Open Access Article
Open Access ArticleChemical inhibition of cell surface modification sensitizes bacteria to phage infection†
Marian Aba
Addo
,
Zhiyu
Zang
and
Joseph P.
Gerdt
 *
*
Department of Chemistry, Indiana University, Bloomington, IN 47405, USA. E-mail: jpgerdt@iu.edu
First published on 13th September 2024
Abstract
Many bacteriophages that infect Gram-positive bacteria rely on the bacterial cell surface polymer wall teichoic acid (WTA) as a receptor. However, some bacteria modulate their cell wall with D-alanine residues, which can disrupt phage adsorption. The prevalence and significance of WTA alanylation as an anti-phage defense is unknown. A chemical inhibitor of WTA D-alanylation could be employed to efficiently screen phage-host combinations for those that exhibit alanylation-dependent infections. Since the incorporation of D-alanine residues into the cell wall requires the activity of D-alanine:alanyl carrier protein ligase (DltA), a DltA inhibitor was employed as this tool. Herein, we found that a chemical probe inhibiting DltA activity impeded bacterial cell wall alanylation and enhanced infectivity of many phages against Bacillus subtilis, including phages Phi29, SPP1, SPO1, SP50, and Goe2. This finding reveals the breadth of immunity conferred by WTA alanylation in B. subtilis, which was previously known to impact only phages Phi29 and SPP1, but not SPO1, SP50, or Goe2. DltA inhibition selectively promoted infection by several phages that bind WTA, having no impact on the flagellotropic phage PBS1. Unexpectedly, DltA inhibition also had no effect on phage SP10, which binds to WTA. This selective chemical tool has the potential to unravel bacteriophage interactions with bacteria, leading to improved phage therapies in the future.
Introduction
Bacteriophages, also known as phages, are viruses that specifically infect bacteria.1,2 They are likely the most abundant organisms on the planet, with a count of approximately 1031,3 and they are estimated to cause 20–40% of bacterial deaths globally.4 The ability of phages to selectively kill bacteria and transmit DNA across bacteria populations has made them useful in biotechnology and healthcare.5 However, bacteria have evolved a range of antiphage mechanisms to defend themselves from viral infections.6 For example, most bacteria selectively degrade phage DNA via restriction-modification and/or CRISPR-Cas systems.7 Many bacteria also employ “abortive-infection” systems that trigger cell suicide before phages are released from the infected cell to infect neighboring bacteria.8 Perhaps the most effective defense is the modification of the bacterial cell surface to prevent initial adsorption of the phage altogether.9New variations on these themes of anti-phage defense are frequently discovered.10–13 For many of these defenses, we still poorly understand their mechanisms of action, their prevalence, and their individual impact on immunity against bacteriophages. Chemical probes that inhibit these defenses will help dissect the defense mechanisms, survey how prevalent the defenses are across bacterial species, and determine how significant each defense is against various phages under different environmental conditions. Therefore, we aim to develop inhibitors of anti-phage defenses. In this work, we targeted a specific mechanism of cell wall modification.
