Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multilevel quantum mechanical calculations show the role of promoter molecules in the dehydration of methanol to dimethyl ether in H-ZSM-5†
Joe
Crossley-Lewis
 a,
Josh
Dunn
a,
Josh
Dunn
 a,
Isabel F.
Hickman
a,
Isabel F.
Hickman
 a,
Fiona
Jackson
b,
Glenn J.
Sunley
a,
Fiona
Jackson
b,
Glenn J.
Sunley
 b,
Corneliu
Buda
*c,
Adrian J.
Mulholland
b,
Corneliu
Buda
*c,
Adrian J.
Mulholland
 *a and
Neil L.
Allan
*a and
Neil L.
Allan
 *a
*a
aCentre for Computational Chemistry, School of Chemistry, University of Bristol, Cantock's Close, Bristol, BS8 1TS, UK. E-mail: neil.allan@bristol.ac.uk
bApplied Sciences, bp Innovation and Engineering, BP plc, Saltend, Hull, HU12 8DS, UK
cApplied Sciences, bp Innovation and Engineering, BP plc, 30 South Wacker Drive, Chicago, IL 60606, USA
First published on 20th May 2024
Abstract
Methyl carboxylate esters promote the formation of dimethyl ether (DME) from the dehydration of methanol in H-ZSM-5 zeolite. We employ a multilevel quantum method to explore the possible associative and dissociative mechanisms in the presence, and absence, of six methyl ester promoters. This hybrid method combines density functional theory, with dispersion corrections (DFT-D3), for the full periodic system, with second-order Møller–Plesset perturbation theory (MP2) for small clusters representing the reaction site, and coupled cluster with single, double, and perturbative triple substitution (CCSD(T)) for the reacting molecules. The calculated adsorption enthalpy of methanol, and reaction enthalpies of the dehydration of methanol to DME within H-ZSM-5, agree with experiment to within chemical accuracy (∼4 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1). For the promoters, a reaction pathway via an associative mechanism gives lower overall reaction enthalpies and barriers compared to the reaction with methanol only. Each stage of this mechanism is explored and related to experimental data. We provide evidence that suggests the promoter's adsorption to the Brønsted acid site is the most important factor dictating its efficiency.
mol−1). For the promoters, a reaction pathway via an associative mechanism gives lower overall reaction enthalpies and barriers compared to the reaction with methanol only. Each stage of this mechanism is explored and related to experimental data. We provide evidence that suggests the promoter's adsorption to the Brønsted acid site is the most important factor dictating its efficiency.
Introduction
Sustainable materials providing alternative, lower energy, greener routes to useful hydrocarbons have an increasingly important role to play in reducing emissions of carbon dioxide. There is recent significant interest in dimethyl ether (DME) due to its potential as a renewable liquefied petroleum gas (LPG) replacement.1–4 It can be made via green methanol; which can be produced from many sources of carbon, including carbon dioxide.1,5–9 Relatively high reaction temperatures (275–350 °C) are required to effectively produce DME commercially using solid catalysts. However, zeolites offer a lower temperature route (110–150 °C) as they are very active and selective for making DME under these conditions where methanol-to-gasoline (MTG) and methanol-to-olefin (MTO) chemistry can be largely avoided.Zeolites, crystalline aluminosilicate porous materials, are among the most commercially important classes of heterogenous catalysts, with large-scale applications in the petrochemical industry and environmental catalysis.10,11 They are formed from corner-sharing (SiO4) and (AlO4)− tetrahedral units, assembled into ordered porous frameworks with large internal surface areas. Interactions at these internal interfaces confine and stabilise reactants and transition states; this well-known confinement effect is a key factor in zeolite catalysis.12–14 Acidic protons at catalytic centres, the Brønsted acid sites (BAS), i.e. bridging Si(OH)Al hydroxyl groups, initiate many well-established reactions in zeolites. The acid-catalysed dehydration of methanol produces dimethyl ether (DME), a commercially important intermediate in MTG and MTO chemistry.15–23
Two distinct mechanisms have been proposed for the dehydration of methanol to DME, associative (concerted) or dissociative (stepwise), as illustrated in Scheme 1 and debated in the literature.24–28 Both mechanisms involve the adsorption of one methanol molecule at a BAS. The dissociative mechanism involves the formation of a surface methoxy species [Si(OMe)Al], followed by reaction with a second methanol molecule to form DME and water. In contrast, in the associative mechanism, a second methanol molecular is adsorbed adjacent to the BAS and a dimer species is formed before conversion to DME and water. A recent combined kinetic data24 and density functional theory (DFT) study25 indicates that the associative mechanism dominates26,27 except at low methanol pressures.28,29
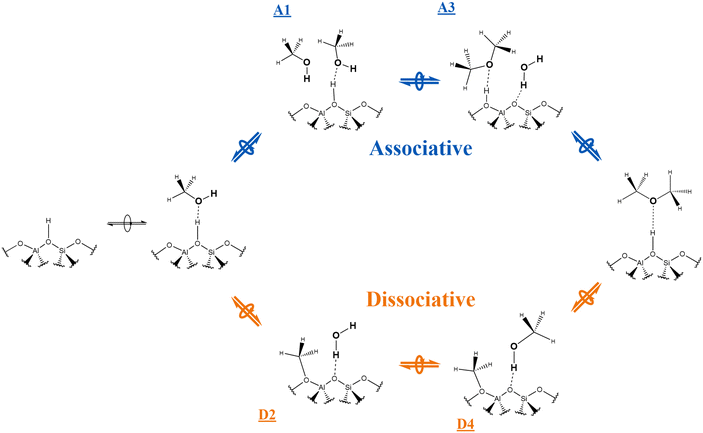 | ||
Scheme 1 The associative and dissociative mechanisms, top path and bottom path, respectively, for the dehydration of methanol to DME reaction. Labels ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/b_char_0031_0332.gif) , , ![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/b_char_0033_0332.gif) , , ![[D with combining low line]](https://www.rsc.org/images/entities/b_char_0044_0332.gif) ![[2 with combining low line]](https://www.rsc.org/images/entities/b_char_0032_0332.gif) , and , and ![[D with combining low line]](https://www.rsc.org/images/entities/b_char_0044_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/b_char_0034_0332.gif) refer to structures in Fig. 3 and 4 (energy minima). refer to structures in Fig. 3 and 4 (energy minima). | ||
Recently, it has been shown30–32 that methyl esters (Fig. 1) can act as potent promoters for the low-temperature (110–150 °C) dehydration of methanol to DME. Preliminary kinetic and spectroscopic studies suggest a mechanism in which the promoters react with methanol to form DME via an associative mechanism,30,32,33 except possibly for methyl formate which partially dissociates into formic acid and a surface methoxy species.32 FT-IR studies show that these promoters rapidly adsorb at BASs through the carbonyl group, binding more strongly than methanol;32 while both species are adsorbed to the BAS through hydrogen bonds, the promoters are further stabilised by van der Waals interactions with the pore walls. As the chain length increases, so too does this increased stabilisation. The shortest alkyl chain length promoter, methyl formate, is the only promoter for which surface methoxy species are observed experimentally, in low-temperature NMR studies by Yang et al.32 These findings suggest that the promotion of DME formation does not necessarily require promoter dissociation into methoxy species. Instead, a bimolecular associative mechanism, which we outline later, has been suggested by some of us to dominate DME formation.30 We use multi-level quantum mechanical methods to explore more fully the proposed reaction pathway and mechanism.
An atomistic understanding of catalyst performance requires comprehensive information regarding active sites and elementary reaction steps. This is challenging through experiment alone as complex reaction pathways with multiple chemical transformations are likely. Application of computational methods is constrained by the size and complexity of the systems and the accuracy of the methods themselves presents challenges. While DFT is a powerful tool, it is not a systematically improvable method, and its predictions vary in accuracy depending on the approximate treatment of correlation and dispersion.34–39 Higher-level methods, such as MP2, can be employed to combat these deficiencies, but at a computational cost; calculations on systems as large as the zeolites in this paper are unfeasible. Furthermore, the “Gold-standard” in quantum chemistry, coupled cluster with single, double, and perturbative triple excitations (CCSD(T)), provides accurate bond energies and molecular properties40–45 but is limited to small molecules and clusters and cannot be applied to periodic systems such as zeolites.34 Thus, quantum embedding (QE) methods have been developed which divide up a system into smaller subsystems that can be modelled at different levels of theory.46 Here, the different subsystems consist of the periodic zeolite framework, the reaction site, and the reacting molecules.
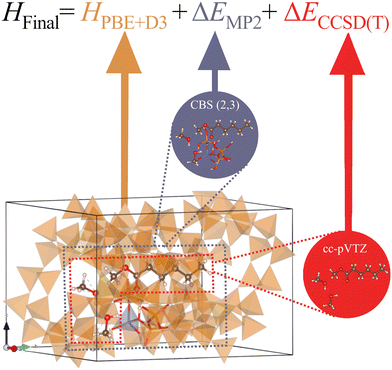
In this work, we apply a hybrid multilevel quantum method to investigate the conversion of methanol to DME in H-ZSM-5 with and without the addition of six promoter molecules. We model the zeolite framework with periodic DFT-D3, the reaction site with MP2, and the reacting molecules with CCSD(T). Specifically, we identify the key elementary steps and analyse how this leads to increased methanol conversion to DME.
