Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhancing protein stability under stress: osmolyte-based deep eutectic solvents as a biocompatible and robust stabilizing medium for lysozyme under heat and cold shock†
Anja
Damjanović‡
a,
Marijan
Logarušić‡
 a,
Lidija-Marija
Tumir
a,
Lidija-Marija
Tumir
 b,
Thanos
Andreou
b,
Thanos
Andreou
 c,
Marina
Cvjetko Bubalo
c,
Marina
Cvjetko Bubalo
 *a and
Ivana
Radojčić Redovniković
a
*a and
Ivana
Radojčić Redovniković
a
aFaculty of Food Technology and Biotechnology, University of Zagreb, Croatia. E-mail: mcvjetko@pbf.hr
bRuđer Bošković Institute, Zagreb, Croatia
cVIO Chemicals AG, Thessaloniki, Greece
First published on 15th July 2024
Abstract
In biomedical and biotechnological domains, liquid protein formulations are vital tools, offering versatility across various fields. However, maintaining protein stability in a liquid form presents challenges due to environmental factors, driving research to refine formulations for broader applications. In our recent study, we investigated the relationship between deep eutectic solvents (DESs) and the natural presence of osmolytes in specific combinations, showcasing the effectiveness of a bioinspired osmolyte-based DES in stabilizing a model protein. Recognizing the need for a more nuanced understanding of osmolyte-based DES stabilization capabilities under different storage conditions, here we broadened the scope of our osmolyte-based DES experimental screening, and delved deeper into structural changes in the enzyme under these conditions. We subjected lysozyme solutions in DESs based on various kosmotropic osmolytes (TMAO, betaine, sarcosine, DMSP, ectoine, GPC, proline, sorbitol and taurine) paired either with another kosmotropic (glycerol) or with chaotropic osmolyte urea to rigorous conditions: heat shock (at 80 °C) and repetitive freeze–thaw cycles (at −20 and −80 °C). Changes in enzyme activity, colloidal stability, and conformational alterations were then monitored using bioassays, aggregation tests, and spectroscopic techniques (FT-IR and CD). Our results demonstrate the remarkable effectiveness of osmolyte-based DES in stabilizing lysozyme under stress conditions, with sarcosine- and betaine-based DESs containing glycerol as a hydrogen bond donor showing the highest efficacy, even at high enzyme loadings up to 200 mg ml−1. Investigation of the individual and combined effects of the DES components on enzyme stability confirmed the synergistic behavior of the kosmotrope–urea mixtures and the cumulative effects in kosmotrope–glycerol mixtures. Additionally, we have shown that the interplay between the enzyme's active and stable (but inactive) states is highly influenced by the water content in DESs. Finally, toxicity assessments of osmolyte-based DESs using cell lines (Caco-2, HaCaT, and HeLa) revealed no risks to human health.
Introduction
Within biomedical and biotechnological domains, liquid protein formulations have emerged as invaluable tools due to their inherent versatility and practical advantages. They play pivotal roles across diverse fields, spanning from the development of biopharmaceuticals and protein-based drugs to biotechnological processes like enzyme production, diagnostic assays, and biocatalysis, which all rely on stable proteins throughout all stages of manufacturing (expression, purification, and formulation), shelf-life management, and product administration/application.1–4 One notable characteristic of liquid protein formulations is their capacity to ensure superior sample homogeneity, as proteins in solution are uniformly distributed, thus guaranteeing consistent sample quality. Additionally, the liquid form facilitates seamless mixing, dilution, and aliquoting of proteins, eliminating the need for cumbersome reconstitution steps and enhancing operational efficiency.2However, for proteins in a liquid form, the delicate balance of their structure can be easily disrupted by environmental factors such as changes in temperature and pH, as well as mechanical stress. These disruptions often lead to denaturation, inactivation, and aggregation of proteins, resulting in a loss of functionality and product quality.5,6 To evaluate protein formulation stability, heat- and freezing-induced protein denaturation has emerged as rapid and reliable methods. These processes disrupt the native structure of proteins when exposed to extreme temperatures, allowing researchers to quickly observe changes in protein stability and gain insights into potential degradation pathways. Additionally, this type of denaturation simulates conditions that proteins may encounter during storage or processing, enhancing its relevance for evaluating formulation stability.1,7,8
To aid in the folding or refolding of proteins in liquid protein formulations, especially under stressful conditions, chemical chaperones are frequently used.9,10 Naturally occurring osmolytes, including methylamines, sugars, alcohols, and amino acids, have been demonstrated as excellent chaperons of proteins in numerous cases.11 When incorporated into formulations, these molecules mimic their natural role in vivo, which is to protect biological systems (such as extremophilic bacteria, marine organisms, sporulating microorganisms, and plants) from stressful environmental conditions by providing thermodynamic stability to biomacromolecules, particularly proteins, without compromising their natural functionality.12 Another relatively new class of chemical chaperones that also effectively mimic the natural environment of biomolecules to stabilize them are non-toxic and highly versatile systems known as deep eutectic solvents (DESs).13,14 Originally, the term DES was coined to describe a physical mixture of two or more components, usually from natural sources, that solidifies at a single temperature lower than the crystallization point of any individual component.15 With time, these solvents/systems have evolved to include mixtures of two or more components that demonstrate properties similar to a eutectic system,16 with one fixed criterion: remaining in a liquid state at a specified temperature, even if one of its components would normally be solid and unsuitable for use as a solvent.17 The chaperon-like activity of DESs for various proteins, such as lysozyme,18–20 lipases,21,22 collagen peptide,23 α-chymotrypsin,24 laccases,25,26 bovine serum albumin,27 various peroxidases,28 cellulases,29 human interferon,30 β-galactosidase,31 immunoglobulin G,32 and ubiquitin,33 has been reported so far.
In our recent study,34 for the first time, a connection was established between two previously parallel approaches to protein stabilization: osmolytes- and DES-assisted protein stabilization. Intrigued by the structural similarity between osmolytes and common DES components, as well as the fact that osmolytes are typically present in cells and tissues in certain combinations and molar ratios, we investigated a set of natural osmolytes and patterns of their natural distribution, preparing new bioinspired two-, three-, and multi-component DESs based on osmolytes. The newly prepared bioinspired solvents were further assessed as a protein stabilization medium at room temperature. The results indicate superior stabilization compared to conventional DESs and the standard buffer used for protein storage. Building upon that research, in this study we subjected lysozyme solution in various DESs to rigorous conditions, including heat shock at 80 °C and multiple freeze–thaw cycles at −20 °C and −80 °C. The stabilizing effect of DESs was examined using bioassays and spectroscopic methods. Finally, as osmolytes accumulate in times of stress, the results are discussed within the framework of osmolyte-based presence in vivo, suggesting a universal mechanism of action used by living systems during periods of stress.