Because phages must first adsorb to a bacterial cell before they infect it, bacterial cell surface modification is a highly effective strategy to avert phage infection.2 The cell surfaces of Gram-positive bacteria consist of polymers such as peptidoglycan and teichoic acids.14 Teichoic acids are anionic glycopolymers composed of poly(glycerol phosphate), poly(ribitol phosphate), or more rarely poly(mannitol phosphate), and they can be attached to the cell membrane (lipoteichoic acid, LTA) or anchored into the peptidoglycan (wall teichoic acid, WTA).15–17 Many phages exploit these cell surface polymers to use them as adsorption sites.18,19 Electrostatic forces play a major role in phage adsorption to both inorganic substrates20,21 and target hosts.22–24 The Lactobacillus delbrueckii phage LL-H has been shown to require negatively charged LTA for adsorption.22,23 Decreasing the LTA surface charge by removing alanines improved LL-H adsorption 100-fold.23 In contrast, the Klebsiella pneumoniae CRKP phage adsorbed better to cell surfaces with less negative charge.24
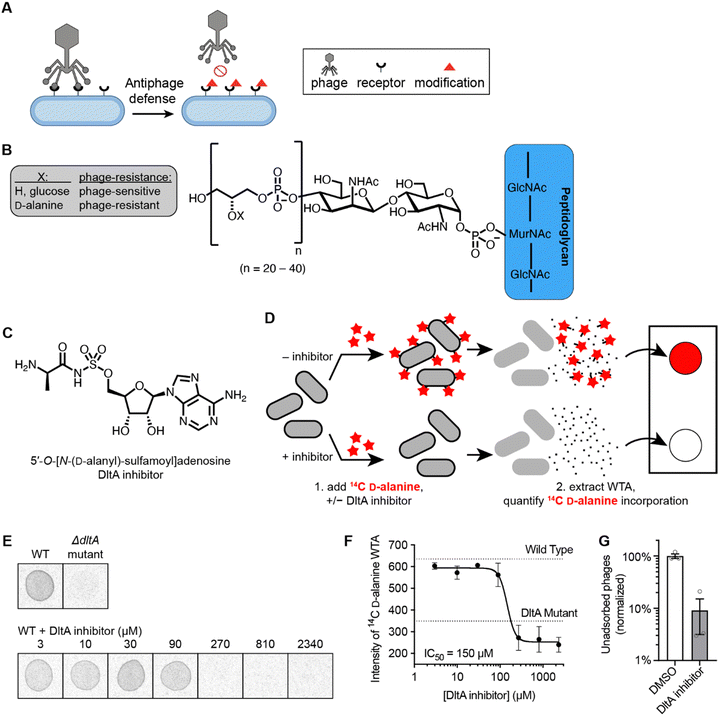
Since these cell surface receptors must be accessible and harbor the proper charge in order for phages to initiate infection, bacteria have developed strategies to modify the receptor structure to prevent phage binding.9 Incorporation of D-alanine residues onto bacterial cell wall teichoic acid is one such receptor modification mechanism that bacteria have evolved against phages.25 In many Gram-positive bacteria such as Bacillus subtilis, the dlt operon that encodes the DltA, DltB, DltC, and DltD proteins is responsible for D-alanylation of the cell WTA.26,27 It was recently found that during phage infection, B. subtilis cells signal neighboring cells to D-alanylate their cell walls, which changes the WTA charge and blocks phage binding sites.25 We predicted that a small molecule inhibitor of the D-alanylation pathway could prevent cell wall modification and re-sensitize bacteria to phage infection. Such an inhibitor would be a useful tool to determine the prevalence and significance of cell wall D-alanylation as a defense mechanism.
In order to arrest the D-alanylation defense, we investigated a previously described inhibitor of the DltA enzyme.28 After synthesizing the inhibitor, we first confirmed its ability to inhibit WTA D-alanylation in cells. We subsequently found that the inhibitor promoted phage adsorption and prevented B. subtilis from resisting phage infection. We finally employed this inhibitor to reveal that cell wall D-alanylation is an effective anti-phage defense strategy for multiple strains of B. subtilis against diverse phages. Since many Gram-positive bacteria (including pathogens like Staphylococcus aureus) have this D-alanylation pathway,29 chemical inhibition of D-alanylation may even improve the efficacy of phage therapies by sensitizing pathogens to phage infections.
Results and discussion
Inhibition of D-alanylation in Bacillus subtilis cells by 5′-O-[N-(D-alanyl)-sulfamoyl]adenosine promotes adsorption of Phi29 phage
Addition of D-alanine esters to wall teichoic acid (WTA) is a strategy by which bacteria regulate surface charge and block phage adsorption (Fig. 1A and B).14,30 We synthesized a DltA inhibitor 5′-O-[N-(D-alanyl)-sulfamoyl]adenosine (Fig. 1C), which has been validated biochemically to block DltA-catalyzed transfer of D-alanine to DltC in vitro.28 We tested whether it inhibited the incorporation of 14C-D-alanine into WTA in live B. subtilis cells. Bacteria were treated with increasing concentrations of 5′-O-[N-(D-alanyl)-sulfamoyl]adenosine in the presence of 14C-D-alanine (Fig. 1D). Phosphor imaging of extracted WTA revealed a complete inhibition of D-alanylation by the chemical inhibitor (IC50 ∼150 μM, Fig. 1E and F) in a dose-dependent fashion. Therefore, this inhibitor effectively arrests D-alanylation of WTA in live B. subtilis cells.Since WTA D-alanylation is believed to inhibit adsorption of certain phages (like Phi29),25 we hypothesized that phage Phi29 would adsorb better to WTA from cells treated with the DltA inhibitor. Indeed, 10× fewer phages remained unadsorbed to purified cell wall material from inhibitor-treated cells compared to non-treated cells (Fig. 1G). Therefore, chemical inhibition of WTA D-alanylation improves adsorption of a phage that preferentially adsorbs to non-alanylated WTA.
