 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
NMR analysis of 15N-labeled naphthyridine carbamate dimer (NCD) to contiguous CGG/CGG units in DNA†
Takeshi
Yamada‡
 a,
Shuhei
Sakurabayashi
a,
Noriaki
Sugiura
a,
Hitoshi
Haneoka
b and
Kazuhiko
Nakatani
a,
Shuhei
Sakurabayashi
a,
Noriaki
Sugiura
a,
Hitoshi
Haneoka
b and
Kazuhiko
Nakatani
 *a
*a
aDepartment of Regulatory Bioorganic Chemistry, SANKEN, Osaka University, Mihogaoka 8-1, Ibaraki, Osaka 567-0047, Japan. E-mail: nakatani@sanken.osaka-u.ac.jp
bComprehensive Analysis Center, SANKEN, Osaka University, Mihogaoka 8-1, Ibaraki, Osaka 567-0047, Japan
First published on 22nd February 2024
Abstract
The structure of the complex formed by naphthyridine carbamate dimer (NCD) binding to CGG repeat sequences in DNA, associated with fragile X syndrome, has been elucidated using 15N-labeled NCD and 1H–15N HSQC. In a fully saturated state, two NCD molecules consistently bind to each CGG/CGG unit, maintaining a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding stoichiometry.
2 binding stoichiometry.
The aberrant expansion of trinucleotide repeat (TNR) sequences in the human genome leads to over 40 serious neurodegenerative diseases, exemplified by fragile X syndrome (FXS, caused by CGG repeat), Huntington's disease (HD, CAG), and myotonic dystrophy type 1 (DM1, CTG).1–3 Unfortunately, there is currently no effective therapy for TNR diseases. This genomic instability results in both contraction and expansion of the repeats.4–6 Aberrantly expanded repeats tend to form metastable slip-outs, comprising hairpin structures with repetitive CXG/CXG units, where X–X mismatches are flanked by two C–G base pairs during replication and transcription reactions.7 The chemical stability of the slip-outs is a contributing factor to the genomic instability of TNR. Recent studies from our group have demonstrated that perturbing slip-out structures with small external molecules can modulate the chemical stability of the hairpin structure and, consequently, genomic instability,8,9 highlighting the importance of developing such small molecules and elucidating their binding modes.10–13
We have reported a variety of small molecules binding to TNR sequences, represented by naphthyridine carbamate dimer (NCD, Fig. 1A),14 which showed a remarkable binding to the CGG repeat DNA. Recently, we reported the NMR structural determination of the complex comprised of NCD and a model DNA containing a single CGG/CGG triad15 (Fig. 1B, PDB: 7YVW). The NMR structure elucidated the distinctive binding pattern of NCD. Two NCD molecules bind to the CGG/CGG triad, and the four naphthyridine carbamate (NC) moieties form hydrogen bonds with four guanines in the CGG/CGG. This interaction leads to the flip-out of two cytosines. To gain insight into the mode of NCD binding to the CGG repeat, we measured the mass spectrometry of a CGG repeat DNA (5′-d(CGG)10-3′) in the presence of NCD, and confirmed that even numbers of NCD molecules bound to the CGG repeat. Based on these experimental results, we have postulated that the complex of NCD molecules and CGG repeat DNA would adopt repetitive structures consisting of one CGG/CGG unit with two NCD molecules, as depicted in Fig. 1C and D.