Methodology
Zeolite structure and periodic DFT calculations
The MFI structure, shown on the left in Fig. 2, was obtained from the database of the international zeolite association (IZA),47 with orthorhombic unit cell parameters a = 20.09 Å, b = 19.74 Å and c = 13.14 Å. There are twelve symmetry-inequivalent T sites, i.e., metal (Al/Si) atom positions, in the unit cell. The BAS incorporated is the so-called Al1–O1(H)–Si5 site:48–51 an aluminium occupies the T1 position, silicon T5, and a proton is attached to the oxygen at the O1 position which links the Al to a neighbouring Si, this is consistent with previous work from collaborators.33 This BAS lies at the intersection of the straight and zig-zag pores, and was chosen as it maximises the dispersion interactions between the promoter molecule and the pore wall.52Periodic plane-wave density functional theory (DFT) calculations were performed on one unit cell of approximately 300 atoms, as implemented in the Vienna ab initio simulation package (VASP) version 5.4.4,53–56 with PAW pseudopotentials57 and the PBE exchange–correlation functional.58 Dispersion energies were calculated with Grimme's D3 dispersion term including Becke and Johnson damping (D3BJ).59,60 The plane-wave basis set kinetic energy cutoff was 520 eV and the SCF energy convergence criterion 10−6 eV. Structural optimisations of reactants and products were considered converged when the maximum atomic force is less than 0.05 eV![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Å−1.
Å−1.
VASPKIT61 was used to compute vibrational frequencies (within the harmonic approximation) to calculate the zero-point vibrational energies, ΔEZPVE, and vibrational contributions to thermal energies62 at 423 K, ΔEtherm. The enthalpy at 423 K is given by:
| HPBE+D3 = ΔEelec + ΔEZPVE + ΔEtherm + pΔV, | (1) |
Zeolites are flexible materials. Metastable states, involving multiple local minima separated by low barriers in the energy landscape, are associated with minor structural changes. The associated energy change to these structural rearrangements has been calculated with periodic DFT and found to be as large as 156 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 for the functional and system size used in this work.63 To avoid problems associated with structural reorganisation,63 we optimised the zeolite framework using the sequential three-step geometry optimisation methodology of Hoffman et al.:63 (i) initial DFT optimisation of both unit cell parameters and all basis atom positions of the bare zeolite obtained from the IZA, (ii) a second full optimisation with methyl n-hexanoate, the largest promoter, as an absorbate, and (iii) a final full optimisation of the zeolite after removal of the absorbate. After the methyl n-hexanoate was removed, the zeolite did not optimise back to the original structure, but to a restructured framework calculated to be 39 kJ
mol−1 for the functional and system size used in this work.63 To avoid problems associated with structural reorganisation,63 we optimised the zeolite framework using the sequential three-step geometry optimisation methodology of Hoffman et al.:63 (i) initial DFT optimisation of both unit cell parameters and all basis atom positions of the bare zeolite obtained from the IZA, (ii) a second full optimisation with methyl n-hexanoate, the largest promoter, as an absorbate, and (iii) a final full optimisation of the zeolite after removal of the absorbate. After the methyl n-hexanoate was removed, the zeolite did not optimise back to the original structure, but to a restructured framework calculated to be 39 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 lower in energy. The reduction in energy associated with the restructuring is consistent with the findings of Hoffman et al.,63 and this structure was used in all subsequent calculations.
mol−1 lower in energy. The reduction in energy associated with the restructuring is consistent with the findings of Hoffman et al.,63 and this structure was used in all subsequent calculations.
To sample the reaction paths, the nudged elastic band (NEB)64,65 method with a minimum of four intermediate images was used with a force convergence criterion of 0.2 eV![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Å−1. The climbing image algorithm66 was applied to identify transition states and convergence set to 0.1 eV
Å−1. The climbing image algorithm66 was applied to identify transition states and convergence set to 0.1 eV![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Å−1. All minima and transition state structures were confirmed by their vibrational frequencies. During optimisation of the transition states, the reacting molecules and four TO4 tetrahedra, i.e., the BAS and neighbouring framework atoms, were allowed to relax, while the rest of the zeolite framework was frozen.
Å−1. All minima and transition state structures were confirmed by their vibrational frequencies. During optimisation of the transition states, the reacting molecules and four TO4 tetrahedra, i.e., the BAS and neighbouring framework atoms, were allowed to relax, while the rest of the zeolite framework was frozen.
Molecular clusters
For the systematically improvable calculations at higher levels of theory, we use Gaussian67 on the systems shown in the middle and right of Fig. 2. We use the hierarchical approach employed by Sauer et al.41–44,52,68–81 as shown in eqn (2):| HFinal = HPBE+D3 + ΔEMP2 + ΔECCSD(T). | (2) |
This involves a scheme of wavefunction-based electron correlation method corrections, which can be capable of obtaining reaction energies and barriers to within chemical accuracy (∼4 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1), applied to the periodic DFT calculations.
mol−1), applied to the periodic DFT calculations.
The system used for the Møller–Plesset perturbation theory (MP2) corrections is shown in the top middle of Fig. 2 and highlighted in purple. It consists of a promoter, two methanol molecules, and a 4T cluster model of the zeolite. The cluster model extracted from the periodic calculations creates dangling bonds which we saturate with hydrogen atoms with O–H bond lengths of 95.3 pm and the same angle as the previous bond (O–Si). Single point energies were obtained at both lower and higher levels of theory, and the difference between the energies is the correction applied. We employ DFT+D3 with the Def2-TZVP basis set for the lower level of theory. The higher level of theory consisted of MP2 energies extrapolated to the complete basis set (CBS) limit using a two-point extrapolation scheme, with cc-pVXZ basis sets (X = D,T).82,83 Previous work by Sauer et al.71 has compared different extrapolation schemes with varying cardinal numbers and found CBS/CPC(3,4) yielded energy barriers and adsorption energies within 1 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 of their best estimate for the complete basis set limit values. However, due to the number of calculations needed and the associated computational cost, we employ CBS/CPC(2,3) which yields results very close to the CBS limit. These corrections aim to combat the key deficiency of periodic DFT calculations, the self-interaction error (SIE).68,71,72,84,85 Uncorrected, SIE results in the systematic underprediction of energy barriers, particularly for charge transfer reactions, such as the proton transfer steps important in zeolite catalysis.71,72,84 For example, Goncalves et al. found that zwitterion intermediates in the MTH reaction, formed by protonation steps at active sites,86 were too low in energy by as much as 40 kJ
mol−1 of their best estimate for the complete basis set limit values. However, due to the number of calculations needed and the associated computational cost, we employ CBS/CPC(2,3) which yields results very close to the CBS limit. These corrections aim to combat the key deficiency of periodic DFT calculations, the self-interaction error (SIE).68,71,72,84,85 Uncorrected, SIE results in the systematic underprediction of energy barriers, particularly for charge transfer reactions, such as the proton transfer steps important in zeolite catalysis.71,72,84 For example, Goncalves et al. found that zwitterion intermediates in the MTH reaction, formed by protonation steps at active sites,86 were too low in energy by as much as 40 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. For the CCSD(T) corrections we extract the reacting atoms only shown on the right of Fig. 2 and highlighted red, i.e., the two methanol molecules, the BAS hydrogen, and the promoter. Single-point energy calculations were performed using the cc-pVTZ basis set87,88 at the MP2 and CCSD(T) levels of theory, where the difference is the correction applied. Of course, such models only take into consideration a portion of the zeolite framework and there are associated complications due to edge effects and lack of proper account of the environment. The counterpoise method was used in both corrections to account for basis set superposition error.89
mol−1. For the CCSD(T) corrections we extract the reacting atoms only shown on the right of Fig. 2 and highlighted red, i.e., the two methanol molecules, the BAS hydrogen, and the promoter. Single-point energy calculations were performed using the cc-pVTZ basis set87,88 at the MP2 and CCSD(T) levels of theory, where the difference is the correction applied. Of course, such models only take into consideration a portion of the zeolite framework and there are associated complications due to edge effects and lack of proper account of the environment. The counterpoise method was used in both corrections to account for basis set superposition error.89
Reaction mechanisms
Within the zeolite, complex reaction mixtures form containing different ratios of methanol, water, DME, and promoters. Experimental kinetic data show dependence on methanol and methyl n-hexanoate partial pressure, with the reaction becoming zero order in methanol at a methanol partial pressure of 1.1 bar and a methyl n-hexanoate partial pressure of 0.11 bar.30 Overall, these kinetic studies reveal the dependence of both methanol and methyl ester partial pressures on the yield of DME. This behaviour is consistent with a bimolecular reaction mechanism involving both methanol and methyl ester adsorbed at the catalyst's active site. Spectroscopic analysis demonstrates ester molecules interact with BAS via the carbonyl group.30,90 Furthermore, recent FT-IR studies involving one of us show that methyl n-hexanoate is rapidly adsorbed at BAS,32 binds more strongly than methanol, and remains adsorbed at the BAS during the reaction. Thus, instead of methanol molecules reacting to form DME at a BAS, the promoter is adsorbed at a BAS. The conformation adopted by the adsorbed promoter involves hydrogen bonding between the carbonyl group and the BAS, with the alkyl chain on the carboxylate carbon pointing along the straight channel,30 as shown in Fig. 2, and the methoxy group of the promoter sitting in a pore intersection. Previous experimental work of some of us has suggested the dehydration of methanol to form dimethyl ether proceeds via an extended catalytic cycle in the presence of a promoter.30There are two possible mechanisms. In the first, the associative mechanism, protonation of the promoter is followed by nucleophilic attack of methanol on the methoxy group, forming DME. A subsequent reaction involving methanol reforms the promoter. The full associative mechanism is illustrated in Scheme 2.
