,148–150Evonetix is the representative company for heat-controlled chip-based DNA synthesis technology. Andrew J. Ferguson et al. managed to control the synthesis process by precisely adjusting the temperature of reaction sites, combined with a microfluidic system. They developed silicon chips by the semiconductor MEMS (micro-electromechanical system) technology to provide thousands of independent temperature-controlled reaction sites for high-throughput parallel DNA synthesis, including error checking to improve yields and the eventual assembly of the as-obtained dsDNA.136,148 The entire reaction processes are carried out at the reaction sites (called “virtual wells”) in a continuously flowing liquid system with thermosensitive reagents. Each heating site on the chip has a diameter of 100 μm and a space of 300 μm resulting in approximately 10 heaters per square millimeter.
Under the control of computer programs, thousands of sites can be independently activated and warmed to start the independent DNA synthesis cycles, respectively. The closed-loop thermal control system allows liquid in each virtual well to reach different temperatures within the same circulation system and avoids the thermal diffusion on each site that happens with an array of conventional heaters. Temperature sensors at the sites feed the actual temperature back to the computer system, and, then, an algorithm compares it with the target temperature to determine whether it needs to be warmed up or cooled down. This requires very precise scaling circuitry and algorithmic programming. To achieve both “warming & cooling” functions, the material with controlled thermal resistance is installed underneath the site, which draws heat from the site to achieve a cooling effect.149 As shown in Fig. 8a, firstly, the circuitry controls the generation of heat at the activated sites. The heat transfers to the liquid above and, as a result, the temperature-sensitive protecting groups are removed.137 Subsequently, a new monomer can be added to each oligonucleotide strand at the activated sites. The cycle of heating and extension is repeated until the target oligonucleotide strands are synthesized. After that, with the help of precise flow pumps and electromagnetic fields, the short ssDNA fragments are selectively released by heating and are transferred to the partner strands with complementary base sequences immobilized on the substrate. In this way, long dsDNA can be automatically assembled on the chip. In addition, mis-matched double strands are identified, once the oligos are annealed because they have a lower denaturation temperature than the desired DNA. Subsequently, unwanted DNA strands are removed by applying precise, sequence-dependent temperature followed by flushing liquid (Fig. 8b). The error correction and purification processes can minimize polluted fragments in the product and help to provide a higher yield. Finally, the successfully matched oligos continue to assemble into longer dsDNA by complementary pairing at the terminal (Fig. 8c).
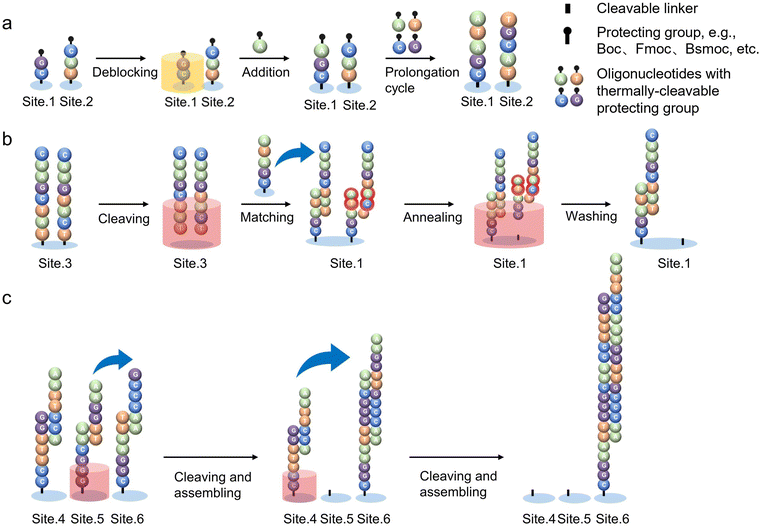 | ||
| Fig. 8 Schematic of thermally controlled oligonucleotide synthesis.103 (a) Thermally controlled strand extension process. The temperature-sensitive protecting group is removed by heating the selected site (site 1). The protecting group may alternatively be Boc, Fmoc, Bsmoc, and more examples could be found in ref. 137. Then, free oligonucleotide monomers are added onto the strand terminal. The cycles of heating and extension are repeated until the desired ssDNA fragments are achieved. (b) Thermally controlled cleavage and error-correction process. Deprotection and cleaving occur at different temperatures. The ssDNAs are released from site 3 by heating and then migrate toward partner strands with complementary base sequences which are immobilized on site 1; the mis-matches can be cleaved by applying a precise temperature during annealing and eventually washed away with the flowing liquid. (c) Thermally controlled assembly process. By heating site 5, the short dsDNAs are released and combined with another dsDNA (site 6) by the principle of complementary base pairing to assemble a longer strands; Heating site 4, short-stranded DNA continues to assemble at site 6. Those processes continue to produce desired long dsDNAs with high yield. Reproduced from ref. 103 with permission from Copyright 2023 Springer Nature. | ||
It is claimed that this technology platform is compatible with chemical and enzymatic DNA synthesis methods. However, it also faces some challenges. For example, appropriate protecting groups are selected according to the type of activating agent used in the heating step. When the activator is acidic (e.g., trifluoroacetic acid), tert-butyloxy carbonyl (Boc) or trityl (Trt) is mostly used. When the activator is basic (e.g., morpholine or piperidine), (1,1-dioxobenzo[b]thiophene-2-ylmethyloxycarbonyl (Bsmoc)) is preferable.137 It is challenging to develop highly temperature-sensitive protecting functional groups. Another serious difficulty is how to independently and precisely control the thermal behavior of micron reaction sites on the chip. To ensure the efficiency of synthesis, it is important to consider the approach that can help the generated heat dispersed evenly around the reaction site without conducting to the gap region or the adjacent sites. To efficiently conduct heat, Evonetix has developed a cooling system that consists of fluid flowing coolant, a thermoelectric cooler, and a copper substrate glued to the back side of the chip. Besides, there are other technical difficulties, such as: which microfluidic system to choose and what is the optimal flow rate? How to control the behavior of DNA under different thermal conditions? How to manufacture precisely assembled silicon wafer modules and avoid the risk of wafer explosion at the weakly bonded area during heating? How to prevent the chip from corroding when it is immersed in the strong acid/alkali reagent at a high temperature?
4.3. Photochemical synthesis
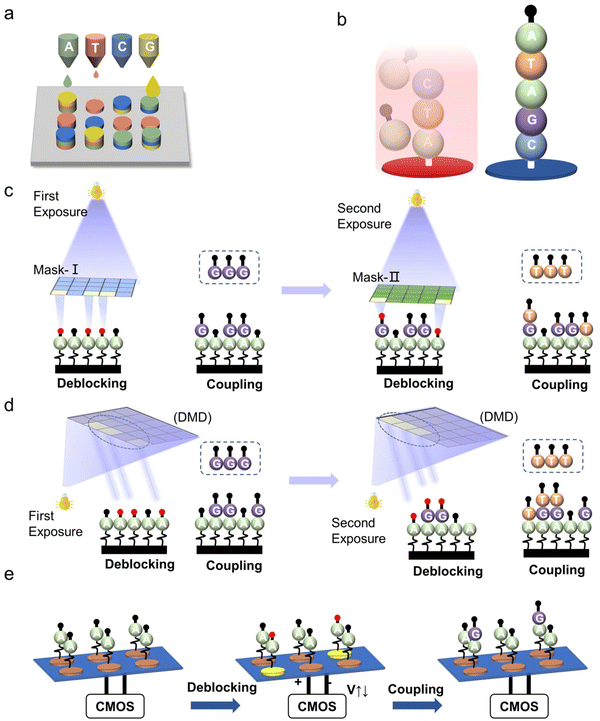
Photochemical synthesis is realized as follows: firstly, a laser with a specific wavelength is precisely projected onto the selected sites of the array substrate. On the irradiated sites, the protecting groups at the 5′ end of the nucleotides are removed. Subsequently, a series of chemical reactions including coupling and capping are performed in a tiling manner, while the nucleotide strands extension only occurs on the irradiated sites. According to the mechanism of deprotection, there are two types of synthesis methods: photo-acids and photo-degration.130,138,151,152 The principle of photo-acids is to decompose the photocatalyst by light exposure, generating acid to remove the protecting group (e.g., DMT).134,135 The photo-degradation approach, on the other hand, is based on direct decomposition of the photolabile protecting groups (e.g., 2-(2-nitrophenyl)-propoxycarbonyl (NPPOC) or benzoyl-2-(2-nitrophenyl) propoxycarbonyl (Bz-NPPOC)) caused by the projected light.138,140,152,153 According to the optical control system, photochemical synthesis is divided into two types, which are mask-based photolithography (used by Affymetrix) and maskless photolithography (used by Roche, LC Sciences), respectively.Mask-based photolithography synthesis refers to the transmission of light through specifically designed physical masks placed over the synthesis surface. Light is only allowed to pass through the transparent area of the mask, and be projected onto the substrate at certain locations.154 Affymetrix's commercial product GeneChipTM represents a mask-based photolithographic in situ synthesis (Fig. 9a).155 This technique typically produces 20–25 nt oligonucleotide strands and more than 106 feature sites per chip. With the development of photolithography process, the feature size of each chip has evolved from 50 μm to 20 μm, 18 μm, 11 μm and eventually down to 5 μm on a 1.28 cm × 1.28 cm chip in 2005.139,156 Subsequently, it was found that a further reduction of feature size of the chip to 1 μm with densities up to 1 × 108 cm−2 was proven to be promising by simulation with a reasonable control of the diffraction.139 However, a unique mask is needed for almost each cycle of nucleotide strands extension. For long sequence synthesis, the mask photolithography method requires a large number of custom-made mask plates, which dramatically increases the cost of synthesis.
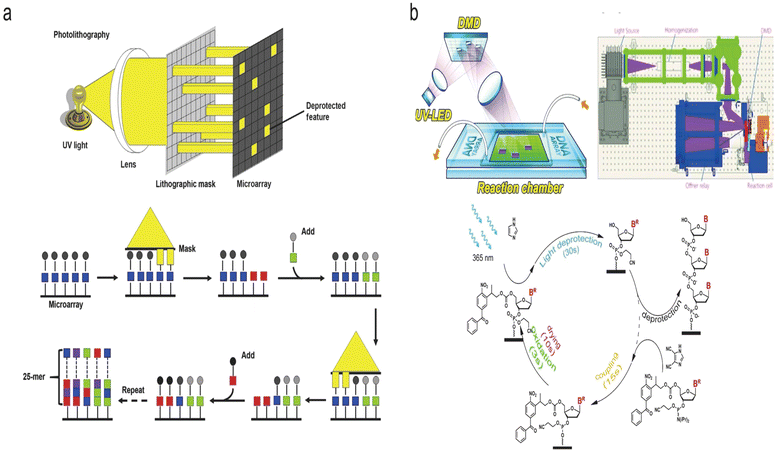 | ||
| Fig. 9 Schematic diagram of two photochemical methods of DNA synthesis. (a) Mask-based photolithography synthesis.155 Top: Mask-based photolithography. UV light passes through a lithographic mask that acts as a filter to either transmit or block the light from the chemically protected microarray surface (wafer). The sequential application of specific lithographic masks determines the order of sequence synthesis on the surface. Bottom: Chemical synthesis cycle. UV light removes the protecting groups (squares) from the array surface, allowing the addition of one nucleotide. The sequential synthesis cycles result in multiple 25-mer probes on the array surface. Reproduced from ref. 155 with permission from Copyright 2015 Elsevier. (b) Maskless photolithography synthesis.152 Top: DMD. The 365 nm UV light from an LED is uniformly projected onto the DMD. Digital micromirrors in the “ON” state reflect the light onto the surface of selected synthesis sites. Bottom: The cycles of phosphoramidite synthesis with a Bz-NPPOC protecting group at the 5′ end that is used in this method. Reproduced from ref. 152 with permission from Copyright 2021 Oxford University Press. | ||
Similar to modern projection techniques, maskless lithography synthesis uses a programmable and addressable digital micromirror device (DMD) instead of chrome masks (Fig. 9b).152 The light path is reflected to the designated synthesis sites for the deprotection reaction by adjusting the angles of each of micromirrors.106,109,135,157 Roche has developed a series of commercial products with densities exceeding 106 oligonucleotides per cm2 using DMD to control the angle of aluminum lenses. LC Sciences' μParafloTM synthesizer uses DMD to irradiate the photocatalyst to produce acid, followed by the removal of the acid-sensitive protecting group DMT. Their microfluidic platform delivers extremely low amounts of reagents. In a single 1.4 cm2 chip that contains 4000 reaction chambers corresponding density is 2857 chambers per cm2, and each of them requires only 270 pl of reagents, remarkably reducing reagent consumption.158,159 Plus, compared with mask-based photolithography synthesis, digital photolithography synthesis does not require expensive, specifically designed and case-sensitive masks so that the synthesis cost can be reduced.
