Highlights from the Biocatalysis Faraday Discussion, May 2024, London, United Kingdom
Agata
Raczyńska
 *abc
*abc
aTunneling Group, Biotechnology Centre, Silesian University of Technology, ul. Krzywoustego 8, 44-100 Gliwice, Poland. E-mail: agata.raczynska@polsl.pl
bToulouse Biotechnology Institute, TBI, Université de Toulouse, CNRS, INRAE, INSA, 135 avenue de Rangueil, F-31077 Toulouse Cedex 04, France
cFaculty of Chemistry, Silesian University of Technology, ul. Strzody 9, 44-100 Gliwice, Poland
First published on 6th September 2024
Abstract
The Biocatalysis Faraday Discussion was held from May 22 to 24, 2024, at the Royal Society of Chemistry in Burlington House, London. This meeting brought together established and early-career scientists, PhD students, and industrial researchers from around the world to engage in rigorous scientific dialogue on the latest advancements in biocatalysis. The conference featured a unique format, where speakers submitted full papers in advance and presented concise summaries, sparking in-depth discussions among participants. This report summarises the event, the presented results, and the concluding remarks, underscoring the collaborative and intellectually stimulating atmosphere of the Faraday Discussions.
Introduction
The Faraday Discussions, named in honor of Michael Faraday, pay homage to his legacy by fostering open dialogue and collaboration among scientists worldwide. Faraday, celebrated for his pioneering experiments in electromagnetism and chemistry, is best remembered for his engaging lectures, including the famed Christmas Lectures at the Royal Institution, which captivated young audiences with demonstrations of scientific principles. His innovative approach not only educated but also inspired generations of scientists. Just as Faraday bridged theory and practice in his experiments, the Faraday Discussions continue to transcend interdisciplinary boundaries in pursuit of scientific advancement. This tradition of interactive and accessible scientific discourse remains a hallmark, embodying Faraday’s commitment to sharing knowledge with the wider community.Faraday Discussions maintain a century-old format, where each speaker submits a full paper in advance and presents a concise summary of their research at the conference. Evolving over time, the current structure involves a 5-minute summary presentation that initiates in-depth discussions among delegates who have reviewed the papers beforehand. This unique format blends traditional conference elements with real-time, interactive peer review, facilitating profound cross-disciplinary engagement and providing a platform to explore cutting-edge research with experts and emerging scholars alike. Unique to the Faraday Discussions, this format ensures that every participant’s voice is heard, and every idea is rigorously evaluated in the pursuit of scientific excellence.
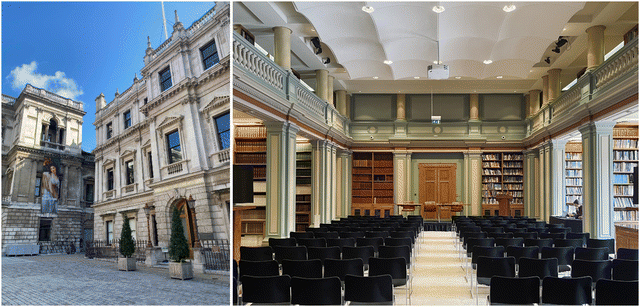
The Biocatalysis Faraday Discussions, held from May 22 to 24, 2024, at the Royal Society of Chemistry in Burlington House, Piccadilly, London, exemplified this format. Situated amidst other esteemed institutions, such as the Royal Academy of Arts and the Royal Astronomical Society, the venue provided an ideal setting for intellectual exchange (Fig. 1). Burlington House’s atmosphere, adorned with historical documents, portraits of past presidents, and modern artworks related to chemistry, underscored the Society’s rich heritage and ongoing dedication to scientific advancement. Busts of scientific luminaries, including Faraday himself, served as a poignant reminder of their lasting influence on future generations.
The lecture hall provided a fitting atmosphere for the Biocatalysis Faraday Discussion, where participants engaged in three days of lively and impactful scientific dialogue. Entering the Library room, delegates were surrounded by scientific journals and books, a testament to the cumulative knowledge amassed over centuries (Fig. 1). Here, amidst the quiet anticipation of participants from diverse corners of the globe, the stage was set for three days of rigorous scientific discourse.
