Open Access Article
Open Access ArticlePhotophysics of a nucleic acid–protein crosslinking model strongly depends on solvation dynamics: an experimental and theoretical study†
Gabriele
Iuzzolino
 ab,
Fulvio
Perrella
ab,
Fulvio
Perrella
 a,
Mohammadhassan
Valadan
a,
Mohammadhassan
Valadan
 cd,
Alessio
Petrone
cd,
Alessio
Petrone
 abd,
Carlo
Altucci
abd,
Carlo
Altucci
 cde and
Nadia
Rega
cde and
Nadia
Rega
 *abd
*abd
aScuola Superiore Meridionale, Largo San Marcellino 10, Napoli I-80138, Italy
bDipartimento di Scienze Chimiche, Università degli Studi di Napoli Federico II, via Cintia 21, Napoli I-80126, Italy. E-mail: nadia.rega@unina.it
cDipartimento di Scienze Biomediche Avanzate, Università degli Studi di Napoli Federico II, via Pansini 5, Napoli I-80131, Italy
dIstituto Nazionale di Fisica Nucleare, Unità di Napoli, via Cintia 21, Napoli I-80126, Italy
eIstituto di Scienze Applicate e Sistemi Intelligenti “Eduardo Caianiello”, URT UNINA, via Cintia 21, Napoli I-80126, Italy
First published on 15th March 2024
Abstract
We present a combined experimental and theoretical study of the photophysics of 5-benzyluracil (5BU) in methanol, which is a model system for interactions between nucleic acids and proteins. A molecular dynamics study of 5BU in solution through efficient DFT-based hybrid ab initio potentials revealed a remarkable conformational flexibility – allowing the population of two main conformers – as well as specific solute–solvent interactions, which both appear as relevant factors for the observed 5BU optical absorption properties. The simulated absorption spectrum, calculated on such an ensemble, enabled a molecular interpretation of the experimental UV-Vis lowest energy band, which is also involved in the induced photo-reactivity upon irradiation. In particular, the first two excited states (mainly involving the uracil moiety) both contribute to the 5BU lowest energy absorption. Moreover, as a key finding, the nature and brightness of such electronic transitions are strongly influenced by 5BU conformation and the microsolvation of its heteroatoms.
1 Introduction
Interactions between nucleic acids and proteins on the molecular scale govern the most basic processes of life, from DNA replication and repair to protein expression and synthesis.1–11 However, it is challenging to isolate nucleoprotein complexes with null or small perturbations, where these last ones in their turn might disrupt the very nature of these complexes. Moreover, very intricate, and indeed not fully understood, is the role of light (i.e. UV irradiation) with protein–nucleic acid complexes. Light can cause the crosslinking of the protein to the nucleic acid, resulting in a covalent modification that can drastically change (usually for the worst) the biological roles of such compounds.12–15 Thus, it is very important to understand how light interacts with nucleoprotein complexes, starting from their optical properties and moving towards the study of the photo-induced crosslinking reaction mechanism.The complete comprehension of protein–nucleic acid interactions and their interplay with light has been limited because, upon irradiation, extensive protein and nucleic acid damage is often observed and the involved reactions are not very efficient. Advanced spectroscopic techniques and model compounds have been developed (and sometimes combined together) to better study this matter. Nucleic acid–protein laser-based photo-crosslinking is, for example, a promising technique to isolate nucleoprotein complexes in their biological conformation. It employs UV femtosecond-stimulated lasers to induce in vivo the formation of covalent bonds – named crosslinks – between a nucleobase and an amino acid residue. In this way, it is easier to stabilize the biological conformation of the complex and allow it to survive the following isolation. In addition, the employment of ultra-short irradiation times minimizes side reactions and allows characterisation of even transient interactions.16–21
The resulting complex reaction mixtures and conditions also make it difficult to identify specific aminoacid/base cross-links.22,23 A common proposed strategy is to attach a pyrimidine base to tyrosine, phenylalanine, and tryptophan side chains, using a short linker to mimic the proximity and orientation in nucleoprotein complexes.24 Furthermore, in this study, we focus on the photophysics of 5-benzyluracil (5BU, see Fig. 1). Such a compound is useful to investigate the photoinduced crosslinking reaction that leads to the formation of 1,2-indaneuracil, allowing to model the photo-addition of phenylalanine to thymine, with the additional advantage of having high reaction yields.24 This reaction has been already shown to be possible in methanol, by photoexcitation between 254 and 266 nm,24–26 corresponding to the maximum of the first band of the UV-Vis spectrum.
This model compound has also been previously investigated theoretically via both wave-function and DFT based approaches, further proving that 5BU is a very good candidate as a model compound for studying the optical properties of nucleoprotein complexes and their photo-reactivity.25,27,28 The 5BU photo-cyclization pathway appears to be influenced by multiple factors: Sun et al.24 proposed a mechanism relying on electron transfer between the chromophores and proton exchange with the solvent; according to Valadan et al.,26 the absorption involves exclusively the S1 ← S0 transition, but the following reactive pathway is determined by a strong coupling among low-lying states and the reciprocal orientation of the two aromatic rings.
However, the photophysics of such a model compound is still far from being fully understood. It is still debated which one of the low-lying excited states of 5BU is responsible for absorption preceding the photoreaction, given the different results of the previous theoretical studies, relying on minimum energy structures.25–28 Indeed, gas-phase simulations27,28 considered the S2 ← S0 transition to be the only bright one; on the contrary, in the methanol solution, the absorption is either attributed entirely to the S1 ← S0 transition,26 or can involve different states depending on the level of theory.28 These different findings reflect on the hypothesized evolution of the system in the excited state, where single and multiple crossings among electronic states can be predicted. Such results can be further influenced by the role of the solvent and how this one is modelled, as well as by the thermal effects, that are not fully taken into account when few minimum energy structures are analyzed.29
In this work, we propose to accurately describe the optical absorption of 5BU in methanol solution at room temperature by including both the explicit solute–solvent interactions and the finite temperature effects with a combined computational and experimental approach. For this aim, we performed a complete study of both the conformational space accessible to the solute and the dynamics of the solute–solvent interactions. In such a way, we were able to have an exhaustive knowledge, gauging our prediction with the experiments, of the ground-state equilibrium of the system in solution at room temperature to provide molecular insights for the absorption spectrum. We collected a room temperature ab initio molecular dynamics (AIMD) trajectory using a hybrid explicit/implicit solvation scheme, combining quantum/molecular mechanics (QM/MM) and non-periodic boundary conditions (NPBCs).30–33 Then, we simulated the optical absorption from configurations extracted from the AIMD29 through linear response time-dependent DFT (LR TD-DFT).34–36 This methodology, taking into account several conformations and solute–solvent interactions, allowed us to unveil the role of different excited states in the overall 5BU observed photophysical properties.
The article is organized as follows: in Sections 2.1 and 2.2, we describe the methodology of the S0 QM/MM/continuum MD; in Sections 2.3 and 2.4, we detail, respectively, the experimental and computational setup for the study of the photophysics; Section 3.1 describes the photophysics of the system from minimum energy structures; in Section 3.2, we characterize the ground-state conformational and solvation equilibrium of the system; in Section 3.3, we describe the optical properties of 5BU as depicted by our experiment and AIMD analysis; in Section 4 we discuss these results.