Photochemical synthesis method ensures that each reaction chamber can synthesize nucleotides independently by controlling the light accurately, which brings great possibilities for increasing the throughput. So far, a standard high-definition DMD (1920 × 1080 dpi) can synthesize up to 786![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 432 sequences, simultaneously.160 The higher resolution enables more precise control of light to reduce diffraction and cross-contamination, contributing to a significant boost in synthetic throughput. However, the maskless lithography synthesis method has some other drawbacks that may diminish its corresponding efficiency, yields, and fidelity. The physical properties of light limit the minimum size and spacing of individual synthesis sites, hindering further miniaturization of the synthesis system. For example, light diffraction and scattering from micromirrors may cause loss of contrast at pixel edges, resulting in unintentional exposure and cross-contamination of the neighboring sites. Unfortunately, diffraction is an inevitable problem in all optical systems. This limits the gap size of pixels to the micron scale. In the case of micromirror array, when the micromirror spacing is reduced to around 1 μm, the average total error rate increases sharply to 21.8% per bp.152,160 Another defect is the local flare caused by refraction along the light path, bubbles in the solvent, or the interface between solvent and surface. They may all lead to unintended exposure.161 Furthermore, high-resolution systems are more sensitive to global scattering, and require higher complexity of the equipment which increases the equipment size and development cost. Efforts on optimizing the micromirror distribution, light irradiation time, capping time, protecting groups and solvents may help enhance the quality of photochemical synthesis.154,162
432 sequences, simultaneously.160 The higher resolution enables more precise control of light to reduce diffraction and cross-contamination, contributing to a significant boost in synthetic throughput. However, the maskless lithography synthesis method has some other drawbacks that may diminish its corresponding efficiency, yields, and fidelity. The physical properties of light limit the minimum size and spacing of individual synthesis sites, hindering further miniaturization of the synthesis system. For example, light diffraction and scattering from micromirrors may cause loss of contrast at pixel edges, resulting in unintentional exposure and cross-contamination of the neighboring sites. Unfortunately, diffraction is an inevitable problem in all optical systems. This limits the gap size of pixels to the micron scale. In the case of micromirror array, when the micromirror spacing is reduced to around 1 μm, the average total error rate increases sharply to 21.8% per bp.152,160 Another defect is the local flare caused by refraction along the light path, bubbles in the solvent, or the interface between solvent and surface. They may all lead to unintended exposure.161 Furthermore, high-resolution systems are more sensitive to global scattering, and require higher complexity of the equipment which increases the equipment size and development cost. Efforts on optimizing the micromirror distribution, light irradiation time, capping time, protecting groups and solvents may help enhance the quality of photochemical synthesis.154,162
4.4. Electrochemical synthesis
Multiple electrode arrays developed by micro/nanofabrication technology can be highly miniaturized, independently controlled as well as addressed in parallel. This offers an exciting opportunity in DNA synthesis: when a potential is applied to the selected electrode in a liquid environment, it initiates a redox reaction on the reagent which creates proton acid. The acid environment removes the pH-sensitive protecting groups at the 5′ end of the nucleotide strands, making the strands available for coupling next nucleotide monomer. Similar to the above approaches, the synthesis cycles are repeated until an oligonucleotide strand of target sequence is obtained.35,102,116,133,163CustomArray, now part of GenScript, developed independently controllable microelectrode arrays based on complementary metal–oxide–semiconductor (CMOS) integrated circuit chips. These microelectrodes are treated with a porous reaction layer (sucrose) to improve the quality of nucleotides synthesis.164 To confine the diffusion of proton acid from the activated electrode sites to the neighboring ones, an opposite potential is applied to the electrodes around the synthesis sites to trigger a reduction electrochemical reaction that produces bases to neutralize the excessive acid.165 Their 12 K microarray chip product has a circular electrode diameter of 44 μm and can synthesize 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 oligos. The 90 K chip offers synthesis throughput of 92
472 oligos. The 90 K chip offers synthesis throughput of 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 918 and oligonucleotide libraries up to 170 nt in length with an error rate of less than 0.5%, and the electrode size is further reduced to 22 μm. On the company's website, it is announced that this is the highest density commercial oligo-synthetic chip at present, with a throughput of 8 million oligos per chip, and the number may reach 200 billion, potentially.166 In addition, the cost is affordable at less than $0.2 per base and the yield of each oligo is up to 1 fmol.166,167 Their chip products are starting to be used in DNA data storage research, which may bring the cost of data storage down to $50 per TB.24,168
918 and oligonucleotide libraries up to 170 nt in length with an error rate of less than 0.5%, and the electrode size is further reduced to 22 μm. On the company's website, it is announced that this is the highest density commercial oligo-synthetic chip at present, with a throughput of 8 million oligos per chip, and the number may reach 200 billion, potentially.166 In addition, the cost is affordable at less than $0.2 per base and the yield of each oligo is up to 1 fmol.166,167 Their chip products are starting to be used in DNA data storage research, which may bring the cost of data storage down to $50 per TB.24,168
Similar approaches were recently studied by Microsoft Research and University of Washington. They have achieved a parallel synthesis of arbitrary sequences of DNA at submicron scale, increasing the synthesis density by three orders of magnitude compared with existing products. The electrodes are 650 nm in diameter and the corresponding pitch length is 2 μm. According to the density of electrodes, 2.5 × 107 oligonucleotide strands are theoretically synthesized in parallel on a 1 cm2 area, which meets the electrode density required for data storage speeds of megabytes per second that we estimated in Fig. 5. In addition, the synthesis length is up to 180 nt, tripled than previous electrochemical microarray-based DNA synthesis methods.116,132 Furthermore, the total cumulative error rate including deletions, insertions, substitutions ranges from 4% to 8%, which is still within the 15% tolerance of DNA data storage technology employing an error-correcting system.14,100,133,169 They designed a special electrode array (Fig. 10c) to resolve the crosstalk problem among adjacent reactors. The synthesis sites (circular-shaped anodes) are at the bottom of a nanowell structure, where deprotection and coupling steps occur. One anode electrode is surrounded by four cathode electrodes (diamond-shaped) applying an opposite potential. As reported, the deprotection step involved the addition of methanol to acetonitrile in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, resulting in the generation of alkaline species that consumed the protonic acid at the cathodes and completed the electrochemical half-reaction. The alkaline methoxide anion chemically confines the acid within the synthesis sites region effectively, preventing unwanted deprotection at the sites which are supposed to be “off” during a synthesis cycle. Additionally, the deep nanowell also provides a physical barrier to limit the acid cross-contamination.
9, resulting in the generation of alkaline species that consumed the protonic acid at the cathodes and completed the electrochemical half-reaction. The alkaline methoxide anion chemically confines the acid within the synthesis sites region effectively, preventing unwanted deprotection at the sites which are supposed to be “off” during a synthesis cycle. Additionally, the deep nanowell also provides a physical barrier to limit the acid cross-contamination.