The Biocatalysis Faraday Discussion began with welcoming remarks from Professors Adrian Mulholland of the University of Bristol and Nicholas Turner of the University of Manchester. Robert Hinde and Samuel Oldknow, Royal Society of Chemistry Editors, then explained the outline of the discussion format, setting the stage for an innovative approach aimed at engaging early career scientists and PhD students, fostering a dynamic and inclusive atmosphere.
Professor Donald Hilvert of ETH Zurich set the tone with an enlightening introductory Spiers Memorial Lecture (https://doi.org/10.1039/D4FD00139G), offering insights into the current state of biocatalysis, its challenges, and future directions. Established to honor Frank Spiers’ significant contributions to the development and advancement of the Faraday Society, the Spiers Memorial Lecture serves as the introductory lecture at each Faraday Discussions meeting, setting the stage for the topics to be discussed. Professor Hilvert presented findings from his groundbreaking work, where a zinc-binding peptide was transformed through design and laboratory evolution into a globular enzyme. This engineered enzyme demonstrated exceptional capabilities, accelerating ester cleavage with precise enantiospecificity and remarkable catalytic efficiency.1 His comprehensive overview helped frame the subsequent discussions on the evolution and engineering of enzymes.
Session 1 “Enzyme evolution, engineering and design: mechanism and dynamics”
After the introductory lecture, the first session, focused on the mechanisms and dynamics of enzyme evolution, engineering, and design, commenced. The first part of the session was chaired by Professor Adrian Mulholland.As the first speaker, Joelle Pelletier from the University of Montreal, Canada, delivered an insightful talk on streamlining the screening process for cytochrome P450 BM3 enzyme variants with high hydroxylation activity (https://doi.org/10.1039/d4fd00017j). Pelletier and coworkers proposed using indigo production as a high-throughput method, where bacterial colonies expressing active variants turn blue for easy identification. The variants were tested for hydroxylation of 12 aromatic compounds using the 4-aminoantipyrine colorimetric assay. Combining indigo (+) variants in recombinant libraries revealed that high hydroxylation activity often correlated with indigo production. This approach simplifies and accelerates the screening process, aiding the development of efficient enzyme variants for industrial and biotechnological applications.
The second talk of the session, delivered by Zhiqi Cong from the Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, China, focused on tuning the peroxidase activity of artificial P450 peroxygenase by engineering redox-sensitive residues (https://doi.org/10.1039/d4fd00008k). By mutating redox-sensitive residues such as methionines, tryptophans, cysteines, and phenylalanines, the team significantly improved the peroxidase activity of P450BM3. The study identified six beneficial mutations that, when combined, enhanced peroxidase activity by over 100-fold, rivaling natural peroxidases. This work offers new strategies for regulating the catalytic promiscuity of cytochrome P450 enzymes for various oxidative functions.
The last talk of the first part of this session was given by Professor Jeremy Harvey, of KU Leuven, Belgium, on exploring the selectivity of cytochrome P450 for enhanced novel anticancer agent synthesis (https://doi.org/10.1039/d4fd00004h). Cytochrome P450 monooxygenases are remarkable enzymes known for their ability to regio- and stereo-selectively oxidise hydrocarbons and have been implicated in a biosynthetic pathway that produces highly potent antitumour compounds. Harvey’s team’s research employed computational techniques to delve into the factors influencing the relative yields of these products. By using conformational search algorithms, the team analysed substrate stereochemistry. Molecular docking on a machine-learned 3D protein structure helped them identify favorable substrate–protein interaction poses. Further molecular dynamics simulations of these poses allowed them to pinpoint interactions that favor specific products, providing insights into the product selectivity of cytochrome P450 enzymes.
Following these presentations, a lively discussion ensued (http://doi.org/10.1039/D4FD90022G), allowing delegates to ask questions and delve deeper into the presented research. After a break, the session continued with Professor Fraser Armstrong chairing the second part, beginning with a talk by Professor Florian Hollfelder of the University of Cambridge, UK, on the synergy between ultrahigh throughput screening and machine learning in biocatalyst engineering.
Professor Hollfelder’s presentation explored the intersection of protein design, directed evolution, and computational methods in biocatalyst engineering (https://doi.org/10.1039/d4fd00065j). Highlighting the complementary nature of these approaches, he discussed how ultrahigh throughput screening, coupled with next-generation sequencing and machine learning, offers new avenues for protein engineering. By miniaturising functional tests into picoliter-scale water-in-oil emulsion droplets capable of rapid generation and analysis, researchers can screen vast libraries exceeding 107 variants in a single day. Sequencing technologies then decode selected clones, generating extensive sequence-function datasets that facilitate the extrapolation of directed evolution experiments to predict and design improved protein mutants. Hollfelder’s work also examined experimental strategies for mapping ‘fitness landscapes’ in sequence space using ultrahigh throughput droplet microfluidics and discussed the current integration of artificial intelligence and machine learning in enhancing biocatalyst engineering outcomes.
