Open Access Article
Open Access ArticleSequence-function space of radical SAM cyclophane synthases reveal conserved active site residues that influence substrate specificity†
Chin-Soon
Phan
 ab and
Brandon I.
Morinaka
ab and
Brandon I.
Morinaka
 *a
*a
aDepartment of Pharmacy, National University of Singapore, Singapore 117544, Singapore. E-mail: phambi@nus.edu.sg
bLatvian Institute of Organic Synthesis, Aizkraukles Street 21, LV-1006 Riga, Latvia
First published on 23rd October 2024
Abstract
Radical SAM cyclophane synthases catalyze C–C, C–N, and C–O crosslinking reactions in the biosynthesis of bioactive peptide natural products. Here, we studied an uncharacterized rSAM enzyme, HtkB from Pandoraea sp., and found this enzyme to catalyze the formation of a HisC2-to-LysCβ crosslink. We used a combination of ColabFold and mutagenesis studies to show that residues D214 in HtkB and H204 in HaaB (another cyclophane synthase) are important for substrate specificity. Mutation of these residues changes the specificity and lowers substrate recognition on the wild-type motifs. This result opens opportunities to alter the specificity and promiscuity for rSAM peptide modifying enzymes.
Introduction
Ribosomally-synthesized and post-translationally modified peptides (RiPPs) are a bioactive, structurally diverse, and rapidly growing natural product class.1,2 Radical SAM (rSAM) cyclophane synthases are involved in the biosynthesis of cyclophane RiPPs, which are characterized by a crosslink formed between an aromatic ring and the side chain of three to five residue motifs.3 The transformations by these enzymes include C–C, C–O, and C–N bond formation to create cyclophane rings.4–22 Similar transformations have been observed in fungi and plants,23–27 but those from rSAM enzymes can incorporate all aromatic residues into three to five residue cyclophanes. The studies on rSAM cyclophane synthases have been focused on characterizing the enzyme and natural products, biochemical characterization, and studying the enzyme promiscuity and mechanism.4–22,28–30 Engineering of rSAM cyclophane synthases for altered function has not been demonstrated, and engineering of radical enzymes in general has been limited.31Our group has focused on a subclass of cyclophane RiPPs termed triceptides. Triceptides are defined by three-residue cyclophanes containing an aromatic to Cβ crosslink formed on Ω1–X2–X3 motifs (Ω1 = aromatic) (Fig. 1A).3 The rSAM enzymes that catalyze this transformation can be referred to as triceptide synthases or 3-residue cyclophane forming enzymes (3-CyFEs).3 To date, we have identified ∼3900 putative triceptide biosynthetic pathways and characterized ten maturases of various families to show that triceptides represent the largest subclass of cyclophane rSAM-RiPPs (Fig. S1, ESI†).19–22 Previous studies on the active site profiling of rSAM cyclophane synthases suggest conserved residues within the active site and may help predict enzyme function.32 Thus, we were interested in whether active site residues in cyclophane synthases can be mutated to alter specificity or promiscuity.
 | ||
| Fig. 1 (A) Examples of cyclophane RiPP macrocycles. (B) SSN of cyclophane synthases (n = 5369; AS = 75; 40% representative nodes). Nodes are colored based on maturase family. | ||
To address this question, we studied a triceptide synthase, HtkB, from new sequence-function space in our sequence similarity network (SSN) of cyclophane synthases. We characterized the activity for HtkB to show that it catalyzes the characteristic triceptide crosslink on an HTK motif. We then show that using a combination of ColabFold and sequence comparison to another triceptide synthase, HaaB, we could identify critical residues proposed to influence selectivity. Mutagenesis at these residues demonstrate the activity of HtkB and HaaB can be altered. This represents a promising step to engineering cyclophane synthases and rSAM-RiPP modifying enzymes.
Results and discussion
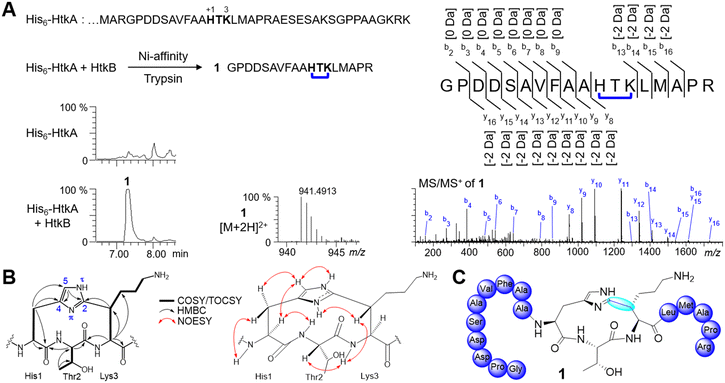
As part of ongoing efforts to explore cyclophane synthase sequence-function space, we focused on the maturases in cluster C09 from our SSN (Fig. 1B). The sequences from C09 are mostly derived from Betaproteobacteria with two main genera Pandoraea (15) and Ralstonia (8) and 11 other bacterial genera. Our previous analysis identified 10 putative precursor sequences.20 However, using RODEO33 to extract precursors associated with C09 we have identified 24 additional putative precursors (Fig. 2A). Two prominent precursor families (P43 and P156) were identified and show conserved Ω1–X2–X3 motifs (Fig. 2B). P43 precursors contain one HxK-motif, while P156 precursors contain 2–3 Hxx-motifs. The remaining precursor families all encode multiple His-containing Ω1–X2–X3 motifs.To understand the reaction catalyzed by the rSAM enzymes from C09, we initiated functional studies on the precursor/rSAM enzyme pair (HtkA/HtkB) from Pandoraea sp. XY-2 (Fig. 2B). The precursor gene htkA was inserted in pET28a(+) to encode an N-terminal His6-tag and the cognate rSAM enzyme gene htkB was inserted into MCS2 of pCDFDuet-1 (Table S1, ESI†). The precursor peptide (His6-HtkA) was expressed in vivo in E. coli NiCo21 (DE3) alone or coexpressed with HtkB, purified by Ni-affinity chromatography, and digested with trypsin. The residues within the precursor are numbered ‘+1’ from the start of the core peptide. The start of the core peptide is arbitrarily assigned to Ω1 of the precursor's first (N-terminal) cyclophane. Comparative analysis of the digests on LC-MS led to identifying a peak 1 with concomitant loss of −2 Da (Fig. 3A). The tandem mass spectrometry (MSMS) analysis localized the −2 Da mass loss to the HTK motif. With activity detected from HtkB we pursued characterization of this fragment by NMR. The trypsin digest prepared from 6 L of culture was subjected to preparative reversed-phase HPLC to give ∼3 mg of fragment 1.
NMR data including COSY, TOCSY, NOESY, edited HSQC, and HMBC were obtained for product fragment 1 (Fig. S2–S14 and Tables S6 and S7, ESI†). The 2D NMR (in D2O) indicated that modified His1 had only a single protonated heteroaromatic carbon His1-H5 (δH 7.39, δC 121.5), which showed HMBC correlations to two quaternary carbons His1-C2 (δC 147.1) and His1-C4 (δC 130.4). In addition, His1-Hβ (δH 3.40) showed HMBC correlations to His1-C5 (δC 121.5) and His1-C4 (δC 130.4). This indicated that His1 was substituted at C2. Thr2 was assigned as unmodified, and we focused our attention on Lys3. The spin system for Lys3 showed Cβ was now a substituted methine (δH 3.50, δC 43.4). This suggested that the cross-link was formed between His1-C2 and Lys3-Cβ. The HMBC correlations of Lys3-Hβ to His1-C2 further supported this assignment (Fig. 3B).
We next turned toward assigning the conformation of the HTK ring. NOESY correlations (DMSO-d6 + 0.15% TFA-d) showed that His1-Hα (δH 4.50), His1-Hβa (δH 3.15), His1-H5 (δH 7.45), His1-NHτ (δH 14.40) and Thr2-NH (δH 8.09) lie on the same face, while His1-NHπ (δH 13.79), Thr2-Hα (δH 4.19), Lys3-NH (δH 8.08) and Lys3-Hβ (δH 3.36) lie on the opposite face. The substitution of the HTK ring is analogous to the HAA and HNR cyclophanes created by triceptide synthases HaaB and YhhB, respectively (Fig. 1 and 3B and C).20 The absolute configuration of the macrocycle was assigned by Marfey's analysis, which showed that all Cα positions are in the L-configuration (Table S8, ESI†). Importantly, the observation of NOESY correlations of His1-NHτ and His1-NHπ allowed the assignment of the orientation of the imidazole ring and leads to the assignment of planar chirality to be 2P or 2Rp when using Lys3-Cα as the pilot atom.
To determine the promiscuity of HtkB at His1, we constructed two variant precursor peptides (His6-HtkA H1F and H1W). His6-HtkA H1F and H1W were coexpressed with HtkB in vivo in E. coli, followed by Ni-affinity purification, and digestion with trypsin. Analysis of the trypsin digest by LC-MS/MS showed that modified fragments consistent with cyclophane formation (−2 Da) were observed for FTK and WTK fragments (Fig. S15 and S16, ESI†). While the modified WTK fragment yield was low, the modified FTK fragment (S1a) was significant. Subsequently, the FTK fragment (S1a) was subjected to large-scale protein expression, Ni-affinity purification, trypsin digest, and preparative reversed-phase HPLC to yield purified fragment S1a. 2D NMR analysis (Fig. S17–S25, ESI†) of fragment S1a showed the modified FTK motif was a paracyclophane crosslinked to Lys3-Cβ. Generally, triceptide synthases (XncB, MscB, OscB) that modify Phe-containing motifs have been observed to catalyze formation of a paracyclophane.19 One exception was the metacyclophane formed by the triceptide synthase ChlB on a nonnative FDR (native = YDR) motif.22 The results here suggest that the topology of Phe-containing cyclophanes may, in part, be substrate-controlled. We tested the double mutant (His6-HtkA H1F/K3R) coexpressed with HtkB and detected activity but low % conversion for this variant prevented characterization by NMR (Fig. S26, ESI†).
Characterizing several triceptide synthases allowed us to start interrogating the active site residues that may alter their activity. We envisioned two engineering experiments: (1) to engineer a triceptide synthase to accept a nonbasic residue at X3, and (2) to engineer a triceptide synthase to accept a basic residue at X3. HtkB was a suitable candidate for the experiment 1. For the second experiment we selected the His-modifying triceptide synthase, HaaB, that catalyzes a His1-C2 to Ala3-Cβ crosslink on an HAA motif. We then analyzed the precursor/enzyme pairs HtkA/HtkB and HaaA/HaaB to identify residues that may be responsible for selectivity. We used ColabFold34 to model the structures of the two enzyme pairs and identified D214 in HtkB and H204 in HaaB, which are both located near Lys3 and Ala3 of the precursor peptides (Fig. 4A and Fig. S27, S28, ESI†). These observations allowed the hypothesis that the D214 and H204 might be associated with the selectivity of HtkB and HaaB, respectively. Sequence alignment showed that D214 in HtkB was encoded at the position analogous to H204 in HaaB (Fig. 4A), which prompted us to exchange these residues for engineering experiments.
For engineering experiment 1, we first tested whether HtkB could accept the precursor peptide variant His6-HtkA K3A. When His6-HtkA K3A was coexpressed in vivo with HtkB we observe a trace conversion of this substrate and showed that HtkB is selective for Lys and suitable for the engineering experiment. Next, we targeted D204 for a directed engineering experiment to alter the specificity to accept Ala instead of Lys on the precursor peptide HtkA. We coexpressed HtkB D214H with His6-HtkA K3A followed by Ni-affinity purification, digest with trypsin, and LC-MS analysis. To our satisfaction, the engineered cyclophane synthase HtkB D214H showed improved cyclophane formation for this variant tested based on increased % conversion to cyclophane product compared to wild-type HtkB (Fig. 4B and Fig. S29, ESI†). Also, HtkB D214H showed a lower conversion when coexpressed with the wild-type His6-HtkA. (Fig. S30, ESI†). These experiments showed that the substrate preference for a cyclophane synthase can be changed.
Next, we moved to engineering experiment 2. We previously showed that HaaB does not accept the HaaA A3R variant and would be a suitable target for engineering the cyclophane synthase HaaB to accept a basic residue at X3.22 We initiated efforts for targeted engineering of a cyclophane synthase to accept a basic residue. We designed HaaB H204D and hypothesized it could lead to a better cyclophane conversion for a motif when A3 is replaced with a basic residue (Arg or Lys). Three different engineered precursors (His6-HaaA A3R, A3K, and H1F/A3R) were designed. Coexpression of all three variants followed by Ni-affinity purification, digest with trypsin, and LC-MS analysis led to an improved cyclophane conversion by HaaB H204D compared to wild-type HaaB (Fig. 4C and Fig. S31–S33, ESI†). Similar to HtkB D214H, a lower yield in cyclophane conversion on native HAA motif was observed by HaaB H204D compared to HaaB (Fig. S34, ESI†). This finding shows the residue at D214 in HtkB that correlated to H204 in HaaB could influence substrate specificity at the X3 position for cyclophane formation.
The two engineering experiments are significant for rSAM enzymes, which are still largely unexplored with regard to engineering. The reasons for this include the difficulty in working with rSAM enzymes in vitro under anaerobic conditions,31 the subtleties in manipulating the active site leads to loss of control of the radical species and undesired side reactions,18 and ability to screen the products from rSAM enzymes in an efficient manner. Previous engineering of rSAM enzymes has been limited to the tryptophan lyase NosL involved in the biosynthesis of nosiheptide.35 NosL catalyzes the formation of 3-methylindole-2-carboxylic acid and 3-methylindole from L-tryptophan. Mutations (R323K, Y90A, S340A) in NosL expanded its substrate scope, allowing the enzyme to process different tryptophan analogs.36,37 This led to the formation of products, 3-methyl-2-indolic acid and 1,3-dimethyl-1H-indole. While NosL uses tryptophan as a substrate, the engineering of HtkB and HaaB represent examples to engineer peptide modifying rSAM enzymes.
Functional study of the remaining gene, HtkC was coexpressed in vivo with His6-HtkA + HtkB, we could not observe any additional changes on the full-length precursor peptide or digested core peptide fragment (Fig. S35, ESI†). The SDS-PAGE result showed HtkC is pulled down with the His6-precursor, and indicated HtkC is expressed and associated with the precursor (Fig. S36, ESI†). The role of HtkC remains unknown at this time.
Conclusions
In conclusion, we further explore and functionally validate an uncharacterized region of rSAM cyclophane synthases. To our knowledge, the natural products from the maturase systems in C09 have not been described. HtkB was characterized as a triceptide synthase catalyzing a His1-C2 to Lys3-Cβ crosslink to give the signature triceptide macrocycle. Our protein engineering efforts show the residue at D214 in HtkB and H204 in HaaB influence the substrate specificity at the X3 position for cyclophane formation. This represents the successful engineering of a peptide-modifying rSAM enzyme and opens the door for engineering this important class of enzymes.Data availability
The Experimental section, figures and tables; 1D and 2D NMR spectra; gene sequences and constructs used in this study.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors acknowledge financial support from the Ministry of Education (R148-000-257-133, R148-000-271-114, R148-000-291-133, and R148-000-340-112) to B. I. M. Part of this work was funded by EU project no. 101087181 (Natural Products Research at Latvian Institute of Organic Synthesis as a Driver for Excellence in Innovation).Notes and references
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis and J. Clardy, et al. , Nat. Prod. Rep., 2013, 30, 108–160, 10.1039/c2np20085f.
- M. Montalbán-López, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud and Y. Goto, et al. , Nat. Prod. Rep., 2021, 38, 130–239, 10.1039/d0np00027b.
- C.-S. Phan and B. I. Morinaka, Nat. Prod. Rep., 2024, 41, 708–720, 10.1039/D3NP00030C.
- I. Barr, J. A. Latham, A. T. Iavarone, T. Chantarojsiri, J. D. Hwang and J. P. Klinman, J. Biol. Chem., 2016, 291, 8877–8884, DOI:10.1074/jbc.C115.699918.
- I. Barr, T. A. Stich, A. S. Gizzi, T. L. Grove, J. B. Bonanno, J. A. Latham, T. Chung, C. M. Wilmot, R. D. Britt, S. C. Almo and J. P. Klinman, Biochemistry, 2018, 57, 1306–1315, DOI:10.1021/acs.biochem.7b01097.
- B. Khaliulin, R. Ayikpoe, M. Tuttle and J. A. Latham, J. Biol. Chem., 2017, 292, 13022–13033, DOI:10.1074/jbc.M117.795682.
- B. Khaliulin, P. Aggarwal, M. Bubas, G. R. Eaton, S. S. Eaton and J. A. Latham, FEBS Lett., 2016, 590, 2538–2548, DOI:10.1002/1873-3468.12249.
- N. A. Bruender and V. Bandarian, Biochemistry, 2016, 55, 2813–2816, DOI:10.1021/acs.biochem.6b00355.
- B.-B. He, Z. Cheng, Z. Zhong, Y. Gao, H. Liu and Y.-X. Li, Angew. Chem., Int. Ed., 2022, 61, e202212447, DOI:10.1002/anie.202212447.
- K. R. Schramma and L. B. Bushin, Nat. Chem., 2015, 7, 431–437, DOI:10.1038/nchem.2237.
- K. M. Davis, K. R. Schramma, W. A. Hansen, J. P. Bacik, S. D. Khare, M. R. Seyedsayamdost and N. Ando, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10420–10425, DOI:10.1073/pnas.1703663114.
- L. B. Bushin, K. A. Clark, I. Pelczer and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2018, 140, 17674–17684, DOI:10.1021/jacs.8b10266.
- A. Caruso, R. J. Martinie, L. B. Bushin and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2019, 141, 16610–16614, DOI:10.1021/jacs.9b09210.
- Y. Imai, K. J. Meyer, A. Iinishi, Q. Favre-Godal, R. Green, S. Manuse, M. Caboni, M. Mori, S. Niles and M. Ghiglieri, et al. , Nature, 2019, 576, 459–464, DOI:10.1038/s41586-019-1791-1.
- R. D. Miller, A. Iinishi, S. M. Modaresi, B.-K. Yoo, T. D. Curtis, P. J. Lariviere, L. Liang, S. Son, S. Nicolau and R. Bargabos, et al. , Nat. Microbiol., 2022, 7, 1661–1672, DOI:10.1038/s41564-022-01227-4.
- K. A. Clark and M. R. Seyedsayamdost, J. Am. Chem. Soc., 2022, 144, 17876–17888, DOI:10.1021/jacs.2c06497.
- L. B. Bunshin, B. C. Covington, K. A. Clark, A. Caruso and M. R. Seyedsayamdost, Nat. Chem. Biol., 2022, 18, 1135–1143, DOI:10.1038/s41589-022-01090-8.
- S. Ma, H. Chen, H. Li, X. Ji, Z. Deng, W. Ding and Q. Zhang, Angew. Chem., Int. Ed., 2021, 60, 19957–19964, DOI:10.1002/anie.202107192.
- T. Q. N. Nguyen, Y. W. Tooh, R. Sugiyama, T. P. D. Nguyen, M. Purushothaman, L. C. Leow, K. Hanif, R. H. S. Yong, I. Agatha and F. R. Winnerdy, et al. , Nat. Chem., 2020, 12, 1042–1053, DOI:10.1038/s41557-020-0519-z.
- R. Sugiyama, A. F. L. Suarez, Y. Morishita, T. Q. N. Nguyen, Y. W. Tooh, M. N. H. B. Roslan, J. Lo Choy, Q. Su, W. Y. Goh and G. A. Gunawan, et al. , J. Am. Chem. Soc., 2022, 144, 11580–11593, DOI:10.1021/jacs.2c00521.
- C.-S. Phan and B. I. Morinaka, ACS Chem. Biol., 2022, 17, 3284–3289, DOI:10.1021/acschembio.2c00621.
- A. F. L. Suarez, T. Q. N. Nguyen, L. Chang, Y. W. Tooh, R. H. S. Yong, L. C. Leow, I. Y. F. Koh, H. Chen, J. W. H. Koh and A. Selvanayagam, et al. , ACS Chem. Biol., 2024, 19, 774–783, DOI:10.1021/acschembio.3c00795.
- N. Nagano, M. Umemura, M. Izumikawa, J. Kawano, T. Ishii, M. Kikuchi, K. Tomii, T. Kumagai, A. Yoshimi, M. Machida, K. Abe, K. Shin-ya and K. Asai, Fungal Genet. Biol., 2016, 86, 58–70, DOI:10.1016/j.fgb.2015.12.010.
- W. Ding, W.-Q. Liu, Y. Jia, Y. Li, W. A. van der Donk and Q. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3521–3526, DOI:10.1073/pnas.1522907113.
- D. N. Chigumba, L. S. Mydy, F. de Waal, W. Li, K. Shafiq, J. W. Wotring, O. G. Mohamed, T. Mladenovic, A. Tripathi, J. Z. Sexton, S. Kautsar, M. H. Medema and R. D. Kersten, Nat. Chem. Biol., 2022, 18, 18–28, DOI:10.1038/s41589-021-00892-6.
- R. D. Kersten, L. S. Mydy, T. R. Fallon, F. de Waal, K. Shafiq, J. W. Wotring, J. Z. Sexton and J.-K. Weng, J. Am. Chem. Soc., 2022, 144, 7686–7692, DOI:10.1021/jacs.2c00014.
- S. T. Lima, B. G. Ampolini, E. B. Underwood, T. N. Graf, C. E. Earp, I. C. Khedi, M. A. Pasquale and J. R. Chekan, Angew. Chem., Int. Ed., 2023, 62, e202218082, DOI:10.1002/anie.202218082.
- A. M. Woodard, F. Peccati, C. D. Navo, G. Jimėnez-Osės and D. A. Mitchell, J. Am. Chem. Soc., 2024, 146, 14328–14340, DOI:10.1021/jacs.4c03994.
- A. R. Balo, A. Caruso, L. Tao, D. J. Tantillo, M. R. Seyedsayamdost and R. D. Britt, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2101571118, DOI:10.1073/pnas.2101571118.
- K. R. Schramma, C. C. Forneris, A. Caruso and M. R. Seyedsayamdost, Biochemistry, 2018, 57, 461–468, DOI:10.1021/acs.biochem.7b01147.
- C. M. Jäger and A. K. Croft, Biochemistry, 2023, 62, 241–252, DOI:10.1021/acs.biochem.2c00376.
- T. W. Precord, S. Ramesh, S. R. Dommaraju, L. A. Harris, B. L. Kille and D. A. Mitchell, ACS Bio Med. Chem. Au, 2023, 3, 240–251, DOI:10.1021/acsbiomedchemau.2c00085.
- J. I. Tietz, C. J. Schwalen, P. S. Patel, T. Maxson, P. M. Blair, H.-C. Tai, U. I. Zakai and D. A. Mitchell, Nat. Chem. Biol., 2017, 13, 470–479, DOI:10.1038/nchembio.2319.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, Nat. Methods, 2022, 19, 679–682, DOI:10.1038/s41592-022-01488-1.
- X. Ji, Y. Li, W. Ding and Q. Zhang, Angew. Chem., Int. Ed., 2015, 54, 9021–9024, DOI:10.1002/anie.201503976.
- D. M. Bhandari, H. Xu, Y. Nicolet, J. C. Fontecilla-Camps and T. P. Begley, Biochemistry, 2015, 54, 4767–4769, DOI:10.1021/acs.biochem.5b00764.
- D. M. Bhandari, D. Fedoseyenko and T. P. Begley, J. Am. Chem. Soc., 2016, 138, 16184–16187, DOI:10.1021/jacs.6b06139.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00227j |
| This journal is © The Royal Society of Chemistry 2024 |