Open Access Article
Open Access ArticleAn integrated approach towards extracting structural characteristics of chlorosomes from a bchQ mutant of Chlorobaculum tepidum†
Lolita
Dsouza
 a,
Xinmeng
Li
a,
Xinmeng
Li
 ab,
Vesna
Erić
c,
Annemarie
Huijser
ab,
Vesna
Erić
c,
Annemarie
Huijser
 d,
Thomas L. C.
Jansen
d,
Thomas L. C.
Jansen
 c,
Alfred R.
Holzwarth
c,
Alfred R.
Holzwarth
 e,
Francesco
Buda
e,
Francesco
Buda
 a,
Donald A.
Bryant
a,
Donald A.
Bryant
 f,
Salima
Bahri
f,
Salima
Bahri
 g,
Karthick Babu Sai Sankar
Gupta
g,
Karthick Babu Sai Sankar
Gupta
 a,
G. J. Agur
Sevink
a,
G. J. Agur
Sevink
 *a and
Huub J. M.
de Groot
*a and
Huub J. M.
de Groot
 *a
*a
aLeiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2300 RA, Leiden, The Netherlands. E-mail: ssnmr@chem.leidenuniv.nl
bDepartment of Chemistry and Hylleraas Centre for Quantum Molecular Sciences, University of Oslo, 0315, Oslo, Norway
cZernike Institute of Advanced Materials, University of Groningen, Nijenborgh 4, 9747 AG, The Netherlands
dMESA+ Institute for Nanotechnology, University of Twente, 7500 AE, The Netherlands
eMax Planck Institute for Chemical Energy Conversion, Stiftstraße 34-36, 45470, Mülheim an der Ruhr, Germany
fDepartment for Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, Pennsylvania 16802, USA
gNMR Spectroscopy, Bijvoet center for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH, Utrecht, The Netherlands
First published on 28th March 2024
Abstract
Chlorosomes, the photosynthetic antenna complexes of green sulfur bacteria, are paradigms for light-harvesting elements in artificial designs, owing to their efficient energy transfer without protein participation. We combined magic angle spinning (MAS) NMR, optical spectroscopy and cryogenic electron microscopy (cryo-EM) to characterize the structure of chlorosomes from a bchQ mutant of Chlorobaculum tepidum. The chlorosomes of this mutant have a more uniform composition of bacteriochlorophyll (BChl) with a predominant homolog, [8Ethyl, 12Ethyl] BChl c, compared to the wild type (WT). Nearly complete 13C chemical shift assignments were obtained from well-resolved homonuclear 13C–13C RFDR data. For proton assignments heteronuclear 13C–1H (hCH) data sets were collected at 1.2 GHz spinning at 60 kHz. The CHHC experiments revealed intermolecular correlations between 132/31, 132/32, and 121/31, with distance constraints of less than 5 Å. These constraints indicate the syn–anti parallel stacking motif for the aggregates. Fourier transform cryo-EM data reveal an axial repeat of 1.49 nm for the helical tubular aggregates, perpendicular to the inter-tube separation of 2.1 nm. This axial repeat is different from WT and is in line with BChl syn–anti stacks running essentially parallel to the tube axis. Such a packing mode is in agreement with the signature of the Qy band in circular dichroism (CD). Combining the experimental data with computational insight suggests that the packing for the light-harvesting function is similar between WT and bchQ, while the chirality within the chlorosomes is modestly but detectably affected by the reduced compositional heterogeneity in bchQ.
1. Introduction
Green sulfur bacteria are nonmotile, Gram-negative bacteria that thrive in strictly anaerobic environments. They are predominantly found in hot springs, sediments, and mats, and in stratified aquatic environments.1–3 The unique feature of these bacteria is that they can survive in extremely low light environments and can harvest light very efficiently.4,5 The ability of these bacteria to survive in low light and efficiently transfer energy is due to their use of efficient chlorosome antenna structures.6,7Chlorosomes harvest light and subsequently transfer the excitation energy to the baseplate, then to the FMO bacteriochlorophyll (BChl) a-binding protein, and ultimately to the reaction center facilitating charge separation.8–11 The self-assembly of chlorosomes is due to aggregates of BChls that form without involving proteins.8,12 The distinctiveness of BChl c, the major pigment in chlorosomes, lies in its 31 hydroxyl group and the absence of a methyl carboxyl group at the 131 position, see Fig. 1. The limited bulkiness of the functionalities at the 31 and 131 positions enables the formation of large supramolecular aggregates by favorable modulation of steric hindrance.13 Self-assembly is a thermodynamically favorable process and for chlorosomes, it is enabled by four main types of intermolecular interactions, (i) coordination of the central magnesium ion to the hydroxyl group, (ii) hydrogen bond formation between the carbonyl group and the hydroxyl group, (iii) pi–pi stacking of the chromophores and (iv) hydrophobic interactions of the tails.14,15 Hydrogen bonding was previously considered to play a dominant role in the emergence of two spectral components.16 With a major component consisting of hydrogen bond donors that support Mg–OH⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C motifs connecting BChls within and between syn–anti stacks, and a minor fraction for which the Mg–OH coordination between BChls in a stack is present, H-bonds between stacks cannot be formed due to the partial rotation of BChls in the plane of their macrocycle.16 The BChls of the second fraction were therefore regarded as non-donors for hydrogen bonds.17 The presence of two fractions agrees with the observed dispersion of the optical absorption profile and the splitting of the NMR signals.14,17,18 The aliphatic functionalities at the 8 and 12 positions are thought to assist in optimizing the size of the BChls and produce steric hindrance for tuning their absorption properties. They provide an overall broadening of the optical absorption profile by side chain heterogeneity.13
C motifs connecting BChls within and between syn–anti stacks, and a minor fraction for which the Mg–OH coordination between BChls in a stack is present, H-bonds between stacks cannot be formed due to the partial rotation of BChls in the plane of their macrocycle.16 The BChls of the second fraction were therefore regarded as non-donors for hydrogen bonds.17 The presence of two fractions agrees with the observed dispersion of the optical absorption profile and the splitting of the NMR signals.14,17,18 The aliphatic functionalities at the 8 and 12 positions are thought to assist in optimizing the size of the BChls and produce steric hindrance for tuning their absorption properties. They provide an overall broadening of the optical absorption profile by side chain heterogeneity.13
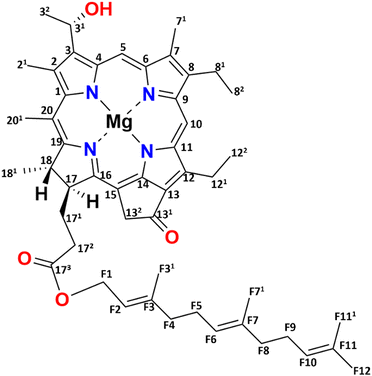 | ||
| Fig. 1 Chemical structure of [8Et, 12Et] BChl c, which is the most abundant (95%) BChl c homolog for the bchQ mutant of Chlorobaculum tepidum. | ||
One chlorosome organelle can contain up to several hundred thousand BChl pigments. Although every chlorosome is unique in both size and composition, for the bchQ mutant they are 95–163 nm in length and 31–57 nm in diameter on average.5,19 Structuring BChl molecules in chlorosomes is a natural solution to overcome the rapid loss of excitonic energy that otherwise would occur for this high pigment density. Naturally self-assembled BChl aggregates like chlorosomes thus provide a paradigm for the bottom–up chemical engineering of biomimetic dye materials for use in optoelectronic devices.20,21 Significant advancements have been made in the analysis of the BChl biosynthesis pathway, leading to the construction of numerous mutations affecting BChl c/d/e.13,22 Inactivation of CT1777, also known as bchQ, causes the accumulation of 5% [8Et, 12Me] and 95% [8Et, 12Et]-BChl c homologs, see Fig. 1, unlike WT chlorosomes, that are comprised of four homologs: 5% [Et, Me], 60% [Et, Et], 30% [Pr, Et] and 5% [Bu, Et].23 Because the homolog [8Et, 12Et] is also the major component in WT chlorosomes, it was taken as the representative pigment component in recent physical modelling studies.16,17,20,24,25
In this paper, we have combined three different methods to study the structural and optical characteristics of chlorosomes of the bchQ mutant of Cba. tepidum. While the side chain variability at the 8 and 12 positions is not anticipated to play a significant role, it is known to tune structural features and absorption properties.23 Because of its nearly homogeneous composition, chlorosomes of the bchQ mutant are being studied experimentally in the present work. The use of mutants and the integration of different techniques has been a successful approach in determining the structure of chlorosomes in Cba. tepdium.26–29 Following Ganapathy et al., we combine two methods, solid-state magic angle spinning (MAS) nuclear magnetic resonance (NMR) spectroscopy and cryo-EM, to first resolve important structural features of BChl c packing in bchQ chlorosomes.26 Additionally, we performed optical measurements. The obtained data were subsequently discussed in relation to earlier findings of WT chlorosomes and employed to propose a chiral angle for bchQ. In addition to the structure and optical properties, we used the dynamic spectral editing technique (DYSE) with solid-state NMR to study the dynamics between WT and bchQ.