The last presentation was by Professor Mikael Widersten of Uppsala University, Sweden, on using phage display to select new alkyl halide hydrolases from a library of mutated epoxide hydrolases (https://doi.org/10.1039/d4fd00001c). He discussed the potato epoxide hydrolase StEH1, structurally similar to haloalkane dehalogenase DhlA, which modestly hydrolyses (2-chloro)- and (2-bromo) ethanebenzene to 2-phenylethanol. By mutating five active-site residues, Widersten’s team created a library of variants and screened them using monovalent phage display. They identified several StEH1-derived enzymes with significantly enhanced dehalogenase activities, demonstrating the potential of this method for improving enzyme functions for industrial and environmental use.
After an ensuing discussion (http://doi.org/10.1039/D4FD90022G), the first session concluded to allow for the subsequent poster session.
Session 2 “Biocatalytic pathways, cascades, cells and systems”
The second session of the Faraday Discussion on biocatalytic pathways, cascades, cells, and systems commenced on the second day and extended through to noon, under the chairmanship of Professor Uwe Bornscheuer.The session commenced with an engaging speech from Professor Pimchai Chaiyen of Vidyasirimedhi Institute of Science and Technology, Thailand, on enhancing essential cofactors for in vivo biocatalysis (https://doi.org/10.1039/d4fd00013g). Chaiyen’s team addressed cofactor scarcity in enzymatic reactions within engineered cells, presenting a novel approach using xylose reductase and lactose to boost intracellular NAD(P)H, ATP, and acetyl coenzyme A. Compared to previous methods, this strategy significantly enhanced the production of sugar alcohols and sugar phosphates. The study introduced the “user-pool” model, showing dynamic cofactor augmentation patterns and using untargeted metabolomics to reveal rapid improvements in key cofactors and intermediates, positively influencing fatty-alcohol production. This early-stage analysis highlighted the potential of the xylose reductase and lactose system in enhancing cofactor capacities in microbial cell factories.
The next speaker, Professor Dominic Campopiano of the University of Edinburgh, UK, presented on developing deprotectase biocatalysts for synthesis (https://doi.org/10.1039/d4fd00016a). Organic synthesis often requires protecting functional groups to control reactivity, which must later be removed. Conventional deprotection methods can be inefficient, prompting exploration of enzymatic approaches as sustainable alternatives. Campopiano’s team focused on biocatalysts capable of selectively removing these protective groups. They used Bacillus BS2 esterase to remove tert-butyloxycarbonyl groups from amino acids, and screened Sphingomonas Cbz-ase (amidohydrolase) for N-carbobenzyloxy group removal. Combining both enzymes enabled efficient two-step deprotection of doubly protected amino acids. Molecular docking studies provided insights into substrate recognition and catalytic mechanisms, suggesting potential enhancements for chemical synthesis methodologies.
After a discussion (http://doi.org/10.1039/D4FD90023E) and a coffee break, the session resumed with a presentation by Clare Megarity from the University of Manchester, UK, chaired by Dr Bruce Lichtenstein. Megarity’s talk focused on the electrochemical leaf’s capabilities (https://doi.org/10.1039/d4fd00020j), which enable precise control and electrification of multi-enzyme cascades. This innovative approach leverages two key discoveries: firstly, the electrical activation of ferredoxin NADP+ reductase within a porous metal oxide electrode to convert NADP+ to NADPH, and secondly, the co-entrapment of additional enzymes within electrode pores. These cascades are extended through NADP(H) recycling facilitated by ferredoxin NADP+ reductase, engineered by mutating a critical active-site tyrosine to serine to alter cofactor specificity, now accepting NAD(H), albeit with a reduced flavin reduction potential. Electrochemical analyses of this variant revealed insights into its catalytic behaviour, including a trapped intermediate state influenced by the applied potential, crucial for overcoming catalytic barriers. The study also highlighted how tightly bound NADP+ inhibits NAD(H) turnover, a phenomenon modulated by electrochemical conditions, emphasising the intricate interplay between enzyme activity and electrode potential in the electrochemical leaf system. These findings are pivotal for advancing practical applications in biocatalysis and synthetic biology.