2 Methods
Multilayered quantum/classical/continuum MD simulations are necessary to accurately describe the ensemble averages, microsolvation dynamics and absorption spectra of chromophores in solution.29,33,37–49 This is particularly true for 5BU in methanol because 5BU is a flexible solute with two interacting aromatic rings and four heteroatoms, while methanol is a flexible, polar and protic solvent: they thus require, at the same time, the accurate treatment of the solute degrees of freedom and the efficient description of the solvent viscosity and mobility; moreover, we have to take into account the specific, polarization and electrostatic solute–solvent interactions.33,50–562.1 Multilayered ONIOM/NPBCs partition scheme
For the aforementioned reasons, the 5BU–methanol system was simulated by a multilayered QM/MM/implicit partition, combining the N-layered integrated molecular orbital and molecular mechanics (ONIOM)57–59 extrapolative scheme with non-periodic boundary conditions.30–33,39,41,60–68 The solute, treated at the B3LYP69–72/6-31G(d,p) level of theory, was put at the center of a spherical cavity with a radius of 20 Å, including 356 methanol molecules, described using the General AMBER Force Field (GAFF) model73 (see Fig. S1 in the ESI†). Using DFT potentials appears in this context the method of choice because of their good compromise between cost and accuracy, for both organic/biological33,74–86 and inorganic/materials systems.29,87–106 The cavity radius was determined using the equilibration procedure detailed in Section 2.2. NPBCs were enforced on the cavity by the conductor-like polarizable continuum model (C-PCM) self-consistent reaction field107–110 (via the ONIOM/PCM-X111–113 formalism) and a dispersion–repulsion potential Wdisp–rep,30–33,39,41,64,66–68 specifically designed by some of us for flexible solvents.For a NVT ensemble, the constant-volume free energy A of the simulated system can be expressed as:
| A(R) = E(P,R) + W(P,R) | (1) |
| W(R) = Wdisp–rep(R) + Welec(R) + Wcav(R) | (2) |
 | (3) |
The energy of the atomistic system – E(P,R) in eqn (1) – can be expressed, according to the ONIOM scheme, as:
| E(P,R) = Emodel,QM(P,R) + Ereal,MM(R) − Emodel,MM(R) | (4) |
The combination of ONIOM and NPBCs provides a well defined, single valued and differentiable potential to perform AIMD simulations in explicit solvents. Such a hybrid solute–solvent QM/MM approach offers a good compromise between accuracy and computational cost for the equilibrium and non-equilibrium simulation of medium-large size systems in solution, including specific solute–solvent interactions.33,75,101,115–123 In fact, a fully correlated, wavefunction-based QM approach for both solute and solvent, although giving in principle a more accurate modelling of solute–solvent hydrogen bonds compared to a QM/MM scheme, is, in practice, not feasible, due to the large dimensions of the simulated system (5BU solute in a solvent box of 356 methanol molecules).
2.2 AIMD simulation
As aforementioned, we collected a ground-state AIMD trajectory of 5BU in methanol at room temperature, with the hybrid ONIOM/NPBCs partition scheme described in Section 2.1, by using a development version of the Gaussian electronic structure software package.124 We exploited the atom-centered density matrix propagation (ADMP)125–129 formalism, which avoids the density matrix convergence at each step by propagating it alongside the nuclear degrees of freedom. A (fictitious) mass is required in the density matrix equation of motion. A tensorial mass weighting scheme was employed:125 the mass of valence orbitals was set to 0.1 amu bohr2, whereas core orbitals were weighted according to the scheme described in ref. 125. The ONIOM/ADMP/NPBCs MD was carried out for 20 ps, with a time step of 0.2 fs, in a NVT ensemble. A 298 K temperature was kept constant by rescaling velocity every 1 ps. A preliminary equilibration was carried out before production AIMD simulation. 5BU was placed into a cubic box of 9582 methanol molecules. A MM Langevin dynamics study was performed (using NAMD software130) in the NpT ensemble (T = 298 K, p = 1 atm) with periodic boundary conditions, applying a Langevin piston to also keep the pressure constant.131,132 GAFF was employed: GAFF atom types were assigned to solute atoms, as well as HF/6-31G(d) RESP charges. Solvent equilibration (with the solute coordinates fixed) was first performed for 10 ns. Then solute degrees of freedom were also allowed to propagate for 1 ns. Then, solvent molecules within 20 Å from the solute center of mass were selected. An AIMD equilibration of the selected system was performed with NPBCs in the NVT ensemble for 1 ps, applying velocity rescaling every 250 fs, using the same partition scheme and level of theory of the following production simulation.2.3 Experimental details
5BU powder was purchased from Giotto Biotech S.r.l. Italia and it was dissolved in 99.9% HPLC grade methanol. A UV-Vis spectrophotometer was used to obtain an absorbance spectrum of the sample in a quartz cuvette with a 1 cm optical path length. The spectrum was acquired in the 200–700 nm range at a 5BU concentration of 10 μg mL−1 (≈50 μM), where we had already verified its linear response.2.4 UV-Vis spectrum simulation
The intensity of the electronic absorption spectrum L(ω) can be calculated (assuming a classical nuclear motion) as:39,133 | (5) |
 | (6) |
 | (7) |
This approach is able to simulate absorption spectra of systems even of significant size including thermal broadening, but neglecting the vibronic fine structure.39,133
In order to simulate the 5BU spectrum in the methanol solution, 2001 uncorrelated configurations were extracted from the AIMD trajectory at regular time intervals (10 fs). The latter was chosen as a compromise between the computational cost of excited states calculation and the accurate sampling of 5BU torsional degrees of freedom.29 The first 8 excited states were calculated for each configuration through linear response time-dependent DFT.34–36 An ONIOM hybrid scheme was used: 5BU and the four methanol molecules closest to the solvation sites (possibly forming hydrogen bonds) were modeled at the QM TD-CAM-B3LYP/6-31+G(d,p) level,69–72,134 and the remaining ones were modeled at the MM level through the GAFF (see Fig. S2 in the ESI†). In this regard, the CAM-B3LYP functional appears suitable to describe the photophysics of pyrimidines (also substituted ones)25,28,135–138 and especially their charge-transfer states. Electrostatic interactions by the bulk solvent were treated using the C-PCM implicit solvent model, placed at the boundary of the spherical solvent box.
3 Results
3.1 Optical absorption data from minimum energy structures
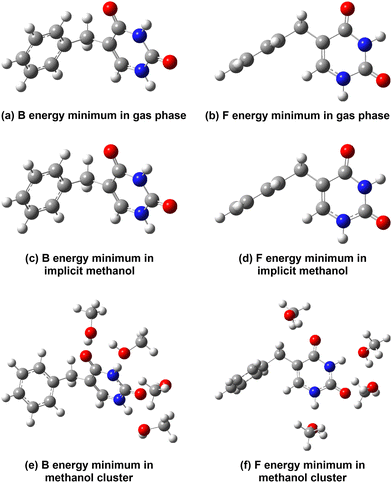
We first describe the optical properties of 5BU in methanol by adopting a static picture, i.e. by performing TD-DFT calculations on ground-state minimum energy structures, differing in 5BU geometries, solvation schemes and levels of theory. First, we chose two conformations that, according to the literature,26 are adopted by the 5BU molecule: the former has the rings arranged in an approximately orthogonal manner (flag-like structure, shortened to “F”), while the latter has the rings approximately facing each other (butterfly-like structure, shortened to “B”) – see Fig. 3. It is expected that the relative orientation of the two rings could influence the amount of orbital interaction and so the excited states character. Moreover, to evaluate the role of solvent effects, three different solvation schemes were employed: first, we simulated the isolated molecule; then, we added the effect of the methanol bulk solvent by an implicit description (C-PCM potential); finally, we included both the implicit solvent and four explicit methanol molecules, each one interacting with a solute hydrogen-bond site. All the optimizations were performed at the B3LYP/6-31G(d,p) level of theory, also employed for AIMD simulations (see also the following discussion). Structures were also optimized at the CAM-B3LYP/6-31+G(d,p) level for comparison because this density functional was previously adopted in the literature for this system.25,26 For each ground-state minimum energy structure, time-dependent calculations were performed at the TD-CAM-B3LYP/6-31+G(d,p) level, with the same solvation scheme used in the relative optimization. All calculations were performed using the Gaussian 16 software package.139The minimum energy structures optimized at the B3LYP/6-31G(d,p) level are shown in Fig. 3: the left and the right columns show the butterfly-like and the flag-like energy minima, respectively; the three rows show instead the structures optimized respectively in the gas phase (Fig. 3a and b), in the methanol treated implicitly (Fig. 3c and d), and in methanol described in a hybrid manner, i.e. by four methanol molecules and bulk implicit solvent (Fig. 3e and f). The two dihedral angles ϑ and φ mainly represent the 5BU conformation: the former is defined by the carbon atoms 5, 7, 8 and 9 and describes the rotation of the phenyl ring, while the latter is defined by the carbon atoms 6, 5, 7 and 8 and describes the rotation of the uracil ring (see Fig. 2). Their values for each B3LYP/6-31G(d,p) optimized structure are reported in Table 1. The structures optimized at the CAM-B3LYP/6-31+G(d,p) level are shown and described instead in the ESI† – please refer to Fig. S3 and Table S1.
 | ||
| Fig. 2 (a) The numbering adopted in the article. (b) The two rotations described by the dihedrals ϑ and φ. | ||
| B structure | F structure | |||
|---|---|---|---|---|
| φ | ϑ | φ | ϑ | |
| 5BU | 93.75 | −102.98 | 0.02 | −89.58 |
| 5BU/C-PCM | 96.63 | −95.78 | 0.01 | −89.80 |
| 5BU + 4MeOH/C-PCM | 107.75 | −60.69 | −21.57 | −79.94 |
In the following discussion, we compare the results of optimization of the structures at the B3LYP/6-31G(d,p) level in different solvation schemes. We also compare the results with those obtained at the CAM-B3LYP/6-31+(d,p) level, and with those reported in the literature.
