Open Access Article
Open Access ArticleExamination of photoinduced radicals in two crystal forms of triphenylamine bis-urea macrocycles†
Md Faizul
Islam
 ,
Gamage Isuri P.
Wijesekera
,
Gamage Isuri P.
Wijesekera
 ,
Ammon J.
Sindt
,
Mark D.
Smith
,
Ammon J.
Sindt
,
Mark D.
Smith
 and
Linda S.
Shimizu
and
Linda S.
Shimizu
 *
*
Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC 29208, USA. E-mail: shimizls@mailbox.sc.edu
First published on 3rd October 2024
Abstract
This study probes two solvates of triphenylamine (TPA) bis-urea macrocycle 1 and their activated structures to evaluate their maximum photoinduced radicals (PIRs), the subsequent decay of the radicals, and their regeneration. The hierarchical assembly of TPAs shows promise in stabilizing less substituted derivatives, potentially expanding the utility of TPAs that lack stabilizing para-substituents. Single crystal structure analysis reveals that host 1 adopts a planar conformation with the two ureas pointing in opposite directions when dimethoxyethane (DME) is encapsulated within the channel. Whereas previously, 1 adopted a bowl-shaped conformation with the two ureas pointing in the same direction (syn) with dimethylsulfoxide (DMSO) bound within the channels. Removal of the guests gives identical activated structures. The bulk materials of 1 are characterized by powder X-ray diffraction (PXRD), thermogravimetric analysis (TGA), and differential scanning calorimetry (DSC). Finally, the process of radical generation under UV-irradiation, decay, and regeneration of radicals was monitored by electron paramagnetic resonance (EPR) spectroscopy. While macrocycle conformation and extended structure are important, the presence of guests was most significant for PIR percentages.
1. Introduction
Supramolecular synthons have been widely used to organize molecular building blocks into desired hierarchical structures to afford solid crystalline forms and polymorphs.1,2 The rational design of these molecular building blocks can yield crystalline solids with permanent cavities.3 Uptake of guest species inside these porous solids can modify the framework's structure, promoting reorganization or recrystallization processes that modulate their properties. For example, incorporating redox-active guests can induce conductivity in metal–organic frameworks (MOFs),4 while the encapsulation of pyrene guests can trigger structural changes in a porphyrin prism.5Supramolecular strategies have been applied to stabilize triphenylamine (TPA) derivatives and their radical cations as generated by chemical, electrochemical, or photochemical methods.6 TPA is an excellent electron donor with favorable redox properties and has been widely incorporated into electronic materials7 and fluorescent probes.8,9 Hierarchical structures that organize TPA with π–π stacking interactions are thought to aid the delocalization of the spin density of their radical cations, ultimately enhancing their stability.10,11 Relative orientation of the TPA neighbors upon assembly can further modulate its photophysical properties. For example, Giuseppone's group studies on TPA supramolecular polymers showed that TPAs could stack with its two limiting enantiomers of TPA alternating between the neighbors or with TPA cores of similar chirality stacked on top of each other.6 These different orientations can influence the hyperfine interactions between the TPA N-centers and increase photoinduced radical (PIR) formation.11,12 PIRs are increasingly recognized due to their exceptional redox properties which enable them to be utilized in photocatalysis and charge/electron transfer events for conductive applications.13
While most TPAs are para-substituted to prevent degradation,10 the Shimizu group has shown that urea-directed assembly of TPAs can enhance the stability of TPAs and enable their PIRs to decay, presumably through a reverse electron transfer, without degradation.14–17 Both host 1 (X = H) and host 2 (X = Br) have been reported, (Fig. 1A) and exhibit columnar assembly. However, host 2 is fully substituted at its para-positions and expected to be more stable. Thus, prior PIR studies focused extensively on this material. Individual columns of 2 pack into hexagonal arrays affording porous crystalline materials that contain the solvent of crystallization (dimethoxyethane, DME).16 Interestingly, heating/activation removes the DME from host 2, and new guests can be introduced, which alters the number of PIRs observed up to 0.85% for 2·0.56benzene. For example, host 2 complexes with less polar guests formed more PIRs upon UV irradiation than host 2 crystals with more polar guests.17 In each case, the PIRs decay slowly to return to starting materials without degradation and can be regenerated simply upon re-exposure to light.
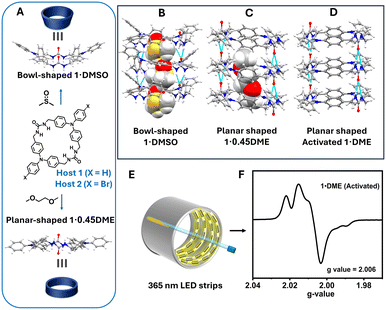 | ||
| Fig. 1 A) TPA bis-urea macrocycles 1 and 2 assemble into columns facilitated by hydrogen bonding of the urea groups, B) prior work on 1·DMSO showed it adopts a bowl-shaped structure in DMSO where the two urea groups are parallel,17 C) 1·0.45DME adopts a planar structure with anti-parallel urea groups, D) heating 1·0.45DME gives an activated structure, E) crystalline materials were UV-irradiation under Ar (g) and then transferred to EPR to measurements, F) photoinduced radical (PIR) formation examined by EPR. | ||
Given our observations that assembly can stabilize TPAs, we were interested to see if the parent bis-urea macrocycle 1, despite having unsubstituted para-positions, would exhibit enhanced stability, enabling us to probe multiple crystallization methods as well as thoroughly examine its PIRs. Herein, we scale the synthesis of this material to investigate how different crystallization environments influence the columnar organization of TPA 1. A new solvate 1·0.45DME and its solvent-free (activated) form are compared with the resynthesized 1·DMSO (Fig. 1 and 2). Intriguingly, macrocycle 1 adopts a planar structure with its two ureas pointing in opposite directions (anti) when crystallized by vapor diffusion of DME into DMSO solutions of 1. In contrast, 1 adopts a bowl-shaped structure with its ureas aligned in parallel (syn-conformation) upon vapor diffusion of H2O into the same solution (Fig. 1A).17 Further screening of crystallization conditions for 2 did not uncover guest-induced changes. While 2·0.72DMSO was structurally characterized herein, it retained its planar shape observed in the previously reported structure 2·0.5DME (Fig. 2G). Slight structural differences in the assemblies of 1 in the presence/absence of solvent guests were found to influence both the concentration and stability of their PIRs. Surprisingly, maximum PIR values for activated host 1 obtained from either solvate was ∼1.4%, which currently represents the highest reported values for crystalline TPAs.
