Open Access Article
Open Access ArticleEffects of sulfoxide and sulfone sidechain–backbone hydrogen bonding on local conformations in peptide models†
Dayi
Liu
 a,
Sylvie
Robin
a,
Sylvie
Robin
 ab,
Eric
Gloaguen
ab,
Eric
Gloaguen
 c,
Valérie
Brenner
c,
Valérie
Brenner
 d,
Michel
Mons
d,
Michel
Mons
 *e and
David J.
Aitken
*e and
David J.
Aitken
 *a
*a
aUniversité Paris-Saclay, CNRS, ICMMO, Orsay 91400, France. E-mail: david.aitken@universite-paris-saclay.fr
bUniversité Paris Cité, Faculté de Pharmacie, Paris 75006, France
cUniversité Paris-Saclay, CNRS, ISMO, Orsay 91400, France
dUniversité Paris-Saclay, CEA, DRF, Gif-sur-Yvette 91191, France
eUniversité Paris-Saclay, CEA, LIDYL, Gif-sur-Yvette 91191, France. E-mail: michel.mons@cea.fr
First published on 15th January 2024
Abstract
We examine peptide model systems designed to probe short-range N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S sidechain–backbone hydrogen bonding involving amino acid residues with sidechain sulfoxide or sulfone functional groups and its effects on local conformations. A strong 7-membered ring hydrogen bond of this type accompanies an intra-residue N–H⋯O
S sidechain–backbone hydrogen bonding involving amino acid residues with sidechain sulfoxide or sulfone functional groups and its effects on local conformations. A strong 7-membered ring hydrogen bond of this type accompanies an intra-residue N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C interaction and stabilizes an extended backbone conformation in preference to classical folded structures.
C interaction and stabilizes an extended backbone conformation in preference to classical folded structures.
Sulfoxides and sulfones are immensely important in chemistry.1 They play useful roles as reagents or additives in organic synthesis,2a–e and in the development of new materials,3a–c and they appear ubiquitously in numerous small molecule therapeutics with wide-ranging activities against human pathologies such as cancer, diabetes, Alzheimer's disease and leprosy.4a–d
Sulfoxide groups appear in an array of natural products, notably in the oxidized sidechains of residues present in some macrocyclic peptide families, including cyclolinopeptides,5 cycloleonuripeptides,6 axinellasins,7 and phakellistatins.8 An oxidized tryptathionine bridge is a unique structural feature of the notorious amatoxin bicyclic octapeptides present in death cap mushrooms.9 While not part of the proteinogenic machinery, methionine sulfoxide is considered highly important in biological systems,10 and the redox system relating it with methionine may be involved in protein activity regulation and serve as a protective antioxidant.11,12 The related compound S-methylcysteine sulfoxide is a phytoalexin with beneficial effects on human health.13
The occurrence of sidechain sulfone moieties in native peptides is rarer, and may in some cases be an artefact of isolation or storage.14 Nonetheless, such residues are important in synthetic peptides, for example in the activation of the anti-cancer prodrug canfosfamide,15 in the modulation of peptide secondary structure16 and bioactivity,17,18 or in substrate discrimination by the enzyme neutrophil elastase.19
Sidechain–backbone interactions, notably via hydrogen bonds (H-bonds), are of considerable importance in determining secondary and tertiary structures.20–23 Despite their prevalence in native and synthetic peptides, very little is known about the conformational effects imparted by sulfoxides or sulfones in peptides. In an isolated case, the oxidation of a macrocyclic thioether derived from a tripeptide was shown to induce a β-turn conformation stabilized by a transannular N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S H-bond implicating a sulfoxide or sulfone motif.24 However, no studies have been designed to assess the intrinsic propensity of amino acid residues bearing sidechain sulfoxide or sulfone groups to form short-range sidechain–backbone interactions with local NH H-bond donors.
S H-bond implicating a sulfoxide or sulfone motif.24 However, no studies have been designed to assess the intrinsic propensity of amino acid residues bearing sidechain sulfoxide or sulfone groups to form short-range sidechain–backbone interactions with local NH H-bond donors.
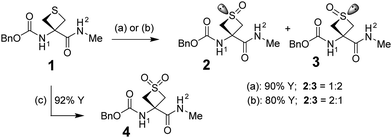
The peptide model Cbz-Attc-NHMe 1 (Attc = 3-aminothietane-3-carboxylic acid) featuring a 4-membered thiacycle appeared to be an attractive platform for the display of a sulfoxide group with a specifically cis or trans geometry (where these terms refer to the relative disposition of the nitrogen and the sulfoxide oxygen on the 4-membered ring) as well as a sulfone group. It was previously observed for compound 1 that a combination of C6γ (sidechain) and C5 (backbone) H-bonds stabilized a predominant extended C5–C6γ local conformation,25 whereas the 5- or 6-membered thiacyclic ring homologs preferred folded δ conformations with π-amide interactions,26 or C7 (backbone) H-bonds typical of γ-turns.27
Compound 1 was subjected to mild oxidation using either hydrogen peroxide or mCPBA to provide a mixture of the trans-sulfoxide Cbz-Attc(trans-O)–NHMe 2 and the cis-sulfoxide Cbz-Attc(cis-O)–NHMe 3 in good yield (Scheme 1). The low diastereoselectivity was of no consequence for our purposes, since both 2 and 3 were required for study and were easily separated by chromatography. Sulfone Cbz-Attc(O,O)–NHMe 4 was prepared from 1 using 2 equiv. of mCPBA.
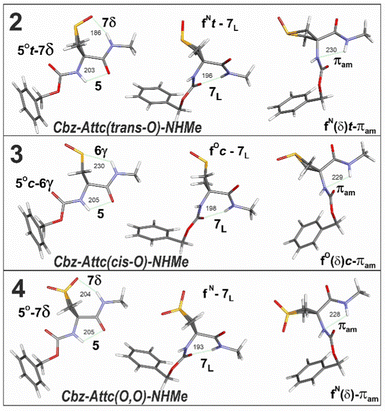
Quantum chemistry structural modelling studies of molecules 2–4 (Fig. 1) were carried out in the gas phase at DFT-D level and in chloroform solution accounted for by a polarizable continuum (for details see ESI,† Section S2.1). The structures were only marginally influenced by solvation (see Fig. S2.2 and S2.3 for comparisons, ESI†), although the energetics (Fig. 2) showed a significant dependence on the environment.
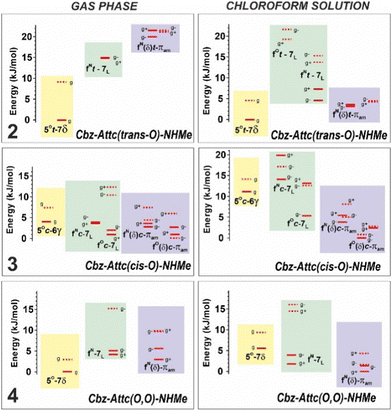 | ||
| Fig. 2 Energetics of the significant conformations of 2–4 in the gas phase (left panels) and in chloroform solution (right panels) at 300 K. See Fig. 1 caption for conformational terminology and ESI,† Section S2.2 for details. Plain and dotted levels refer to trans and cis conformations of the carbamate respectively; labels g+/− stand for gauche+/− orientations of the Cbz moiety. | ||
trans-Sulfoxide 2 showed one low-energy extended conformer, labelled C5/C7δ, characterized by an intra-residue C5 backbone H-bond interaction for NH1 and a sidechain C7 H-bond interaction between NH2 and the sulfoxide oxygen, with H-bonding distances in the gas phase that are short for N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (d = 202 pm) and remarkably short for N–H⋯O
C (d = 202 pm) and remarkably short for N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S (d = 183 pm). Two alternative folded conformers, devoid of an H-bonding role for the sulfoxide and characterized by a free NH1 and either a backbone C7 H-bond or a π-amide interaction for NH2, were higher (solution) or much higher (gas phase) in energy.
S (d = 183 pm). Two alternative folded conformers, devoid of an H-bonding role for the sulfoxide and characterized by a free NH1 and either a backbone C7 H-bond or a π-amide interaction for NH2, were higher (solution) or much higher (gas phase) in energy.
In contrast, cis-sulfoxide 3 showed three stable conformer types. The first was extended, labelled C5/C6γ, with an intra-residue C5 backbone H-bond for NH1 and a sidechain C6 H-bond interaction between NH2 and the sulfoxide sulfur. The gas phase H-bonding distances, N–H⋯S (d = 240 pm) and N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (d = 204 pm), were very close to those found previously for the C5/C6γ conformer of the non-oxygenated derivative 1.25 The second and third conformers were folded, similarly to those detected for 2, with a C7 or a π-amide interaction for NH2 and no H-bonding role for the sulfoxide. The folded conformers of 3 were slightly more stable than the extended conformer in the gas phase; the difference in energies was increased in solution with a stability order π-amide > C7 > C5/C6γ.
C (d = 204 pm), were very close to those found previously for the C5/C6γ conformer of the non-oxygenated derivative 1.25 The second and third conformers were folded, similarly to those detected for 2, with a C7 or a π-amide interaction for NH2 and no H-bonding role for the sulfoxide. The folded conformers of 3 were slightly more stable than the extended conformer in the gas phase; the difference in energies was increased in solution with a stability order π-amide > C7 > C5/C6γ.
Sulfone 4 showed three stable conformer types: C5/C7δ, C7, and π-amide. In the C5/C7δ conformer the N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S gas phase distance was perceptibly longer (d = 196 pm) than in the corresponding conformer of 2. Although it was the most stable form in the gas phase, this conformer was more energetic than the folded forms in solution.
S gas phase distance was perceptibly longer (d = 196 pm) than in the corresponding conformer of 2. Although it was the most stable form in the gas phase, this conformer was more energetic than the folded forms in solution.
It is striking that the oxygen atom of the sulfoxide or sulfone group is well inclined to form a C7 H-bond with NH2 in 2 (particularly) and in 4, but not to form a C6 H-bond with NH1 in 3 nor 4. Indeed, in compound 3 the only implication of the sulfoxide function involves the sulfur atom, akin to the behavior observed previously for 1. This behavior contrasts with that of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O motif of asparagine sidechains, which is able to form both intra-residue C6γ and inter-residue C7δ H-bonds with neighboring backbone NH atoms.28
O motif of asparagine sidechains, which is able to form both intra-residue C6γ and inter-residue C7δ H-bonds with neighboring backbone NH atoms.28
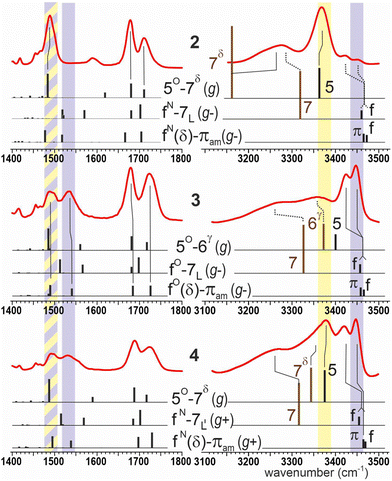
Experimental assessment of the solution state behavior of 2–4 was carried out using IR spectroscopy in chloroform solutions (5 mM). The absorption bands in the amide I, II and A regions are shown in Fig. 3. No concentration effects were observed (see ESI,† Section S3), indicating the intramolecular nature of the non-covalent interactions. Interpretation of the data was in good agreement with the quantum chemistry calculations, which facilitated assignments, as previously demonstrated.26
trans-Sulfoxide 2 showed two significant amide A (NH stretch) bands, at 3274 and 3368 cm−1, indicating the presence of the dominant C5/C7δ conformer. The latter band, well reproduced by theory, was assigned to NH1; its C5 H-bond was perceptibly stronger than usual for an α,α-disubstituted amino acid, whose diagnostic C5 absorbance band (Fig. 3, yellow region) habitually appears around 3380 cm−1.29 The former band was remarkably red-shifted and was assigned to NH2 engaged in a strong C7δ N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S H-bond, despite its red-shift being seemingly overestimated by theory, presumably because its calibration was originally carried out on N–H⋯O
S H-bond, despite its red-shift being seemingly overestimated by theory, presumably because its calibration was originally carried out on N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C H-bonds.26 Small bands at 3421 and 3452 cm−1 were assigned to minor contributions from C7 and/or π-amide conformers. The amide I bands at 1678 and 1710 cm−1 were in good agreement with calculations for the C5/C7δ conformer, while the amide II region showed a single intense band at 1488 cm−1, considered to be a benchmark for a C5 interaction.30
C H-bonds.26 Small bands at 3421 and 3452 cm−1 were assigned to minor contributions from C7 and/or π-amide conformers. The amide I bands at 1678 and 1710 cm−1 were in good agreement with calculations for the C5/C7δ conformer, while the amide II region showed a single intense band at 1488 cm−1, considered to be a benchmark for a C5 interaction.30
cis-Sulfoxide 3 showed a more complex set of amide A bands, best explained in terms of contributions from the three conformer families found by quantum chemistry. The most intense bands, at 3424 and 3447 cm−1, were assigned to NH1 and NH2 respectively, in a dominant folded π-amide conformation (or a completely free form, i.e. non H-bonded). The diagnostic πam absorbance band for α,α-disubstituted amino acids (Fig. 3; violet region) was recently reported to appear around 3450 cm−1.26 A modest contribution from the extended C5/C6γ conformer results in the plateau at 3350–3380 cm−1, reminiscent of the C5/C6γ conformer known for compound 1. Similarly, the broad feature around 3280 cm−1 can be ascribed to a weakly populated C7 conformer. The amide I bands at 1679 and 1723 cm−1 of 3 were in good agreement with calculations for the π-amide conformer, as were the two bands in the amide II region at 1490 and 1534 cm−1 which are thus revealed as the spectroscopic signature in this region for a peptide that adopts the classical δ conformation featuring a π-amide interaction.26
Sulfone 4 showed an IR absorption spectrum that suggested the presence of the three conformers found by quantum chemistry in balanced populations. As was the case for compound 3, the high frequency amide A bands at 3418 and 3449 cm−1 arose from NH1 and NH2 respectively, in a π-amide (or a free) conformation, while the C7 conformer gave rise to the broad red band at c. 3260 cm−1 from H-bonded NH2 and contributed to the 3418 cm−1 band from free NH1. The remaining amide A feature, a band with a maximum at 3378 cm−1 (yellow region) and a broadened red-side shoulder around 3360 cm−1 was assigned to the C5/C7δ conformer, by comparison with 2. The appearance of broadened or overlapping peaks in the amide I and II regions likewise indicated that several conformations were present.
To complement solution phase assignments, a gas phase conformation-selective laser study was also conducted using UV and IR/UV double-resonance spectroscopies.31 The first study was carried out on the sulfoxide 2/3 mixture (ratio 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) obtained by synthesis (Scheme 1). The UV spectrum showed one main band and three minor bands that were all assigned to the same conformer A (see ESI,† Section S4). The conformer-selective IR absorption spectrum obtained (Fig. 4, center) showed two bands in the NH stretch region, at 3262 and 3377 cm−1. Comparison with the calculated spectra for the stable forms of 2 and 3 led us to identify A as the C5/C7δ conformer of 2, with a red-shifted NH2 in a strong N–H⋯O
2) obtained by synthesis (Scheme 1). The UV spectrum showed one main band and three minor bands that were all assigned to the same conformer A (see ESI,† Section S4). The conformer-selective IR absorption spectrum obtained (Fig. 4, center) showed two bands in the NH stretch region, at 3262 and 3377 cm−1. Comparison with the calculated spectra for the stable forms of 2 and 3 led us to identify A as the C5/C7δ conformer of 2, with a red-shifted NH2 in a strong N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S interaction and NH1 in a C5 N–H⋯O
S interaction and NH1 in a C5 N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C interaction. The predominance of the C5/C7δ conformer of 2 was in complete agreement with the calculations and the solution state studies.
C interaction. The predominance of the C5/C7δ conformer of 2 was in complete agreement with the calculations and the solution state studies.
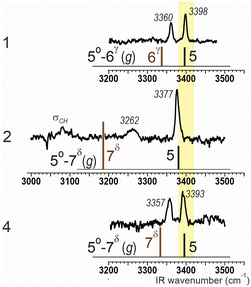 | ||
| Fig. 4 Gas-phase conformation-selective NH stretch IR spectra for the main gas phase conformation of compounds 2 (center panel) and 4 (lower), together with the calculated spectra for the most stable C5 conformations. For comparison, the corresponding IR data for compound 1 are also given (top), adapted from ref. 25. | ||
The trend of theory to overestimate the red-shift of C6γ or C7δ bands was confirmed by the gas phase results, supporting the present solution assignment. In contrast, the absence of spectral data for 3 should be seen as a caveat for the gas phase technique (see ESI,† Section S4 for further details).
The gas phase study of sulfone 4 likewise revealed only one conformation type (Fig. 4, bottom). The conformer-selective IR absorption spectrum exhibited two intense bands in the 3350–3400 cm−1 region, a signature that was very similar to that of the principal C5/C6γ conformer of compound 1 (Fig. 4, top). This similarity, together with the calculated spectra, indicated that this was the C5/C7δ conformer of 4. Its C5 interaction is slightly weaker than that observed for compound 2, and more in line with the C5 band in other α,α-disubstituted amino acids such as 1 (Fig. 4, top). Its N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S C7δ interaction, however, is much weaker than that of in trans-sulfoxide 2, in qualitative agreement with the theoretical structures (Fig. 1) and the energetics (Fig. 2), leading to the C5/C7δ conformer of 4 being challenged by the alternative folded forms in solution.
S C7δ interaction, however, is much weaker than that of in trans-sulfoxide 2, in qualitative agreement with the theoretical structures (Fig. 1) and the energetics (Fig. 2), leading to the C5/C7δ conformer of 4 being challenged by the alternative folded forms in solution.
The solution-state behavior of compounds 2–4 in chloroform was examined by 1H NMR spectroscopy (see ESI†). The NH chemical shifts and DMSO-d6 titration results are in good agreement with a predominant C5/C7δ conformer for compound 2 and a mixed conformational landscape for 3 and 4 in which folded C7 and π-amide interactions make significant contributions. Further supportive evidence was obtained from qualitative NOESY experiments. In compound 2, a strong correlation was observed between NH2 and the syn protons of the 4-membered ring, as expected for a C5/C7δ conformer. In contrast, the corresponding correlation in compounds 3 and 4 was much weaker, whereas a strong correlation between NH1 and the syn protons was in evidence, in agreement with folded conformations for these two compounds.
In summary, peptide model systems demonstrate that sulfoxide and sulfone functional groups may form sidechain–backbone N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S hydrogen bonds with a neighboring NH amide atom, leading to a stabilized local geometry with an extended backbone and an intra-residue C5 H-bond. This type of secondary structure is quite rare,30,32 and has been proposed as a basis for molecular spacers.33 Amino acids with oxidized thioether sidechains appear to be useful building blocks for the design and control of peptide architectures.
S hydrogen bonds with a neighboring NH amide atom, leading to a stabilized local geometry with an extended backbone and an intra-residue C5 H-bond. This type of secondary structure is quite rare,30,32 and has been proposed as a basis for molecular spacers.33 Amino acids with oxidized thioether sidechains appear to be useful building blocks for the design and control of peptide architectures.
Support from the French National Research Agency (Grant ANR-17-CE29-0008) and from “Investissements d’Avenir” programs (LabEx PALM; grant ANR-10-LABX-0039-PALM; DIRCOS) is acknowledged. D. L. was the awardee of a China Scholarship Council PhD grant.
Conflicts of interest
There are no conflicts to declare.Notes and references
- The Chemistry of Sulphones and Sulphoxides, ed., S. Patai, Z. Rappoport, C. Stirling, Wiley, Chichester, UK, 1988 Search PubMed.
- (a) A. N. R. Alba, X. Companyó and R. Rios, Chem. Soc. Rev., 2010, 39, 2018 RSC; (b) N. S. Simpkins, Sulphones in Organic Synthesis, Pergamon, Oxford, 1993 Search PubMed; (c) M. C. Carreno, Chem. Rev., 1995, 95, 1717 CrossRef CAS; (d) I. Fernández and N. Khiar, Chem. Rev., 2003, 103, 3651 CrossRef PubMed; (e) X. Salom-Roig and C. Bauder, Synthesis, 2020, 964 CAS.
- (a) J. Yuan, Z. Xu and M. O. Wolf, Chem. Sci., 2022, 13, 5447 RSC; (b) W. Y. Wu, Y. Bai, X. R. Wang and C. Wu, Chin. Chem. Lett., 2021, 32, 1309 CrossRef CAS; (c) M. Geven, R. D'Arcy, Z. Y. Turhan, F. El-Mohtadi, A. Alshamsan and N. Tirelli, Eur. Polym. J., 2021, 149, 110387 CrossRef CAS.
- (a) M. H. Feng, B. Q. Tang, S. H. Liang and X. F. Jiang, Curr. Top. Med. Chem., 2016, 16, 1200 CrossRef CAS PubMed; (b) E. Wojaczynska and J. Wojaczynski, Curr. Opin. Chem. Biol., 2023, 76, 102340 CrossRef CAS PubMed; (c) A. Regueiro-Ren, Adv. Heterocycl. Chem., 2021, 134, 1 CrossRef; (d) M. Mustafa and J. Y. Winum, Expert Opin. Drug Discovery, 2022, 17, 501 CrossRef CAS PubMed.
- H. Morita, A. Shishido, T. Matsumoto, H. Itokawa and K. Takeya, Tetrahedron, 1999, 55, 967 CrossRef CAS.
- H. Morita, A. Gonda, K. Takeya and H. Itokawa, Bioorg. Med. Chem. Lett., 1996, 6, 767 CrossRef CAS.
- Y. Wu, Z. M. Wu, S. S. Zhang, L. Y. Liu, F. Sun, W. H. Jiao, S. P. Wang and H. W. Lin, Org. Lett., 2022, 24, 934 CrossRef CAS PubMed.
- O. S. Kwon, C. K. Kim, W. S. Byun, J. Oh, Y. J. Lee, H. S. Lee, C. J. Sim, D. C. Oh, S. K. Lee, K. B. Oh and J. Shin, J. Nat. Prod., 2018, 81, 1426 CrossRef CAS PubMed.
- J. Garcia, V. M. Costa, A. Carvalho, P. Baptista, P. G. de Pinho, M. D. Bastos and F. Carvalho, Food Chem. Toxicol., 2015, 86, 41 CrossRef CAS PubMed.
- N. Brot and H. Weissbach, in The Chemistry of Sulphones and Sulphoxides, ed. S. Patai, Z. Rappoport, C. Stirling, Wiley, Chichester, UK, 1988, pp. 851 Search PubMed.
- L. Aussel and B. Ezraty, Front. Mol. Biosci., 2021, 8, 665492 CrossRef CAS PubMed.
- J. M. Lim, G. Kim and R. L. Levine, Neurochem. Res., 2019, 44, 247 CrossRef CAS PubMed.
- W. M. B. Edmands, N. J. Gooderham, E. Holmes and S. C. Mitchell, Toxicol. Res., 2013, 2, 11 CrossRef CAS.
- C. O. Miles, J. E. Melanson and A. Ballot, Environ. Sci. Technol., 2014, 48, 13307 CrossRef CAS PubMed.
- K. D. Tew, Expert Opin. Invest. Drugs, 2005, 14, 1047 CrossRef CAS PubMed.
- H. L. Schenck, G. P. Dado and S. H. Gellman, J. Am. Chem. Soc., 1996, 118, 12487 CrossRef CAS.
- G. T. Perell, R. L. Staebell, M. Hairani, A. Cembran and W. C. K. Pomerantz, BioChemComm, 2017, 18, 1836 CAS.
- N. Shinohara, H. Itoh, S. Matsuoka and M. Inoue, ChemMedChem, 2012, 7, 1770 CrossRef CAS PubMed.
- D. Leahy, C. Grant, A. Jackson, A. Duff, N. Tardiota, J. Van Haeften, X. C. Chen, J. M. Peake, M. D. Kruppa, E. T. Smith, D. A. Johnson, W. B. Lott and J. M. Harris, Molecules, 2021, 26, 5344 CrossRef CAS PubMed.
- C. L. Worth and T. L. Blundell, Proteins, 2009, 75, 413 CrossRef CAS PubMed.
- N. Eswar and C. Ramakrishnan, Protein Eng., 2000, 13, 227 CrossRef CAS PubMed.
- M. Vijayakumar, H. Qian and H. X. Zhou, Proteins, 1999, 34, 497 CrossRef CAS.
- D. Bordo and P. Argos, J. Mol. Biol., 1994, 243, 504 CrossRef CAS PubMed.
- L. Y. Jiang and K. Burgess, J. Am. Chem. Soc., 2002, 124, 9028 CrossRef CAS PubMed.
- Z. Imani, V. R. Mundlapati, G. Goldsztejn, V. Brenner, E. Gloaguen, R. Guillot, J. P. Baltaze, K. Le Barbu-Debus, S. Robin, A. Zehnacker, M. Mons and D. J. Aitken, Chem. Sci., 2020, 11, 9191 RSC.
- Z. Imani, V. R. Mundlapati, V. Brenner, E. Gloaguen, K. Le Barbu-Debus, A. Zehnacker-Rentien, S. Robin, D. J. Aitken and M. Mons, Chem. Commun., 2023, 59, 1161 RSC.
- M. De Zotti and J. Clayden, Org. Lett., 2019, 21, 2209 CrossRef CAS PubMed.
- P. G. Vasudev, M. Banerjee, C. Ramakrishnan and P. Balaram, Proteins, 2012, 80, 991 CrossRef CAS PubMed.
- V. R. Mundlapati, Z. Imani, V. C. D'Mello, V. Brenner, E. Gloaguen, J. P. Baltaze, S. Robin, M. Mons and D. J. Aitken, Chem. Sci., 2021, 12, 14826 RSC.
- C. Peggion, A. Moretto, F. Formaggio, M. Crisma and C. Toniolo, Biopolymers, 2013, 100, 621 CrossRef CAS PubMed.
- E. Gloaguen, M. Mons, K. Schwing and M. Gerhards, Chem. Rev., 2020, 120, 12490 CrossRef CAS PubMed.
- M. Crisma, F. Formaggio, C. Alemán, J. Torras, C. Ramakrishnan, N. Kalmankar, P. Balaram and C. Toniolo, Pept. Sci., 2018, 110, e23100 CrossRef.
- C. Toniolo, M. Crisma, F. Formaggio, C. Peggion, Q. B. Broxterman and B. Kaptein, Biopolymers, 2004, 76, 162 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc05933b |
| This journal is © The Royal Society of Chemistry 2024 |