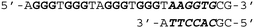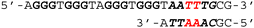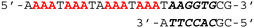Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A dual DNA-binding conjugate that selectively recognizes G-quadruplex structures†
Mitsuharu
Ooga
a,
Vinodh J.
Sahayasheela
 a,
Yuki
Hirose
a,
Yuki
Hirose
 a,
Daisuke
Sasaki
a,
Kaori
Hashiya
a,
Toshikazu
Bando
*a and
Hiroshi
Sugiyama
a,
Daisuke
Sasaki
a,
Kaori
Hashiya
a,
Toshikazu
Bando
*a and
Hiroshi
Sugiyama
 *b
*b
aDepartment of Chemistry, Graduate School of Science, Kyoto University, Kitashirakawa-Oiwakecho, Sakyo-ku, Kyoto 606-8502, Japan. E-mail: bando@kuchem.kyoto-u.ac.jp
bInstitute for Integrated Cell-Material Sciences (iCeMS), Kyoto University, Yoshida-ushinomiyacho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: hs@kuchem.kyoto-u.ac.jp
First published on 22nd July 2024
Abstract
G-quadruplex (G4) structures play roles in various biological processes, but the challenge lies in specific targeting. To address this, we synthesized a conjugate capable of recognizing the G4 structure and its proximal duplex. Our conjugate can enable recognition of specific G4s in the human genome to understand and target those structures.
Nucleic acids are known to form structures apart from the canonical Watson–Crick double-helix.1 Among them, G-quadruplexes (G4s) are well-known secondary structures that arise from guanine-rich sequences through stacked G tetrads by Hoogsteen hydrogen bonding and are further stabilized by monovalent cations (K+, Na+) between the tetrad.2 G4 structures arising from the telomere repeat sequences and oncogene promoter regions are thought to play important biological roles in the processing of telomere-ends3 and the regulation of gene expression, respectively.4 Therefore, various small molecules have been developed to stabilize the G4 structure, making it a promising drug target.5–7 However, the genome-wide analysis using a G4 ligand identified more than 700
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 G4-forming sequences,8 and targeting a specific G4-forming sequence still remains a challenge given the topological similarity among diverse G4-forming sequences.
000 G4-forming sequences,8 and targeting a specific G4-forming sequence still remains a challenge given the topological similarity among diverse G4-forming sequences.
A promising approach to overcome this challenge is the simultaneous recognition of both G4 and its proximal DNA regions. Earlier using this approach our lab9,10 and other groups11,12 were able to achieve success in obtaining G4 selectivity. With the advancement of G4-mapping techniques13 and reports of crystal structures of a G4-duplex complex,14,15 it is possible to design molecules that can recognize both G4 and its proximal DNA regions to reveal its function by selective targeting. More recently, while this manuscript was under preparation, the G4 ligand conjugated with oligonucleotide was also reported.16
However, to achieve a higher-level recognition retaining affinity towards a selective G4 site for biological studies, it is necessary to identify diverse candidates. Herein, we synthesized a conjugate consisting of a PyPDS, a well-known G4 ligand,17,18 and pyrrole–imidazole polyamide (PIP), a programmable sequence-specific DNA minor-groove binder.19,20 We demonstrated the selective G4 recognition of the conjugate, thereby illustrating its potential as a tool to delineate the function of selective G4 structures and target them.
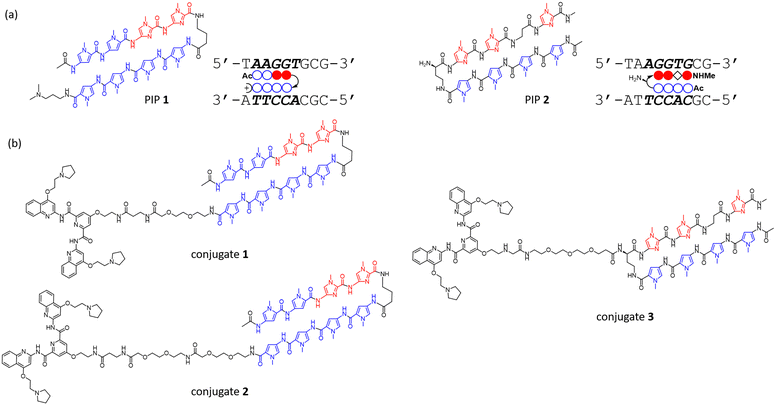
To investigate the binding ability and sequence selectivity, we chose the c-MYC G4 forming region and its adjacent duplex sequence in the nuclease hypersensitive element (NHE) III1 as a dual-target (Fig. S1, ESI†). PIPs 1 and 2 (Fig. 1a) targeting the flanking region proximal to G4 in the c-MYC promoter were utilized as the duplex binder. The PIPs obtained by solid-phase synthesis were conjugated with PyPDS via linkers to produce conjugates 1–3 (Fig. 1b and Schemes S1–S3, ESI†). Conjugates 1 and 2 were designed using PIP 1, whereas conjugate 3 was designed using PIP 2.