2. Materials and methods
2.1. Sample preparation
The cells for bchQ and WT were grown at 40 °C in a growth chamber according to standard procedures.1 For uniform 13C labeling, 1.0 L of the Wahlund medium was inoculated with 50 mL freshly grown cell culture without ammonium acetate and grown for 1 day for light adaptation. 1.0 L of the fresh medium containing 99.5% enriched NaH13CO3 was inoculated with 50 mL of light-adapted culture grown without ammonium acetate.15 Cells were grown for 2 days, then harvested at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g by centrifugation. The supernatant was discarded, and the pellet was resuspended in 35 mL of 20 mM Tris-HCl (pH 8.0), 10 mM sodium ascorbate, and 2 M sodium thiocyanate. The pellet was completely dissolved by keeping it at 4 °C for 30 minutes. The cells were disrupted using a French pressure cell at 8.27 MPa at 4 °C. Then, 1.5 mg of DNAse-1 was added to the suspension and stirred for 30 minutes. The cells were passed through the French pressure cell again to ensure complete disruption. The suspension was centrifuged for 20 minutes at 23
000 × g by centrifugation. The supernatant was discarded, and the pellet was resuspended in 35 mL of 20 mM Tris-HCl (pH 8.0), 10 mM sodium ascorbate, and 2 M sodium thiocyanate. The pellet was completely dissolved by keeping it at 4 °C for 30 minutes. The cells were disrupted using a French pressure cell at 8.27 MPa at 4 °C. Then, 1.5 mg of DNAse-1 was added to the suspension and stirred for 30 minutes. The cells were passed through the French pressure cell again to ensure complete disruption. The suspension was centrifuged for 20 minutes at 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 × g to remove large cell debris in a Beckman ultra-centrifuge using a Ti-70 rotor. After centrifugation, the chlorosomes were isolated by subjecting them to centrifugation on a linear 20–50% sucrose density gradient. The separation process was carried out by subjecting the mixture to sucrose density gradient centrifugation in a swing-out rotor at 4 °C for 18 h at 133
200 × g to remove large cell debris in a Beckman ultra-centrifuge using a Ti-70 rotor. After centrifugation, the chlorosomes were isolated by subjecting them to centrifugation on a linear 20–50% sucrose density gradient. The separation process was carried out by subjecting the mixture to sucrose density gradient centrifugation in a swing-out rotor at 4 °C for 18 h at 133![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 668 × g. The sample was dialyzed overnight against 20 mM Tris-HCl buffer to remove sucrose and cyanate.30 After dialysis, the optical absorption profile was collected using UV-Vis measurement. The sample was stored at 4 °C until further use. For solid-state NMR studies, the sample was further concentrated using an ultracentrifuge at 200
668 × g. The sample was dialyzed overnight against 20 mM Tris-HCl buffer to remove sucrose and cyanate.30 After dialysis, the optical absorption profile was collected using UV-Vis measurement. The sample was stored at 4 °C until further use. For solid-state NMR studies, the sample was further concentrated using an ultracentrifuge at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 hour, and the obtained pellet was used to fill the NMR rotor in the form of a paste.
000 × g for 1 hour, and the obtained pellet was used to fill the NMR rotor in the form of a paste.
2.2. Solid-state NMR measurements
Central to this work is the use of MAS NMR, which has been developed over five decades into a versatile tool for studying biological systems, starting with the pioneering work on bacteriorhodopsin by Herzfeld et al.31MAS-NMR data were collected from freshly prepared 13C-labeled chlorosomes from bchQ and WT cells. The measurements were performed with the sample in a 3.2 mm rotor spinning at 11 kHz at the magic angle in a Bruker AV-750 (17.6 T) spectrometer equipped with a triple resonance MAS probe head by using a 13C radio frequency of 188.6 MHz. One-dimensional, direct polarization (DP), cross polarization (CP) and insensitive nuclei enhanced by polarization transfer (INEPT) measurements were performed with 1024 scans.32 The recycle delay was set to 2 s for CP and the delay period was set to 1.75 ms and 1.15 ms for INEPT. The proton π/2 pulse length for the CP was 2.5 μs, corresponding to 100 kHz rf amplitude. Two-dimensional dipolar correlation spectra were recorded by using 13C–13C radiofrequency driven recoupling (RFDR) at mixing times of 9.2 ms and 3.2 ms to probe long and short-range correlations.33 The NMR data were collected in 256 scans for every trace in the t1 dimension with phase-sensitive detection in ω1. Two-dimensional dipolar correlation spectra were recorded by using the 13C–13C CHHC/CP3 sequence to probe the indirect detection of 1H–1H contacts at proton diffusion times of 350 μs.34,35 The dipolar hCH spectra were collected with a 1.3 mm probe in a 1.2 GHz spectrometer operating at a magnetic field strength of 28.2 Tesla. The sample was spun at 60 kHz at a set temperature of 245 K, which corresponds to a sample temperature of 277 K.36 To assign the proton chemical shifts we made use of 13C chemical shifts. The 1H chemical shift was internally calibrated using the water signal at 4.7 ppm. In the 2D dipolar hCH spectra, 90° pulses were set at 105 kHz rf-field amplitude for 1H and at 51 kHz rf-field amplitude for 13C.37,381H–13C forward cross-polarization (CP) was achieved using a contact time of 1.2 ms. 13C–1H back CP was achieved using a contact time of 0.3 ms, and PISSARRO decoupling was used for both 1H and 13C with 15 kHz RF amplitude.39 The MISSISSIPPI water suppression was used to suppress the water signal, with a 17 kHz irradiation for 120 ms.40 All the spectra were acquired and processed using the states-TPPI procedure. See ESI† Table S4 for more information on the set up of solid state 2D NMR.
The data were processed using a Bruker TopSpin 4.1.4 and analyzed using MestReNova.