Chemical inhibition of D-alanylation sensitizes B. subtilis to infection by Phi29 and SPP1 phages
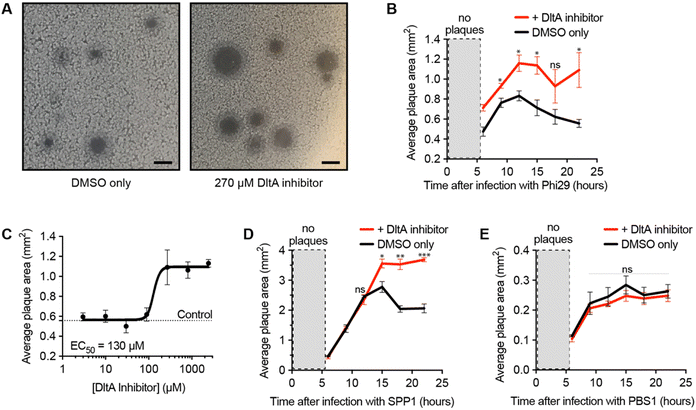
After validating the inhibitor's activity on WTA D-alanylation and phage adsorption, we hypothesized that the chemical inhibitor would sensitize the bacteria to phage infection. To test this hypothesis, we used a phage/host combination previously shown to exhibit DltA-dependent phage defense: phage Phi29 and B. subtilis PY79.25 We infected the bacteria with the phage in the presence of different chemical inhibitor concentrations and monitored the plaque area over time (Fig. 2A and B). As previously reported,25 Phi29 plaques contracted—presumably due to the re-growth of cells with D-alanylated WTA. The DltA inhibitor mitigated this effect, affording significantly larger plaques. Furthermore, this plaque expansion effect was dose-dependent: the chemical inhibitor promoted phage infection with an EC50 ∼ 130 μM (Fig. 2C and Fig. S3, ESI†), which was comparable to its potency of inhibiting D-alanine incorporation into WTA (Fig. 1F). At very high concentrations (>2 mM), the compound was toxic (Fig. S4, ESI†). Previous work also indicated that phage SPP1 was susceptible to DltA-mediated host defense.25 As predicted, the DltA inhibitor also prevented plaque restriction of SPP1 (Fig. 2D). Thus, our hypothesis was supported that chemical inhibition of D-alanylation sensitizes B. subtilis to WTA-binding phages.DltA inhibitor does not promote infection of phage that uses flagellum as the receptor
To ensure that the DltA inhibitor does not generally promote the infectivity of all Bacillus phages, we tested its impact on a phage known to adsorb to the flagellum of Bacillus (PBS1).31 Since DltA does not D-alanylate flagella, the DltA inhibitor should not increase the plaque size of the phage PBS1. As expected, the inhibitor had no impact on the plaque size of the flagellotropic PBS1 (Fig. 2E). Therefore, the DltA inhibitor is selective for its intended activity: promoting plaquing of phages that require adsorption to non-alanylated WTA.DltA inhibitor reveals the significance of D-alanylation for protection from phage SPO1 but not phage SP10
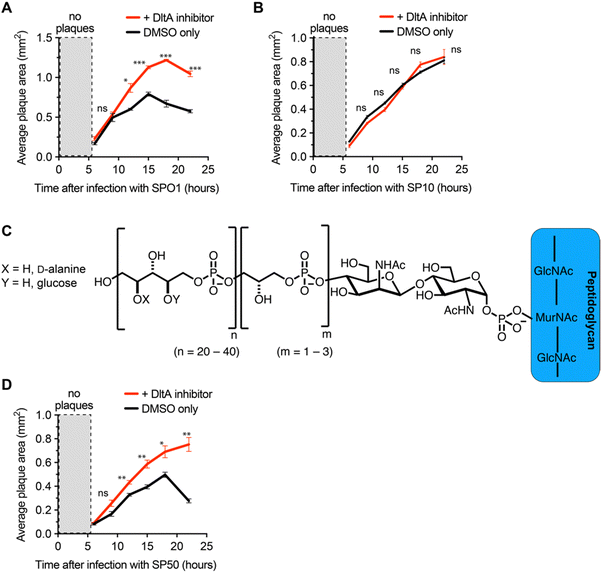
We next employed the inhibitor as a tool to investigate the impact of D-alanylation on the infectivity of other bacteriophages. We tested SPO1 and SP10, two lytic phages that are known to require glucosylated WTA (gWtA) for adsorption to their Bacillus host.30,32 The impact of WTA D-alanylation on SPO1 and SP10 is currently unknown. Since the phages must bind gWTA, we hypothesized that D-alanylation would inhibit their adsorption. Indeed, the DltA inhibitor increased the plaque areas of cells infected with SPO1 phage (Fig. 3A). However, SP10 plaques were not enlarged by DltA inhibition (Fig. 3B). Therefore, the DltA inhibitor has proven to be a useful tool to discover that SPO1 is sensitive to cell wall D-alanylation but SP10 is not. We hypothesize that some (but not all) of the other Bacillus phages that require gWTA for adsorption are likely also impacted by cell wall D-alanylation.DltA inhibitor reveals that B. subtilis strains with alternative WTA structure can also resist phage infection via cell wall D-alanylation
To further employ the DltA inhibitor as a chemical tool to study phage interaction with bacteria cell surfaces, we tested the SP50 phage that infects B. subtilis strain W23. This B. subtilis strain has a different WTA structure than PY79. The WTA of B. subtilis W23 consists of a ribitol-5-phosphate polymer functionalized with glucose and D-alanine residues (Fig. 3C).33 Despite its altered WTA, we found that the W23 strain was also sensitized to SP50 phage infection by the DltA inhibitor (Fig. 3D). Therefore, B. subtilis W23 likely also employs D-alanylation as a phage defense mechanism, and SP50 is susceptible to this defense.DltA inhibitor promotes infection of the newly isolated phage Goe2
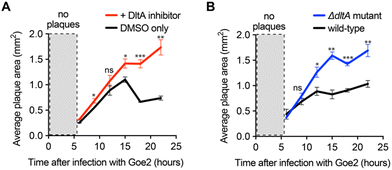
As a final implementation of the DltA inhibitor, we used it to determine the significance of WTA D-alanylation for a newly discovered phage with an unknown cell surface receptor—Bacillus VB_BsuM-Goe2 (Goe2) phage.34 If Goe2 requires non-D-alanylated WTA for phage adsorption, then inhibition of D-alanylation by the DltA inhibitor should significantly promote phage adsorption and subsequent replication. To test this hypothesis, we infected B. subtilis PY79 cells with Goe2 phage in the absence and presence of the chemical inhibitor. The DltA inhibitor significantly increased the area of Goe2 plaques (Fig. 4A), revealing that phage Goe2 requires non-D-alanylated WTA for adsorption. To confirm this result, we tested if a ΔdltA mutant was sensitized to the Goe2 phage. Indeed, it was (Fig. 4B), affording further confidence that WTA alanylation is an effective defense against the new phage Goe2.Inhibition of D-alanylation promotes antibacterial activity of both antibiotics and bacteriophages
It is noteworthy that D-alanylation also plays a role in antibiotic resistance—particularly to positively charged antibiotics. For example, WTA D-alanylation in Lactococcus affords resistance to nisin,35 and LTA D-alanylation in Streptococcus provides resistance to polymyxin B, colistin, and peptides.36 Therefore, cell wall D-alanylation is a bacterial mechanism of protection against both antibiotics and bacteriophages. Inhibition of D-alanylation has been shown to sensitize bacteria to antibiotics such as imipenem and vancomycin.28,37 We are unaware of any prior work sensitizing bacteria to phages through chemical inhibition of D-alanylation. Collectively with prior antibiotic efforts, our work show that inhibition of D-alanylation may sensitize bacteria to chemical antibiotics, bacteriophage therapies, and even synergistic antibiotic-phage therapies.Conclusion
Bacterial cell surface modification is a common anti-phage defense strategy that bacteria have evolved against bacteriophages.9,25 Our study indicates that chemical inhibition of surface modification (specifically, WTA D-alanylation) is an effective means of re-sensitizing bacteria to many phages. The specificity of the chemical tool to block this anti-phage system led us to discover that several phages were susceptible to cell wall D-alanylation—including a novel phage with a yet-uncharacterized receptor. Although the current work was limited to strains of B. subtilis, this inhibitor (or the general strategy) should be applicable to other bacteria, as well. Notably, it could be employed to sensitize DltA-containing pathogens to phage therapies38 (e.g., Staphylococci,39Streptococci,40Enterococci,41Listeria,42Clostridia,43 Fig. S5, ESI†). It can also be used as a tool to dissect teichoic acid D-alanylation defenses in bacteria without easy genetic tools for mutation experiments.Materials and methods
Strains and phages
All bacterial strains and phages used in this study are listed in the ESI.†General growth conditions
Bacterial cultures were inoculated to ∼0.2–0.4 OD600nm from an overnight culture grown in LB broth at 37 °C.Synthesis of DltA inhibitor
Following the synthesis protocol outlined by May et al. with slight modification, the chemical inhibitor was achieved over three steps.28 The final product was achieved from HPLC purification. 1H NMR, 13C NMR, and HRMS were used to validate the product identity (find details in ESI†).Alanylation assay in bacterial cells
Adapted from a protocol by Pasquina et al.,44 bacteria culture (B. subtilis 168) was inoculated to 0.4 OD600nm from an overnight culture grown in LB broth. The culture (0.05mL) was incubated with D-cycloserine (200 μg mL−1) and the DltA inhibitor (3-fold dilution from 2340 μM to 3 μM) for 30 mins at 30 °C. Subsequently, 14C-D-alanine (0.005 mCi mL−1) was added, and the culture was incubated for another 30 minutes. The cells were then pelleted by centrifugation and washed with buffer 1 (50 mM MES, pH 6.5). The cell pellet was boiled 1 hour in SDS containing buffer 2 (4% [wt/vol] SDS, 50 mM MES, pH 6.5) to remove all LTA and contaminating lipids. The pellet was washed sequentially in buffer 1, buffer 2, buffer 3 (2% NaCl, 50 mM MES, pH 6.5), and buffer 1 to remove all lipids and residual SDS. Proteins were removed by digestion with proteinase K for 4 hours at 50 °C, in digestion buffer (0.5% [w/v] SDS, 20 mM Tris–HCl, pH 8.0). The pellet was washed in buffer 3 and then three times in MilliQ water. The sample was then incubated 16 hours in 0.1 M NaOH, to release the WTA from the peptidoglycan.45 The 14C-D-alanylated WTA released into the supernatant was spotted on a Whatman filter paper and phosphor imaging was performed using Typhoon FLA 9500 Imager.Adsorption to cell wall material assay