The next speaker, Professor Neil Marsh from the University of Michigan, USA, discussed aromatic decarboxylations catalysed by prenylated-flavin dependent enzymes (https://doi.org/10.1039/d4fd00006d). These UbiD-like enzymes use a modified flavin cofactor to facilitate decarboxylation of unsaturated carboxylic acids via a unique 1,3-dipolar cyclo-addition mechanism, making them valuable for biocatalysis. Marsh and coworkers highlighted their broad substrate tolerance and the potential of approximately 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 annotated protein sequences within this enzyme family. Their study used enzyme-catalysed solvent deuterium exchange to screen potential substrates, focusing on ferulic acid decarboxylase. The research revealed that ferulic acid decarboxylase catalyses hydrogen/deuterium atom exchange into various small aromatic molecules, indicating substrate promiscuity but selective decarboxylation efficiency. This work provides insights into exploring novel UbiD-like enzymes for expanding (de)carboxylation reactions in biocatalytic applications.
000 annotated protein sequences within this enzyme family. Their study used enzyme-catalysed solvent deuterium exchange to screen potential substrates, focusing on ferulic acid decarboxylase. The research revealed that ferulic acid decarboxylase catalyses hydrogen/deuterium atom exchange into various small aromatic molecules, indicating substrate promiscuity but selective decarboxylation efficiency. This work provides insights into exploring novel UbiD-like enzymes for expanding (de)carboxylation reactions in biocatalytic applications.
The session concluded with a presentation by Dr Francesca Valetti from the University of Torino, Italy. Her talk focused on oxygen-resistant [FeFe]hydrogenases and their potential as biocatalysts for clean energy and cascade reactions (https://doi.org/10.1039/d4fd00010b). These enzymes, unlike their oxygen-sensitive counterparts, exhibit a natural defense against oxygen damage, making them promising for applications such as biofuel cells and CO2 conversion. Valetti highlighted the discovery of clostridial hydrogen-producing enzymes, their protein engineering strategies, and their potential for enhanced stability and efficiency in biotechnological processes, paving the way for sustainable hydrogen-driven biocatalysis.
Session 3 “Artificial, biomimetic and hybrid enzymes”
After lunch in the beautiful Council Room (Fig. 2), the afternoon session commenced with the third session of the meeting, focused on artificial, biomimetic, and hybrid enzymes, chaired by Dr Amanda Jarvis. The first presentation by Professor Gerard Roelfes from the University of Groningen, Netherlands, elucidated the computation-guided engineering of distal mutations in an artificial enzyme (https://doi.org/10.1039/d4fd00069b). Roelfes demonstrated how the catalytic performance of an already engineered artificial enzyme could be enhanced by introducing distal mutations that affect the protein’s conformational equilibrium. Using an innovative algorithm, the team identified 73 potential variants, of which two, with mutations more than 11 Å from the active site, exhibited increased catalytic activity for a new-to-nature hydrazone formation reaction. These variants showed a 66% higher turnover number and improved thermostability by 14 °C. Molecular dynamics simulations revealed that these improvements resulted from a shift in the distribution of productive enzyme conformations initiated by the introduced mutations.The second speaker of this session, Dr Ivana Drienovská of VU Amsterdam, Netherlands, presented her work on designing Michaelases, exploring novel protein scaffolds for iminium biocatalysis (https://doi.org/10.1039/d4fd00057a). Drienovská highlighted the potential of biocatalysis as a sustainable alternative for asymmetric catalysis, though often limited by the inherent reactivity of natural enzymes. To address this, her team focused on protein engineering to introduce new-to-nature functional groups into proteins, creating artificial enzymes. They investigated various protein scaffolds for the Michael addition of nitromethane to E-cinnamaldehyde, a model of iminium-ion catalysis. They selected scaffolds with open hydrophobic pockets and known binding sites for the substrate, expressing and analysing variants with functional amine groups such as lysine, p-aminophenylalanine, or N6-(D-prolyl)-L-lysine.
