 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cupin-domain containing protein is not essential for the alkyl salicylaldehyde formation in Aspergillus ustus†
Marlies
Peter
and
Shu-Ming
Li
 *
*
Institut für Pharmazeutische Biologie und Biotechnologie, Fachbereich Pharmazie, Philipps-Universität Marburg, Marburg 35037, Germany. E-mail: shuming.li@pharmazie.uni-marburg.de
First published on 18th September 2024
Abstract
Previous studies demonstrated the requirement of four enzymes including a cupin-domain containing protein for the formation of alkyl salicylaldehydes and derivatives. Heterologous expression of three biosynthetic genes from Aspergillus ustus resulted in the formation of such compounds in high-yields without involvement of a cupin analogue.
Alkyl salicylaldehydes and derivatives are polyketides and widely distributed in nature, especially in fungi. They differ from each other in their oxidation states as alcohols, aldehydes or carboxylic acids, in the length and saturation grade of their alkyl chains as well as further modifications.1 Differing from other aromatic polyketides, which are mainly biosynthesised by non-reducing polyketide synthases (NR-PKSs),2 the members of this group are products of highly-reducing PKSs (HR-PKSs).3 HR-PKSs are a large group of enzymes with multiple domains to generate usually non-aromatic products like lovastatin.4 Some aromatic polyketides require the collaboration of both HR-PKS and NR-PKS for their biosynthesis.5 For the HR-PKS-catalysed formation of alkyl salicylaldehydes and derivatives in fungi, additional enzymes are necessary. These include usually two short-chain dehydrogenases (SDRs) and one cupin-domain containing protein (Fig. 1).3,6–8 It was proposed that the HR-PKSs release linear hydroxylated polyketide products, which are successively oxidised to ketones by the SDRs.6,7 In the case of the annullatin biosynthesis, deletion of the SDR anuD did not change the product formation, whereas deletion of anuF led to a significantly reduced productivity of annullatins. Therefore, AnuF can be also speculated to have a crucial function during the biosynthesis of the core skeleton.9 The resulting ketones enable subsequent aldol condensation and further aromatization.7 The cupin-domain containing proteins function as key enzymes during the aromatisation and enhance significantly the yields of the aromatic products.3,10 Deletion of the cupin-domain containing protein fogC from the flavoglaucin gene cluster led to the formation of a linear polyketide, which undergoes spontaneous rearrangements to different products.8In vitro assay demonstrated that only traces of aromatic products are formed in the absence of the cupin-domain containing protein StrC.3 A very recent publication described StrC-catalysed water elimination of a hydroxylated cyclohexenone and proved its function as an aromatase (Fig. 1).10
 | ||
| Fig. 1 Examples of HR-PKS-catalysed formation of alkyl salicylaldehydes and alcohols with involvement of two SDRs and a cupin-domain containing protein. | ||
Here, we report the formation of an alkylated salicylaldehyde and derivatives after expression of genes coding for a HR-PKS and two SDRs in Aspergillus nidulans in high yields without requirement of a cupin-domain containing protein.
Genome mining revealed the presence of a putative biosynthetic gene cluster, termed psa for propyl salicylaldehyde hereafter, in Aspergillus ustus 3.3904 (A. ustus). This cluster likely consists of 12 genes coding for a transcription factor PsaTF (KIA75503), a HR-PKS PsaPS (KIA75504), two SDRs (PsaOX1, PsaOx2 (KIA75505)), a cupin-domain containing protein PsaCP (KIA75508) and additional tailoring enzymes like cytochrome P450 enzyme (CYP), prenyltransferase (PT), and flavin-containing monooxygenase (FMO) (Fig. 2A). The genomic sequence of PsaOX1 is located downstream of KIA75503 and upstream of KIA75504 and was not annotated prior to this study. It spans bp 11996–13388 of JOMC01000132 and comprises 9 exons of 128, 53, 169, 8, 153, 73, 137, 137, 45 bp, interrupted by 8 introns of 52, 54, 65, 70, 55, 58, 75, 61 bp. PsaPS, PsaOX1, PsaOX2, and PsaCP with deduced polypeptide chains of 2387, 300, 277, and 189 amino acids share sequence identities of 41–65%, 54–63%, 24–49%, and 49–75% with the homologues mentioned in Fig. 1 (Table S1, ESI†), respectively.