To evaluate whether the hybrid molecules could maintain their G4-stabilizing effect on the c-MYC quadruplex, we monitored the thermal denaturing profiles of the G4 formation using circular dichroism (CD) in the presence or absence of the hybrid compounds. We utilized the target DNA sequence with G-to-T modifications at the G4-duplex interface to facilitate the formation of G4 and prevent the alternative hybridization of the shorter strand based on our previous research9 (Table 1). The CD spectrum of the target DNA showed combined signals derived from both G4 and B-form structures (Fig. S2a, ESI,† and Table 1). To accurately measure the melting temperature from the G4 structures alone, it is necessary to use a wavelength where the duplex effect is minimal. From our previous study using hairpin duplex DNA segments,9 we found that 267 nm was the ideal wavelength for measuring the signal derived from the G4 structure. To validate this, the signal of the duplex segment at that specific wavelength was measured, and the results revealed little change (Fig. S3, ESI†), indicating that the duplex region had minimal impact on the target sequence at this wavelength. Moving forward, the signal at 267 nm was measured to obtain the melting curves of the G4 moiety in the presence or absence of each compound (Fig. 2 and Fig. S4, ESI†). The melting curves indicated that all the conjugates stabilized the G4 structure, and from the comparison of the melting temperature increase (ΔTm), conjugate 3 showed higher values (ΔTm = 7.6 °C) than the other two conjugates. Interestingly, conjugate 3 also showed better G4 stabilizing ability than PyPDS alone (ΔTm = 6.5 °C) under the condition in which one equivalent of the compound was added to DNA. These results validate that the conjugates were able to stabilize the G4 structures comparable to PyPDS.
Next, the stabilization of the duplex region by the hybrid compounds was evaluated through UV melting assays. The absorption-based melting curve of the target DNA segment detected at 260 nm revealed two inflection points and it was estimated to undergo melting in the following order: (i) the duplex and (ii) the G4 structure (Fig. S5a, ESI†). To clarify the inflection point corresponding to duplex melting, several validations were carried out. First, the CD signals of the target DNA and the DNA lacking the complementary strand of the duplex region were measured (Fig. S5b, ESI†). Measurements using CD melting at 10 °C and 55 °C revealed that the CD signals were very similar in both DNA sequences, suggesting that the duplex signal largely disappeared from the CD signal of the target DNA (Fig. S5b and c, ESI†). Additionally, a comparison of CD spectra in the presence or absence of the conjugates at 10 °C and 55 °C showed that the signal derived from parallel G4 remained, while the signal derived from PIP binding in the minor groove (330 nm) disappeared with increasing temperature (Fig. S6, ESI†). These results indicated that the increase in absorbance from 10 to 55 °C was largely due to the dissociation of the duplex region, and the binding of PIP towards the target sequence increased the melting temperature. Note that the addition of PyPDS had no substantial impact on the waveform of the CD spectrum of the target DNA, and similarly to other G4-stabilizing ligands, a slight decrease in G4-derived signals, such as those at 260 and 240 nm, was also detected21 (Fig. S7, ESI†).
To assess the thermal stability of the conjugates to their target duplex region, we calculated the positive shifts of the melting temperature by the addition of compounds (Table 2 and Fig. S8, ESI†). The increased ΔTm values of the conjugates compared to PIP or PyPDS alone confirmed their enhanced double-strand stabilizing ability. The slightly higher ΔTm values for conjugate 3 (ΔTm = 26.8 °C) compared to PIP 2 (ΔTm = 24.3 °C) could be attributed to the high ΔTm values of PIP 2 itself. The stabilization of the double-stranded DNA by PyPDS (ΔTm = 11.0 °C) may be likely due to its binding in the region between G4 and the duplex segment and indeed a NMR-based model of PyPDS binding at the interface of the G-quartet and the double-stranded loop structure has been reported.22 Additionally, we performed the FRET melting assay utilizing DNA oligomers labeled with a fluorescein and a tetramethylrhodamine at the opposite end of G4 (Fig. S9a, ESI†). This method also demonstrated the duplex stabilizing ability of the conjugates, and the order of the ΔTm values was consistent with that of UV melting at 260 nm, except for a reversal due to a slight difference between conjugates 1 and 2 (Table S1 and Fig. S9b–g, ESI†).
| PyPDS | PIP 1 | Conjugate 1 | Conjugate 2 | PIP 2 | Conjugate 3 | |
|---|---|---|---|---|---|---|
| ΔTm [°C] | 11.0 ± 0.3 | 0.4 ± 0.3 | 17.2 ± 0.4 | 16.8 ± 0.6 | 24.3 ± 0.5 | 26.8 ± 0.3 |
The results of the melting studies demonstrated that the conjugates were capable of stabilizing both the G4 and duplex regions of the target DNA, indicating their dual recognition abilities. In sum, conjugate 3 exhibited high thermal stability in both G4 and duplex regions. The energy-minimized structure of the DNA–conjugate 3 complex suggested a binding model in which the PyPDS moiety was inserted between the G4 and duplex regions (Fig. S10, ESI†).
To investigate the selectivity of the conjugates, we performed native gel electrophoretic mobility shift assays (EMSA) using the target DNA sequence, the non-target DNA sequence, and the non-G4 DNA sequence (Fig. 3 and Fig. S11, ESI†). The DNA sequences used in this assay were initially confirmed to form G4 structures by CD spectrometry (Fig. S2, ESI†). As shown in Fig. 3a, in the case of the target DNA sequence, all conjugates including PyPDS showed an upward shift of the DNA band, with the intensity of the upshifted bands in the order of conjugate 1 < 2 < 3. The increase in band intensity by conjugate 3 from our EMSA can be explained by its high thermal stability demonstrated from our melting experiments. In contrast, in the presence of the non-target DNA sequence, only PyPDS showed a band shift, validating the selective binding of the conjugates to the target sequence alone (Fig. 3b). Finally, the conjugates showed no band shift with the non-G4 forming sequence containing duplex binding sites (Fig. 3c). Taken together, our EMSA assays indicate that efficient binding of conjugates requires the presence of both G4 and target duplex regions.
To further assess the selectivity of conjugates, fluorescent indicator displacement assays using thioflavin T (ThT) or ethidium bromide (EtBr) were carried out. ThT is a fluorescent probe that emits enhanced fluorescence upon binding to G4 structures, and its fluorescence intensity decreases upon displacement by G4 ligands, allowing for the identification of selective G4 binding compounds.23,24 We first confirmed that ThT showed enhanced fluorescence intensity when bound to the different G4 sequences, including the dual binding sites compared to double-stranded or non-G4 forming sequences. Additionally, EtBr, a commonly used fluorescent probe for double-stranded DNA, showed enhanced fluorescence when bound to DNA sequences containing duplex regions, compared to G4-only sequences (Fig. S12, ESI,† and Table 1). By using target and non-target DNA sequences, we then examined the concentration-dependent displacement of ThT or EtBr by each compound, resulting in the generation of displacement curves (Fig. S13 and S14, ESI†). The evaluation of the binding ability of the compounds was based on the DC50 value, which is the concentration required to displace 50% of the fluorescent indicator.25 With the DC50 values for both DNA sequences, the S value (DC50 for the non-target sequence/DC50 for the target sequence) was calculated as the measure of selectivity. For the ThT displacement assay, which evaluated G4 binding, PyPDS showed a value of 1 for the S value, indicating no difference between the sequences. In contrast, all conjugates had S values greater than 1, indicating preferential binding to the target sequences (Table 3) and conjugate 3 had the highest value (S = 2.3). The use of EtBr as a double-stranded fluorescent indicator demonstrated that the displacement rate for PyPDS reached saturation at 60%, with an S value of 0.67 (Fig. S14a, ESI,† and Table 4). In contrast, the S value for conjugates was above 1. These results suggest that the selectivity of the duplex region in conjugates contributes to the selectivity of G4.
| DC50 [μM] | S | ||
|---|---|---|---|
| Target | Non-target | ||
| PyPDS | 0.54 | 0.56 | 1.0 |
| Conjugate 1 | 1.1 | 2.1 | 1.9 |
| Conjugate 2 | 1.2 | 2.0 | 1.7 |
| Conjugate 3 | 0.88 | 2.0 | 2.3 |
| DC50 [μM] | S | ||
|---|---|---|---|
| Target | Non-target | ||
| PyPDS | 2.1 | 1.4 | 0.67 |
| Conjugate 1 | 1.8 | 3.5 | 1.9 |
| Conjugate 2 | 1.5 | 3.4 | 2.3 |
| Conjugate 3 | 1.3 | 2.9 | 2.2 |
In summary, the PyPDS–PIP hybrid compounds we developed exhibit the ability to recognize both G4 and its proximal duplex regions concurrently. This allows for the selective recognition of a specific G4, thereby suggesting that the conjugation of G4 ligands with PIP could be an effective strategy for designing ligands targeting specific G4-forming sequences. In the future, we will test our approach to visualize and modulate selective G4 structures for biological application.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Notes and references
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- H. J. Lipps and D. Rhodes, Trends Cell Biol., 2009, 19, 414–422 CrossRef CAS PubMed.