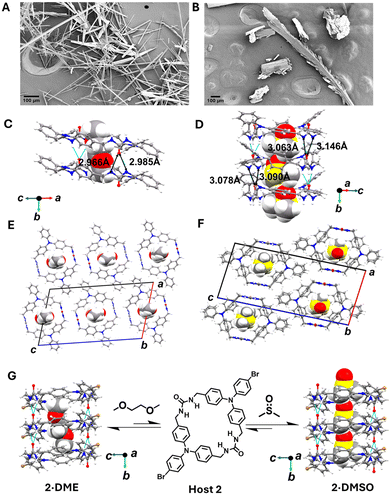 | ||
| Fig. 2 FESEM images and views from the crystal structures. A) FESEM image of 1·0.45DME complex showing long needle-like crystals, B) FESEM image of 1·DMSO complex with multifaceted cylindrical morphology, C) urea hydrogen bonding distances between neighboring molecules in 1·0.45DME, D) urea hydrogen bonding distances in the 1·DMSO shows the bowl shape,15 E) packing motif of 1·0.45DME complex, F) packing motif of 1·DMSO complex, G) comparison of prior reported host 2·0.5DME,14 which also showed similar planar structure and columnar organization with 2·0.72DMSO. | ||
2. Experimental section
2.1 Synthesis and crystallization of the host
Both hosts were resynthesized in five steps according to the literature procedures.17 Two different solvated forms of 1 were obtained (Fig. 2A and B). Both are colorless with an acicular morphology. 1·0.45DME was crystallized by vapor diffusion of DME into the DMSO solution (10 mg mL−1) of host 1 at room temperature. 1·DMSO was crystallized by the vapor diffusion of H2O into the DMSO solution (10 mg mL−1) of host 1, as observed before.17 Macrocycle 2 was crystallized by the vapor diffusion of H2O or DME into the DMSO solution of 2 (2.5 mg mL−1), affording 2·DMSO and 2·0.5DME, respectively. The 1·0.45DME complex was activated by heating and diffracted for SCXRD. The three new structures were deposited into the Cambridge Structural Database (CSD), CCDC No. 2344711, 2344712, 2344713.2.2 Single-crystal X-ray diffraction (SCXRD)
X-ray intensity data were collected using a Bruker D8 QUEST diffractometer with a PHOTON-II area detector and an Incoatec Microfocus Source (Mo Kα radiation, λ = 0.71073 Å). Data for 1·0.45DME were collected at 301(2) while data for 1·DME (activated) and 2·DMSO were collected at 100(2) K. The raw area detector data frames were reduced, scaled, ‘detwinned’ and corrected for absorption effects using the Bruker APEX3, Cell_Now, SAINT+, SADABS and TWINABS (for 1·DMSO) programs.18,19 The structures were solved with SHELXT.20,21 Subsequent difference Fourier calculations and full-matrix least-squares refinement against F2 were performed with SHELXL-2018 using OLEX2.22 SCXRD data for the three new structures were deposited in the database, CCDC no. 2344711–2344713, while the fourth structure matched that previously deposited CCDC no. 1961246. See ESI† for full details.2.3 Thermogravimetric analysis (TGA)
TGA was carried out using TA instruments SDT-Q600 simultaneous DTA/TGA instrument. TGA was measured at a rate of 2 °C min−1 from 25–200 °C. All measurements were done with a 5 minute isotherm before temperature increase, followed by a 30 minute isotherm.2.4 Differential scanning calorimetry analysis (DSC)
DSC was carried out using TA instruments Q2000 with a nitrogen atmosphere and a heating/cooling rate of 10 °C min−1.2.5 Power X-ray diffraction (PXRD)
PXRD data were collected on a Rigaku MiniFlex 6G powder X-ray diffractometer using a Bragg–Brentano geometry with CuKα radiation. The step scans covered the angular range 2–40° 2θ in steps of 1° per minute with accelerating voltage and current of 40 kV and 15 mA, respectively.2.6 Scanning electron microscopy (SEM)
SEM measurements were collected in Zeiss Gemini500 thermal field emission scanning electron microscope (FESEM). The samples were spread over a conductive carbon tape attached to an Al pin pan mounted on the specimen holder. Images were captured with the secondary electron detector (SE2) at a working distance of ∼10 mm with an acceleration voltage of ∼10 kV. The image size is 1024 × 768 pixels with line average scanning mode with a Zeiss setting of 8.2.7 Electron paramagnetic resonance (EPR) measurements
EPR measurements were carried out on a Bruker EMX plus equipped with a Bruker X-band microwave bridgehead and Xenon software (v 1.1b.66). All spectra were recorded at room temperature and a power of ∼1.589 mW with a modulation amplitude of 2.0 G. The double integration to obtain peak areas was performed in the Xenon software. Samples were sealed under argon and UV-irradiated in Norell Suprasil Quartz EPR tubes, then transferred to the EPR for measurements. 365 nm LEDs were used as light source. Dark decay studies were carried out after irradiating samples up to their maximum radical generation and storing them in dark under argon. EPR spectra were recorded over 7 days. Radical signals were regenerated by irradiating the sample overnight and monitoring their decay for 2 days. All the spectra were doubly integrated to obtain the area under the curve and plotted against time after UV irradiation.3. Results and discussion
3.1 Crystal morphology and crystal structure analysis
Given the literature precedents that TPAs with unsubstituted para-positions are less stable, we had previously prepared only small amounts of host 1. However, assembled urea-tethered TPA dimers also lacked para-substituents and exhibited good stability,15,17 which emboldened us to scale the synthesis of 1 to probe its crystalline forms, and extensively characterize its PIRs with different guests. Macrocycle 1 was sparingly soluble in DMF and DMSO and gave crystals by vapor diffusion of a poor solvent into 1 in DMSO (10 mg mL−1). Initially, the morphology of the crystals of 1·0.45DME and 1·DMSO complexes were analysed using field emission scanning electron microscopy (FESEM). The FESEM images of the complexes revealed that while the single crystals of both complexes show conventional needle-shaped morphology of the bis-urea crystals, their fine structure vary significantly. 1·0.45DME formed typical columnar crystals without any visible defects (Fig. 2A), while 1·DMSO complex displays multifaceted cylindrical morphology with defects along c-axis (Fig. 2B). Further analysis of the single crystal structure of the 1·0.45DME complex revealed that the macrocycle crystallizes in the monoclinic space group P21/c. The macrocycle adopts a planar conformation with its two urea groups aligned in opposite directions to minimize the dipole interactions.23 The asymmetric unit of the 1·0.45DME complex consists of half of one crystallographically independent host molecule located on a crystallographic inversion center and disordered DME guest molecules with a host–guest ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.45 (Fig. 2C). The macrocycles organize into a columnar structure through the characteristic bifurcated urea hydrogen bonds (d(N⋯O) = 2.966(5) Å, 2.985(5) Å). This creates infinite hydrogen-bonded tubes along the crystallographic b-axis with a macrocycle to macrocycle repeat distance of 4.712 Å (Fig. S9†). Additional stabilization occurs through intracolumnar π-stacking of the TPA groups of the building blocks (Fig. 2E, Table S2†). Further analysis of the structure shows the N⋯N distances in one independent macrocycle unit is 10.776 Å (Fig. S8†). The repeating TPA units with similar chirality are stacked on each other with N⋯N distance of 4.712 Å (Fig. 3C). Investigations of the propeller arrangement of the TPA aromatic rings within the same macrocycle show that they are in symmetrical orientation (Fig. S13†).
0.45 (Fig. 2C). The macrocycles organize into a columnar structure through the characteristic bifurcated urea hydrogen bonds (d(N⋯O) = 2.966(5) Å, 2.985(5) Å). This creates infinite hydrogen-bonded tubes along the crystallographic b-axis with a macrocycle to macrocycle repeat distance of 4.712 Å (Fig. S9†). Additional stabilization occurs through intracolumnar π-stacking of the TPA groups of the building blocks (Fig. 2E, Table S2†). Further analysis of the structure shows the N⋯N distances in one independent macrocycle unit is 10.776 Å (Fig. S8†). The repeating TPA units with similar chirality are stacked on each other with N⋯N distance of 4.712 Å (Fig. 3C). Investigations of the propeller arrangement of the TPA aromatic rings within the same macrocycle show that they are in symmetrical orientation (Fig. S13†).
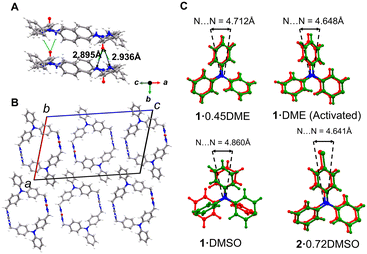 | ||
| Fig. 3 A) Crystal structure of 1·DME (activated), B) packing structure of the 1·DME (activated), C) comparison of the N⋯N distances in 1·0.45DME, 1·DME (activated), 1·DMSO and 2·0.72DMSO complex. | ||
In the case of 1·DMSO complex,17 the compound crystallized in the monoclinic space group P21/c. The asymmetric unit consists of one complete host molecule and one DMSO molecule, with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest ratio (Fig. 2D). The guest DMSO is disordered over two closely spaced orientations. Surprisingly, the host adopts an atypical syn-conformation, where two urea groups in a single macrocycle are oriented in the same direction. The macrocycles assemble into similar columnar structures along the crystallographic b-axis with DMSO inside using the three-centered urea hydrogen bonding motif (d(N⋯O) = 3.090(5) Å, 3.063(5) Å, 3.147(5) Å, 3.078(5) Å). The macrocycle-to-macrocycle distance within a column is 4.843 Å, slightly larger than the 1·0.45DME complex (Fig. S9†). The stacking is further stabilized by the edge-to-face π-stacking of the TPA groups (Fig. 3C, Table S2†). The packing shows that the neighboring columns are oriented oppositely to minimize the macrodipoles (Fig. 2F and S11†). The N⋯N distance in a single macrocycle unit is found to be 10.24 Å, which is slightly shorter than the 1·0.45DME complex (Fig. S8†). The TPA units with similar chirality stack on each other with N⋯N center distance of 4.860 Å (Fig. 3C). In contrast to the 1·0.45DME complex, 1·DMSO complex shows unsymmetrical propeller arrangement of the aromatic rings of the TPA unit (Fig. S13†).
1 host–guest ratio (Fig. 2D). The guest DMSO is disordered over two closely spaced orientations. Surprisingly, the host adopts an atypical syn-conformation, where two urea groups in a single macrocycle are oriented in the same direction. The macrocycles assemble into similar columnar structures along the crystallographic b-axis with DMSO inside using the three-centered urea hydrogen bonding motif (d(N⋯O) = 3.090(5) Å, 3.063(5) Å, 3.147(5) Å, 3.078(5) Å). The macrocycle-to-macrocycle distance within a column is 4.843 Å, slightly larger than the 1·0.45DME complex (Fig. S9†). The stacking is further stabilized by the edge-to-face π-stacking of the TPA groups (Fig. 3C, Table S2†). The packing shows that the neighboring columns are oriented oppositely to minimize the macrodipoles (Fig. 2F and S11†). The N⋯N distance in a single macrocycle unit is found to be 10.24 Å, which is slightly shorter than the 1·0.45DME complex (Fig. S8†). The TPA units with similar chirality stack on each other with N⋯N center distance of 4.860 Å (Fig. 3C). In contrast to the 1·0.45DME complex, 1·DMSO complex shows unsymmetrical propeller arrangement of the aromatic rings of the TPA unit (Fig. S13†).
Prior work crystallized 2 by the vapor diffusion of DME into a DMSO solution of 2 and showed that the macrocycle adopted the anti-conformation with bound DME in the pore.16 Given the changes observed with host 1, we next examined the vapor diffusion of H2O into the DMSO solution of 2 to see if alternative bowl-shaped structures could be observed. Colorless needles were obtained and subjected to SCXRD analysis. The macrocycle crystallizes in the space group P21/c, with a framework nearly isostructural to the 2·0.72DME structure.16 The asymmetric unit consists of half of one host located on a crystallographic inversion center and a partially occupied, disordered DMSO molecule with host–guest ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.72. The macrocycle assembles into the columnar structure guided by the three-centered hydrogen bonding motif (d(N⋯O) = 2.954(5) Å, 2.877(5) Å) in anti-conformation with further stability coming from the offset π-stacking interaction.
0.72. The macrocycle assembles into the columnar structure guided by the three-centered hydrogen bonding motif (d(N⋯O) = 2.954(5) Å, 2.877(5) Å) in anti-conformation with further stability coming from the offset π-stacking interaction.
Next, both the crystalline forms of 1 were subjected to thermogravimetric analysis (TGA), heating up to 200 °C (2 °C min−1) to remove the guest molecules. TGA for both the compounds showed 1-step desorption curves between 25–120 °C (Fig. 4A, B, S24, and S25†). For the 1·0.45DME complex, a 5.1% weight loss was observed by TGA, which corresponds to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.45 host–guest ratio. In comparison, the 1·DMSO complex displayed a weight loss of 13%, which corresponds to 1
0.45 host–guest ratio. In comparison, the 1·DMSO complex displayed a weight loss of 13%, which corresponds to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.3 host–guest ratio. Interestingly, the 1·0.45DME crystals retained their crystallinity upon activation and were subjected to SCXRD analysis. However, upon activation, 1·DMSO transitions to a powder-like solid, which crumbles upon further handling.
1.3 host–guest ratio. Interestingly, the 1·0.45DME crystals retained their crystallinity upon activation and were subjected to SCXRD analysis. However, upon activation, 1·DMSO transitions to a powder-like solid, which crumbles upon further handling.
The crystal structure of activated 1·DME revealed an effectively unchanged host structure but with complete removal of DME guests (Fig. 3 and S12†). The asymmetric unit comprises half of one host molecule on a crystallographic inversion center with no significant residual electron density from the guest observed in the channel. The individual macrocycles retain the anti-conformation and are still organized into a columnar structure with only slight changes in the urea hydrogen bond distances (d(N⋯O) = 2.895(8) Å, 2.93(8) Å). The assembled host is further stabilized by offset π-stacking of the phenyl rings with slight changes in the stacking distances (Table S2†). The N⋯N distance is slightly shorter (10.68 Å) than the DME solvated one (Fig. S8†). The TPAs retain their conformation6 with the repeating unit with N⋯N distance between two subsequent macrocycles along b-axis is 4.648 Å (Fig. 3C).
To further analyse the interactions that promoted the packing of the TPA units and guide the assembly, Hirshfeld surface analysis24,25 and fingerprint plots were generated for 1·0.45DME, 1·DMSO, and activated 1·DME crystalline materials (Fig. S18–S20†). Prior Hirshfeld analysis of 2·DME (activated) showed that the exterior bromine formed Br⋯Caryl interactions, which increased the intercolumnar interaction and could contribute to the increased stability of the crystalline complex.16 Similar Hirshfeld analysis and further fingerprint plots of the three TPA host 1 complexes, which lack the halide, showed that all complexes primarily assemble through the three centered hydrogen bonding interactions. Further, packing is stabilized through the Caryl–Caryl and Caryl⋯H stacking interactions between the neighboring units.
To compare the interaction energies in the complexes, the energy framework calculations were performed using B3LYP/6-31G(d,p) energy model in the Crystal Explorer software.24 Inspection of the energy table, suggests that the interaction energies along the column axis are higher due to the hydrogen bonding and aryl–aryl interaction between the macrocycles. Interestingly, the packing energies corresponding to H–Caryl interaction energies between the neighboring macrocycles in 1·DME is slightly higher than the 1·DMSO (Fig. S21–S23†). This suggests that the 1·0.45DME has stronger intercolumnar interactions than 1·DMSO, which likely results in its higher stability and retention of crystallinity upon activation.
Differential scanning calorimetry (DSC) measurements were carried out to evaluate the thermal stability of the crystals. The DSC profile for the 1·DMSO complex up to 200 °C (Fig. 4A, blue) displays an endothermic broad peak up to ∼100 °C, which corresponds to the loss of the DMSO guest. Interestingly, appearance of a second endothermic peak at ∼140 °C can be attributed to the phase transition, which might be another polymorph. Further cooling in the first and second heating–cooling cycles (Fig. S26†) shows no significant changes, suggesting this phase change is irreversible. In contrast, the DSC profile of 1·0.45DME complex (Fig. 4B, blue) shows an endothermic peak around 60 °C corresponding to the removal of the DME guest. A second broad endothermic peak around 160 °C may be due to a minor structural transition. No transition was observed upon first cooling and second heating–cooling cycle (Fig. S26†).
For further insight into the phase transitions of the crystals upon guest removal, PXRD analysis on all the complexes was performed. Fig. 4C shows the PXRD data for the 1·0.45DME and 1·DMSO complexes before and after activation. The experimental PXRD pattern closely matches the theoretical PXRD spectra predicted from the single crystal data for 1·0.45DME, activated 1·DME, and 1·DMSO (Fig. S15–S17†), suggesting that these complexes are single phase. As expected, removal of the solvent from complexes induces changes that can been observed by PXRD. Surprisingly, activation of 1·DMSO complex gives a PXRD pattern that matches the activated 1·DME pattern, suggesting that the two activated materials are isostructural (Fig. 4C and S45†). These activated species' Fourier transform infrared (FTIR) spectra are also nearly identical (Fig. S27†), consistent with two structures being the same.
3.2 EPR experiments for radical generation study
Prior studies on 2 showed that UV irradiation with a Hannovia medium pressure Hg lamp or 365 nm LEDs generated radicals without degradation.17 The activated crystals of host 2 form a maximum of 0.69% PIRs upon 20 h of irradiation with 365 nm light-emitting diodes (LEDs). These radicals displayed a half-life of ∼24 h, decaying without degradation. The radicals can be regenerated to their maximum concentration upon UV irradiation. Thus, we were curious to see how the different structural motifs of 1 would modulate the amount of PIRs and to determine if these radicals would be stable and decay without degradation.Fresh triply recrystallized samples (∼7 mg) were filtered, dried under Ar (g) at room temperature in the dark, and loaded inside quartz EPR tubes for radical generation study. X-band EPR spectra were recorded before and after UV irradiation with 365 nm LEDs. EPR spectra were taken with incrementally increasing irradiation time (2 h) to monitor the maximum radical generation over (1 to 40 h). Radical formation was estimated by plotting the double integration of the EPR spectra over time and comparing it with a standard. Commercially available magic blue, which contains a TPA radical cation, was used as a standard to prepare a calibration curve (Fig. S35†). Next, the radical formation in each sample was compared with the calibration curve to get the approximate radical concentration.
Fig. 5 shows the EPR spectra for the four crystalline samples of host 1 before and after UV irradiation. Before irradiation, little to no radical signal was observed. After irradiation, broad axial powder pattern-shaped EPR signals were observed, similar to 2. The line shapes of the powder patterns differed between the four samples. The asymmetry of the powder pattern observed in all the samples may arise from the anisotropic nature of the crystalline complexes.17 The corresponding g-values for the TPA host are 2.006 (1·0.45DME, 1·DME (activated), 1·DMSO (activated)) and 2.007 (1·DMSO), which are close to the g-value of the TPA radical cations in solution (2.002–2.005).26 Hyperfine splitting is observed in the 1·0.45DME, 1·DME (activated), and 1·DMSO (activated) due to through-space hyperfine coupling of the nearby N centers, which can differ based on the TPA conformation and its relative packing.27
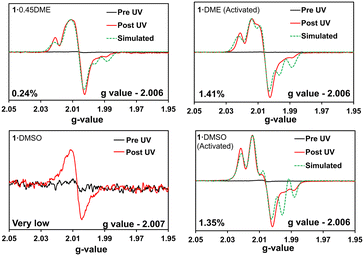 | ||
| Fig. 5 EPR spectra of the 1·0.45DME complex, 1·DMSO complex, activated 1·DME and activated 1·DMSO. EPR signals are shown for the pre and post UV irradiation. Additionally, maximum radical concentration and g-values are given for each complex. Best simulation of the EPR spectra of 1·DME, 1·DME (activated), 1·DMSO (activated) using EasySpin are plotted with dashed lines. See Table S4† for simulation parameters. | ||
Next, simulations of the EPR spectra of the 1·0.45DME, 1·DME (activated), and 1·DMSO (activated) were carried out using EasySpin.28 Two different radical components were required to fit the spectrum for all the complexes reasonably. The best fits are plotted on the corresponding spectra in Fig. 5. The first radical contains N hyperfine interactions, and the second one is featureless without any hyperfine interactions. The N hyperfine interactions varied with increased N-hyperfine interaction observed for the activated complexes. Similar line shapes and hyperfine couplings have been observed in triarylamines and their derivatives, which show a high propensity for assembly upon photoinduced radical formation.6,12,27,29
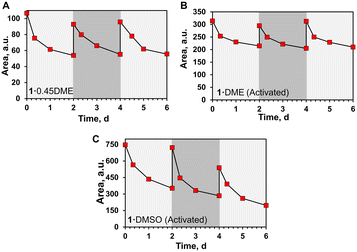
Maximum radical concentrations and radical concentrations after 6 h for all the crystalline complexes are summarized in Table 1. Upon increasing the UV irradiation time from 0 to 7 days, 1·0.45DME (7.1 mg) shows an evolution of hyperfine splitting from a broad one-line to a five-line pattern. The plot of the double integration of the EPR spectra versus time shows the number of radicals reach a plateau (Fig. S30B†). Through comparison to the Magic Blue calibration curve, we estimate that 1·0.45DME complex forms a maximum of ∼0.24% radicals upon 32 h of UV irradiation. Hyperfine splitting in the 1·0.45DME complex is more pronounced than the previously reported 2·0.5DME complex,17 although the maximum radical concentration is similar (0.28% in 2·0.5DME).
In contrast, activated 1·DME reaches a plateau faster at ∼8 h of UV-irradiation and forms more PIRs ∼1.41%, which is also higher than prior reports of activated host 2 (0.69%). Hyperfine splitting patterns are also different in the activated 1·DME complex. The characteristic five-line pattern in the EPR spectrum is more intense upon activation. Interestingly, 1·DMSO showed no or minimal amounts of radicals, below the detection limit, suggesting that the polar DMSO guest hinders the radical formation pathway.30
As the activated structures are nearly identical although their size distributions are different, we expect that PIRs in 1·DMSO (activated) should be very similar to 1·DME (activated). As expected, the removal of DMSO from the channel resulted in a dramatic increase in the radical concentration of the host. Irradiation of three samples of the microcrystalline powder, 1·DMSO (activated), generates a maximum PIRs of 1.35% with a standard deviation of 0.06%. Gratifyingly, crystalline 1·DME (activated) also gives a maximum PIR of 1.41%, consistent with them being isostructural. In comparison, the highest reported PIRs in TPA films is 6%, observed for TPAs in conjugated donor–acceptor molecules. Those systems afforded up to 16–19% PIRs in CHCl3 solution, although with lower stability as compared to the solid-state.31
Next, the persistence of the radicals in the complexes was monitored in the dark (Fig. S30–S33†). The samples were each UV-irradiated until they reached the maximum radical concentration, then stored in the dark, and monitored periodically by EPR to estimate the decay of radical species. Dark decay was followed for 7 days for all the samples. 1·0.45DME sample shows a radical decay of ∼58% in seven days with a half-life of ∼5 days. Both activated species showed a decrease in radical concentration in 7 days with 1·DMSO (activated) decaying slightly faster than 1·DME (activated) ∼79% versus 50% radical decay in seven days with a half-life of ∼2 days. In each case, there appears to be an initial faster decay of radicals followed by a slower process. This suggests the presence of two pathways and/or two types of radicals in the system. Prior observations on 2 and its complexes also exhibited both faster and slower decaying species.17
Next, the reversibility of radical generation and decay was investigated by exposure of the samples to UV light after initial decay. Fig. 6 illustrates the process of radical generation, decay, and regeneration of radicals for 1·0.45DME and the two activated complexes. In each graph, the double integration of the EPR signals was plotted versus time. Samples were initially UV-irradiated to their maximum radical generation, then stored in the dark, and the decay of the radicals was monitored by EPR at time t = 8 h, 24 h, and 48 h. After 48 h, the samples were re-irradiated overnight to restore the initial radical signal. This process is repeated two times and suggests that the formation and decay of the radicals are remarkably reversible in the crystalline samples.
Finally, the stability of the complexes upon UV-irradiation was checked by 1H NMR and IR spectra. Samples used for the regeneration study were taken after the third cycle and dissolved in DMSO-d6 for 1H NMR and compared with the 1H NMR of fresh samples. No significant changes were observed for any of the complexes (Fig. S36–S39†). Similarly, the IR spectra of all the samples after the regeneration study were recorded and compared with fresh samples. Again, no observable changes occurred for any of the complexes (Fig. S40–S43†). This study suggests that the crystalline complexes exhibit good stability even after multiple irradiation events despite the presence of unsubstituted para-positions.
4. Conclusion
In summary, TPA host 1 crystallizes in two forms depending on whether DMSO or DME guests are encapsulated. Upon binding the DME guests, the TPAs adopt a planar conformation with the urea groups pointing in opposite directions. The host–DME guest ratio is 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. X-ray structural analysis revealed that the hosts assemble into columns through hydrogen bonding with further stabilization from π-stacking interactions. In comparison, the 1·DMSO complex adopts a bowl-shaped conformation where the urea groups are aligned in the same direction. The single crystal structure shows that the urea hydrogen bonding distances are slightly larger than the 1·0.45DME complex. The two solvates form similar columnar structures through hydrogen bonding along with edge-to-face interactions. Both forms pack through C–H⋯π interactions with neighboring units. Hirshfeld analysis suggests greater C–H⋯π interaction in 1·0.45DME than 1·DMSO. This correlates well with the observed stability of the crystal forms upon removal of the guests from the pore. The 1·0.45DME complex remains single crystalline upon activation and can be further analysed by SCXRD. In comparison, 1·DMSO loses its single crystallinity upon activation, forming microcrystalline powder similar in structure to activated 1·DME. Given the observed bowl-shaped structure of 1·DMSO, we also revisited the TPA host 2, screening further crystallization conditions; however, while a new crystal 2·DMSO was obtained, its structure was identical to those previously observed.
1. X-ray structural analysis revealed that the hosts assemble into columns through hydrogen bonding with further stabilization from π-stacking interactions. In comparison, the 1·DMSO complex adopts a bowl-shaped conformation where the urea groups are aligned in the same direction. The single crystal structure shows that the urea hydrogen bonding distances are slightly larger than the 1·0.45DME complex. The two solvates form similar columnar structures through hydrogen bonding along with edge-to-face interactions. Both forms pack through C–H⋯π interactions with neighboring units. Hirshfeld analysis suggests greater C–H⋯π interaction in 1·0.45DME than 1·DMSO. This correlates well with the observed stability of the crystal forms upon removal of the guests from the pore. The 1·0.45DME complex remains single crystalline upon activation and can be further analysed by SCXRD. In comparison, 1·DMSO loses its single crystallinity upon activation, forming microcrystalline powder similar in structure to activated 1·DME. Given the observed bowl-shaped structure of 1·DMSO, we also revisited the TPA host 2, screening further crystallization conditions; however, while a new crystal 2·DMSO was obtained, its structure was identical to those previously observed.
Our goal of probing the photophysical properties of host 1 and characterizing its PIRs was achieved. The macrocycle was surprisingly robust when assembled into crystals, despite its two unsubstituted para-positions, suggesting that other structurally simple TPAs may also be stabilized by assembly. The different crystal forms (1·DMSO, 1·0.45DME, and the activated structure) varied in PIRs produced as determined by EPR and underwent the reverse electron transfer without degradation. The presence of guests modulated the shape of the EPR spectra upon radical formation. The complex with the most polar guest, DMSO, did not form significant quantities of radicals. In contrast, the 1·DME complex exhibited maximum PIRs of 0.24% upon 32 h irradiation. Activated hosts obtained from the two solvates reproducibly exhibited higher number of PIRs with very similar behaviour and radical percentage. Interestingly, the activated 1 reaches its maximum radical concentration of ∼1.4% in a shorter period (8 h). To our knowledge, this is the highest PIR percentage for crystalline TPA materials reported. The stability of TPA radical cations is known to impact their electronic properties and utility in OLEDs32 and perovskite solar cells.33 Crystalline TPAs deposited in the CSD and their associated PIRs offer well-defined structural characteristics that might be used to develop algorithms to predict radical percentages simply from the molecular structure and its assembly. We are currently employing machine-learning strategies to predict radical generation based on structure and hope to report on this shortly.
Data availability
The three new structures in this manuscript have been deposited in the Cambridge Structural Database managed by the Cambridge Crystallographic Data Centre (CCDC No. 2344711, 2344712, 2344713). The ESI† contains characterization of macrocycle 1 and its crystalline assemblies including DSC, FTIR, FESEM, NMR, PXRD, SCXRD, TGA, and measurement of its PIRs by EPR. In addition, the ESI† contains calculations of interactions by Hirshfeld analysis.Author contributions
L. S. S. and M. F. I. initiated the study. M. F. I., G. I. P. W., A. J. S. and L. S. S. planned experiments, analyzed data, and contributed to the manuscript. M. F. I., G. I. P. W., A. J. S. and M. D. S. performed experiments. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the National Science Foundation (NSF) CHE-2203830 and by an ASPIRE grant from the Office of the Vice President for Research at the University of South Carolina. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect those of the NSF.Notes and references
- S. Bhattacharya and B. K. Saha, Guest-Induced Isomerization of Net and Polymorphism in Trimesic Acid–Arylamine Complexes, Cryst. Growth Des., 2011, 11(6), 2194–2204, DOI:10.1021/cg101531h.
- B. A. Nogueira, S. M. M. Lopes, A. Milani, V. André, J. A. Paixão, M. E. S. Eusébio, T. M. V. D. Pinho e Melo, M. T. Duarte, C. Castiglioni and R. Fausto, Color Polymorphs of ROY-Ol, Cryst. Growth Des., 2022, 22(9), 5375–5389, DOI:10.1021/acs.cgd.2c00462.
- J. T. A. Jones, T. Hasell, X. Wu, J. Bacsa, K. E. Jelfs, M. Schmidtmann, S. Y. Chong, D. J. Adams, A. Trewin, F. Schiffman, F. Cora, B. Slater, A. Steiner, G. M. Day and A. I. Cooper, Modular and Predictable Assembly of Porous Organic Molecular Crystals, Nature, 2011, 474(7351), 367–371, DOI:10.1038/nature10125.
- L. S. Xie, G. Skorupskii and M. Dinca, Electrically Conductive Metal–Organic Frameworks, Chem. Rev., 2020, 120(16), 8536–8580 CrossRef PubMed.
- N. Fujita, K. Biradha, M. Fujita, S. Sakamoto and K. Yamaguchi, A Porphyrin Prism: Structural Switching Triggered by Guest Inclusion, Angew. Chem., Int. Ed., 2001, 40(9), 1718–1721, DOI:10.1002/1521-3773(20010504)40:9%3C1718::AID-ANIE17180%3E3.0.CO;2-7.
- E. Moulin, J. J. Armao and N. Giuseppone, Triarylamine-Based Supramolecular Polymers: Structures, Dynamics, and Functions, Acc. Chem. Res., 2019, 52(4), 975–983, DOI:10.1021/acs.accounts.8b00536.
- P. Cias, C. Slugovc and G. Gescheidt, Hole Transport in Triphenylamine Based OLED Devices: From Theoretical Modeling to Properties Prediction, J. Phys. Chem. A, 2011, 115(50), 14519–14525, DOI:10.1021/jp207585j.
- V. Parthasarathy, S. Fery-Forgues, E. Campioli, G. Recher, F. Terenziani and M. Blanchard-Desce, Dipolar versus Octupolar Triphenylamine-Based Fluorescent Organic Nanoparticles as Brilliant One- and Two-Photon Emitters for (Bio)imaging, Small, 2011, 7(22), 3219–3229, DOI:10.1002/smll.201100726.
- A. Ito, H. Ino, K. Tanaka, K. Kanemoto and T. Kato, Facile Synthesis, Crystal Structures, and High-Spin Cationic States of All-para-Brominated Oligo(N-phenyl-m-aniline)s, J. Org. Chem., 2002, 67(2), 491–498, DOI:10.1021/jo0160571.
- G. F. Huo, X. Shi, Q. Tu, Y. X. Hu, G. Y. Wu, G. Q. Yin, X. Li, L. Xu, H. M. Ding and H. B. Yang, Radical-Induced Hierarchical Self-Assembly Involving Supramolecular Coordination Complexes in Both Solution and Solid States, J. Am. Chem. Soc., 2019, 141(40), 16014–16023, DOI:10.1021/jacs.9b08149.
- E. Moulin, F. Niess, M. Maaloum, E. Buhler, I. Nyrkova and N. Giuseppone, The Hierarchical Self-Assembly of Charge Nanocarriers: A Highly Cooperative Process Promoted by Visible Light, Angew. Chem., Int. Ed., 2010, 49, 6974–6978, DOI:10.1002/anie.201001833.
- J. J. I. Armao, M. Maaloum, T. Ellis, G. Fuks, M. Rawiso, E. Moulin and N. Giuseppone, Healable Supramolecular Polymers as Organic Metals, J. Am. Chem. Soc., 2014, 136(32), 11382–11388, DOI:10.1021/ja5044006.
- Y. Chen, S. Xu, C. Fang Wen, H. Zhang, T. Zhang, F. Lv, Y. Yue and Z. Bian, Unravelling the Role of Free Radicals in Photocatalysis, Chem. – Eur. J., 2024, 30(29), e202400001, DOI:10.1002/chem.202400001.
- A. J. Sindt, B. A. DeHaven, D. F. McEachern, D. M. M. M. Dissanayake, M. D. Smith, A. K. Vannucci and L. S. Shimizu, UV-Irradiation of Self-Assembled Triphenylamines Affords Persistent and Regenerable Radicals, Chem. Sci., 2019, 10(9), 2670–2677, 10.1039/C8SC04607G.
- M. S. Hossain, A. J. Sindt, R. L. Goodlett, D. J. Shields, C. J. O'Connor, A. Antevska, S. G. Karakalos, M. D. Smith, S. Garashchuk, T. D. Do, A. D. Gudmundsdottir and L. S. Shimizu, Effects of Self-Assembly on the Photogeneration of Radical Cations in Halogenated Triphenylamines, J. Phys. Chem. C, 2021, 125(36), 19991–20002, DOI:10.1021/acs.jpcc.1c04933.
- A. J. Sindt, M. D. Smith, S. Berens, S. Vasenkov, C. R. Bowers and L. S. Shimizu, Single-Crystal-to-Single-Crystal Guest Exchange in Columnar Assembled Brominated Triphenylamine Bis-Urea Macrocycles, Chem. Commun., 2019, 55(39), 5619–5622, 10.1039/C9CC01725A.
- A. J. Sindt, B. A. Dehaven, D. W. Goodlett, J. O. Hartel, P. J. Ayare, Y. Du, M. D. Smith, A. K. Mehta, A. M. Brugh, M. D. E. Forbes, C. R. Bowers, A. K. Vannucci and L. S. Shimizu, Guest Inclusion Modulates Concentration and Persistence of Photogenerated Radicals in Assembled Triphenylamine Macrocycles, J. Am. Chem. Soc., 2020, 142(1), 502–511, DOI:10.1021/jacs.9b11518.
- SAINT+ Version 8.37 A, Bruker AXS Madison Wis. USA, 2016 Search PubMed.
- L. Krause, R. Herbst-Irmer, G. M. Sheldrick and D. Stalke, J. Appl. Crystallogr., 2015, 48, 3–10 CrossRef PubMed.
- G. M. Sheldrick, SHELXT–Integrated Space-Group and Crystal-Structure Determination, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71(1), 3–8 CrossRef PubMed.
- G. Sheldrick, Using Phases to Determine the Space Group, Acta Crystallogr., Sect. A: Found. Adv., 2018, 74, a353 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. Howard and H. Puschmann, OLEX2: A Complete Structure Solution, Refinement and Analysis Program, J. Appl. Crystallogr., 2009, 42(2), 339–341 CrossRef.
- Functional Materials from Self-Assembled Bis-urea Macrocycles|Accounts of Chemical Research, https://pubs.acs.org/doi/full/10.1021/ar500106f (accessed 2024-03-03) Search PubMed.
- P. R. Spackman, M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, D. Jayatilaka and M. A. Spackman, CrystalExplorer: A Program for Hirshfeld Surface Analysis, Visualization and Quantitative Analysis of Molecular Crystals, J. Appl. Crystallogr., 2021, 54(3), 1006–1011, DOI:10.1107/S1600576721002910.
- M. A. Spackman and D. Jayatilaka, Hirshfeld Surface Analysis, CrystEngComm, 2009, 11(1), 19–32, 10.1039/B818330A.
- A. R. Forrester, J. M. Hay and R. H. Thomson, Organic Chemistry of Stable Free Radicals, 1968 Search PubMed.
- T. Liang, D. Collin, M. Galerne, G. Fuks, A. Vargas Jentzsch, M. Maaloum, A. Carvalho, N. Giuseppone and E. Moulin, Covalently Trapped Triarylamine-Based Supramolecular Polymers, Chem. – Eur. J., 2019, 25(63), 14341–14348, DOI:10.1002/chem.201902404.
- S. Stoll and A. Schweiger, EasySpin, a Comprehensive Software Package for Spectral Simulation and Analysis in EPR, J. Magn. Reson., 2006, 178(1), 42–55, DOI:10.1016/j.jmr.2005.08.013.
- J. Liu, X. Rong, J. Wu, B. Chen, Z. Lu and Y. Huang, Air-Stable Organic Radicals in Solid State from a Triphenylamine Derivative by UV Irradiation, Tetrahedron Lett., 2023, 115, 154259, DOI:10.1016/j.tetlet.2022.154259.
- D. A. Caminos, M. Puiatti, J. I. Bardagí and A. B. Peñéñory, Anions Involved in the Initiation of the Thermally Induced SRN1 Reaction for α-Arylation of Ketones, RSC Adv., 2017, 7(50), 31148–31157, 10.1039/C7RA05156E.
- X. Rong, J. Liu, J. Wu, C. Li, K. Wang, Z. Lu, Y. Liu, M. Gu and Y. Huang, A Feasible Strategy to Obtain Air-Stable Triphenylamine Radicals in the Solid State by Introducing Conjugated Donor–Acceptor Modules, J. Mater. Chem. C, 2023, 11(28), 9640–9648, 10.1039/D3TC01187A.
- J. Zhao, Z. Yang, X. Chen, Z. Xie, T. Liu, Z. Chi, Z. Yang, Y. Zhang, M. P. Aldred and Z. Chi, Efficient Triplet Harvesting in Fluorescence–TADF Hybrid Warm-White Organic Light-Emitting Diodes with a Fully Non-Doped Device Configuration, J. Mater. Chem. C, 2018, 6(15), 4257–4264, 10.1039/C8TC01210E.
- A. Farokhi, H. Shahroosvand, G. D. Monache, M. Pilkington and M. K. Nazeeruddin, The Evolution of Triphenylamine Hole Transport Materials for Efficient Perovskite Solar Cells, Chem. Soc. Rev., 2022, 51(14), 5974–6064, 10.1039/D1CS01157J.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis, characterization of macrocycle 1, its crystalline assemblies including DSC, FTIR, FESEM, NMR, PXRD, SCXRD, TGA, and measurement of its PIRs by EPR and calculations of interactions by Hirshfeld analysis. CCDC 2344711–2344713. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ce00839a |
| This journal is © The Royal Society of Chemistry 2024 |