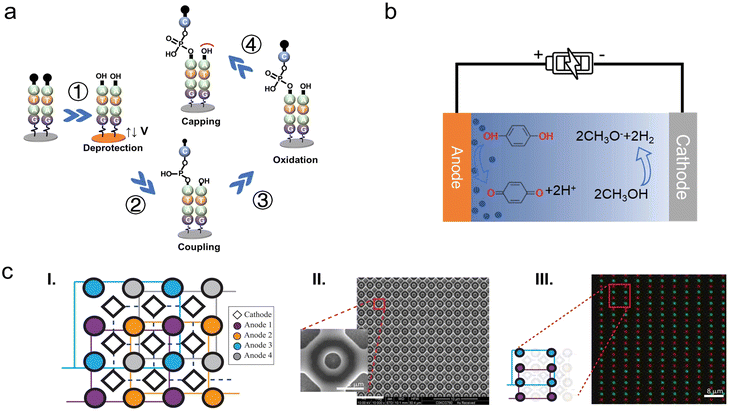 | ||
| Fig. 10 An overview of the electrochemical DNA synthesis. (a) Schematic diagram of the electrochemical synthesis of nucleotide strand on an electrode. ① A positive potential is applied to the electrode, producing a protonic acid to remove the DMT protecting group and exposing the “–OH” to start the next cycle. ② A free phosphoramidite monomer with a protecting group (DMT) at its 5′ end is coupled to the “–OH” on the electrode/previous nucleotide. ③ The newly formed phosphite backbone linkage is oxidized to the more stable phosphate by an oxidizing agent. ④ The capping reagents seal off “–OH” groups that are not coupled to the monomer, making them unavailable for subsequent reactions. (b) An example of reaction of redox pairs at electrodes during the electrochemical deblock step. The anode undergoes an oxidation reaction to generate protons; the cathode undergoes a reduction reaction that consumes protons. Reproduced from ref. 133 with permission from Copyright 2021 AAAS. (c) (I) Cathodes (diamond-shapes) are connected together (dashed line) while four anodes (circle-shapes) of the same color connected together (solid line). (II) SEM image of a nanoscale electrode array. The 650-nm anodes with the pitch length of 2 μm are sunk in a 200-nm deep well and surrounded by four counter electrodes. (III) A fluorescent image of the array in (II) after parallel synthesis of two different sequences with different fluorophores. The clear demarcation of the different fluorescence proves that the acid generated by the electrodes is strictly confined and demonstrates independently controlled parallel synthesis. Reproduced from ref. 133 with permission from Copyright 2021 AAAS. | ||
Further shrinking electrode feature size and shortening electrode pitch are effective solutions to greatly increase synthesis density and throughput. Typically, electrode sizes based on advanced semiconductor manufacturing technologies can reach submicron or even nanometer scale. It is relatively feasible to prepare ultra-dense micro/nanoelectrode arrays. In other applications, researchers have succeeded in narrowing down the diameter of micro-electrodes to 100–200 nm or even 10 nm, and the pitch of electrodes to 750 nm.170–172
However, the risk that the acid diffuses to neighboring electrodes raises at a higher density of electrodes.165,173 This results in unwanted deprotection on the surface of adjacent electrodes, which increases error rates and reduces synthesis yields. Currently, the biggest technical challenge in electrochemical DNA synthesis is to strictly confine the acid produced around the activated microelectrodes and prevent it from diffusing to the adjacent electrodes. A compromise between the synthesis density and the ion diffusion must be studied before a breakthrough technology that can solve the conflict appears.116,132,163,174
Although the aforementioned array-based DNA synthesis technologies have improved the throughput by several orders of magnitude over the traditional column-based synthesis, their capabilities are still not yet ready for applications such as DNA data storage. Each of these technologies faces its own challenges to substantially increase throughput, reduce costs and speed up synthesis while ensuring appropriate coupling efficiency: (1) for inkjet printing, the size, complexity and cost of piezo printheads are considered as the crucial limit. (2) The size and spacing of heater units and precise control of heat become an obstacle for thermal synthesis techniques. (3) Photochemical methods struggle with the inherent diffraction and refraction of light. Novel developments in applied physics, such as the plasmonics, may bring disruptive technological innovation to overcome the physical constrain of the chip size. It has been demonstrated that metallic nanostructures can generate localized surface plasmons when they couple to electromagnetic waves, resulting in thermoplasmonics effect like localized heating,175 or subdiffraction-limited spatial resolution in optics,176 which might shine a light in further reducing the working units of chips for thermal or photochemical DNA synthesis. (4) Electrochemical synthesis has to overcome the proton acid diffusion and crosstalk effects between electrodes (Table 1). For example, technologists have proposed new ways in generating ions or protons, e.g., by using ion-releasing materials as a working electrode that releases protons directly instead of via the redox reaction in the solution, which may hold potential to further localize the protons and enable even higher synthesis density.177
| Synthesis method | Merit | Challenge | Ref. |
|---|---|---|---|
| Inkjet printing | • Low volume reagent and cost; | • Relatively low throughput; | 109, 143–145 and 148 |
| • Parallel synthesis without sacrificing fidelity; | • Limited positioning accuracy; | ||
| • Highly commercialized. | • Large surface tension of a liquid droplet. | ||
| Thermal | • High throughput up to 100 million; | • Emerging heat-sensitive reaction system; | 32, 104, 137 and 138 |
| • Difficulty in controlling local heat transfer; | |||
| • Highly complex process; | |||
| • Simultaneous assembly and error correction. | • Relatively immature, more R & D is expected. | ||
| Photolithography | • The photolithography resolution is high; | • Inherent light diffraction, scattering and other phenomena; | 94, 135, 136, 153, 166, 186 and 207 |
| • Highest commercialized synthesis density; | • Difficult to reduce the size further; | ||
| • Technology is relatively mature. | • Expensive masks and complex system. | ||
| Electrochemical | • Utilize highly integrated circuit manufacturing techniques; | • Acid diffusion crosstalk; | 26, 92, 133, 134 and 164 |
| • Ultra-high synthesis throughput; | • High integration and process requirements. | ||
| • Further miniaturization possibilities. | |||
5. Enzymatic synthesis: the new opportunity
Several companies including DNA Script, Camena Bioscience, Molecular Assemblies, Nuclera, Ansa Biotechnologies, etc., have already explored the commercialization of enzymatic DNA synthesis, enabling the synthesis of long single-stranded oligonucleotides. Their strategies are similar: TdT mediates the progressive extension of the nucleotides, while the extension is controlled by using modified nucleotides or nucleotide-enzyme conjugates that can block the subsequential coupling until further treatment.34,92,94,101,123,178 However, native TdT is highly selective to the modification of monomers, e.g., the 3′-blocked dNTPs, therefore, the enzyme often needs to be engineered so as to enhance its binding with the monomer.122,125The initial research focus was on the modification of the 3′-O-end of the nucleotides. A reversible “terminator” group is added to the nucleotide monomer, which interrupts the synthesis and prevents the next nucleotide from being coupled (Fig. 11a).91,119,122,152,179 A variety of reversible terminators are available, such as 3′-O-NH2-dNTP (DNA Script and Nuclera),180–183 3′-O-azidomethyl-dNTP184,185 (Molecular Assemblies), 3′-O-(2-nitrobenzyl)-dNTP (Camena Bioscience),186 3′-phosphate187 (Codexis188) or photo-cleavable nucleotides189,190 and other protecting groups.191,192
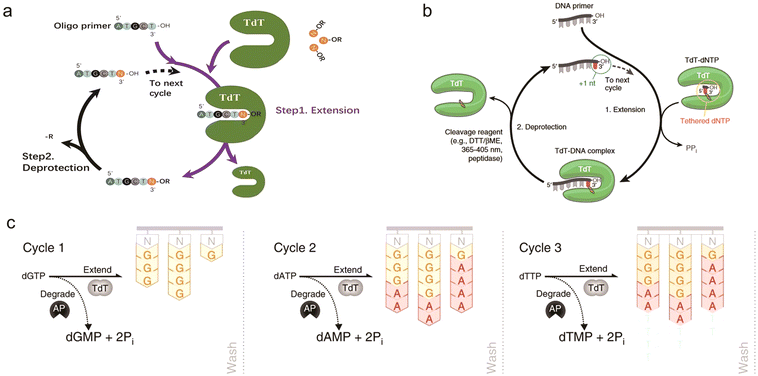 | ||
| Fig. 11 Three typical TdT-based enzymatic DNA synthesis routes. (a) The use of TdT to mediate synthesis of modified dNTP.91 Normal dNTP is modified with 3′-O-blocking group. Step 1: extension: TdT catalyzes the specific dNTP to couple with the primer. The blocking group can effectively prevent the next cycle of extension. Step 2: deprotection: the 3′-O blocking group of the primer that has been extended is removed to start the next nucleotide coupling. Reproduced from ref. 91 with permission from Copyright 2021 Frontiers. (b) Scheme for TdT–dNTP conjugate mediated two-step oligonucleotide extension.92 A short DNA primer is immobilized to a solid-phase support for the conjugate to begin synthesis. The TdT–dNTP conjugate consists of a TdT tethered with a dNTP via a cleavable linker. Step 1: extension, the TdT–dNTP conjugate incorporate into a primer. After incorporation of the tethered dNTP, the 3′ end of the primer remains covalently bound to TdT and is inaccessible to other TdT–dNTP molecules. Step 2: deprotection, the linkage between TdT and the incorporated nucleotide is cleaved, releasing the primer and allows subsequent extension. Reproduced from ref. 92 with permission from Copyright 2018 Springer Nature. (c) Schematic depiction of competitive reaction between TdT and apyrase (AP) enzymes. First, oligonucleotide initiators (N, gray) are tethered to a solid support. During synthesis, TdT catalyzes nucleoside triphosphate to the 3′ end of the initiators, whereas apyrase terminates the coupling cycle to prevent excessive extension. A wash step after synthesis is necessary to remove byproducts.127 Reproduced from ref. 127 with permission from Copyright 2019 Springer Nature. | ||
In 2020, DNA Script launched the world's first desktop DNA printer “SYNTAX” based on enzymatic synthesis, enabling automated synthesis, purification and quantification. The platform uses resin as a solid-phase carrier, and a cleavable linker DNA molecule (called “initiator DNA (iDNA)”) is attached to it. The entire synthesis process is completed in two steps. Firstly, the engineered TdT adds a modified monomer (3′-O-NH2 dNTP) to the iDNA.180,181 Then, the terminator group at the 3′ end of the monomer is removed to initiate the next DNA extension. The two steps are repeated until the target sequence is synthesized. In detail, 3′-OH modified monomers (also known as reversible terminators-dNTP) are employed to prevent enzymes from conjugating multiple monomers to extend DNA strands at each cycle. The reversible terminators can be removed by a mild acid buffer reagent without contaminating the final product. Meanwhile, the TdT is engineered in vitro by DNA Script to catalyze rapid and selective addition of monomers to iDNA, maintaining high fidelity and coupling efficiency. In addition, it allowed for longer synthetic length. They successfully synthesized 360 nt oligonucleotide strands and the stepwise efficiency was up to 99.5%. However, the engineered polymerase currently takes about 5 minutes to add a modified monomer, while native TdT is efficient with unblocked nucleotides and is capable of adding a monomer within 10–20 seconds.119,193 Better engineered enzyme variants for blocked nucleotides are thus desired for improving the efficiency of controlled enzymatic synthesis. Recently, Verardo et al. reported the first demonstration of multiplex enzymatic synthesis of DNA with single-base resolution, showing the possibility of microspatial control of nucleic acid sequence and length and a longer length than nonmultiplexing studies. Besides, they used a silicon MEMS platform to synthesize 50 nt sequences with spot density up to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 560 on a 75 mm × 25 mm slide, and the cycling efficiency was estimated to be 98.9% based on 21 nt long synthesized sequence.129
560 on a 75 mm × 25 mm slide, and the cycling efficiency was estimated to be 98.9% based on 21 nt long synthesized sequence.129
Molecular assemblies developed a novel enzymatic method of DNA synthesis. In their pioneering fully enzymatic synthesis™ (or FES™) technology, a high-performance engineered TdT, i.e., calf thymus TdT sourced from engineered E. coli was used for synthesis.194,195 More than 25% of the enzyme's amino acid sequence is altered, enabling industry-leading coupling efficiency, accuracy and speed at high temperature.194,195 This new enzymatic synthesis technology allows for rapid synthesis of ultra-long sequence-specific DNA on demand, with an in-process purification process that selectively removes erroneous sequences rather than waiting until the entire reaction is completed, ensuring yields while saving time. Michael J. Kamdar, President and CEO of molecular assemblies announced on website in 2018 that they were the first industrial group to complete DNA data storage by enzymatic DNA synthesis.196,197,198 They recently delivered the first enzymatically synthesized oligonucleotides using the new technique FES™.199
Nuclera' eDropTM platform is based on thin film transistor (TFT) materials powered by electrical wetting effects. The software-controlled electronic signals modify the interaction between droplets and surfaces. Their high-resolution device provides precise control of droplet manipulation, enabling a high level of parallelization and generation of small volumes (tens to hundreds of nanoliters) of droplets. They have utilized the nitrite-mediated deprotection of the 3′-O-aminooxy reversible terminator to convert the aminooxy moiety (–ONH2) to the hydroxyl moiety (–OH) in the presence of acid, combined with engineered TdT. At the basal pH of the system, the nitrite deprotection solution is inactive while the modified TdT is active. When the system pH is lowered to the point where the deprotection solution is active and the reversible terminator is removed, the modified TdT is inactive. Subsequently, when the system pH is back to basal, protected monomers are added to the 3′-terminal of the oligo strands at the selected sites by the active modified TdT. This strategy prevents extension of the released “–OH” groups prior to addition of the next nucleotide.182 Based on the advantages of this platform, Nuclera is investigating novel enzymatic DNA synthesis techniques that promise to automate efficient DNA synthesis. Similar to DNA Script's monomer deprotection strategy, the reaction is fast and highly specific because of the enhanced nucleophilicity of the 3′-O-aminooxy group. Although there is a risk of unwanted side effects such as acid-induced depurination, it can be suppressed by pH adjustment and addition of certain salts (e.g., Mg2+, Na+, spermine).114 Another concern is that the nonspecific nitrosation of base leads to a much longer time for deprotection,180 which would affect the speed of synthesizing long DNA strands. This can be solved by using non-nitrosation methods instead.129,200
Ansa Biotechnologies aims to commercialize artificial enzymatic DNA synthesis by controlling the release of TdT enzyme from the 3′ end of the DNA based on their published technique.92 Instead of modifying the enzyme itself, they conjugate each TdT with a single dNTP using a reversible linker.201 This offers a much faster synthesis than several other enzymatic synthesis methods involving free monomers: the coupling time of using TdT with reversible terminator is around 60 min179,189 while for template-dependent polymerase mediated oligo synthesis it is 1 min.184 The utilization of TdT–dNTP conjugates allows for a reduction in reaction time to 10–20 seconds.92 During synthesis, the conjugate adds the monomer to the exposed 3′ end of the primer nucleotide, and the enzyme remains attached to the 3′ end to prevent the addition of other monomers (Fig. 11b).92,125 This strategy requires a higher amount of enzyme, but typically only consumes ∼11 μM of the enzyme–substrate conjugates in each cycle as stated, while the concentration of the monomers need for phosphoramidite method is 9000 times higher.92 In this sense, it is still cost-effective compared with other techniques that require high concentrations of expensive modified nucleotides.125 The company recently announced the successful de novo synthesis of the world's longest DNA oligonucleotide whose length can reach 1005 nt. The proportion of full-length products reached 28%, indicating an industry-leading average stepwise yield of approximately 99.9% during synthesis.202
Due to enzyme's general nature of high catalytic activity, the native TdTs are able to add unblocked nucleotides extremely quickly. Therefore, it's a huge challenge to control the synthesis of individual bases. For specific long sequence synthesis, enzyme engineering and modification monomer strategies are necessary to ensure coupling efficiency. However, for applications with high fault tolerance (e.g., data storage), a modification-free strategy is feasible. Kern Systems, founded in 2019, used an enzyme competition strategy:127,203 Two enzymes, TdT and apyrase, bring two opposite effects on nucleotides (Fig. 11c).127 TdT adds new nucleotides to the DNA strand, while apyrase degrades the remaining nucleotides, preventing them from being added to the existing strand redundantly. TdT, apyrase, and short oligonucleotide initiators are mixed in different concentrations into a synthesis system. By tuning the ratio of the concentration of the two enzymes, the TdT adds a limited number of the nucleotides with the same base to the extending strand in each synthesis round, until this batch of nucleotides are degraded by apyrase. At the same time, the company uses a non-exact synthesis co-coding technology that relies on redundancy and error correction mechanisms. This strategy suits the application of DNA data storage well, because in this way the validity of the data is not merely dependent on the exact sequence, which relieves the pressure in ensuring the synthesis accuracy. Although this strategy results in longer synthesis lengths that may lead to a compromise of volumetric storage density, and it puts higher demands on the coding system as consecutively identical bases are not permitted in coding, it is nevertheless an inspiring solution for the application of the enzymatic synthesis in DNA data storage.