Two different ground-state energy minima were actually found for each solvation scheme, in accordance with the previous works.26 The orthogonal (φ ≈ 0° and ϑ ≈ 90°, Table 1) arrangement of the two rings is retained by flag-like structures in almost all solvation models (Fig. 3b and d) and for both levels of theory (see Fig. S3b, S3d and S3f in the ESI†). A slight rotation of the uracil ring (φ = −21.57°) is observed for the B3LYP/6-31G(d,p) optimized F conformer in the methanol cluster (Fig. 3f).
The butterfly-like conformer, on the other hand, displays a more symmetric arrangement of the two rings, with both dihedral angles at around 90° (Table 1). This is consistent with the gas-phase structures optimized at different levels of theory (such as M06/cc-pVTZ and SA-CASSCF(20,14)/DZP)27,28 proposed in recent literature studies. In gas phase and implicit methanol, this symmetric arrangement is indeed highly preserved in the B3LYP/6-31G(d,p) optimized B conformer (Fig. 3a and c). However, the addition of explicit solvent molecules induces a slight phenyl ring rotation (ϑ = −60.69°, see Fig. 3e), a finding also confirmed by the CAM-B3LYP optimized B conformer with the same solvation scheme (ϑ = −56.96°, see Fig. S3e, ESI†).
We then discuss the main features of 5BU lowest energy excited states in solution, computed at the TD-CAM-B3LYP/6-31+G(d,p) level from B3LYP/6-31G(d,p) minimum energy structures. Their vertical excitation energies and oscillator strengths are reported in Table 2, while the natural transition orbital (NTO) pairs140 describing the first two excitations are displayed in the ESI† (see Section SI-4 and Fig. S4–S9). Excited states from the CAM-B3LYP/6-31+G(d,p) optimized structures were also computed for further comparison and confirmation, and reported in the ESI† (Table S2).
| Transition | V.E.E. (f) | ||
|---|---|---|---|
| 5BU | 5BU/C-PCM | 5BU + 4MeOH/C-PCM | |
| F structure | |||
| S1 ← S0 | 5.12 (0.000) | 5.16 (0.218) | 5.04 (0.226) |
| S2 ← S0 | 5.26 (0.155) | 5.31 (0.000) | 5.39 (0.005) |
| S3 ← S0 | 5.41 (0.000) | 5.42 (0.000) | 5.45 (0.003) |
| S4 ← S0 | 5.99 (0.000) | 6.04 (0.024) | 5.98 (0.010) |
| B structure | |||
| S1 ← S0 | 5.10 (0.004) | 5.09 (0.265) | 5.04 (0.294) |
| S2 ← S0 | 5.14 (0.189) | 5.28 (0.002) | 5.38 (0.001) |
| S3 ← S0 | 5.38 (0.012) | 5.39 (0.003) | 5.43 (0.003) |
| S4 ← S0 | 5.74 (0.020) | 5.84 (0.056) | 5.87 (0.066) |
For each conformer optimized in the gas phase, S2 is the only low-lying bright state (f = 0.155 and f = 0.189 in F and B conformers, respectively, see Table 2) and is dominated by a uracil Uππ* transition. In the B conformer, such transition is mixed with a charge-transfer transition from a phenyl ring π orbital to a uracil π* orbital (PπUπ*) (see Fig. S5, ESI†). In contrast, for both conformers, S1 has a dark uracil nπ* character, where the n orbital is localized on oxygen #4 (Fig. S4 ESI,† please refer to Fig. 2a for atoms labels). Interestingly, the inclusion of solvent effects (either implicitly or through cluster models) increases the energy of the dark Unπ* transition and stabilizes the bright Uππ* state. Therefore, in the methanol solution, the S1 state has a main Uππ* character (Fig. S6 and S8, ESI†) and becomes the bright state (Table 2). Conversely, the S2 state acquires a dark Unπ* character (including an implicit solvent model, Fig. S7, ESI†), or a dark Pππ* one with a minimal Unπ* contribution in a cluster/implicit solvent model (Fig. S9, ESI†), while the Unπ* character (S1 in gas phase) is shown instead by the S3 state (NTOs not shown).
In particular, specific solvation effects (included in the cluster/implicit solvent model) induce a relevant increase (0.24 eV for the B conformer) of the adiabatic S2 state energy, while S1 energy is slightly reduced. This results in an increase of the S2–S1 energy gap, as well as a decrease of the S3–S2 gap, both for the B3LYP/6-31G(d,p) and CAM-B3LYP/6-31+G(d,p) optimized structures.
For each solvation scheme, we also observe that the transitions from the B minimum are generally brighter and red-shifted with respect to the corresponding ones from the F minimum; the same conformational effect is observed for the CAM-B3LYP/6-31+G(d,p) structures.
Comparing transitions calculated on structures optimized at B3LYP/6-31G(d,p) and CAM-B3LYP/6-31+G(d,p) levels, we observe overall very similar features of the excited states, both in terms of energy and brightness. The former level of theory was also employed in ADMP/ONIOM/NPBCs ab initio molecular dynamics to sample the conformation space for the following excited states and spectrum simulations (Section 3.3).
3.2 Conformational and solvation equilibrium in the ground state
In order to better characterize and systematically explore 5BU conformational space and, in particular, other possible minimum energy structures, an energy scan in methanol with respect to ϑ and φ dihedral angles (defined in Section 3.1) was performed. In particular, the energy was evaluated in the whole domain ϑ,φ ∈ [−180°![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180°] at the B3LYP/6-31G(d,p)/C-PCM level of theory, starting the scan from the (−89.8, 0.01) optimized F conformer.
180°] at the B3LYP/6-31G(d,p)/C-PCM level of theory, starting the scan from the (−89.8, 0.01) optimized F conformer.
The total energy of the system, relative to the scan minimum energy (in the following referred to as “R.T.E”, for “relative total energy”), is reported in Fig. 4 as a function of the dihedrals ϑ and φ. Because of the C2 symmetry of the phenyl ring, the scan can be divided into two congruent halves: the upper one, where ϑ > 0, and the lower one, where ϑ < 0; we can thus restrict our analysis of the scan to the region −180° < ϑ < 0°. Three wells (R.T.E. < 1 kcal mol−1) can be recognized: (i) the well on the left, hereafter called “L”, lies approximately between −160° < ϑ < −50° and −120° < φ < −70° and its lowest point has ϑ = −89.8°, φ = −100.0° and R.T.E. = 0.118 kcal mol−1; (ii) the well at the center, hereafter named “C”, ranges approximately between −130° < ϑ < −60° and −30° < φ < 50° and its lowest point has ϑ = −89.8°, φ = 0.0° and R.T.E. = 0 kcal mol−1 (the absolute minimum of the scan); (iii) the well on the right, named “R”, lies approximately between −130° < ϑ < −10° and 70° < φ < 120° and its lowest point has ϑ = −89.8°, φ = 100.0° and R.T.E. = 0.118 kcal mol−1.
Conformers belonging to L and R wells are actually mirror images (enantiomers). Thus, the energy scan shows that 5BU in methanol has just two distinct conformers: the flag-like minimum energy structure (Fig. 3d), corresponding to the C well, and the butterfly-like minimum energy structure (Fig. 3c), located in the R (or L) well, both previously characterized in Section 3.1.
At the same time, the saddle point between the C and R wells (ϑ = −109.8°, φ = 60°) has a R.T.E. = 0.961 kcal mol−1, suggesting a low activation barrier for the interconversion of the two conformers: the ground-state equilibrium of 5BU does not appear to be dominated by rigid structures, but rather by a plethora of energetically close configurations: a dynamical analysis is thus necessary to achieve a complete understanding of the excited states, overcoming the bias associated with the study of few minimum energy structures.
