Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Organometallic modification confers oligonucleotides new functionalities
Tharun K.
Kotammagari
 ,
Lange Yakubu
Saleh
,
Lange Yakubu
Saleh
 and
Tuomas
Lönnberg
and
Tuomas
Lönnberg
 *
*
Department of Chemistry, University of Turku, Henrikinkatu 2, 20500 Turku, Finland. E-mail: tuanlo@utu.fi
First published on 16th February 2024
Abstract
To improve their properties or to introduce entirely new functionalities, the intriguing scaffolds of nucleic acids have been decorated with various modifications, most recently also organometallic ones. While challenging to introduce, organometallic modifications offer the potential of expanding the field of application of metal-dependent functionalities to metal-deficient conditions, notably those of biological media. So far, organometallic moieties have been utilized as probes, labels and catalysts. This Feature Article summarizes recent efforts and predicts likely future developments in each of these lines of research.
1. Introduction
The programmable target recognition based on the simple rules of Watson–Crick base pairing makes nucleic acids attractive scaffolds for diverse applications, ranging from therapeutic agents1–3 to nanoelectronic components.4–6 The other properties of nucleic acids, however, leave much to be desired from the point of view of many of these applications. Natural oligonucleotides tend to accumulate in the liver and kidneys and get rapidly excreted in urine.7–10 They also do not readily penetrate cell membranes and, once inside, are promptly degraded by nucleases.11,12 The hybridization affinity is insufficient for targets with long double-helical regions, notably miRNAs.13–16 While strongly chromophoric, oligonucleotides do not provide a signal that would stand out against the background of the intracellular medium. Nucleic acid catalysts (ribozymes and DNAzymes) and aptamers would both benefit from a wider range of functional groups than what is provided by the canonical nucleotides.17–19 Electron transfer along DNA double helices has been detected but is insufficient to make DNA useful as a molecular wire.20 All of these issues have been addressed by chemical modifications, with varying degrees of success.21–231.1. Organic vs. metal-carrying modifications
Simple organic modifications are often sufficient for altering the physicochemical properties or enzymatic stability of oligonucleotides. Cellular uptake can be improved by conjugation of hydrophobic groups or cell-penetrating peptides.24–28 Nuclease resistance, in turn, can be achieved through modification of the sugar or phosphate moieties, such as 2′-O-alkylation or replacement by phosphorothioate, respectively.29,30 Glycoclusters show promise in targeting the liver and similar strategies for other organs, utilizing other conjugate groups, can be envisaged.31–33 Organic fluorophores are widely employed in oligonucleotide-based hybridization probes and for visualization in biological samples.34–36 Hybridization affinity can be enhanced through either backbone modifications, such as LNA37 or PNA, or augmenting the base moieties with expanded stacking surface38,39 or additional hydrogen bond donors and acceptors.40–48 Similar strategies have also met with considerable success in increasing the binding affinity and specificity of aptamers.49–57 Chemical modification of ribozymes and DNAzymes17,18 has largely focused on biostability but recently rationally designed changes based on high-resolution 3D structures have been shown to also improve the catalytic activity.58 As long as the modifications are confined to the sugar moieties, such engineered nucleic acid catalysts still usually retain their reliance on metal ion cofactors. Modifications of the base moieties, on the other hand, have afforded a number of metal-independent DNAzymes.59–62Renal clearance can be retarded by increasing the size of the molecule. With oligonucleotides, this concept was first realized by immobilization of thiol-functionalized strands onto gold nanoparticles, giving rise to a spherical nucleic acid.63,64 A metal core is not indispensable, however, and more recently spherical nucleic acids have also been synthesized with various non-metallic cores, such as fullerene or cubic silsesquioxane.65 Even completely coreless ones have been prepared by dissolving the gold nanoparticle core after assembly of the spherical nucleic acid and cross-linking the constituent strands.66
In certain applications, metal complexes offer clear advantages over purely organic modifications. Lanthanide chelates offer longer luminescence lifetime, less concentration quenching and larger Stokes shift than organic fluorophores.67 Analogously, while a number of redox active organic compounds potentially useful as electrochemical labels have been reported,68–72 ferrocene is by far the most widely employed moiety for this purpose.73–77 Artificial ribonucleases bearing a transition metal ion at the catalytic core tend to be more active than their purely organic counterparts.78–83 For converting DNA into a nanowire, coordination of an array of metal ions appears all but indispensable.84–88
1.2. Self-assembly vs. covalent metalation
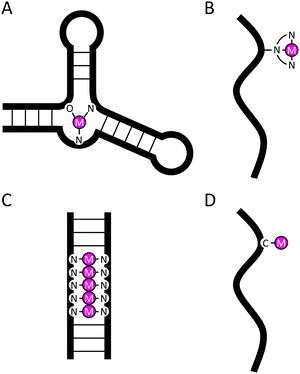
Coordinative complexes between oligonucleotides and metal ions are obtained conveniently through self-assembly provided that the former presents an appropriate high-affinity binding site and the latter is sufficiently labile kinetically. In ribozymes and DNAzymes, such a binding site is formed on folding of the oligonucleotide into a specific tertiary structure, bringing donor atoms from nucleobase and phosphate moieties that are often separated by several residues to converge on a common metal center (Fig. 1(A)).89,90 Artificial nucleases and oligonucleotides labelled with a radiometal or a fluorescent lanthanide ion, in turn, usually exploit conjugation with high-affinity chelating groups (Fig. 1(B)).91,92 Long arrays of metal ions have been formed within double-helical DNA through metal-mediated base pairing (Fig. 1(C)), paving the way to rational design and bottom-up synthesis of well-defined nanowires.93–95 A complementary approach to self-assembly for functionalizing oligonucleotides with metal ions is covalent metalation, i.e. using a stable organometallic bond to “pin down” a metal ion at a predetermined site (Fig. 1(D)).96–100 The merits and limitations of this approach are discussed below. | ||
| Scheme 1 Preparation of oligonucleotides bearing organometallic modification through (A) automated synthesis with organometallic phosphoramidite building blocks,103 (B) post-synthetic metalation of an oligonucleotide in solution105 and (C) on-support conjugation with a previously synthesized organometallic complex.117Reagents and conditions: (a) conventional phosphoramidite coupling with 3-chloroperoxybenzoic acid as the oxidant and no capping step; (b) NH3, MeNH2, H2O, 55 °C, 30 min; (c) Hg(OAc)2, H2O, 60 °C, 16 h; (d) CH2Cl2, 25 °C, 12 h; (e) NH3, MeNH2, H2O, 65 °C, 10 min. | ||
Metalation strategies involving coupling of a previously synthesized organometallic species offer unparalleled control over the site of metalation as long as the oligonucleotide presents a single “handle” with unique reactivity, such as the 5′-hydroxy group in Scheme 1(A) or the aminooxy group in Scheme 1(C). Ligand-directed cyclopalladation can also be carried out with high site-selectivity as the necessary arrangement of an aromatic carbon atom and a proximal soft Lewis base is not found on natural nucleic acids. Similarly, Ag(0) has been deposited exclusively on aldehyde-bearing DNA by a Tollens reaction but the precise structure and extent of metalation remained obscure.122 With mercuration, the situation is different because the C5 atoms of both cytosine and uracil bases readily react with various Hg(II) salts.104 Some artificial nucleobase analogues110 are much more reactive than the natural pyrimidine bases but even so cytosines should be replaced by 5-methylcytosines and uracils by thymines to prevent off-target mercuration. This approach has been successful with short oligonucleotides105,110,123–127 but has never been tested with very long ones.