Experimental part
Materials
Lysozyme extracted from hen egg white and Micrococcus lysodeikticus (ATCC No. 4698) was purchased from Sigma-Aldrich (St. Louis, MI, USA). All components of the DESs were also procured from Sigma-Aldrich. All chemicals were of at least 99% purity and were used without additional purification.DES preparation and characterization
DESs were prepared in the way that two or more components in specific ratios with a certain amount of water (20, 40, 60 or 80% of water (w/w)) were placed in a glass flask, stirred, and heated at 50 °C until a clear homogeneous liquid was formed (Table 1).| DES | Abbreviationsa | Molar ratio | pH (20 °C) | E NR [kcal mol−1] | ρ (20 °C) [g cm−3] | EC50 (mg ml−1) | ||
|---|---|---|---|---|---|---|---|---|
| HaCaT | Caco-2 | HeLa | ||||||
| a Choline–chloride (ChCl), betaine (Bet), sarcosine (Sar), ectoine (Ect), trimethylamine N-oxide (TMAO), proline (Pro), dimethylsulfoniopropionate (DMSP), urea (U), glycerol (Gly), sorbitol (Sor), taurine (Tau), glycerophosphocholine (GPC). | ||||||||
| Choline-based DES | ChCl:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
7.7 | 48.98 | 1.13 | ND | ND | ND |
| ChCl:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.2 | 49.60 | 1.14 | ND | ND | ND | |
| Betaine-based DES | Bet:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
8.1 | 49.96 | 1.14 | ND | ND | ND |
| Bet:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.0 | 49.72 | 1.15 | ND | ND | ND | |
| Sarcosine-based DES | Sar:U | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
5.6 | 49.75 | 1.19 | >1000 | >1000 | >1000 |
| Sar:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
5.2 | 49.48 | 0.88 | >1000 | >1000 | >1000 | |
| Ectoine-based DES | Ect:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.8 | 48.92 | 1.20 | >1000 | >1000 | >1000 |
| Ect:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.3 | 49.78 | 1.15 | >1000 | >1000 | >1000 | |
| TMAO-based DES | TMAO:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
9.6 | 49.79 | 1.09 | >1000 | >1000 | >1000 |
| TMAO:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
9.1 | 50.43 | 1.14 | >1000 | >1000 | >1000 | |
| Proline-based DES | Pro:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
7.2 | 49.81 | 1.16 | >1000 | >1000 | >1000 |
| Pro:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
6.2 | 49.93 | 1.18 | >1000 | >1000 | >1000 | |
| DMSP-based DES | DMSP:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
5.6 | 48.67 | 1.19 | >1000 | >1000 | >1000 |
| DMSP:Gly | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1.0 | 48.54 | 1.18 | 310.2 | 335.5 | 380.9 | |
| Bioinspired multicomponent DES | Bet:Sor:Tau:GPC:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.1 3.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.8 2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.1 7.1 |
7.0 | 49.24 | 1.18 | >1000 | >1000 | >1000 |
| TMAO:Bet:Tau:U | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6:0.5 6:0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 15 |
6.8 | 49.34 | 1.16 | >1000 | >1000 | >1000 | |
Upon cooling to room temperature, the mixture was left on a bench for a week to observe possible solidification or precipitation. Before use, DES forming compounds choline chloride, TMAO and sarcosine were dried in a vacuum drier (Memmert GmbH + Co. KG) at 40 °C and 100 mbar for 24 h. The pH of all DESs was measured at room temperature using a pH electrode (InLab® Micro Pro-ISM pH electrode, Mettler-Toledo). The density at room temperature was measured by using a pycnometer (V = 1 ml). To determine the polarity and molar transition energy (ENR) of the DESs Nile red was used as a solvatochromatic probe.35
Extent of lysozyme aggregation
Heat-induced aggregation of lysozyme solution (5 mg ml−1) in DESs and the referent solvent (50 mM potassium phosphate buffer, pH 6.4) was studied using a temperature-controlled UV/vis spectrophotometer (Lambda 35, PerkinElmer). A sample was placed in a quartz cuvette and covered with mineral oil to retard evaporation. Turbidity changes (λ = 600 nm) were monitored at a temperature of 80 °C.Lysozyme activity assay
Lysozyme activity was determined according to the method of Shugar et al.36 Briefly, 30 μl of Micrococcus lysodeikticus bacteria suspension in sterile PBS buffer (7 mg ml−1) and 30 μl of the lysozyme solution were added to 525 μl of 50 mM potassium phosphate buffer solution (pH 6.4) in a cuvette. Immediately after mixing, the cuvette was placed in a UV/vis spectrophotometer (Thermo Fisher Scientific, GenesysTM10S) and the absorbance was measured at a wavelength of 450 nm over a period of linear turbidity decline. Relative activity was calculated as a ratio of activity measured in the DES and the activity measured in the reference buffer.Residual activity of lysozyme after heat shock and freeze–thaw conditions
Lysozyme solutions at a concentration of 5 mg ml−1 (0.1 mg ml−1 for preliminary screening on the influence of the water content on DES ability to stabilize lysozyme; 50, 100 and 200 mg ml−1 for experiments related to the stability of highly concentrated lysozyme solutions) were prepared in different DES and in 50 mM potassium phosphate buffer solution (pH 6.4). Enzyme solutions were subjected to different storage conditions: (i) incubation for 1 hour at 80 °C; and (ii) freeze–thaw conditions: 24-hour freeze–thaw cycle at −20 °C or −80 °C, total of 5 cycles.Residual lysozyme activity after each storage condition was determined according to the method of Shugar et al.36 described above. In brief, an aliquot of lysozyme solution in DES was withdrawn, and the residual activity was measured. The residual activity (%) was calculated from the initial reaction rate obtained by the enzyme after incubation, compared to the one obtained without previous exposure.
Circular dichroism (CD) measurements
CD spectra of lysozyme dissolved in 50 mM potassium phosphate buffer solution (pH 6.4) or relevant DESs (final protein concentration of 0.2–0.75 mg ml−1) were recorded on a JASCO J815 spectrophotometer (Jasco Coorp., Tokyo, Japan), with a Peltier cell holder for temperature control.CD spectra in the near UV region (250–350 nm) were obtained at a fixed temperature (20 °C, 80 °C or 95 °C) with a scan rate of 200 nm min−1, a bandwidth of 1 nm and a response time of 1 s. Near UV CD spectra were recorded using appropriate 1 cm path quartz cuvettes, except for Ect:Gly solution (path of 0.5 cm). Concentrations of lysozyme were 0.75 mg ml−1 (52 μM). Each spectrum was accumulated for 2 scans. Baselines taken with DES or buffer under the same conditions were subtracted from each spectrum.
CD spectra in the far UV region (190–250 nm) were measured at room temperature in the wavelength range of 200–250 nm with a scan rate of 200 nm min−1, bandwidth of 1 nm and a response time of 1 s. Due to the high absorbance of DES,37,38 far UV CD spectra were recorded using appropriate 0.1 mm path quartz glasses to avoid HT voltage above 600 V. The concentration of lysozyme was 0.2 mg ml−1 (15 μM). The study of far-UV spectra of lysozyme after storage in the DES was performed by hydration:39 diluting of DES with buffer, to achieve a final DES content of 0.5% (w/w). The spectra after hydration were recorded using 1 cm path quartz cuvettes.
Thermal CD-scans were collected at a fixed wavelength (227 ± 5 nm) in a temperature range 20 °C to 95 °C in appropriate 1 mm path quartz cuvettes at a heating rate of 1 °C per minute. The concentration of lysozyme was 0.3–0.4 mg ml−1 (20–26 μM). Ellipticities ([θ]) were expressed in observed mdeg and in units of deg cm2 dmol−1, using the protein concentration.
Fourier transform infrared spectroscopy (FT-IR)
FT-IR spectra of the mixtures, spectra of the pure starting compounds, and spectra of lysozyme solutions in DESs (20 mg ml−1) and 50 mM potassium phosphate buffer solution (pH 6.4) were recorded on an FT-IR spectrometer equipped with an attenuated total reflection module (Tensor II, Bruker, Ettlingen, Germany) with diamond ATR crystal from 4000–400 cm−1, with a resolution of 4 cm−1. The spectra are the average of 16 scans. For measurements of protein solutions (20 mg ml−1), the samples were left to equilibrate for 1 h prior to measurements, with all measurements being made in the next 1 h. FT-IR spectra of each solvent, without the protein present, were acquired under the same conditions and used for solvent subtraction.Cytotoxicity assay
The cytotoxicity of DESs was tested against three human adherent cell lines: cancer cells derived from the colorectal adenocarcinoma (Caco-2 – ATCC No. HTB-37™), normal human keratinocyte cells (HaCaT – CVCL No. 0038), and epithelial cells from cervical carcinoma (HeLa – ATCC No. CCL-2™). Cells were maintained in DMEM – Dulbecco's modified Eagle's medium (Capricorn Scientific GmbH), supplemented with 5% (v/v) FBS – fetal bovine serum (GIBCO by Life Technologies) in an incubator with 5% CO2 and a humidified atmosphere at 37 °C. The impact of synthesized DESs on cellular proliferation was investigated in vitro utilizing the CellTiter96® AQueous One Solution Cell Proliferation assay (Promega), also known as the MTS assay. Briefly, cells were seeded in 96-well plates at a density of 3 × 104 cells per well in 100 μl of media. Following overnight incubation, cells were exposed to the prepared DESs at three concentrations spanning from 100 to 500, and ultimately 1000 mg ml−1, and then incubated for 72 hours. A 10 μl volume of MTS reagent was added to each well and the absorbance was measured at 492 nm on the microplate reader after the 3-hour incubation period. Cell viability percentage was calculated by comparing the absorbance of treated cells to that of untreated control cells. The experiments were conducted in triplicate with four replicates for each concentration. The corresponding EC50 values, defined as the concentration of tested compounds leading to 50% growth inhibition, were calculated from the dose–response curves.Results and discussion
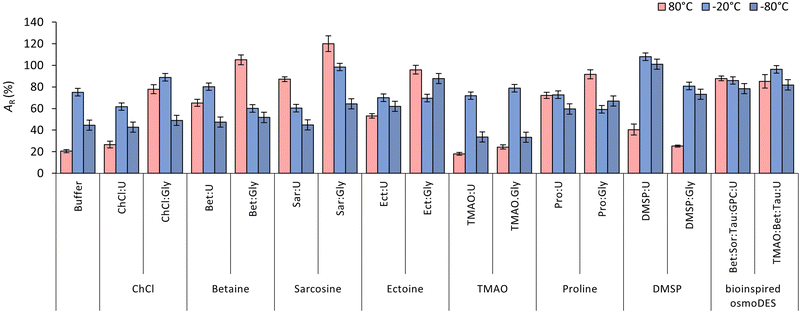
In our previous work, we showcased the ability of novel osmolyte-based DESs based on kosmotropes sarcosine, ectoine and DMSP (all HBAs), when paired with glycerol as a HBD, to stabilize a model protein (lysozyme) at 25 °C to a greater extent than the conventional choline chloride-based DES (ChCl:Gly) and the reference buffer.34 This was also confirmed for bioinspired multicomponent DES replicated from an osmolyte cocktail in mammalian kidneys comprising chaotrope urea. To gain a deeper understanding of novel osmolyte-based DESs’ capacity to stabilize lysozyme, this study expanded both the range of DESs utilized and the incubation conditions applied to enzyme solutions in DESs. Lysozyme was once again chosen for its cost-effectiveness, the accessibility of rapid and reliable spectrophotometric methods for activity measurement, and the wealth of information available on its behaviour under various conditions in vitro.40 In this research, we first prepared and characterized various osmolyte-based DESs in terms of their physicochemical properties and cytotoxicity (Table 1). Following this, lysozyme solutions in DESs were subjected to stressful conditions, including heat shock or repeated freezing–thawing cycles (Fig. 1), whereby the stabilizing effect of DESs was examined using bioassays and spectroscopic methods.Osmolyte-based DESs preparation and characterization
Versatile DESs were prepared for analysis by combining a range of components, such as kosmotropic osmolytes (trimethylamine-N-oxide (TMAO), betaine, sarcosine, dimethylsulfoniopropionate (DMSP), ectoine, glycerophosphocholine (GPC), proline, glycerol, sorbitol, and taurine) and chaotrope urea (Table 1). It is important to note that the osmolytes utilized in this study span across all kingdoms of life, ranging from deep-sea fish (TMAO, sarcosine, and urea), plants (betaine), fungi (ectoine and glycerol), algae (DMSP), to mammals (GPC and urea). DESs based on choline chloride and the osmolyte betaine, which have already been established as effective lysozyme stabilizers41–44 were prepared and tested as well.Preliminary experiments investigating the impact of water content on the solvent's ability to stabilize lysozyme at 80 °C, conducted on a limited selection of DESs (ChCl:Gly, Sar:Gly and Ect:Gly), revealed that lysozyme was either more effectively or equally stabilized in DESs containing 40% water (w/w) compared to those with 20% water (with a difference of ±10%; data not shown). A water content of 40% was also considered beneficial from a practical standpoint: water helps reduce the solvent's viscosity45 making it easier to handle and potentially suitable for larger-scale applications.46 At this point, dilutions of DESs beyond 40% were not considered, as additional water would result in a solvent behaving more like a solution of its components in water.47
In accordance with the above, a total of fourteen two-component osmolyte-based DESs and two bioinspired multicomponent DESs, all containing 40% water (w/w), were prepared and characterized (Table 1). The pH value of the prepared DESs ranged widely: from 1.0 (DMSP:Gly) to 9.8 (TMAO:U). In general, the most acidic DESs were those with DMSP (pH 1.0 and 5.6) and sarcosine (pH 5.2 and 5.6) as HBA, while TMAO-based DESs were shown to be basic (pH 9.1 and 9.6). This is not surprising, as it is well-established that the acidity/basicity of a DES originates from its starting components, which in this case significantly differ from one another (e.g., the pKa values of sarcosine, ectoine, TMAO, glycerol, and urea are 2.2, 3.1, 7.7, 13.5, and 13.9 respectively). Furthermore, DMSP was utilized in its HCl salt form, and its acidity predominantly originates from the protonation of the sulfonate group (–SO3H). All the tested DES were more polar than ethanol (ENR < 52.17 kcal mol−1) and less polar (or of similar polarity) than water (ENR < 48.20 kcal mol−1).48 The measured densities of the DESs were in the range from 0.9 and 1.2 g cm−3, with the majority falling in the 1.1–1.2 g cm−3 range at room temperature. Finally, we utilized FT-IR analysis for the chemical characterization of the prepared DESs, aiming to confirm the presence of strong interactions within the components. As anticipated, the FT-IR spectra of pure methylamines (betaine, TMAO and sarcosine) and amino acids (ectoine and proline) showed characteristic bands: the NH bond stretching (ν(NH)) between 3300–3500 cm−1 and the carbonyl bond stretching (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)) between 1600–1800 cm−1 (Fig. S1–S2, ESI†). In the DESs spectra, we observed signals reminiscent of the starting materials, but with distinct features suggesting the formation of mixtures where hydrogen bonding predominates. Specifically, when methylamines or amino acids were paired with HBD (glycerol and urea) we noticed a broad, strong peak spanning from 3650 to 3000 cm−1, indicating the formation of robust NH–O
O)) between 1600–1800 cm−1 (Fig. S1–S2, ESI†). In the DESs spectra, we observed signals reminiscent of the starting materials, but with distinct features suggesting the formation of mixtures where hydrogen bonding predominates. Specifically, when methylamines or amino acids were paired with HBD (glycerol and urea) we noticed a broad, strong peak spanning from 3650 to 3000 cm−1, indicating the formation of robust NH–O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C hydrogen bonds between the components. Additionally, a shift towards higher wavenumbers in the carbonyl stretching (ν(C
C hydrogen bonds between the components. Additionally, a shift towards higher wavenumbers in the carbonyl stretching (ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)) band in the DESs spectra further supported the occurrence of hydrogen bonding.49
O)) band in the DESs spectra further supported the occurrence of hydrogen bonding.49
The heat shock: quantitative monitoring of lysozyme colloidal and conformational stability in osmolyte-based DESs
As can be seen in Fig. S3 (ESI†), in the reference buffer, a rapid increase in optical intensity signifies aggregation of the enzyme. Conversely, in DESs tested (including reference cholinium- and betaine-based DESs) no time-dependent aggregation of lysozyme was observed, except for TMAO:U (only data for buffer, TMAO:U, and a bioinspired multicomponent DES are shown). This chaperone-like activity of conventional cholinium-based DESs containing urea and polyols has already been observed for lysozyme18 and immunoglobulin G.32 Remarkably, even in DMSP:Gly with a pH value as low as 1.0, no aggregation was observed. This is noteworthy considering that, according to the literature, at temperatures of 80 °C aggregation in lysozyme solutions typically initiates immediately at pH levels below 4.0.52
This simple and rapid aggregation test gave us valuable hints on osmolyte-based DESs’ potential to act as chemical chaperons. Furthermore, the recovery of enzymatic activity after heat treatment was measured upon dilution in the buffer.34 It is crucial to emphasize herein that in all bioactivity experiments, lysozyme solutions were incubated in DESs, and aliquots were subsequently withdrawn to measure the residual enzyme activity in the buffer.39 This procedure was applied both at the initial time point (“zero”), where each enzyme solution in DES served as its own reference for subsequent activity measurements, and after treatments such as heat or cold shock. Thus, any potential effects of osmolytes on enzyme activity were minimized using this approach. Furthermore, enzyme activities measured immediately after dissolving lysozyme in DES (initial lysozyme activity in DES) were comparable to the initial activity of lysozyme in the reference sample. This suggests that the concentrations of osmolytes present during the enzyme activity measurements had no significant effect.
Fig. 1 shows the residual activity (AR) of lysozyme after the heat treatment. In the reference buffer, AR decreased to approximately 20%, a value like the one observed for TMAO-based DESs. However, the remaining DESs exhibited greater enzyme stabilization. This effect was particularly notable in three DESs variants, all incorporating glycerol as the HBD: Bet:Gly, Sar:Gly, and Ect:Gly, where enzyme activity was fully preserved (AR ≥ 96%). Here, we have demonstrated that Sar:Gly (AR = 120%), functioning as a DES with superior lysozyme stabilization ability, significantly surpasses ChCl:Gly (AR = 78%), which has previously been indicated to stabilize lysozyme during thermal treatment at 80 °C.44 The observation that glycerol-based DESs function as excellent chemical chaperones aligns with previous studies by Delorme et al.25 and Toledo et al.53 that highlighted a positive relationship between higher counts of OH-groups in the HBD and improved thermostability through hydrogen bond formation between the OH-groups on the HBD and the enzyme's amino acids. In glycerol-based DESs the same authors also observed an increase in AR values above 100%. To adequately explain this phenomenon, a more in-depth analysis is necessary. Although glycerol-based DESs herein demonstrated a higher stabilization ability compared to their urea-based counterparts, the latter still exhibited improved stabilization of lysozyme compared to the reference system (except for TMAO-based DES).
Additionally, even though strongly acidic DES, specifically DMSP:Gly, demonstrated lower lysozyme stabilization potential compared to other glycerol-based DES, it unexpectedly exhibited a comparable stabilization ability to the reference buffer (AR ∼ 25%). This occurrence might be attributed to the abundance of HBA/HBD-acting groups, such as hydroxyl and carboxyl, which contribute to buffering the system through the formation of dense hydrogen bonds.54 From the thermostability results, it appears that the pH value of a DES is not a critical factor in its ability to stabilize lysozyme. For example, despite having similar pH values, Sar:Gly, Sar:U and DMSP:U (ranging from 5.2 to 5.6), exhibited significantly different levels of lysozyme residual activity after heat shock (120%, 87%, and 40%, respectively). This observation is intriguing and suggests that other mechanisms may be at play, such as direct interactions between DES components and proteins, or interactions with water molecules in close proximity to the protein, thereby altering the water activity of the medium.
Finally, two bioinspired multicomponent DESs, both based on urea as an HBD, also showed excellent chaperon-like activity with AR > 85%. Similar to what was observed in our previous work,34 two-component DESs consisting of betaine and TMAO with urea as the HBD demonstrated a lesser ability to stabilize lysozyme compared to their multicomponent bioinspired DES counterpart. This suggests once again that including additional osmolytes in the cocktail, in molar ratios replicated from a natural context, significantly enhances the DES's ability to stabilize lysozyme.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.34 It has been proposed that such kosmotrope counterbalancing is achieved through the attraction of urea molecules, preventing their interaction with the protein surface, or by hindering the mobility of urea molecules through hydrogen bonding with kosmotrope, thereby restricting their access to protein domains.56,57
2.34 It has been proposed that such kosmotrope counterbalancing is achieved through the attraction of urea molecules, preventing their interaction with the protein surface, or by hindering the mobility of urea molecules through hydrogen bonding with kosmotrope, thereby restricting their access to protein domains.56,57
In DESs where glycerol acted as the HBD, this kind of synergistic effect was only observed for Sar:Gly. Specifically, lysozyme retained its activity completely in the glycerol–water mixture (AR ∼ 100%), whereas in Sar:Gly, the enzyme was “overactivated” with an AR = 120%. However, in the other DESs, a cumulative effect was noted: all glycerol-based DESs exhibited superior enzyme stabilization capability compared to their HBA counterparts.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg ml−1). This knowledge gap hinders potential applications of DESs in protein stabilization, as storing or formulating proteins in such dilute conditions would demand impractical volumes of solvent.39 Namely, the primary hurdle in formulating protein solutions at elevated concentrations lies in navigating the intricate degradation pathway driven by concentration-dependent aggregation.58 To the best of our knowledge, the efficacy of DESs, specifically ChCl:Gly, in maintaining the physical integrity of lysozyme at high protein concentrations up to 143 mg ml−1 has been demonstrated only in the study by Sanchez-Fernandez et al.39 Here, we evaluated the effectiveness of the three most promising DES candidates identified during the initial screening, Sar:Gly, Bet:Gly, and bioinspired multicomponent DES Bet:Sor:Tau:GPC:U, in stabilizing lysozyme across a broad concentration range, specifically from 5 to 200 mg ml−1, while ensuring the protein remained soluble. As shown in Fig. 2B, in the bioinspired multicomponent DES there is a significant decrease in AR value after heat shock, correlating with the enzyme loading (AR = 45% at a loading of 200 mg ml−1). Additionally, above a lysozyme concentration of 5 mg ml−1 protein aggregates were visibly formed in this DES after the heat shock. However, in Sar:Gly and Bet:Gly, lysozyme activity was fully preserved at loadings up to 50 mg ml−1. At loadings of 100 and 200 mg ml−1, a slight reduction in activity was observed in both DESs, but still with excellent recovery (AR > 75%) and no observable aggregation.
mg ml−1). This knowledge gap hinders potential applications of DESs in protein stabilization, as storing or formulating proteins in such dilute conditions would demand impractical volumes of solvent.39 Namely, the primary hurdle in formulating protein solutions at elevated concentrations lies in navigating the intricate degradation pathway driven by concentration-dependent aggregation.58 To the best of our knowledge, the efficacy of DESs, specifically ChCl:Gly, in maintaining the physical integrity of lysozyme at high protein concentrations up to 143 mg ml−1 has been demonstrated only in the study by Sanchez-Fernandez et al.39 Here, we evaluated the effectiveness of the three most promising DES candidates identified during the initial screening, Sar:Gly, Bet:Gly, and bioinspired multicomponent DES Bet:Sor:Tau:GPC:U, in stabilizing lysozyme across a broad concentration range, specifically from 5 to 200 mg ml−1, while ensuring the protein remained soluble. As shown in Fig. 2B, in the bioinspired multicomponent DES there is a significant decrease in AR value after heat shock, correlating with the enzyme loading (AR = 45% at a loading of 200 mg ml−1). Additionally, above a lysozyme concentration of 5 mg ml−1 protein aggregates were visibly formed in this DES after the heat shock. However, in Sar:Gly and Bet:Gly, lysozyme activity was fully preserved at loadings up to 50 mg ml−1. At loadings of 100 and 200 mg ml−1, a slight reduction in activity was observed in both DESs, but still with excellent recovery (AR > 75%) and no observable aggregation.
First, both buffered and DES solutions of lysozyme were monitored at a single wavelength in the far UV region. Upon heating, the negative CD signal intensity decreased. The melting curve displayed a sigmoidal shape, indicating a cooperative, two-state denaturation process for lysozyme in the buffered aqueous solution (Fig. 3). The transition temperature (thermal melting, Tm), which signifies the temperature at which the protein undergoes structural transition or denaturation, resulting in the unfolding of its secondary and tertiary structure, was determined to be 75.9 °C. A similar cooperative shape was observed for lysozyme solutions in DESs. All DESs tested (Bet:Gly, Sar:Gly, Pro:Gly, and bioinspired multicomponent DES Bet:Sor:Tau:GPC:U) exhibited a strong stabilizing influence on the protein's secondary structure. The Tm value of lysozyme in Pro:Gly exceeded that in the buffered aqueous solution by 5 °C, and by more than 10 °C in Bet:Gly, Sar:Gly, and Bet:Sor:Tau:GPC:U. The highest Tm value of 91.5 °C was observed in Bet:Sor:Tau:GPC:U (Fig. S4 and Table S1, ESI†).
While the far UV CD spectral region reflects changes in secondary structure, the near UV CD region (250–350 nm) serves as a fingerprint of the protein's tertiary structure. The near UV CD spectrum is influenced by aromatic amino acids and the rigidity of their surroundings, including intramolecular interactions such as hydrogen bonding.38 Thus, following the CD research conducted by Esquembre et al.,44 we recorded the near CD spectra of the enzyme's solutions during thermal treatment. This allowed us to determine whether renaturation of lysozyme after heat shock occurs in DESs upon cooling, or later upon dilution in buffer. At room temperature, the near CD spectra of lysozyme in the buffer and DES exhibited triplet-like signals attributed to tyrosine, tryptophan, and disulfide residues,38,62 indicating a comparable tertiary structure of lysozyme in these solvents (Fig. 4 and Fig. S5, ESI†). The heating of the solution led to a featureless spectrum in all tested solutions, attributable to the unfolding of the protein and the exposure of the aromatic side chains of amino acids to the isotropic environment.41,43,44 It is noteworthy that in DES solutions the tertiary structure was partially maintained even at 80 °C, with complete protein unfolding occurring only at 95 °C. Overall, CD studies confirmed that in osmolyte-based DESs with glycerol as the HBD and in multicomponent bioinspired DES lysozyme shows excellent thermostability and is able to entirely regain its folded structure after cooling to room temperature.
The cold shock: quantitative monitoring of lysozyme stability in osmolyte-based DESs
To assess whether osmolyte-based DESs protect proteins during freeze–thaw cycles in a similar way, lysozyme solutions (5 mg ml−1) underwent five consecutive cycles of freezing (−80 °C and −20 °C) and thawing at room temperature. The residual enzyme activity (AR) was measured after each cycle, and the results are presented in Fig. 1 and Fig. S6 and S7 (ESI†).Overall, a decrease in enzyme activity was observed in all DESs tested across consecutive freeze–thaw cycles at both storage temperatures, except for Sar:Gly and DMSP:U at −80 °C, and DMSP:U at both temperatures. Notably, DMSP:U exhibited the best ability to stabilize lysozyme under cold shock, with AR = 100% after five freeze–thaw cycles at both storage temperatures. In comparison, lysozyme in the reference buffer exhibited a reduction in AR to 45% and 75% at −80 °C and −20 °C, respectively. Moreover, osmolyte-based DESs again demonstrated chaperon-like activity superior to conventional choline chloride-based DESs. In heat-induced stress experiments, glycerol emerged as the most effective HBD among those tested. However, in scenarios involving cold-induced stress, DESs based on urea have proven to be equally effective candidates for enzyme stabilization as glycerol-based ones. The observation that DESs containing glycerol are effective in cryoprotecting proteins comes as no surprise, considering glycerol's well-established reputation as a protein cryoprotectant.63 However, since urea has previously been demonstrated to promote enzyme inactivation during freeze-thawing,64 this discovery is rather exciting. The results suggest that kosmotropes, such as sarcosine, ectoine, DMSP, and proline, effectively counteract the detrimental effects of urea on lysozyme during cold shock in deep eutectic environments: these mixtures provide more efficient protection for the enzyme compared to the reference buffer. On top of that, urea-based bioinspired multicomponent DESs, especially TMAO:Bet:Tau:U, again showed excellent chaperon-like activity with AR > 85%, much higher than their two-component counterparts TMAO:U and Bet:U. This observation aligns well with natural phenomena: to reduce the cytoplasmic freezing point and fend off frost, psychrophilic bacteria, diapausing insects, amphibians, and reptiles accumulate urea, alongside other osmolytes such as betaine, sugars, sugar alcohols, and amino acids.34
Balancing enzyme activity and stability in DES–water mixtures: could transitions between dormant and active states of enzymes in vivo be explained through the formation of a DES?