A conformational analysis of the solute from ADMP/ONIOM/NPBCs MD was performed to understand the 5BU conformations explored at the equilibrium. The (ϑ,φ) two-dimensional distribution from the AIMD is plotted in Fig. 5. We clearly distinguish two regions: one on the left, approximately located at −130° < ϑ < −30°, −60° < φ < 50°, the other one on the right, approximately at −170° < ϑ < −50°, 50° < φ < 130°. These two regions correspond, respectively, to the aforementioned C and R wells of the energy scan (Fig. 4) and so to the population of the flag-like and the butterfly-like conformers. Therefore, a sufficient sampling of both 5BU distinct conformers in methanol at room temperature is provided by the collected trajectory, as well as structural fluctuations within each conformer.
Solvent structuring and 5BU microsolvation were then studied, through radial distribution functions (RDFs) between solvent oxygen atoms and 5BU solvation sites, i.e. uracil O2, O4 (as hydrogen bond acceptors) and N1, N3 (as hydrogen bond donors, please see Fig. 2a for atoms numbering). Clear 5BU–methanol specific interactions can be recognized. Looking at the O2 RDF (Fig. 6a), the first peak at 2.75 Å suggests a strong interaction with 1–2 methanol molecules (according to g(r) radial integral) in the first solvation shell. The second peak at 3.55 Å is instead actually due to the methanol molecules solvating the other three heteroatoms: the g(r) integral (between 2 and 3) is compatible with the coordination numbers of N1, N3 and O4. The O2 second solvation shell is represented by the peak at 7.25 Å, including, on an average, about 14 solvent molecules. A third shell at 11.45 Å is moreover recognized. While the O4 first solvation shell is located at the same distance as that of O2 (2.75 Å, Fig. 6b), it is instead less populated (between 0 and 1 methanol molecule). This effect can be explained by the steric hindrance of the benzyl group, closer to O4. Moreover, a reduced exchange between first and outer shells of O4 is observed, as suggested by g(r) minimum values between the peaks (0.338 for O4, 0.797 for O2), likely due to the lower exposure to the solvent. The two low peaks at 3.75 and 4.75 Å actually reflect O2, N1 and N3 solvation. The O4 second solvation shell (at 6.75 Å) includes, on average, 9–10 solvent molecules. The third shell is located instead at 11.55 Å. N1 and N3 first solvation shells are at the same average distance (2.85 Å, Fig. 6c and d, respectively) and show a comparable population (1 methanol molecule). Nevertheless, the first minimum in N3 RDF is much deeper and wider (0.295 and 0.580 g(r) minimum values for N3 and N1, respectively). This suggests that the number of solvent molecules moving between the first two shells of N3 is, on average, less than that of N1. This is likely due to the position of N3 between the two oxygen atoms, where the interplay of three solvation sites causes a stronger local structuring of the solvent. The intense second peaks at 4.95 Å include the methanol molecules solvating O2 (N1 RDF) or O2 and O4 (N3 RDF). The N3 second solvation shell is represented by the peak at 6.65 Å (Fig. 6d) and includes, on average, about 18 methanol molecules. The N1 RDF in the same region (Fig. 6c) instead appears to have already converged to 1, suggesting a lower structuration.
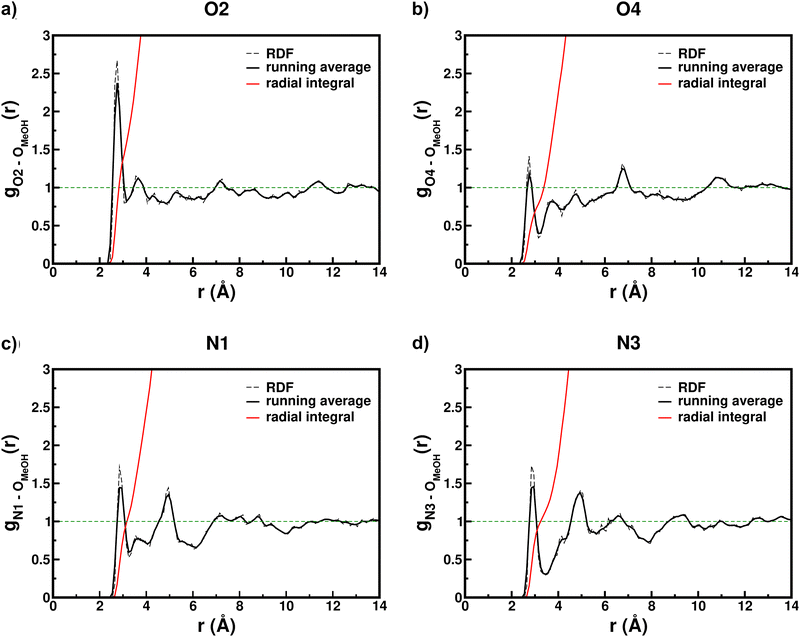 | ||
| Fig. 6 RDFs (dashed black) of methanol oxygen atoms – 5BU heteroatoms distances (please refer to Fig. 2a for atoms labels). The solid black line represents the 3-point g(r) running average and the red line represents its radial integral; the green dashed straight line is set to 1, the value of g(r) for a homogeneous distribution. | ||
In addition to the average information provided by the RDFs, we further investigated the microsolvation of 5BU heteroatoms to observe the occurrence of strong hydrogen bonds. A structural criterion was employed (based on a donor–acceptor distance ≤4 Å and hydrogen-donor–acceptor angle ≤40°). For each heteroatom and for each AIMD frame, the distance from the oxygen of the closest methanol molecule having an orientation suitable for hydrogen bonding (i.e., verifying the angle criterion) was evaluated. Distributions of such distance from the AIMD for each heteroatom are shown in Fig. 7. These distributions reveal a mobile solvation equilibrium, exploring both fully solvated configurations and partially solvated ones. Indeed, we can consider a heteroatom unsolvated when the closest protic molecule with an adequate angle is too distant (>4 Å). Therefore, O4 and N1 appear unsolvated in a significant fraction of the configurations explored by the AIMD because their distributions (Fig. 7b and c, respectively) show a significant population of >4 Å distances.
 | ||
| Fig. 7 Distributions of the distance of each 5BU heteroatom from the closest solvent oxygen verifying HMeOH–OMeOH–O or H–N–OMeOH < 40°. Such a criterion allows to select solvent molecules potentially able to participate in hydrogen bonds. Please refer to Fig. 2a for atoms labels. | ||
According to the previous discussion, the collected 5BU ADMP/ONIOM/NPBCs MD simulation therefore provided a complete picture of both conformational and solvation equilibria in methanol at room temperature. Such an ensemble allowed a detailed prediction of 5BU excited states and absorption spectra under such conditions, as shown in the following section (Section 3.3).
3.3 Optical absorption spectrum: experimental and simulation results from AIMD sampling
The experimental UV-Vis absorption spectrum of 5BU in methanol was recorded according to the procedure detailed in Section 2.3. As shown in Fig. 8a, it exhibits two distinctive absorption bands, located at approximately 210 and 265 nm, corresponding to 5.90 and 4.68 eV, respectively. Our interest was to disentangle the main electronic transitions and the structural features determining the observed absorption. To this end, we simulated the optical absorption spectrum of 5BU in methanol by performing TD-DFT calculations on an ensemble of configurations extracted from the AIMD, according to the protocol detailed in Section 2.4. As shown in Fig. 8a, the band shape of the experimental spectrum is reproduced with a systematic blue shift, suggesting that the energy spacing among the excited states is correctly estimated. By recognizing the individual contributions of the different states, we attribute the lowest-energy band, which is the one excited during photo-cyclization,25,26 mainly to the transition towards S1, with contributions also from S2 and S3, the latter only found beyond the band maximum (see Fig. 8b).The different conformations and solvation states sampled by the QM/MM/NPBCs trajectory in methanol solution at room temperature (Section 3.2) suggest that the adiabatic S1 and S2 lowest-energy states may actually exchange their character based on such parameters. We thus investigated the character of the first two excitations (through a NTO analysis) on a small subset of representative configurations extracted from the QM/MM/NPBCs MD, differing in the conformation and solvation state (please refer to Fig. S10 in the ESI† for their structures). The features of S1 and S2 states calculated for such configurations are correlated to the structural parameters as shown in Table 3. NTO pairs isosurfaces can be instead found in Fig. S11–S15 in the ESI.†
| Configuration | Conformer | O4 | N1 | S1 ← S0 | S2 ← S0 | ||
|---|---|---|---|---|---|---|---|
| V.E.E. (f) | Character | V.E.E. (f) | Character | ||||
| α | B | H-bonded | H-bonded | 4.92 (0.217) | Uππ* | 5.25 (0.019) | Unπ* + PπUπ* |
| β | F | H-bonded | H-bonded | 5.05 (0.179) | Uππ* | 5.27 (0.017) | Unπ* |
| γ | B | Not H-bonded | H-bonded | 4.83 (0.158) | Uππ* + PπUπ* | 5.13 (0.054) | Unπ* |
| δ | F | Not H-bonded | H-bonded | 5.12 (0.170) | Uππ* | 5.30 (0.006) | Unπ* |
| ε | F | Not H-bonded | Not H-bonded | 5.03 (0.024) | Unπ* | 5.14 (0.102) | Uππ* |
Consistently with the calculations on optimized structures with either implicit or explicit solvation (Section 3.1), S1 is a bright Uππ* state, while S2 has a dark Unπ* character (being the n orbital located on O4), due to specific methanol–5BU interactions (especially on N1). Interestingly, in the ε configuration, where neither of the two heteroatoms O4 and N1 are solvated, S1 and S2 characters are swapped: the former is therefore a dark Unπ* state, while the latter has instead a bright Uππ* character (see Fig. S15, ESI†). We also observe that S1 and S2 of the butterfly-like representative configurations (α and γ) also show a small PπUπ* charge-transfer character, which is absent in the corresponding transitions of flag-like configurations (see Fig. S11 and S12, ESI†).