2. Metal-mediated base pairing of organometallic nucleobase surrogates
While metal-mediated base pairing of organometallic nucleobase analogues was first reported with small molecule model compounds already in the mid-1990s,148 to the best of our knowledge we remain the only group to study such interactions within oligonucleotides. Initially, these studies were motivated by the desire to enhance the hybridization affinity of therapeutic oligonucleotides through metal coordination. Increased melting temperatures have indeed been observed for many short double-105,108,110,125,127,149–151 and triple-helical123,147 oligonucleotides incorporating a single organometallic residue and in one case a palladacyclic splice-switching oligonucleotide modestly outperformed its unmodified counterpart in a human cell line.152 The connection between these two observations, however, remains uncertain. As higher hybridization affinity often comes at the cost of lower sequence selectivity, we were surprised to find that in some cases an organometallic nucleobase surrogate not only increased the melting temperature of a double helix but also robustly discriminated between canonical nucleobases.110,125,127 Inspired by these results, the focus of our studies on oligonucleotides bearing organometallic base modifications has recently been on potential applications in single nucleotide polymorphism (SNP) genotyping.1532.1. Hg(II)-mediated base pairing
The T–Hg(II)–T homo base pair was the first metal-mediated base pair reported and arguably the most thoroughly investigated one.154–157 Coordination of Hg(II) between the N3 atoms of two thymine residues results in a structure with similar dimensions as the canonical Watson–Crick base pairs,155 serving as a good starting point for the design of other Hg(II)-mediated base pairs. Given the relative ease of mercuration of the C5 atom of pyrimidine bases,104 5-mercuricytosine was a natural candidate to be tested first.105 In the syn conformation of pyrimidine nucleosides, C5 assumes the position normally occupied by N3 and thus in a double helix points the Hg(II) ion towards the opposite nucleobase, analogous to the T–Hg(II)–T base pair. This first organometallic nucleoside within an oligonucleotide was later followed by several artificial C-nucleoside analogues retaining the same position of the Hg(II) ion but varying the other substituents or expanding the aromatic ring system.Fig. 2 summarizes the melting temperatures of various 11-mer double-helical oligodeoxyribonucleotides featuring a single organomercury nucleobase in the middle of one strand and any of the four canonical nucleobases in the middle of the other strand, the rest of the sequences being identical in all cases. For reference, respective results on duplexes with thymine in place of the organometallic nucleobase are also included. As expected, with the unmodified duplexes the highest melting temperature was observed with adenine opposite to thymine and all of the mispairs were destabilizing by more than 10 °C (Fig. 2(A)). The addition of 1 equivalent of Hg(II) to the samples led to a significant increase in the melting temperatures of the duplexes containing a T–G or a T–T mispair, consistent with formation of a Hg(II)-mediated base pair (Fig. 2(B)).105 The T–A-containing duplex was still the most stable one and only the T–C mispair led to a sufficiently different (lower) melting temperature to allow reliable SNP identification. In other words, coordinative Hg(II)-mediated base pairing offered no real advantage over Watson–Crick base pairing in this case.
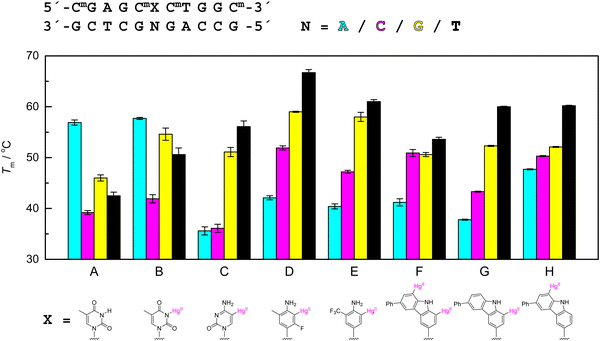 | ||
| Fig. 2 UV melting temperatures of 11-mer oligodeoxyribonucleotide duplexes pairing either (A) thymine through hydrogen bonding, (B) thymine through Hg(II) coordination, (C) 5-mercurycytosine,105 (D) 3-fluoro-2-mercuri-6-methylaniline,110 (E) 2-mercuri-6-trifluoromethylaniline,127 (F) 1,8-dimercuri-6-phenylcarbazole,124 (G) 1-mercuri-6-phenylcarbazole or (H) 8-mercuri-6-phenylcarbazole125 with adenine (cyan), cytosine (magenta), guanine (yellow) or thymine (black). For experimental conditions, see the original publications. | ||
At monomer level, 5-mercuricytidine pairs weakly with adenosine and cytidine and strongly with any nucleoside that can undergo deprotonation on coordination of Hg(II).105 Hybridization properties of the respective 11-mer oligodeoxyribonucleotide reflected these preferences, the most stable duplexes being the ones with thymine or guanine opposite to the organometallic residue (Fig. 2(C)). Pairing with adenine or cytosine, on the other hand, was even more destabilizing than any of the mispairs between canonical nucleobases. Thus a hybridization probe based on Hg(II)-mediated base pairing of 5-mercuricytosine would be able to distinguish reliably between guanine and adenine or thymine and cytosine but not between adenine and cytosine. Comparable (but opposite) discrimination of thymine and cytosine through Ag(I)-mediated base pairing with imidazophenanthroline has been reported within model sequences relevant for the development of breast or pancreatic cancer.158,159
Interestingly, replacing the cytosine ligand with a 3-fluoro-6-methylaniline ligand while retaining the position of the mercuri substituent ortho to the amino substituent markedly increased the melting temperature of all of the corresponding duplexes (Fig. 2(D)).110 Furthermore, the duplex placing cytosine opposite to the organometallic residue was stabilized considerably more than the others, leading to discrimination between all canonical nucleobases in the order of A < C < G < T and with at least 5 °C difference between the melting temperatures. Comparable results have been obtained exploiting coordinative Ag(I)-mediated base pairing between cytosine and any of the canonical nucleobases but in that case, two probes were needed for reliable discrimination, one with α and the other one with β anomer of the cytidine residue at the recognition site.160 Removal of the fluoro substituent from the Watson–Crick face and replacing the methyl group with a trifluoromethyl group did not change the stability order although the melting temperature of the duplex pairing the organometallic residue with thymine decreased somewhat (Fig. 2E).127 Encouraged by these results, we employed 3-fluoro-2-mercuri-6-methylaniline in a molecular beacon-type Förster resonance energy transfer (FRET) probe and were gratified to find a clear correlation between fluorescence emission and melting temperature for each of the nucleobases at the polymorphic site.126
Qualitatively, the stability order of A < C < G < T for the organometallic Hg(II)-mediated base pairs discussed above (Fig. 2(C)–(E)) can be explained in terms of the preference of Hg(II) for anionic ligands and the better fit of the smaller pyrimidine bases within the double-helical environment (Fig. 3). With guanine and thymine, hydrogen bonding between the oxo and amino substituents may provide additional stabilization. To explore Hg(II)-mediated base pairing beyond this paradigm, we expanded the aniline ring to a carbazole ring, with Hg(II) ions at both carbon atoms ortho to the nitrogen atom.124 Melting temperatures of duplexes placing adenine or cytosine opposite to this dimercurated residue (Fig. 2(F)) were comparable to those of the respective duplexes incorporating one of the aniline derivatives (Fig. 2(D) and (E)) but with guanine and, especially, thymine considerable destabilization was observed. In principle, the 1,8-dimercuri-6-phenylcarbazole should be able to form the same kind of Hg(II)-mediated base pairs with guanine and thymine as depicted in Fig. 3 but the large drops in melting temperature suggested a different binding mode. In the case of thymine, DFT optimization yielded a dinuclear base pair with each of the Hg(II) ions coordinated to one of the oxo substituents of thymine, rather than the usual N3. Such a base pair is stronger than one having only one of the Hg(II) ions coordinated to N3 but less compatible with the geometry of the double helix, consistent with the observed decrease in melting temperature. A dinuclear Hg(II)-mediated base pair has also been reported to form between thymine and 1,N6-ethenoadenine within a parallel-stranded double helix.161,162
The carbazole C-nucleoside bearing a Hg(II) ion only at the C1 atom (Fig. 2(G)) structurally resembles its aniline counterparts (Fig. 2(D) and (E)) so it was not unexpected to once again observe a similar pattern of hybridization preferences for the corresponding modified oligonucleotide.125 Curiously, switching the Hg(II) from C1 to C8 had no effect on pairing with guanine or thymine but pairing with adenine and cytosine became much more favorable. The most interesting results, however, were obtained with duplexes placing either 2- or 4-thiothymine opposite to the organometallic residue.125 All of the organomercury carbazole derivatives showed very high affinity towards these rare nucleobases and the C1- and C8-monomercurated ones preferred pairing with 2- and 4-thiothymine, respectively.
2.2. Pd(II)-mediated base pairing
The very first metal-mediated base pair between artificial nucleoside analogues featured Pd(II) as the bridging metal ion.163 Like Pt(II), Pd(II) also exhibits a high affinity towards nitrogen donor atoms of natural nucleic acids.164 The square planar coordination geometry of Pd(II) should be compatible with base stacking within a double helix, at least as long as steric crowding of the ligands is kept to a minimum.165 Finally, aromatic rings bearing suitable directing ligands undergo cyclopalladation under conditions tolerated by nucleic acids, providing relatively easy access to oligonucleotides furnished with organopalladium modifications. All of these factors make Pd(II) a promising candidate for metal-mediated base pairing between organometallic oligonucleotides and their natural counterparts. Compared to Hg(II), the realization of this promise has, however, proven much more challenging.Our first organopalladium oligonucleotide featured a palladacyclic phenylpyridine C-nucleoside analogue (Fig. 4(A)) in the middle of the same sequence as the one used with Hg(II)-mediated base pairing (see above).108 UV melting studies on corresponding duplexes revealed a problem encountered since with most duplexes incorporating a putative Pd(II)-mediated base pair, namely multiphasicity of the melting curves. In the best case the analysis of such curves yields two melting temperatures, with the higher one presumably associated with dissociation of the two strands. Often, however, the overlapping of several melting events prevents reliable determination of the melting temperatures altogether. Furthermore, while Pd(II) is kinetically orders of magnitude more labile than Pt(II),166 in many cases hysteresis between the denaturation and renaturation curves is observed,150,151 indicating that when these processes involve changes in Pd(II)-mediated base pairing, they are too slow to be studied by conventional UV melting temperature measurements. We have explored FRET-based competition assays151 and 15N NMR spectroscopy167 as alternative methods, with moderate success.
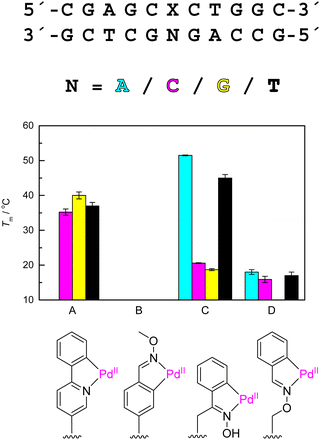 | ||
| Fig. 4 UV melting temperatures of 11-mer oligodeoxyribonucleotide duplexes pairing either (A) phenylpyridine palladacycle108 or (B)–(D) benzaldoxime palladacycles of varying flexibility167,168 with adenine (cyan), cytosine (magenta), guanine (yellow) or thymine (black). Missing columns indicate cases where a sigmoidal melting curve was not observed. In case of multiphasic melting curves, the highest melting temperature is given. For experimental conditions, see the original publications. | ||
Our earlier studies on coordinative Pd(II)-mediated base pairing had revealed that the same base pair could be stabilizing at the end of a double helix but destabilizing in the middle, suggesting suboptimal geometry that is better tolerated outside of the base stack than within.169,170 This turned out to be the case also with the palladacyclic phenylpyridine – 11-mer duplexes incorporating this modification in the middle melted at a lower temperature than their unmodified counterparts108 while the opposite was true for duplexes bearing the same modification at either end.151 Interestingly, base-pairing preferences of the palladacyclic C-nucleoside analogue were also somewhat different depending on whether it was incorporated at the 3′- or 5′-terminus.
Besides placing it at the end of the double helix, the strain caused by a geometrically incompatible metal-mediated base pair could also be alleviated by making the metalated nucleoside analogue more flexible. We tested this approach with three isomeric oligonucleotides bearing oxime palladacyles of varying rigidity as the base moiety of their central nucleoside analogue (Fig. 4(B)–(D)).167,168 For duplexes incorporating the most rigid version (Fig. 4(B)), sigmoidal melting curves were not observed so their stabilities remain obscure. With the most flexible version (Fig. 4(D)), melting temperatures could be determined and were found to be very low. The acetophenone oxime palladacycle (Fig. 4(C)), representing an isomer of intermediate flexibility, was tested as both α and β anomers, with markedly different results. The α anomer favored pairing with thymine and guanine but the resulting duplexes were still less stable than unmodified counterparts featuring a single mismatch in the middle. The β anomer, in turn, was stabilizing when placed opposite to an adenine or thymine residue, the melting temperatures of the corresponding duplexes approaching those of fully matched unmodified duplexes.
3. Oligonucleotides bearing organometallic labels
The properties of some organometallic complexes make them useful as labels to facilitate imaging of oligonucleotides or elucidation of their secondary structures. The field is dominated by studies on ferrocene-labeled electrochemical probes and sensors73–77 but recently luminescent organometallic labels119,171 have also received attention.Ferrocene-labeled oligonucleotides can be detected in femtomolar quantities.111,172 Depending on the intended application, the signal can be rendered either insensitive or sensitive to secondary structure by either separating the label and the oligonucleotide by a long and flexible linker73 or by incorporating the label as part of the backbone.173–175 Changes in secondary structure, in turn, can be induced by hybridization with another nucleic acid,176,177 coordination of metal ions140,178 or aptamer-type interaction with a variety of analytes.179,180 The strong correlation between the oxidation current of a ferrocene label and its proximity to the electrode can be easily harnessed to create electrochemical sensors by immobilizing the labeled oligonucleotide on the surface of the electrode.181 Finally, the oxidation potential of ferrocene can be tweaked by changing the substituents of the cyclopentadiene rings, allowing simultaneous detection on multiple channels.182
Tridentate platinacyclic chelates of 1-(1,2,3-triazol-4-yl)-6-phenylpyridine represent an interesting alternative to organic fluorophores and lanthanide chelates as luminescent labels, combining some of the desirable characteristics of both.183,184 Their luminescence lifetimes are relatively long (although not quite on par with lanthanide chelates), allowing time-resolved experiments. In contrast to the bulky lanthanide chelates, on the other hand, planar platinacycles readily intercalate within the base stack of a double helix, a useful feature for many applications involving nucleic acids. As a step towards such applications, the platinacyclic chelate was recently incorporated into short double-helical oligodeoxynucleotides through either DNA- or GNA-type scaffolds pointing it towards the center of the double helix or through uridine-C5, placing it in the major groove.119 Oligonucleotides placing the platinacycle within the base stack had longer fluorescence lifetimes than those placing it in the major groove and this difference was attributed to shielding from water and triplet oxygen. This interpretation was further borne out by the fact that the shortest lifetimes were observed with corresponding single-stranded oligonucleotides.