 | ||
| Fig. 2 Biosynthetic gene cluster encoding propyl salicylaldehydes and derivatives in A. ustus (A) and proposed biosynthetic pathway of 1–4 (B). | ||
To prove their functions, we firstly carried out heterologous expression of the four neighbouring genes, psaTF, psaOX1, psaPS, and psaOX2, in Aspergillus nidulans LO8030 (A. nidulans). For this purpose, the genomic sequence of a 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 572 bp segment was amplified from A. ustus by PCR in three fragments and assembled with pJN01711via homologous recombination in Saccharomyces cerevisiae.12 The sequence in the obtained expression construct pMP024 is directly cloned after a strong gpdA promoter. The SwaI-linearised pMP024 was introduced into A. nidulans protoplasts via a PEG-mediated protocol13 to get the expression strain A. nidulans MP13. After selection with riboflavin autotrophy and PCR verification of the correct integrity of the A. ustus sequence into the genome of A. nidulans (Fig. S1, ESI†), the transformant MP13 was cultivated in PDB medium for 14 days under static conditions, by using A. nidulans BK06 as an isogenic control strain. LC-MS analysis of the EtOAc extracts of the fungal cultures revealed the presence of four new peaks (1–4) in strain MP13, with a [M+Na]+ ion at m/z 189.0893 for 1 and [M+H]+ ions at m/z 181.0856, 165.0909, and 185.1164 for 2, 3, and 4, respectively. These peaks were not detected in the control strain BK06 (Fig. 3). For structure elucidation, compounds 1–4 were isolated from a fermentation broth of strain MP13 in 6 L PDB medium and subjected to MS and NMR analyses including 1H, 13C, 1H–1H COSY, HSQC and HMBC spectra (Fig. S3–19, ESI†). In the 1H NMR spectra of 1–3, signals in the high field (δH 0.9–3 ppm) can be easily assigned to a propyl group and signals between δH 6.7 and 7.5 ppm are resonances of three vicinal aromatic protons (Fig. S3–S7, ESI†). Interpretation of the MS and NMR data and comparison with those described in the literature confirmed compound 1 to be 6-propyl salicyl alcohol,14 which was isolated from two endophytic fungi Phomopsis sp. E02011 and Aspergillus sp. ZJ-68.14,15 Compounds 2 and 3 were identified as the corresponding acid and aldehyde of 1 by comparison of their NMR spectra (Fig. S8–S14, ESI†) and MS data. 2 was isolated from a secretion of the Crematogaster ant during a defense reaction before.16 The signals for a hydroxymethyl group at δH 4.91 and δc 60.2 ppm in the NMR spectra of 1 have been replaced by signals at δH 11.94 and δc 195.4 ppm for an aldehyde in 3. Further analysis of HMBC correlations proved 3 to be 6-propyl salicylaldehyde. Compound 4 is a dihydro-γ-pyrone derivative (see ESI† for detailed structure elucidation, Fig. S15–19, ESI†), an analogue reported in a previous study during the biosynthesis of stachysalicyloid C.3 Compounds 3 and 4 have not been described before.