2.3. cryo-EM measurements
cryo-EM was performed with a Titan Krios microscope (FEI). Aliquots of purified chlorosomes were applied onto a holey carbon grid with a thin layer of carbon and were plunge frozen in liquid ethane at 83 K with a Vitrobot vitrification system (FEI). Experiments were performed with a Falcon 3 camera operated in a linear mode with a defocus of −0.5 μm. A magnification of 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 was used which corresponds to a pixel size of 0.878 Å per pixel. The image processing and analysis was performed using Fiji software.41
000 was used which corresponds to a pixel size of 0.878 Å per pixel. The image processing and analysis was performed using Fiji software.41
2.4. Optical measurements
UV-Vis spectra were measured using a Shimadzu 1800 UV-Vis spectrometer, with the sample contained in a 1 mm optical path length micro cuvette (Hellma, QS absorption cuvette). CD spectra were measured using a JASCO J-815 CD spectrometer, with the sample contained in a 1 mm optical path length micro cuvette (Hellma, QS absorption cuvette).2.5. cryo-EM mimics
A pre-assembled BChl c tube structure with a helical angle of δ = 90° was taken from our previous set of structures.20 The tube structure was 15 nm in diameter and 60 nm in length and was thermalised through 10 ns of molecular dynamics (MD) simulations. Using the EMAN2 package, we generated 6 Å resolution electron density maps (e2pdb2mrc.py) and obtained 2D projections (e2project3d.py). We performed Fourier Transform analysis over the 2D density maps using the Fiji software.413. Results
3.1. Structure of chlorosomes of bchQ based on MAS NMR and cryo-EM
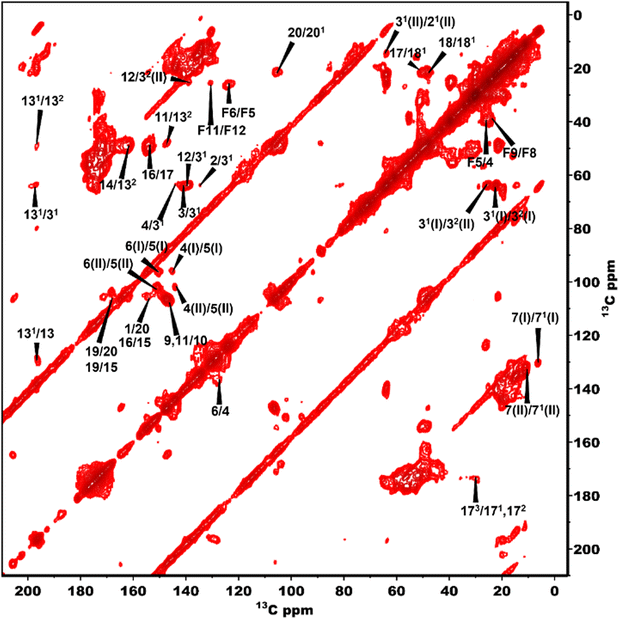
The 13C NMR chemical shift assignments for the BChl c molecules in the chlorosomes of the bchQ mutant of Cba. tepidum were obtained from 2D 13C–13C magic angle dipolar correlation spectra. Assignments are summarized in the ESI,† Table S1. Fig. 2 is an RFDR homonuclear dipolar correlation dataset collected with a mixing time of 9.2 ms. The measurements were also done at 3.2 ms which is used to predict the short-range correlations, shown in Fig. S4, in the ESI.† While the assignments closely match the assignments of the WT, the resonances were better resolved for the chlorosomes of the bchQ mutant. Like WT chlorosomes, a doubling of signals is observed for the mutant, for 13C resonances 21-C, 31-C, 32-C, 4-C, 5-C, 6-C, 7-C, and 71-C of the BChl ring, revealing two different BChl components within the system.16,17 The 13C chemical shifts were used to calculate the aggregation shifts (see the ESI† Fig. S3). The BChl aggregates tend to shift upfield or downfield relative to monomeric BChl in solution due to the macrocycle overlap between neighboring BChl molecules that can give rise to significant ring current shifts.21,42 The solution data for the 13C were taken for the [8Et, 12Et] homolog as it is the most abundant homolog in the bchQ mutant.15 The 21, 3, 32, 5, 7, 71, 82, 12, 121, 122, 19 and F3 carbons are shifted by >|1.5| ppm or more relative to the data for monomeric BChl c in solution.Fig. 3 shows a 2D dipolar hCH spectrum obtained at 60 kHz spinning frequency under a 28.2 Tesla magnetic field which gives rise to well-resolved cross peaks. The specific aim of this experiment is to resolve the 1H shifts correlating with 13C for the chlorosomes of the bchQ mutant. Protons have a high gyromagnetic ratio and are abundant spins.
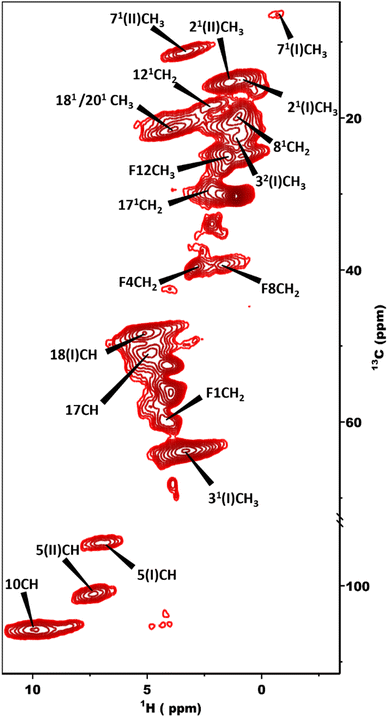 | ||
| Fig. 3 2D dipolar hCH spectrum obtained at 1.2 GHz with the sample in a 1.3 mm rotor spinning at 60 kHz at a sample temperature of 277 K. | ||
Due to strong homonuclear dipolar couplings and little shift dispersion for 1H, it is not easy to obtain well-resolved spectra.
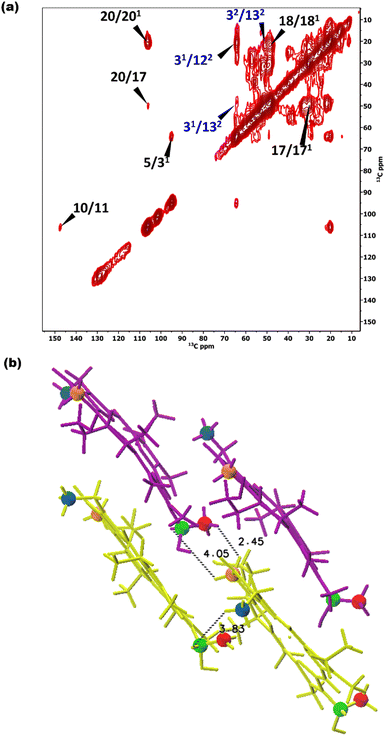
To overcome this problem, the use of a high magnetic field in combination with faster spinning and inverse detection is beneficial.43 Proton assignments are provided in Table S2 in the ESI.† The doubling for 5 and 71 that is observed for 13C is also seen for protons. Upfield shifts >−1.5 ppm are detected for 21(I and II), 31, 5 (I and II), 71, 81, 121, and downfield shifts >1.5 ppm for 181 proton resonances compared to BChl c in solution.14 The 1H chemical shifts were used to calculate the aggregation shifts (see the ESI,† Fig. S3). To proceed from chemical shift assignments and aggregation shift calculations to distance constraints, we collected homonuclear 13C–13C dipolar correlation data for the bchQ mutant, which gave us three unambiguous intermolecular correlations, namely 132/31, 132/32 and 121/31, correlating carbons from ring 1 and ring 3 (Fig. 4).