The next speaker, Professor Anthony Green from the University of Manchester, UK, presented on the development of an efficient pyrrolysyl-tRNA synthetase for the economical production of MeHis-containing enzymes (https://doi.org/10.1039/d4fd00019f). By incorporating mutations from Methanomethylophilus alvus into Methanogenic archaeon ISO4-G1, they created a highly efficient aminoacyl tRNA synthetase for encoding MeHis into proteins, which functions at low MeHis concentrations (∼0.1 mM). This advancement allows for cost-effective production of MeHis-containing enzymes and the incorporation of various non-canonical amino acids, enhancing enzyme engineering.
After a lively discussion (http://doi.org/10.1039/D4FD90024C) and a refreshments break, the third session continued, chaired by Professor Jeremy Harvey, with a talk from Professor Sílvia Osuna from the Universitat de Girona & ICREA, Spain. Her presentation focused on harnessing conformational dynamics in enzyme catalysis to achieve nature-like catalytic efficiencies using the shortest path map tool for computational enzyme redesign (https://doi.org/10.1039/d3fd00156c). By constructing graphs based on residue distances and correlated motions from molecular dynamics simulations, the shortest path map method identifies key mutations that enhance specific conformational changes, even at distal sites. The study showcased the integration of the shortest path map with a template-based AlphaFold2 approach and short molecular dynamics simulations, successfully estimating conformational landscapes and identifying crucial hotspots for computational enzyme engineering. Using the beta subunit of tryptophan synthase as a model, the efficacy of the method was demonstrated, highlighting its potential in rapidly capturing enzyme dynamics and facilitating laboratory evolution of enzymes.
The penultimate speaker, Professor Vicent Moliner of Universitat Jaume I, Spain, presented findings from a computational study of the mechanism of a polyurethane esterase A (PueA) from Pseudomonas chlororaphis in the hydrolysis of a polyester polyurethane, Impranil DLN (https://doi.org/10.1039/d4fd00022f). Highlighting the urgent need for effective plastic waste management, Professor Moliner’s team explored PueA’s potential in enzymatic biodegradation. Using AlphaFold2 to model the enzyme’s 3D structure and QM/MM molecular dynamics simulations, the study identified the enzyme’s active site and protein–substrate binding interactions. The free energy landscape analysis revealed that wild-type PueA can degrade polyurethane chains, following a four-step serine hydrolase mechanism.
The last speaker, Professor Lynn Kamerlin of the Georgia Institute of Technology, USA, gave an online presentation shedding light on conformational regulation across orthologues and evolutionary lineages using KIF and KIN tools (https://doi.org/10.1039/d4fd00018h). These tools analyse protein structures via noncovalent interaction networks, covering static and dynamic conformational ensembles. Kamerlin explored their application in understanding substrate specificity evolution in class A beta-lactamases, focusing on transitions from generalist to specialist catalytic functions. The study suggests targets for experimental protein engineering, providing insights into manipulating catalytic functions through targeted modifications.
Following the discussion (http://doi.org/10.1039/D4FD90024C), the second day of presentations concluded with brief lightning talks by selected poster-presenting delegates, followed by a poster session. The day wrapped up with a conference dinner, providing attendees with an opportunity to network in a more relaxed setting.
Session 4 “Biocatalysis for industry, medicine and the circular economy”
The final fourth session on biocatalysis for industry, medicine, and the circular economy, began on the third and concluding day of the Biocatalysis Faraday Discussion, chaired by Dr Meilan Huang. Professor Per-Olof Syrén from KTH Royal Institute of Technology, Sweden, opened the session with a presentation on the degradation of polyethylene terephthalate (PET) microplastic particles to monomers in human serum using PETase (https://doi.org/10.1039/d4fd00014e). Highlighting the environmental and health risks posed by billions of tons of plastic waste, his research demonstrates PETase’s capability to break down PET into substances like terephthalic acid and mono(2-hydroxyethyl)terephthalate within human serum at 37 °C, without affecting cell viability in vitro. This study explores biocatalysis as a potential solution to microplastic pollution in biomedical contexts.The next speech was given by Dr Louis Luk from Cardiff School of Chemistry, UK, who presented research on controlling the activity of a peptide asparaginyl ligase through lumazine synthetase compartmentalisation (https://doi.org/10.1039/d4fd00002a). These ligases are prized for their efficient kinetics and wide substrate compatibility in protein bioconjugation. Luk’s team explored encapsulating OaAEP1-C247A, a representative enzyme of this group, within protein containers inspired by natural organelles, using engineered lumazine synthases. Initially, they attempted to use a super-positively charged GFP(+36) tag with AaLS-13, which resulted in enzyme truncation. A successful alternative involved fusing OaAEP1-C247A with a circularly permutated variant of lumazine synthetases, effectively compartmentalising the ligase within a composite protein structure. Despite reduced activity compared to its free form, this study highlights the feasibility of lumazine synthase-based encapsulation in E. coli, paving the way for optimising stable peptide-ligating artificial organelles for efficient protein modifications.