The strain PY79 (PIPTG-sigX)25 was subcultured into fresh LB broth 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 from an overnight culture in LB broth and grown in the presence of DltA inhibitor (300 μM) or DMSO vehicle for 5 hours at 37 °C with shaking at 220 rpm. The expression of sigX was then induced by the addition of 0.5 mM IPTG before growing the cells overnight at 37 °C with shaking at 220 rpm. Cells were pelleted by centrifugation, washed with buffer 1 (50 mM MES, pH 6.5) and incubated with buffer 2 (4% [wt/vol] SDS, 50 mM MES, pH 6.5) for 1 hour. Cell wall material was washed first with buffer 3 (2% NaCl, 50 mM MES, pH 6.5), then with buffer 1, and finally resuspended in LB broth. After normalizing cell wall material to OD600nm = 4, 8 × 109 PFU of Phi29 phages were added to 100 μL of each normalized cell wall suspension and incubated for 10 minutes. The samples were centrifuged to pellet cell wall material and adsorbed phages. The supernatants (containing unadsorbed phages) were serially diluted and spotted in 10 μL aliquots onto LB-agar plates containing a lawn of PY79 cells. Plates were incubated overnight at 37 °C. Plaque forming units (PFUs) were counted, and the number of PFUs were normalized the average number remaining unadsorbed to untreated cell wall material (100%).
100 from an overnight culture in LB broth and grown in the presence of DltA inhibitor (300 μM) or DMSO vehicle for 5 hours at 37 °C with shaking at 220 rpm. The expression of sigX was then induced by the addition of 0.5 mM IPTG before growing the cells overnight at 37 °C with shaking at 220 rpm. Cells were pelleted by centrifugation, washed with buffer 1 (50 mM MES, pH 6.5) and incubated with buffer 2 (4% [wt/vol] SDS, 50 mM MES, pH 6.5) for 1 hour. Cell wall material was washed first with buffer 3 (2% NaCl, 50 mM MES, pH 6.5), then with buffer 1, and finally resuspended in LB broth. After normalizing cell wall material to OD600nm = 4, 8 × 109 PFU of Phi29 phages were added to 100 μL of each normalized cell wall suspension and incubated for 10 minutes. The samples were centrifuged to pellet cell wall material and adsorbed phages. The supernatants (containing unadsorbed phages) were serially diluted and spotted in 10 μL aliquots onto LB-agar plates containing a lawn of PY79 cells. Plates were incubated overnight at 37 °C. Plaque forming units (PFUs) were counted, and the number of PFUs were normalized the average number remaining unadsorbed to untreated cell wall material (100%).
General plaque assay conditions
The indicated strains were grown in LB broth to 0.2 OD600nm after subculturing from an overnight culture. Cells (200 μL) were infected with a dilution of phage lysate in LB broth (50 μL) in the presence of 300 μM chemical inhibitor or DMSO in 4 mL LB broth containing 0.5% agar (supplemented with 5 mM MgCl2, 5 mM CaCl2 and 0.5 mM MnCl2). The mixture was then poured onto an empty Petri dish (100 mm) and incubated overnight at 37 °C.Dose-dependent DltA inhibitor with phage assay
The bacteria strain B. subtilis PY79 was grown in LB broth to 0.2 OD600nm after subculturing from an overnight culture. Next, to 15 mL centrifuge tubes containing prewarmed LB broth plus 0.5% agar (2.5 mL supplemented with 5 mM MgCl2 and 0.5 mM MnCl2) were each treated with chemical inhibitor (3-fold dilution from 2340 μM to 3 μM). The cells were infected with phages (Phi29, 10−7 PFU mL−1) for 10 mins and added to the prewarmed agar–DMSO or compound mixture. The mixture was then poured onto an empty Petri dish (60 mm) and incubated overnight at 37 °C.Plaque area determination
The plaque area was measured after taking pictures of Petri dishes with plaques after each incubation time. The plaque areas were measured using Fiji with ViralCounter_0.8.ijm macro.46 The area of plaques was output in an MS excel document, which was then plotted with Prism Graph Pad data analysis software.Construction of ΔdltA mutant of PY79 via transduction