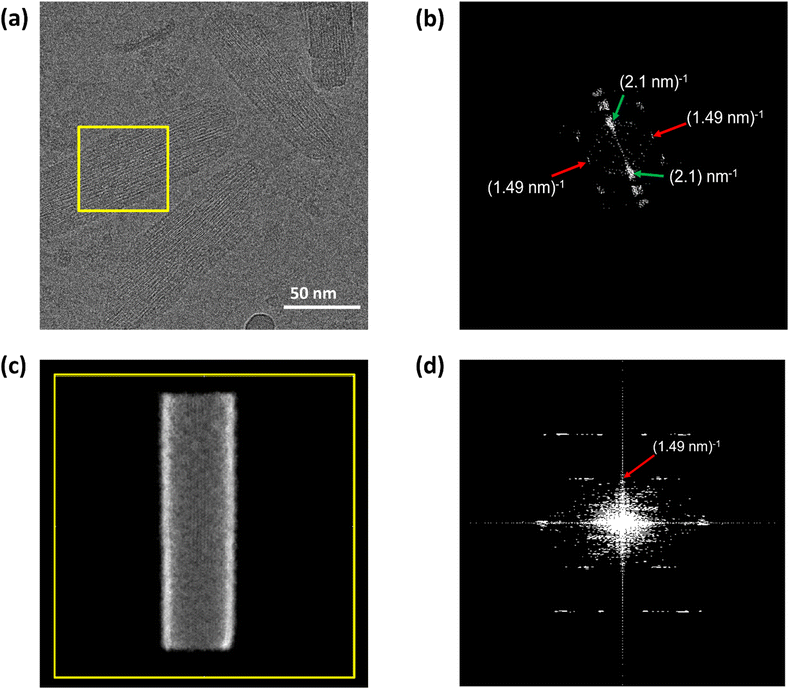
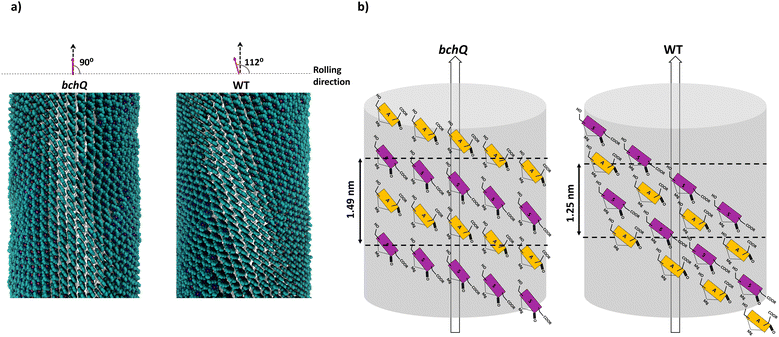
cryo-EM observations reveal that bchQ chlorosomes are tubular in nature and are similar to WT chlorosomes.27 A side-on view of isolated chlorosomes shows striped patterns, Fig. 5(a), which, after fast Fourier transform (FFT) of the image, yielded layer lines indicative of helicity/chirality of the pigment stacking within tubes, with an axial repeat of 1.49 nm, Fig. 5(b). The strongest pair of equatorial reflections relate to a 2.1 nm inter-tube distance. While the latter is in complete agreement with the WT case and pertains to the presence of the same aliphatic tail in the BChl pigment, the axial repeat of bchQ is significantly increased compared to the 1.25 nm for WT. This points to a difference in the chiral angle between bchQ and WT.
3.2. Optical characteristics of the chlorosomes
Optical responses, such as UV-Vis absorption and circular dichroism (CD), have been measured for chlorosomes of both WT and bchQ. For WT chlorosomes, absorption, linear dichroism and CD spectra have been collected before from preparations with variable heterogeneity and homolog composition.8 In particular the CD appears to depend strongly on the composition. We therefore performed independent measurements for the WT chlorosomes grown in well-controlled high light conditions and found the measured spectra to reproduce well between preparations.Chlorosomes are quasi one-dimensional, and due to the syn–anti parallel stacking of the BChl c molecules there are both positive and negative couplings.44 As a result, the Qy absorbance bands of BChl c were observed at 750 nm for the WT and 735 nm for the bchQ. The overall shift towards longer wavelength in both WT and bchQ in comparison to monomeric BChl c in solution originates from strong excitonic coupling that leads to excitonic delocalization.11,17
Among UV and CD, it has been reported that CD is sensitive to chirality.20,24,45 We find psi shaped bands featuring a negative/positive/negative (−/+/−) signature in the CD spectra of both chlorosomes. Yet, the details clearly show a difference: for instance, the very intense negative band in the 750–800 region for the WT is almost absent for bchQ. In addition to this, the bands for bchQ are shifted in absolute terms and have different rotational strengths relative to WT (Fig. 6).
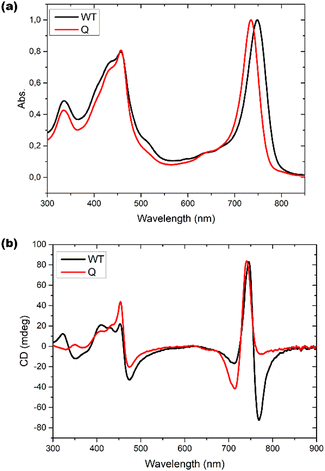 | ||
| Fig. 6 (a) UV-Vis absorption spectra and (b) CD spectra for WT (black) and bchQ (red), respectively, measured at room temperature. | ||
From the absorption spectra, one may identify a clear 15 nm red shift in the absorption maxima of chlorosomes of WT compared to the bchQ mutant, from 750 nm for the WT to 735 nm for the bchQ mutant. It has been noted that a small change in the absorption maxima from 750 nm to a lower wavelength can result in the attenuation of the longest wavelength negative band while strengthening the band at 720 nm.46 In the current context, it is noticeable that the absorption maximum of bchQ chlorosomes is at approximately 735 nm, whereas it is around 750 nm in WT chlorosomes.