The results presented in this study demonstrate: (i) the remarkable effectiveness of novel osmolyte-based DESs in stabilizing the model protein under stress conditions, and (ii) observable changes in the protein structure during stress in DESs, which can nonetheless be fully restored upon returning the enzyme to ambient conditions. However, in line with prior findings,39,44 we observed low or negligible enzyme activity in several of the most promising osmolyte-based DESs in terms of stabilization efficacy (<20% compared to the activity measured in the reference buffer) (Fig. S8, ESI†) that could be completely recovered after dilution of the enzyme in the buffer (see the section ‘Heat induced aggregation and inactivation of lysozyme in DESs’).Hence, we delved deeper into the intriguing relationship between enzyme activity and stability, focusing specifically on the bioinspired DES (Bet:Sor:Tau:GPC:U), and the dependence of these two enzyme's properties on water content (20, 40, 60 and 80% of water in DES, w/w). Again, lysozyme showed slight or no activity in the DES containing ≤40% of water (w/w), while the activity increased upon the addition of water, peaking in the highly diluted mixture (80% of water, w/w), however, the value was still much lower than that observed in the buffer (about 40% of the activity observed in the buffer) (Fig. 5). Conversely, enzyme stability (assessed by subjecting lysozyme to heat shock to elicit a rapid response), was notably better preserved in highly “concentrated” DES, reaching its peak at a water concentration of 20% (w/w) (Fig. 5). Temperature-dependent CD spectra of lysozyme solution in the DES in the temperature range 20–95 °C confirmed the results: the stabilization effect of DES with lower water content (≤40%, w/w) is so strong that the full unfolding of the protein does not occur under the measurement conditions, as seen by the lack of a plateau in the melting curve (Fig. 6A and Fig. S9, ESI†). On the other hand, when approaching higher water concentrations, the unfolding of the enzyme follows a trend more similar to that in the buffer. Fig. 6B further highlights the observed dependence of lysozyme's transition temperatures on water content, showing a peak at 20% water in DES (w/w) with Tm = 93 °C.
To investigate whether changes in the enzyme's behaviour in DES are associated with specific alterations in secondary structure, and whether these alterations can be reversed upon enzyme dilution in buffer, we initially recorded the far UV CD and FT-IR spectra of lysozyme in the “concentrated” Bet:Sor:Tau:GPC:U solution. Subsequently, we recorded the far UV CD spectrum after diluting (rehydrating) lysozyme in buffer. Both the far UV CD and FT-IR spectra of the enzyme in the “concentrated” DES revealed noticeable peak wavelength shifts in the Bet:Sor:Tau:GPC:U solution, indicating a greater presence of β-sheets (a structure more resistant to unfolding than α-helices65,66) compared to lysozyme in its native state in buffer, where α-helices predominate (Fig. 7A and B). Furthermore, the far UV spectra confirmed that upon rehydration of lysozyme preincubated in Bet:Sor:Tau:GPC:U into buffer, the native lysozyme secondary structure was virtually fully restored (Fig. 7A and Fig. S10, ESI†).
Overall, these findings suggest that the interplay between the enzyme's active and stable (but inactive) states is highly influenced by the water content in DES, supporting our hypothesis46 that osmolyte mixtures form DES at the nano/microscale in vivo, preserving enzymes in a catalytically inactive state during stressful conditions such as cryoprotection, drought resistance, and germination, with the eutectic systems becoming diluted upon overcoming these stressors and the subsequent entry of water into the cell, leading to enzyme activation. Such readily reversible transitions between inactive and active enzyme states (dormancy and wake-up states) in response to alterations in its microenvironment (reactivation of lysozyme occurs within a minute following dilution from a “concentrated” DES, as observed during stability tests) would be energetically beneficial since metabolic regulation at the level of enzyme degradation or synthesis would be avoided.67 Most importantly, the in vivo fluctuations of water content in the protein microenvironment and the resulting reversible interplay between active/dormant states would open a new window through which information flows in natural systems could be understood, investigated or even monitored.
Finally, it is crucial to acknowledge that the mechanisms by which protein structure is influenced by its environment are far more intricate than what is presented here, where only solvent–protein interactions are considered. In vivo, protein folding/unfolding occurs within the crowded cellular milieu, characterized by volume exclusion due to neighbouring soluble macromolecules, and within confined spaces, influenced by rigid or fixed structures. These environments are distinctly different. Models of crowding examine the effects of varied concentrations of soluble crowding agents, such as polymers, while confinement models involve molecular meshes, pores, and channels, simulating cytoskeletal and extracellular matrix structures.68,69 Given these considerations, it is reasonable to assume that proteins encounter more complex landscapes in vivo compared to those described here. These differences likely play a significant role in the protein folding and unfolding puzzle in DES environments and warrant further exploration. In the context of using DES for protein stabilization, an intriguing approach could involve designing solvent media that combine macromolecular crowders27 or confining proteins into nanopores70 with DES technology. This hybrid strategy could offer novel solutions for enhancing protein stability.
Cytotoxicity assessment of osmolyte-based DESs
Finally, to assess DESs biocompatibility, the newly synthesized osmolyte-based DESs were tested at three different concentrations (100 mg ml−1, 500 mg ml−1, and 1000 mg ml−1) on three cell lines: Caco-2, HaCaT, and HeLa. These cell lines were selected to cover various potential applications of the tested DES, ranging from oral administration (intestinal, Caco-2), to topical (skin, HaCaT), to possible antiproliferative effects (tumor cell line, HeLa). This evaluation serves as a preliminary assessment of DES ecotoxicity as well as proof of their cytocompatibility with the used cells and whether they can proceed to further application. Because none of the tested DES caused 50% growth inhibition in any of the used cell lines, the EC50 values were declared to be higher than 1000 mg L−1 and DESs were considered to have low cytotoxicity and non-antiproliferative activity (EC50 >2000 mg L−1i.e., >5 mM) (Table 1). The only exception is DMPS:Gly, which significantly inhibited the growth of all three cell lines at concentrations of 500 mg ml−1 and 1000 mg ml−1. This is probably due to the extremely low pH value of DMPS:Gly (1.0) which can denature membrane proteins and cause cell death.71 Despite the observed decrease in cell survival associated with DMPS:Gly, it is important to note that the tested concentrations ranging from 100 mg ml−1 to 1000 mg ml−1, are exceptionally high. By testing these high concentrations, we took into account the estimation that cell lines are approximately ten times less sensitive than in vivo tests. Therefore, the overall conclusion is that the tested DESs pose no environmental hazards or risks to human health. However, additional investigations on other model organisms are certainly desirable for final confirmation of the safety of these newly synthesized solvents.Conclusion
In conclusion, our study demonstrates that DESs based on naturally occurring osmolytes, in all their variety, are excellent biocompatible chemical chaperones promoting the correct refolding of unfolded lysozyme in vitro after exposing the enzyme to stressful conditions, such as heat shock and repeated freezing–thawing cycles. Glycerol-based DESs with betaine, sarcosine, and ectoine as HBA stood out as the best candidates for stabilizing lysozyme under heat shock conditions, while the urea-based DES, specifically DMSP, provided the greatest stabilization of lysozyme under freezing conditions. Interestingly multicomponent bioinspired DESs (Bet:Sor:Tau:GPC:U and TMAO:Bet:Tau:U) were excellent stabilizing medium for lysozyme under both storage conditions, showing excellent chaperon-like activity, much higher than their two-component counterparts Bet:U and TMAO:U. Through an analysis of the intriguing relationship between enzyme activity and stability in DES, with a focus on a bioinspired DES (Bet:Sor:Tau:GPC:U) and the dependency of these enzyme properties on water content, we have demonstrated that highly concentrated DES efficiently stabilize lysozyme in its inactive form. Furthermore, lysozyme activation can be easily achieved by diluting the enzyme–DES mixture in water, supporting our hypothesis that the formation of DES in vivo could facilitate rapid and reversible transitions between inactive (dormant) and active (wake-up) states in response to changes in their microenvironment. We propose that by mimicking the natural landscape of a specific protein through tracking osmolyte cocktails and replicating them in vitro through DES, it is feasible to engineer the ideal medium for this specific protein to be stored for a long term and resistant to fluctuations in storage temperature. The present study suggests that a template protein stability can be enhanced by carefully tailored DES, and that further experiments and careful optimization of DES formulations will be needed to elucidate working combinations for additional proteins beyond lysozyme. The ability to selectively tune a protein microenvironment has the potential to foster manufacturing sustainability “by design” across diverse industries.The current study primarily focused on identifying and reporting the observed stabilization effects by screening a large number of DESs to showcase the concept of osmolyte-based DESs as excellent protein stabilizers against heat and freeze shock, rather than delving into the detailed mechanisms underlying these behaviors. Therefore, designing a comprehensive study that combines various experimental and computational techniques would provide detailed mechanistic insight into how osmolyte-based DESs stabilize proteins. This approach would help in understanding the specific interactions and environmental factors that contribute to protein stabilization, thereby guiding the development of more effective DES formulations for biotechnological applications. Addressing this aspect is a key goal for our future research endeavors.