4. Oligonucleotides bearing organometallic catalytic moieties
Given the programmable sequence recognition properties, the most obvious and also the most extensively studied use for oligonucleotide catalysts is the cleavage of other nucleic acids.185–191 Efficient artificial nucleases are accessible through the simple and modular approach of covalently tethering a cleaving agent to a guiding oligonucleotide strand. Alternatively, catalytic oligonucleotides can be found through an iterative process known as systematic evolution of ligands by exponential enrichment (SELEX).192,193 In the latter case, the scope of the catalyzed reactions is not limited to cleavage (or formation) of RNA or DNA phosphodiester linkages. Regardless of the design and application, most oligonucleotide catalysts require metal ion cofactors.As discussed above in Section 1.2.3., dissociation under dilute conditions makes most coordinative metal complexes unsuitable for applications in biological media. Organometallic complexes could offer a solution to this problem and we recently set out to test this hypothesis on simple RNA cleavage model systems. The requirement for a hydrolytically stable organometallic complex ruled out the metal ions most extensively studied in the context of RNA cleavage and led us to choose Hg(II) instead. The potential of this overlooked metal ion was first tested in the free form and found to be sufficient to warrant further experiments.194 Replacing free Hg(II) by an arylmercury compound led to lower apparent catalytic activity but the loss was mostly attributable to lower affinity for the scissile phosphodiester, an issue that could be addressed by the inclusion of an appropriate guiding element, such as an oligonucleotide.195
Two oligonucleotide conjugates, differing in the arylmercury group as well as the linker between this moiety and the guiding sequence (Scheme 2), were tested for their ability to catalyze the cleavage of the sole RNA phosphodiester linkage within a complementary 2′-O-methyl-RNA strand.196 Both conjugates were active and the observed rates indicated a significant proximity effect on tethering the cleaving agent to a guiding oligonucleotide. The linker length and flexibility also played a major role, with the conjugate incorporating a triethylene glycol linker being approximately 6 times as active as the one incorporating the shorter and more rigid ethylene glycol oxime linker. While the cleavage efficiency fell short of what has been achieved with the best metal-dependent artificial ribonucleases, we were nevertheless able to demonstrate the feasibility of furnishing oligonucleotides with organometallic catalytic functions.
 | ||
| Scheme 2 Cleavage of the single RNA phosphodiester linkage within a target oligonucleotide by oligonucleotide conjugates of two different arylmercury complexes. | ||
5. Conclusion and outlook on future progression
Redox labeling of oligonucleotides by ferrocene is an established method that has already been exploited in myriad sensor applications. Apart from these, the most successful use of oligonucleotides bearing organometallic modifications has been as hybridization probes for SNP genotyping. Replacing the base moiety at the recognition site by an arylmercury moiety appears particularly promising, allowing robust discrimination of all canonical nucleobases with a single probe. Inexpensive point-of-care SNP genotyping tools based on Hg(II)-mediated base pairing by arylmercury nucleobase surrogates could become reality in near future. Owing to their more complicated coordination chemistry and slower ligand-exchange kinetics, palladacycles are less suitable for this role but have instead shown some promise as modifications of therapeutic oligonucleotides.Interesting properties of organometallic modifications as labels are not limited to redox chemistry, as exemplified by the recent report on oligonucleotides bearing phosphorescent platinacyclic chelates. Many more studies have been published on related nucleoside, nucleotide and dinucleotide analogues,171,197–199 predicting this to be an active field in the coming years. The atomic weights and isotope distributions of many metals forming stable organometallic complexes, notably mercury and the platinum group metals, are very distinct from those of the biogenic elements, possibly allowing laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) or nanoscale secondary ion mass spectrometry (NanoSIMS) imaging of correspondingly modified oligonucleotides.
The field of oligonucleotides featuring organometallic catalytic groups is still in its infancy but the concept has been proven in the case of artificial ribonucleases. The catalytic activity was modest but can likely be improved considerably by optimizing the linker and the overall architecture of the oligonucleotide conjugate. Organometallic modifications could expand the catalytic repertoire of DNAzymes and ribozymes to reactions that have so far remained elusive. Stereoselective C–C cross-couplings appear a particularly attractive application given the ubiquity of palladacycles as pre-catalysts for such reactions.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
L. Y. S. expresses sincere gratitude for the financial support received from the Finnish National Agency for Education (grant number TM-19-11029), Turku University Foundation (decision numbers 080836 and 081007), Otto A. Malm Foundation, and the University of Turku Graduate School. T. K. K. gratefully acknowledges the support from Turku Collegium for Science, Medicine and Technology (TCSMT) and Turku University Foundation (decision number 26005842).Notes and references
- C. I. E. Smith and R. Zain, Annu. Rev. Pharmacol. Toxicol., 2019, 59, 605–630 CrossRef CAS PubMed.
- J. Talap, J. Zhao, M. Shen, Z. Song, H. Zhou, Y. Kang, L. Sun, L. Yu, S. Zeng and S. Cai, J. Pharm. Biomed. Anal., 2021, 206, 114368 CrossRef CAS PubMed.
- H. Adachi, M. Hengesbach, Y. T. Yu and P. Morais, Biomedicines, 2021, 9, 550 CrossRef CAS PubMed.
- S. R. K. Pandian, C. J. Yuan, C. C. Lin, W. H. Wang and C. C. Chang, Adv. Phys. X, 2017, 2, 22–34 CAS.
- L. Hui, R. Bai, H. Liu, L. Hui, R. Bai and H. Liu, Adv. Funct. Mater., 2022, 32, 2112331 CrossRef CAS.
- X. Dai, Q. Li, A. Aldalbahi, L. Wang, C. Fan and X. Liu, Nano Lett., 2020, 20, 5604–5615 CrossRef CAS PubMed.
- H. Takakusa, N. Iwazaki, M. Nishikawa, T. Yoshida, S. Obika and T. Inoue, Nucleic Acid Ther., 2023, 33, 83–94 CrossRef CAS PubMed.
- R. S. Geary, Expert Opin. Drug Metab. Toxicol., 2009, 5, 381–391 CrossRef CAS PubMed.
- P. Andersson and C. Den Besten, Drug Discovery, Royal Society of Chemistry, 2019, vol. 2019, pp. 474–531 Search PubMed.
- L. Weidolf, A. Björkbom, A. Dahlén, M. Elebring, P. Gennemark, M. Hölttä, D. Janzén, X. Q. Li and S. Andersson, Drug Discovery Today, 2021, 26, 2244–2258 CrossRef CAS PubMed.
- M. Gagliardi and A. T. Ashizawa, Biomedicines, 2021, 9, 433 CrossRef CAS PubMed.
- J. P. Bost, H. Barriga, M. N. Holme, A. Gallud, M. Maugeri, D. Gupta, T. Lehto, H. Valadi, E. K. Esbjörner, M. M. Stevens and S. El-Andaloussi, ACS Nano, 2021, 15, 13993–14021 CrossRef CAS PubMed.
- J. Stenvang and S. Kauppinen, Expert Opin. Biol. Ther., 2008, 8, 59–81 CrossRef CAS PubMed.
- E. van Rooij and S. Kauppinen, EMBO Mol. Med., 2014, 6, 851–864 CrossRef CAS PubMed.
- J. Stenvang, A. Petri, M. Lindow, S. Obad and S. Kauppinen, Silence, 2012, 3, 1 CrossRef CAS PubMed.
- Z. Liu, A. Sall and D. Yang, Int. J. Mol. Sci., 2008, 9, 978–999 CrossRef CAS PubMed.
- P. J. Huang and J. Liu, ChemistryOpen, 2020, 9, 1046–1059 CrossRef CAS PubMed.
- M. Hollenstein, Curr. Opin. Chem. Biol., 2019, 52, 93–101 CrossRef CAS PubMed.
- K. Y. Chan, A. B. Kinghorn, M. Hollenstein and J. A. Tanner, ChemBioChem, 2022, 23, e202200006 CrossRef CAS PubMed.
- F. D. Lewis, T. Wu, Y. Zhang, R. L. Letsinger, S. R. Greenfield and M. R. Wasielewski, Science, 1979, 1997(277), 673–676 Search PubMed.
- A. Khvorova and J. K. Watts, Nat. Biotechnol., 2017, 35, 238–248 CrossRef CAS PubMed.
- Y. Kawamoto, Y. Wu, Y. Takahashi and Y. Takakura, Adv. Drug Delivery Rev., 2023, 199, 114872 CrossRef CAS PubMed.
- L. K. McKenzie, R. El-Khoury, J. D. Thorpe, M. J. Damha and M. Hollenstein, Chem. Soc. Rev., 2021, 50, 5126–5164 RSC.
- M. Hawner and C. Ducho, Molecules, 2020, 25, 5963 CrossRef CAS PubMed.
- K. Klabenkova, A. Fokina and D. Stetsenko, Molecules, 2021, 26, 5420 CrossRef CAS PubMed.
- S. Anwar, F. Mir and T. Yokota, Pharmaceutics, 2023, 15, 1130 CrossRef CAS PubMed.
- C. Fàbrega, A. Aviñó, N. Navarro, A. F. Jorge, S. Grijalvo and R. Eritja, Pharmaceutics, 2023, 15, 320 CrossRef PubMed.
- P. Tran, T. Weldemichael, Z. Liu and H. Y. Li, Pharmaceutics, 2022, 2022(14), 342 CrossRef PubMed.
- G. Clavé, M. Reverte, J. J. Vasseur and M. Smietana, RSC Chem. Biol., 2021, 2, 94–150 RSC.
- A. I. Lamond and B. S. Sproat, FEBS Lett., 1993, 325, 123–127 CrossRef CAS PubMed.
- H. Cui, X. Zhu, S. Li, P. Wang and J. Fang, ACS Omega, 2021, 6, 16259–16265 CrossRef CAS PubMed.
- V. Kumar and W. B. Turnbull, Chem. Soc. Rev., 2023, 52, 1273–1287 RSC.
- A. J. Debacker, J. Voutila, M. Catley, D. Blakey and N. Habib, Mol. Ther., 2020, 28, 1759–1771 CrossRef CAS PubMed.
- S. Munan, Y.-T. Chang and A. Samanta, Chem. Commun., 2024, 60, 501–521 RSC.
- B. Juskowiak, Anal. Bioanal. Chem., 2010, 399, 3157–3176 CrossRef PubMed.
- B. Y. Michel, D. Dziuba, R. Benhida, A. P. Demchenko and A. Burger, Front. Chem., 2020, 8, 511052 Search PubMed.
- P. H. Hagedorn, R. Persson, E. D. Funder, N. Albæk, S. L. Diemer, D. J. Hansen, M. R. Møller, N. Papargyri, H. Christiansen, B. R. Hansen, H. F. Hansen, M. A. Jensen and T. Koch, Drug Discovery Today, 2018, 23, 101–114 CrossRef CAS PubMed.
- A. T. Krueger, H. Lu, A. H. F. Lee and E. T. Kool, Acc. Chem. Res., 2006, 40, 141–150 CrossRef PubMed.
- K. L. Seley, Recent Advances in Nucleosides: Chemistry and Chemotherapy, Elsevier, 2002, pp. 299–326 Search PubMed.
- M. B. Danielsen and J. Wengel, Beilstein J. Org. Chem., 2021, 17, 1828–1848 CrossRef CAS PubMed.
- K. Y. Lin and M. D. Matteucci, J. Am. Chem. Soc., 1998, 120, 8531–8532 CrossRef CAS.
- K. H. Scheit and H. R. Rackwitz, Nucleic Acids Res., 1982, 10, 4059–4069 CrossRef CAS PubMed.
- C. J. Wilds, M. A. Maier, V. Tereshko, M. Manoharan and M. Egli, Angew. Chem., Int. Ed., 2002, 41, 115–117 CrossRef CAS PubMed.
- F. Wojciechowski and R. H. E. Hudson, J. Am. Chem. Soc., 2008, 130, 12574–12575 CrossRef CAS PubMed.
- A. M. Varizhuk, T. S. Zatsepin, A. V. Golovin, E. S. Belyaev, Y. I. Kostyukevich, V. G. Dedkov, G. A. Shipulin, G. V. Shpakovski and A. V. Aralov, Bioorg. Med. Chem., 2017, 25, 3597–3605 CrossRef CAS PubMed.
- J. Brzezinska, Z. Gdaniec, L. Popenda and W. T. Markiewicz, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 1163–1170 CrossRef CAS PubMed.
- K. Yamada, Y. Masaki, H. Tsunoda, A. Ohkubo, K. Seio and M. Sekine, Org. Biomol. Chem., 2014, 12, 2255–2262 RSC.
- C. Lou, A. Dallmann, P. Marafini, R. Gao and T. Brown, Chem. Sci., 2014, 5, 3836–3844 RSC.
- J. P. Elskens, J. M. Elskens and A. Madder, Int. J. Mol. Sci., 2020, 21, 4522 CrossRef CAS PubMed.
- D. Ji, H. Feng, S. W. Liew and C. K. Kwok, Trends Biotechnol., 2023, 41, 1360–1384 CrossRef CAS PubMed.
- R. Oliveira, E. Pinho, A. L. Sousa, J. J. DeStefano, N. F. Azevedo and C. Almeida, Trends Biotechnol., 2022, 40, 549–563 CrossRef CAS PubMed.
- B. N. Gawande, J. C. Rohloff, J. D. Carter, I. Von Carlowitz, C. Zhang, D. J. Schneider and N. Janjic, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 2898–2903 CrossRef CAS PubMed.
- A. Lozoya-Colinas, Y. Yu and J. C. Chaput, J. Am. Chem. Soc., 2023, 145, 25789–25796 CrossRef CAS PubMed.
- B. Majumdar, D. Sarma, Y. Yu, A. Lozoya-Colinas and J. C. Chaput, RSC Chem. Biol., 2024, 5, 41–48 RSC.
- Y. W. Cheung, P. Röthlisberger, A. E. Mechaly, P. Weber, F. Levi-Acobas, Y. Lo, A. W. C. Wong, A. B. Kinghorn, A. Haouz, G. Paul Savage, M. Hollenstein and J. A. Tanner, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 16790–16798 CrossRef CAS PubMed.
- M. Kimoto, H. P. Tan, K. I. Matsunaga, N. A. Binte Mohd Mislan, G. Kawai and I. Hirao, J. Am. Chem. Soc., 2023, 145, 20432–20441 CrossRef CAS PubMed.
- K. I. Matsunaga, M. Kimoto, V. W. Lim, H. P. Tan, Y. Q. Wong, W. Sun, S. Vasoo, Y. S. Leo and I. Hirao, Nucleic Acids Res., 2021, 49, 11407–11424 CrossRef CAS PubMed.
- K. Chiba, T. Yamaguchi and S. Obika, Chem. Sci., 2023, 14, 7620–7629 RSC.
- S. Paul, A. A. W. L. Wong, L. T. Liu and D. M. Perrin, ChemBioChem, 2022, 23, e202100600 CrossRef CAS PubMed.
- Y. Wang, E. Liu, C. H. Lam and D. M. Perrin, Chem. Sci., 2018, 9, 1813–1821 RSC.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, ACS Comb. Sci., 2013, 15, 174–182 CrossRef CAS PubMed.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, ChemBioChem, 2009, 10, 1988–1992 CrossRef CAS PubMed.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- C. A. Mirkin and S. H. Petrosko, ACS Nano, 2023, 17, 16291–16307 CrossRef CAS PubMed.
- H. Li, B. Zhang, X. Lu, X. Tan, F. Jia, Y. Xiao, Z. Cheng, Y. Li, D. O. Silva, H. S. Schrekker, K. Zhang and C. A. Mirkin, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4340–4344 CrossRef CAS PubMed.
- J. I. Cutler, K. Zhang, D. Zheng, E. Auyeung, A. E. Prigodich and C. A. Mirkin, J. Am. Chem. Soc., 2011, 133, 9254–9257 CrossRef CAS PubMed.