3.3. Structural dynamics of bchQ chlorosomes in comparison to WT
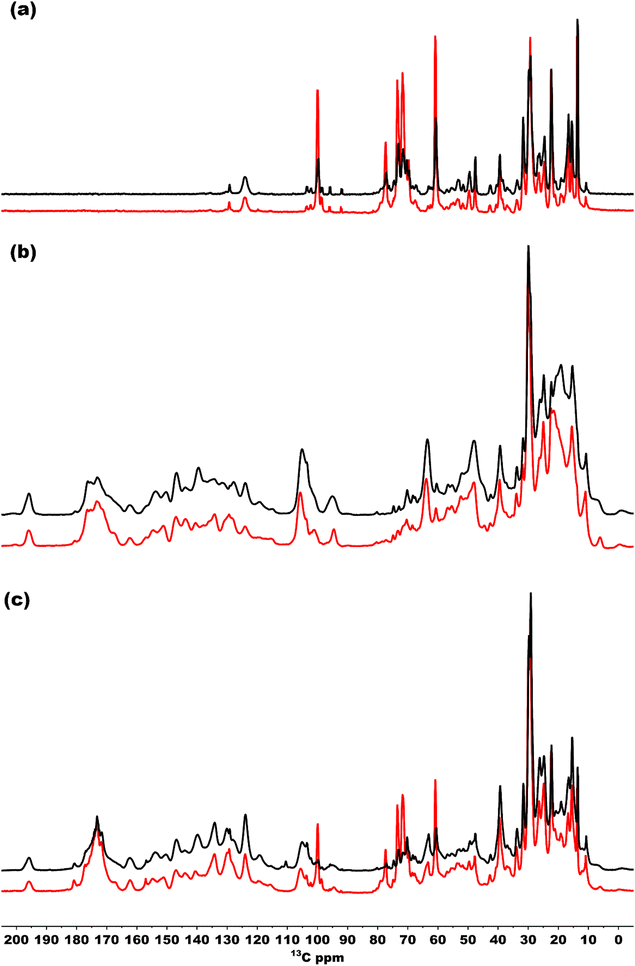
To also study the structural dynamics features in our sample, we employed a dynamic spectral editing (DYSE) approach. We performed three distinct types of experiments: direct polarization (DP), cross polarization (CP) and Insensitive nucleus enhanced by polarization transfer (INEPT).32 DP provides all the 13C signals present in the sample, and it is only moderately affected by mobility in the system whereas CP and INEPT act as dynamic filters. CP probes motion around the millisecond time scale and is most effective if the system is rigid.47 When there is significant isotropic motion in the system, dipolar couplings are averaged to zero, making CP inefficient, resulting in CP acting as a dynamic filter to probe rigid parts of the system. On the other hand, INEPT is effective only if the coherence lifetimes (T2) of carbons are long. Because this is not the case for rigid solids, INEPT selectively probes dynamic phenomena within the system.32,48Fig. 7(a) shows a comparison of the INEPT spectra, highlighting the flexible domains in the system for the bchQ (red) and WT (black) chlorosomes. The spectra for the bchQ chlorosomes show increased signal intensity from galactolipids that resonate between 65–80 ppm and at 100 ppm. Each BChl contains fifty carbon atoms, out of which sixteen are quaternary carbons. Quaternary carbons which resonate above 130 ppm do not show any signal in the INEPT. The signals originating from the tail 13C resonances are F1-CH2, F4-CH2, F5-CH2, F10-CH, F31-CH3, and side chains 17-CH, 18-CH, 171-CH2, 172-CH2, 32-C, 21-C, 71-C, 81-C, 82-CH3. The table with the position and chemical shifts is given in the ESI,† Table S3. INEPT-enhanced signals may appear less intense in CP, and the opposite may also occur, see Fig. S1 (ESI†). This observation suggests the existence of distinct subpopulations of resonances in the sample, each with varying mobilities.49
Fig. 7(b) shows the CP spectra of bchQ (red) and WT (black) chlorosomes. The 13C-5 signal at 102 ppm of the major fraction is better resolved for bchQ in comparison to WT chlorosomes. The CP signals for the rigid 13C resonances resolved in the spectra are 131-C, 173-C, 14-C, 16-C, 1-C, 6-C, 9-C, 4-C, 3-C, 2-C, 10-C, 15-C, 5-CH, 31-CH, F1-CH2, 17-CH, 18-CH, 171-CH2, 172-CH2, F8-CH2, F5-CH2, 32-CH3, F31-CH3, 21-CH2, 71(II)-CH3. The 20-C, 15-C, and 5-C(II) methine signals overlap with the 10-C signal. The signals located in the aliphatic region in the CP spectra are broad, which can be attributed to sample heterogeneity.
Fig. 7(c) presents an overlaid DP spectrum of bchQ (red) and WT (black) chlorosomes showing signals for all the 13C resonances present in the sample irrespective of the mobility. The signal for bchQ shows a significant increase in intensity around 60–80 ppm and 100 ppm. Strong signals are detected in these regions from the sugar headgroups of the galactolipids and the anomeric carbons at 100 ppm surrounding the chlorosomes.15 The increase in relative intensity for these signals for chlorosomes from the bchQ mutant indicates that there may be an increase of the lipid to BChl ratio for bchQ compared to the WT, which would be in line with smaller chlorosomes in bchQ compared to WT observed with TEM.23
4. Discussion
By combining solid-state NMR, cryo-EM, and optical methods, we were able to retrieve information about chlorosomes of the bchQ mutant that is usually gathered separately from bacteria that may have been grown under varying conditions. Moreover, we made sure that we reproduced known results for the WT chlorosomes, a system that serves as a reference, using the same procedures. Solid-state NMR is a widely used technique for resolving structure and dynamics, for systems such as proteins, amyloids, supramolecular aggregates, polymers.50–55 We employed the two-dimensional 13C–13C RFDR, dipolar hCH to get the 13C and 1H assignments. CHHC spectra were used to probe the distance constraints. It showed 1H–1H distances of less than 5 Å. This agrees well with the proton transfer range previously established for this experiment. The data reveals the presence of parallel stacking motifs, as seen also for the WT chlorosomes.1D solid-state NMR experiments CP, INEPT, and DP were also applied to provide insight into the global dynamics properties. Recent progress on MAS NMR has demonstrated the importance of using both dipolar and J-based pulse sequences for probing rigid and dynamic components in the system and in turn elucidating the structure and dynamics of a system under study.32,56 On a higher structural level, cryo-EM can be employed to obtain insight into molecular packing and supra-molecular structuring.26,27,57 As chlorosomes self-assemble into an array of stacks without incorporating proteins into the stacks, this makes chlorosomes a less complex system to study the relationship between structure and function compared to their protein-pigment-based counterparts. On the other hand, chlorosomes are intrinsically disordered systems, featuring both static and dynamic disorder,28 and the fine structure of chlorosomes has long eluded our characterisation efforts. Comparing the composition of WT and bchQ chlorosomes, i.e., the degree of static disorder, the WT chlorosome is considerably more heterogenous, with variable degrees of methylation at 82-C and 121-C, see Fig. S7 (ESI†). Yet, one may conclude from the 13C–13C RFDR and CHHC data that bchQ and WT chlorosomes both exhibit alternate parallel stacking of syn–anti monomers. On the other hand, the optical response measured for the two chlorosome types show remarkable differences. Because of the syn–anti parallel stacking, both are redshifted by 60–80 nm compared to monomeric BChl c in solution. The absorption spectra of bchQ showed a narrower and 15 nm blue shifted Qy band compared to WT. The 15-nm red-shift for WT chlorosomes compared to bchQ mutant chlorosomes suggests that WT has better potential for light harvesting because WT has a larger window to utilize light in the far-red region that can be utilized for light harvesting.23 The red shift of 15 nm in the absorption maximum for the WT in comparison to bchQ chlorosomes can be attributed to several factors, including compositional differences between WT and bchQ, or total number of BChl molecules in the assembly, as discussed in the literature.23 Also, chirality can provide a contribution. Model calculations show that a different overall chirality can shift the position of the absorption maximum by as much as 7 nm.20 The red-shift is also reflected in the corresponding CD spectra. Apparently, an increased number of homologs, e.g., for the WT, gives rise to the growth of larger chlorosomes.13
Because CD spectra are sensitive to supramolecular chirality, one may use this relation for determining chirality. The internal chirality of a molecule results in psi shaped bands in the CD spectra of large chiral aggregates.58 For chlorosomes, four types of CD bands were reported: type 1, positive negative (+/−), type II, negative positive (−/+), and two mixed types, negative positive negative (−/+/−) and positive negative positive (+/−/+).8 This signature also changes for tubes that are very short compared to the size of natural chlorosomes.59 Previously, Li et al. developed a computational procedure that directly relates the signature of the CD signal to the chiral angle. It shows that each signature can be attributed to regions of chiral angles, albeit that domain boundaries are affected by spectral broadening.24 Moreover, the CD signals undergo a sign inversion if the direction of rolling is switched, i.e., when the inside and outside of a tube are interchanged. While the CD spectra of chlorosomes for WT and the bchQ mutant exhibit rather similar signatures, the details differ significantly. Combined with layer line information, the clear −/+/− bands for the WT narrow down the number of commensurate chiral angles to a few distinct values, including a chiral angle of 112, see Fig. S5 (ESI†).20 On the other hand, the reduction of the compositional heterogeneity for bchQ chlorosomes relative to WT induces a change to a −/+ type pattern, since the negative band at ca. 720 strongly increases relative to WT, while the strong negative band between 750–800 nm for WT virtually disappears for the bchQ chlorosomes with predominantly 95% [8Et, 12Et]-BChl c. This change in the signature reflects a change in the angle between the stacks and the tube axis for bchQ relative to WT. A chiral angle of 90° is both relatively close to the proposed values of 112° for WT and consistent with this signature.