Author contributions
The concept for the article was jointly conceived and developed by M. C. B. and T. A., while M. C. B. designed the experiments, wrote the first draft and provided direction. I. R. R. and L. T. helped refine the concept. M. C. B., L. T., A. D. and M. L. performed the experiments. The paper was reviewed and edited by all authors.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Croatian Science Foundation (IPS-2022-02-3938, IP-2019-04-7712 and IP-2018-01-4694). We gratefully acknowledge undergraduate students Marija Karin and Petra Lacić for their contributions to the experiments.Notes and references
- V. A. Borzova, T. B. Eronina, V. V. Mikhaylova, S. G. Roman, A. M. Chernikov and N. A. Chebotareva, Int. J. Mol. Sci., 2023, 24, 10298 CrossRef CAS PubMed.
- R. J. Falconer, Biotechnol. Adv., 2019, 37, 107412 CrossRef CAS PubMed.
- M. C. Rodríguez, J. Villarraza, M. B. Tardivo, S. Antuña, D. Fontana, N. Ceaglio and C. Prieto, J. Pharm. Sci., 2023, 112, 2756–2765 CrossRef PubMed.
- M. M. C. H. van Schie, J. D. Spöring, M. Bocola, P. Domínguez de María and D. Rother, Green Chem., 2021, 23, 3191–3206 RSC.
- M. Akbarian and S.-H. Chen, Pharmaceutics, 2022, 14, 2533 CrossRef CAS PubMed.
- S. R. Wlodarczyk, D. Custódio, A. Pessoa and G. Monteiro, Eur. J. Pharm. Biopharm., 2018, 131, 92–98 CrossRef CAS PubMed.
- L. Zhang, R. Zhou, J. Zhang and P. Zhou, Int. Dairy J., 2021, 123, 105175 CrossRef CAS.
- J. W. Bye, L. Platts and R. J. Falconer, Biotechnol. Lett., 2014, 36, 869–875 CrossRef CAS PubMed.
- G. Lentzen and T. Schwarz, Appl. Microbiol. Biotechnol., 2006, 72, 623–634 CrossRef CAS PubMed.
- T. Ueda, M. Nagata and T. Imoto, J. Biochem., 2001, 130, 491–496 CrossRef CAS PubMed.
- S. Khan, S. Siraj, M. Shahid, M. M. Haque and A. Islam, Int. J. Biol. Macromol., 2023, 234, 123662 CrossRef CAS PubMed.
- P. H. Yancey, J. Exp. Biol., 2005, 208, 2819–2830 CrossRef CAS PubMed.
- N. Zhang, P. Domínguez de María and S. Kara, Catalysts, 2024, 14, 84 CrossRef CAS.
- A. M. Curreri, S. Mitragotri and E. E. L. Tanner, Adv. Sci., 2021, 8, 2004819 CrossRef CAS PubMed.
- A. P. Abbott, G. Capper, D. L. Davies, R. K. Rasheed and V. Tambyrajah, Chem. Commun., 2003, 70–71 RSC.
- Y. Zhou, W. Xu, Y. Pan, F. Wang, X. Hu, Y. Lu and M. Jiang, BioResources, 2022, 17, 5485–5509 Search PubMed.
- D. O. Abranches and J. A. P. Coutinho, Annu. Rev. Chem. Biomol. Eng., 2023, 14, 141–163 CrossRef CAS PubMed.
- F. Niknaddaf, S. S. Shahangian, A. Heydari, S. Hosseinkhani and R. H. Sajedi, ChemistrySelect, 2018, 3, 10603–10607 CrossRef CAS.
- P. Xu, Y. Wang, J. Chen, X. Wei, W. Xu, R. Ni, J. Meng and Y. Zhou, Talanta, 2019, 202, 1–10 CrossRef CAS PubMed.
- Z. Takalloo, F. Niknaddaf, S. S. Shahangian, A. Heydari, S. Hosseinkhani and R. H. Sajedi, Biotechnol. Appl. Biochem., 2020, 67, 330–342 CAS.
- S. H. Kim, S. Park, H. Yu, J. H. Kim, H. J. Kim, Y. H. Yang, Y. H. Kim, K. J. Kim, E. Kan and S. H. Lee, J. Mol. Catal. B Enzym., 2016, 128, 65–72 CrossRef CAS.
- I. Juneidi, M. Hayyan, M. A. Hashim and A. Hayyan, Biochem. Eng. J., 2017, 117, 129–138 CrossRef CAS.
- M. Shehata, A. Unlu, U. Sezerman and E. Timucin, J. Phys. Chem. B, 2020, 124, 8801–8810 CrossRef CAS PubMed.
- N. Yadav, K. Bhakuni, M. Bisht, I. Bahadur and P. Venkatesu, ACS Sustainable Chem. Eng., 2020, 8, 10151–10160 CrossRef CAS.
- A. E. Delorme, J. M. Andanson and V. Verney, Int. J. Biol. Macromol., 2020, 163, 919–926 CrossRef CAS PubMed.
- H. Sun, R. Xin, D. Qu and F. Yao, J. Biotechnol., 2020, 323, 264–273 CrossRef CAS PubMed.
- K. Bhakuni, N. Yadav and P. Venkatesu, Phys. Chem. Chem. Phys., 2020, 22, 24410–24422 RSC.
- B. P. Wu, Q. Wen, H. Xu and Z. Yang, J. Mol. Catal. B: Enzym., 2014, 101, 101–107 CrossRef CAS.
- A. A. N. Gunny, D. Arbain, E. M. Nashef and P. Jamal, Bioresour. Technol., 2015, 181, 297–302 CrossRef CAS PubMed.
- M. S. Lee, K. Lee, M. W. Nam, K. M. Jeong, J. E. Lee, N. W. Kim, Y. Yin, S. Y. Lim, D. E. Yoo, J. Lee and J. H. Jeong, J. Ind. Eng. Chem., 2018, 65, 343–348 CrossRef CAS.
- J. Hoppe, R. Drozd, E. Byzia and M. Smiglak, Int. J. Biol. Macromol., 2019, 136, 296–304 CrossRef CAS PubMed.
- D. Dhiman, A. S. C. Marques, M. Bisht, A. P. M. Tavares, M. G. Freire and P. Venkatesu, Green Chem., 2023, 25, 650–660 RSC.
- I. Gomes and N. Galamba, J. Chem. Phys., 2023, 159, 235101 CrossRef CAS PubMed.
- M. Cvjetko Bubalo, T. Andreou, M. Panić, M. Radović, K. Radošević and I. R. Redovniković, Green Chem., 2023, 25, 3398–3417 RSC.
- K. M. Jeong, J. Ko, J. Zhao, Y. Jin, D. E. Yoo, S. Y. Han and J. Lee, J. Cleaner Prod., 2017, 151, 87–95 CrossRef CAS.
- D. Shugar, Biochim. Biophys. Acta, 1952, 8, 302–309 CrossRef CAS PubMed.
- P. Angsantikul, K. Peng, A. M. Curreri, Y. Chua, K. Z. Chen, J. Ehondor and S. Mitragotri, Adv. Funct. Mater., 2021, 31, 2002912 CrossRef CAS.
- S. M. Kelly, T. J. Jess and N. C. Price, Biochim. Biophys. Acta, Proteins Proteomics, 2005, 1751, 119–139 CrossRef CAS PubMed.
- A. Sanchez-Fernandez, S. Prevost and M. Wahlgren, Green Chem., 2022, 24, 4437–4442 RSC.
- R. Swaminathan, V. K. Ravi, S. Kumar, M. V. S. Kumar and N. Chandra, Adv. Protein Chem. Struct. Biol., 2011, 84, 63–111 CAS.
- A. Sanchez-Fernandez, K. J. Edler, T. Arnold, D. Alba Venero and A. J. Jackson, Phys. Chem. Chem. Phys., 2017, 19, 8667–8670 RSC.
- K. Park, B. Y. Ham, K. Li, S. Kang, D. Jung, H. Kim, Y. Liu, I. Hwang and J. Lee, J. Mol. Liq., 2022, 349, 118143 CrossRef CAS.
- C. X. Zeng, S. J. Qi, R. P. Xin, B. Yang and Y. H. Wang, J. Mol. Liq., 2016, 219, 74–78 CrossRef CAS.
- R. Esquembre, J. M. Sanz, J. G. Wall, F. Del Monte, C. R. Mateo and M. L. Ferrer, Phys. Chem. Chem. Phys., 2013, 15, 11248–11256 RSC.
- A. Mitar, M. Panić, J. Prlić Kardum, J. Halambek, A. Sander, K. Zagajski Kučan, I. Radojčić Redovniković and K. Radošević, Chem. Biochem. Eng. Q., 2019, 33, 1–18 CrossRef CAS.
- D. Rente, M. Cvjetko Bubalo, M. Panić, A. Paiva, B. Caprin, I. Radojčić Redovniković and A. R. C. Duarte, J. Cleaner Prod., 2022, 380, 135147 CrossRef CAS.
- L. Cicco, G. Dilauro, F. M. Perna, P. Vitale and V. Capriati, Org. Biomol. Chem., 2021, 19, 2558–2577 RSC.
- M. A. Karadendrou, I. Kostopoulou, V. Kakokefalou, A. Tzani and A. Detsi, Catalysts, 2022, 12, 249 CrossRef CAS.
- B. Ozturk, C. Parkinson and M. Gonzalez-Miquel, Sep. Purif. Technol., 2018, 206, 1–13 CrossRef CAS.
- M. Hirai, S. Arai, H. Iwase and T. Takizawa, J. Phys. Chem. B, 1998, 102, 1308–1313 CrossRef CAS.
- D. Shah, A. R. Shaikh, X. Peng and R. Rajagopalan, Biotechnol. Prog., 2011, 27, 513–520 CrossRef CAS PubMed.
- L. N. Arnaudov and R. De Vries, Biophys. J., 2005, 88, 515 CrossRef CAS PubMed.
- M. L. Toledo, M. M. Pereira, M. G. Freire, J. P. A. Silva, J. A. P. Coutinho and A. P. M. Tavares, ACS Sustainable Chem. Eng., 2019, 7, 11806–11814 CrossRef CAS.
- A. Salvi, J. M. Quillan and W. Sadee, AAPS PharmSci, 2002, 4, 1–8 CrossRef PubMed.
- J. P. Bittner, N. Zhang, L. Huang, P. Domínguez De María, S. Jakobtorweihen and S. Kara, Green Chem., 2022, 24, 1120–1131 RSC.
- H. Monhemi, M. R. Housaindokht, A. A. Moosavi-Movahedi and M. R. Bozorgmehr, Phys. Chem. Chem. Phys., 2014, 16, 14882–14893 RSC.
- S. Sarkar, S. Ghosh and R. Chakrabarti, RSC Adv., 2017, 7, 52888–52906 RSC.
- S. J. Shire, Z. Shahrokh and J. Liu, J. Pharm. Sci., 2004, 93, 1390–1402 CrossRef CAS PubMed.
- L. Z. Wu, B. L. Ma, Y. B. Sheng and W. Wang, J. Mol. Struct., 2008, 891, 167–172 CrossRef CAS.
- T. Knubovets, J. J. Osterhout, P. J. Connolly and A. M. Klibanov, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 1262–1267 CrossRef CAS PubMed.
- A. Esposito, L. Comez, S. Cinelli, F. Scarponi and G. Onori, J. Phys. Chem. B, 2009, 113, 16420–16424 CrossRef CAS PubMed.
- F. Tanaka, L. S. Forster, P. K. Pal and J. A. Rupley, J. Biol. Chem., 1975, 250, 6977–6982 CrossRef CAS PubMed.
- S. K. Singh and S. Nema, in Formulation and Process Development Strategies for Manufacturing Biopharmaceuticals, ed. F. Jameel and S. Hershenson, John Wiley & Sons, New Jersey, 2010, vol. 26, pp. 625–675 Search PubMed.
- J. F. Carpenter and J. H. Crowe, Cryobiology, 1988, 25, 244–255 CrossRef CAS PubMed.
- R. Arunkumar, C. J. Drummond and T. L. Greaves, Front. Chem., 2019, 7, 74 CrossRef CAS PubMed.
- D. E. Otzen, Biophys. J., 2002, 83, 2219–2230 CrossRef CAS PubMed.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Science, 1982, 217, 1214–1222 CrossRef CAS PubMed.
- D. Lucent, V. Vishal and V. S. Pande, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 10430–10434 CrossRef CAS PubMed.
- L. W. Simpson, T. A. Good and J. B. Leach, Biotechnol. Adv., 2020, 42, 107573 CrossRef CAS PubMed.
- D. L. Z. Caetano, R. Metzler, A. G. Cherstvy and S. J. de Carvalho, Phys. Chem. Chem. Phys., 2021, 23, 27195–27206 RSC.
- L. Lomba, M. P. Ribate, E. Zaragoza, J. Concha, M. P. Garralaga, D. Errazquin, C. B. García and B. Giner, Appl. Sci., 2021, 11, 10061 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp02275k |
| ‡ These authors contributed equally to this work. |
| This journal is © the Owner Societies 2024 |