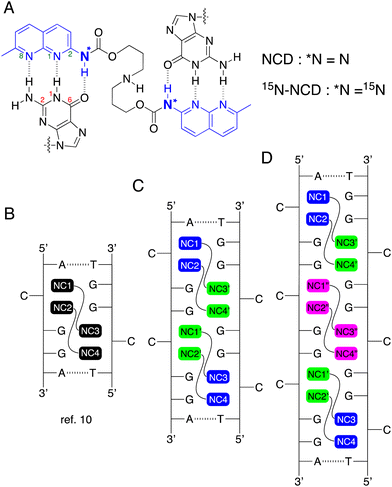 | ||
| Fig. 1 (A) Chemical structures of NCD and 15N-NCD. The partial numberings of guanine and naphthyridine rings are shown in red and green, respectively. (B) Illustration of complexes composed of two NCD molecules and a CGG/CGG unit, as elucidated by NMR structural determination (ref. 15). The proposed structures of complexes involving NCD molecules and consecutive (C) two or (D) three repetitive CGG/CGG units. ‘NC’ refers to the naphthyridine carbamate moiety of NCD. | ||
In the proposed binding model of NCD to the CGG repeat DNA, one question arises as to whether NCD binding to the CGG repeats followed the nearest-neighbor exclusion principle as observed for the binding of DNA intercalators.16 When an intercalator binds to a duplex DNA, the two adjacent intercalation sites would remain unoccupied due to the perturbation of the base pair structure by intercalation. When CGG repeat DNA formed the hairpin secondary structure, there are contiguous CGG/CGG units, and these CGG/CGG units are in different environments depending on the position, e.g., the end or the middle of the hairpin structure. Furthermore, NCD-binding to one CGG/CGG unit should perturb the structure of the neighboring CGG/CGG unit and, hence, the binding of NCD. Although MS data revealed an even number of NCD molecules bound to the CGG repeat DNAs, our understanding of the NCD binding mode to CGG repeat DNA remains limited. To address the question raised above, we synthesized 15N-labeled NCD (15N-NCD, Fig. 1A) and measured the 15N NMR spectrum to see how the 15N signal changed as the number of CGG/CGG units increased. In the proposed model of (CGG/CGG)2 with four NCD molecules (Fig. 1C), the NC moieties (NC1, NC2, NC3, and NC4) shown in blue has likely a similar chemical environment to those in the complex of (CGG/CGG)3 with six NCD molecules (Fig. 1D). Likewise, NC moieties shown in green should be in similar environments but different from those in blue. In addition, the NC moieties colored in cyan in Fig. 1D would be in different environments from those colored in blue and green, although the difference could be small. The different chemical environments of NC moieties could be observed by the difference in the chemical shift in NMR analysis. Here, we report that 1H–15N HSQC spectra of 15N-NCD with DNA involving one, two, and three units of CGG/CGG clearly indicated that NCD could bind to the contiguous CGG/CGG sites.
2-Amino-1,8-naphthyridine labeled with 15N at the amino group was synthesized by reacting 2-chloro-1,8-naphthyridine with 22% aqueous 15N ammonia in the presence of 1.0 equivalent copper in 40% yield. Boc-protected 15N-NCD and 15N-NCD were prepared following the reported procedure17 (Scheme S1 in ESI†). The 1H-NMR of Boc-protected 15N-NCD in CDCl3 is presented in Fig. S1 in the ESI.† The amide protons of 15N-NCD appeared at 7.69 ppm as a superposition of two doublets due to the heteronuclear coupling between 1H and 15N.
Next, 15N-NCD was used to assign amide protons in the complexes involving NCD and a CGG/CGG unit. The dsDNA GG1, composed of DNA1: 5′-d(CTAA CGG AATG)-3′ and DNA2: 5′-d(CATT CGG TTAG)-3′, was used for the NMR structural determination of the NCD-CGG/CGG complex in our previous study15 (Fig. 2A). The 1H–15N HSQC spectrum of GG1 (200 μM) in the presence of 15N-NCD (400 μM) is depicted in Fig. 2B. For clarity, the 1H-NMR spectrum of the NCD-GG1 complex is overlaid above the HSQC spectrum. In the 1H–15N HSQC, four cross peaks were observed at (1H, 15N) coordinates: (11.02, 118.4), (11.09, 119.6), (11.18, 119.8), and (11.63, 119.6), and identified as amide protons of NC1, NC2, NC3, and NC4, respectively (Fig. 2C). As two amide protons would be anticipated for one NCD binding, the HSQC spectra clearly indicated the binding of two NCD molecules to GG1.
 | ||
| Fig. 2 (A) dsDNAs used in this study, denoted as GGn (n = 1 to 3) based on the number of CGG/CGG units. The green numbers indicate residue numbering in GG1. (B) 1H–15N HSQC of GG1 (200 μM) in 5% D2O in sodium phosphate buffer (20 mM, pH = 6.8) containing 100 mM NaCl in the presence of 15N-NCD (400 μM). The imino-proton region (10.5 to 12.5 ppm) of the 1H and 15N HSQC spectra is displayed. The 1H-NMR of the NCD-GG1 complex is presented above the HSQC. Amide protons of NCD are labeled as NCn (n = 1 to 4), and the imino-protons of guanines are denoted as Gn (n =numbering). (C) Schematic illustration of the binding of two 15N-NCD molecules to the CGG/CGG unit in GG1. Amide protons next to 15N are colored in blue and observed in the HSQC shown in Fig. 3B. | ||
Continuing, we employed another model DNA named GG2, comprising two consecutive CGG/CGG units (Fig. 2A, n = 2) for titration experiments using 15N-NCD. The sequences of GG1 and GG2 are identical except for the number of CGG/CGG units. The titration results are presented in Fig. 3A. In the 1H-NMR spectrum of GG2, the imino-protons of thymidine were observed between 13.5 and 14.0 ppm, while those of guanine were between 12.5 and 13.0 ppm. Upon titration with 15N-NCD, the signal initially became complex (e.g., GG2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15N-NCD = 1
15N-NCD = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), but as the titration proceeded, the spectrum simplified. Eventually, the shifts of signals saturated upon the addition of 6 equivalents of 15N-NCD. In Fig. 3B, the 1H–15N HSQC of the GG2 and 15N-NCD complex under saturated conditions is displayed. The 1H-NMR spectrum shown above the HSQC spectrum is the GG2 and unlabeled NCD complex under the saturated conditions. At a glance, seven cross peaks in the HSQC spectrum were identified at (1H, 15N) coordinates: (10.89, 118.6), (10.94, 119.8), (11.03, 119.1), (11.03, 119.2), (11.03, 119.8), (11.11, 118.8), and (11.48, 119.6). Upon further analysis using a 3D view (Fig. 3C), the peak at (11.11, 118.8), marked with a red star, displayed approximately twice the intensity compared to other peaks, indicating a superposition of two peaks. Therefore, eight cross peaks corresponding to the 1H–15N coupling were identified, suggesting that four NCD molecules bound to GG2.
3), but as the titration proceeded, the spectrum simplified. Eventually, the shifts of signals saturated upon the addition of 6 equivalents of 15N-NCD. In Fig. 3B, the 1H–15N HSQC of the GG2 and 15N-NCD complex under saturated conditions is displayed. The 1H-NMR spectrum shown above the HSQC spectrum is the GG2 and unlabeled NCD complex under the saturated conditions. At a glance, seven cross peaks in the HSQC spectrum were identified at (1H, 15N) coordinates: (10.89, 118.6), (10.94, 119.8), (11.03, 119.1), (11.03, 119.2), (11.03, 119.8), (11.11, 118.8), and (11.48, 119.6). Upon further analysis using a 3D view (Fig. 3C), the peak at (11.11, 118.8), marked with a red star, displayed approximately twice the intensity compared to other peaks, indicating a superposition of two peaks. Therefore, eight cross peaks corresponding to the 1H–15N coupling were identified, suggesting that four NCD molecules bound to GG2.
 | ||
| Fig. 3 (A) One-dimensional 1H spectra of GG2 (500 μM) in sodium phosphate buffer (pH 6.8, 20 mM) containing 100 mM NaCl in the presence of 15N-NCD with various GG2–15N-NCD ratios at 283K. The ratio displayed in the left side of each spectrum represents the proportion of GG2 and 15N-NCD. (B) 1H–15N HSQC of GG2 (500 μM) in 5% D2O in sodium phosphate buffer (20 mM, pH = 6.8) containing 100 mM NaCl in the presence of 15N-NCD (3 mM). The 1H-NMR spectrum of the NCD-GG2 complex is presented above the HSQC. Dashed guidelines illustrate the correlation of 1H-NMR and HSQC. A red star highlights a superposition of two peaks. (C) The 3D view of Fig. 2B. The peak highlighted with a red star has approximately twice the intensity of the other peaks. | ||
In a similar manner, the model DNA GG3, comprising three consecutive CGG/CGG units, was subjected to titration with 15N-NCD (Fig. 4A). Corresponding to the results of Fig. 3A, a complex spectrum was observed at the early stages of titration, but it simplified as the titration progressed. Ultimately, with the addition of 15 equivalents of 15N-NCD, the changes in the peaks reached saturation. In Fig. 4B, the 1H–15N HSQC spectrum, capturing the complex of GG3 and 15N-NCD under saturated conditions, is presented. At a glance, nine peaks are visible at (1H, 15N) coordinates: (10.90, 118.6), (10.92, 119.2), (10.94, 119.8), (11.03, 118.9), (11.04, 119.1), (11.04, 119.2), (11.04, 119.8), (11.09, 118.9), (11.48, 119.6). Through a 3D view analysis (Fig. 4C), it was determined that the peaks at (10.92, 119.2), (11.03, 118.9), and (11.09, 118.9) were superpositions of the two peaks. Consequently, twelve cross peaks are observed as amide-protons, indicating the binding of six NCD molecules to GG3.
In these studies, three different DNAs containing one (GG1), two (GG2), and three units (GG3) of CGG/CGG were employed to measure 1H–15N HSQC spectra of the complexes with 15N-labeled NCD. In GG1, the CGG/CGG unit was sandwiched between Watson–Crick base pairs. GG2 had two adjacent CGG/CGG units at one side with Watson–Crick base pairs, and GG3 additionally included an “inner” CGG/CGG unit, sandwiched by two “outer” CGG/CGG units.
Comparison of the cross peaks observed in the HSQC spectra for GG1, GG2, and GG3 with 15N-NCD was conducted (Fig. 5). In the HSQC spectra of GG1, four cross peaks were identified, exhibiting different 1H and 15N chemical shifts (Fig. 5A). These cross peaks were assigned based on prior NMR studies.15 A notable difference in the 1H and 15N chemical shifts between NC1 and NC4 amide N–H was attributed to the adjacent nucleotide bases, such as adenine for NC1 and thymine for NC4.
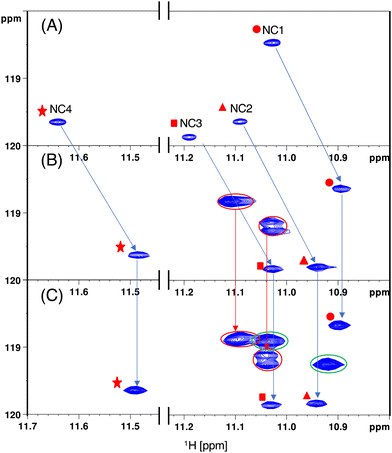 | ||
| Fig. 5 Comparison of 1H–15N HSQC. (A) GG1 and 15N-NCD, (B) GG2 and 15N-NCD, and (C) GG3 and 15N-NCD complex. | ||
In the HSQC spectra of GG2 with 15N-NCD, these four cross peaks shifted upfield in terms of the 1H chemical shift, while no significant changes in the 15N chemical shift were observed. Additionally, three new cross peaks, totaling four N–H, appeared (Fig. 5B, circled with a red line). These seven cross peaks in the GG2–NCD complex displayed almost the same 1H and 15N chemical shifts in the HSQC spectra of the GG3–NCD complex, where two new cross peaks, each containing two N–H, emerged (Fig. 5C, circled with a green line).
The remarkable similarity in the 1H and 15N chemical shifts of the eight NC moieties in both GG2 and GG3 complexes indicated that the chemical environment of these NC moieties remained consistent. This suggests that NCD-binding to the two “outer” CGG/CGG units in GG3 is similar to that in GG2. The two new cross peaks in GG3 are evidently due to NCD binding to the “inner” CGG/CGG unit and have different 1H and 15N chemical shifts, indicating a distinct chemical environment compared to the NC moieties bound to the “outer” unit. These observations strongly indicate that NCD can bind to the contiguous CGG/CGG units in the CGG repeat hairpin, even though the “inner” unit may experience structural perturbations due to NCD binding to neighboring units.
Another crucial observation was made in these studies. In the titration experiments with both GG2 and GG3, the spectrum initially became complicated but eventually converged. Such complexity was not observed in the titration of NCD to GG1.15 The binding pathway of NCD in the presence of contiguous CGG/CGG units likely involves intermediates but converges to a specific structure under equilibrium, suggesting that NCD can likely bind to CGG repeat DNA, even if the repeat is substantial in length.
In conclusion, our investigation focused on the binding mode of NCD to repetitive CGG/CGG units using 15N-labeled NCD and 1H–15N HSQC NMR techniques. By comparing the 1H–15N HSQC spectra of complexes with three DNA models containing one, two, and three CGG/CGG units, we confirmed that NCD can bind to all three CGG/CGG units in GG3. This implies the plausible scenario of NCD binding to long CGG repeat DNA.
Conflicts of interest
There are no conflicts to declare.Notes and references
- A. M. Gacy, G. Goellner, N. Juranić, S. Macura and C. T. McMurray, Cell, 1995, 81, 533–540 CrossRef CAS PubMed.
- Y. Nadel, P. Weisman-Shomer and M. Fry, J. Biol. Chem., 1995, 270, 28970–28977 CrossRef CAS PubMed.