Although in this study sample variability has been minimized via improved preparation protocols, other effects on the CD spectra collected from ensembles of chlorosomes, in particular a varying signal with length in the lower size range, has long been considered to seriously challenge the direct interpretation of optical information for ensembles of chlorosomes.60,61 Moreover, calculations indicate that CD spectra for model chlorosomes become length-independent beyond 60 nm, which sets a more precise limit for this relation.24 We note that measured dimensions for both WT and bchQ chlorosomes are greater than 90 nm according to TEM, thereby sufficiently exceeding this theoretical lower limit.23
As discussed, the observation of layer lines in the Fourier transform cryo-EM data of bchQ chlorosomes is crucial for their structural characterization, as it corroborates evidence for their helical nature. The layer line position in reciprocal space directly relates to the distance between subunits along the direction of the tube axis in real space.62 A repeat of 1.25 nm was previously identified for WT, meaning that the repeat of 1.49 nm determined for bchQ, reflects a significant difference in BChl stacking (Fig. 8).
Using a geometrical analysis developed by Li et al.20,24 This measured layer line position for the bchQ mutant relates to chiral angles δ = 57°, 89°, 90°, 123°, 160°, 172° depending on the chiral family that one considers, see Fig. S6 (ESI†), with the direction of rolling making no difference. Among these, only four angles, namely δ = 57°, 90°, 123° and 160°, also match the signature of the CD spectra. We note that δ = 90° gives rise to the previously proposed stacking mode parallel to the tube axis.26,27 It is important to note that although CD lacks the precision to resolve pigment arrangements, it does provide information about the interaction of electronic transition dipoles within pigments. Small changes may lead to a shift of absorption maximum and alter the position of the negative and positive CD bands.25,46 Combining data from different characterization methods, i.e. CD and cryo-EM, enables us to narrow down commensurate chiral angles, using the protocol outlined by Li et al.20,24 To compare beyond the feature of the layer line, we performed Fourier analysis on an atomically resolved chlorosome model for the commensurate chiral angles associated with the reverse rolling direction, which is the one associated with δ = 112° proposed for the WT chlorosomes. Direct comparison of simulated scattering features indeed shows distinct similarities for δ = 90° (Fig. 5c and d), while matching to the other angles 57°, 123°, 160° is difficult. Since the CD signals for δ = 90° (bchQ) and δ = 112° (WT) are both from the helical H(0,1) family, it is tempting to conclude that the two distinct chiral angles explain the available information for the two chlorosome types. The small shoulder around 750–800 in the CD spectrum for the bchQ chlorosomes is then due to the remaining 5% methylation at C12 and heterogeneity within the system, including distributions in tube lengths. The role of overall chirality of the BChl stacks on the particulars of exciton transfer was recently evaluated computationally.25
A global perspective on the dynamics was obtained by performing one-dimensional NMR spectroscopy. For the spectra acquired using direct polarization, the differences between bchQ and WT chlorosomes are primarily in the region between 65–80 ppm, which corresponds to galactosyl lipid groups present in the system. It appears that bchQ contains a larger amount of lipids than WT. This is due to the fact that chlorosomes are smaller in size as shown by Gomez Maqueo Chew et al. using Transmission Electron Microscopy (TEM) images.23 This smaller size in chlorosomes leads to increased lipid to BChl ratio in bchQ compared to WT.63
Most of the NMR signals of BChl c appear in the CP-based spectrum for both WT and bchQ chlorosomes. However, the signals for chlorosomes from the bchQ mutant are better resolved compared to WT. This confirms that the BChl c aggregates in chlorosomes from the bchQ mutant are more ordered and homogenous because of less methylation and the presence of a single homolog.23 The INEPT works like a solution state experiment which gives signals if there is through bond coupling and probes flexible regions in the system. The INEPT spectra show a major increase in signal intensity of the bchQ chlorosomes for the lipid region and the fact that both WT and bchQ chlorosomes samples show signals in INEPT around that region, points simply to increased lipids to BChl ratio present in chlorosomes of the bchQ mutant. The other signals observed in INEPT, which belong to side groups and tails, show similar dynamic trends. Overall, we may conclude that chlorosomes of the bchQ mutant and WT display equivalent dynamical properties. This suggests the idea that chlorosomes can be considered as plastic crystals. In this model, the rigid macrocycle core is crystalline while the tails attached to it are dynamic. However, a property that characterizes plastic crystals – rotational mode – has not yet been observed. This is because any distinguishing dynamics related to pigment rotation would be too fast for our current NMR setup to detect.20
5. Conclusions
By modifying the BChl biosynthetic pathway, a mutant chlorosome with reduced compositional and structural heterogeneity compared to WT has been produced.23 The bchQ mutant, containing 95% [8Et, 12Et], closely matches the entirely homogeneous setup in existing molecular dynamics (MD) simulations, setting the stage for a more thorough understanding of the structure–function properties of chlorosomes through MAS NMR, cryo-EM, and optical data. Proton and carbon chemical shift assignments have been obtained using 2D RFDR and dipolar hCH experiments. Distance constraints obtained from CHHC spectra showed the same alternate parallel stacking of syn–anti monomers in bchQ as obtained earlier for WT chlorosomes. cryo-EM data revealed the characteristic tube chirality of chlorosomes, but with an axial repeat of 1.49 nm and a separation of 2.1 nm between tubes. Optical data was also obtained for chlorosomes from the bchQ mutant. Apart from showing that the absorption and CD spectra are substantially different from the WT spectra, in combination with cryo-EM data they provide us the means for further narrowing down matching chiral angles for bchQ.Our measurements reveal differences between bchQ and WT chlorosomes. On the smallest molecular scale, the unit cell containing a syn–anti dimer is conserved between the WT and the bchQ mutant. The modification of the composition upon mutation with limited side chain heterogeneity leads to a blue-shifted absorption maximum for the bchQ chlorosomes, and a significantly different layer line position and CD spectra type. The increased molecular homogeneity results in better resolved half-bandwidth in the absorption spectra, which may explain the need for homolog heterogeneity in WT chlorosomes.
The protocol developed in this study is applicable to artificial systems with tubes or multistart helices, to extract the overall chirality of the assemblies formed by synthetic molecules. The syn–anti parallel stacking of BChl in chlorosomes26,27 is known to be different from the stacking in most artificial analogues, in which the farnesyl chain is replaced.64 Molecular modifications will most often lead to antiparallel packing.65 Very recently also parallel stacked analogs have been presented, albeit that bulky side chains instead of farnesyl chain change the overall morphology.64 It can be noted, however, that for parallel stacks and sheets, chirality does not necessarily translate into functional information, as exemplified by the conservation of function for different chiralities in chlorosomes.