572 bp segment was amplified from A. ustus by PCR in three fragments and assembled with pJN01711via homologous recombination in Saccharomyces cerevisiae.12 The sequence in the obtained expression construct pMP024 is directly cloned after a strong gpdA promoter. The SwaI-linearised pMP024 was introduced into A. nidulans protoplasts via a PEG-mediated protocol13 to get the expression strain A. nidulans MP13. After selection with riboflavin autotrophy and PCR verification of the correct integrity of the A. ustus sequence into the genome of A. nidulans (Fig. S1, ESI†), the transformant MP13 was cultivated in PDB medium for 14 days under static conditions, by using A. nidulans BK06 as an isogenic control strain. LC-MS analysis of the EtOAc extracts of the fungal cultures revealed the presence of four new peaks (1–4) in strain MP13, with a [M+Na]+ ion at m/z 189.0893 for 1 and [M+H]+ ions at m/z 181.0856, 165.0909, and 185.1164 for 2, 3, and 4, respectively. These peaks were not detected in the control strain BK06 (Fig. 3). For structure elucidation, compounds 1–4 were isolated from a fermentation broth of strain MP13 in 6 L PDB medium and subjected to MS and NMR analyses including 1H, 13C, 1H–1H COSY, HSQC and HMBC spectra (Fig. S3–19, ESI†). In the 1H NMR spectra of 1–3, signals in the high field (δH 0.9–3 ppm) can be easily assigned to a propyl group and signals between δH 6.7 and 7.5 ppm are resonances of three vicinal aromatic protons (Fig. S3–S7, ESI†). Interpretation of the MS and NMR data and comparison with those described in the literature confirmed compound 1 to be 6-propyl salicyl alcohol,14 which was isolated from two endophytic fungi Phomopsis sp. E02011 and Aspergillus sp. ZJ-68.14,15 Compounds 2 and 3 were identified as the corresponding acid and aldehyde of 1 by comparison of their NMR spectra (Fig. S8–S14, ESI†) and MS data. 2 was isolated from a secretion of the Crematogaster ant during a defense reaction before.16 The signals for a hydroxymethyl group at δH 4.91 and δc 60.2 ppm in the NMR spectra of 1 have been replaced by signals at δH 11.94 and δc 195.4 ppm for an aldehyde in 3. Further analysis of HMBC correlations proved 3 to be 6-propyl salicylaldehyde. Compound 4 is a dihydro-γ-pyrone derivative (see ESI† for detailed structure elucidation, Fig. S15–19, ESI†), an analogue reported in a previous study during the biosynthesis of stachysalicyloid C.3 Compounds 3 and 4 have not been described before.
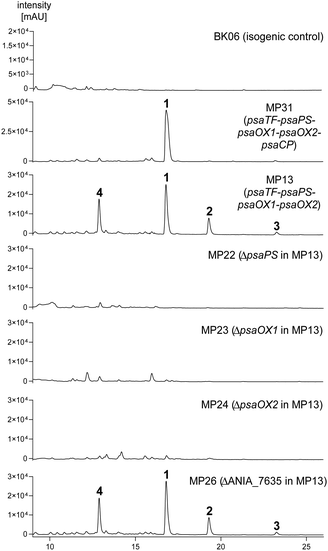 | ||
| Fig. 3 LC-MS analysis of the crude extracts of A. nidulans transformants with elution profile A. Illustrated are UV absorptions at 270–340 nm. | ||
Single gene deletion of psaOX1, psaPS or psaOX2 in the expression strain MP13 did not lead to the detection of any aromatic compounds (Fig. 3). These results confirmed the essential role of the three enzymes in the formation of 1–3, but are somewhat surprised, because a cupin-domain containing protein is required for the formation of alkyl salicylaldehydes and derivatives.3,6–8 As mentioned above, non-aromatic derivatives were detected as predominant products in the absence of cupin-domain containing proteins.3,8 We quantified therefore the yields of 1–3 in MP13 after cultivation for 14 days in PDB medium and compared them with that of 4. The total yield of 1–3 with 0.73 ± 0.07 mmol (0.11 ± 0.02 g) per litre culture is significantly higher than 0.22 ± 0.03 mmol (0.04 ± 0.01 g) for 4 and is comparable to the reported value for annullatin F at 0.82 ± 0.06 mmol, the main product of the anu cluster.17
Since PsaPS, PsaOX1, and PsaCP share the highest sequence identities with AnuA, AnuB, and AnuC from the anu cluster (Table S1, ESI†), we decided to prove the importance of the cupin-domain containing protein AnuC for the formation of annullatin F. Therefore, we deleted its genomic sequence in the A. nidulans expression strain BK08 containing the whole anu cluster. For this purpose, fragments of 5′-UTR and 3′-UTR of anuC with approximately 600 bp were amplified from genomic DNA of P. roqueforti, the origin of the anu cluster, and assembled them with sequences of the selection markers URA3, AmpR, and AfpyrG, amplified from pYH-wA-pyrG13 in Saccharomyces cerevisiae12 (Fig. S1, ESI†). Transformation, selection, and verification led to the A. nidulans deletion strain MP15, which was cultivated for metabolite detection. LC-MS analysis of the culture extract revealed clearly the abolishment of the annullatin production after cultivation for 14 days (Fig. 4). This proved unequivocally the essential role of AnuC from P. roqueforti for the formation of alkyl salicylaldehyde derivatives, as also reported for other cupin-domain containing proteins in the biosynthetic pathways mentioned in Fig. 1.3,6–8 These results provide evidence for the psa gene cluster from A. ustus as an exception for the formation of alkyl salicylaldehyde and derivatives which does not require a cupin-domain containing protein.