- R. Al-Jarf, A. G. C. de Sá, D. E. V. Pires and D. B. Ascher, J. Chem. Inf. Model., 2021, 61, 3314–3322 CrossRef CAS PubMed.
- S. Kang, A. Jaworski, K. Ohshima and R. D. Wells, Nat. Genet., 1995, 10, 213–218 CrossRef CAS PubMed.
- K. Ohshima and R. D. Wells, J. Biol. Chem., 1997, 272, 16798–16806 CrossRef CAS PubMed.
- A. J. M. H. Verkerk, M. Pieretti, J. S. Sutcliffe, Y. H. Fu, D. P. A. Kuhl, A. Pizzuti, O. Reiner, S. Richards, M. F. Victoria, F. Zhang, B. E. Eussen, G. J. B. van Ommen, L. A. J. Blonden, G. J. Riggins, J. L. Chastain, C. B. Kunst, H. Galjaard, C. Thomas Caskey, D. L. Nelson, B. A. Oostra and S. T. Warran, Cell, 1991, 65, 905–914 CrossRef CAS PubMed.
- C. E. Pearson, K. N. Edamura and J. D. Cleary, Nat. Rev. Genet., 2005, 6, 729–742 CrossRef CAS PubMed.
- M. Nakamori, G. B. Panigrahi, S. Lanni, T. Gall-Duncan, H. Hayakawa, H. Tanaka, J. Luo, T. Otabe, J. Li, A. Sakata, M.-C. Caron, N. Joshi, T. Prasolava, K. Chiang, J.-Y. Masson, M. S. Wold, X. Wang, M. Y. W. T. Lee, J. Huddleston, K. M. Munson, S. Davidson, M. Layeghifard, L.-M. Edward, R. Gallon, M. Santibanez-Koref, A. Murata, M. P. Takahashi, E. E. Eichler, A. Shlien, K. Nakatani, H. Mochizuki and C. E. Pearson, Nat. Genet., 2020, 52, 146–159 CrossRef CAS PubMed.
- Y. Hasuike, H. Tanaka, T. Gall-Duncan, M. Mehkary, K. Nakatani, C. E. Pearson, S. Tsuji, H. Mochizuki and M. Nakamori, Neurobiol. Dis., 2022, 163, 105604 CrossRef CAS PubMed.
- A. Granzhan, N. Kotera and M. P. Teulade-Fichou, Chem. Soc. Rev., 2014, 43, 3630–3665 RSC.
- C. M. Chien, P. C. Wu, R. Satange, C. C. Chang, Z. L. Lai, L. D. Hagler, S. C. Zimmerman and M. H. Hou, J. Am. Chem. Soc., 2020, 142, 11165–11172 CrossRef CAS PubMed.
- W. H. Tseng, C. K. Chang, P. C. Wu, N. J. Hu, G. H. Lee, C. C. Tzeng, S. Neidle and M. H. Hou, Angew. Chem., Int. Ed., 2017, 56, 8761–8765 CrossRef CAS PubMed.
- M. Krafcikova, S. Dzatko, C. Caron, A. Granzhan, R. Fiala, T. Loja, M. P. Teulade-Fichou, T. Fessl, R. Hänsel-Hertsch, J. L. Mergny, S. Foldynova-Trantirkova and L. Trantirek, J. Am. Chem. Soc., 2019, 141, 13281–13285 CrossRef CAS PubMed.
- T. Peng and K. Nakatani, Angew. Chem., Int. Ed., 2005, 44, 7280–7283 CrossRef CAS PubMed.
- T. Yamada, K. Furuita, S. Sakurabayashi, M. Nomura, C. Kojima and K. Nakatani, Nucleic Acids Res., 2022, 50, 9621–9631 CrossRef CAS PubMed.
- W. Saenger, Principles of Nucleic Acid Structure, Springer, New York, New York, NY, 1984 Search PubMed.
- K. Nakatani, H. He, S. Uno, T. Yamamoto and C. Dohno, Curr. Protoc. Nucleic Acid Chem., 2008, 1–21 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cc00544a |
| ‡ Current address: Nucleotide and Peptide Drug Discovery Center, Tokyo Medical and Dental University, Yushima 1-5-45, Bunkyo-ku, Tokyo 113-8519, Japan. |
| This journal is © The Royal Society of Chemistry 2024 |