 | ||
| Scheme 2 Reaction scheme for the associative mechanism of the formation of DME in H-ZSM-5 with methanol(s) and a promoter. The promoter is coloured purple and the reaction steps labelled in orange are those listed in Table 1. | ||
In the alternative dissociative mechanism, the promoter first rotates 90° about the carbonyl bond, allowing the formation of a surface methoxy species. Another methanol molecule reacts with this species forming DME and water. NMR studies by some of us observed that methyl formate more easily dissociated to form surface methoxy species compared to the longer alkyl chain promoters. Therefore, a bimolecular associative mechanism plays an important role in the methyl ester promoted methanol dehydration to DME. However, at low methanol coverages, and for the methyl formate promoted reaction, we do not rule out the dissociative mechanism.32
All elementary steps for both associative and dissociative mechanisms in the presence and absence of promoters are listed in Table 1. It is worth noting the competing nature of the mechanisms, i.e., the surface methoxy species formed in the dissociative mechanism deactivates that catalytic site until a second methanol molecule reacts with the surface methoxy species forming DME.
| Adsorption | CH3OH(g) + H–Z ↔ CH3OH⋯H–Z | (M1) |
| ROOCH3(g) + H–Z + 2CH3OH ↔ ROOCH3⋯H–Z + 2CH3OH | (P1) | |
| Dissociative mechanism (methanol) | CH3OH⋯H–Z ↔ H2O + CH3–Z | (M2) |
| CH3OH(g) + CH3–Z ↔ CH3OCH3⋯H–Z | (M3) | |
| Associative mechanism (methanol) | CH3OH(g) + CH3OH⋯H–Z ↔ CH3OH⋯CH3OH⋯H–Z | (M4) |
| CH3OH⋯CH3OH⋯H–Z ↔ CH3OCH3⋯H2O⋯H–Z | (M5) | |
| Dissociative mechanism (promoter) | ROOCH3⋯H–Z ↔ ROOH + CH3–Z | (P2) |
| CH3OH(g) + CH3–Z ↔ CH3OCH3⋯H–Z | (M3) | |
| Associative mechanism (promoter) | CH3OH⋯CH3OH⋯ROOCH3⋯H–Z ↔ CH3OH⋯CH3OCH3⋯ROOH⋯Z–H | (P3) |
| CH3OH⋯CH3OH⋯ROOH⋯Z–H ↔ ROOCH3⋯H–Z⋯CH3OH⋯H2O | (P4) | |
The full description of each mechanism requires the localization of stationary points on the potential energy surface. The adsorption enthalpy of a promoter molecule is given by:
| Hads = HR-a − HR | (4) |
| H‡intr = HTS − HR-a | (5) |
Results and discussion
Methanol dehydration to dimethyl ether
The calculated energetics and full reaction pathways for both the associative and dissociative mechanisms for the dehydration of methanol to DME in the absence of promoters are illustrated in Fig. 3. The structures involved in each pathway are displayed in Fig. 4. The calculated reaction enthalpy is −28 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1.
mol−1.
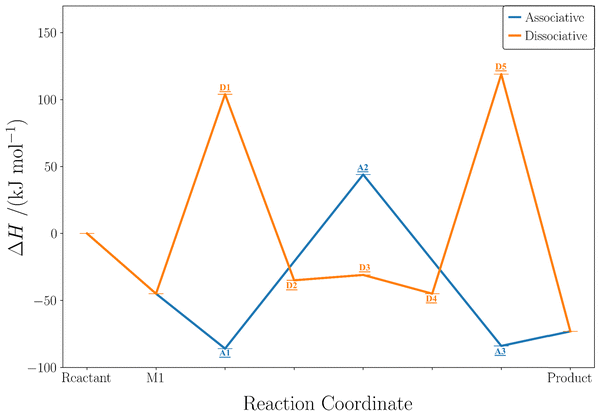 | ||
Fig. 3 Reaction coordinate diagram showing the enthalpies (ΔH; kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) calculated with the hybrid QM method, for the dehydration of methanol to DME in the competing associative and dissociative mechanisms. Since the reaction pathway differs for each mechanism only Reactant, M1, and Product are labelled on the x-axis, axis and individual labels can be found on the diagram. Labels refer to reaction coordinate structures in Fig. 4. mol−1) calculated with the hybrid QM method, for the dehydration of methanol to DME in the competing associative and dissociative mechanisms. Since the reaction pathway differs for each mechanism only Reactant, M1, and Product are labelled on the x-axis, axis and individual labels can be found on the diagram. Labels refer to reaction coordinate structures in Fig. 4. | ||
 | ||
| Fig. 4 Structures involved in the reaction coordinate for each mechanism, labels in each box refer to their corresponding enthalpy in Fig. 3, A1, A3, D2, and D4 are minima shown also in Scheme 1, the others are transition states. Colour key for boxes: blue = associative; orange = dissociative. Colour key for structures: red = oxygen; brown= carbon; pink = hydrogen. Structures made using VESTA.91 | ||
In both mechanisms, the first stage involves the adsorption of a methanol molecule to the BAS with an enthalpy of −45 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. This value is ∼40 kJ
mol−1. This value is ∼40 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 less negative than others have reported in the literature using similar methodologies;44,84,92–94 this is a consequence of taking account of the structural rearrangement discussed above and following the methodology of Hoffman et al.63 Our zeolite framework is 39 kJ
mol−1 less negative than others have reported in the literature using similar methodologies;44,84,92–94 this is a consequence of taking account of the structural rearrangement discussed above and following the methodology of Hoffman et al.63 Our zeolite framework is 39 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 lower in energy compared to those typically used in the earlier literature. In the associative mechanism, a methanol dimer is formed at the BAS with an enthalpy of −86 kJ
mol−1 lower in energy compared to those typically used in the earlier literature. In the associative mechanism, a methanol dimer is formed at the BAS with an enthalpy of −86 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. The rate-determining step—in the associative mechanism—involves DME formation from the protonated dimer intermediate with an intrinsic barrier of 130 kJ
mol−1. The rate-determining step—in the associative mechanism—involves DME formation from the protonated dimer intermediate with an intrinsic barrier of 130 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. After desorption of the water by-product, the overall reaction enthalpy is −28 kJ
mol−1. After desorption of the water by-product, the overall reaction enthalpy is −28 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 and the formation enthalpy is −73 kJ
mol−1 and the formation enthalpy is −73 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. In the dissociative mechanism, methylation of the zeolite surface takes place. The barrier height is 149 kJ
mol−1. In the dissociative mechanism, methylation of the zeolite surface takes place. The barrier height is 149 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 and the reaction enthalpy 10 kJ
mol−1 and the reaction enthalpy 10 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. Subsequently, water desorbs and a new methanol molecule is adsorbed. The final step is the methylation of methanol forming DME with a barrier of 164 kJ
mol−1. Subsequently, water desorbs and a new methanol molecule is adsorbed. The final step is the methylation of methanol forming DME with a barrier of 164 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. Our barrier for the associative mechanism is lower than those in the dissociative mechanism.
mol−1. Our barrier for the associative mechanism is lower than those in the dissociative mechanism.
Our calculated intrinsic barriers using PBE+D3 alone give similar results to those calculated in the literature.94,95 For example, using PBE+D3 we calculate an intrinsic barrier of 128 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 for the formation of a surface methoxy species, Smith et al. calculates an intrinsic barrier of 143 kJ
mol−1 for the formation of a surface methoxy species, Smith et al. calculates an intrinsic barrier of 143 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1,95 whilst Kilburn et al. calculates an intrinsic barrier of 119 kJ
mol−1,95 whilst Kilburn et al. calculates an intrinsic barrier of 119 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1.96
mol−1.96
Di Iorio et al. have investigated the same dehydration reaction in a different zeolite,25 CHA. They report results for mechanisms involving an SN2 like step involving concerted backside attack. We have also modelled the associative and dissociative mechanisms in ZSM-5 with this geometry. The results can be found in the ESI.† The calculated intrinsic enthalpy barriers for the formation of the surface methoxy species in the dissociative and associative mechanisms are 154 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 and 214 kJ
mol−1 and 214 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 respectively, values considerably higher than those for our proposed mechanisms. The structures for the transition states we propose above are similar to those reported for ZSM-5 by Nastase et al.97 on the basis of QM/MM calculations.
mol−1 respectively, values considerably higher than those for our proposed mechanisms. The structures for the transition states we propose above are similar to those reported for ZSM-5 by Nastase et al.97 on the basis of QM/MM calculations.
Tables 2 and 3 show the importance of our hybrid multilevel QM correction scheme in analysing the mechanisms for methanol dehydration to DME. All the transition states contain charged species, which are over-stabilised by SIE – inherent in DFT+D3 only. In the absence of this QM scheme, every transition state is considerably more stable and lower in energy. However, the corrections applied to the transition states in the dissociative mechanism are much larger than those in the associative mechanism due to increased charge polarisation, since an extra water molecule stabilises the charge polarisation in the associative pathway.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) of intermediates and transition states involved in the dissociative mechanism of methanol dehydration to DME, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
mol−1) of intermediates and transition states involved in the dissociative mechanism of methanol dehydration to DME, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
| Dissociative | Reactant | M1 | TS1 | M2 | Int | M3 | TS2 | Product |
|---|---|---|---|---|---|---|---|---|
| ΔHFinal | 0 | −45 | 104 | −35 | −31 | −45 | 119 | −73 |
| ΔHPBE+D3 | 0 | −41 | 87 | −7 | 7 | −9 | 100 | −61 |
| ΔEMP2 | 0 | 0 | 33 | −11 | −19 | −12 | 41 | −5 |
| ΔECCSD(T) | 0 | −4 | −16 | −17 | −19 | −24 | −22 | −7 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration to DME, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration to DME, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
| Associative | Reactant | M1 | M4 | TS | M5 | Product |
|---|---|---|---|---|---|---|
| ΔHFinal | 0 | −45 | −86 | 44 | −84 | −73 |
| ΔHPBE+D3 | 0 | −41 | −65 | 38 | −74 | −61 |
| ΔEMP2 | 0 | 0 | −18 | 21 | −7 | −5 |
| ΔECCSD(T) | 0 | −4 | −3 | −15 | −3 | −7 |
Experimental enthalpies of methanol adsorption and dehydration to DME in H-ZSM-5 reported in the literature vary widely due to the varying methods employed, the zeolite structure, and the coverage of molecules at the BAS. For example, enthalpies of adsorption of −115 and −45 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 have been reported respectively by Lee et al.98 in 1997 and by Ortega et al. in 2021.99 In Table 4 we compare our results for the associative mechanism with those from a recent experimental study.99
mol−1 have been reported respectively by Lee et al.98 in 1997 and by Ortega et al. in 2021.99 In Table 4 we compare our results for the associative mechanism with those from a recent experimental study.99
| Methanol adsorption (kJ mol−1) | Reaction enthalpy (kJ mol−1) | Ref. |
|---|---|---|
| −45 | −28 | This study |
| −44.5 ± 0.7 | −24 | Ortega et al. Expt.99 |
Our results for methanol adsorption and reaction enthalpies agree with those in Table 4 within the threshold of chemical accuracy (∼4 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1), although we caution that any such direct comparison with experiment is problematic for several reasons including the variation of adsorption enthalpies with coverage at active sites.99 Experimental heats of desorption of methanol from ZSM-5 catalysts range from 47–107 kJ
mol−1), although we caution that any such direct comparison with experiment is problematic for several reasons including the variation of adsorption enthalpies with coverage at active sites.99 Experimental heats of desorption of methanol from ZSM-5 catalysts range from 47–107 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 depending on the coverage and experimental method.100–102
mol−1 depending on the coverage and experimental method.100–102
Methyl formate: associative vs. dissociative mechanisms
Experimentally, the promoters are added after several hours of methanol feed and competitively adsorb to the BAS. The full reaction pathways, for both the associative and dissociative mechanisms for the dehydration of methanol to DME in the presence of methyl formate are reported in Tables 5 and 6 and illustrated in Fig. 5. The structures along the reaction coordinate in the dissociative mechanism are displayed in Fig. 4 (but the methanol molecule in step![[D with combining low line]](https://www.rsc.org/images/entities/b_char_0044_0332.gif)
![[1 with combining low line]](https://www.rsc.org/images/entities/b_char_0031_0332.gif) is now methyl formate) and in the associative mechanism in Fig. 6. The higher-order corrections can be found in the ESI.† The addition of a promoter molecule reveals a new reaction pathway via the associative mechanism. Instead of DME being formed from two methanol molecules, the methyl group of the promoter undergoes nucleophilic attack from one methanol molecule whilst another methanol molecule acts as a proton shuttler. As we have seen, the first stage of both mechanisms involves the adsorption of a methanol molecule to the BAS, with an adsorption enthalpy of 45 kJ
is now methyl formate) and in the associative mechanism in Fig. 6. The higher-order corrections can be found in the ESI.† The addition of a promoter molecule reveals a new reaction pathway via the associative mechanism. Instead of DME being formed from two methanol molecules, the methyl group of the promoter undergoes nucleophilic attack from one methanol molecule whilst another methanol molecule acts as a proton shuttler. As we have seen, the first stage of both mechanisms involves the adsorption of a methanol molecule to the BAS, with an adsorption enthalpy of 45 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1. The formation reaction enthalpies for the associative and dissociative mechanisms are −92 kJ
mol−1. The formation reaction enthalpies for the associative and dissociative mechanisms are −92 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 and −85 kJ
mol−1 and −85 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, respectively; these are not the same because the promoter is left adsorbed to the BAS in the associative mechanism, whereas DME is left adsorbed in the dissociative mechanism.
mol−1, respectively; these are not the same because the promoter is left adsorbed to the BAS in the associative mechanism, whereas DME is left adsorbed in the dissociative mechanism.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) of intermediates and transition states involved in the dissociative mechanism of methanol dehydration too DME with methyl formate, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
mol−1) of intermediates and transition states involved in the dissociative mechanism of methanol dehydration too DME with methyl formate, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
| Dissociative mechanism | Reaction coordinate | |||||||
|---|---|---|---|---|---|---|---|---|
| Reactant | P1 | PD1‡ | PD3 | D3 | D4 | D5 | Product | |
| ΔHfinal | 0 | −30 | 192 | −59 | −22 | −42 | 113 | −85 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration too DME with methyl formate, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration too DME with methyl formate, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
| Associative mechanism | Reaction coordinate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adsorption | Reaction 1 | Reaction 2 | Product | ||||||||
| Reactant | P1 | P1‡ | P1* | P1*‡ | P3 | P4 | P4‡ | P5* | P5*‡ | ||
| ΔHfinal | 0 | −73 | −6 | −55 | −28 | −90 | −104 | −49 | −90 | −22 | −92 |
 | ||
Fig. 5 Reaction coordinate diagram, ΔH (kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1), involving intermediates and transition states involved in methanol dehydration to DME in the presence of methyl formate in the competing associative and dissociative mechanisms. Note that the reaction coordinate is different for each mechanism so only Reactant, M1, P1, and Product are shown on the x-axis and individual labels can be found on the diagram. Structures for the dissociative mechanism are displayed in Fig. 4, while the structures for the associative are shown in Fig. 6. mol−1), involving intermediates and transition states involved in methanol dehydration to DME in the presence of methyl formate in the competing associative and dissociative mechanisms. Note that the reaction coordinate is different for each mechanism so only Reactant, M1, P1, and Product are shown on the x-axis and individual labels can be found on the diagram. Structures for the dissociative mechanism are displayed in Fig. 4, while the structures for the associative are shown in Fig. 6. | ||
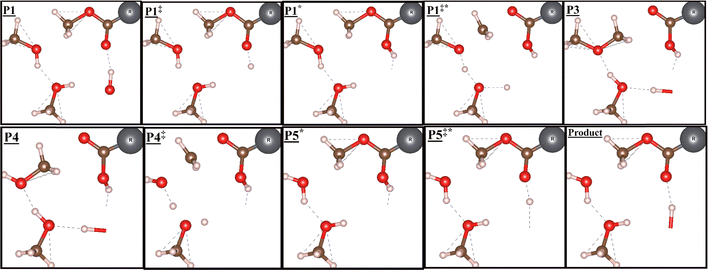 | ||
Fig. 6 Structures involved in the reaction coordinate for the associative mechanism with a promoter, labels in each box refer to their corresponding reaction coordinate in Fig. 5 and 8. Reaction coordinate ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) — —![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) describes reaction (1), whereas reaction (2) describes reaction coordinate describes reaction (1), whereas reaction (2) describes reaction coordinate ![[P with combining low line]](https://www.rsc.org/images/entities/char_0050_0332.gif) ![[4 with combining low line]](https://www.rsc.org/images/entities/char_0034_0332.gif) —Product. Colour key for structures: red = oxygen; brown = carbon; pink = hydrogen; grey = R (which refers to the alkyl chain in the promoter molecule). Structures made using VESTA.91 —Product. Colour key for structures: red = oxygen; brown = carbon; pink = hydrogen; grey = R (which refers to the alkyl chain in the promoter molecule). Structures made using VESTA.91 | ||
In the associative mechanism, the promoter points along the straight pore with an adsorption enthalpy of −73 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 to the BAS, a value larger than that of methanol itself. This is consistent with the experimental observation that the promoter displaces methanol from the BAS. The promoter is then protonated from the BAS with an intrinsic barrier and reaction enthalpy of 67 and 18 kJ
mol−1 to the BAS, a value larger than that of methanol itself. This is consistent with the experimental observation that the promoter displaces methanol from the BAS. The promoter is then protonated from the BAS with an intrinsic barrier and reaction enthalpy of 67 and 18 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, respectively. Subsequent nucleophilic attack by one of the methanol molecules on the methyl group of the promoter produces DME, formic acid, and water; the intrinsic barrier and reaction enthalpy are 27 and −35 kJ
mol−1, respectively. Subsequent nucleophilic attack by one of the methanol molecules on the methyl group of the promoter produces DME, formic acid, and water; the intrinsic barrier and reaction enthalpy are 27 and −35 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, respectively. The protonation of the promoter is the rate-determining step. Once the water has desorbed and methanol has adsorbed to the reaction site, a second reaction takes place. Re-esterification of the promoter occurs, with an intrinsic barrier of 55 kJ
mol−1, respectively. The protonation of the promoter is the rate-determining step. Once the water has desorbed and methanol has adsorbed to the reaction site, a second reaction takes place. Re-esterification of the promoter occurs, with an intrinsic barrier of 55 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, and deprotonation of the promoters to re-form the BAS with an intrinsic barrier of 68 kJ
mol−1, and deprotonation of the promoters to re-form the BAS with an intrinsic barrier of 68 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 and a calculated reaction enthalpy of −19 kJ
mol−1 and a calculated reaction enthalpy of −19 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1.
mol−1.
In the dissociative mechanism, to form the surface methoxy species the promoter must first rotate 90° about the carbonyl bond (Fig. 7) after adsorption to the BAS, since the methyl group of the promoter must be aligned to the zeolitic surface oxygen to allow dissociation. This rotation is accompanied by a decrease in the magnitude of the adsorption enthalpy by 43 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1; strikingly in this conformation the promoter adsorbs more weakly than methanol. Subsequent dissociation produces a surface methoxy species and formic acid, with a barrier and reaction enthalpy of 222 and −29 kJ
mol−1; strikingly in this conformation the promoter adsorbs more weakly than methanol. Subsequent dissociation produces a surface methoxy species and formic acid, with a barrier and reaction enthalpy of 222 and −29 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, respectively. Formic acid is then desorbed and methanol adsorbed once more. From this point on, the dissociative mechanism proceeds as in the absence of the promoter.
mol−1, respectively. Formic acid is then desorbed and methanol adsorbed once more. From this point on, the dissociative mechanism proceeds as in the absence of the promoter.
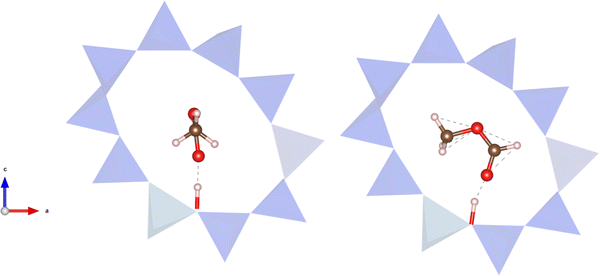 | ||
| Fig. 7 Left: Conformation of adsorbed methyl formate necessary for the associative mechanism. Right: Conformation of adsorbed methyl formate necessary for the dissociative mechanism. The BAS is at the bottom of the 10-ring zeolite structure below methyl formate. Colour key for structures: red = oxygen; brown= carbon; pink = hydrogen; blue polyhedral = TO4 tetrahedra. Structures made using VESTA.91 | ||
Although more barriers exist in the associative pathway, they are much smaller than those in the dissociative pathway; 67, 27, 55, 68 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 in the associative vs. 222 and 155 kJ
mol−1 in the associative vs. 222 and 155 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 in the dissociative. Nevertheless, the possibility of some dissociation is consistent with the experimental observation of surface methoxy species. The higher-order corrections listed in the ESI,† are similar to the corresponding values in the mechanisms with methanol only. Specifically, the corrections applied to the transition state energies in the dissociative mechanism are much greater than those applied in the associative.
mol−1 in the dissociative. Nevertheless, the possibility of some dissociation is consistent with the experimental observation of surface methoxy species. The higher-order corrections listed in the ESI,† are similar to the corresponding values in the mechanisms with methanol only. Specifically, the corrections applied to the transition state energies in the dissociative mechanism are much greater than those applied in the associative.
Larger promoters: the associative mechanism
The full reaction pathways for the associative mechanism for the remaining promoters are illustrated in Fig. 8 with corresponding values reported in Table 7, and structures for each step of the reaction coordinate are displayed in Fig. 6.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration too DME with each promoter, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
mol−1) of intermediates and transition states involved in the associative mechanism of methanol dehydration too DME with each promoter, calculated with the hybrid multilevel QM scheme relative to the enthalpy of the reactants
| Promoter | Reaction coordinate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adsorption | Reaction 1 | Reaction 2 | Product | ||||||||
| Reactant | P1 | P1‡ | P1* | P1*‡ | P3 | P4 | P4‡ | P5* | P5*‡ | ||
| Methyl acetate | 0 | −82 | −33 | −99 | −50 | −100 | −114 | −64 | −119 | −48 | −120 |
| Methyl propionate | 0 | −94 | −48 | −123 | −58 | −124 | −131 | −87 | −132 | −71 | −138 |
| Methyl n-butyrate | 0 | −104 | −60 | −132 | −72 | −130 | −136 | −95 | −137 | −84 | −147 |
| Methyl n-pentanoate | 0 | −109 | −66 | −137 | −76 | −136 | −148 | −104 | −150 | −97 | −152 |
| Methyl n-hexanoate | 0 | −119 | −71 | −145 | −80 | −144 | −156 | −118 | −161 | −106 | −166 |
The formation reaction enthalpies, i.e., relative to the reactants, and calculated reaction enthalpies, i.e., relative to the adsorbed promoter, are listed in Table 8, where methyl formate is also included for completeness. There is a significant correlation between the formation reaction enthalpies for the promoters via the associative mechanism and their experimental DME STY (Fig. 1). This is not so for the calculated reaction enthalpies. This suggests the enthalpy of reaction is highly dependent on the adsorption of the promoter. In a PBE+D3 calculation alone, a protonated species is the lowest in enthalpy, whereas the hybrid multilevel method shows the lowest enthalpy structure is the final product itself, revealing the thermodynamic driving force toward the overall product.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) for the associative mechanism for each promoter obtained by the hybrid multilevel QM method
mol−1) for the associative mechanism for each promoter obtained by the hybrid multilevel QM method
| Promoter | Methyl formate | Methyl acetate | Methyl propionate | Methyl n-butyrate | Methyl n-pentanoate | Methyl n-hexanoate |
|---|---|---|---|---|---|---|
| Formation reaction enthalpy | −92 | −120 | −138 | −147 | −152 | −166 |
| Calculated reaction enthalpy | −19 | −38 | −44 | −43 | −43 | −47 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1) for each promoter at the T1 site obtained by the hybrid multilevel QM method
mol−1) for each promoter at the T1 site obtained by the hybrid multilevel QM method
| Methyl formate | Methyl acetate | Methyl propionate | Methyl n-butyrate | Methyl n-pentanoate | Methyl n-hexanoate | |
|---|---|---|---|---|---|---|
| H ads | −73 | −82 | −94 | −104 | −109 | −119 |
Every promoter adsorbs to the BAS through a strong hydrogen bond interaction at a length of 1.49 Å between its carbonyl group and the BAS proton. The adsorption enthalpies increase with alkyl chain length due to attractive dispersion interactions between the chain and the zeolite pore walls. An extra –CH2 group in the chain increases the adsorption enthalpy by approximately 10 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1, except between methyl n-butyrate and methyl n-pentanoate. We speculate that here, the extra –CH2 group lies in an adjacent pore intersection unlike the other promoters, so it does not receive the same stabilisation from the pore walls. This effect in turn disappears with a further –CH2 group in the chain, as with methyl n-hexanoate, because the increased chain length is now sufficiently long for attractive dispersion interactions with the pore wall after the intersection. We note this provides a static view of the most stable conformation of the adsorbed molecule rather one based on the temperature-dependent spatial distribution of adsorbed molecules. Molecular dynamics sampling of MP2 quality adsorption enthalpies could be achieved using parameterisation from MP2: DFT-D potential energy surfaces, as was done recently by Berger, Rybicki, and Sauer but is beyond the scope of this work.73 The PBE+D3 functional by itself systematically predicts too strong adsorption. This is consistent with the work of Goncalves et al.86 and Sauer et al.77 on the adsorption energies of a wide range of molecules.
mol−1, except between methyl n-butyrate and methyl n-pentanoate. We speculate that here, the extra –CH2 group lies in an adjacent pore intersection unlike the other promoters, so it does not receive the same stabilisation from the pore walls. This effect in turn disappears with a further –CH2 group in the chain, as with methyl n-hexanoate, because the increased chain length is now sufficiently long for attractive dispersion interactions with the pore wall after the intersection. We note this provides a static view of the most stable conformation of the adsorbed molecule rather one based on the temperature-dependent spatial distribution of adsorbed molecules. Molecular dynamics sampling of MP2 quality adsorption enthalpies could be achieved using parameterisation from MP2: DFT-D potential energy surfaces, as was done recently by Berger, Rybicki, and Sauer but is beyond the scope of this work.73 The PBE+D3 functional by itself systematically predicts too strong adsorption. This is consistent with the work of Goncalves et al.86 and Sauer et al.77 on the adsorption energies of a wide range of molecules.
The correlation between the formation reaction enthalpy and promoter adsorption enthalpy highlights the importance of binding. The bound conformation is crucial for protonation of the promoter, i.e., the initial stage of the reaction, and promoters that adsorb more strongly to the BAS are less susceptible to inhibition of the catalytic site from by-products.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1
mol−1
| Promoter | Proton affinity | (i) Protonation | (ii) DME formation | ||
|---|---|---|---|---|---|
| H ‡intr | Reaction enthalpy | H ‡intr | Reaction enthalpy | ||
| Methyl acetate | 822103 | 49 | −17 | 49 | −14 |
| Methyl propionate | 830,103 832104 | 46 | −29 | 65 | −9 |
| Methyl n-butyrate | 836103 | 44 | −28 | 60 | −4 |
| Methyl n-pentanoate | 839,103 839104 | 43 | −28 | 61 | −10 |
| Methyl n-hexanoate | 840104 | 48 | −26 | 65 | −8 |
The transition states for both reactions are shown in Fig. 9 and relevant bond distances between labelled atoms are given in the ESI.† During protonation of the promoter molecule, O1–H1, and H1–O2 distances are ∼1.39 Å, and ∼1.26 Å, respectively, for all promoters. The proton is not equidistant between O1 and O2 but instead lies closer to the promoter molecule. Even though these bond distances do not change, the barrier and reaction enthalpy for protonation are significantly different for methyl acetate from the remaining promoters, due to the different proton affinities of the promoters (Table 10).
 | ||
| Fig. 9 The transition state found for (i) protonation of the promoter molecule, and (ii) formation of DME. The atoms involved in these steps are labelled and referred to in the ESI.† Colour key for structures: red = oxygen; brown = carbon; pink = hydrogen; grey = alkyl chain; blue polyhedral = TO4 tetrahedra. Figure made using VESTA.91 | ||
While our results support the widely accepted notion that a variety of molecules readily deprotonate the BAS, we calculate much higher barriers—up to 49 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1—than other previous computational studies of reactions in zeolites.105–109 The difference is likely due to SIE in DFT which causes the artificial stabilisation of charged structures. We find that barriers are underpredicted by up to 26 kJ
mol−1—than other previous computational studies of reactions in zeolites.105–109 The difference is likely due to SIE in DFT which causes the artificial stabilisation of charged structures. We find that barriers are underpredicted by up to 26 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 without the higher-level corrections. This is consistent with the findings of Sauer et al.71 who found that proton exchange barriers for alkanes in H-ZSM-5 were underpredicted by 36–40 kJ
mol−1 without the higher-level corrections. This is consistent with the findings of Sauer et al.71 who found that proton exchange barriers for alkanes in H-ZSM-5 were underpredicted by 36–40 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1 by DFT-D alone. Accurate calculations of proton attachment energies are crucial when modelling reactions in zeolites. A comparison of the methods and the corrections for each promoter can be found in the ESI.†
mol−1 by DFT-D alone. Accurate calculations of proton attachment energies are crucial when modelling reactions in zeolites. A comparison of the methods and the corrections for each promoter can be found in the ESI.†
The formation of DME is exothermic for every promoter and every transition state involves multiple molecules in a cyclic transition state involving proton shuttling and hydrogen bonding. Initially, a nucleophilic attack of methanol on the methoxy group of the protonated promoter molecule occurs; subsequent proton transfer from DME to another methanol molecule takes place with no barrier.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1
mol−1
| Promoter | (i) Re-esterification | (ii) Deprotonation | ||
|---|---|---|---|---|
| H ‡intr | Reaction enthalpy | H ‡intr | Reaction enthalpy | |
| Methyl acetate | 50 | −5 | 71 | −2 |
| Methyl propionate | 44 | −6 | 61 | −6 |
| Methyl n-butyrate | 41 | −1 | 53 | −10 |
| Methyl n-pentanoate | 44 | −2 | 53 | −2 |
| Methyl n-hexanoate | 38 | −5 | 55 | −5 |
The re-esterification of the promoter molecule involves a cyclic, concerted, transition state with three molecules; it is exothermic for each promoter. Umbrella inversion occurs as the –CH3 group of the methanol molecule undergoes nucleophilic attack from the oxygen atom of the promoter. Simultaneous proton transfer, from the protonated methanol to the transient hydroxide, takes place and maintains charge neutrality. Table 11 shows that the intrinsic barriers decrease as the length of the alkyl chain in the promoter increases. These data are consistent with the earlier discussion of proton affinities reported in Table 10.
Conclusions
We have employed a hybrid multilevel quantum method to explore the competing associative and dissociative mechanisms underpinning the formation of DME from methanol in H-ZSM-5 in the presence, and absence, of six methyl ester promoters. For the dehydration of methanol to DME in the absence of promoters, most of our results are in agreement with experimental values to within chemical accuracy (∼4 kJ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol−1). We show a reaction mechanism (not previously modelled) involving a promoter and two methanol molecules reacting to form DME. The promoters provide a more exothermic route to DME formation because the promoter is left adsorbed at the BAS, the intrinsic activation enthalpy barriers are also reduced. Highlighted in this work are the inaccuracies prevalent in common theoretical methodologies employed to investigate zeolite catalysis. Specifically, SIE over-stabilises polar species leading to inaccuracies in the calculation of reaction barriers within key steps of pathways, especially protonation steps. The results demonstrate the importance of high level (ab initio) calculations for drawing quantitative mechanistic conclusions, to overcome limitations of DFT. We show a thermodynamic driving force toward the final product by revealing the lowest enthalpy structure is the final product – a conclusion that would not be possible without employing the hybrid multilevel QM methodology. Overall, our results show why the promoter molecules increase the formation of DME from methanol, in H-ZSM-5, in agreement with experiment and rationalise the extent of promotion with promoter size. The promoter's adsorption to the BAS is a key factor, associated with lower overall reaction enthalpies (compared to methanol only) and lower barriers along the reaction pathway.
mol−1). We show a reaction mechanism (not previously modelled) involving a promoter and two methanol molecules reacting to form DME. The promoters provide a more exothermic route to DME formation because the promoter is left adsorbed at the BAS, the intrinsic activation enthalpy barriers are also reduced. Highlighted in this work are the inaccuracies prevalent in common theoretical methodologies employed to investigate zeolite catalysis. Specifically, SIE over-stabilises polar species leading to inaccuracies in the calculation of reaction barriers within key steps of pathways, especially protonation steps. The results demonstrate the importance of high level (ab initio) calculations for drawing quantitative mechanistic conclusions, to overcome limitations of DFT. We show a thermodynamic driving force toward the final product by revealing the lowest enthalpy structure is the final product – a conclusion that would not be possible without employing the hybrid multilevel QM methodology. Overall, our results show why the promoter molecules increase the formation of DME from methanol, in H-ZSM-5, in agreement with experiment and rationalise the extent of promotion with promoter size. The promoter's adsorption to the BAS is a key factor, associated with lower overall reaction enthalpies (compared to methanol only) and lower barriers along the reaction pathway.
Data availability
All structure files can be found online (https://doi.org/10.5281/zenodo.10261595; https://doi.org/10.5281/zenodo.10260386) and are provided as open-source material.Conflicts of interest
The ester promoters described in this manuscript are the subject of two patent filings (WO2019/037764 and WO2019037768) by BP plc and BP (China) Holdings Limited. There are no other conflicts to declare.References
- M. Tomatis, A. Mahmud Parvez, M. T. Afzal, S. Mareta, T. Wu, J. He and T. He, Fuel, 2019, 254, 115627 CrossRef CAS
.
- M. Marchionna, R. Patrini, D. Sanfilippo and G. Migliavacca, Fuel Process. Technol., 2008, 89, 1255–1261 CrossRef CAS
.
- R. Anggarani, M. Muchar, C. S. Wibowo and R. Sukaraharja, Energy Proc., 2015, 65, 274–281 CrossRef CAS
.
- M. Himabindu and R. V. Ravikrishna, J. Renewable Sustainable Energy, 2010, 2, 52701 CrossRef
.
- M. Martín, J. CO2 Utilization, 2016, 13, 105–113 CrossRef
.
- R. G. Santiago, J. A. Coelho, S. M. P. de Lucena, A. P. S. Musse, M. de, F. Portilho, E. Rodriguez-Castellón, D. C. S. de Azevedo and M. Bastos-Neto, Front. Chem., 2022, 10, 1–11 Search PubMed
.
- S. P. Naik, T. Ryu, V. Bui, J. D. Miller, N. B. Drinnan and W. Zmierczak, Chem. Eng. J., 2011, 167, 362–368 CrossRef CAS
.
- S. Le Goff, Electric Hybrid Rail Technol., 2022, 2021, 36–37 CrossRef
.
- F. Pontzen, W. Liebner, V. Gronemann, M. Rothaemel and B. Ahlers, Catal. Today, 2011, 171, 242–250 CrossRef CAS
.
- S. Ernst, Angew. Chem., Int. Ed., 2011, 50, 5425–5426 CrossRef CAS
.
- J. Liang, Z. Liang, R. Zou and Y. Zhao, Adv. Mater., 2017, 29, 1701139 CrossRef PubMed
.
- G. Sastre and A. Corma, J. Mol. Catal. A: Chem., 2009, 305, 3–7 CrossRef CAS
.
- A. B. Grommet, M. Feller and R. Klajn, Nat. Nanotechnol., 2020, 15, 256–271 CrossRef CAS PubMed
.
- E. Grifoni, G. M. Piccini, J. A. Lercher, V. A. Glezakou, R. Rousseau and M. Parrinello, Nat. Commun., 2021, 12, 2630 CrossRef CAS PubMed
.
- J. F. Haw, W. Song, D. M. Marcus and J. B. Nicholas, Acc. Chem. Res., 2003, 36, 317–326 CrossRef CAS PubMed
.
- M. Stöcker, Microporous Mesoporous Mater., 1999, 29, 3–48 CrossRef
.
- U. Olsbye, S. Svelle, K. P. Lillerud, Z. H. Wei, Y. Y. Chen, J. F. Li, J. G. Wang and W. B. Fan, Chem. Soc. Rev., 2015, 44, 7155–7176 RSC
.
- B. E. Langner, Appl. Catal., 1982, 2, 289–302 CrossRef CAS
.
- A. D. Chowdhury, A. L. Paioni, K. Houben, G. T. Whiting, M. Baldus and B. M. Weckhuysen, Angew. Chem., Int. Ed., 2018, 57, 8095–8099 CrossRef CAS PubMed
.
- T. Mole, G. Bett and D. Seddon, J. Catal., 1983, 84, 435–445 CrossRef CAS
.
- I. Lezcano-Gonzalez, E. Campbell, A. E. J. Hoffman, M. Bocus, I. V. Sazanovich, M. Towrie, M. Agote-Aran, E. K. Gibson, A. Greenaway, K. De Wispelaere, V. Van Speybroeck and A. M. Beale, Nat. Mater., 2020, 19, 1081–1087 CrossRef CAS PubMed
.
- I. M. Dahl and S. Kolboe, Catal. Lett., 1993, 20, 329–336 CrossRef CAS
.
- S. Kolboe, Stud. Surf. Sci. Catal., 1988, 36, 189–193 CrossRef CAS
.
- A. J. Jones, S. I. Zones and E. Iglesia, J. Phys. Chem. C, 2014, 118, 17787–17800 CrossRef CAS
.
- J. R. Di Iorio, A. J. Hoffman, C. T. Nimlos, S. Nystrom, D. Hibbitts and R. Gounder, J. Catal., 2019, 380, 161–177 CrossRef CAS
.
- R. T. Carr, M. Neurock and E. Iglesia, J. Catal., 2011, 278, 78–93 CrossRef CAS
.
- A. Ghorbanpour, J. D. Rimer and L. C. Grabow, ACS Catal., 2016, 6, 2287–2298 CrossRef CAS
.
- A. J. Jones and E. Iglesia, Angew. Chem., Int. Ed., 2014, 53, 12177–12181 CrossRef CAS PubMed
.
- J. Park, J. Cho, M. J. Park and W. B. Lee, Catal. Today, 2021, 375, 314–323 CrossRef CAS
.
- B. J. Dennis-Smither, Z. Yang, C. Buda, X. Liu, N. Sainty, X. Tan and G. J. Sunley, Chem. Commun., 2019, 55, 13804–13807 RSC
.
- J. Crossley-Lewis, J. Dunn, C. Buda, G. J. Sunley, A. M. Elena, I. T. Todorov, C. W. Yong, D. R. Glowacki, A. J. Mulholland and N. L. Allan, J. Mol. Graph. Model., 2023, 125, 108606 CrossRef CAS PubMed
.
- Z. Yang, B. J. Dennis-Smither, Z. Xu, Z. Zhao, M. Guo, N. Sainty, G. Hou, X. Liu and G. J. Sunley, Chemistry, 2023, 5, 511–525 CrossRef CAS
.
- Z. Yang, B. J. Dennis-smither, C. Buda, A. Easey, F. Jackson, G. A. Price, N. Sainty, X. Tan, Z. Xu and G. J. Sunley, Catal. Sci. Technol., 2023, 13, 3590–3605 RSC
.
- F. Neese, A. Hansen, F. Wennmohs and S. Grimme, Acc. Chem. Res., 2009, 42, 641–648 CrossRef CAS PubMed
.
- Y. Zhao and D. G. Truhlar, J. Phys. Chem. A, 2005, 109, 5656–5667 CrossRef CAS PubMed
.
- S. Mallikarjun Sharada, T. Bligaard, A. C. Luntz, G. J. Kroes and J. K. Nørskov, J. Phys. Chem. C, 2017, 121, 19807–19815 CrossRef CAS
.
- E. Ruiz, D. R. Salahub and A. Vela, J. Phys. Chem., 1996, 100, 12265–12276 CrossRef CAS
.
- J. Klime, D. R. Bowler and A. Michaelides, Phys. Rev. B: Condens. Matter Mater. Phys., 2011, 83, 1–29 Search PubMed
.
- B. Vlaisavljevich, J. Huck, Z. Hulvey, K. Lee, J. A. Mason, J. B. Neaton, J. R. Long, C. M. Brown, D. Alfè, A. Michaelides and B. Smit, J. Phys. Chem. A, 2017, 121, 4139–4151 CrossRef CAS PubMed
.
- F. Claeyssens, J. N. Harvey, F. R. Manby, R. A. Mata, A. J. Mulholland, K. E. Ranaghan, M. Schütz, S. Thiel, W. Thiel and H.-J. Werner, Angew. Chem., Int. Ed., 2006, 45, 6856–6859 CrossRef CAS PubMed
.
- J. Sauer, Acc. Chem. Res., 2019, 52, 3502–3510 CrossRef CAS PubMed
.
- P. Nachtigall and J. Sauer, Stud. Surf. Sci. Catal., 2007, 168, 701–736 CrossRef
.
- G. Piccini, M. Alessio and J. Sauer, Angew. Chem., Int. Ed., 2016, 55, 5235–5237 CrossRef CAS PubMed
.
- G. Piccini, M. Alessio and J. Sauer, Phys. Chem. Chem. Phys., 2018, 20, 19664–19670 RSC
.
- K. E. Ranaghan, D. Shchepanovska, S. J. Bennie, N. Lawan, S. J. Macrae, J. Zurek, F. R. Manby and A. J. Mulholland, J. Chem. Inf. Model., 2019, 59, 2063–2078 CrossRef CAS PubMed
.
- L. O. Jones, M. A. Mosquera, G. C. Schatz and M. A. Ratner, J. Am. Chem. Soc., 2020, 142, 3281–3295 CrossRef CAS PubMed
.
- Database of Zeolite Structures, https://www.iza-structure.org/databases/, (accessed October 2022).
- R. Grau-Crespo, A. G. Peralta, A. R. Ruiz-Salvador, A. Gómez and R. López-Cordero, Phys. Chem. Chem. Phys., 2000, 2, 5716–5722 RSC
.
- A. Palčić and V. Valtchev, Appl. Catal., A, 2020, 606, 117795 CrossRef
.
- M. Trachta, O. Bludský, J. Vaculík, R. Bulánek and M. Rubeš, Sci. Rep., 2023, 13, 12380 CrossRef CAS PubMed
.
- M. Boronat and A. Corma, ACS Catal., 2019, 9, 1539–1548 CrossRef CAS PubMed
.
- H. Windeck, F. Berger and J. Sauer, Angew. Chem., Int. Ed., 2023, 62, e202303204 CrossRef CAS PubMed
.
- G. Kresse and J. Hafner, Phys. Rev. B: Condens. Matter Mater. Phys., 1993, 47, 558–561 CrossRef CAS PubMed
.
- G. Kresse and J. Furthmüller, Comput. Mater. Sci., 1996, 6, 15–50 CrossRef CAS
.
- G. Kresse and D. Joubert, J. Phys. Chem. A, 2020, 124, 4053–4061 CrossRef PubMed
.
- J. Kresse and G. Furthmuller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169–11184 CrossRef PubMed
.
- G. Kresse and D. Joubert, Phys. Rev. B: Condens. Matter Mater. Phys., 1999, 59, 1758–1775 CrossRef CAS
.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed
.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed
.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456–1465 CrossRef CAS PubMed
.
- V. Wang, N. Xu, J. C. Liu, G. Tang and W. T. Geng, Comput. Phys. Commun., 2021, 267, 108033 CrossRef CAS
.
-
C. J. Cramer, Essentials of Computational Chemistry: Second Edition, J Wiley, New York, 2004 Search PubMed
.
- A. Hoffman, M. Deluca and D. Hibbitts, J. Phys. Chem. C, 2019, 123, 6572–6585 CrossRef CAS
.
- G. Henkelman and H. Jónsson, J. Chem. Phys., 2000, 113, 9978–9985 CrossRef CAS
.
-
H. Jonsson, G. Mills and K. W. Jacobsen, Classical and Quantum Dynamics in Condensed Phase Simulations, World Scientific, Singapore, 1998 Search PubMed
.
- G. Henkelman, B. P. Uberuaga and H. Jonsson, J. Chem. Phys., 2000, 113, 9901–9904 CrossRef CAS
.
-
M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, Williams, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16 (Revision A.03), Gaussian Inc., Pittsburgh, PA, 2016 Search PubMed
.
- F. Berger, M. Rybicki and J. Sauer, J. Catal., 2021, 395, 117–128 CrossRef CAS
.
- F. Berger and J. Sauer, Angew. Chem., Int. Ed., 2021, 60, 3529–3533 CrossRef CAS PubMed
.
- G. Piccini and J. Sauer, J. Chem. Theory Comput., 2014, 10, 2479–2487 CrossRef CAS PubMed
.
- M. Rybicki and J. Sauer, J. Am. Chem. Soc., 2018, 140, 18151–18161 CrossRef CAS PubMed
.
- C. Tuma and J. Sauer, Chem. Phys. Lett., 2004, 387, 388–394 CrossRef CAS
.
- F. Berger, M. Rybicki and J. Sauer, ACS Catal., 2023, 13, 2011–2024 CrossRef CAS
.
- U. Eichler, C. M. Kölmel and J. Sauer, J. Comput. Chem., 1997, 18, 463–477 CrossRef CAS
.
- S. Svelle, C. Tuma, X. Rozanska, T. Kerber and J. Sauer, J. Am. Chem. Soc., 2009, 131, 816–825 CrossRef CAS PubMed
.
- K. P. Schröder, J. Sauer, M. Leslie, C. Richard and C. R. A. Catlow, Zeolites, 1992, 12, 20–23 CrossRef
.
- N. Hansen, T. Kerber, J. Sauer, A. T. Bell and F. J. Keil, J. Am. Chem. Soc., 2010, 132, 11525–11538 CrossRef CAS PubMed
.
- K. P. Schröder, J. Sauer, M. Leslie, C. Richard, C. R. A. Catlow and J. M. Thomas, Chem. Phys. Lett., 1992, 188, 320–325 CrossRef
.
- G. Piccini, M. Alessio, J. Sauer, Y. Zhi, Y. Liu, R. Kolvenbach, A. Jentys and J. A. Lercher, J. Phys. Chem. C, 2015, 119, 6128–6137 CrossRef CAS
.
- C. Campbell, R. Van Santen, M. Stamatakis, N. Collis, H. J. Freund, C. Plaisance, J. Sauer, B. Garrett, E. Gross, A. Kotarba, B. Weckhuysen, W. Ueda, C. Reece, C. R. A Catlow, A. Trunschke, L. Briquet, T. Ho, I. Panas, D. Willock, C. Friend, M. Bowker, N. Robinson, A. Bruix, S. Gupta, B. Gates, A. Corma, R. Madix, D. Manganas, A. Roldan, A. O’malley, J. Staszak-Jirkovsky and J. Jirkovsky, Faraday Discuss., 2016, 188, 279–307 RSC
.
- P. Selvam, J. Sauer, B. Garrett, C. Campbell, R. Van Santen, P. Davies, A. L. Miller, M. Bowker, G. Hutchings, D. Wotton, H.-J. Freund, M. Howard, M. Feaviour, R. Burch, A. Maria, R. Galletti, E. Gross, F. Ivars Barcelo, A. Kotarba, S. Kondrat, B. Weckhuysen, B. Majumdar, A. Bruix, N. Fischer, B. Gates, J. Moulijn, A. Roldan, N. Novak Tusar, T. Jakubek, D. Willock, M. Craven, K. Sethu, C. R. A. Catlow, R. Madix, H. Manyar, C. Friend, A. Corma, P. Wells, W. Ueda, A. Trunschke and R. Palmer, Faraday Discuss., 2016, 188, 131 RSC
.
- F. Weigend, Phys. Chem. Chem. Phys., 2006, 8, 1057–1065 RSC
.
- F. Weigend and R. Ahlrichs, Phys. Chem. Chem. Phys., 2005, 7, 3297–3305 RSC
.
- S. Svelle, C. Tuma, X. Rozanska, T. Kerber and J. Sauer, J. Am. Chem. Soc., 2009, 131, 816–825 CrossRef CAS PubMed
.
- P. N. Plessow and F. Studt, J. Phys. Chem. Lett., 2020, 11, 4305–4310 CrossRef CAS PubMed
.
- T. J. Goncalves, P. N. Plessow and F. Studt, ChemCatChem, 2019, 11, 4368–4376 CrossRef CAS
.
- T. H. Dunning, J. Chem. Phys., 1989, 90, 1007–1023 CrossRef CAS
.
- D. E. Woon and T. H. Dunning, J. Chem. Phys., 1993, 98, 1358–1371 CrossRef CAS
.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553–566 CrossRef CAS
.
- O. Mowla, E. Kennedy and M. Stockenhuber, Fuel, 2018, 232, 12–26 CrossRef CAS
.
- K. Momma and F. Izumi, J. Appl. Crystallogr., 2011, 44, 1272–1276 CrossRef CAS
.
- J. Van Der Mynsbrugge, K. Hemelsoet, M. Vandichel, M. Waroquier and V. Van Speybroeck, J. Phys. Chem. C, 2012, 116, 5499–5508 CrossRef CAS
.
- L. Kilburn, M. DeLuca, A. J. Hoffman, S. Patel and D. Hibbitts, J. Catal., 2021, 400, 124–139 CrossRef CAS
.
- A. Ghorbanpour, J. D. Rimer and L. C. Grabow, Catal. Commun., 2014, 52, 98–102 CrossRef CAS
.
- A. T. Smith, P. N. Plessow and F. Studt, J. Phys. Chem. C, 2021, 125, 20373–20379 CrossRef CAS
.
- L. Kilburn, M. DeLuca, A. J. Hoffman, S. Patel and D. Hibbitts, J. Catal., 2021, 400, 124–139 CrossRef CAS
.
- S. A. F. Nastase, C. R. A. Catlow and A. J. Logsdail, Phys. Chem. Chem. Phys., 2021, 23, 2088–2096 RSC
.
- C. C. Lee, R. J. Gorte and W. E. Farneth, J. Phys. Chem. B, 1997, 101, 3811–3817 CrossRef CAS
.
- C. Ortega, M. Rezaei, V. Hessel and G. Kolb, Chem. Eng. J., 2018, 347, 741–753 CrossRef CAS
.
- T. Omojola, N. Cherkasov, A. I. McNab, D. B. Lukyanov, J. A. Anderson, E. V. Rebrov and A. C. van Veen, Catal. Lett., 2018, 148, 474–488 CrossRef CAS
.
- G. Mirth, J. A. Lercher, M. W. Anderson and J. Klinowski, J. Chem. Soc., Faraday Trans., 1990, 86, 3039–3044 RSC
.
- C. G. Pope, J. Chem. Soc., Faraday Trans., 1993, 89, 1139–1141 RSC
.
- S. G. Lias, J. E. Bartmess, J. F. Liebman, J. L. Holmes, R. D. Levin and W. G. Mallard, J. Phys. Chem. Ref. Data, 1988, 17, 1–861 CrossRef
.
- J. L. Holmes, N. A. van Huizen and P. C. Burgers, Eur. J. Mass Spectrosc., 2017, 23, 341–350 CrossRef CAS PubMed
.
- P. Liu and D. Mei, J. Phys. Chem. C, 2020, 124, 22568–22576 CrossRef CAS
.
- N. S. Gould, S. Li, H. J. Cho, H. Landfield, S. Caratzoulas, D. Vlachos, P. Bai and B. Xu, Nat. Commun., 2020, 11, 1–13 CrossRef PubMed
.
- P. Liu, Z. Yan and D. Mei, Phys. Chem. Chem. Phys., 2021, 23, 10395–10401 RSC
.
- J. H. Hack, J. P. Dombrowski, X. Ma, Y. Chen, N. H. C. Lewis, W. B. Carpenter, C. Li, G. A. Voth, H. H. Kung and A. Tokmakoff, J. Am. Chem. Soc., 2021, 143, 10203–10213 CrossRef CAS PubMed
.
- P. Deshlahra and E. Iglesia, Chem. Commun., 2020, 56, 7371–7398 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cp05987a |
| This journal is © the Owner Societies 2024 |