- T. Tian, Y.-Q. Chen, S.-R. Wang and X. Zhou, Chemistry, 2018, 4, 1314–1344 CrossRef CAS.
- N. Kosiol, S. Juranek, P. Brossart, A. Heine and K. Paeschke, Mol. Cancer, 2021, 20, 40 CrossRef CAS PubMed.
- S. Asamitsu, S. Obata, Z. Yu, T. Bando and H. Sugiyama, Molecules, 2019, 24, 429 CrossRef PubMed.
- Y. Ma, K. Iida and K. Nagasawa, Biochem. Biophys. Res. Commun., 2020, 531, 3–17 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. Di Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef CAS PubMed.
- S. Asamitsu, S. Obata, A. T. Phan, K. Hashiya, T. Bando and H. Sugiyama, Chemistry, 2018, 24, 4428–4435 CrossRef CAS PubMed.
- S. Mandal, Y. Kawamoto, Z. Yue, K. Hashiya, Y. Cui, T. Bando, S. Pandey, M. E. Hoque, M. A. Hossain, H. Sugiyama and H. Mao, Nucleic Acids Res., 2019, 47, 3295–3305 CrossRef CAS PubMed.
- E. Cadoni, L. De Paepe, G. Colpaert, R. Tack, D. Waegeman, A. Manicardi and A. Madder, Nucleic Acids Res., 2023, 51, 4112–4125 CrossRef CAS PubMed.
- J.-H. Yuan, J.-L. Tu, G.-C. Liu, X.-C. Chen, Z.-S. Huang, S.-B. Chen and J.-H. Tan, Nucleic Acids Res., 2022, 50, 4246–4257 CrossRef CAS PubMed.
- S. Galli, G. Flint, L. Růžičková and M. Di Antonio, RSC Chem. Biol., 2024, 5, 426–438 RSC.
- I. Russo Krauss, S. Ramaswamy, S. Neidle, S. Haider and G. N. Parkinson, J. Am. Chem. Soc., 2016, 138, 1226–1233 CrossRef CAS PubMed.
- R. C. Monsen, E. Y. D. Chua, J. B. Hopkins, J. B. Chaires and J. O. Trent, Nucleic Acids Res., 2023, 51, 1943–1959 CrossRef CAS PubMed.
- A. Berner, R. N. Das, N. Bhuma, J. Golebiewska, A. Abrahamsson, M. Andréasson, N. Chaudhari, M. Doimo, P. P. Bose, K. Chand, R. Strömberg, S. Wanrooij and E. Chorell, J. Am. Chem. Soc., 2024, 146, 6926–6935 CrossRef CAS PubMed.
- S. Müller, D. A. Sanders, M. Di Antonio, S. Matsis, J.-F. Riou, R. Rodriguez and S. Balasubramanian, Org. Biomol. Chem., 2012, 10, 6537–6546 RSC.
- M. Di Antonio, A. Ponjavic, A. Radzevičius, R. T. Ranasinghe, M. Catalano, X. Zhang, J. Shen, L.-M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed.
- C. L. Kielkopf, E. E. Baird, P. B. Dervan and D. C. Rees, Nat. Struct. Biol., 1998, 5, 104–109 CrossRef CAS PubMed.
- P. B. Dervan and B. S. Edelson, Curr. Opin. Struct. Biol., 2003, 13, 284–299 CrossRef CAS PubMed.
- E. Palma, C. Içhedef, C. Fernandes, A. Belchior, P. Raposinho, L. Gano, A. Miranda, D. Moreira, P. Lourenço, C. Cruz, A. S. Pires, M. F. Botelho and A. Paulo, Chem. – Eur. J., 2024, 30, e202400285 CrossRef CAS PubMed.
- L.-Y. Liu, T.-Z. Ma, Y.-L. Zeng, W. Liu and Z.-W. Mao, J. Am. Chem. Soc., 2022, 144, 11878–11887 CrossRef CAS PubMed.
- A. Renaud de la Faverie, A. Guédin, A. Bedrat, L. A. Yatsunyk and J.-L. Mergny, Nucleic Acids Res., 2014, 42, e65 CrossRef CAS PubMed.
- J. Jamroskovic, M. Livendahl, J. Eriksson, E. Chorell and N. Sabouri, Chemistry, 2016, 22, 18932–18943 CrossRef CAS PubMed.
- D. Monchaud and M.-P. Teulade-Fichou, Methods Mol. Biol., 2010, 608, 257–271 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc01572j |
| This journal is © The Royal Society of Chemistry 2024 |