In addition, Camena Bioscience uses a proprietary enzyme combination to achieve template-free DNA synthesis with trinucleotide building blocks. Their novel de novo synthesis and gene assembly technology-gSynthTM are able to synthesize 300 nt DNA strands and successfully produce a 2.7 kb plasmid. For the synthesis of 300 nt fragments, on average, the yield of gSynthTM full-length product is 85.3%, approximately indicating a coupling efficiency of higher than 99.9%. While for phosphoramidite synthesis, the yield is only 22.3%, which corresponds to a coupling efficiency of only 99.5%.126,186,204
An overview of the present technologies of DNA artificial synthesis is summarized in Table 2. Compared with phosphoramidite chemistry method, enzymatic synthesis technology uses aqueous reagents most of which are non-toxic, requires less purification process, and enables faster synthesis and large-scale synthesis of longer strands. Some reagents, such as the cacodylate buffer, still require careful disposal as they could be toxic for organisms at a high dose. Nevertheless, enzymatic synthesis generally offers a more cost-effective and environmentally friendly approach. Although enzymatic synthesis is still at its early stage where the “high-throughput” is mostly realized at multi-well plate scale, it is foreseeable that further optimization of enzymatic methods combined with the semiconductor microchip technology will likely drive the large-scale oligo synthesis technology into the next generation. There are already pioneer works: Light-mediated template-free TdT enzymatic synthesis provides a new strategy for accurate stacking of single nucleotides;179,189 Lee et al.205 proposed a strategy for DNA synthesis using array chips based on the enzymatic principle, which achieved point control of TdT enzymatic activity by coupling synthesis on a chip with a DMD platform. In 2023, Smith et al. reported a polymerase-nucleotide conjugate which works as a protecting group and is electrochemically cleavable. When the conjugate detaches from the surface at mild oxidative voltages, the oligonucleotide with an extendable 3′-end is spatially available for subsequent nucleotide coupling catalyzed by TdT. They preliminarily demonstrated controllable single-base enzymatic synthesis on a microelectrode array.206 These works is an important step forward toward high-throughput enzymatic DNA synthesis using high-density array chips platform. Han Sae Jung et al. from Harvard University has developed a control system for real-time pH monitoring, utilizing a CMOS integrated circuit system that applies voltages independently to electrode sites in a “concentric circle” layout, confining electrochemically-generated acid within the vicinity of the anode to prevent diffusion and enabling real-time monitoring of acid distribution on and around the activation site. This system enables parallelized pH-regulated enzymatic oligonucleotide extension and potentially can be extended to high-throughput enzymatic DNA synthesis in dense arrangements and mild aqueous media.207 However, it is not clear whether this system can properly develop pH-controlled effects in the high-density arrays required for DNA array-based synthesis, and further validation is needed. In addition, rapid amplification using biological cells, their enzymes or organelles is also the trend to achieve fast and accurate DNA synthesis. The booming revolution of synthetic biology as well as biotechnology will surely promote the development of large-scale, high-throughput DNA synthesis for data storage.
| Synthesis method | Platform | Technique | Control mechanism of the synthesis cycle | Core components | Reagent | Addictives | Equipment | Representative company |
|---|---|---|---|---|---|---|---|---|
| Solid-phase phosphoramidite | Column | Columnar liquid flow | Each oligo is synthesized in a separate column having an independent synthesis cycle | pH-sensitive 5′ end terminal protected dNTP | Organic based | NA | Solenoid/pneumatic valve liquid circuit | Twist Bioscience; IDT |
| Micro-array | Inkjet printing | Oligos share a synthesis cycle, while monomer and catalyst are applied to individual sites all over the array in order by an inkjet printer | NA | Piezo printheads and precision step-shift mechanical system | Agilent; Twist Bioscience | |||
| Thermal synthesis | Oligos share a synthesis cycle, while the deprotection step is controlled synchronously on individual sites by temperature change | Temperature sensitive 5′ end terminal protected dNTP | NA | Addressable heating control circuit system | Evonetix | |||
| Photo-degration | Oligos share a synthesis cycle, while the deprotection step is controlled individually by light irradiation | Photoliable 5′ end terminal protected dNTP | NA | Lithographic mask and optical control system, or digital | Affymetrix; LC Sciences; Roche | |||
| Photo-acids | Oligos share a synthesis cycle, while the deprotection step is controlled individually by light irradiation, light-induced acidogenesis and a pH-sensitive protecting group on the monomer | pH-sensitive 5′ end terminal protected dNTP | Light sensitive | Micromirror control system | ||||
| Electrochemical-acids | Oligos share a synthesis cycle, while the deprotection step is controlled individually by voltage, electrochemical acidogenesis and a pH-sensitive protecting group on the monomer | Redox | Integrated circuit-controlled addressable electrode arrays | CustomArray; Twist Bioscience; Intel Corporation; Microsoft | ||||
| Enzymatic synthesis | Column/array | pH/thermal/photographic/kinetics | Column: similar to column based platform for solid-phase phosphoramidite method; array: similar to the micro-array based platform for solid-phase phosphoramidite method. | pH/thermal/photon sensitive 3′ end terminal protected dNTP, engineered TdT; or dNTP–TdT conjugate; or TdT and apyrase | Aqueous | Redox, or temperature/light sensitive | Workstations with solenoid valves or pipettes | Molecular Assemblies; DNA Script; Thermo Fisher; Nuclera |
6. Summary and prospect
In summary, in the context of the data-exploding digital information age, high-throughput DNA synthesis technology is expected to be the next promising solution for digital data storage. Accelerating the development and scaling up of DNA synthesis technology will greatly alleviate the current problems in data storage and significantly reduce the consumption of electronic materials and energy. Similar to the efforts in integrated circuit semiconductor manufacturing to reduce chip process nodes and increase processing power, low cost and high throughput are the goals of current technological innovation in DNA synthesis. For DNA data storage to be actually used in a commercial application, large-scale automated synthesis and sequencing are the primary prerequisites. Currently, there still exists a constraint on the length for precisely synthesizing longer sequences. However, for DNA data storage applications, strand length is not paramount. As we encode large amounts of data in high-density oligonucleotide arrays, it is more effective to further improve the density, reliability, accuracy, and speed of synthesis, as well as to lower the synthesis cost per base. DNA sequencing technology has developed rapidly, and several sequencing schemes based on different mechanisms have greatly enhanced the sequencing scale.208–210 Next-generation sequencing technologies (NGS) have increased throughput from the KB level to GB and TB, and the costs are decreasing at a rate predicted to surpass the Moore's law.211 The third-generation nanopore single-molecule sequencing technologies will further increase sequencing length, accelerate sequencing speed, and reduce costs. The development of biotechnologies, including enzyme engineering specific for DNA synthesis or sequencing, as well as next-generation sequencing will likely continue to lead to overall cost reductions for DNA data storage, but these costs are still expected to be several orders of magnitude higher than existing long-term data storage solutions.28 In order to improve the practicality of DNA data storage and make it a promising competitor of the state-of-the-art archival tape technology, both the speed of its data storing and retrieving processes, and the cost of synthesis, sequencing and reading still require significant breakthroughs.Novel ideas and approaches are expected to further boost the development of technological platforms for DNA data storage. Optical or electrical recognition methods, which are utilized in DNA sequencing, can be integrated into the DNA synthesis process to achieve error correction during synthesis. For example, Xu et al. achieved synthesis and sequencing on the common electrode. The fluorescence spectra after the coupling of the expected bases are analyzed in real-time to check whether they are consistent with the expected spectra. A polymerase is used to catalyze primer extension during sequencing, which in turn causes charge redistribution, resulting in measurable current spikes.80 Combining real-time sequencing with in situ synthesis on a chip is expected to reduce the errors as well as reagent consumption. However, there may be a mismatch between synthesis and detection time. Furthermore, gene synthesis routes in the field of synthetic biology may further inspire the array-based synthesis technology. The synthesis process may be improved by monitoring various indicators such as electrical properties, temperature, magnetism or light influenced by the changes in the structure and length of ssDNA through physical or chemical analysis techniques. It is promising that in the future, the collaborative innovation of both academia and industry will give birth to new technological solutions, and drive the DNA data storage technology to become mature gradually and move further towards commercialization.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support from the National Key Research and Development Program (no. 2021YFF1200300), the National Natural Science Foundation of China (T2188102 and 22025404), the Shanghai Pujiang Program (no. 21PJ1422900 and 22PJ1422900), the Science and Technology Commission of Shanghai Municipality (no. 23J21900200), the Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZLCX20212602), and GuangCi Deep Mind Project of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. We thank Dr Fan Wu from Photonic View Technology Co., Ltd and Dr Long Qian from Peking University for the fruitful discussion during the revision of the manuscript.Notes and references
- S. Shrivastava and R. Badlani, Int. J. Electr. Energy, 2014, 2, 119–124 CrossRef.