 | ||
| Fig. 4 LC-MS results of the crude extracts of A. nidulans transformants BK08 and MP15 with elution profile A. Illustrated are UV absorptions at 270 nm. | ||
In a previous study, an endogenous enzyme from A. nidulans was speculated as a cupin analogue for the formation of 5-deoxyaurocitrin.7 To exclude this possibility for the formation of 1–3 in the expression strain MP13, we used the sequences of AnuC, VirC, and StrC as queries for search of cupin homologues. An uncharacterized protein encoded by ANIA_7635 (acc. no. XP_680904) shares 38–43% identities on the amino acid level with the above mentioned cupin-domain containing proteins (Table S1, ESI†). The sequence of ANIA_7635 was deleted in the A. nidulans expression strain MP13 by using the aforementioned strategy with afpyro18 as a selection marker. As shown in Fig. 2A, no changes in the secondary metabolite profile of the obtained ΔANIA_7635 mutant MP26 were detected after cultivation in PDB medium for 14 days (Fig. 3). The product yields of 1–4 in MP26 remain the same as in MP13. This proved that ANIA_7635 is not involved in the formation of the alkyl salicylaldehyde and derivatives in MP13.
Accumulation of 1–3 in strains MP13 and MP26 indicates strongly that a cupin-domain containing protein seems not essential for their biosynthesis. Although replacement of its function by an unknown endogenous enzyme cannot be completely excluded, however, the results from deletion experiments for the biosynthesis of flavoglaucin8 and annullatin (Fig. 4) did not support such a hypothesis. This raises the question about the role of psaCP in the putative cluster (Fig. 2A), which is separated from psaOX2 by 6 kbp. To explore the gene function, psaCP was cloned into pMP024 described above and introduced into A. nidulans protoplasts. After selection, verification (Fig. S1, ESI†) and cultivation in PDB medium for 14 days, compound 1 was detected as the predominant peak and 2–4 as minor ones. The product yield was calculated to be 0.90 ± 0.04 mmol (0.15 ± 0.03 g) for 1 in strain MP31, almost the total amount of 1–4 in MP13. This indicates that the PKS product was directed effectively to the alcohol 1, instead for the formation of the shunt products 2 and 4, as also demonstrated for StrC.3 In that study, the presence of the cupin-domain containing protein strongly support the pathway for main product formation. PsaCP is very likely responsible for the conversion of 3 to 1, which is now under investigation.
It can be speculated that the HR-PKS PsaPS releases a linear polyketide with a double bond by water elimination on the polyketide assembling for ring formation and therefore water elimination after ring formation catalysed by a cupin-domain containing protein would be redundant (see below for details).
As illustrated in Fig. 1, the products of the four enzymes can be alkyl salicylaldehydes or alcohols and their oxidation states can also be altered by the host enzymes.6 To prove the influence of the endogenous enzymes for the detected products in MP13, we did cultivation of A. nidulans LO8030 with the isolated compounds 1–3. The compounds were added to 2-days old cultures in 10 mL PDB medium at 25 °C to final concentrations of 25 μM. HPLC analysis revealed the complete conversion of compound 3 to 1 after one day (Fig. S2, ESI†). No consumption of 1 and 2 was detected, even after cultivation for 5 days (data not shown). This proved that the proposed function of PsaCP can be complemented by an endogenous enzyme, at least partially in strains MP13 and MP26.
Based on these results, we propose the biosynthetic pathways for the compounds 1–4 (Fig. 2B). The HR-PKS PsaPS generates the linear polyketide with three hydroxyl groups at the last three iterations. Most likely, the DH domain is responsible for a dehydration and introduction of a double bond. The polyketide chain is either directly released by hydrolysis or by a KR domain-catalysed reductive mechanism, as reported before.3 These intermediates are used by the SDRs PsaOX1 and PsaOX2 as substrates for successive oxidations of the two remaining hydroxyl groups. A subsequent aldol condensation provides the aromatic products 2 and 3. Conversion of 3 to 1 is very likely catalysed by PsaCP or by a host enzyme from A. nidulans in its absence. Compound 4 can be considered as a shunt product and is formed by ineffective participation of the DH domain. After the reductive polyketide chain release, PsaOX1 or PsaOX2, and an endogenous enzyme conduct two oxidations, followed by keto-enol tautomerism and water elimination of two hydroxyl groups to provide product 4.
