Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
4-(Ammoniomethyl)benzoate, a protic zwitterionic bifunctional linker in uranyl ion carboxylate complexes†
Youssef
Atoini
a,
Jack
Harrowfield
 *b and
Pierre
Thuéry
*b and
Pierre
Thuéry
 *c
*c
aTechnical University of Munich, Campus Straubing, Schulgasse 22, 94315 Straubing, Germany
bUniversité de Strasbourg, ISIS, 8 allée Gaspard Monge, 67083 Strasbourg, France. E-mail: harrowfield@unistra.fr
cUniversité Paris-Saclay, CEA, CNRS, NIMBE, 91191 Gif-sur-Yvette, France. E-mail: pierre.thuery@cea.fr
First published on 11th June 2024
Abstract
4-(Aminomethyl)benzoic acid (Hamb) has been found to complex the uranyl ion in its neutral, zwitterionic form in a series of mixed-ligand complexes synthesized under solvo-hydrothermal conditions with several dicarboxylate coligands. [UO2(tdc)(Hamb)]·5H2O (1), [UO2(pda)(Hamb)]·CH3CN (2), and [UO2(cam)(Hamb)]·CH3CN (3), where tdc2−, pda2− and cam2− are 2,5-thiophenedicarboxylate, 1,2-phenylenediacetate and (1R,3S)-(+)-camphorate, respectively, crystallize as simple chains in which the dicarboxylate ligand is bridging and Hamb is terminal, all carboxylates being κ2O,O′-chelating. [UO2(pht)(Hamb)]·0.5H2O (4), where pht2− is phthalate, is a ribbonlike chain in which both ligands are bridging, while the presence of the coordinated solvent N,N-dimethylacetamide (dma) prevents polymerization in [(UO2)4(O)2(pht)2(Hamb)2(dma)2]·2H2O (5), a bis(μ3-oxo)-bridged tetranuclear assembly. 1,2-Phenylenedioxydiacetate (pdda2−) gives [UO2(pdda)(Hamb)]·0.5CH3CN (6), another ribbonlike chain in which only pdda2− is bridging. Finally, [UO2(pim)(Hamb)] (7), involving the pimelate ligand (pim2−) is a double-stranded, ribbonlike chain in which two UO2(pim) linear polymers are bridged by the Hamb ligands. In all these complexes, the ammonium group of Hamb is involved in extended hydrogen bonding giving rise to weakly bonded assemblies of higher periodicity. All complexes except 5 and 6 are emissive, with photoluminescence quantum yields between 2 and 11%, and with the exception of a broad signal associated with 1, all display the usual vibronic fine structure, with peak positions clearly related to the uranium coordination numbers.
Introduction
The fact that the presence of a positively charged centre adjacent to a carboxylate-donor group, as in the well-known ligand “betaine” (trimethylammonioacetate), appears to have negligible influence on M–O bond lengths in comparison to those in anionic carboxylate donors1,2 has been the basis of our efforts to characterize mixed zwitterionic/anionic ligand uranyl ion complexes.3,4 The intention there was to determine how the structure of uranyl ion coordination polymers based on polycarboxylate ligands might be modified by the co-coordination of neutral oligozwitterionic carboxylates. To avoid possible complications in synthesis due to additional acid–base equilibria and coordination modes, the oligozwitterions employed in this work5–11 were those with quaternary nitrogen centres, thus lacking dissociable protons. This has the disadvantage of eliminating usage of the positively charged centre for strong hydrogen bonding interactions, a factor which can strongly influence the role of a zwitterionic ligand as a structure determinant.12–23 The best known protic zwitterions are the amino acids but the known structural chemistry of their uranyl ion complexes, while showing this importance of ammonium-centre hydrogen bonding, is very limited, involving only glycine,24–26 alanine25 and proline.27 This paucity of information is due, in our experience, to reactions, both under solvothermal or mild ambient conditions and in the presence or absence of light, of most amino acids in the presence of uranyl ion which result in their decomposition. Hence, to obtain broader indications of the influence of a protic zwitterion on uranyl ion coordination polymer structures, we have turned to the chemically more robust species 4-(ammoniomethyl)benzoate (Hamb, Scheme 1), for which, in uranyl ion complexes where it is the sole carboxylate ligand apart from solvent-derived formate, some information useful for comparison is available.28 Mixed-ligand complexes with several carboxylate ligands derived from well-studied dicarboxylic acids, 2,5-thiophenedicarboxylic acid (H2tdc), 1,2-phenylenediacetic acid (H2pda), (1R,3S)-(+)-camphoric acid (H2cam), phthalic acid (H2pht), 1,2-phenylenedioxydiacetic acid (H2pdda), and pimelic (heptanedioic) acid (H2pim) (Scheme 1) have here proven to be readily obtained under solvo-hydrothermal conditions. All these complexes have been characterized by their crystal structure and emission properties in the solid state.Experimental
Synthesis
Caution! Uranium is a radioactive and chemically toxic element, and uranium-containing samples must be handled with suitable care and protection. Small quantities of reagents and solvents were employed to minimize any potential hazards arising both from the presence of uranium and the use of pressurized vessels for the syntheses.[UO2(NO3)2(H2O)2]·4H2O (RP Normapur, 99%) was purchased from Prolabo; 4-(aminomethyl)benzoic acid and all dicarboxylic acids were from Aldrich. Elemental analyses were performed by MEDAC Ltd. For all syntheses, the solutions were placed in 10 mL tightly closed glass vessels (Pyrex culture tubes with SVL15 stoppers and Teflon-coated seals, provided by VWR) and heated at 140 °C in a sand bath (Harry Gestigkeit ST72). The crystals were grown in the hot, pressurized solutions and not as a result of a final return to ambient conditions.