- D. Reinsel, J. Gantz and J. Rydning, Data age 2025: The Digitization of the World From Edge to Core, https://www.seagate.com/files/www-content/our-story/trends/files/idc-seagate-dataage-whitepaper.pdf, (accessed July 16, 2023).
- DNA Data Storage Alliance, Preserving our Digital Legacy: An Introduction to DNA Data Storage, https://dnastoragealliance.org/dev/publications/, (accessed July 16, 2023).
- D. Mytton and M. Ashtine, Joule, 2022, 6, 2032–2056 CrossRef.
- D. Mytton, How much energy do data centers use?, https://davidmytton.blog/how-much-energy-do-data-centers-use/, (accessed July 16, 2023).
- E. Masanet, A. Shehabi, N. Lei, S. Smith and J. Koomey, Science, 2020, 367, 984–986 CrossRef CAS PubMed.
- A. Andrae, Projecting the chiaroscuro of the electricity use of communication and computing from 2018 to 2030, 2019.
- D. Carmean, L. Ceze, G. Seelig, K. Stewart, K. Strauss and M. Willsey, Proc. IEEE, 2019, 107, 63–72 CAS.
- IBM Inc, IBM Unveils World's First 2 Nanometer Chip Technology, Opening a New Frontier for Semiconductors, https://newsroom.ibm.com/2021-05-06-IBM-Unveils-Worlds-First-2-Nanometer-Chip-Technology,-Opening-a-New-Frontier-for-Semiconductors, (accessed October 24, 2023).
- L. Heineck, First to Market, Second to None: The World's First 232-Layer NAND, https://www.micron.com/about/blog/2022/july/first-to-market-second-to-none-the-worlds-first-232-layer-nand, (accessed 10 December, 2023).
- B. Li, N. Y. Song, L. Ou and D. Du, Can We Store the Whole World's Data in DNA Storage?, 2020.
- Y. Dong, F. Sun, Z. Ping, Q. Ouyang and L. Qian, Natl. Sci. Rev., 2020, 7, 1092–1107 CrossRef PubMed.
- G. M. Church, Y. Gao and S. Kosuri, Science, 2012, 337, 1628 CrossRef CAS PubMed.
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos and E. Birney, Nature, 2013, 494, 77–80 CrossRef CAS PubMed.
- R. N. Grass, R. Heckel, M. Puddu, D. Paunescu and W. J. Stark, Angew. Chem., Int. Ed., 2015, 54, 2552–2555 CrossRef CAS PubMed.
- G. M. Mortuza, J. Guerrero, S. Llewellyn, M. D. Tobiason, G. D. Dickinson, W. L. Hughes, R. Zadegan and T. Andersen, BMC Bioinf., 2023, 24, 160 CrossRef CAS PubMed.
- L. Organick, Y. J. Chen, S. Dumas Ang, R. Lopez, X. Liu, K. Strauss and L. Ceze, Nat. Commun., 2020, 11, 616 CrossRef CAS PubMed.
- A. Extance, Nature, 2016, 537, 22–24 CrossRef CAS PubMed.
- V. Zhirnov, R. M. Zadegan, G. S. Sandhu, G. M. Church and W. L. Hughes, Nat. Mater., 2016, 15, 366–370 CrossRef CAS PubMed.
- L. Song, F. Geng, Z. Y. Gong, X. Chen, J. Tang, C. Gong, L. Zhou, R. Xia, M. Z. Han, J. Y. Xu, B. Z. Li and Y. J. Yuan, Nat. Commun., 2022, 13, 5361 CrossRef CAS PubMed.
- Z. Ping, D. Ma, X. Huang, S. Chen, L. Liu, F. Guo, S. J. Zhu and Y. Shen, Gigascience, 2019, 8, 1–10 CrossRef CAS PubMed.
- K. Chen, J. Kong, J. Zhu, N. Ermann, P. Predki and U. F. Keyser, Nano Lett., 2019, 19, 1210–1215 CrossRef CAS PubMed.
- Y. Erlich and D. Zielinski, Science, 2017, 355, 950–954 CrossRef CAS PubMed.
- Y. Choi, T. Ryu, A. C. Lee, H. Choi, H. Lee, J. Park, S. H. Song, S. Kim, H. Kim, W. Park and S. Kwon, Sci. Rep., 2019, 9, 6582 CrossRef PubMed.
- J. Bornholt, R. Lopez, D. M. Carmean, L. Ceze and K. Strauss, IEEE Micro, 2016, 44, 637–649 Search PubMed.
- W. D. Chen, A. X. Kohll, B. H. Nguyen, J. Koch, R. Heckel, W. J. Stark, L. Ceze, K. Strauss and R. N. Grass, Adv. Funct. Mater., 2019, 29, 1901672 CrossRef.
- J. Koch, S. Gantenbein, K. Masania, W. J. Stark, Y. Erlich and R. N. Grass, Nat. Biotechnol., 2020, 38, 39–43 CrossRef CAS PubMed.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS PubMed.
- Y. Zhang, Y. Ren, Y. Liu, F. Wang, H. Zhang and K. Liu, ChemPlusChem, 2022, 87, e202200183 CrossRef CAS PubMed.
- M. H. Raza, S. Desai, S. Aravamudhan and R. Zadegan, Biotechnol. Adv., 2023, 66, 108155 CrossRef CAS PubMed.
- L. C. Meiser, B. H. Nguyen, Y. J. Chen, J. Nivala, K. Strauss, L. Ceze and R. N. Grass, Nat. Commun., 2022, 13, 352 CrossRef CAS PubMed.
- S. Crosby, Chim. Oggi, 2020, 38, 22–24 CAS.
- A. Doricchi, C. M. Platnich, A. Gimpel, F. Horn, M. Earle, G. Lanzavecchia, A. L. Cortajarena, L. M. Liz-Marzán, N. Liu, R. Heckel, R. N. Grass, R. Krahne, U. F. Keyser and D. Garoli, ACS Nano, 2022, 16, 17552–17571 CrossRef CAS PubMed.
- E. Yoo, D. Choe, J. Shin, S. Cho and B. K. Cho, Comput. Struct. Biotechnol. J., 2021, 19, 2468–2476 CrossRef CAS PubMed.
- C. Xu, C. Zhao, B. Ma and H. Liu, Nucleic Acids Res., 2021, 49, 5451–5469 CrossRef CAS PubMed.
- R. P. Feynman, Eng. Sci., 1960, 23, 22–36 Search PubMed.
- N. Wiener, US News World Rep., 1964, 56, 84–86 Search PubMed.
- M. S. Neiman, Radiotekhnika, 1964, 1, 3–12 Search PubMed.
- J. Davis, Art J., 1996, 55, 70–74 CrossRef.
- N. Yachie, K. Sekiyama, J. Sugahara, Y. Ohashi and M. Tomita, Biotechnol. Prog., 2007, 23, 501–505 CrossRef CAS PubMed.
- N. G. Portney, Y. Wu, L. K. Quezada, S. Lonardi and M. Ozkan, Langmuir, 2008, 24, 1613–1616 CrossRef CAS PubMed.
- M. Ailenberg and O. Rotstein, Biotechniques, 2009, 47, 747–754 CrossRef CAS PubMed.
- R. Lopez, Y. J. Chen, S. Dumas Ang, S. Yekhanin, K. Makarychev, M. Z. Racz, G. Seelig, K. Strauss and L. Ceze, Nat. Commun., 2019, 10, 2933 CrossRef PubMed.
- L. Anavy, I. Vaknin, O. Atar, R. Amit and Z. Yakhini, Nat. Biotechnol., 2019, 37, 1229–1236 CrossRef CAS PubMed.
- L. Organick, S. D. Ang, Y. J. Chen, R. Lopez, S. Yekhanin, K. Makarychev, M. Z. Racz, G. Kamath, P. Gopalan, B. Nguyen, C. N. Takahashi, S. Newman, H. Y. Parker, C. Rashtchian, K. Stewart, G. Gupta, R. Carlson, J. Mulligan, D. Carmean, G. Seelig, L. Ceze and K. Strauss, Nat. Biotechnol., 2018, 36, 242–248 CrossRef CAS PubMed.
- J. Bornholt, R. Lopez, D. M. Carmean, L. Ceze, G. Seelig and K. Strauss, IEEE Micro, 2017, 37, 98–104 Search PubMed.
- Z. Ping, S. Chen, G. Zhou, X. Huang, S. J. Zhu, H. Zhang, H. H. Lee, Z. Lan, J. Cui, T. Chen, W. Zhang, H. Yang, X. Xu, G. M. Church and Y. Shen, Nat. Comput. Sci., 2022, 2, 234–242 CrossRef PubMed.
- B. W. A. Bögels, B. H. Nguyen, D. Ward, L. Gascoigne, D. P. Schrijver, A. M. Makri Pistikou, A. Joesaar, S. Yang, I. K. Voets, W. J. M. Mulder, A. Phillips, S. Mann, G. Seelig, K. Strauss, Y. J. Chen and T. F. A. de Greef, Nat. Nanotechnol., 2023, 18, 912–921 CrossRef PubMed.
- J. L. Banal, T. R. Shepherd, J. Berleant, H. Huang, M. Reyes, C. M. Ackerman, P. C. Blainey and M. Bathe, Nat. Mater., 2021, 20, 1272–1280 CrossRef CAS PubMed.
- C. Bee, Y. J. Chen, M. Queen, D. Ward, X. Liu, L. Organick, G. Seelig, K. Strauss and L. Ceze, Nat. Commun., 2021, 12, 4764 CrossRef CAS PubMed.
- K. J. Tomek, K. Volkel, A. Simpson, A. G. Hass, E. W. Indermaur, J. M. Tuck and A. J. Keung, ACS Synth. Biol., 2019, 8, 1241–1248 CrossRef CAS PubMed.
- F. Farzadfard, N. Gharaei, Y. Higashikuni, G. Jung, J. Cao and T. K. Lu, Mol. Cell, 2019, 75(769–780), e764 Search PubMed.
- S. Mohan, S. Vinodh and F. R. Jeevan, Int. J. Comput. Appl., 2013, 69, 53–57 Search PubMed.
- F. Liu, J. Li, T. Zhang, J. Chen and C. L. Ho, ACS Synth. Biol., 2022, 11, 3583–3591 CrossRef CAS PubMed.
- M. Hao, H. Qiao, Y. Gao, Z. Wang, X. Qiao, X. Chen and H. Qi, Commun. Biol., 2020, 3, 416 CrossRef CAS PubMed.
- W. Chen, M. Han, J. Zhou, Q. Ge, P. Wang, X. Zhang, S. Zhu, L. Song and Y. Yuan, Natl. Sci. Rev., 2021, 8, nwab028 CrossRef CAS PubMed.