- U. Cho and J. K. Chen, Cell Chem. Biol., 2020, 27, 921–936 CrossRef CAS PubMed.
- H. Cahová, L. Havran, P. Brázdilová, H. Pivoňková, R. Pohl, M. Fojta and M. Hocek, Angew. Chem., Int. Ed., 2008, 47, 2059–2062 CrossRef PubMed.
- A. Simonova, J. Balintová, R. Pohl, L. Havran, M. Fojta and M. Hocek, ChemPlusChem, 2014, 79, 1703–1712 CrossRef CAS.
- J. Balintová, M. Plucnara, P. Vidláková, R. Pohl, L. Havran, M. Fojta and M. Hocek, Chem. – Eur. J., 2013, 19, 12720–12731 CrossRef PubMed.
- J. Balintová, J. Špaček, R. Pohl, M. Brázdová, L. Havran, M. Fojta and M. Hocek, Chem. Sci., 2015, 6, 575–587 RSC.
- N. A. Whittemore, A. N. Mullenix, G. B. Inamati, M. Manoharan, P. D. Cook, A. A. Tuinman, D. C. Baker and J. Q. Chambers, Bioconjugate Chem., 1999, 10, 261–270 CrossRef CAS PubMed.
- T. Ihara, Y. Maruo, S. Takenaka and M. Takagi, Nucleic Acids Res., 1996, 24, 4273–4280 CrossRef CAS PubMed.
- M. Ortiz, M. Jauset-Rubio, O. Trummer, I. Foessl, D. Kodr, J. L. Acero, M. L. Botero, P. Biggs, D. Lenartowicz, K. Trajanoska, F. Rivadeneira, M. Hocek, B. Obermayer-Pietsch and C. K. O’Sullivan, ACS Cent. Sci., 2023, 9, 1591–1602 CrossRef CAS PubMed.
- M. Ortiz, M. Jauset-Rubio, D. Kodr, A. Simonova, M. Hocek and C. K. O’Sullivan, Biosens. Bioelectron., 2022, 198, 113825 CrossRef CAS PubMed.
- M. Ortiz, M. Jauset-Rubio, V. Skouridou, D. Machado, M. Viveiros, T. G. Clark, A. Simonova, D. Kodr, M. Hocek and C. K. O’Sullivan, ACS Sens., 2021, 6, 4398–4407 CrossRef CAS PubMed.
- C. Figazzolo, Y. Ma, J. H. R. Tucker and M. Hollenstein, Org. Biomol. Chem., 2022, 20, 8125–8135 RSC.
- M. Murtola, M. Wenska and R. Strömberg, J. Am. Chem. Soc., 2010, 132, 8984–8990 CrossRef CAS PubMed.
- M. Murtola, A. Ghidini, P. Virta and R. Strömberg, Molecules, 2017, 22, 1856 CrossRef PubMed.
- D. Desbouis, I. P. Troitsky, M. J. Belousoff, L. Spiccia and B. Graham, Coord. Chem. Rev., 2012, 256, 897–937 CrossRef CAS.
- J. Suh, Comprehensive Inorganic Chemistry II: From Elements to Applications, Elsevier, 2nd edn, 2013, vol. 6, pp. 779–803 Search PubMed.
- J. R. Morrow and O. Iranzo, Curr. Opin. Chem. Biol., 2004, 8, 192–200 CrossRef CAS PubMed.
- M. Diez-Castellnou, G. Salassa, F. Mancin and P. Scrimin, Front. Chem., 2019, 7, 469 CrossRef CAS PubMed.
- P. C. Chang, C. Y. Chang, W. Bin Jian, C. J. Yuan, Y. C. Chen and C. C. Chang, Nano Res., 2019, 12, 1293–1300 CrossRef CAS.
- S. Hensel, K. Eckey, P. Scharf, N. Megger, U. Karst and J. Müller, Chem. – Eur. J., 2017, 23, 10244–10248 CrossRef CAS PubMed.
- H.-A. Wagenknecht, Angew. Chem., Int. Ed., 2003, 42, 3204–3206 CrossRef CAS PubMed.
- A. Pérez-Romero, A. Domínguez-Martín, S. Galli, N. Santamaría-Díaz, O. Palacios, J. A. Dobado, M. Nyman and M. A. Galindo, Angew. Chem., Int. Ed., 2021, 60, 10089–10094 CrossRef PubMed.
- F. Linares, E. García-Fernández, F. J. López-Garzón, M. Domingo-García, A. Orte, A. Rodríguez-Díeguez and M. A. Galindo, Chem. Sci., 2019, 10, 1126–1137 RSC.
- K. Hwang, P. Hosseinzadeh and Y. Lu, Inorg. Chim. Acta, 2016, 452, 12–24 CrossRef CAS PubMed.
- W. Zhou and J. Liu, Metallomics, 2018, 10, 30–48 CrossRef CAS PubMed.
- Z. J. Lesnikowski, Curr. Org. Chem., 2007, 11, 355–381 CrossRef CAS.
- Y. Liu, G. Liu and D. J. Hnatowich, Materials, 2010, 3, 3204 CrossRef CAS.
- A. Ono, H. Kanazawa, H. Ito, M. Goto, K. Nakamura, H. Saneyoshi and J. Kondo, Angew. Chem., Int. Ed., 2019, 58, 16835–16838 CrossRef CAS PubMed.
- J. Kondo, Y. Tada, T. Dairaku, Y. Hattori, H. Saneyoshi, A. Ono and Y. Tanaka, Nat. Chem., 2017, 9, 956–960 CrossRef CAS PubMed.
- G. H. Clever and T. Carell, Angew. Chem., Int. Ed., 2007, 46, 250–253 CrossRef CAS PubMed.
- D. Ukale, S. Maity, M. Hande and T. Lönnberg, Synlett, 2019, 1733–1737 CAS.
- T. A. Lönnberg, M. A. Hande and D. U. Ukale, Comprehensive Organometallic Chemistry IV, Elsevier, 2022, pp. 146–182 Search PubMed.
- J.-L. H. A. Duprey and J. H. R. Tucker, Chem. Lett., 2013, 43, 157–163 CrossRef.
- K. Kowalski, Coord. Chem. Rev., 2021, 432, 213705 CrossRef CAS.
- A. Collado, M. Gómez-Gallego and M. A. Sierra, Eur. J. Org. Chem., 2018, 1617–1623 CrossRef CAS.
- T. S. Zatsepin, S. Y. Andreev, T. Hianik and T. S. Oretskaya, Russ. Chem. Rev., 2003, 72, 537–554 CrossRef CAS.
- R. C. Mucic, M. K. Herrlein, C. A. Mirkin and R. L. Letsinger, Chem. Commun., 1996, 555–557 RSC.
- H. Dougan, J. B. Hobbs, J. I. Weitz and D. M. Lyster, Nucleic Acids Res., 1997, 25, 2897–2901 CrossRef CAS PubMed.
- R. M. K. Dale, E. Martin, D. C. Livingston and D. C. Ward, Biochemistry, 1975, 14, 2447–2457 CrossRef CAS PubMed.
- D. Ukale, V. S. Shinde and T. Lönnberg, Chem. – Eur. J., 2016, 22, 7917–7923 CrossRef CAS PubMed.
- D. Ukale and T. Lönnberg, ChemBioChem, 2021, 22, 1733–1739 CrossRef CAS PubMed.
- A. C. Cope and R. W. Siekman, J. Am. Chem. Soc., 1965, 87, 3272–3273 CrossRef CAS.
- S. K. Maity and T. Lönnberg, Chem. – Eur. J., 2018, 24, 1274–1277 CrossRef CAS PubMed.
- M. Martín-Ortíz, M. Gómez-Gallego, C. Ramírez de Arellano and M. A. Sierra, Chem. – Eur. J., 2012, 18, 12603–12608 CrossRef PubMed.
- A. Aro-Heinilä, T. Lönnberg and P. Virta, Bioconjugate Chem., 2019, 30, 2183–2190 CrossRef PubMed.
- S. Takenaka, Y. Uto, H. Kondo, T. Ihara and M. Takagi, Anal. Biochem., 1994, 218, 436–443 CrossRef CAS PubMed.
- S. C. Hillier, C. G. Frost, A. T. A. Jenkins, H. T. Braven, R. W. Keay, S. E. Flower and J. M. Clarkson, Bioelectrochemistry, 2004, 63, 307–310 CrossRef CAS PubMed.
- A. Anne and C. Demaille, J. Am. Chem. Soc., 2006, 128, 542–557 CrossRef CAS PubMed.
- A. Anne, A. Bouchardon and J. Moiroux, J. Am. Chem. Soc., 2003, 125, 1112–1113 CrossRef CAS PubMed.
- E. E. Ferapontova, E. M. Olsen and K. V. Gothelf, J. Am. Chem. Soc., 2008, 130, 4256–4258 CrossRef CAS PubMed.
- D. Ge and R. Levicky, Chem. Commun., 2010, 46, 7190–7192 RSC.
- S. K. Maity and T. A. Lönnberg, ACS Omega, 2019, 4, 18803–18808 CrossRef CAS PubMed.
- J. Moreau, N. Dendane, B. Schöllhorn, N. Spinelli, C. Fave and E. Defrancq, Bioorg. Med. Chem. Lett., 2013, 23, 955–958 CrossRef CAS PubMed.
- F. Boisten, I. Maisuls, T. Schäfer, C. A. Strassert and J. Müller, Chem. Sci., 2023, 14, 2399–2404 RSC.
- C. Ligeour, A. Meyer, J.-J. Vasseur and F. Morvan, Eur. J. Org. Chem., 2012, 1851–1856 CrossRef CAS.
- K. Tanaka, G. H. Clever, Y. Takezawa, Y. Yamada, C. Kaul, M. Shionoya and T. Carell, Nat. Nanotechnol., 2006, 1, 190–194 CrossRef CAS PubMed.
- G. A. Burley, J. Gierlich, M. R. Mofid, H. Nir, S. Tal, Y. Eichen and T. Carell, J. Am. Chem. Soc., 2006, 128, 1398–1399 CrossRef CAS PubMed.
- D. U. Ukale and T. Lönnberg, Angew. Chem., Int. Ed., 2018, 57, 16171–16175 CrossRef CAS PubMed.
- D. U. Ukale, P. Tähtinen and T. Lönnberg, Chem. – Eur. J., 2020, 26, 2164–2168 CrossRef CAS PubMed.
- T. K. Kotammagari, P. Tähtinen and T. Lönnberg, Chem. – Eur. J., 2022, 28, e202202530 CrossRef CAS PubMed.
- A. Aro-Heinilä, T. Lönnberg and P. M. Virta, ChemBioChem, 2020, 22, 354–358 CrossRef PubMed.
- A. Aro-Heinilä, A. Lepistö, A. Äärelä, T. A. Lönnberg and P. Virta, J. Org. Chem., 2022, 87, 137–146 CrossRef PubMed.
- D. Liu, Q. Xia, D. Ding and W. Tan, Front. Bioeng. Biotechnol., 2022, 10, 986412 CrossRef PubMed.
- O. Luige, K. Karalė, P. P. Bose, M. Bollmark, U. Tedebark, M. Murtola and R. Strömberg, RSC Adv., 2022, 12, 5398–5406 RSC.
- O. Luige, P. P. Bose, R. Stulz, P. Steunenberg, O. Brun, S. Andersson, M. Murtola and R. Strömberg, Chem. Commun., 2021, 57, 10911–10914 RSC.
- S. W. Santoro, G. F. Joyce, K. Sakthivel, S. Gramatikova and C. F. Barbas, J. Am. Chem. Soc., 2000, 122, 2433–2439 CrossRef CAS PubMed.
- S. Taherpour, O. Golubev and T. Lönnberg, Inorg. Chim. Acta, 2016, 452, 43–49 CrossRef CAS.
- E. Mutti, M. Hunger, S. Fedosov, E. Nexo and B. Kräutler, ChemBioChem, 2017, 18, 2280–2291 CrossRef CAS PubMed.
- M. Hunger, E. Mutti, A. Rieder, B. Enders, E. Nexo and B. Kräutler, Chem. – Eur. J., 2014, 20, 13103–13107 CrossRef CAS PubMed.
- A. B. Olejniczak, R. Kierzek, E. Wickstrom and Z. J. Lesnikowski, J. Organomet. Chem., 2013, 747, 201–210 CrossRef CAS.
- A. B. Olejniczak, J. Plešek, O. Křiž and Z. J. Lesnikowski, Angew. Chem., Int. Ed., 2003, 42, 5740–5743 CrossRef CAS PubMed.
- A. M. Debela, M. Ortiz, V. Beni, S. Thorimbert, D. Lesage, R. B. Cole, C. K. O’Sullivan and B. Hasenknopf, Chem. – Eur. J., 2015, 21, 17721–17727 CrossRef CAS PubMed.
- M. Ortiz, A. M. Debela, M. Svobodova, S. Thorimbert, D. Lesage, R. B. Cole, B. Hasenknopf and C. K. O’Sullivan, Chem. – Eur. J., 2017, 23, 10597–10603 CrossRef CAS PubMed.
- W. Zhou, R. Saran and J. Liu, Chem. Rev., 2017, 117, 8272–8325 CrossRef CAS PubMed.
- M. Jarczewska, A. Szymczyk, J. Zajda, M. Olszewski, R. Ziółkowski and E. Malinowska, Molecules, 2022, 27, 7481 CrossRef CAS PubMed.
- J. Xu, R. Jiang, Y. Feng, Z. Liu, J. Huang, C. Ma and K. Wang, Coord. Chem. Rev., 2022, 459, 214453 CrossRef CAS.
- X. Mao, M. Liu, Q. Li, C. Fan and X. Zuo, JACS Au, 2022, 2, 2381–2399 CrossRef CAS PubMed.
- Y. Takezawa, L. Hu, T. Nakama, M. Shionoya and M. Shionoya, Angew. Chem., Int. Ed., 2020, 59, 21488–21492 CrossRef CAS PubMed.
- T. Nakama, Y. Takezawa, D. Sasaki and M. Shionoya, J. Am. Chem. Soc., 2020, 142, 10153–10162 CrossRef CAS PubMed.
- Y. Takezawa, K. Mori, W. E. Huang, K. Nishiyama, T. Xing, T. Nakama and M. Shionoya, Nat. Commun., 2023, 14, 1–10 Search PubMed.
- M. H. Heddinga and J. Müller, Org. Biomol. Chem., 2022, 20, 4787–4793 RSC.
- D. U. Ukale and T. Lönnberg, ChemBioChem, 2018, 19, 1096–1101 CrossRef CAS PubMed.
- F. Zamora, M. Sabat and B. Lippert, Inorg. Chem., 1996, 35, 4858–4864 CrossRef CAS PubMed.
- M. Hande, S. Maity and T. Lönnberg, Int. J. Mol. Sci., 2018, 19, 1588 CrossRef PubMed.
- H. Räisälä and T. Lönnberg, Chem. – Eur. J., 2019, 25, 4751–4756 CrossRef PubMed.
- M. Hande, S. Maity and T. Lönnberg, J. Inorg. Biochem., 2021, 222, 111506 CrossRef CAS PubMed.
- M. Hande, O. Saher, K. E. Lundin, C. I. E. Smith, R. Zain and T. Lönnberg, Molecules, 2019, 24, 1180 CrossRef PubMed.
- T. Lönnberg, Metal Ions Life Sci., 2023, 25, 211–227 Search PubMed.
- S. Katz, Biochim. Biophys. Acta, Spec. Sect. Lipids Relat. Subj., 1963, 68, 240–253 CrossRef CAS.
- J. Kondo, T. Yamada, C. Hirose, I. Okamoto, Y. Tanaka and A. Ono, Angew. Chem., Int. Ed., 2014, 53, 2385–2388 CrossRef CAS PubMed.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa, T. Fujimoto, T. Machinami and A. Ono, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed.
- Y. Tanaka, S. Oda, H. Yamaguchi, Y. Kondo, C. Kojima and A. Ono, J. Am. Chem. Soc., 2007, 129, 244–245 CrossRef CAS PubMed.
- B. Jash, P. Scharf, N. Sandmann, C. Fonseca Guerra, D. A. Megger and J. Müller, Chem. Sci., 2017, 8, 1337–1343 RSC.
- B. Jash and J. Müller, Eur. J. Inorg. Chem., 2017, 3857–3861 CrossRef CAS.
- X. Guo and F. Seela, Chem. – Eur. J., 2017, 23, 11776–11779 CrossRef CAS PubMed.
- S. Mandal, M. Hebenbrock and J. Müller, Angew. Chem., Int. Ed., 2016, 55, 15520–15523 CrossRef CAS PubMed.
- J. Bachmann, I. Schönrath, J. Müller and N. L. Doltsinis, Molecules, 2020, 25, 4942 CrossRef CAS PubMed.
- K. Tanaka and M. Shionoya, J. Org. Chem., 1999, 64, 5002–5003 CrossRef CAS PubMed.
- R. B. Martin, Acc. Chem. Res., 1985, 18, 32–38 CrossRef CAS.
- J. Müller, E. Freisinger, P. Lax, D. A. Megger and F.-A. Polonius, Inorg. Chim. Acta, 2007, 360, 255–263 CrossRef.
- F. Basolo, H. B. Gray and R. G. Pearson, J. Am. Chem. Soc., 1960, 82, 4200–4203 CrossRef CAS.
- M. Toiviala, V. Kleemola, S. Maity and T. Lönnberg, Eur. J. Org. Chem., 2023, e202201443 CrossRef CAS.
- S. K. Maity, M. A. Hande and T. Lönnberg, ChemBioChem, 2020, 21, 2321–2328 CrossRef CAS PubMed.
- S. Taherpour, H. Lönnberg and T. Lönnberg, Org. Biomol. Chem., 2013, 11, 991–1000 RSC.
- O. Golubev, G. Turc and T. Lönnberg, J. Inorg. Biochem., 2016, 155, 36–43 CrossRef CAS PubMed.
- J. Skiba, A. Kowalczyk, A. Gorski, N. Dutkiewicz, M. Gapińska, J. Stróżek, K. Woźniak, D. Trzybiński and K. Kowalski, Dalton Trans., 2023, 52, 1551–1567 RSC.
- I. Magriñá, A. Toldrà, M. Campàs, M. Ortiz, A. Simonova, I. Katakis, M. Hocek and C. K. O’Sullivan, Biosens. Bioelectron., 2019, 134, 76–82 CrossRef PubMed.
- G. Chatelain, C. Chaix, H. Brisset, C. Moustrou, F. Fages and B. Mandrand, Sens. Actuators, B, 2008, 132, 439–442 CrossRef CAS.
- G. Chatelain, H. Brisset and C. Chaix, New J. Chem., 2009, 33, 1139–1147 RSC.
- T. Ihara, D. Sasahara, M. Shimizu and A. Jyo, Supramol. Chem., 2009, 21, 207–217 CrossRef CAS.
- M. Trotter, N. Borst, R. Thewes and F. von Stetten, Biosens. Bioelectron., 2020, 154, 112069 CrossRef CAS PubMed.
- E. E. Ferapontova, Annu. Rev. Anal. Chem., 2018, 11, 197–218 CrossRef CAS PubMed.
- A.-E. Radi and C. K. O’Sullivan, Chem. Commun., 2006, 3432–3434 RSC.
- S. Arimoto, K. Shimono, T. Yasukawa, F. Mizutani and T. Yoshioka, Anal. Sci., 2016, 32, 469–472 CrossRef PubMed.
- K. Han, T. Liu, Y. Wang and P. Miao, Rev. Anal. Chem., 2016, 35, 201–211 CrossRef CAS.
- C. Fan, K. W. Plaxco and A. J. Heeger, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9134 CrossRef CAS PubMed.
- A. Simonova, I. Magriñá, V. Sýkorová, R. Pohl, M. Ortiz, L. Havran, M. Fojta, C. K. O’Sullivan and M. Hocek, Chem. – Eur. J., 2020, 26, 1286–1291 CrossRef CAS PubMed.
- M. Hebenbrock, D. González-Abradelo, A. Hepp, J. Meadowcroft, N. Lefringhausen, C. A. Strassert and J. Müller, Inorg. Chim. Acta, 2021, 516, 119988 CrossRef CAS.
- M. Hebenbrock, L. Stegemann, J. Kösters, N. L. Doltsinis, J. Müller and C. A. Strassert, Dalton Trans., 2017, 46, 3160–3169 RSC.
- Y. Staroseletz, S. Gaponova, O. Patutina, E. Bichenkova, B. Amirloo, T. Heyman, D. Chiglintseva and M. Zenkova, Molecules, 2021, 26, 1732 CrossRef CAS PubMed.
- A. Kuzuya and M. Komiyama, Curr. Org. Chem., 2007, 11, 1450–1459 CrossRef CAS.
- L. Koroleva, I. Serpokrylova, V. Vlassov and V. Silnikov, Protein Pept. Lett., 2007, 14, 151–163 CrossRef CAS PubMed.
- A. Ghidini, M. Murtola and R. Strömberg, DNA in Supramolecular Chemistry and Nanotechnology, 2015, pp. 158–171 Search PubMed.
- T. Niittymaki and H. Lönnberg, Org. Biomol. Chem., 2006, 4, 15–25 RSC.
- Y. Aiba, J. Sumaoka and M. Komiyama, Chem. Soc. Rev., 2011, 40, 5657–5668 RSC.
- P. Muangkaew and T. Vilaivan, Bioorg. Med. Chem. Lett., 2020, 30, 127064 CrossRef CAS PubMed.
- S. K. Silverman, Trends Biochem. Sci., 2016, 41, 595–609 CrossRef CAS PubMed.
- M. Hollenstein, Molecules, 2015, 20, 20777–20804 CrossRef CAS PubMed.
- L. Y. Saleh, M. Ora and T. Lönnberg, Catalysts, 2020, 10, 219 CrossRef CAS.
- L. Y. Saleh, M. Ora and T. Lönnberg, ChemBioChem, 2021, 22, 1761–1764 CrossRef CAS PubMed.
- L. Y. Saleh, M. Ora and T. Lönnberg, J. Inorg. Biochem., 2023, 247, 112331 CrossRef CAS PubMed.
- M. Valencia, M. Martín-Ortiz, M. Gómez-Gallego, C. Ramírez de Arellano and M. A. Sierra, Chem. – Eur. J., 2014, 20, 3831–3838 CrossRef CAS PubMed.
- C. Lorenzo-Aparicio, S. Moreno-Blázquez, M. Oliván, M. A. Esteruelas, M. Gómez Gallego, P. García-Alvarez, J. A. Cabeza and M. A. Sierra, Inorg. Chem., 2023, 62, 8232–8248 CrossRef CAS PubMed.
- C. Lorenzo-Aparicio, M. Gómez Gallego, C. Ramírez de Arellano and M. A. Sierra, Dalton Trans., 2022, 51, 5138–5150 RSC.
| This journal is © The Royal Society of Chemistry 2024 |