Crystallography
Data collections were performed at 100(2) K on a Bruker D8 Quest diffractometer using an Incoatec Microfocus Source (IμS 3.0 Mo) and a PHOTON III area detector, and operated with APEX4.29 The data were processed with SAINT,30 and empirical absorption corrections were made with SADABS.31,32 The structures were solved by intrinsic phasing with SHELXT,33 and refined by full-matrix least-squares on F2 with SHELXL,34 using the ShelXle interface.35 The hydrogen atoms bound to oxygen and nitrogen atoms were retrieved from residual electron density maps and they were refined either freely (1 and 7) or with geometric restraints (all other compounds). All other hydrogen atoms in all compounds were introduced at calculated positions and treated as riding atoms with an isotropic displacement parameter equal to 1.2 times that of the parent atom (1.5 for CH3). For compound 3, the SQUEEZE36 software was used to subtract the contribution of disordered solvent molecules to the structure factors, the number of electrons added corresponding to approximately 0.5 water molecule per formula unit. In 4, the water molecule was given an occupancy factor of 0.5 in order to retain an acceptable displacement parameter, and its hydrogen atoms were treated as riding atoms after improvement of the geometry. The dma molecule in 5 is disordered over two positions sharing the oxygen and two carbon atoms; these positions were refined with occupancy parameters constrained to sum to unity and restraints on some bond lengths and displacement parameters. Several parts of the structure of 6 are affected by disorder, in particular the two oxo groups and two carboxylate groups bound to U2; the two positions have been refined with occupancy parameters constrained to sum to unity and restraints on bond lengths and displacement parameters. Crystal data and structure refinement parameters are given in Table 1. Drawings were made with ORTEP-337,38 and VESTA.39| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| Chemical formula | C14H21NO13SU | C20H20N2O8U | C20H28N2O9U | C16H14NO8.5U | C40H48N4O26U4 | C19H18.5N1.5O10U | C15H19NO8U |
| M/g mol−1 | 681.41 | 654.41 | 678.47 | 594.31 | 1952.94 | 665.88 | 579.34 |
| Crystal system | Monoclinic | Monoclinic | Monoclinic | Triclinic | Monoclinic | Orthorhombic | Triclinic |
| Space group | C2/c | P21/c | P21 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P21/c | Pca21 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a/Å | 16.8957(7) | 11.5459(8) | 8.9441(4) | 8.8256(6) | 8.3349(3) | 13.3970(4) | 8.4357(4) |
| b/Å | 16.0165(7) | 7.6285(5) | 15.7697(8) | 9.8335(7) | 12.2484(3) | 13.1905(4) | 10.8597(5) |
| c/Å | 16.5508(7) | 23.9919(18) | 18.0045(10) | 10.6502(8) | 24.3610(8) | 23.2768(6) | 10.9300(5) |
| α/° | 90 | 90 | 90 | 69.704(3) | 90 | 90 | 107.615(2) |
| β/° | 111.2954(17) | 93.933(3) | 102.834(2) | 77.981(3) | 94.5772(15) | 90 | 98.577(2) |
| γ/° | 90 | 90 | 90 | 74.084(2) | 90 | 90 | 110.9835(19) |
| V/Å3 | 4173.0(3) | 2108.2(3) | 2476.0(2) | 827.09(10) | 2479.06(14) | 4113.3(2) | 852.93(7) |
| Z | 8 | 4 | 4 | 2 | 2 | 8 | 2 |
| Reflections collected | 170![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 683 |
51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 067 067 |
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 652 652 |
43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 034 034 |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 731 731 |
83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 254 254 |
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 111 111 |
| Independent reflections | 5406 | 3988 | 9381 | 3129 | 4694 | 7797 | 5202 |
| Observed reflections [I > 2σ(I)] | 5117 | 3847 | 9172 | 2951 | 4501 | 7602 | 5046 |
| R int | 0.043 | 0.043 | 0.059 | 0.036 | 0.049 | 0.042 | 0.050 |
| Parameters refined | 317 | 290 | 616 | 253 | 377 | 652 | 238 |
| R 1 | 0.015 | 0.044 | 0.032 | 0.027 | 0.028 | 0.025 | 0.014 |
| wR2 | 0.041 | 0.095 | 0.071 | 0.068 | 0.061 | 0.061 | 0.034 |
| S | 1.070 | 1.443 | 1.165 | 1.154 | 1.336 | 1.066 | 1.106 |
| Δρmin/e Å−3 | −1.01 | −2.78 | −1.16 | −2.31 | −1.63 | −0.80 | −0.73 |
| Δρmax/e Å−3 | 2.38 | 1.53 | 1.16 | 2.51 | 1.59 | 1.97 | 2.17 |
Luminescence measurements
Emission spectra were recorded on solid samples using an Edinburgh Instruments FS5 spectrofluorimeter equipped with a 150 W CW ozone-free xenon arc lamp, dual-grating excitation and emission monochromators (2.1 nm mm−1 dispersion; 1200 grooves mm−1) and an R928P photomultiplier detector. The powdered compounds were pressed to the wall of a quartz tube, and the measurements were performed using the right-angle mode in the SC-05 cassette. An excitation wavelength of 420 nm was used in all cases and the emission was monitored between 450 and 600 nm. The quantum yield measurements were performed by using a Hamamatsu Quantaurus C11347 absolute photoluminescence quantum yield spectrometer and exciting the samples between 300 and 400 nm.Results and discussion
Context of the work
A crystal structure determination of the monohydrate of Hamb40 has shown it to be present in its zwitterionic form, i.e. as 4-(ammoniomethyl)benzoate, not 4-(aminomethyl)benzoic acid, and illustrates the hydrogen bonding capacity of the ammonio group in that all three protons are involved in interactions with separate carboxylate-O atoms (disorder of these carboxylate-O atoms gives rise to relatively large uncertainty in the H⋯O distances). The benzene rings form slipped-stack arrays with a relatively large interplanar separation of 3.66 Å and a centroid⋯centroid distance of 3.8602(18) Å, that appear from the Hirshfeld surface41,42 to involve dispersion interactions only, though this surface also indicates weak interactions beyond dispersion of the methylene CH atoms with the disordered water molecule oxygen atom. Several crystal structures have been reported of this molecule in its cationic, non-zwitterionic form, H2amb+,43–47 with examples in particular of multiple NH⋯O bonds with crown ethers44 and of the use of this cation as a structure-directing species for the building of layered structures,46 those being nice illustrations of the potential of the ammonium group for supramolecular organization. Only two cases of metal ion complexes (not including the uranyl ion) formed either by Hamb with Cd2+,48 or by amb− with Ag+ (with both carboxylate and amine groups coordinated)49 have been reported. The related 4-ammoniobenzoate zwitterion is not commonly found as a ligand either, with only some complexes reported in the Cambridge Structural Database (CSD, Version 5.45).50In the one known complex with uranyl ion where Hamb is present exclusively as the zwitterion,28 [UO2(Hamb)(OH)(HCOO)(H2O)]2·2CB6·2dmf·14H2O, the ligand is involved in various weak interactions largely involving the cucurbit[6]uril (CB6) also present in the crystal as well as being coordinated to uranyl ion in a κ1O mode. All three ammonio group protons act as hydrogen bond donors, one to solvent dmf (H⋯O, 1.99 Å), the two others to cucurbituril-O (H⋯O, 2.04 and 2.17 Å), while both methylene group protons act as hydrogen bond donors again to cucurbituril-O (H⋯O, 2.48 and 2.44 Å). Cucurbituril-H atoms also interact to each side of the carboxylate-C atom (H⋯C, 2.58 and 2.56 Å), the Hamb ligand thus being rather encumbered by CB6 units, perhaps explaining its limitation to unidentate coordination to UVI. In the tetranuclear complex involving CB8 impurity, [(UO2)4(Hamb)2(amb)(O)2(OH)2(H2O)4](NO3)·2CB6·0.5CB8·Hamb·20H2O, the coordinated carboxylate is present both as zwitterionic Hamb and as its conjugate base, amb−. Unfortunately, because of the low quality and complicated composition of the crystal, amino hydrogen atoms could not be located, so hydrogen bonding of the amino groups could not be defined in detail but a significant feature of this structure is that both the Hamb and amb− ligands coordinate to UVI in a κ2O,O′-chelating mode.