- S. L. Shipman, J. Nivala, J. D. Macklis and G. M. Church, Science, 2016, 353, aaf1175 CrossRef PubMed.
- S. L. Shipman, J. Nivala, J. D. Macklis and G. M. Church, Nature, 2017, 547, 345–349 CrossRef CAS PubMed.
- S. S. Yim, R. M. McBee, A. M. Song, Y. Huang, R. U. Sheth and H. H. Wang, Nat. Chem. Biol., 2021, 17, 246–253 CrossRef CAS PubMed.
- J. Bonnet, P. Subsoontorn and D. Endy, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8884–8889 CrossRef CAS PubMed.
- T. Ham, S. Lee, J. Keasling and A. Arkin, PLoS One, 2008, 3, e2815 CrossRef PubMed.
- K. N. Lin, K. Volkel, J. M. Tuck and A. J. Keung, Nat. Commun., 2020, 11, 2981 CrossRef CAS PubMed.
- Merck Inc, Oligonucleotide Handling & Stability, https://www.sigmaaldrich.cn/CN/en/technical-documents/protocol/genomics/dna-and-rna-purification/oligonucleotide-handling-and-stability, (accessed October 24, 2023).
- T. J. Anchordoquy and M. C. Molina, Cell Preserv. Technol., 2007, 5(4), 180–188 CrossRef CAS.
- B. Röder, K. Frühwirth, C. Vogl, M. Wagner and P. Rossmanith, J. Clin. Microbiol., 2010, 48, 4260–4262 CrossRef PubMed.
- N. Pazdernik, How to store oligonucleotides for greatest stability, https://www.idtdna.com/pages/education/decoded/article/storing-oligos-7-things-you-should-know?cUSS#:~:text=Real%2Dtime%20oligo%20stability%20depends,free%20water%20(Figure%201A), (accessed 10 December, 2023).
- K. Matange, J. M. Tuck and A. J. Keung, Nat. Commun., 2021, 12, 1358 CrossRef CAS PubMed.
- X. Tan, T. Zhang, L. Ge and Z. Lu, Russ. Chem. Rev., 2021, 90, 280 CrossRef CAS.
- A. X. Kohll, P. L. Antkowiak, W. D. Chen, B. H. Nguyen, W. J. Stark, L. Ceze, K. Strauss and R. N. Grass, Chem. Commun., 2020, 56, 3613–3616 RSC.
- Y. Liu, Y. Ren, J. Li, F. Wang, F. Wang, C. Ma, D. Chen, X. Jiang, C. Fan, H. Zhang and K. Liu, Sci. Adv., 2022, 8, eabo7415 CrossRef CAS PubMed.
- F. Farzadfard and T. K. Lu, Science, 2014, 346, 1256272 CrossRef PubMed.
- H. H. Nguyen, J. Park, S. J. Park, C.-S. Lee, S. Hwang, Y.-B. Shin, T. H. Ha and M. Kim, Polymers, 2018, 10, 28 CrossRef PubMed.
- F. Sun, Y. Dong, M. Ni, Z. Ping, Y. Sun, Q. Ouyang and L. Qian, Adv. Sci., 2023, 10, e2206201 CrossRef PubMed.
- R. Veneziano, T. R. Shepherd, S. Ratanalert, L. Bellou, C. Tao and M. Bathe, Sci. Rep., 2018, 8, 6548 CrossRef PubMed.
- T. L. Schmidt, B. J. Beliveau, Y. O. Uca, M. Theilmann, F. Da Cruz, C. T. Wu and W. M. Shih, Nat. Commun., 2015, 6, 8634 CrossRef CAS PubMed.
- S. Kosuri, N. Eroshenko, E. M. Leproust, M. Super, J. Way, J. B. Li and G. M. Church, Nat. Biotechnol., 2010, 28, 1295–1299 CrossRef CAS PubMed.
- S. Newman, A. P. Stephenson, M. Willsey, B. H. Nguyen, C. N. Takahashi, K. Strauss and L. Ceze, Nat. Commun., 2019, 10, 1706 CrossRef CAS PubMed.
- D. R. Bentley, S. Balasubramanian, H. P. Swerdlow, G. P. Smith, J. Milton, C. G. Brown, K. P. Hall, D. J. Evers, C. L. Barnes, H. R. Bignell, J. M. Boutell, J. Bryant, R. J. Carter, R. Keira Cheetham, A. J. Cox, D. J. Ellis, M. R. Flatbush, N. A. Gormley, S. J. Humphray, L. J. Irving, M. S. Karbelashvili, S. M. Kirk, H. Li, X. Liu, K. S. Maisinger, L. J. Murray, B. Obradovic, T. Ost, M. L. Parkinson, M. R. Pratt, I. M. Rasolonjatovo, M. T. Reed, R. Rigatti, C. Rodighiero, M. T. Ross, A. Sabot, S. V. Sankar, A. Scally, G. P. Schroth, M. E. Smith, V. P. Smith, A. Spiridou, P. E. Torrance, S. S. Tzonev, E. H. Vermaas, K. Walter, X. Wu, L. Zhang, M. D. Alam, C. Anastasi, I. C. Aniebo, D. M. Bailey, I. R. Bancarz, S. Banerjee, S. G. Barbour, P. A. Baybayan, V. A. Benoit, K. F. Benson, C. Bevis, P. J. Black, A. Boodhun, J. S. Brennan, J. A. Bridgham, R. C. Brown, A. A. Brown, D. H. Buermann, A. A. Bundu, J. C. Burrows, N. P. Carter, N. Castillo, E. C. M. Chiara, S. Chang, R. Neil Cooley, N. R. Crake, O. O. Dada, K. D. Diakoumakos, B. Dominguez-Fernandez, D. J. Earnshaw, U. C. Egbujor, D. W. Elmore, S. S. Etchin, M. R. Ewan, M. Fedurco, L. J. Fraser, K. V. Fuentes Fajardo, W. Scott Furey, D. George, K. J. Gietzen, C. P. Goddard, G. S. Golda, P. A. Granieri, D. E. Green, D. L. Gustafson, N. F. Hansen, K. Harnish, C. D. Haudenschild, N. I. Heyer, M. M. Hims, J. T. Ho, A. M. Horgan, K. Hoschler, S. Hurwitz, D. V. Ivanov, M. Q. Johnson, T. James, T. A. Huw Jones, G. D. Kang, T. H. Kerelska, A. D. Kersey, I. Khrebtukova, A. P. Kindwall, Z. Kingsbury, P. I. Kokko-Gonzales, A. Kumar, M. A. Laurent, C. T. Lawley, S. E. Lee, X. Lee, A. K. Liao, J. A. Loch, M. Lok, S. Luo, R. M. Mammen, J. W. Martin, P. G. McCauley, P. McNitt, P. Mehta, K. W. Moon, J. W. Mullens, T. Newington, Z. Ning, B. Ling Ng, S. M. Novo, M. J. O'Neill, M. A. Osborne, A. Osnowski, O. Ostadan, L. L. Paraschos, L. Pickering, A. C. Pike, A. C. Pike, D. Chris Pinkard, D. P. Pliskin, J. Podhasky, V. J. Quijano, C. Raczy, V. H. Rae, S. R. Rawlings, A. Chiva Rodriguez, P. M. Roe, J. Rogers, M. C. Rogert Bacigalupo, N. Romanov, A. Romieu, R. K. Roth, N. J. Rourke, S. T. Ruediger, E. Rusman, R. M. Sanches-Kuiper, M. R. Schenker, J. M. Seoane, R. J. Shaw, M. K. Shiver, S. W. Short, N. L. Sizto, J. P. Sluis, M. A. Smith, J. Ernest Sohna Sohna, E. J. Spence, K. Stevens, N. Sutton, L. Szajkowski, C. L. Tregidgo, G. Turcatti, S. Vandevondele, Y. Verhovsky, S. M. Virk, S. Wakelin, G. C. Walcott, J. Wang, G. J. Worsley, J. Yan, L. Yau, M. Zuerlein, J. Rogers, J. C. Mullikin, M. E. Hurles, N. J. McCooke, J. S. West, F. L. Oaks, P. L. Lundberg, D. Klenerman, R. Durbin and A. J. Smith, Nature, 2008, 456, 53–59 CrossRef CAS PubMed.
- Y. Choi, H. J. Bae, A. C. Lee, H. Choi, D. Lee, T. Ryu, J. Hyun, S. Kim, H. Kim, S. H. Song, K. Kim, W. Park and S. Kwon, Adv. Mater., 2020, 32, e2001249 CrossRef PubMed.
- C. Xu, B. Ma, Z. Gao, X. Dong, C. Zhao and H. Liu, Sci. Adv., 2021, 7, eabk0100 CrossRef CAS PubMed.
- M. Dimopoulou, M. Antonini, P. Barbry and R. Appuswamy, Signal Process. Image Commun., 2021, 97, 116331 CrossRef.
- J. Bonnet, M. Colotte, D. Coudy, V. Couallier, J. Portier, B. Morin and S. Tuffet, Nucleic Acids Res., 2010, 38, 1531–1546 CrossRef CAS PubMed.
- D. Bennet, T. Vo-Dinh and F. Zenhausern, Nano Select, 2022, 3, 883–902 CrossRef.
- P. L. Antkowiak, J. Koch, B. H. Nguyen, W. J. Stark, K. Strauss, L. Ceze and R. N. Grass, Small, 2022, 18, 2107381 CrossRef CAS PubMed.
- G. D. Dickinson, G. M. Mortuza, W. Clay, L. Piantanida, C. M. Green, C. Watson, E. J. Hayden, T. Andersen, W. Kuang, E. Graugnard, R. Zadegan and W. L. Hughes, Nat. Commun., 2021, 12, 2371 CrossRef CAS PubMed.
- C. Fan, Q. Deng and T. F. Zhu, Nat. Biotechnol., 2021, 39, 1548–1555 CrossRef CAS PubMed.
- J. Chen, M. Chen and T. F. Zhu, Nat. Biotechnol., 2022, 40, 1601–1609 CrossRef CAS PubMed.
- L. Pasteur, Researches on the Molecular Asymmetry of Natural Organic Products, 1905 Search PubMed.
- M. G. T. A. Rutten, F. W. Vaandrager, J. A. A. W. Elemans and R. J. M. Nolte, Nat. Rev. Chem., 2018, 2, 365–381 CrossRef.
- B. Nguyen, J. Sinistore, J. A. Smith, P. S. Arshi, L. M. Johnson, T. Kidman, T. J. Dicaprio, D. Carmean and K. Strauss, Architecting Datacenters for Sustainability: Greener Data Storage using Synthetic DNA, IEEE, 2020 Search PubMed.
- L. F. Song, Z. H. Deng, Z. Y. Gong, L. L. Li and B. Z. Li, Front. Bioeng. Biotechnol., 2021, 9, 689797 CrossRef PubMed.
- S. Palluk, D. H. Arlow, T. de Rond, S. Barthel, J. S. Kang, R. Bector, H. M. Baghdassarian, A. N. Truong, P. W. Kim, A. K. Singh, N. J. Hillson and J. D. Keasling, Nat. Biotechnol., 2018, 36, 645–650 CrossRef CAS PubMed.
- C. N. Takahashi, B. H. Nguyen, K. Strauss and L. Ceze, Sci. Rep., 2019, 9, 4998 CrossRef PubMed.
- S. Kosuri and G. M. Church, Nat. Methods, 2014, 11, 499–507 CrossRef CAS PubMed.
- R. Heckel, G. Mikutis and R. N. Grass, Sci. Rep., 2019, 9, 9663 CrossRef PubMed.
- J. M. Ruijter, C. Ramakers, W. M. Hoogaars, Y. Karlen, O. Bakker, M. J. van den Hoff and A. F. Moorman, Nucleic Acids Res., 2009, 37, e45 CrossRef CAS PubMed.
- C. M. Hommelsheim, L. Frantzeskakis, M. Huang and B. Ülker, Sci. Rep., 2014, 4, 5052 CrossRef CAS PubMed.
- K. A. Wetterstrand, DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP), https://www.genome.gov/sequencingcostsdata, (accessed July 16, 2023).
- C. K. Lim, S. Nirantar, W. S. Yew and C. L. Poh, Trends Biotechnol., 2021, 39, 990–1003 CrossRef CAS PubMed.
- P. L. Antkowiak, J. Lietard, M. Z. Darestani, M. M. Somoza, W. J. Stark, R. Heckel and R. N. Grass, Nat. Commun., 2020, 11, 5345 CrossRef CAS PubMed.
- R. A. Hughes and A. D. Ellington, Cold Spring Harbor Perspect. Biol., 2017, 9 Search PubMed.
- P. Kuhn, K. Wagner, K. Heil, M. Liss and N. Netuschil, Eng. Life Sci., 2017, 17, 6–13 CrossRef CAS PubMed.
- A. Hoose, R. Vellacott, M. Storch, P. S. Freemont and M. G. Ryadnov, Nat. Rev. Chem., 2023, 7, 144–161 CrossRef CAS PubMed.
- A. M. Michelson and A. R. Todd, J. Chem. Soc., 1955, 0, 2632–2638 RSC.
- P. T. Gilham and H. G. Khorana, J. Am. Chem. Soc., 2002, 80, 6212–6222 CrossRef.
- R. L. Letsinger and K. K. Ogilvie, J. Am. Chem. Soc., 1969, 91, 3350–3355 CrossRef CAS.
- S. L. Beaucage and M. H. Caruthers, Tetrahedron Lett., 1981, 22, 1859–1862 CrossRef CAS.
- H. Li, Y. Huang, Z. Wei, W. Wang, Z. Yang, Z. Liang and Z. Li, Sci. Rep., 2019, 9, 5058 CrossRef PubMed.
- M. Septak, Nucleic Acids Res., 1996, 24, 3053–3058 CrossRef CAS PubMed.
- B. Xia, S.-J. Xiao, D.-J. Guo, J. Wang, J. Chao, H.-B. Liu, J. Pei, Y.-Q. Chen, Y.-C. Tang and J.-N. Liu, J. Mater. Chem., 2006, 16, 570–578 RSC.
- M. Jensen, L. Roberts, A. Johnson, M. Fukushima and R. Davis, J. Biotechnol., 2014, 179, 76–81 CrossRef PubMed.
- J. Y. Cheng, H. H. Chen, Y. S. Kao, W. C. Kao and K. Peck, Nucleic Acids Res., 2002, 30, e93 CrossRef PubMed.
- J. Tian, K. Ma and I. Saaem, Mol. BioSyst., 2009, 5, 714–722 RSC.
- R. An, Y. Jia, B. Wan, Y. Zhang, P. Dong, J. Li and X. Liang, PLoS One, 2014, 9, e115950 CrossRef PubMed.
- E. M. LeProust, B. J. Peck, K. Spirin, H. B. McCuen, B. Moore, E. Namsaraev and M. H. Caruthers, Nucleic Acids Res., 2010, 38, 2522–2540 CrossRef CAS PubMed.
- R. D. Egeland and E. M. Southern, Nucleic Acids Res., 2005, 33, e125 CrossRef PubMed.
- M. H. Caruthers, J. Biol. Chem., 2013, 288, 1420–1427 CrossRef CAS PubMed.
- B. L. Simmons, N. D. McDonald and N. G. Robinett, Front. Bioeng. Biotechnol., 2023, 11 Search PubMed.
- E. A. Motea and A. J. Berdis, Biochim. Biophys. Acta, 2010, 1804, 1151–1166 CrossRef CAS PubMed.
- F. J. Bollum, J. Biol. Chem., 1960, 235, 2399–2403 CrossRef CAS PubMed.
- F. J. Bollum, J. Biol. Chem., 1959, 234, 2733–2734 CrossRef CAS PubMed.
- M. A. Jensen and R. W. Davis, Biochemistry, 2018, 57, 1821–1832 CrossRef CAS PubMed.
- S. M. Minhaz Ud-Dean, Syst. Synth. Biol., 2008, 2, 67–73 CrossRef CAS PubMed.
- F. J. Bollum, J. Biol. Chem., 1962, 237, 1945–1949 CrossRef CAS PubMed.
- M. Eisenstein, Nat. Biotechnol., 2020, 38, 1113–1115 CrossRef CAS PubMed.
- N. M. Bell, S. A. Mankowska, S. A. Harvey and D. L. Stemple, A highly accurate, enzymatic, de novo synthesis and gene assembly technology, https://www.camenabio.com/assets/media/2019-10-24-pplication-note.pdf, (accessed July 16, 2023) Search PubMed.
- H. H. Lee, R. Kalhor, N. Goela, J. Bolot and G. M. Church, Nat. Commun., 2019, 10, 2383 CrossRef PubMed.
- X. Lu, J. Li, C. Li, Q. Lou, K. Peng, B. Cai, Y. Liu, Y. Yao, L. Lu, Z. Tian, H. Ma, W. Wang, J. Cheng, X. Guo, H. Jiang and Y. Ma, ACS Catal., 2022, 12, 2988–2997 CrossRef CAS.
- D. Verardo, B. Adelizzi, D. A. Rodriguez-Pinzon, N. Moghaddam, E. Thomée, T. Loman, X. Godron and A. Horgan, Sci. Adv., 2023, 9, eadi0263 CrossRef CAS PubMed.
- A. C. Pease, D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes and S. P. Fodor, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 5022–5026 CrossRef CAS PubMed.
- S. P. Fodor, J. L. Read, M. C. Pirrung, L. Stryer, A. T. Lu and D. Solas, Science, 1991, 251, 767–773 CrossRef CAS PubMed.
- R. D. Egeland, F. Marken and E. M. Southern, Anal. Chem., 2002, 74, 1590–1596 CrossRef CAS PubMed.
- B. H. Nguyen, C. N. Takahashi, G. Gupta, J. A. Smith, R. Rouse, P. Berndt, S. Yekhanin, D. P. Ward, S. D. Ang, P. Garvan, H. Y. Parker, R. Carlson, D. Carmean, L. Ceze and K. Strauss, Sci. Adv., 2021, 7, eabi6714 CrossRef CAS PubMed.
- B. Y. Chow, C. J. Emig and J. M. Jacobson, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15219–15224 CrossRef CAS PubMed.
- X. Gao, E. LeProust, H. Zhang, O. Srivannavit, E. Gulari, P. Yu, C. Nishiguchi, Q. Xiang and X. Zhou, Nucleic Acids Res., 2001, 29, 4744–4750 CrossRef CAS PubMed.
- A. J. Ferguson, M. J. Hayes, B. C. Kirkpatrick, Y. C. Lin, V. Narayan and A. Prak, Eng. biol., 2019, 3, 20–23 CrossRef.
- S. R. Crosby, M. Jennison and J. Brennan, WIPO Pat., WO2018189546A1, 2018 Search PubMed.
- S. Bühler, I. Lagoja, H. Giegrich, K.-P. Stengele and W. Pfleiderer, Helv. Chim. Acta, 2004, 87, 620–659 CrossRef.
- A. R. Pawloski, G. McGall, R. G. Kuimelis, D. Barone, A. Cuppoletti, P. Ciccolella, E. Spence, F. Afroz, P. Bury, C. Chen, C. Chen, D. Pao, M. Le, B. McGee, E. Harkins, M. Savage, S. Narasimhan, M. Goldberg, R. Rava and S. P. A. Fodor, J. Vac. Sci. Technol., B, 2007, 25, 2537–2546 CrossRef CAS.
- G. H. McGall, A. D. Barone, M. Diggelmann, S. P. A. Fodor, E. Gentalen and N. Ngo, J. Am. Chem. Soc., 1997, 119, 5081–5090 CrossRef CAS.
- A. P. Blanchard, R. J. Kaiser and L. E. Hood, Biosens. Bioelectron., 1996, 11, 687–690 CrossRef CAS.
- P. K. Wolber, P. J. Collins, A. B. Lucas, A. De Witte and K. W. Shannon, in DNA Microarrays Part A: Array Platforms and Wet-Bench Protocols, ed. A. Kimmel and B. Oliver, 2006, vol. 410, pp. 28–57 Search PubMed.
- N. Tang, S. Ma and J. Tian, in Synthetic Biology, ed. H. Zhao, Academic Press, Boston, 2013, pp. 3–21 Search PubMed.
- T. R. Hughes, M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend and P. S. Linsley, Nat. Biotechnol., 2001, 19, 342–347 CrossRef CAS PubMed.
- E. Leproust, Ind. Biotechnol., 2021, 17, 107–108 CrossRef.
- P. F. Indermuhle, E. P. Marsh and A. Fernandez, US Pat., US2020/0299322A1, 2020 Search PubMed.
- Twist Bioscience Inc, Twist Bioscience Reports Fourth Quarter and Full Year Fiscal 2021 Financial Results, https://investors.twistbioscience.com/news-releases/news-release-details/twist-bioscience-reports-fourth-quarter-and-full-year-fiscal-1, (accessed July 16, 2023).
- M. J. Hayes and A. J. Ferguson, WIPO Pat., WO2018104698A1, 2018 Search PubMed.
- M. J. Hayes, R. M. Sanches-Kuiper and D. A. Bygrave, WIPO Pat., WO2019064006A1, 2019 Search PubMed.
- S. R. Crosby, M. Jennison and J. Brennan, US Pat., US11161869B2, 2021 Search PubMed.
- X. Gao, E. Gulari and X. Zhou, Biopolymers, 2004, 73, 579–596 CrossRef CAS PubMed.
- J. Lietard, A. Leger, Y. Erlich, N. Sadowski, W. Timp and M. M. Somoza, Nucleic Acids Res., 2021, 49, 6687–6701 CrossRef CAS PubMed.
- A. Hasan, K.-P. Stengele, H. Giegrich, P. Cornwell, K. R. Isham, R. A. Sachleben, W. Pfleiderer and R. S. Foote, Tetrahedron, 1997, 53, 4247–4264 CrossRef CAS.
- S. Singh-Gasson, R. D. Green, Y. Yue, C. Nelson, F. Blattner, M. R. Sussman and F. Cerrina, Nat. Biotechnol., 1999, 17, 974–978 CrossRef CAS PubMed.
- S. Miller, U. Karaoz, E. Brodie and S. Dunbar, in Current and Emerging Technologies for the Diagnosis of Microbial Infections, ed. A. Sails and Y. W. Tang, 2015, vol. 42, pp. 395–431 Search PubMed.