In conclusion, we identified a biosynthetic gene cluster from A. ustus with significant sequence similarities to those involved in the biosynthesis of alkyl salicylaldehydes and derivatives. Heterologous expression in A. nidulans revealed that the formation of the aromatic products in high yields requires only three enzymes, i.e. a HR-PKS and two SDRs. In contrast to other studies published before, a cupin-domain containing protein was not necessary for the formation of products described in this study. Therefore, we provide here an example for the formation of alkyl salicylaldehydes and derivatives without the requirement of a cupin-domain containing protein.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- H. Ran and S.-M. Li, Nat. Prod. Rep., 2021, 38, 240–263 RSC.
- Y. H. Chooi and Y. Tang, J. Org. Chem., 2012, 77, 9933–9953 CrossRef CAS PubMed.
- R. Yang, J. Feng, H. Xiang, B. Cheng, L.-D. Shao, Y.-P. Li, H. Wang, Q.-F. Hu, W.-L. Xiao, Y. Matsuda and W.-G. Wang, J. Am. Chem. Soc., 2023, 145, 11293–11300 CrossRef CAS PubMed.
- R. J. Cox, Org. Biomol. Chem., 2007, 5, 2010–2026 RSC.
- E. Skellam, Nat. Prod. Rep., 2022, 39, 754–783 RSC.
- Z. Zhao, Y. Ying, Y. S. Hung and Y. Tang, J. Nat. Prod., 2019, 82, 1029–1033 CrossRef CAS PubMed.
- L. Liu, M. C. Tang and Y. Tang, J. Am. Chem. Soc., 2019, 141, 19538–19541 CrossRef CAS PubMed.
- J. Nies, H. Ran, V. Wohlgemuth, W. B. Yin and S.-M. Li, Org. Lett., 2020, 22, 2256–2260 CrossRef CAS PubMed.
- P. Xiang, B. Kemmerich, L. Yang and S.-M. Li, Org. Lett., 2022, 24, 6072–6077 CrossRef CAS PubMed.
- H. Wang, C. Peng, X.-X. Chen, H. Y. Wang, R. Yang, H. Xiang, Q. F. Hu, L. Liu, L.-W. Chung, Y. Matsuda and W.-G. Wang, ACS Catal., 2024, 14, 10796–10805 CrossRef CAS.
- F. Kindinger, J. Nies, A. Becker, T. Zhu and S.-M. Li, ACS Chem. Biol., 2019, 14, 1227–1234 CrossRef CAS PubMed.
- L. Mojardín, M. Vega, F. Moreno, H. P. Schmitz, J. J. Heinisch and R. Rodicio, Fungal Genet. Biol., 2018, 111, 16–29 CrossRef PubMed.
- W. B. Yin, Y. H. Chooi, A. R. Smith, R. A. Cacho, Y. Hu, T. C. White and Y. Tang, ACS Synth. Biol., 2013, 2, 629–634 CrossRef CAS PubMed.
- D. Weber, S. Gorzalczany, V. Martino, C. Acevedo, O. Sterner and T. Anke, Z. Naturforsch., 2005, 60c, 467–477 CrossRef PubMed.
- R. Cai, H. Jiang, Z. Zang, C. Li and Z. She, Mar. Drugs, 2019 Search PubMed.
- T. H. Jones, S. R. Brunner, A. A. Edwards, D. W. Davidson and R. R. Snelling, J. Chem. Ecol., 2005, 31, 407–417 CrossRef CAS PubMed.
- J. Zhou, X. L. Chen and S. M. Li, Appl. Microbiol. Biotechnol., 2024, 108, 427 CrossRef CAS PubMed.
- D. J. Janzen, H. Wang and S.-M. Li, J. Nat. Prod., 2023, 86, 1779–1785 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc04227a |
| This journal is © The Royal Society of Chemistry 2024 |