Crystal structures
All the seven complexes presently obtained are true mixed-ligand species, with both anionic dicarboxylate and Hamb (always in its zwitterionic form) bound to uranium, which confirms the ease with which true heteroleptic complexes can be generated from mixtures of anionic and zwitterionic carboxylates.3 Six complexes, [UO2(tdc)(Hamb)]·5H2O (1), [UO2(pda)(Hamb)]·CH3CN (2), [UO2(cam)(Hamb)]·CH3CN (3), [UO2(pht)(Hamb)]·0.5H2O (4), [UO2(pdda)(Hamb)]·0.5CH3CN (6) and [UO2(pim)(Hamb)] (7), crystallize as monoperiodic coordination polymers, while [(UO2)4(O)2(pht)2(Hamb)2(dma)2]·2H2O (5), involving the same carboxylate ligands as 4, contains an additional coordinated solvent (dma) and is a discrete, tetranuclear species. Prior to the examination of the coordination differences and supramolecular arrangement in these compounds, an interesting point to address is that of the relative strength of the anionic dicarboxylate and zwitterionic carboxylate ligands. The uranium atom environment in the series is either pentagonal-bipyramidal (4–7) or hexagonal-bipyramidal (1–3), the latter being obtained when all carboxylate groups are κ2O,O′-chelating, the uranium coordination number (CN) being thus 7 or 8. Table 2 gives the mean values of U–O bond lengths and of bond valence parameters,51 calculated with PLATON,52 for individual carboxylate oxygen atoms (BVAC for anionic carboxylates, BVZI for Hamb). Comparable values are those for equal CNs and for identically monodentate or chelating carboxylate donors. Among the three complexes 1–3, with a CN of 8, the mean BV values are slightly less for Hamb than for the AC ligand, but the large dispersion makes the difference statistically insignificant. In complexes 4 and 5, the BV is larger for bridging monodentate Hamb than for monodentate pht2−, while the reverse is true for pim2− in 7. These results thus show no clear general trend, in contrast to previous findings showing more neatly a slight decrease of BVs and hence of donor strength for zwitterionic ligands in self-sorted, homoleptic complexes10 or in other mixed-ligand complexes.8 It may be that the rather large separation between the carboxylate group and the positive charge in Hamb reduces the effect of the latter in comparison with that in betaine or the pyridinium-based zwitterions used in the previous studies.| Complex | CN | U–Ooxo | U–OAC (monodentate) | U–OAC (chelating) | U–OZI (monodentate) | U–OZI (chelating) | BVAC (monodentate) | BVAC (chelating) | BVZI (monodentate) | BVZI (chelating) |
|---|---|---|---|---|---|---|---|---|---|---|
| a The esds on mean values measure the dispersion of individual values; no esd is given for single BV values. | ||||||||||
| 1 | 8 | 1.7818(10) | 2.46(2) | 2.47(4) | 0.45(2) | 0.44(4) | ||||
| 2 | 8 | 1.778(5) | 2.45(2) | 2.494(2) | 0.454(15) | 0.420(1) | ||||
| 3 | 8 | 1.773(13) | 2.47(2) | 2.479(15) | 0.440(16) | 0.433(13) | ||||
| 4 | 7 | 1.761(2) | 2.369(4) | 2.46(3) | 2.327(7) | 0.536 | 0.45(3) | 0.582(9) | ||
| 5 | 7 | 1.787(5) | 2.407(5) | 2.52(2) | 2.355(6) | 0.498 | 0.40(2) | 0.553(7) | ||
| 6 | 7 | 1.75(2) | 2.35(7) | 2.43(2) | 0.56(8) | 0.47(2) | ||||
| 7 | 7 | 1.7787(6) | 2.3009(14) | 2.474(2) | 2.323(3) | 0.613 | 0.437(2) | 0.587(3) | ||
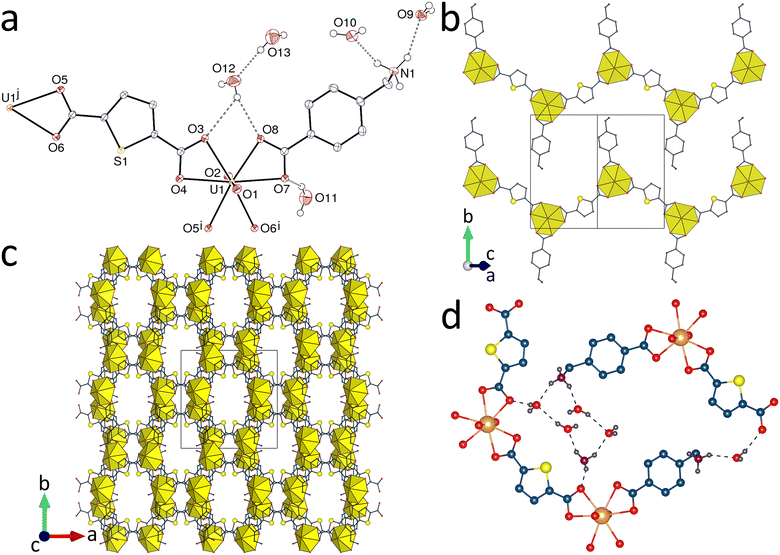
The three complexes 1–3 crystallize as simple chains in which the dicarboxylate ligand is bridging and Hamb is a simple decorating, pendent group. As shown in Fig. 1–3, the shape of the polymer is slightly different in the three compounds, with the Hamb ligands located on both edges of serpentine chains in 1 and 3, or on only one side of a linear chain in 2. The tdc2− ligand found in 1 has frequently been employed in uranyl chemistry,5,7,53–61 and complexes involving also aprotic dizwitterions are known but an unusual feature is the formation of crystals in which there is separation of the ligands into anionic and cationic polymer units,5,7 which is not observed here. The polymer chains in 1 can be considered to lie side-by-side in sheets parallel to (10![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) ), with the Hamb ligands alternating in orientation relative to the mean polymer axis and projecting, along with water molecules, into dips in the adjacent strands. Hydrogen-bonding chains NH⋯OH(water)⋯O(carboxylate) cross-link chains within a sheet, while the projection of all ammonio groups of one sheet to the same side of that sheet means that further hydrogen bonding by the ammonio groups to carboxylate oxygen atoms of an adjacent sheet leads to sheet pairing. These double sheets are further linked into a triperiodic array by hydrogen bonding involving water molecules as well as stacking of tdc2− units involving S⋯S interactions [3.522(2) Å] exceeding dispersion. The packing displays channels