- D. D. Dalma-Weiszhausz, J. Warrington, E. Y. Tanimoto and G. Miyada, in DNA Microarrays Part A: Array Platforms and Wet-Bench Protocols, ed. A. Kimmel and B. Oliver, 2006, vol. 410, pp. 3–28 Search PubMed.
- J. P. Pellois, X. Zhou, O. Srivannavit, T. Zhou, E. Gulari and X. Gao, Nat. Biotechnol., 2002, 20, 922–926 CrossRef CAS PubMed.
- H. Ailing, Z. Qi, Z. Yulu, Z. Xiaochuan, E. Christoph and G. Xiaolian, PepArray™ - Epitope Mapping on a Flexible High-Density Microfluidic Chip, https://lcsciences.com/documents/PEGS_Poster_2010%20final_small.pdf, (accessed July 16, 2023) Search PubMed.
- X. Zhou, S. Cai, A. Hong, Q. You, P. Yu, N. Sheng, O. Srivannavit, S. Muranjan, J. M. Rouillard, Y. Xia, X. Zhang, Q. Xiang, R. Ganesh, Q. Zhu, A. Matejko, E. Gulari and X. Gao, Nucleic Acids Res., 2004, 32, 5409–5417 CrossRef CAS PubMed.
- C. Agbavwe, C. Kim, D. Hong, K. Heinrich, T. Wang and M. M. Somoza, J. Nanobiotechnol., 2011, 9, 57 CrossRef CAS PubMed.
- N. Kretschy, A. K. Holik, V. Somoza, K. P. Stengele and M. M. Somoza, Angew. Chem., Int. Ed., 2015, 54, 8555–8559 CrossRef CAS PubMed.
- M. Sack, N. Kretschy, B. Rohm, V. Somoza and M. M. Somoza, Anal. Chem., 2013, 85, 8513–8517 CrossRef CAS PubMed.
- K. Maurer, J. Cooper, M. Caraballo, J. Crye, D. Suciu, A. Ghindilis, J. A. Leonetti, W. Wang, F. M. Rossi, A. G. Stover, C. Larson, H. Gao, K. Dill and A. McShea, PLoS One, 2006, 1, e34 CrossRef PubMed.
- A. Gindilis, US Pat., US20190255504A1, 2019 Search PubMed.
- M. Karl, J. J. Cooper and F. H. Sho, US Pat., US20200086290A1, 2020 Search PubMed.
- CustomArray Inc., Miniature Semiconductor Technology for Nucleic Acid Synthesis, https://www.customarrayinc.com/msc-technology, (accessed July 16, 2023).
- Gen Script Inc., Precise Synthetic Oligo Pools:A perfect solution for cost-effective library construction and efficient high-throughput screening, https://www.genscript.com/location.php?href=/gsfiles/techfiles/Oligo_pool_folder_2019Q2.pdf, (accessed July 16, 2023).
- CustomArray Inc., MSC DNA Cold Storage Solutions, https://www.customarrayinc.com/dna-cold-storage, (accessed July 16, 2023).
- W. H. Press, J. A. Hawkins, S. K. Jones, Jr., J. M. Schaub and I. J. Finkelstein, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18489–18496 CrossRef CAS PubMed.
- Y. Liu, X. Li, J. Chen and C. Yuan, Front. Chem., 2020, 8, 573865 CrossRef CAS PubMed.
- R. Liu, R. Chen, A. T. Elthakeb, S. H. Lee, S. Hinckley, M. L. Khraiche, J. Scott, D. Pre, Y. Hwang, A. Tanaka, Y. G. Ro, A. K. Matsushita, X. Dai, C. Soci, S. Biesmans, A. James, J. Nogan, K. L. Jungjohann, D. V. Pete, D. B. Webb, Y. Zou, A. G. Bang and S. A. Dayeh, Nano Lett., 2017, 17, 2757–2764 CrossRef CAS PubMed.
- N. Karimian and P. Ugo, Curr. Opin. Electrochem., 2019, 16, 106–116 CrossRef CAS.
- M. Chen, L. Zheng, B. Santra, H. Y. Ko, R. A. DiStasio, Jr., M. L. Klein, R. Car and X. Wu, Nat. Chem., 2018, 10, 413–419 CrossRef CAS PubMed.
- X. Ji, C. E. Banks, D. S. Silvester, A. J. Wain and R. G. Compton, J. Phys. Chem. C, 2007, 111, 1496–1504 CrossRef CAS.
- L. Jauffred, A. Samadi, H. Klingberg, P. M. Bendix and L. B. Oddershede, Chem. Rev., 2019, 119, 8087–8130 CrossRef CAS PubMed.
- K. A. Willets, A. J. Wilson, V. Sundaresan and P. B. Joshi, Chem. Rev., 2017, 117, 7538–7582 CrossRef CAS PubMed.
- P. Vereecken and B. Put, US Pat., US20190355964A1, 2019 Search PubMed.
- K. Ramadan, I. V. Shevelev, G. Maga and U. Hubscher, J. Mol. Biol., 2004, 339, 395–404 CrossRef CAS PubMed.
- A. S. Mathews, H. Yang and C. Montemagno, Curr. Protoc. Nucleic Acid Chem., 2017, 71, 13.17.1–13.17.38 Search PubMed.
- D. Hutter, M.-J. Kim, N. Karalkar, N. A. Leal, F. Chen, E. Guggenheim, V. Visalakshi, J. Olejnik, S. Gordon and S. A. Benner, Nucleosides, Nucleotides Nucleic Acids, 2010, 29, 879–895 CrossRef CAS PubMed.
- DNA Script Inc., Enzymatic DNA Synthesis Technical Note, https://www.dnascript.com/wpcontent/uploads/2021/04/DNAScript_EDS_3Pillars_TN900100.pdf, (accessed July 16, 2023).
- M. C. Chen and G. R. Mcinroy, WIPO Pat., WO2020178603A1, 2020 Search PubMed.
- M. C. Chen, R. A. Lazar and J. Huang, US Pat., US20180201968A1, 2018 Search PubMed.
- K. Hoff, M. Halpain, G. Garbagnati, J. S. Edwards and W. Zhou, ACS Synth. Biol., 2020, 9, 283–293 CrossRef CAS PubMed.
- J. W. Efcavitch and J. L. Tubbs, WIPO Pat., WO2016064880A1, 2016 Search PubMed.
- A. G. Fraser, S. Mankowska and N. Bell, WIPO Pat., WO2020150143, 2020 Search PubMed.
- M. Flamme, S. Hanlon, I. Marzuoli, K. Püntener, F. Sladojevich and M. Hollenstein, Commun. Chem., 2022, 5, 68 CrossRef CAS PubMed.
- D. Entwistle, Revolutionizing Nucleic Acid Synthesis with Engineered Enzymes, https://d1io3yog0oux5.cloudfront.net/_7aaee8ad8f1cff46ac1b18e728d64cf1/codexis/db/1165/11842/pdf/CDXS+TIDES+EU+Presentation+November+2023.pdf, (accessed Feb 07, 2024).
- A. S. Mathews, H. Yang and C. Montemagno, Org. Biomol. Chem., 2016, 14, 8278–8288 RSC.
- L. Takeshita, Y. Yamada, Y. Masaki and K. Seio, J. Org. Chem., 2020, 85, 1861–1870 CrossRef CAS PubMed.
- M. L. Metzker, R. Raghavachari, S. Richards, S. E. Jacutin, A. Civitello, K. Burgess and R. A. Gibbs, Nucleic Acids Res., 1994, 22, 4259–4267 CrossRef CAS PubMed.
- M. Flamme, S. Hanlon, H. Iding, K. Puentener, F. Sladojevich and M. Hollenstein, Bioorg. Med. Chem. Lett., 2021, 48, 128242 CrossRef CAS PubMed.
- J. W. Efcavitch and J. L. Tubbs, WIPO Pat., WO2022063835A1, 2022 Search PubMed.
- J. W. Efcavitch and J. L. Tubbs, WIPO Pat., WO2018175436A1, 2018 Search PubMed.
- S. Agarwalla, WIPO Pat., WO2021207158A1, 2021 Search PubMed.
- Molecular assemblies Inc, Molecular Assemblies Announces First Storage and Retrieval of Information with Enzymatic DNA Synthesis, https://molecularassemblies.com/press-releases/molecular-assemblies-announces-first-storage-and-retrieval-of-information-with-enzymatic-dna-synthesis/, (accessed July 16, 2023).
- Molecular assemblies Inc., The First Fully Enzymatic Synthesis™ Technology for Synthetic DNA, https://molecularassemblies.com/technology/, (accessed July 16, 2023).
- Molecular assemblies Inc., Codexis and Molecular Assemblies Announce Results of First Collaboration on a Proprietary High Performing DNA Polymerase to Supercharge Fully Enzymatic DNA Synthesis, https://molecularassemblies.com/press-releases/codexis-and-molecular-assemblies-announce-results-of-first-collaboration-on-a-proprietary-high-performing-dna-polymerase-to-supercharge-fully-enzymatic-dna-synthesis/, (accessed July 16, 2023).
- Molecular assemblies Inc, Molecular Assemblies Ships First Enzymatically Synthesized Oligonucleotides, https://molecularassemblies.com/press-releases/molecular-assemblies-ships-first-enzymatically-synthesized-oligonucleotides/, (accessed July 16, 2023).
- D. L. H. Williams, Nitrosation reactions & the chemistry of nitric oxide, 2004 Search PubMed.
- D. Arlow and S. Palluk, US Pat., US20220251617A1, 2022 Search PubMed.
- Ansa Biotechnoloies Inc., Ansa Biotechnologies Announces Successful de novo Synthesis of World's Longest Oligonucleotide at 1005 Bases, https://ansabio.com/news, (accessed July 16, 2023).
- E. Hultin, M. Kaller, A. Ahmadian and J. Lundeberg, Nucleic Acids Res., 2005, 33, e48 CrossRef PubMed.
- S. A. Mankowska and S. A. Harvey, WIPO Pat., WO2018152323, 2018 Search PubMed.
- H. Lee, D. J. Wiegand, K. Griswold, S. Punthambaker, H. Chun, R. E. Kohman and G. M. Church, Nat. Commun., 2020, 11, 5246 CrossRef CAS PubMed.
- J. A. Smith, B. H. Nguyen, R. Carlson, J. G. Bertram, S. Palluk, D. H. Arlow and K. Strauss, ACS Synth. Biol., 2023, 12, 1716–1726 CrossRef CAS PubMed.
- H. S. Jung, W. B. Jung, J. Wang, J. Abbott, A. Horgan, M. Fournier, H. Hinton, Y. H. Hwang, X. Godron, R. Nicol, H. Park and D. Ham, Sci. Adv., 2022, 8, eabm6815 CrossRef PubMed.
- M. Escalona, S. Rocha and D. Posada, Nat. Rev. Genet., 2016, 17, 459–469 CrossRef CAS PubMed.
- E. R. Mardis, Nature, 2011, 470, 198–203 CrossRef CAS PubMed.
- S. Goodwin, J. D. McPherson and W. R. McCombie, Nat. Rev. Genet., 2016, 17, 333–351 CrossRef CAS PubMed.
- M. Jain, I. T. Fiddes, K. H. Miga, H. E. Olsen, B. Paten and M. Akeson, Nat. Methods, 2015, 12, 351–356 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |