 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembling short peptide amphiphiles as versatile delivery agents: a new frontier in antibacterial research
Ramesh
Singh†
 *,
Shruti
Sharma
*,
Shruti
Sharma
 ,
Aanand
Kautu
,
Aanand
Kautu
 and
Khashti Ballabh
Joshi
and
Khashti Ballabh
Joshi
 *
*
Department of Chemistry, School of Chemical Science and Technology, Dr Harisingh Gour Vishwavidyalaya (A Central University), Sagar, 470003, Madhya Pradesh, India. E-mail: kbjoshi77@gmail.com; kbjoshi@dhsgsu.edu.in
First published on 3rd July 2024
Abstract
Self-assembling short peptide amphiphiles, crafted through a minimalistic approach, spontaneously generate well-ordered nanostructures, facilitating the creation of precise nanostructured biomaterials for diverse biomedical applications. The seamless integration of bioactive metal ions and nanoparticles endows them with the potential to serve as pioneering materials in combating bacterial infections. Nanomanipulation of these molecules’ binary structures enables effective penetration of membranes, forming structured nanoarchitectures with antibacterial properties. Through a comprehensive exploration, we attempt to reveal the innovative potential of short peptide amphiphiles, particularly in conjugation with metal cations and nanoparticles, offering insights for future research trajectories.
The pressing issue of antibiotic resistance underscores the imperative for novel antibacterial therapeutics.1 Conventional antibiotics are becoming increasingly ineffective against a burgeoning array of resistant bacterial strains, underscoring the critical need for alternative approaches.2 The widespread misuse and overuse of antibiotics have expedited the emergence of multidrug-resistant pathogens,3 exemplified by notorious strains like methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococcus (VREF).4,5 This proliferation of resistant strains has severely curtailed the efficacy of conventional treatments for various infections. In light of these challenges, the development of novel antibacterial agents exhibiting heightened efficacy against bacteria while minimizing host toxicity assumes paramount importance.6
In the ongoing battle against antibacterial resistance, peptide nanotechnology has emerged as a promising avenue, offering novel solutions through the self-assembling capability of multifunctional short peptides.7–11 Short peptides, capable of self-assembly, have garnered considerable attention due to their distinct attributes and versatile applications.11,12 Their spontaneous organization into well-defined nanostructures lays the groundwork for the production of antibacterial nanomaterials, ranging from basic fibrils and sheets to intricate micellar and vesicular assemblies.13 These nanostructures confer unique physicochemical properties upon the peptides, enhancing their efficacy as broad-spectrum antimicrobial agents in combating bacterial resistance (Scheme 1).14–18
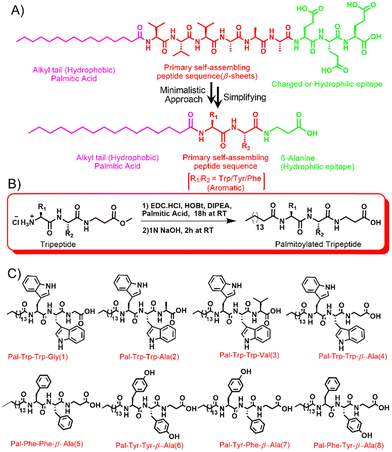 | ||
| Scheme 1 This figure illustrates (A) the minimalist engineering of short peptide amphiphiles (sPAs), akin to established peptide amphiphiles (PAs).35 sPAs feature a hydrophobic palmitic acid, aromatic core for self-assembly, and linear amino acid sequence for hydrophilicity. (B) A representative synthesis scheme17 and (C) structural adjustments yielding uniquely different sPA architectures, enabling self-assembly into various nanostructures driven by secondary structures like beta sheets and alpha helices. This advancement holds promise for diverse applications, especially in antibacterial strategies and nanomaterial design. Scheme (B) reproduced/adapted from reference17 with permission of the Royal Society of Chemistry. | ||
The intrinsic antibacterial attributes and drug delivery proficiency of self-assembling peptides augment the efficacy of conventional antibiotics against drug-resistant pathogens.15,16,18 Co-assembling these nanomaterials proves effective in combating drug-resistant bacterial strains.19 For instance, amalgamating antimicrobial nanofibers with self-assembling peptides, such as H-Lys–βAla–βAlaLys–βAla–OEt, exhibits potential against both Gram-positive and Gram-negative bacterial strains.20 In a separate study, a tripeptide (D-Leu–Phe–Phe)-based hydrogel demonstrated the ability to encapsulate and regulate the release of antibiotic drugs like ciprofloxacin.21 In our previous investigation, we observed that delivering the commercial antibiotic levofloxacin via H2S-responsive peptide nanostructures enhances its efficacy, effectively inhibiting bacterial growth, including methicillin-resistant S. aureus (MRSA).22
Alternatively, the amalgamation of metal ions with self-assembling short peptides yields hybrid supramolecular nanostructures with promising antibacterial attributes.23 These nanostructures, formed through the fusion of metal ions with peptides possessing commendable antibacterial properties, exemplify notable advancements. For instance, Zn(II)-coordinated dipeptide, tryptophan–phenylalanine (Trp–Phe), based self-assembling nanostructures have garnered attention for their ability to disrupt bacterial membranes.24 Similarly, nanostructures derived from gold and silver ion-conjugated di-proline dipeptides exhibit inhibitory effects against both Gram-positive and Gram-negative bacterial strains.25 Furthermore, our research group discovered that co-assembling zinc with tyrosine amino acids over the diatom surface impedes bacterial growth by delivering zinc ions to bacterial cells.26,27
The attachment of fatty acid chains to self-assembling short peptides stands as a strategy to augment their antimicrobial efficacy while preserving their intrinsic properties.28 This covalent linkage significantly enhances the biomedical functionality of the peptides, endowing them with enhanced amphiphilic characteristics, heightened thermal stability, and resistance to enzymatic degradation, thereby prolonging their effectiveness in biological environments. Furthermore, the hydrophobic nature of the lipid chain facilitates interactions with lipid membranes, potentially modulating membrane dynamics and facilitating traversal through biological barriers. The fusion of lipids and peptides engenders multifaceted alterations in peptide self-assembly.29,30
Extensive research has unveiled numerous short lipopeptides manifesting potent antibacterial and antifungal activities, efficacious against biofilms and multidrug-resistant bacteria.31,32 PAs offer several distinct advantages over traditional lipidic nanoparticles. While both PAs and lipidic nanoparticles possess amphiphilic properties, PAs exhibit superior biocompatibility with cell membranes due to their peptide-based nature.15 This enhanced biocompatibility translates into reduced cytotoxicity and improved stability, making PAs particularly well-suited for biomedical applications.30 Moreover, the self-assembling mechanism of PAs allows for precise control over the formation of nanostructures, leading to uniform and stable assemblies with tunable properties. This level of control is often challenging to achieve with lipidic nanoparticles, which can exhibit variability in size, shape, and stability. Furthermore, PAs offer the versatility to incorporate a wide range of bioactive molecules, including metal ions or nanoparticles, within their nanostructures. This capability enables the design of multifunctional delivery systems with tailored therapeutic payloads, enhancing their potential as antibacterial agents.33 In summary, while lipidic nanoparticles have been widely utilized as drug delivery vehicles, PAs offer distinct advantages in terms of biocompatibility, structural control, and versatility in payload incorporation. By elucidating these differences, we aim to provide a clearer understanding of the unique benefits offered by PAs as delivery agents in anti-bacterial research.34 This review primarily delves into our group's research on the self-assembly of short peptide amphiphiles and their amalgamation with bioactive metal ions and metal nanoparticles, elucidating their applications in antibacterial endeavors.
Self-assembly of peptide amphiphiles
Amphiphilic biomolecules, integral components of biological systems, possess dual hydrophilic and lipophilic characteristics, enabling interaction with both aqueous and lipid environments. This property facilitates their self-assembly, a phenomenon observed prominently in cellular membranes where lipid bilayers spontaneously form.35 Peptide amphiphiles, a crucial subset of these biomolecules, are achieved either by conjugating hydrophilic peptide sequences to lipophilic groups or by structuring amino acids within a peptide sequence to have hydrophobic and hydrophilic ends. This classification includes amphiphilic peptides, lipidated peptide amphiphiles, and supramolecular peptide amphiphile conjugates.36Amphiphilic peptides typically feature sequences with hydrophilic amino acids at one end and hydrophobic amino acids at the other, as studied extensively by researchers such as Zhang et al. and Schneider et al., with peptides like EAK8, EAK12, and EAK16.37–39 Conversely, lipidated amphiphiles comprise hydrophobic lipid chains attached to hydrophilic amino acid groups, making them biomimetic and known as lipopeptides. The self-assembly behavior of these molecules is dictated by their arrangement and composition, primarily driven by the interplay between hydrophilic and hydrophobic interactions, as elucidated by seminal works by Stupp et al. and Banerjee et al.35,40,41
A novel category of peptide amphiphiles, termed supramolecular peptide amphiphile conjugates, has emerged through the integration of traditional amphiphiles with supramolecular chemistry. This category involves the formation of pseudopeptidic bonds between amphiphilic peptides or lipopeptides and separate molecules that regulate supramolecular interactions. Researchers such as Kros et al. and Das et al. have synthesized pseudopeptide amphiphiles using various molecules like FMOC protection, pyrene, naphthalimide, naphthalene, and arylenediimides. Furthermore, Halder et al. and Ma Su et al. have contributed to the development of peptidomimetic amphiphiles, particularly focusing on their antibacterial efficacy (Fig. 1).36,42,43
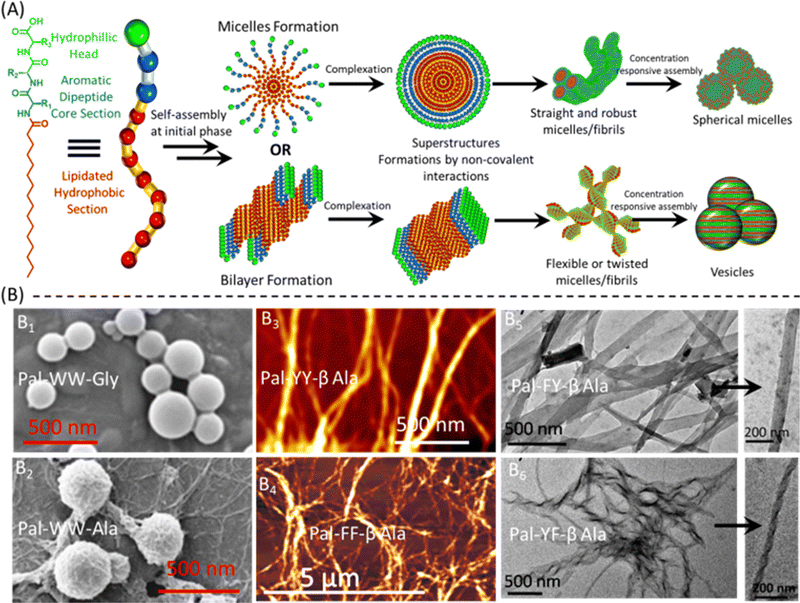 | ||
| Fig. 1 Formation and characterization of self-assembled sPA nanostructures. (A) The proposed model illustrates the formation of various nanostructures of short peptide amphiphiles (sPAs) via lipid bilayer or micelle formation, followed by various electrostatic interactions leading to the conversion into vesicles, straight or twisted fibers, and spherical micelles. The resulting supramolecular species exhibit structural diversity dependent on their aromatic or hydrophilic core. (B) SEM, AFM, and TEM characterization of self-assembled sPA nanostructures. (B1) and (B2) FE-SEM images depict the formation of vesicles and spherical micelle structures of Pal–WW–Gly and Pal–WW–Ala, respectively. (B3) and (B4) AFM images reveal robust and needle-like fibrous structures of Pal–YY–βAla and Pal–FF–βAla. (B5) and (B6) TEM images illustrate sheeted straight and twisted fibrous structures of Pal–FY–βAla and Pal–YF–βAla, respectively, and the arrow directs the intact images providing insights into the morphology of these nanostructures. The various sections within the image have been reproduced or adapted with permission as follows: B1 from ref. 66 with permission from Elsevier; B2 from ref. 45 with permission from Wiley; B3 from ref. 23 with permission from Wiley; B4 from ref. 17 with permission from the Royal Society of Chemistry; and B5 and B6 from ref. 46 with permission from Wiley. | ||
The amphiphilicity of these molecules serves as a crucial determinant in shaping their self-assembly characteristics. It dictates the formation of a wide range of nano- to microstructures, including micelles, spherical micelles, micro-emulsions, cylindrical fibers, and vesicles.44–46 This diversity arises from the delicate balance between different types of molecular forces, such as hydrogen bonding and hydrophobic interactions. By adjusting the length and composition of the hydrophobic chains, researchers can finely tune these interactions, thus modulating the morphology and properties of the assembled structures.35,36,47
The self-assembly of amphiphilic peptides in polar solvents is a sophisticated process driven by the intricate balance of molecular interactions. When suspended in a polar solvent, these peptides undergo a remarkable organizational shift where the hydrophobic segments minimize their exposure to the solvent, while the polar portions remain exposed, leading to the spontaneous formation of ordered aggregates through non-covalent forces, resulting in various nanostructures.48 The molecular mechanisms governing the self-assembly of short peptides, including lipidated peptides, are multifaceted and influenced by several factors. These include the specific amino acid sequences, the distribution of hydrophobic and hydrophilic residues along the peptide chain, and the interplay of non-covalent forces such as hydrogen bonding, hydrophobic interactions, electrostatic interactions, and van der Waals forces. The delicate equilibrium between these forces ultimately dictates the arrangement and stability of the resulting assemblies.49
A seminal study by Stupp et al. elucidated that the dominance of hydrophobic interactions in a system leads to the formation of micellar structures, while an increase in hydrogen bonding can transform these structures into elongated fibers, demonstrating the profound impact of these molecular forces on the self-assembly process.35,48 Consequently, controlling the interplay of hydrophobic forces and hydrogen bonding is pivotal in dictating the morphology and properties of the self-assembled peptides. This control can be achieved by judiciously selecting the appropriate polar head and hydrophilic tail in the peptide structure. In essence, peptide self-assembly represents a dynamic process where peptides spontaneously organize into ordered aggregates, driven by a complex interplay of molecular forces. Understanding and manipulating these forces offers profound insights into the design and development of functional peptide-based materials for diverse biomedical and nanotechnological applications.49
Antibacterial action mechanism
Short-peptide-based self-assembled nanomaterials exhibit unique characteristics that significantly augment their antibacterial efficacy.10,11 Their large surface area and dynamic capability to encapsulate and transport antimicrobial agents enhance their activity against bacterial pathogens. Furthermore, the self-assembly of peptides into nanostructures enables targeted delivery of antimicrobial components, thereby improving efficacy while mitigating the risk of resistance development.19Research has delved into the antibacterial mechanisms of short peptide and lipidated short peptide-based nanostructures, elucidating their disruptive effects on bacterial cell walls, leading to cell lysis and demise.44 Additionally, these nanostructures can impede crucial intracellular processes, including DNA, RNA, and protein synthesis, further contributing to bacterial eradication. Unlike traditional antibiotics, short peptides interact directly with bacterial cell membranes, lowering the likelihood of bacterial drug resistance.31 Their rapid germ-killing ability, low bactericidal concentration, and synergistic effects with conventional antibiotics render them highly effective against endotoxins and antibiotic-resistant strains.32 Moreover, short peptides demonstrate excellent biocompatibility, low toxicity, and non-irritating properties, underscoring their potential for safe and efficacious utilization in clinical settings. These attributes position short-peptide-based self-assembled nanomaterials as promising candidates for combating bacterial infections and advancing biomedical applications.
Additionally, it's important to elucidate the proposed mechanism of action of PA-metal ion or PA-metal nanoparticle complexes. These complexes may exert antibacterial activity through various mechanisms, including disruption of bacterial cell membranes, generation of reactive oxygen species (ROS), interference with essential cellular processes, or induction of bacterial cell death pathways.40,41 By detailing these mechanisms, we aim to provide clarity on how PA-metal complexes exert their antibacterial effects. Furthermore, we recognize the importance of addressing selectivity for bacteria versus mammalian cells (Fig. 2B, cytotoxicity data), a crucial aspect in the development of antibacterial agents. PAs offer the potential for enhanced selectivity through several strategies. For instance, the structural design of PAs can be tailored to specifically target bacterial membranes or cell wall components while sparing mammalian cell membranes. Additionally, the incorporation of specific metal ions or nanoparticles with inherent antibacterial activity can further enhance selectivity towards bacterial cells. Moreover, the development of multifunctional PAs capable of targeting specific bacterial strains or antibiotic-resistant pathogens while minimizing cytotoxicity to mammalian cells represents an area of active research.46,50–54
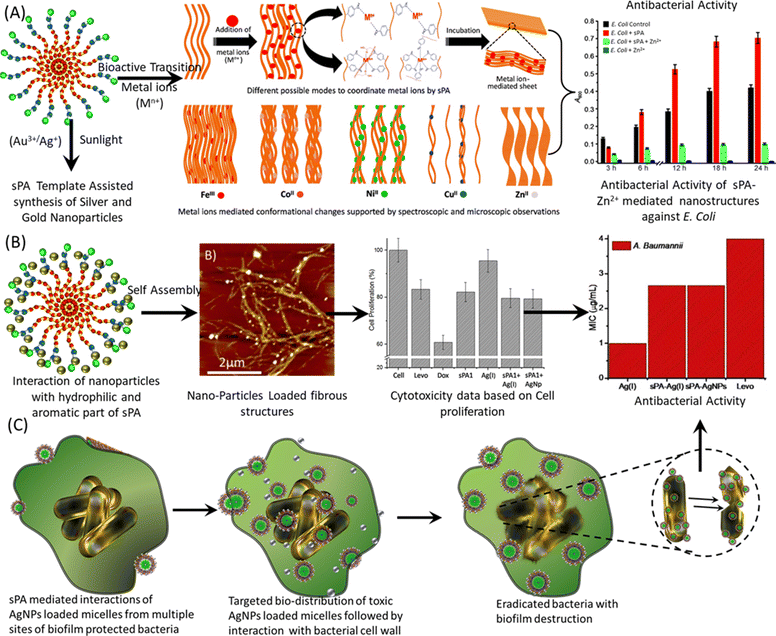 | ||
| Fig. 2 Nano-manipulation of sPAs nanostructures via bioactive transition and novel metal ions for enhanced antibacterial activity. (A) Proposed model illustrating the primary aggregation of short peptide amphiphiles (sPAs) in polar solvents, leading to micelle formation followed by vertical stacking into fibril structures. This nanostructure is then manipulated through structural and conformational transformations upon the addition of different metal ions, positively impacting the antibacterial activity of sPA-metal conjugates, as depicted in a bar graph. (B) Addition of novel metal ions such as Au3+ and Ag+ results in the decoration of sPA nanostructures with AuNPs and AgNPs, respectively, due to the reduction of metal ions and oxidation of the aromatic core of sPAs. The resulting metal nanoparticle-loaded sPA fibers exhibit high biocompatibility and reduced cytotoxicity, leading to enhanced antibacterial activity, as demonstrated in the bar graphs. (C) Proposed model illustrating the antibacterial activity of sPA-metal ion or sPA-metal nanoparticle conjugates. Images have been reproduced or adapted with permission as follows: (A) from ref. 23 with permission from Wiley; (B) from ref. 46,50 and 55 with permission from Wiley; and (C) from ref. 56 with permission from Wiley. | ||
Innovations in peptide amphiphile design
The modification of peptides through lipidation constitutes a strategic approach that influences their hydrophobicity, thereby exerting significant impacts on their absorption, distribution, metabolism, and excretion. Leveraging this modification, engineers can tailor nanostructures capable of releasing antibacterial agents upon encountering pathogens.55,56 Lipidation of short peptides introduces hydrophobicity, enhancing membrane interaction, permeability, stability, and protection against enzymatic proteolysis.44 The supramolecular assembly of lipidated peptides often adopts a β-sheet secondary structure, crucial for the formation of nanofibrils and the antimicrobial activity of the peptides.35 Natural lipopeptide antibiotics like polymyxin B16 and daptomycin57 have demonstrated efficacy against a broad spectrum of bacteria, including multidrug-resistant strains. Notably, their activity relies on the presence of the lipid chain, as its removal renders them inactive. Exploiting this natural phenomenon, researchers have synthesized biocompatible antibacterial nanomaterials.Studies have unveiled the antimicrobial potential of various lipopeptides, including single amino acid derivatives. Certain dipeptides, particularly those derived from polar amino acids such as Ser and His, exhibit significant antimicrobial activity against Gram-positive bacteria, with some displaying remarkable selectivity.58 Short peptides containing arginine and lysine, with their net positive charge, also demonstrate antibacterial activity.59,60 For instance, the lipopeptide (C10)2–KKKK–NH2 has displayed potent antibacterial properties against biofilms on surfaces like polystyrene and contact lenses, surpassing conventional antibiotics.61 However, despite their efficacy, cationic amphiphiles like these pose concerns regarding their potential harm to human cells, reflecting the broader challenges faced by antimicrobial peptides (AMPs) and their derivatives.62 High production costs, unfavorable pharmacokinetic profiles, and limited effectiveness in animal studies are among the obstacles encountered by these compounds.16,57–60
Moreover, prolonged use of cationic AMPs may lead to bacterial resistance, highlighting the imperative for ongoing research aimed at developing more efficient and safer antibacterial peptides. Addressing these challenges will be pivotal in harnessing the full potential of peptide-based antimicrobial agents in clinical settings.62
Antibacterial activity by manipulation with metal ions and nanoparticles
Non-covalent interactions are fundamental drivers of peptide self-assembly, with their manifestation influenced by amino acid sequence, environmental factors, and the presence of metal ions.63 Metal ions, pivotal in the self-assembly of natural proteins and the functioning of biological systems, exhibit specific recognition of amino acids, facilitating connections and rearrangement of non-covalent forces among peptide molecules, thereby instigating peptide self-assembly. Peptides serve as electron-donating ligands and protective agents,64,65 thus finding utility as reducing agents and protectants in the synthesis of metal nanoparticles such as gold and silver.45,66,67 Our research endeavors extensively explore the impact of metal ions on self-assembling peptide amphiphiles (sPAs), elucidating alterations in secondary structures, overall morphology, and their potential in combating bacterial infections.17,68 The delivery of metal ions possessing antibacterial properties, such as zinc, copper, silver, and gold, induces significant perturbations in bacterial cell homeostasis, contributing to antimicrobial efficacy.Our investigation unveiled that short peptide amphiphiles (sPAs) incorporating the Phe–Phe aromatic segment manifest robust π–π stacking interactions, effectively shielding the core constituents—the palmitoyl lipid chain and aromatic dipeptides—within the micellar or fibrillar structure of the sPA.17 The core components remain largely unaffected by most divalent metal ions, including Cu(II), Co(II), Ni(II), and Zn(II). However, metal ions possessing higher positive charges, such as Fe(III), can attract the negatively charged π-cloud, thereby promoting the formation of β-sheet structures. Moreover, these metal ions interact with the outer hydrophilic residues (β-alanine) of the sPA, triggering the nucleation of fibers into diverse configurations, encompassing twists, random coils, and aggregations. The multi-mode versatility of these fiber structures of sPA has been exploited for synthesizing antibacterial nanostructures embedded with metal cations or novel metal nanoparticles.17,23,68
The antibacterial efficacy of these nanosystems arises from two primary mechanisms: the generation of reactive oxygen species (ROS) and the high drug payload capacity of silver nanoparticles, inducing oxidative stress and perturbation of bacterial cell membranes. Notably, sPA nanostructures, such as Pal–Phe–Phe–β–Ala, facilitate the reduction of gold56 and silver salts55 under sunlight exposure, leading to nucleation into nanoparticles. These nanoparticles, when embedded over the nanofibers, exhibit potent antibacterial effects against bacteria. Specifically, the sPA–AuNP hybrid structures demonstrated antibacterial effects against Escherichia coli, evident from a substantial inhibition zone.25 Of note, the minimum inhibitory concentration (MIC) values of sPA–AgNPs hybrids displayed comparable potency against both Gram-positive Staphylococcus aureus and Gram-negative bacterial pathogens, including drug-resistant strains such as Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii.45,46,50
A notable modification in our study involved substituting the central Phe–Phe segment of short peptide amphiphiles (sPAs) with slightly hydrophilic aromatic dipeptides Tyr–Tyr, characterized by metal-chelating hydroxyl groups.23 This alteration was particularly significant as it allowed for a fascinating morphological transformation in these metal-chelating nanofibers, transitioning their structure from fibrous to sheet-like nanostructures. The process of this transformation was multifaceted and depended on the specific metal ions present. For instance, Fe(III) ions were observed to prompt the development of elongated β-sheet fiber structures, which gradually transformed into robust and orderly sheet-like assemblies. In contrast, the introduction of Cu(II) ions led to the disintegration of fibers. Similarly, Co(II) and Ni(II) ions induced the coiling and twisting of fibers, eventually resulting in the formation of sheet-like nanostructures. On the other hand, Zn(II) ions prompted the folding of nanosheets into needle-like shapes.23 These newly formed sheet-like structures served a crucial function as reservoirs for metal ions, releasing toxic ions upon contact with bacterial cells. For example, the zinc ion-conjugated Pal–Tyr–Tyr–β–Ala self-assembled nanostructures were observed to efficiently transport zinc ions into Escherichia coli bacterial strains upon contact.17,68
The integration of metal ions into self-assembling sPAs introduced an alternative interaction pathway with bacterial cell walls, enhancing the antibacterial mechanisms of these nanostructures. Metal ions acted as cationic bridges between the nanostructures and the negatively charged components of microbial membranes, thus amplifying their antibacterial efficacy. Further insights into the structure–function relationships of these sPAs were gained by modifying their structure, specifically by removing the hydroxyl group from one of the tyrosine residues. This structural manipulation resulted in the synthesis of two constitutional isomers: Palmitoyl–Phe–Tyr–β–Ala and Palmitoyl–Tyr–Phe–β–Ala. These modifications disrupted or preserved the binary hydrophobic–hydrophilic nature of the amphiphiles, respectively, influencing the formation of their nanostructures. In Pal–Tyr–Phe–β–Ala–OH, the disruption of the binary nature led to a sequence of hydrophobic (Pal), hydrophilic (Tyr), hydrophobic (Phe), and hydrophilic (β-Ala) components, which impacted the formation of regularly stacked sheet-like structures. Conversely, in Palmitoyl–Tyr–Phe–β-Ala–OH, the preserved binary nature facilitated the formation of regular structures. This intricate interplay between peptide structure and metal ion interactions provides valuable insights into the design and engineering of peptide-based nanomaterials with tailored antibacterial properties, offering promising avenues for future research and development in the field of antimicrobial materials.46,51,57
Additionally, the combination of these nanofibers with silver nanoparticles results in the production of antibacterial nanofibers, thereby further enhancing their antibacterial effectiveness. The inherent characteristics of these sPAs, particularly their favorable hydropathy index, facilitate efficient binding to cell membranes, enabling the effective transport of toxic silver nanoparticles (AgNPs) as cargo to target sites. Cytotoxicity assays conducted against healthy eukaryotic cells have demonstrated that both sPAs and their silver hybrids exhibit negligible toxicity and display higher biocompatibility compared to conventional drugs such as levofloxacin and doxorubicin. This targeted delivery approach holds promise for combating neurodegenerative diseases associated with microbiota.46,50,56
The antibacterial efficacy of these nanosystems was thoroughly evaluated against a spectrum of bacterial strains, including Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. By augmenting interactions with bacterial surfaces or biofilms, these nanosystems enhance biodistribution and pharmacokinetics, ultimately leading to increased antibacterial efficacy. The accumulation of toxic silver nanoparticles at elevated concentrations in specific regions results in more efficient inhibition of bacterial growth and lower minimum inhibitory concentration (MIC) values, highlighting the potential of these nanosystems for combating bacterial infections effectively.46
Structure–activity relationships of a few short peptide conjugates and designed sPAs
Structure–activity relationship (SAR) studies are fundamental in drug discovery and development, particularly in the context of antimicrobial peptides (AMPs) where even subtle changes in molecular structure can significantly impact biological activity.52 Our exploration of SAR, building upon the groundwork laid by established researchers, focuses on short peptide conjugates and their potential as potent antibacterial agents. In our investigation, we delve into understanding how modifications in the molecular structure of these peptides influences their efficacy against a spectrum of bacterial strains, encompassing both Gram-positive and Gram-negative types. Notably, we assess their activity against notorious pathogens such as Staphylococcus aureus (including MRSA and MSSA strains), Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii. This comprehensive approach allows us to gauge the versatility and breadth of action of our designed sPA nanostructures (Table 1).22,52–54| Entry | Short peptide conjugates | Nanostructures | Function | Activity/selectivity/applicationsref. |
|---|---|---|---|---|
| 1. | L-Tyrosine | Diatom fabricated microcontainer | Delivery of bioactive metal ions | Antibacterial activity against E. coli.26 |
| 2. | Di-L-Proline | 2D Nanosheets | Delivery of AuNPs and AgNPs | Antibacterial activity against S. aureus.25 |
| 3. | L-Tryptophan-L-Phenylalanine | Spherical nanoparticles | Delivery of bioactive metal ions (Zn2+) | Antibacterial activity against E. coli24 |
| 4. | L-Phe-L-Phe and more | Nanotubes | Nanostructures as antibacterial agents | Broad-spectrum antibacterial activity9 |
| 5. | H2N–X–Met–Phe–C12H25, X = L–lysine, X = L–histidine, and X = L–leucine | Supramolecular hydrogels | Nanostructures as antibacterial agents | Antibacterial activity against Gram-negative and Gram-positive41 |
| 6. | D-Leu–Phe–Phe | Fibrous hydrogel formation | Delivery of ciprofloxacin | Antibacterial activity against S. aureus, E. coli, K. pneumonia.21 |
| 7. | H2N–Tyr(DNB)–Phe–Phe–OH (PHSC) | Nanospheres/nanovesicles | Delivery of levofloxacin | Antibacterial activity against Gram-negative and Gram-positive and MRSA22 |
| 8. | H-Lys–β-Ala–β-Ala–Lys–β-Ala–OEt | Cationic nanovesicles | Delivery of L-Dopa and curcumin | Antibacterial activity against Gram-negative and Gram-positive20 |
| 9. | Lipidated amino acids [Phe, His, Ser, Cys, Val] | Spherical micelles, worm like micelles | Antibacterial gelators | Antibacterial activity against Gram-negative (E. coli) and Gram-positive (S. aureus)58 |
| 10. | Pal-Trp | Nanorod/nanofibers | Delivery of Au(II) and AuNPs | Antibacterial activity against Gram-negative and Gram-positive54 |
| 11. | Pal–Trp–Trp–Gly | Nanospheres/nanovesicles | Thermoplasmonic nanomaterials | Proposed photothermal activity45,66,67 |
| 12. | Pal–Trp–Trp–Ala | Spherical micelles | Plasmonic welding of AuNPs | Proposed photothermal, catalytic and drug delivery activity45,66,67 |
| 13. | Pal–Phe–Phe–β-Ala | Needle/nanofibers | Delivery of AuNPs, AgNPs, and bioactive metal ions | Antibacterial activity against Gram-negative and Gram-positive17,55,56 |
| 14. | Pal–Tyr–Tyr–β-Ala | Nanofibers | Delivery of, AgNPs, bioactive metal ions (Zn2+), plasmonic welding of AuNPs | Antibacterial activity against Gram-negative and Gram-positive23,46 |
| 15. | Pal–Tyr–Phe–β-Ala | Nanofibers | Delivery of, AgNPs, bioactive metal ions, plasmonic welding of AuNPs | Antibacterial activity against Gram-negative and Gram-positive46,51,68 |
| 16. | Pal–Phe–Tyr–β-Ala | Nanofibers/twisted fibers | Delivery of, AgNPs, bioactive metal ions, plasmonic welding of AuNPs | Antibacterial activity against Gram-negative and Gram-positive46,51,68 |
Beyond merely evaluating their antimicrobial activity, we also scrutinize the morphology of these nanostructures. This examination is crucial as it offers insights into how the physical characteristics of the peptides, such as size, shape, and surface properties, influence their interaction with bacterial membranes and subsequent bactericidal effects.52 Our findings unveil the promising potential of the newly designed sPA nanostructures as a captivating platform for the development of nanoantibacterials. Notably, they demonstrate comparability or even superiority to existing counterparts in various critical aspects. For instance, our nanostructures exhibit ease of synthesis, which is pivotal for scalability and cost-effectiveness in large-scale production. Additionally, their molecular stability at room temperature presents a significant advantage, ensuring prolonged shelf life and ease of storage compared to conventional antibiotics.22
Furthermore, the biocompatibility and biodegradability of our sPA nanostructures underscore their potential as safer alternatives to traditional antibiotics, mitigating concerns regarding toxicity and environmental impact.54 This multifaceted superiority positions them as promising candidates for addressing the urgent challenge of antimicrobial resistance, offering a holistic solution that combines efficacy with sustainability. Therefore, tabulated SAR studies, coupled with extensive evaluation of nanostructure morphology and antibacterial activity, provide compelling evidence for the potential of our designed sPA nanostructures as a next-generation antimicrobial platform. By leveraging insights from both established research69 and our own innovative contributions, we pave the way for the development of effective and sustainable antibacterial agents to combat infectious diseases.54,70
Future direction and prospects
Ongoing research in peptide amphiphile design is poised to lead significant advancements, further enhancing physiological properties and biocompatibility through meticulous atomic-level manipulation of short peptide amphiphiles (sPAs). The integration of sPAs with supramolecular chemistry holds immense potential for developing personalized nanomedicines, fostering the creation of theranostic platforms with integrated diagnostic and therapeutic capabilities, thereby revolutionizing targeted drug delivery and tissue engineering practices. Anticipated future developments may overhaul the integration of nanomaterials into antibacterial interventions, offering innovative approaches to combat bacterial resistance. Continued research endeavors will prioritize enhancing multifunctional hybrid self-assembly materials as nanocarriers, with a focus on refining targeted drug delivery systems for precise therapeutic agent release at infection sites. Such nanomaterials offer unique properties to disrupt biofilm structures, improving bacterial susceptibility to treatment. Recent studies demonstrate the potential of versatile nanostructures in combating bacterial infections, antibiotic resistance, and applications in photothermal therapy and advanced imaging, contributing significantly to nanoscience and biotechnology. Integration of nanomaterials into antibacterial interventions will reshape strategies against bacterial resistance, validated through in vivo or animal models, driving significant progress in biomedicine.Conclusions
The self-assembly of lipid-modified ultra-short peptides represents a compelling frontier in antibacterial research, capitalizing on their distinctive characteristics to bolster antimicrobial efficacy and surmount challenges linked with conventional antibiotics. These nanomaterials, derived from the self-assembly of short peptide amphiphiles, furnish a multifaceted strategy to engage and disrupt bacterial membranes notorious for their resistance to traditional antimicrobial agents. The salient attributes of self-assembling peptide amphiphiles, including potent antibacterial properties and pharmaceutical promise, position them at the vanguard of the fight against antibiotic-resistant bacteria. This comprehensive review elucidates the progressions in peptide amphiphile design, spotlighting the successful creation of novel structures to augment physiological attributes and compatibility. Integration of metal ions and nanoparticles into these peptide architectures demonstrates auspicious antibacterial outcomes against diverse bacterial strains, underscoring their potential in efficaciously combating bacterial infections. Profound morphological and physiological alterations have been elucidated at the atomic level, focusing on modulating these structures to achieve diverse self-assembly morphologies, ranging from linear fibers to spherical vesicles, through adjustments in the hydrophilicity of the head group. The findings of this study underscore the versatility and prospective applications of these pioneering peptide structures in the realms of biomedicine and antibacterial therapeutics.Conflicts of interest
There are no conflicts to declare.Acknowledgements
RS thanks CSIR-UGC for the doctoral research fellowship. SS thanks SERB-DST-India for the project associate-I fellowship. AK thanks DHSGSU for a doctoral fellowship. This work is supported by SERB-DST-India project no. CRG/2021/002526 and DST/TDT/TEC/2022/47(G) to KBJ.Notes and references
- M. A. Salam, M. Y. Al-Amin, M. T. Salam, J. S. Pawar, N. Akhter, A. A. Rabaan and M. A. A. Alqumber, Healthcare, 2023, 11(13), 1946 CrossRef PubMed
.
- C. Ghosh, P. Sarkar, R. Issa and J. Haldar, Trends Microbiol., 2019, 27(4), 323–338 CrossRef CAS PubMed
.
- G. Kalın, E. Alp, A. Chouaikhi and C. Roger, Microorganisms, 2023, 11(10), 2575 CrossRef PubMed
.
- H. F. Chambers and F. R. Deleo, Nat. Rev. Microbiol., 2009, 7(9), 629–641 CrossRef CAS PubMed
.
- G. Li, M. J. Walker and D. M. P. De Oliveira, Microorganisms., 2023, 11(1), 24 CrossRef CAS PubMed
.
- U. Theuretzbacher and L. J. V. Piddock, Cell Host Microbe, 2019, 10 Search PubMed
.
- J. M. Ageitos, A. Sánchez-Pérez, P. Calo-Mata and T. G. Villa, Biochem. Pharmacol., 2017, 133, 117–138 CrossRef CAS PubMed
.
- A. A. Bahar and D. Ren, Pharmaceuticals, 2013, 6(12), 1543–1575 CrossRef PubMed
.
- L. Schnaider, S. Brahmachari, N. W. Schmidt, B. Mensa, S. Shaham-Niv, D. Bychenko, L. A. Abramovich, L. J. W. Shimon, S. Kolusheva, W. F. DeGrado and E. Gazit, Nat. Commun., 2017, 8, 1365 CrossRef PubMed
.
- L. Lombardi, A. Falanga, V. D. Genio and S. Galdiero, Pharmaceutics, 2019, 11(4), 166 CrossRef CAS PubMed
.
- Y. Liu, Y. Yang, C. Wang and X. Zhao, Nanoscale, 2013, 5, 6413–6421 RSC
.
- E. Gazit, Chem. Soc. Rev., 2007, 36, 1263–1269 RSC
.
- A. P. McCloskey, B. F. Gilmore and G. Laverty, Pathogens, 2014, 3(4), 791–821 CrossRef PubMed
.
- M. Abbas, M. Ovais, A. Atiq, T. M. Ansari, R. Xing, E. Spruijt and X. Yan, Coord. Chem. Rev., 2022, 460, 214481 CrossRef CAS
.
- M. Mehta, T. A. Bui, X. Yang, Y. Aksoy, E. M. Goldys and W. Deng, ACS Mater. Au, 2023, 3(6), 600–619 CrossRef CAS PubMed
.
- E. V. K. Ledger, A. Sabnis and A. M. Edwards, Microbiology, 2022, 168(2), 001136 CrossRef CAS PubMed
.
- R. Singh, N. K. Mishra, N. Singh, P. Rawal, P. Gupta and K. B. Joshi, New J. Chem., 2020, 44, 9255–9263 RSC
.
- P. Zou, W.-T. Chen, T. Sun, Y. Gao, L.-L. Li and H. Wang, Biomater. Sci., 2020, 8, 4975–4996 RSC
.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC
.
- R. Goel, C. Garg, H. K. Gautam, A. K. Sharma, P. Kumar and A. Gupta, Int. J. Biol. Macromol., 2018, 111, 880–893 CrossRef CAS PubMed
.
- S. Marchesan, Y. Qu, L. J. Waddington, C. D. Easton, V. Glattauer, T. J. Lithgow, K. M. McLean, J. S. Forsythe and P. G. Hartley, Biomaterials, 2013, 34(14), 3678–3687 CrossRef CAS PubMed
.
- N. Singh, R. Singh, M. Shukla, G. Kaul, S. Chopra, K. B. Joshi and S. Verma, ACS Infect. Dis., 2020, 6(9), 2441–2450 CrossRef CAS PubMed
.
- R. Singh, N. K. Mishra, V. Kumar, V. Vinayak and K. B. Joshi, ChemBioChem, 2018, 19(15), 1630–1637 CrossRef CAS PubMed
.
- S. Anwar, M. B. Khawar, A. Afzal, M. Ovais and Z. Xiang, Appl. Microbiol. Biotechnol., 2023, 107(18), 5775–5787 CrossRef CAS PubMed
.
- R. Singh, V. Suryavashi, V. Vinayak and K. B. Joshi, ChemistrySelect, 2019, 4(19), 5810–5816 CrossRef CAS
.
- R. Singh, M. J. Khan, J. Rane, A. Gajbhiye, V. Vinayak and K. B. Joshi, ChemistrySelect, 2020, 5(10), 3091–3097 CrossRef CAS
.
- B. Mishra, A. Saxena and A. Tiwari, Biotechnol. Rep., 2020, 28, e00571 CrossRef PubMed
.
- R. Menacho-Melgar, J. S. Decker, J. N. Hennigan and M. D. Lynch, J. Controlled Release, 2019, 295, 1–12 CrossRef CAS PubMed
.
- S. Zhang, Acc. Chem. Res., 2012, 45(12), 2142–2150 CrossRef CAS PubMed
.
- C. J. C. Edwards-Gaylea and I. W. Hamley, Org. Biomol. Chem., 2017, 15, 5867–5876 RSC
.
- A. Makovitzki, D. Avrahami and Y. Shai, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(43), 15997–16002 CrossRef CAS PubMed
.
- S. Chou, H. Guo, F. G. Zingl and X. Mou, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(4), e2219679120 CrossRef CAS PubMed
.
- X. Yin, Z. Chen, Y. Chen, Y. Xie, B. Xiong, H. Jiang and J. Zhu, Colloids Surf., B, 2020, 195, 111271 CrossRef CAS PubMed
.
- L. Xu, X. Wang, Y. Liu, G. Yang, R. J. Falconer and C. X. Zhao, Adv. NanoBiomed Res., 2022, 2, 2100109 CrossRef CAS
.
- M. P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50(10), 2440–2448 CrossRef CAS PubMed
.
- A. Dasgupta and D. Das, Langmuir, 2019, 35(33), 10704–10724 CrossRef CAS PubMed
.
- S. Zhang, T. Holmes, C. Lockshin and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3334–3338 CrossRef CAS PubMed
.
- J. P. Schneider, D. J. Pochan, B. Ozbas, K. Rajagopal, L. Pakstis and J. Kretsinger, J. Am. Chem. Soc., 2002, 124, 15030–15037 CrossRef CAS PubMed
.
- D. J. Pochan, J. P. Schneider, J. Kretsinger, B. Ozbas, K. Rajagopal and L. Haines, J. Am. Chem. Soc., 2003, 125, 11802–11803 CrossRef CAS PubMed
.
- S. Basak, I. Singh, A. Banerjee and H. B. Kraatz, RSC Adv., 2017, 7, 14461–14465 RSC
.
- T. Mondal, A. Chatterjee, B. Hansda, B. Mondal, P. Sen and A. Banerjee, Soft Matter, 2024, 20, 1236–1244 RSC
.
- M. M. Konaia, S. Samaddar, G. Bocchinfuso, V. Santucci, L. Stella and J. Haldar, Chem. Commun., 2018, 54, 4943–4946 RSC
.
- Y. Chen, Z. Ye, W. Zhen, L. Zhang, X. Min, Y. Wang, F. Liu and M. Su, Bioorg. Chem., 2023, 140, 106766 CrossRef CAS PubMed
.
- A. Adak, V. Castelletto, A. de Sousa, K.-A. Karatzas, C. Wilkinson, N. Khunti, J. Seitsonen and I. W. Hamley, Biomacromolecules, 2024, 25(2), 1205–1213 CrossRef CAS PubMed
.
- V. Kumar, N. K. Mishra, S. Gupta and K. B. Joshi, ChemistrySelect, 2017, 2(1), 211–218 CrossRef CAS
.
- K. Kesharwani, R. Singh, S. K. Tripathi, G. Kaul, A. Akhir, D. Saxena, V. Kumar, N. K. Mishra, S. Chopra and K. B. Joshi, ChemistrySelect, 2022, 7(35), e202202234 CrossRef CAS
.
- A. Dehsorkhi, V. Castelletto and I. W. Hamley, J. Pept. Sci., 2014, 20(7), 453–467 CrossRef CAS PubMed
.
- H. Cui, Matthew J. Webber and S. I. Stupp, Biopolymers, 2010, 94(1), 1–18 CrossRef CAS PubMed
.
- K. D. Ristroph and R. K. Prud’homme, Nanoscale Adv., 2019, 1(11), 4207–4237 RSC
.
- S. K. Tripathi, K. Kesharwani, D. Saxena, R. Singh, A. Kautu, S. Sharma, A. Pandey, S. Chopra and K. B. Joshi, ChemMedChem, 2023, 18(5), e202200654 CrossRef PubMed
.
- K. Kesharwani, A. Kautu, S. Sharma, R. Singh, V. Kumar, S. K. Tripathi, P. Shukla and K. B. Joshi, Chem. Commun., 2022, 58, 13815–13818 RSC
.
- M. G. Ciulla and F. Gelain, Microb. Biotechnol., 2023, 16(4), 757–777 CrossRef PubMed
.
- P. L. Nyembe, T. Ntombela and M. M. Makatini, Pharmaceutics, 2023, 15, 1506 CrossRef CAS PubMed
.
- N. Swain, S. Sharma, R. Maitra, D. Saxena, A. Kautu, R. Singh, K. Kesharwani, S. Chopra and K. B. Joshi, ChemMedChem, 2024, e202300576 CrossRef CAS PubMed
.
- K. Kesharwani, R. Singh, M. J. Khan, V. Vinayak and K. B. Joshi, ChemistrySelect, 2021, 6(27), 6827–6833 CrossRef CAS
.
- S. K. Tripathi, K. Kesharwani, G. Kaul, A. Akhir, D. Saxena, R. Singh, N. K. Mishra, A. Pandey, S. Chopra and K. B. Joshi, ChemMedChem, 2022, 7(15), e202200251 CrossRef PubMed
.
- A. B. Fernandez, F. Vogt, R. Sodian and F. Weis, Infect Drug Resist., 2010, 3, 95–101 Search PubMed
.
- C. V. García and I. Colomer, Org. Biomol. Chem., 2021, 19, 6797–6803 RSC
.
- D. I. Chan, E. J. Prenner and H. J. Vogel, Biochim. Biophys. Acta, Biomembr., 2006, 1758(9), 1184–1202 CrossRef CAS PubMed
.
- D. Kalafatovic and E. Giralt, Molecules, 2017, 22(11), 1929 CrossRef PubMed
.
- K. Reddersen, K. E. Greber, I. K. Glowniak and C. Wiegand, Antibiotics, 2020, 9(12), 879 CrossRef CAS PubMed
.
- N. H. Z. Baharin, N. F. K. Mokhtar, M. N. M. Desa, B. Gopalsamy, N. N. M. Zaki, M. H. Yuswan, A. Muthanna, N. D. Dzaraly, S. Abbasiliasi, A. M. Hashim, M. S. A. Sani and S. Mustafa, PeerJ, 2021, 14(9), e12193 CrossRef PubMed
.
- A. Levin, T. A. Hakala, L. Schnaider, G. J. L. Bernardes, E. Gazit and T. P. J. Knowles, Nat. Rev. Chem., 2020, 4(11), 615–634 CrossRef CAS
.
- D. E. Shalev, Int. J. Mol. Sci., 2022, 23(24), 15957 CrossRef CAS PubMed
.
- R. Zou, Q. Wang, J. Wu, J. Wu, C. Schmuck and H. Tian, Chem. Soc. Rev., 2015, 44, 5200–5219 RSC
.
- N. K. Mishra, K. B. Joshi and S. Verma, J. Colloid Interface Sci., 2015, 455, 145–153 CrossRef CAS PubMed
.
- N. K. Mishra, V. Kumar and K. B. Joshi, Nanoscale, 2015, 7, 20238–20248 RSC
.
- R. Singh, N. K. Mishra, P. Gupta and K. B. Joshi, Chem. – Asian J., 2020, 15(4), 531–539 CrossRef CAS PubMed
.
- R. Kowalczyk, P. W. R. Harris, G. M. Williams, S. H. Yang and M. A. Brimble., Adv. Exp. Med. Biol., 2017, 1030, 185–227 CrossRef CAS PubMed
.
- S. Sharma, A. Kautu, N. Singh, N. Kumar, V. Kumar, R. Singh, K. Kesharwani, N. Swain, P. Gupta and K. B. Joshi, Next Mater., 2024, 4, 100118 CrossRef
.
Footnote |
| † Current address: Department of Mechanical Engineering, Colorado State University, Fort Collins, CO, USA. E-mail: ramesh.singh@colostate.edu |
| This journal is © The Royal Society of Chemistry 2024 |