The main purpose of our study is to resolve a structure for the primary homolog that was consistently used in modelling studies, tacitly assuming there would be no difference between a mixture of homologs and a pure single ethyl–ethyl homolog. The further we progress in drawing conclusions from the modelling, the more urgent it is to have a structural basis as close as possible to the models. The present study fulfills that need. The main outcomes of this study, which also support earlier findings, indicate that the differences between bchQ and WT are small (Fig. 8). Earlier results indicate that the effects of differences in chirality on exciton transfer are minor.25 Hence the specific chirality is not critical for the function.
With MAS NMR, it has been observed that chlorosomes exhibit significant dynamics despite the presence of large supramolecular aggregates. We observed dynamics in the side chain and tail carbon resonances, which provided a comprehensive view of the NMR time scale dynamics. Our data are in line with the plastic crystalline character proposed by Li et al.20 Which supports the idea that the macrocycle core is crystalline but there is a fast rotational motion in the ps time scale which couples to the exciton energy levels, promoting long-range energy transfer by modulating energy levels and overlap between exciton states. This spectral information is influenced by local fluctuations and should be further investigated in the future.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This publication is part of the project “The molecular mechanism of long-range exciton transfer in chiral self-assembled supramolecular matrices” (with project number 715.018.001) of the 29th research program NWO TOP which is financed by the Dutch Research Council (NWO). The high-field NMR experiments and SB were supported by uNMR-NL, the National Roadmap Large-Scale NMR Facility of the Netherlands (NWO grant 184.032.207) and the uNMR-NL grid (NWO grant 184.035.002). We extend our sincere acknowledgement to the cryo-EM facility Leiden (NeCEN). Studies in the laboratories of D.A.B were supported by grant DE-FG02-94ER20137 from the Photosynthetic Systems Program, Division of Chemical Sciences, Geosciences, and Biosciences (CSGB), Office of Basic Energy Sciences of the U. S. Department of Energy.Notes and references
- T. M. Wahlund, C. R. Woese, R. W. Castenholz and M. T. Madigan, Arch. Microbiol., 1991, 156, 81–90 CrossRef CAS.
- J. T. Beatty, J. Overmann, M. T. Lince, A. K. Manske, A. S. Lang, R. E. Blankenship, C. L. Van Dover, T. A. Martinson and F. G. Plumley, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 9306–9310 CrossRef CAS PubMed.
- L. Andrew Staehelin, J. R. Golecki and G. Drews, Biochim. Biophys. Acta, Bioenerg., 1980, 589, 30–45 CrossRef PubMed.
- J. Dostal and D. Zigmantas, J. Am. Chem. Soc., 2012, 7 Search PubMed.
- G. T. Oostergetel, H. van Amerongen and E. J. Boekema, Photosynth. Res., 2010, 104, 245–255 CrossRef CAS PubMed.
- C. M. Borrego and L. J. Garcia-Gil, Photosynth. Res., 1995, 45, 21–30 CrossRef CAS PubMed.
- C. M. Borrego, P. D. Gerola, M. Miller and R. P. Cox, Photosynth. Res., 1999, 59, 159–166 CrossRef CAS.
- A. R. Holzwarth, K. Griebenow and K. Schaffner, J. Photochem. Photobiol. Chem., 1992, 65, 61–71 CrossRef CAS.
- J. Dostál, J. Pšenčík and D. Zigmantas, Nat. Chem., 2016, 8, 705–710 CrossRef PubMed.
- N. C. M. Magdaong, D. M. Niedzwiedzki, R. G. Saer, C. Goodson and R. E. Blankenship, Biochim. Biophys. Acta, Bioenerg., 2018, 1859, 1180–1190 CrossRef CAS PubMed.
- S. K. Frehan, L. Dsouza, X. Li, V. Eríc, T. L. C. Jansen, G. Mul, A. R. Holzwarth, F. Buda, G. J. A. Sevink, H. J. M. de Groot and A. Huijser, J. Phys. Chem. B, 2023, 127, 7581–7589 CrossRef CAS PubMed.
- N.-U. Frigaard and D. A. Bryant, in Complex Intracellular Structures in Prokaryotes, ed. J. M. Shively, Springer Berlin Heidelberg, Berlin, Heidelberg, 2006, vol. 2, pp. 79–114 Search PubMed.
- N.-U. Frigaard and D. A. Bryant, Arch. Microbiol., 2004, 182, 265–276 CrossRef CAS PubMed.
- B.-J. van Rossum, D. B. Steensgaard, F. M. Mulder, G. J. Boender, K. Schaffner, A. R. Holzwarth and H. J. M. de Groot, Biochemistry, 2001, 40, 1587–1595 CrossRef CAS PubMed.
- T. S. Balaban, A. R. Holzwarth, K. Schaffner, G.-J. Boender and H. J. M. de Groot, Biochemistry, 1995, 34, 15259–15266 CrossRef CAS PubMed.
- X. Li, F. Buda, H. J. M. de Groot and G. J. A. Sevink, J. Phys. Chem. C, 2018, 122, 14877–14888 CrossRef CAS PubMed.
- V. Erić, X. Li, L. Dsouza, S. K. Frehan, A. Huijser, A. R. Holzwarth, F. Buda, G. J. A. Sevink, H. J. M. de Groot and T. L. C. Jansen, J. Phys. Chem. B, 2023, 127, 1097–1109 CrossRef PubMed.
- J. Pšenčík, T. Polívka, P. Němec, J. Dian, J. Kudrna, P. Malý and J. Hála, J. Phys. Chem. A, 1998, 102, 4392–4398 CrossRef.
- G. A. Montaño, B. P. Bowen, J. T. LaBelle, N. W. Woodbury, V. B. Pizziconi and R. E. Blankenship, Biophys. J., 2003, 85, 2560–2565 CrossRef PubMed.
- X. Li, F. Buda, H. J. M. De Groot and G. J. A. Sevink, iScience, 2022, 25, 103618 CrossRef CAS PubMed.
- A. Pandit, K. Ocakoglu, F. Buda, T. van Marle, A. R. Holzwarth and H. J. M. de Groot, J. Phys. Chem. B, 2013, 117, 11292–11298 CrossRef CAS PubMed.
- J. L. Thweatt, D. P. Canniffe and D. A. Bryant, Adv. Bot. Res., 2019, 90, 35–89 CAS.
- A. G. M. Chew, N.-U. Frigaard and D. A. Bryant, J. Bacteriol., 2007, 189, 6176–6184 CrossRef CAS PubMed.
- X. Li, F. Buda, H. J. M. de Groot and G. J. Agur Sevink, J. Phys. Chem. C, 2019, 123, 16462–16478 CrossRef CAS.
- V. Erić, J. L. Castro, X. Li, L. Dsouza, S. K. Frehan, A. Huijser, A. R. Holzwarth, F. Buda, G. J. A. Sevink, H. J. M. de Groot and T. L. C. Jansen, J. Phys. Chem. B, 2023, 127, 7487–7496 CrossRef PubMed.
- S. Ganapathy, G. T. Oostergetel, P. K. Wawrzyniak, M. Reus, A. Gomez Maqueo Chew, F. Buda, E. J. Boekema, D. A. Bryant, A. R. Holzwarth and H. J. M. de Groot, Proc. Natl. Acad. Sci. U.S. A., 2009, 106, 8525–8530 CrossRef CAS PubMed.
- S. Ganapathy, G. T. Oostergetel, M. Reus, Y. Tsukatani, A. Gomez Maqueo Chew, F. Buda, D. A. Bryant, A. R. Holzwarth and H. J. M. de Groot, Biochemistry, 2012, 51, 4488–4498 CrossRef CAS PubMed.
- L. M. Günther, M. Jendrny, E. A. Bloemsma, M. Tank, G. T. Oostergetel, D. A. Bryant, J. Knoester and J. Köhler, J. Phys. Chem. B, 2016, 120, 5367–5376 CrossRef PubMed.
- L. M. Günther, A. Löhner, C. Reiher, T. Kunsel, T. L. C. Jansen, M. Tank, D. A. Bryant, J. Knoester and J. Köhler, J. Phys. Chem. B, 2018, 122, 6712–6723 CrossRef PubMed.
- Y. Tian, R. Camacho, D. Thomsson, M. Reus, A. R. Holzwarth and I. G. Scheblykin, J. Am. Chem. Soc., 2011, 133, 17192–17199 CrossRef CAS PubMed.
- G. S. Harbison and J. Herzfeld, Biochemistry, 1983, 22, 1–5 CrossRef CAS PubMed.
- I. Matlahov and P. C. A. van der Wel, Methods, 2018, 148, 123–135 CrossRef CAS PubMed.
- A. E. Bennett, R. G. Griffin, J. H. Ok and S. Vega, J. Chem. Phys., 1992, 96, 8624–8627 CrossRef CAS.
- A. Lange, S. Luca and M. Baldus, J. Am. Chem. Soc., 2002, 124, 9704–9705 CrossRef CAS PubMed.
- I. De Boer, L. Bosman, J. Raap, H. Oschkinat and H. J. M. De Groot, J. Magn. Reson., 2002, 157, 286–291 CrossRef CAS PubMed.
- K. R. Thurber and R. Tycko, J. Magn. Reson., 2009, 196, 84–87 CrossRef CAS PubMed.
- E. K. Paulson, C. R. Morcombe, V. Gaponenko, B. Dancheck, R. A. Byrd and K. W. Zilm, J. Am. Chem. Soc., 2003, 125, 14222–14223 CrossRef CAS PubMed.
- E. Barbet-Massin, A. J. Pell, J. S. Retel, L. B. Andreas, K. Jaudzems, W. T. Franks, A. J. Nieuwkoop, M. Hiller, V. Higman, P. Guerry, A. Bertarello, M. J. Knight, M. Felletti, T. Le Marchand, S. Kotelovica, I. Akopjana, K. Tars, M. Stoppini, V. Bellotti, M. Bolognesi, S. Ricagno, J. J. Chou, R. G. Griffin, H. Oschkinat, A. Lesage, L. Emsley, T. Herrmann and G. Pintacuda, J. Am. Chem. Soc., 2014, 136, 12489–12497 CrossRef CAS PubMed.
- M. Weingarth, P. Tekely and G. Bodenhausen, Chem. Phys. Lett., 2008, 466, 247–251 CrossRef CAS.
- D. H. Zhou and C. M. Rienstra, J. Magn. Reson., 2008, 192, 167–172 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- R. J. Abraham and K. M. Smith, J. Am. Chem. Soc., 1983, 105, 5734–5741 CrossRef CAS.
- C. M. Quinn, M. Wang and T. Polenova, in Protein NMR, ed. R. Ghose, Springer New York, New York, NY, 2018, vol. 1688, pp. 1–35 Search PubMed.
- J. Knoester, Int. J. Photoenergy, 2006, 1–10 Search PubMed.
- G. Garab and H. van Amerongen, Photosynth. Res., 2009, 101, 135–146 CrossRef CAS PubMed.
- D. C. Brune, P. D. Gerola and J. M. Olson, Photosynth. Res., 1990, 24, 253–263 CrossRef CAS PubMed.
- F. Azadi-Chegeni, C. Schiphorst and A. Pandit, Photosynth. Res., 2018, 135, 227–237 CrossRef CAS PubMed.
- F. Azadi Chegeni, G. Perin, K. B. Sai Sankar Gupta, D. Simionato, T. Morosinotto and A. Pandit, Biochim. Biophys. Acta, Bioenerg., 2016, 1857, 1849–1859 CrossRef CAS PubMed.
- H.-K. Lin, J. C. Boatz, I. E. Krabbendam, R. Kodali, Z. Hou, R. Wetzel, A. M. Dolga, M. A. Poirier and P. C. A. van der Wel, Nat. Commun., 2017, 8, 15462 CrossRef CAS PubMed.
- M. Weingarth and M. Baldus, Acc. Chem. Res., 2013, 46, 2037–2046 CrossRef CAS PubMed.
- P. C. A. van der Wel, Emerg. Top. Life Sci., 2018, 2, 57–67 CrossRef CAS PubMed.
- B. Reif, S. E. Ashbrook, L. Emsley and M. Hong, Nat. Rev. Methods Primers, 2021, 1, 2 CrossRef CAS PubMed.
- P. Schanda and M. Ernst, Prog. Nucl. Magn. Reson. Spectrosc., 2016, 96, 1–46 CrossRef CAS PubMed.
- F. Castellani, B. van Rossum, A. Diehl, M. Schubert, K. Rehbein and H. Oschkinat, Nature, 2002, 420, 99–102 CrossRef PubMed.
- A. Abragam, The Principles of Nuclear Magnetism, Clarendon Press, 1983 Search PubMed.
- O. C. Andronesi, S. Becker, K. Seidel, H. Heise, H. S. Young and M. Baldus, J. Am. Chem. Soc., 2005, 127, 12965–12974 CrossRef CAS PubMed.
- A. Alia, F. Buda, H. J. M. de Groot and J. Matysik, Annu. Rev. Biophys., 2013, 42, 675–699 CrossRef CAS PubMed.
- Y. Miloslavina, P. H. Lambrev, T. Jávorfi, Z. Várkonyi, V. Karlický, J. S. Wall, G. Hind and G. Garab, Photosynth. Res., 2012, 111, 29–39 CrossRef CAS PubMed.
- C. Didraga, J. A. Klugkist and J. Knoester, J. Phys. Chem. B, 2002, 106, 11474–11486 CrossRef CAS.
- C. Didraga and J. Knoester, J. Lumin., 2003, 102–103, 60–66 CrossRef CAS.
- T. Kunsel, L. M. Günther, J. Köhler, T. L. C. Jansen and J. Knoester, J. Chem. Phys., 2021, 155, 124310 CrossRef CAS PubMed.
- G. T. Oostergetel, M. Reus, A. Gomez Maqueo Chew, D. A. Bryant, E. J. Boekema and A. R. Holzwarth, FEBS Lett., 2007, 581, 5435–5439 CrossRef CAS PubMed.
- Y. Tsukatani, J. Harada, T. Mizoguchi and H. Tamiaki, Photochem. Photobiol. Sci., 2013, 12, 2195 CrossRef CAS PubMed.
- A. Löhner, T. Kunsel, M. I. S. Röhr, T. L. C. Jansen, S. Sengupta, F. Würthner, J. Knoester and J. Köhler, J. Phys. Chem. Lett., 2019, 10, 2715–2724 CrossRef PubMed.
- S. Ganapathy, S. Sengupta, P. K. Wawrzyniak, V. Huber, F. Buda, U. Baumeister, F. Würthner and H. J. M. de Groot, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11472–11477 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cp00221k |
| This journal is © the Owner Societies 2024 |