B. subtilis 168 ΔdltA donor cells were grown in LB broth supplemented with 1 μg mL−1 erythromycin, and 10 mM CaCl2. In three separate 1.7 mL tubes, donor cells were infected with 50 μL SPP1 phage 10−5, 10−6, 10−7 PFU mL−1 and each was overlayed on 1.5% agar LB plate (supplemented with 1 μg mL−1 erythromycin and 10 mM CaCl2). The agar plate was incubated overnight at 37 °C. Phage buffer (10 mM Tris pH 7.4, 10 mM MgSO4, 4 g L−1 NaCl) was added to the most confluently lysed plate to extract propagated phages. The recipient cells (B. subtilis PY79) were grown in LB broth and infected with extracted phages diluted 10-fold (10−1–10−3) for 10 mins. Phage adsorption was halted with the addition of prewarmed LB broth supplemented with 20 mM citrate. This mixture was then centrifuged, and the pellet was resuspended in leftover supernatant (∼50 μL) and plated on 1.5% agar LB (supplemented with 1 μg mL−1 erythromycin and 20 mM Sodium citrate). The agar plate was incubated overnight at 37 °C. Colonies obtained were regrown on selection media (1.5% agar LB supplemented with 1 μg mL−1 erythromycin and 20 mM sodium citrate) and verified by polymerase chain reaction (PCR) using primers detailed in Table S4 (ESI†).Author contributions
Conceptualization, J. P. G.; methodology, M. A. A., Z. Z., J. P. G.; investigation, M. A. A.; writing – original draft, M. A. A., J. P. G.; writing – review & editing, M. A. A., Z. Z., J. P. G.; visualization, M. A. A., J. P. G.; supervision, J. P. G.; funding acquisition, J. P. G.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Bacillus Genomic Stock Center (Ohio State University), the Félix d'Hérelle Reference Center for Bacterial Viruses (University of Laval), Robert Hertel (University of Goettingen), and Sigal Ben-Yehuda (Hebrew University) for providing bacteria and phages. The research was supported by a National Science Foundation CAREER award (IOS-2143636) to J. P. G. The Laboratory for Biological Mass Spectrometry was supported by the Indiana University Precision Health Initiative. The 500 MHz NMR and 600 MHz spectrometer of the Indiana University NMR facility were supported by NSF grant CHE-1920026, and the Prodigy probe was purchased in part with support from the Indiana Clinical and Translational Sciences Institute funded, in part, by NIH Award TL1TR002531.References
- M. R. Clokie, A. D. Millard, A. V. Letarov and S. Heaphy, Phages in nature, Bacteriophage, 2011, 1(1), 31–45, DOI:10.4161/bact.1.1.14942.
- E. Stone, K. Campbell, I. Grant and O. McAuliffe, Understanding and Exploiting Phage-Host Interactions, Viruses, 2019, 11(6), 567, DOI:10.3390/v11060567.
- M. B. Dion, F. Oechslin and S. Moineau, Phage diversity, genomics and phylogeny, Nat. Rev. Microbiol., 2020, 18(3), 125–138, DOI:10.1038/s41579-019-0311-5.
- C. A. Suttle, Marine viruses–major players in the global ecosystem, Nat. Rev. Microbiol., 2007, 5(10), 801–812, DOI:10.1038/nrmicro1750.
- P. Rogovski, R. D. Cadamuro, R. da Silva, E. B. de Souza, C. Bonatto, A. Viancelli, W. Michelon, E. M. Elmahdy, H. Treichel and D. Rodríguez-Lázaro, et al., Uses of Bacteriophages as Bacterial Control Tools and Environmental Safety Indicators, Front. Microbiol., 2021, 12, 793135, DOI:10.3389/fmicb.2021.793135.
- H. Georjon and A. Bernheim, The highly diverse antiphage defence systems of bacteria, Nat. Rev. Microbiol., 2023, 21, 686–700, DOI:10.1038/s41579-023-00934-x.
- H. G. Hampton, B. N. J. Watson and P. C. Fineran, The arms race between bacteria and their phage foes, Nature, 2020, 577(7790), 327–336, DOI:10.1038/s41586-019-1894-8.
- A. Lopatina, N. Tal and R. Sorek, Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy, Annu. Rev. Virol., 2020, 7(1), 371–384, DOI:10.1146/annurev-virology-011620-040628.
- K. D. Seed, Battling Phages: How Bacteria Defend against Viral Attack, PLoS Pathog., 2015, 11(6), e1004847, DOI:10.1371/journal.ppat.1004847.
- A. Millman, S. Melamed, A. Leavitt, S. Doron, A. Bernheim, J. Hör, J. Garb, N. Bechon, A. Brandis and A. Lopatina, et al., An expanded arsenal of immune systems that protect bacteria from phages, Cell Host Microbe, 2022, 30(11), 1556–1569.e1555, DOI:10.1016/j.chom.2022.09.017.
- N. Duan, E. Hand, M. Pheko, S. Sharma and A. Emiola, Structure-guided discovery of anti-CRISPR and anti-phage defense proteins, Nat. Commun., 2024, 15(1), 649, DOI:10.1038/s41467-024-45068-7.
- J. Li, R. Cheng, Z. Wang, W. Yuan, J. Xiao, X. Zhao, X. Du, S. Xia, L. Wang and B. Zhu, et al., Structures and activation mechanism of the Gabija anti-phage system, Nature, 2024, 629, 467–473, DOI:10.1038/s41586-024-07270-x.
- A. Deep; Q. Liang; E. Enustun; J. Pogliano and K. D. Corbett, Architecture and infection-sensing mechanism of the bacterial PARIS defense system. bioRxiv 2024, 2024, 2001.2002.573835 DOI:10.1101/2024.01.02.573835.
- S. Brown, J. P. Santa Maria and S. Walker, Wall teichoic acids of Gram-positive bacteria, Annu. Rev. Microbiol., 2013, 67, 313–336, DOI:10.1146/annurev-micro-092412-155620.
- T. J. Silhavy, D. Kahne and S. Walker, The bacterial cell envelope, CSH Perspect. Biol., 2010, 2(5), a000414, DOI:10.1101/cshperspect.a000414.
- C. Weidenmaier and A. Peschel, Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions, Nat. Rev. Microbiol., 2008, 6(4), 276–287, DOI:10.1038/nrmicro1861.
- S. Brown, T. Meredith, J. Swoboda and S. Walker, Staphylococcus aureus and Bacillus subtilis W23 make polyribitol wall teichoic acids using different enzymatic pathways, Chem. Biol., 2010, 17(10), 1101–1110, DOI:10.1016/j.chembiol.2010.07.017.
- J. G. Swoboda, J. Campbell, T. C. Meredith and S. Walker, Wall Teichoic Acid Function, Biosynthesis, and Inhibition, ChemBioChem, 2009, 11(1), 35–45, DOI:10.1002/cbic.200900557.
- J. Bertozzi Silva, Z. Storms and D. Sauvageau, Host receptors for bacteriophage adsorption, FEMS Microbiol. Lett., 2016, 363(4), fnw02, DOI:10.1093/femsle/fnw002.
- J. Zhuang and Y. Jin, Virus retention and transport through Al-oxide coated sand columns: effects of ionic strength and composition, J. Contam. Hydrol., 2003, 60(3–4), 193–209, DOI:10.1016/s0169-7722(02)00087-6.
- A. Armanious, M. Aeppli, R. Jacak, D. Refardt, T. Sigstam, T. Kohn and M. Sander, Viruses at Solid-Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption, Environ. Sci. Technol., 2016, 50(2), 732–743, DOI:10.1021/acs.est.5b04644.
- P. Munsch-Alatossava and T. Alatossava, The extracellular phage-host interactions involved in the bacteriophage LL-H infection of Lactobacillus delbrueckii ssp. lactis ATCC 15808, Front. Microbiol., 2013, 4, 408, DOI:10.3389/fmicb.2013.00408.
- L. Raisanen, C. Draing, M. Pfitzenmaier, K. Schubert, T. Jaakonsaari, S. von Aulock, T. Hartung and T. Alatossava, Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on D-alanyl and alpha-glucose substitution of poly(glycerophosphate) backbones, J. Bacteriol., 2007, 189(11), 4135–4140, DOI:10.1128/JB.00078-07.
- G. Hao, A. I. Chen, M. Liu, H. Zhou, M. Egan, X. Yang, B. Kan, H. Wang, M. Goulian and J. Zhu, Colistin-resistance-mediated bacterial surface modification sensitizes phage infection, Antimicrob. Agents Chemother., 2019, 63(12), e01609-19, DOI:10.1128/AAC.01609-19.
- E. Tzipilevich, O. Pollak-Fiyaksel, B. Shraiteh and S. Ben-Yehuda, Bacteria elicit a phage tolerance response subsequent to infection of their neighbors, EMBO J., 2022, 41(3), e109247, DOI:10.15252/embj.2021109247.
- B. M. Wood, J. P. Santa Maria, L. M. Matano, C. R. Vickery and S. Walker, A partial reconstitution implicates DltD in catalyzing lipoteichoic acid d-alanylation, J. Biol. Chem., 2018, 293(46), 17985–17996, DOI:10.1074/jbc.ra118.004561.
- A. P. Bhavsar, L. K. Erdman, J. W. Schertzer and E. D. Brown, Teichoic acid is an essential polymer in Bacillus subtilis that is functionally distinct from teichuronic acid, J. Bacteriol., 2004, 186(23), 7865–7873, DOI:10.1128/JB.186.23.7865-7873.2004.
- J. J. May, R. Finking, F. Wiegeshoff, T. T. Weber, N. Bandur, U. Koert and M. A. Marahiel, Inhibition of the D-alanine:D-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium's susceptibility to antibiotics that target the cell wall, FEBS J., 2005, 272(12), 2993–3003, DOI:10.1111/j.1742-4658.2005.04700.x.
- B. J. Schultz, E. D. Snow and S. Walker, Mechanism of d-alanine transfer to teichoic acids shows how bacteria acylate cell envelope polymers, Nat. Microbiol., 2023, 8(7), 1318–1329, DOI:10.1038/s41564-023-01411-0.
- F. E. Young, Requirement of glucosylated teichoic acid for adsorption of phage in Bacillus subtilis 168, Proc. Natl. Acad. Sci. U. S. A., 1967, 58(6), 2377–2384, DOI:10.1073/pnas.58.6.2377.
- L. M. Raimondo, N. P. Lundh and R. J. Martinez, Primary Adsorption Site of Phage PBS1: the Flagellum of Bacillus subtilis, J. Virol., 1968, 2(3), 256–264, DOI:10.1128/jvi.2.3.256-264.1968.
- C. Baptista, M. A. Santos and C. São-José, Phage SPP1 Reversible Adsorption to Bacillus subtilis Cell Wall Teichoic Acids Accelerates Virus Recognition of Membrane Receptor YueB, J. Bacteriol., 2008, 190(14), 4989–4996, DOI:10.1128/JB.00349-08.
- A. L. Givan, K. Glassey, R. S. Green, W. K. Lang, A. J. Anderson and A. R. Archibald, Relation between wall teichoic acid content of Bacillus subtilis and efficiency of adsorption of bacteriophages SP 50 and phi 25, Arch. Microbiol., 1982, 133(4), 318–322, DOI:10.1007/bf00521298.
- I. M. Willms, M. Hoppert and R. Hertel, Characterization of Bacillus Subtilis Viruses vB_BsuM-Goe2 and vB_BsuM-Goe3, Viruses, 2017, 9(6), 146, DOI:10.3390/v9060146.
- E. Giaouris, R. Briandet, M. Meyrand, P. Courtin and M. P. Chapot-Chartier, Variations in the degree of D-Alanylation of teichoic acids in Lactococcus lactis alter resistance to cationic antimicrobials but have no effect on bacterial surface hydrophobicity and charge, Appl. Environ. Microbiol., 2008, 74(15), 4764–4767, DOI:10.1128/AEM.00078-08.
- R. Saar-Dover, A. Bitler, R. Nezer, L. Shmuel-Galia, A. Firon, E. Shimoni, P. Trieu-Cuot and Y. Shai, D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B Streptococcus by increasing the cell wall density, PLoS Pathog., 2012, 8(9), e1002891, DOI:10.1371/journal.ppat.1002891.
- D. Coupri, N. Verneuil, A. Hartke, A. Liebaut, T. Lequeux, E. Pfund and A. Budin-Verneuil, Inhibition of d-alanylation of teichoic acids overcomes resistance of methicillin-resistant Staphylococcus aureus, J. Antimicrob. Chemother., 2021, 76(11), 2778–2786, DOI:10.1093/jac/dkab287.
- C. Neuhaus Francis and J. Baddiley, A Continuum of Anionic Charge: Structures and Functions of d-Alanyl-Teichoic Acids in Gram-positive Bacteria, Microbiol. Mol. Biol. Rev., 2003, 67(4), 686–723, DOI:10.1128/mmbr.67.4.686-723.2003.
- A. Nakao, S.-i Imai and T. Takano, Transposon-mediated insertional mutagenesis of the D-alanyl-lipoteichoic acid (dlt) operon raises methicillin resistance in Staphylococcus aureus, Res. Microbiol., 2000, 151(10), 823–829, DOI:10.1016/S0923-2508(00)01148-7.
- M. Kovács, A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck and R. Brückner, A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae, J. Bacteriol., 2006, 188(16), 5797–5805, DOI:10.1128/jb.00336-06.
- D. Wobser, L. Ali, E. Grohmann, J. Huebner and T. Sakinc, A Novel Role for D-Alanylation of Lipoteichoic Acid of Enterococcus faecalis in Urinary Tract Infection, PLoS One, 2014, 9(10), e107827, DOI:10.1371/journal.pone.0107827.
- E. Abachin, C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche and P. Trieu-Cuot, Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes, Mol. Microbiol., 2002, 43(1), 1–14, DOI:10.1046/j.1365-2958.2002.02723.x.
- S. M. McBride and A. L. Sonenshein, The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile, Microbiology, 2011, 157(Pt 5), 1457–1465, DOI:10.1099/mic.0.045997-0.
- L. Pasquina, J. P. Santa Maria, B. Mckay Wood, S. H. Moussa, L. M. Matano, M. Santiago, S. E. S. Martin, W. Lee, T. C. Meredith and S. Walker, A synthetic lethal approach for compound and target identification in Staphylococcus aureus, Nat. Chem. Biol., 2016, 12(1), 40–45, DOI:10.1038/nchembio.1967.
- K. Kho and T. C. Meredith, Extraction and Analysis of Bacterial Teichoic Acids, Bio. Protoc., 2018, 8(21), e3078, DOI:10.21769/BioProtoc.3078.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld and B. Schmid, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682, DOI:10.1038/nmeth.2019.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00070f |
| This journal is © The Royal Society of Chemistry 2024 |