We then compared the average values of energy and oscillator strength of the first four states from the AIMD (see Table 4) with the values calculated from the minimum energy structures optimized in the methanol cluster at the B3LYP/6-31G(d,p) level (i.e., the same level of the AIMD, see the third column of Table 2). This comparison revealed consistent energies and spacing, but fairly different oscillator strength values: S1 is the brightest state from both static and dynamics calculations, but its oscillator strength calculated from AIMD frames is on average only 0.123, compared with the values of 0.294 and 0.226 from the minimum energy structures. S2 and S3, which from static calculations appeared to be completely dark states, instead exhibit an average oscillator strength of 0.042 and 0.013 respectively, based on AIMD frames. The S2 state is therefore not completely dark because it acquires a Uππ* character in a reduced fraction of the sampled configurations. Besides S1, S2 is of particular interest since it contributes, together with S1, to the first band, which is excited to induce 5BU photo-cyclization.25,26
| Transition | V.E.E. (eV) | f |
|---|---|---|
| S1 ← S0 | 5.01 | 0.123 |
| S2 ← S0 | 5.20 | 0.042 |
| S3 ← S0 | 5.34 | 0.013 |
| S4 ← S0 | 5.82 | 0.050 |
| Average of the first band | 5.06 | |
| Experimental peak | 4.68 |
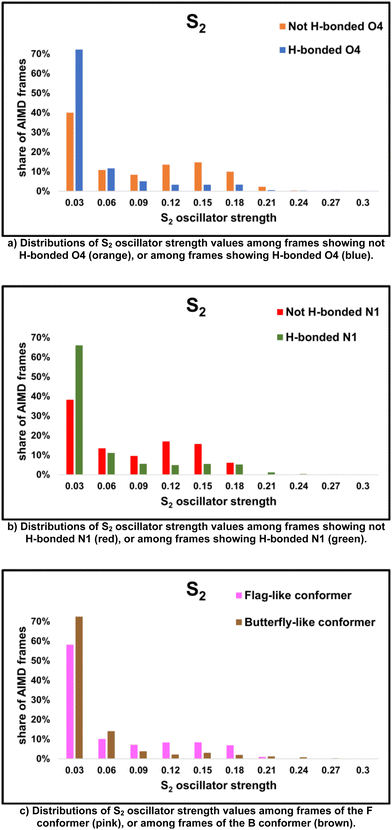
The swap of the character of the first two states depending on 5BU conformation and solvation can explain several trends in their brightness. Regarding the first excited state, the formation of a hydrogen bond between O4 and a solvent molecule appears to be an important feature. As discussed in Section 3.2, some of the sampled configurations have at least one solvent molecule with distance and orientation suitable for a hydrogen bond with O4, while in the other ones, O4 does not form hydrogen bonds (see Fig. 7b). In the former group, the average S1 oscillator strength is 0.146 and its distribution peak lies between 0.15 and 0.18 (see the blue bars in Fig. 9a, please refer to Table 5 for average S1 oscillator strength within each cluster of MD frames); in the latter group, instead, the average S1 oscillator strength is 0.069 and its distribution peak lies between 0.00 and 0.03 (see the orange bars in Fig. 9a). Analogously to O4, N1 microsolvation (see Fig. 7c) also influences the brightness of S1: S1 is mostly a bright state when there is at least one solvent molecule with distance and orientation suitable for a hydrogen bond with N1, while it is darker when N1 does not form hydrogen bonds. In the former group, the average S1 oscillator strength is 0.132 and its distribution peak lies between 0.15 and 0.18 (see the green bars in Fig. 9b); in the latter group, instead, the average S1 oscillator strength is 0.059 and its distribution peak lies between 0.00 and 0.03 (see the red bars in Fig. 9b). Finally, the relative orientation of the uracil and phenyl rings also plays a role: when they are approximately orthogonal to each other (flag-like conformer, occupying the right half of the distribution in Fig. 5, see for instance Fig. 3b), S1 oscillator strength has a bimodal distribution, with a dark peak between 0.00 and 0.03 and a bright peak between 0.15 and 0.18 (see the pink bars in Fig. 9c). When the rings face each other (butterfly-like conformer, occupying the left half of the distribution in Fig. 5, see for instance Fig. 3a), the distribution dark peak disappears and S1 is mostly a bright state, with a distribution peak between 0.15 and 0.18 (see the brown bars in Fig. 9c).
| Cluster | Average f (S1) | Cluster | Average f (S1) |
|---|---|---|---|
| Not H-bonded O4 | 0.069 | H-bonded O4 | 0.146 |
| Not H-bonded N1 | 0.059 | H-bonded N1 | 0.132 |
| F conformer | 0.123 | B conformer | 0.127 |
S2 exhibits an opposite behaviour with respect to S1, even if its variations are more subtle. Like S1, indeed, its brightness mainly depends on the microsolvation of O4, but, contrarily to S1, it is higher in the configurations where O4 does not form hydrogen bonds and decreases in those where a methanol molecule solvates O4. In the first group, S2 oscillator strength has a bimodal distribution, with the highest peak between 0.00 and 0.03 and a lower peak (with a population half of the first one) between 0.15 and 0.18. In the second group instead, the distribution shows no bright peaks (see the blue bars in Fig. 10a) and the second excited state is mostly dark, with an average oscillator strength of 0.0308 (please refer to Table 6 for average S2 oscillator strength within each cluster of MD frames). Again in contrast to the first state, the fraction of frames with a bright second excited state significantly increases when there are no nearby solvent molecules able to form a hydrogen bond with N1, while it is lower when N1 is solvated: in the first group, S2 oscillator strength has a bimodal distribution, with the highest peak between 0.00 and 0.03 and a second one between 0.12 and 0.15, with a relative abundance half of the first one (see the red bars in Fig. 10b). The second group does not show this bright peak in the distribution, but a mostly dark second excited state, with an average oscillator strength of 0.039 (see the green bars in Fig. 10b). Finally, the effect of the two rings arrangement is also reversed with respect to S1: the second excited state is brighter when the two rings are approximately orthogonal (flag-like conformer), while it is darker when they face each other (butterfly-like conformer) (see Fig. 10c).