The final presentation before a coffee break was delivered by Dr Lu Shin Wong of the University of Manchester, UK, on the biocatalytic synthesis of silicone polymers (https://doi.org/10.1039/d4fd00003j). Polysiloxanes, like poly(dimethyl)siloxane, are widely used in industry and consumer products for their Si–O–Si backbone structure. Dr Wong demonstrated a biocatalytic approach using silicatein-a, an enzyme from marine sponges capable of catalysing Si–O bond hydrolysis and condensation. By using dialkoxysilane precursors in non-aqueous conditions, silicatein-a enabled the formation of poly(dimethyl)siloxane with higher molecular weights (approximately 1000–2000 Da) compared to traditional non-enzymatic methods. However, prolonged exposure led to gradual polymer degradation. This study highlights silicatein-a’s role in polysiloxane synthesis, suggesting biocatalysis as a promising approach for sustainable silicone production.
After a discussion (http://doi.org/10.1039/D4FD90025A) and a short break, the session continued with a talk from Dr Stefan Lutz, Chief Scientific Officer at Codexis, USA, on an engineered T7 RNA polymerase designed to enhance co-transcriptional capping and minimise dsRNA byproducts in mRNA synthesis (https://doi.org/10.1039/D4FD00023D). With the surge in mRNA therapies, exemplified by mRNA vaccines against SARS-CoV-2, there’s an urgent need for efficient large-scale mRNA production. The traditional approach using wild-type T7 RNA polymerase for in vitro transcription often results in costly and inefficient incorporation of the essential 5′ 7-methylguanosine cap analog, alongside the formation of unwanted dsRNA byproducts. Dr Lutz’s team addressed these challenges by developing a T7-68 RNA polymerase, which significantly enhances the co-transcriptional incorporation of both di- and tri-nucleotide cap analogs, even at reduced concentrations. Importantly, in vitro transcription products synthesised with T7-68 demonstrate markedly reduced dsRNA content, offering a promising advancement for safer and more cost-effective mRNA manufacturing processes.
The next speaker, Dr Daniel Dourado of Almac Sciences, UK, discussed the application of rational enzyme engineering in developing a new biocatalytic route to etonogestrel and levonorgestrel, vital contraceptive active ingredients (https://doi.org/10.1039/d4fd00011k). This project, funded by the Bill & Melinda Gates Foundation, aimed to lower production costs to enhance access in developing countries where cost is a barrier to family planning. Almac Group utilised their selectAZyme™ panel to select and engineer a carbonyl reductase enzyme from Bacillus weidmannii. Through computational mutant selection, Mutant-75 was developed, demonstrating improved catalytic activity and stability in bioreduction reactions of ethyl secodione, a critical intermediate for active-ingredient synthesis. This advancement allows efficient production under challenging conditions, paving the way for scalable and cost-effective contraceptive manufacturing methods in underserved regions.
In the last presentation of the session, Dr Bruce Lichtenstein from the University of Portsmouth, UK, explored the impact of fusion partners on the enzymatic activity and thermal stability of PET-degrading enzymes (https://doi.org/10.1039/d4fd00067f). Plastics pose significant environmental challenges, and engineered enzymes offer promising solutions for their recycling. Dr Lichtenstein’s team used SpyCatcher003 technology to investigate how thermal stability influences PETase enzymes. Despite significant shifts in thermal stability upon fusion, the study found no clear enhancement in enzyme activity or stability on PET films. This research highlights the complexity of enzyme engineering for plastic degradation and suggests that further detailed analysis is needed to optimise enzyme performance in recycling applications.