directed along the c axis and containing columns of hydrogen bonded water molecules (this compound being the most strongly solvated in the series). The Kitaigorodsky packing index (KPI, evaluated with PLATON52) is 0.71 for the complete structure and 0.55 when water molecules are excluded. The water column does not contain a continuous chain of hydrogen bonded water molecules, but separate groups linking the polymeric chains, in which a ring formed by two ammonium groups and four water molecules, with the graph set descriptor62 R46(12), is particularly conspicuous, together with a much larger R44(40) ring involving two [UO2(tdc)(Hamb)] interdigitated units and two water molecules (Fig. 1d). Three parallel-displaced π-stacking interactions between tdc2− and Hamb units, either with their own kind or with one another, are possibly present [centroid⋯centroid distances, 3.6599(13)–4.0353(12) Å; dihedral angles, 0–6.23(11)°; slippage, 1.26–2.15 Å].
), with the Hamb ligands alternating in orientation relative to the mean polymer axis and projecting, along with water molecules, into dips in the adjacent strands. Hydrogen-bonding chains NH⋯OH(water)⋯O(carboxylate) cross-link chains within a sheet, while the projection of all ammonio groups of one sheet to the same side of that sheet means that further hydrogen bonding by the ammonio groups to carboxylate oxygen atoms of an adjacent sheet leads to sheet pairing. These double sheets are further linked into a triperiodic array by hydrogen bonding involving water molecules as well as stacking of tdc2− units involving S⋯S interactions [3.522(2) Å] exceeding dispersion. The packing displays channels directed along the c axis and containing columns of hydrogen bonded water molecules (this compound being the most strongly solvated in the series). The Kitaigorodsky packing index (KPI, evaluated with PLATON52) is 0.71 for the complete structure and 0.55 when water molecules are excluded. The water column does not contain a continuous chain of hydrogen bonded water molecules, but separate groups linking the polymeric chains, in which a ring formed by two ammonium groups and four water molecules, with the graph set descriptor62 R46(12), is particularly conspicuous, together with a much larger R44(40) ring involving two [UO2(tdc)(Hamb)] interdigitated units and two water molecules (Fig. 1d). Three parallel-displaced π-stacking interactions between tdc2− and Hamb units, either with their own kind or with one another, are possibly present [centroid⋯centroid distances, 3.6599(13)–4.0353(12) Å; dihedral angles, 0–6.23(11)°; slippage, 1.26–2.15 Å].
In complex 2, the chains, with their pendent Hamb molecules all on one side, are only interdigitated in groups of two, and the hydrogen bond links are here directly between ammonium donors and carboxylate acceptors, no solvent water molecule being present. The chain dimers thus formed are stacked slantwise to form layers parallel to (100). No parallel-displaced π-stacking interaction is present, although the packing is quite compact (KPI, 0.70). Together with its positional isomers, pda2− has been relatively well-studied as a ligand for uranyl ion.63–66 While complex 1 provides an example of where the presence of Hamb appears to significantly alter the properties of its coligand, in complex 2 any such influence appears to be minimal and close parallels can be found in previously studied species. Perhaps unsurprisingly, a very similar monoperiodic polymer is found in the uranyl ion complex with the metallazwitterion Zn(pda)(phen)2,64 but other similar polymers are also found where the simple neutral chelates 2,2′-bipyridine and 1,10-phenanthroline or the anionic chelate nitrate accompany pda2−.63,64 Nonetheless, there are in all cases significant differences in the interdigitation patterns of the polymer chains reflecting the differences in the nature of the coligands.
The pure (1R,3S) enantiomer of camphoric acid having been used, complex 3 is chiral and crystallizes in the Sohncke group P21. The chains are interdigitated so as to form layers parallel to (101), but the ammonium groups are directed outwards on both sides and involved in hydrogen bonds with carboxylate groups of adjacent layers and with water and acetonitrile solvent molecules. The triperiodic assembly thus formed displays channels parallel to the c axis occupied by the acetonitrile molecules (KPI, 0.65). Complex 3 provides another example of a monoperiodic polymer with a close parallel in the polymer formed with nitrate ion as a coligand, there necessarily involving a counter cation, tetraphenylphosphonium.67 A significant difference is that whereas in the known complex the polymer is helical and right-handed, in complex 3 there are inequivalent helical polymer chains of opposite handedness.
Compound 4, as well as 6 and 7, crystallizes as a ribbon-like, doubly bridged chain (Fig. 4). While the pht2− ligand has one κ2O,O′-chelating carboxylate group (as are all groups in 1–3) and one monodentate, the Hamb ligand is bridging in the μ2-κ1O:κ1O′ mode. The ammonium groups, located on both edges of the chain, point on opposite sides, as the uncomplexed carboxylate oxygen atoms, and they are hydrogen bonded to carboxylate groups (complexed and uncomplexed) and to the water molecule, the latter being bonded to one uranyl oxo group and to the uncomplexed atom O4. Ammonium hydrogen bonding generates layers parallel to (111), while the water molecules (with partial occupancy and not shown in Fig. 4c for clarity) build links between adjacent layers. Further association is due to parallel-displaced π-stacking interactions between pht2− and Hamb units, here also either with their own kind or with one another [centroid⋯centroid distances, 3.849(4)–3.915(4) Å; dihedral angles, 0–20.7(3)°; slippage, 1.09–1.81 Å], and the resulting packing is very compact (KPI, 0.75). Phthalate is known to give a simple zigzag chain with the uranyl ion when the additional, terminal chelating ligand is nitrate,68 and also diperiodic networks with sql or V2O5 topologies when mixed with the dizwitterions 1,1′-[(2,3,5,6-tetramethylbenzene-1,4-diyl)bis(methylene)]bis(pyridin-1-ium-(3 or 4)-carboxylate)6 or 1,4-bis(4′-carbonylatopyridiniomethyl)benzene,8 but the present mixed-ligand ribbon has seemingly no precedent.
Changing acetonitrile for dma as a cosolvent during the synthesis with phthalate as a coligand produces the very different complex 5, the only discrete species in this series due to dma coordination as a terminal ligand (Fig. 5). This complex pertains to the common family of bis(μ3-oxo)-bridged tetranuclear uranyl species, found in particular with the pht2− ligand alone.68–71 The peculiarity here is that two of the four pht2− ligands found in the previous form are replaced by two Hamb ligands, resulting in an overall neutral, centrosymmetric complex and also making the metal ion environment much more irregular since a pht2− ligand coordinated by its two carboxylate groups is replaced by a single bridging carboxylate. The bonding mode of both ligands is the same as that found in 4, with an additional bridging interaction of the chelating group (μ2-κ2O,O′:κ1O). While the environment of U1 is a rather regular pentagonal bipyramid, that of U2 is a strongly distorted one, with O10–U2–O11 and O10–U2–O12 angles of 95.97(17) and 74.22(19)°, respectively. The oxo (instead of hydroxo) nature of O11 is shown by its bond valence parameter of 1.91 and the near planarity of its environment, with the sum of the three U–O–U angles equal to 353.6°. The ammonium group forms hydrogen bonds with uranyl oxo, bridging oxo and one carboxylate group pertaining to neighbouring molecules, resulting in the formation of layers parallel to (001). Hamb ligands are involved in a parallel-displaced π-stacking interaction [centroid⋯centroid distance, 3.878(4) Å; dihedral angle, 0°; slippage, 1.37 Å], thus generating columns of interacting molecules along the a axis. The packing has a KPI of 0.72.
Complex 6, with the pdda2− ligand, with which several uranyl ion complexes have already been described,6,72,73 differs from 4 by the Hamb ligand being here a terminal, κ2O,O′-chelating ligand, as in 1–3, bridging being due only to pdda2−, which is bound to three metal ions through one μ2-κ1O:κ1O′-bridging and one monodentate carboxylate groups (Fig. 6). Another difference is that all ammonium groups are located on the same side of the ribbon-like polymer. Bilayers parallel to (100) are formed through interlocking of chains related by a binary screw axis, with extensive hydrogen bonding between ammonium groups and carboxylate or ether oxygen atoms. Parallel-displaced π-stacking interactions are formed between pdda2− and Hamb units within columns parallel to the a axis [centroid⋯centroid distances, 3.732(5) and 3.841(6) Å; dihedral angles, 2.9(5) and 5.6(5)°; slippage, 1.43 and 1.61 Å], thus giving a triperiodic assembly (KPI, 0.71). A slightly different chain with also double pdda2− bridges has been found in a complex in which an additional water ligand forms intrachain hydrogen bonds,72 whereas, in its mixed complex with 1,1′-[(2,3,5,6-tetramethylbenzene-1,4-diyl)bis(methylene)]bis(pyridin-1-ium-4-carboxylate) as a coligand,6 pdda2− forms with uranyl a simple zigzag chain with further zwitterion overarching bridges.
The pimelate ligand, for which several uranyl ion complexes are known,74–81 gives also a ribbon-like, double-stranded polymer, but a different geometry results from the elongated shape of pim2− (Fig. 7). Both ligands are bridging, pim2−, in an extended although somewhat irregular conformation, through one chelating and one monodentate carboxylate groups, and Hamb as a μ2-κ1O:κ1O′ linker. Two neutral UO2(pim) strands running along the b axis direction are linked to one another by the Hamb ligands located in between. The ammonium group forms one intrachain hydrogen bond with the carboxylate oxygen atom O6 (another, weak bond with one oxo group of the uranyl ion bound to O6 is possibly significant), and it is also bound to three pim2− oxygen atoms from two different chains, thus generating a triperiodic assembly (KPI, 0.72). Seven hydrogen bonding rings can be discerned, with sizes spanning a large range from R24(8) up to R66(78). Similar chains in which the central bridging ligands are also pim2− anions are known, but in these cases these bridging ligands are bis(κ2O,O'-chelating), and not bound in the μ2-κ1O:κ1O′ mode as here, with the consequence of a larger separation between the two strands.77,81 A case of bridging by oxalate ligands has also been reported.81
In contrast to previous work involving aprotic dizwitterionic dicarboxylates of large size,3–11 which are conducive to the formation of mixed-ligand complexes with periodicities varying from 0 to 3 displaying entanglement in several cases, the present work with the Hamb monozwitterionic ligand has only yielded monoperiodic coordination polymers and one tetranuclear discrete species. The reduced metal ion-bridging ability of Hamb is of course a drawback here, but, due to the presence of the primary ammonium group, its bifunctionality is more obvious than with pyridinium-based zwitterionic carboxylates. Three types of chains are found within the series. In the simple chains found in 1–3, all the carboxylate groups are κ2O,O′-chelating, the dicarboxylates being bridging and Hamb terminal. Generally speaking, when only anionic dicarboxylates are used in uranyl ion complex synthesis, a frequent outcome of uranyl tris-chelation is the formation of honeycomb networks, as is for example very often found with tdc2−.55–59,61 The fact that complex 1 is formed preferentially to a honeycomb network involving only tdc2− ligands may be seen as further indication of the particular ease with which true mixed-ligand species can be obtained through mixing anionic and zwitterionic carboxylates.3 Chains with double bridges between uranyl centres are found in complexes 4 and 6, with either both ligands or only the dicarboxylate, respectively, being bridging, and complex 5 illustrates the effect of a coordinating solvent (deleterious in regard to polymer formation in the present case, but not necessarily so in general82). Complex 7 is a double-stranded chain in which Hamb links the two linear uranyl pimelate polymers. In all these complexes, the pendent ammonium group provides an attachment point through hydrogen bonding, with a minimum of three bonds formed, or more where some are bifurcated, the interaction with neighbouring polymeric units being facilitated by the sideways orientation of this group with respect to the aromatic ring.
While in general the structural influence of Hamb as a ligand on uranyl ion is most obviously related to the interactions of its ammonium group, there are indications of subtle influences on the coordination of dicarboxylate ligands, as seen in complexes 1 and 3, and there is a contrast with aprotic zwitterions in that in none of the present cases does Hamb bind in a unidentate manner. This and the bond valence parameters of oxygen donors (which do not differ significantly from those of the anionic dicarboxylate ligands) are perhaps indicative of a higher Lewis basicity of the carboxylate group of Hamb than of those in known aprotic zwitterions or even of that of betaine itself, where unidentate bonding is quite common in general,4 although one earlier case of unidentate coordination of Hamb is known.28 That in all the present mixed ligand species the dicarboxylic acid is deprotonated while Hamb is not must be in part due to pKa differences but may also reflect the influence of the ammonio group on lattice energy and hence solubility. Such an effect could explain why Hamb partially displaces pht2− in the cluster found in complex 5, where all four UVI centres have pentagonal-bipyramidal coordination, rather than two with hexagonal-bipyramidal coordination as in other cases of the phthalate-bound U4O2 cluster. In relation to the biological chemistry of uranyl ion and its binding to zwitterionic amino-acids, it is clear that there is nothing inherently unstable about a protic zwitterion bound to UVI and that difficulty in the synthesis of amino-acid complexes must relate to the core composition of the amino-acids themselves.
Luminescence properties
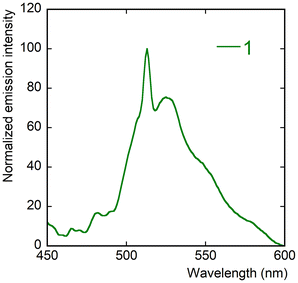
Complexes 1–4 and 7 are emissive in the solid state under excitation at 420 nm, with photoluminescence quantum yields (PLQYs) of 11% for 1, 7% for 2, 11% for 3, 2% for 4, and 4% for 7, whereas 5 and 6 are virtually non-emissive, with PLQYs lower than 1%. The emission spectra for complexes 2–4 and 7, shown in Fig. 8, display the typical vibronic progression due to the S10 → S0ν (ν = 0–4) transitions of the uranyl ion.83,84 These complexes are clearly separated into two groups, with the hexagonal-bipyramidal complexes 2 and 3 having signals blue-shifted with respect to pentagonal-bipyramidal 4 and 7. The maxima positions at 480/479, 500/498, 521/520, 545/544 and 572/569 nm for 2/3, and 491/496, 512/514, 533/537, 558/563 and 586/591 nm for 4/7 match the usual values for O6 and O5 equatorial environments, respectively.85 The low intensity “hot-band” (S11 → S00) due to electron–phonon coupling86 is observed at 466 and 465 nm for 2 and 3, respectively, but its presence is unclear due to the irregular baseline for 4 and it is not seen for 7. In contrast, the emission spectrum of 1 only displays a very broad and irregular signal with a superimposed narrow peak at 513 nm which could correspond to the most intense peak from the usual sequence (Fig. 9). Given that the structure of complex 1 does show some unusual features, in particular stacking of tdc2− ligands involving detectable S⋯S interactions, it is possible that its emission is not uranyl-centred. | ||
| Fig. 8 Emission spectra of complexes 2, 3, 4 and 7 in the crystalline state upon excitation at 420 nm. | ||
Conclusions
We have reported the synthesis, crystal structure and luminescence properties of seven uranyl ion complexes which all involve the monozwitterion 4-(ammoniomethyl)benzoate coupled with different dicarboxylate coligands. True mixed-ligand, heteroleptic complexes are formed in all cases, thus confirming the interest of zwitterionic ligands in the design of such compounds, a promising development in the domain of uranyl-containing coordination polymers and frameworks.87–89 In contrast to pyridinium-based dizwitterions mostly used in our previous work, Hamb combines a moderate assembling ability through metal ion coordination, with only one carboxylate group, and an important hydrogen-bond donor capacity associated with the primary ammonium group. As a consequence, the coordination polymers formed are monoperiodic only (but for one tetranuclear, discrete species), which contrasts with the very diverse architectures of higher periodicity obtained with elongated dizwitterionic dicarboxylates. Extended hydrogen bonding however associates the chains into di- or triperiodic assemblies, making Hamb a true bifunctional ligand. A possible development of this work would be to use partial or pseudo-zwitterionic ligands3 possessing two or more carboxylates and one primary ammonium group, which would however not be neutral but anionic.Conflicts of interest
There are no conflicts of interest to declare.References
- R. S. McEwen, J. Chem. Soc., Chem. Commun., 1973, 68 RSC.
- X. M. Chen, Synthesis and Structural Studies of Some Metal Complexes of Betaines, Doctoral thesis, The Chinese University of Hong Kong, 1992 Search PubMed.
- P. Thuéry and J. Harrowfield, Coord. Chem. Rev., 2024, 510, 215821 CrossRef.
- L. Baklouti and J. Harrowfield, Dalton Trans., 2023, 52, 7772 RSC.
- S. Kusumoto, Y. Atoini, S. Masuda, Y. Koide, J. Y. Kim, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, CrystEngComm, 2022, 24, 7833 RSC.
- S. Kusumoto, Y. Atoini, S. Masuda, J. Y. Kim, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, Inorg. Chem., 2022, 61, 15182 CrossRef CAS PubMed.
- S. Kusumoto, Y. Atoini, S. Masuda, Y. Koide, J. Y. Kim, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, Inorg. Chem., 2023, 62, 3929 CrossRef CAS PubMed.
- S. Kusumoto, Y. Atoini, Y. Koide, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, CrystEngComm, 2023, 25, 5748 RSC.
- S. Kusumoto, Y. Atoini, S. Masuda, Y. Koide, K. Chainok, Y. Kim, J. Harrowfield and P. Thuéry, Inorg. Chem., 2023, 62, 7803 CrossRef CAS PubMed.
- S. Kusumoto, Y. Atoini, Y. Koide, K. Chainok, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, Chem. Commun., 2023, 59, 10004 RSC.
- S. Kusumoto, Y. Atoini, Y. Koide, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, J. Inclusion Phenom. Macrocyclic Chem., 2024, 104, 209 CrossRef CAS.
- G. Bombieri, E. Forsellini, G. Tomat, L. Magon and R. Graziani, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1974, 30, 2659 CrossRef.
- Y. Z. Zheng, M. L. Tong and X. M. Chen, Eur. J. Inorg. Chem., 2005, 4109 CrossRef.
- H. Sopo, J. Sviili, A. Valkonen and R. Sillanpää, Polyhedron, 2006, 25, 1223 CrossRef CAS.
- P. Nockemann, R. Van Deun, B. Thijs, D. Huys, E. Vanecht, K. Van Hecke, L. Van Meervelt and K. Binnemans, Inorg. Chem., 2010, 49, 3351 CrossRef CAS PubMed.
- D. K. Unruh, K. Gojdas, A. Libo and T. Z. Forbes, J. Am. Chem. Soc., 2013, 135, 7398 CrossRef CAS PubMed.
- A. S. Jayasinghe, D. K. Unruh, A. Kral, A. Libo and T. Z. Forbes, Cryst. Growth Des., 2015, 15, 4062 CrossRef CAS.
- A. Riisiö, A. Väisänen and R. Sillanpää, Inorg. Chem., 2013, 52, 8591 CrossRef PubMed.
- X. Hou and S. F. Tang, RSC Adv., 2014, 4, 34716 RSC.
- X. Gao, J. Song, L. X. Sun, Y. H. Xing, F. Y. Bai and Z. Shi, New J. Chem., 2016, 40, 6077 RSC.
- L. Mei, Z. N. Xie, K. Q. Hu, L. Wang, L. Y. Yuan, Z. J. Li, Z. F. Chai and W. Q. Shi, Dalton Trans., 2016, 45, 13304 RSC.
- Z. Wang, X. Hou and S. F. Tang, J. Coord. Chem., 2021, 74, 1693 CrossRef CAS.
- X. Li, J. Lu, W. Mu, B. Chen, D. Luo, B. Liu, Y. Yang, H. Wei and S. Peng, Inorg. Chim. Acta, 2022, 530, 120675 CrossRef CAS.
- N. W. Alcock, D. J. Flanders, T. J. Kemp and M. A. Shand, J. Chem. Soc., Dalton Trans., 1985, 517 RSC.
- J. de Groot, K. Gojdas, D. K. Unruh and T. Z. Forbes, Cryst. Growth Des., 2014, 14, 1357 CrossRef CAS.
- J. de Groot, B. Cassell, M. Basile, T. Fetrow and T. Z. Forbes, Eur. J. Inorg. Chem., 2017, 1938 CrossRef CAS.
- R. N. Shchelokov, Y. N. Mikhailov, G. M. Lobanova, A. S. Kanishcheva, I. M. Orlova, N. B. Generalova and G. V. Podnebesnova, Russ. J. Inorg. Chem., 1982, 27, 2348 CAS.
- P. Thuéry, Cryst. Growth Des., 2012, 12, 499 CrossRef.
- APEX4, ver. 2021.10–0, Bruker AXS, Madison, WI, 2021 Search PubMed.
- SAINT, ver. 8.40A, Bruker Nano, Madison, WI, 2019 Search PubMed.
- SADABS, ver. 2016/2, Bruker AXS, Madison, WI, 2016 Search PubMed.
- L. Krause, R. Herbst-Irmer, G. M. Sheldrick and D. Stalke, J. Appl. Crystallogr., 2015, 48, 3 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3 Search PubMed.
- C. B. Hübschle, G. M. Sheldrick and B. Dittrich, J. Appl. Crystallogr., 2011, 44, 1281 CrossRef PubMed.
- A. L. Spek, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 9 CrossRef CAS PubMed.
- M. N. Burnett and C. K. Johnson, ORTEPIII, Report ORNL-6895, Oak Ridge National Laboratory, TN, 1996 Search PubMed.
- L. J. Farrugia, J. Appl. Crystallogr., 2012, 45, 849 CrossRef CAS.
- K. Momma and F. Izumi, J. Appl. Crystallogr., 2011, 44, 1272 CrossRef CAS.
- A. M. Atria, M. T. Garland and R. Baggio, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2014, 70, 385 CrossRef CAS PubMed.
- P. R. Spackman, M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, D. Jayatilaka and M. A. Spackman, J. Appl. Crystallogr., 2021, 54, 1006 CrossRef CAS PubMed.
- S. K. Wolff, D. J. Grimwood, J. J. McKinnon, M. J. Turner, D. Jayatilaka and M. A. Spackman, CrystalExplorer, University of Western Australia, 2012 Search PubMed.
- T. D. Keene, I. Zimmermann, A. Neels, O. Sereda, J. Hauser, S. X. Liu and S. Decurtins, Cryst. Growth Des., 2010, 10, 1854 CrossRef CAS.
- Y. H. Luo, S. W. Ge, W. T. Song and B. W. Sun, New J. Chem., 2014, 38, 723 RSC.
- A. Direm, A. Altomare, A. Moliterni and N. Benali-Cherif, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2015, 71, 427 CrossRef CAS PubMed.
- M. L. Aubrey, A. S. Valdes, M. R. Filip, B. A. Connor, K. P. Lindquist, J. B. Neaton and H. I. Karunadasa, Nature, 2021, 597, 355 CrossRef CAS PubMed.
- A. Kaiba, F. Al Otaibi, M. H. Geesi, Y. Riadi, T. A. Aljohani and P. Guionneau, J. Mol. Struct., 2021, 1234, 130129 CrossRef CAS.
- Y. V. Kokunov, V. V. Kovalev, Y. E. Gorbunova, G. A. Razgonyaeva and S. A. Kozyukhin, Russ. J. Inorg. Chem., 2018, 63, 333 CrossRef CAS.
- Y. V. Kokunov, V. V. Kovalev, Y. E. Gorbunova, S. A. Kozyukhin and G. A. Razgonyaeva, Russ. J. Coord. Chem., 2018, 44, 722 CrossRef CAS.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171 CrossRef CAS PubMed.
- N. E. Brese and M. O'Keeffe, Acta Crystallogr., Sect. B: Struct. Sci., 1991, 47, 192 CrossRef.
- A. L. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 148 CrossRef CAS PubMed.
- S. G. Thangavelu, M. B. Andrews, S. J. A. Pope and C. L. Cahill, Inorg. Chem., 2013, 52, 2060 CrossRef CAS PubMed.
- S. G. Thangavelu, S. J. A. Pope and C. L. Cahill, CrystEngComm, 2015, 17, 6236 RSC.
- S. G. Thangavelu, R. J. Butcher and C. L. Cahill, Cryst. Growth Des., 2015, 15, 3481 CrossRef CAS.
- H. H. Li, X. H. Zeng, H. Y. Wu, X. Jie, S. T. Zheng and Z. R. Chen, Cryst. Growth Des., 2015, 15, 10 CrossRef CAS.
- P. Thuéry and J. Harrowfield, CrystEngComm, 2016, 18, 1550 RSC.
- S. J. Jennifer and A. K. Jana, Cryst. Growth Des., 2017, 17, 5318 CrossRef CAS.
- P. Thuéry and J. Harrowfield, Inorg. Chem., 2021, 60, 9074 CrossRef PubMed.
- P. Thuéry and J. Harrowfield, Inorg. Chem., 2022, 61, 9725 CrossRef PubMed.
- Y. Atoini, S. Kusumoto, Y. Koide, S. Hayami, Y. Kim, J. Harrowfield and P. Thuéry, Polyhedron, 2024, 250, 116848 CrossRef CAS.
- J. Bernstein, R. E. Davis, L. Shimoni and N. L. Chang, Angew. Chem., Int. Ed., 1995, 34, 1555 CrossRef CAS.
- P. Thuéry, Y. Atoini and J. Harrowfield, Cryst. Growth Des., 2019, 19, 6611 CrossRef.
- P. Thuéry, Y. Atoini and J. Harrowfield, Inorg. Chem., 2019, 58, 6550 CrossRef PubMed.
- P. Thuéry, Y. Atoini and J. Harrowfield, Inorg. Chem., 2020, 59, 2503 CrossRef PubMed.
- P. Thuéry and J. Harrowfield, Eur. J. Inorg. Chem., 2021, 2182 CrossRef.
- P. Thuéry, Y. Atoini and J. Harrowfield, Inorg. Chem., 2019, 58, 870 CrossRef PubMed.
- P. Thuéry and J. Harrowfield, Cryst. Growth Des., 2021, 21, 3000 CrossRef.
- I. A. Charushnikova, N. N. Krot, I. N. Polyakova and V. I. Makarenkov, Radiochemistry, 2005, 47, 241 CrossRef CAS.
- I. Mihalcea, C. Volkringer, N. Henry and T. Loiseau, Inorg. Chem., 2012, 51, 9610 CrossRef CAS PubMed.
- G. Andreev, N. Budantseva, A. Levtsova, M. Sokolova and A. Fedoseev, CrystEngComm, 2020, 22, 8394 RSC.
- P. Thuéry, Cryst. Growth Des., 2011, 11, 347 CrossRef.
- P. Thuéry, Y. Atoini and J. Harrowfield, Inorg. Chem., 2020, 59, 2923 CrossRef PubMed.
- L. A. Borkowski and C. L. Cahill, Cryst. Growth Des., 2006, 6, 2241 CrossRef CAS.
- L. A. Borkowski and C. L. Cahill, Cryst. Growth Des., 2006, 6, 2248 CrossRef CAS.
- P. Thuéry, Cryst. Growth Des., 2011, 11, 2606 CrossRef.
- P. Thuéry and J. Harrowfield, Inorg. Chem., 2016, 55, 2133 CrossRef PubMed.
- P. Thuéry, E. Rivière and J. Harrowfield, Cryst. Growth Des., 2016, 16, 2826 CrossRef.
- P. Thuéry and J. Harrowfield, CrystEngComm, 2016, 18, 3905 RSC.
- P. Thuéry and J. Harrowfield, Cryst. Growth Des., 2017, 17, 2116 CrossRef.
- P. Thuéry, Y. Atoini and J. Harrowfield, Inorg. Chem., 2019, 58, 567 CrossRef PubMed.
- P. Thuéry and J. Harrowfield, Cryst. Growth Des., 2014, 14, 1314 CrossRef.
- A. Brachmann, G. Geipel, G. Bernhard and H. Nitsche, Radiochim. Acta, 2002, 90, 147 CrossRef CAS.
- M. Demnitz, S. Hilpmann, H. Lösch, F. Bok, R. Steudtner, M. Patzschke, T. Stumpf and N. Huittinen, Dalton Trans., 2020, 49, 7109 RSC.
- P. Thuéry and J. Harrowfield, Inorg. Chem., 2017, 56, 13464 CrossRef PubMed.
- D. H. Chen, N. Vankova, G. Jha, X. Yu, Y. Wang, L. Lin, F. Kirschhöfer, R. Greifenstein, E. Redel, T. Heine and C. Wöll, Angew. Chem., Int. Ed., 2024, e202318559 CAS.
- T. Loiseau, I. Mihalcea, N. Henry and C. Volkringer, Coord. Chem. Rev., 2014, 266–267, 69 CrossRef CAS.
- P. Thuéry and J. Harrowfield, Dalton Trans., 2017, 46, 13660 RSC.
- K. Lv, S. Fichter, M. Gu, J. März and M. Schmidt, Coord. Chem. Rev., 2021, 446, 214011 CrossRef CAS.
Footnote |
| † CCDC 2347791–2347797. For crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d4ce00381k |
| This journal is © The Royal Society of Chemistry 2024 |