| Cluster | Average f (S2) | Cluster | Average f (S2) |
|---|---|---|---|
| Not H-bonded O4 | 0.071 | H-bonded O4 | 0.031 |
| Not H-bonded N1 | 0.066 | H-bonded N1 | 0.039 |
| F conformer | 0.042 | B conformer | 0.021 |
4 Conclusion
In this work, new features regarding the photophysics of 5BU in methanol solution at room temperature were found, using an accurate combined computational and experimental approach. From our analysis of the ground-state equilibrium and of the electronic layout of 5BU in methanol, an intricate and subtle correlation between the instantaneous configurations and the photophysics was found. The nature and brightness of the first two excited states were found to be dependent on the conformation of the solute and on the instantaneous microsolvation of its heteroatoms. Indeed, 5BU appears to have a soft conformational landscape, composed of a plethora of relevant conformations and microsolvation configurations at room temperature. As main results, we observed that the character and brightness of these two transitions can swap, depending on the reciprocal orientation of the aromatic rings and the instantaneous microsolvation of the heteroatoms O4 and N1. The dependence of absorption on the conformation and microsolvation may influence the successive photoreaction. The irradiation at different wavelengths may induce different reaction pathways by exciting different initial conformations and solute–solvent configurations, activating either S1 or S2 excited states. This may acquire even greater importance when applied to nucleoprotein complexes, where the involved residues are often blocked in a specific orientation and solvation state and may thus be susceptible or not to photo-crosslinking based on these features. This suggests that the description of the subsequent photorelaxation and photoreaction has to include a non-adiabatic treatment of both the first two excited states, already in its earliest stages. Moreover, given the significant role of the solvent in both the excitation and the photoreaction, excited state dynamics would benefit from the inclusion of an explicit treatment of the several solvent molecules to fully saturate the solvation sites. Our work will help in advancing such analysis, towards the comprehension of photophysics of DNA– and RNA–protein model complexes.Author contributions
N. R. and C. A.: conceptualization, funding acquisition, project administration, supervision, writing – original draft. A. P.: formal analysis, methodology, supervision, writing – original draft, writing – review & editing. M. V.: data curation, formal analysis, methodology, investigation, supervision, writing – original draft. F. P.: formal analysis, methodology, software, validation, writing – original draft, writing – review & editing. G. I.: data curation, investigation, validation, visualization, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
N. R. and A. P. gratefully acknowledge the Italian Ministry of University and Research (projects: PRIN 202082CE3T_002) for financial support. N. R. and A. P. acknowledge IBiSCo (Infrastructure for Big data and Scientific Computing) for HPC resources. N. R., A. P. and F. P. also acknowledge Gaussian Inc. for financial support. A. P. thanks the University of Napoli Federico II for financial support (project: FRA2022-CosmoHab CUP: E65F22000050001). G. I., F. P., A. P. and N. R. also thank Francesco Gambuli for his assistance and support.References
- R. Berisio, F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram and A. Yonath, Nat. Struct. Biol., 2003, 10, 366–370 CrossRef CAS PubMed.
- A. G. Matera, R. M. Terns and M. P. Terns, Nat. Rev. Mol. Cell Biol., 2007, 8, 209–220 CrossRef CAS PubMed.
- T. A. Steitz, Nat. Rev. Mol. Cell Biol., 2008, 9, 242–253 CrossRef CAS PubMed.
- T. Glisovic, J. L. Bachorik, J. Yong and G. Dreyfuss, FEBS Lett., 2008, 582, 1977–1986 CrossRef CAS PubMed.
- L. A. Yates, C. J. Norbury and R. J. Gilbert, Cell, 2013, 153, 516–519 CrossRef CAS PubMed.
- M. W. Hentze, A. Castello, T. Schwarzl and T. Preiss, Nat. Rev. Mol. Cell Biol., 2018, 19, 327–341 CrossRef CAS PubMed.
- G. L. Hager, C. Elbi and M. Becker, Curr. Opin. Genet. Dev., 2002, 12, 137–141 CrossRef CAS PubMed.
- D. J. Patel and Z. Wang, Annu. Rev. Biochem., 2013, 82, 81–118 CrossRef CAS PubMed.
- T. Siggers and R. Gordân, Nucleic Acids Res., 2014, 42, 2099–2111 CrossRef CAS PubMed.
- Q. Du, P. L. Luu, C. Stirzaker and S. J. Clark, Epigenomics, 2015, 7, 1051–1073 CrossRef CAS PubMed.
- C. D. Allis and T. Jenuwein, Nat. Rev. Genet., 2016, 17, 487–500 CrossRef CAS PubMed.
- K. Smith, B. Hodgkins and M. O'Leary, Biochim. Biophys. Acta, Nucleic Acids Protein Synth, 1966, 114, 1–15 CrossRef CAS PubMed.
- J. G. Peak and M. J. Peak, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1991, 246, 187–191 CrossRef CAS PubMed.
- S. Barker, M. Weinfeld and D. Murray, Mutat. Res., Rev. Mutat. Res., 2005, 589, 111–135 CrossRef CAS PubMed.
- T. Nakano, X. Xu, A. M. Salem, M. I. Shoulkamy and H. Ide, Free Radical Biol. Med., 2017, 107, 136–145 CrossRef CAS PubMed.
- L. Zhang, K. Zhang, R. Prändl and F. Schöffl, Biochem. Biophys. Res. Commun., 2004, 322, 705–711 CrossRef CAS PubMed.
- C. Altucci, A. Nebbioso, R. Benedetti, R. Esposito, V. Carafa, M. Conte, M. Micciarelli, L. Altucci and R. Velotta, Laser Phys. Lett., 2012, 9, 234–239 CrossRef CAS.
- A. Nebbioso, R. Benedetti, M. Conte, V. Carafa, F. De Bellis, J. Shaik, F. Matarese, B. Della Ventura, F. Gesuele, R. Velotta, J. H. A. Martens, H. G. Stunnenberg, C. Altucci and L. Altucci, Sci. Rep., 2017, 7, 11725 CrossRef PubMed.
- A. Reim, R. Ackermann, J. Font-Mateu, R. Kammel, M. Beato, S. Nolte, M. Mann, C. Russmann and M. Wierer, Nat. Commun., 2020, 11, 3019 CrossRef CAS PubMed.
- A. Stützer, L. M. Welp, M. Raabe, T. Sachsenberg, C. Kappert, A. Wulf, A. M. Lau, S. S. David, A. Chernev, K. Kramer, A. Politis, O. Kohlbacher, W. Fischle and H. Urlaub, Nat. Commun., 2020, 11, 5250 CrossRef PubMed.
- A. Knörlein, C. P. Sarnowski, T. de Vries, M. Stoltz, M. Götze, R. Aebersold, F. H. Allain, A. Leitner and J. Hall, Nat. Commun., 2022, 13, 2719 CrossRef PubMed.
- H. Morrison, Bioorganic photochemistry, J. Wiley and Sons, Ltd, New York, 1990 Search PubMed.
- M. Shetlar, Int. Congr. Ser., 1993, 67–72 CAS.
- G. Sun, C. J. Fecko, R. B. Nicewonger, W. W. Webb and T. P. Begley, Org. Lett., 2006, 8, 681–683 CrossRef CAS PubMed.
- M. Micciarelli, M. Valadan, B. Della Ventura, G. Di Fabio, L. De Napoli, S. Bonella, U. Röthlisberger, I. Tavernelli, C. Altucci and R. Velotta, J. Phys. Chem. B, 2014, 118, 4983–4992 CrossRef CAS PubMed.
- M. Valadan, E. Pomarico, B. Della Ventura, F. Gesuele, R. Velotta, A. Amoresano, G. Pinto, M. Chergui, R. Improta and C. Altucci, Phys. Chem. Chem. Phys., 2019, 21, 26301–26310 RSC.
- M. Micciarelli, C. Altucci, B. D. Ventura, R. Velotta, V. Tosa, A. B. G. Pérez, M. P. Rodríguez, A. R. de Lera and A. Bende, Phys. Chem. Chem. Phys., 2013, 15, 7161–7173 RSC.
- M. Micciarelli, B. F. E. Curchod, S. Bonella, C. Altucci, M. Valadan, U. Rothlisberger and I. Tavernelli, J. Phys. Chem. A, 2017, 121, 3909–3917 CrossRef CAS PubMed.
- A. Petrone, F. Perrella, F. Coppola, L. Crisci, G. Donati, P. Cimino and N. Rega, Chem. Phys. Rev., 2022, 3, 021307 CrossRef CAS.
- N. Rega, G. Brancato and V. Barone, Chem. Phys. Lett., 2006, 422, 367–371 CrossRef CAS.
- G. Brancato, N. Rega and V. Barone, J. Chem. Phys., 2006, 125, 164515 CrossRef PubMed.
- G. Brancato, N. Rega and V. Barone, J. Phys. Chem., 2008, 128, 144501 CrossRef PubMed.
- U. Raucci, F. Perrella, G. Donati, M. Zoppi, A. Petrone and N. Rega, J. Comput. Chem., 2020, 41, 2228–2239 CrossRef CAS PubMed.
- M. E. Casida, in Recent Advances in Density Functional Methods (Part I), ed. D. P. Chong, World Scientific Publishing Co. Pte. Ltd., 1995, ch. 5, pp. 475–516 Search PubMed.
- R. E. Stratmann, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 1998, 109, 8218–8224 CrossRef CAS.
- A. Dreuw and M. Head-Gordon, Chem. Rev., 2005, 105, 4009–4037 CrossRef CAS PubMed.
- R. Crespo-Otero and M. Barbatti, Theor. Chem. Acc., 2012, 131, 1237 Search PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2015, 119, 958–967 CrossRef CAS PubMed.
- A. Petrone, J. Cerezo, F. J. A. Ferrer, G. Donati, R. Improta, N. Rega and F. Santoro, J. Phys. Chem. A, 2015, 119, 5426–5438 CrossRef CAS PubMed.
- J. Cerezo, F. J. Avila Ferrer, G. Prampolini and F. Santoro, J. Chem. Theory Comput., 2015, 11, 5810–5825 CrossRef CAS PubMed.
- J. Cerezo, A. Petrone, F. J. Avila Ferrer, G. Donati, F. Santoro, R. Improta and N. Rega, Theor. Chem. Acc., 2016, 135, 263 Search PubMed.
- J. Cerezo, G. Mazzeo, G. Longhi, S. Abbate and F. Santoro, J. Phys. Chem. Lett., 2016, 7, 4891–4897 Search PubMed.
- D. Padula, J. Cerezo, G. Pescitelli and F. Santoro, Phys. Chem. Chem. Phys., 2017, 19, 32349–32360 RSC.
- J. M. Milanese, M. R. Provorse, E. J. Alameda and C. M. Isborn, J. Chem. Theory Comput., 2017, 13, 2159–2171 CrossRef CAS PubMed.
- J. Cerezo, D. Aranda, F. J. Avila Ferrer, G. Prampolini, G. Mazzeo, G. Longhi, S. Abbate and F. Santoro, Chirality, 2018, 30, 730–743 CrossRef CAS PubMed.
- Y. Liu, J. Cerezo, N. Lin, X. Zhao, R. Improta and F. Santoro, Theor. Chem. Acc., 2018, 137, 40 Search PubMed.
- G. Donati, A. Petrone, P. Caruso and N. Rega, Chem. Sci., 2018, 9, 1126–1135 RSC.
- S. V. Shedge, T. J. Zuehlsdorff, M. J. Servis, A. E. Clark and C. M. Isborn, J. Phys. Chem. A, 2019, 123, 6175–6184 CrossRef CAS PubMed.
- S. Del Galdo, M. Fusè and V. Barone, J. Chem. Theory Comput., 2020, 16, 3294–3306 CrossRef CAS PubMed.
- C. Reichardt, Chem. Rev., 1994, 94, 2319–2358 CrossRef CAS.
- K. M. Solntsev, D. Huppert, L. M. Tolbert and N. Agmon, J. Am. Chem. Soc., 1998, 120, 7981–7982 CrossRef CAS.
- K. M. Solntsev, D. Huppert and N. Agmon, J. Phys. Chem. A, 1999, 103, 6984–6997 CrossRef CAS.
- H. A. Frank, J. A. Bautista, J. Josue, Z. Pendon, R. G. Hiller, F. P. Sharples, D. Gosztola and M. R. Wasielewski, J. Phys. Chem. B, 2000, 104, 4569–4577 CrossRef CAS.
- C. Adamo, M. Cossi, N. Rega and V. Barone, in Theoretical Biochemistry, ed. L. A. Eriksson, Elsevier, Amsterdam, The Netherlands, 2001, vol. 9 of Theoretical and Computational Chemistry, pp. 467–538 Search PubMed.
- E. Krystkowiak, K. Dobek and A. Maciejewski, J. Photochem. Photobiol., A, 2006, 184, 250–264 CrossRef CAS.
- V. Barone, R. Improta and N. Rega, Acc. Chem. Res., 2008, 41, 605–616 CrossRef CAS PubMed.
- M. Svensson, S. Humbel, R. D. J. Froese, T. Matsubara, S. Sieber and K. Morokuma, J. Phys. Chem., 1996, 100, 19357–19363 CrossRef CAS.
- T. Vreven, K. S. Byun, I. Komáromi, S. Dapprich, J. A. Montgomery Jr, K. Morokuma and M. J. Frisch, J. Chem. Theory Comput., 2006, 2, 815–826 CrossRef CAS PubMed.
- K. Morokuma, Q. Wang and T. Vreven, J. Chem. Theory Comput., 2006, 2, 1317–1324 CrossRef CAS PubMed.
- M. Berkowitz, C. L. Brooks and S. A. Adelman, J. Phys. Chem., 1980, 72, 3889–3898 CrossRef CAS.
- K. Kratky, J. Comput. Phys., 1980, 37, 205–217 CrossRef.
- C. L. Brooks and M. Karplus, J. Phys. Chem., 1983, 79, 6312–6325 CrossRef CAS.
- D. Beglov and B. Roux, J. Chem. Phys., 1994, 100, 9050–9063 CrossRef CAS.
- G. Brancato, A. Di Nola, V. Barone and A. Amadei, J. Chem. Phys., 2005, 122, 154109 CrossRef PubMed.
- E.-S. Riihimäki, J. M. Martínez and L. Kloo, THEOCHEM, 2006, 760, 91–98 CrossRef.
- N. Rega, G. Brancato, A. Petrone, P. Caruso and V. Barone, J. Chem. Phys., 2011, 134, 074504 CrossRef PubMed.
- A. Petrone, G. Donati, P. Caruso and N. Rega, J. Am. Chem. Soc., 2014, 136, 14866–14874 CrossRef CAS PubMed.
- M. G. Chiariello and N. Rega, J. Phys. Chem. A, 2018, 122, 2884–2893 CrossRef CAS PubMed.
- A. D. Becke, Phys. Rev. A, 1988, 38, 3098 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B, 1988, 37, 785 CrossRef CAS PubMed.
- B. Miehlich, A. Savin, H. Stoll and H. Preuss, Chem. Phys. Lett., 1989, 157, 200–206 CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- L. Goerigk, A. Hansen, C. Bauer, S. Ehrlich, A. Najibi and S. Grimme, Phys. Chem. Chem. Phys., 2017, 19, 32184–32215 RSC.
- M. G. Chiariello, G. Donati, U. Raucci, F. Perrella and N. Rega, J. Phys. Chem. B, 2021, 125, 10273–10281 CrossRef CAS PubMed.
- J. Antony and S. Grimme, Phys. Chem. Chem. Phys., 2006, 8, 5287–5293 RSC.
- F. Perrella, F. Coppola, A. Petrone, C. Platella, D. Montesarchio, A. Stringaro, G. Ravagnan, M. P. Fuggetta, N. Rega and D. Musumeci, Biomolecules, 2021, 11, 1048 CrossRef CAS PubMed.
- S. Grimme, J. Antony, T. Schwabe and C. Mück-Lichtenfeld, Org. Biomol. Chem., 2007, 5, 741–758 RSC.
- F. Coppola, F. Perrella, A. Petrone, G. Donati and N. Rega, Front. Mol. Biosci., 2020, 7, 569990 CrossRef CAS PubMed.
- D. Jacquemin, V. Wathelet, E. A. Perpète and C. Adamo, J. Chem. Theory Comput., 2009, 5, 2420–2435 CrossRef CAS PubMed.
- C. A. Guido, D. Jacquemin, C. Adamo and B. Mennucci, J. Phys. Chem. A, 2010, 114, 13402–13410 CrossRef CAS PubMed.
- U. Raucci, M. G. Chiariello, F. Coppola, F. Perrella, M. Savarese, I. Ciofini and N. Rega, J. Comput. Chem., 2020, 41, 1835–1841 CrossRef CAS PubMed.
- C. A. Guido, S. Knecht, J. Kongsted and B. Mennucci, J. Chem. Theory Comput., 2013, 9, 2209–2220 CrossRef CAS PubMed.
- F. Perrella, U. Raucci, M. G. Chiariello, M. Chino, O. Maglio, A. Lombardi and N. Rega, Biopolymers, 2018, 109, e23225 CrossRef PubMed.
- É. Brémond, M. Savarese, N. Q. Su, Á. J. Pérez-Jiménez, X. Xu, J. C. Sancho-García and C. Adamo, J. Chem. Theory Comput., 2016, 12, 459–465 CrossRef PubMed.
- D. Jacquemin, E. A. Perpète, G. E. Scuseria, I. Ciofini and C. Adamo, J. Chem. Theory Comput., 2008, 4, 123–135 CrossRef CAS PubMed.
- M. Steinmetz and S. Grimme, ChemistryOpen, 2013, 2, 115–124 CrossRef CAS PubMed.
- F. Perrella, V. Langellotti, E. Buttarazzi, M. E. Cucciolito, M. Melchiorre, G. Pinto, V. Prokopenko, N. Rega, F. Ruffo, A. Petrone and R. Esposito, ChemCatChem, 2024, 16, e202300945 CrossRef CAS.
- T. Risthaus and S. Grimme, J. Chem. Theory Comput., 2013, 9, 1580–1591 CrossRef CAS PubMed.
- E. Buttarazzi, F. Perrella, N. Rega and A. Petrone, J. Chem. Theory Comput., 2023, 19, 8751–8766 CrossRef CAS PubMed.
- M. A. Iron and T. Janes, J. Phys. Chem. A, 2019, 123, 3761–3781 CrossRef CAS PubMed.
- F. Perrella, F. Coppola, N. Rega and A. Petrone, Molecules, 2023, 28, 3411 CrossRef CAS PubMed.
- D. Nazarian, P. Ganesh and D. S. Sholl, J. Mater. Chem. A, 2015, 3, 22432–22440 RSC.
- F. Perrella, A. Petrone and N. Rega, J. Chem. Theory Comput., 2023, 19, 626–639 CrossRef CAS PubMed.
- M. M. Quintal, A. Karton, M. A. Iron, A. D. Boese and J. M. L. Martin, J. Phys. Chem. A, 2006, 110, 709–716 CrossRef CAS PubMed.
- F. Perrella, X. Li, A. Petrone and N. Rega, JACS Au, 2023, 3, 70–79 CrossRef CAS PubMed.
- C. Latouche, D. Skouteris, F. Palazzetti and V. Barone, J. Chem. Theory Comput., 2015, 11, 3281–3289 CrossRef CAS PubMed.
- F. Coppola, P. Cimino, F. Perrella, L. Crisci, A. Petrone and N. Rega, J. Phys. Chem. A, 2022, 126, 7179–7192 CrossRef CAS PubMed.
- Y. Zhao and D. G. Truhlar, J. Chem. Theory Comput., 2005, 1, 415–432 CrossRef CAS PubMed.
- M. P. Waller, H. Braun, N. Hojdis and M. Bühl, J. Chem. Theory Comput., 2007, 3, 2234–2242 CrossRef CAS PubMed.
- F. Perrella, A. Petrone and N. Rega, Phys. Chem. Chem. Phys., 2021, 23, 22885–22896 RSC.
- A. Jain, Y. Shin and K. A. Persson, Nat. Rev. Mater., 2016, 1, 15004 CrossRef CAS.
- J. Hafner, C. Wolverton and G. Ceder, MRS Bull., 2006, 31, 659–668 CrossRef.
- P. J. Hasnip, K. Refson, M. I. J. Probert, J. R. Yates, S. J. Clark and C. J. Pickard, Phil. Trans. R. Soc. A, 2014, 372, 20130270 CrossRef PubMed.
- C. J. Cramer and D. G. Truhlar, Phys. Chem. Chem. Phys., 2009, 11, 10757–10816 RSC.
- M. Bühl and H. Kabrede, J. Chem. Theory Comput., 2006, 2, 1282–1290 CrossRef PubMed.
- S. Miertuš, E. Scrocco and J. Tomasi, Chem. Phys., 1981, 55, 117–129 CrossRef.
- V. Barone and M. Cossi, J. Phys. Chem. A, 1998, 102, 1995–2001 CrossRef CAS.
- M. Cossi, N. Rega, G. Scalmani and V. Barone, J. Chem. Phys., 2001, 114, 5691–5701 CrossRef CAS.
- M. Cossi, N. Rega, G. Scalmani and V. Barone, J. Comput. Chem., 2003, 24, 669–681 CrossRef CAS PubMed.
- T. Vreven, B. Mennucci, C. O. da Silva, K. Morokuma and J. Tomasi, J. Chem. Phys., 2001, 115, 62–72 CrossRef CAS.
- S. J. Mo, T. Vreven, B. Mennucci, K. Morokuma and J. Tomasi, Theor. Chem. Acc., 2004, 111, 154–161 Search PubMed.
- A. Petrone, P. Caruso, S. Tenuta and N. Rega, Phys. Chem. Chem. Phys., 2013, 15, 20536–20544 RSC.
- A. Ben-Naim, Solvation Thermodynamics, Plenum, New York, 1987 Search PubMed.
- A. Nakayama, G. Arai, S. Yamazaki and T. Taketsugu, J. Chem. Phys., 2013, 139, 214304 CrossRef PubMed.
- S. Komin, C. Gossens, I. Tavernelli, U. Rothlisberger and D. Sebastiani, J. Phys. Chem. B, 2007, 111, 5225–5232 CrossRef CAS PubMed.
- J. Gao and M. Freindorf, J. Phys. Chem. A, 1997, 101, 3182–3188 CrossRef CAS.
- M.-E. Moret, I. Tavernelli and U. Rothlisberger, J. Phys. Chem. B, 2009, 113, 7737–7744 CrossRef CAS PubMed.
- I. Tavernelli, B. F. Curchod and U. Rothlisberger, Chem. Phys., 2011, 391, 101–109 CrossRef CAS.
- M. Nonella, G. Mathias and P. Tavan, J. Phys. Chem. A, 2003, 107, 8638–8647 CrossRef CAS.
- V. Pham, I. Tavernelli, C. Milne, R. van der Veen, P. DAngelo, C. Bressler and M. Chergui, Chem. Phys., 2010, 371, 24–29 CrossRef CAS.
- R. M. Abolfath, P. K. Biswas, R. Rajnarayanam, T. Brabec, R. Kodym and L. Papiez, J. Phys. Chem. A, 2012, 116, 3940–3945 CrossRef CAS PubMed.
- J. Wen, S. Mai and L. González, J. Phys. Chem. A, 2023, 127, 9520–9529 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, B. G. Janesko, F. Lipparini, G. Zheng, J. L. Sonnenberg, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, P. V. Parandekar, N. J. Mayhall, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian Development Version, Revision H.37+, Gaussian Inc., Wallingford CT, 2010 Search PubMed.
- S. S. Iyengar, H. B. Schlegel, J. M. Millam, G. A. Voth, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 2001, 115, 10291–10302 CrossRef CAS.
- H. B. Schlegel, J. M. Millam, S. S. Iyengar, G. A. Voth, A. D. Daniels, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 2001, 114, 9758–9763 CrossRef CAS.
- H. B. Schlegel, S. S. Iyengar, X. Li, J. M. Millam, G. A. Voth, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 2002, 117, 8694–8704 CrossRef CAS.
- S. S. Iyengar, H. B. Schlegel, G. A. Voth, J. M. Millam, G. E. Scuseria and M. J. Frisch, Isr. J. Chem., 2002, 42, 191–202 CrossRef CAS.
- N. Rega, S. S. Iyengar, G. A. Voth, H. B. Schlegel, T. Vreven and M. J. Frisch, J. Phys. Chem. B, 2004, 108, 4210–4220 CrossRef CAS.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- G. J. Martyna, D. J. Tobias and M. L. Klein, J. Phys. Chem., 1994, 101, 4177–4189 CrossRef CAS.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, J. Phys. Chem., 1995, 103, 4613–4621 CrossRef CAS.
- A. Lami and F. Santoro, in Computational Strategies for Spectroscopy: From Small Molecules to Nano Systems, ed. V. Barone, John Wiley & Sons, Ltd, New York, NY, 2011, ch. 10, pp. 475–516 Search PubMed.
- T. Yanai, D. P. Tew and N. C. Handy, Chem. Phys. Lett., 2004, 393, 51–57 CrossRef CAS.
- R. Improta and V. Barone, Top. Curr. Chem., 2015, 355, 329–357 CrossRef CAS PubMed.
- R. Improta, F. Santoro and L. Blancafort, Chem. Rev., 2016, 116, 3540–3593 CrossRef CAS PubMed.
- A. J. Pepino, J. Segarra-Martí, A. Nenov, R. Improta and M. Garavelli, J. Phys. Chem. Lett., 2017, 8, 1777–1783 CrossRef CAS PubMed.
- A. J. Pepino, J. Segarra-Martí, A. Nenov, I. Rivalta, R. Improta and M. Garavelli, Phys. Chem. Chem. Phys., 2018, 20, 6877–6890 RSC.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery, Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16 Revision C.01, Gaussian Inc., Wallingford CT, 2016 Search PubMed.
- R. L. Martin, J. Chem. Phys., 2003, 118, 4775–4777 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cp06254f |
| This journal is © the Owner Societies 2024 |