Poster session and social events
At the end of the first and second days of presentations, a select group of delegates were given the opportunity to present their posters in a format known as a “Lightning Presentation” – a concise presentation involving only one slide and a mere minute to convey their key findings (Fig. 3). Despite its brevity, this format allowed presenters to highlight the essence of their work, directing interested delegates to seek further discussion during the subsequent poster sessions. | ||
| Fig. 3 Selected delegates queuing to present their findings in a one-minute “Lightning Presentation” format. | ||
Following these succinct introductions, the poster sessions commenced. Delegates made their way to the Science and Fish Rooms, where the fifty-five posters were displayed. The air buzzed with conversations, scientific debates, and laughter, fostering an atmosphere of intellectual exchange. The posters’ topics spanned from enhancing chemo-enzymatic oxidation and exploring protein dynamics to developing deep learning tools for predicting biocatalytic activities and reshaping enzymes for biotechnological applications. The posters themselves were of an exceptional standard, with two distinguished for their exemplary presentation and scientific rigour, recognised with Catalysis Science & Technology Poster Prizes, awarded to Evelina Venckute from the University of Edinburgh, UK, and Thomas Lister from the University of Manchester, UK.
After the poster session on the second day, the conference dinner took place. To the delegates’ delight, the Library was transformed into an elegant banquet hall, with round tables draped in white linens. In a place of honour, where presentations usually took place, stood the ‘Faraday Division Loving Cup’ – a silver chalice reputedly crafted by the lady silversmith Heslie Fawdery in 1728 (Fig. 4). Historically, this cup had played a central role in the Loving Cup Ceremony, a cherished tradition of the Faraday Discussions symbolising fellowship, respect, and the sharing of knowledge among scientists. The ceremony had involved passing the silver cup around, with each participant taking a sip of port wine as a gesture of unity.
In the aftermath of the Covid-19 pandemic, this tradition had been adapted to allow participants to sip from their own cups while preserving the ceremony’s historical essence. Professor Dwayne Heard, President of the Faraday Community for Physical Chemistry and Professor of Atmospheric Chemistry at the University of Leeds, introduced the ceremony, elucidating its significance and storied past. Together with Professor Donald Hilvert, he commenced the ceremony with a toast in piam memoriam of G. S. Marlow and Angela and Tony Fish, esteemed contributors to the Faraday Discussions (Fig. 4). The Loving Cup ceremony imbued the conference dinner with a rich historical and cultural dimension, providing a backdrop for informal discussions and the forging of professional relationships, thus connecting modern scientists with the traditions of their predecessors.
Concluding remarks
Professor Uwe Bornscheuer from the University of Greifswald, Germany, delivered the summary and closing talk for the Biocatalysis Faraday Discussion (https://doi.org/10.1039/D4FD00127C) (Fig. 5). His presentation offered profound insights into the evolution of the field, culminating with the observation: “In the past, an enzyme-based process was designed around the limitations of the enzyme; today, the enzyme is engineered to fit the process specifications.” He also highlighted key points from this Faraday Discussion, addressing significant common challenges that remain unresolved. The concluding remarks mainly revolved around current advances and limitations in predicting enzyme properties and functions, assessing the effects of mutations, and making enzyme engineering and discovery more reliable. Professor Bornscheuer also initiated a discussion on how basic biocatalysis knowledge can be integrated into undergraduate education to raise awareness of this increasingly important field.Conclusions
In the view of this report’s author, the Biocatalysis Faraday Discussion was a quintessential example of an ideal scientific meeting. Held at the beautiful Burlington House, the event was an enriching and memorable experience that facilitated fruitful scientific discussions and established new collaborations and friendships. The enthusiasm for the subject matter was palpable, with scientific dialogue continuing seamlessly into coffee breaks and lunch conversations. Participants were keen to exchange thoughts and ideas, creating an atmosphere of collaboration and engagement. The discussions were not only effortless but also deeply interesting, especially when exploring the current limits of our knowledge and strategies to extend these boundaries. Overall, it was an immensely rewarding experience and an exceptionally enjoyable and productive conference.Acknowledgements
The author acknowledges the financial support of the Ministry of Science and Higher Education, Poland, for funding the research project under the ‘Diamond Grant’ program (no. 0169/DIA/2020/49) from the budget for science for the years 2019–2023, and of the French government through Campus France as part of the doctorate cotutelle program. Additionally, the author wishes to thank Marta Dolińska, Stuart Govan and Professor Mulholland for sharing photos from the event.References
- S. Studer, D. A. Hansen, Z. L. Pianowski, P. R. E. Mittl, A. Debon, S. L Guffy, B. S. Der, B. Kuhlman and D. Hilvert, Science, 2018, 362, 1285–1288 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |