Open Access Article
Open Access ArticleProbing the metalloproteome: an 8-mercaptoquinoline motif enriches minichromosome maintenance complex components as significant metalloprotein targets in live cells†
Sean M.
McKenna
ab,
Bogdan I.
Florea
 c,
Daniela M.
Zisterer
d,
Sander I.
van Kasteren
c,
Daniela M.
Zisterer
d,
Sander I.
van Kasteren
 c and
Joanna F.
McGouran
c and
Joanna F.
McGouran
 *ab
*ab
aSchool of Chemistry, Trinity Biomedical Sciences Institute, Trinity College Dublin, 152-160 Pearse St, Dublin 2, Ireland. E-mail: jmcgoura@tcd.ie
bSynthesis and Solid State Pharmaceutical Centre (SSPC), Ireland
cDepartment of Bioorganic Synthesis, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands
dSchool of Biochemistry and Immunology, Trinity Biomedical Sciences Institute, Trinity College Dublin, 152-160 Pearse St, Dublin 2, Ireland
First published on 25th June 2024
Abstract
Affinity-based probes are valuable tools for detecting binding interactions between small molecules and proteins in complex biological environments. Metalloproteins are a class of therapeutically significant biomolecules which bind metal ions as part of key structural or catalytic domains and are compelling targets for study. However, there is currently a limited range of chemical tools suitable for profiling the metalloproteome. Here, we describe the preparation and application of a novel, photoactivatable affinity-based probe for detection of a subset of previously challenging to engage metalloproteins. The probe, bearing an 8-mercaptoquinoline metal chelator, was anticipated to engage several zinc metalloproteins, including the 26S-proteasome subunit Rpn11. Upon translation of the labelling experiment to mammalian cell lysate and live cell experiments, proteomic analysis revealed that several metalloproteins were competitively enriched. The diazirine probe SMK-24 was found to effectively enrich multiple components of the minichromosome maintenance complex, a zinc metalloprotein assembly with helicase activity essential to DNA replication. Cell cycle analysis experiments revealed that HEK293 cells treated with SMK-24 experienced stalling in G0/G1 phase, consistent with inactivation of the DNA helicase complex. This work represents an important contribution to the library of cell-permeable chemical tools for studying a collection of metalloproteins for which no previous probe existed.
Introduction
Affinity-based probes (AfBP) are valuable tools for studying the binding interactions of small molecules in complex and dynamic biological environments.1,2 AfBPs initially engage target proteins through non-covalent interactions, while the presence of a photoactivatable functional group enables covalent labelling to be initiated through UV irradiation (Fig. 1a).2,3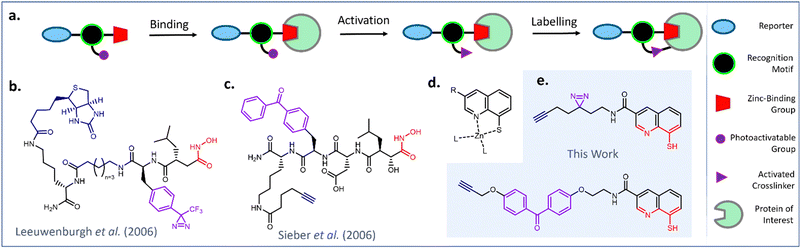 | ||
| Fig. 1 Design and select examples of AfBPs for profiling the metalloproteome (a) schematic representation of protein labelling with a generic AfBP. (b) Diazirine-functionalized hydroxamic acid probe for MMPs reported by Leeuwenburgh et al.4 (c) Benzophenone-functionalized hydroxamic acid probe for MMP13 reported by Sieber et al.5 (d) Proposed binding mode of 8MQ (e) design of novel AfBPs incorporating an 8MQ metal-binding group. | ||
Metal-binding proteins, or metalloproteins, are exciting targets to study using AfBPs, as metal co-factors impart essential structural features or catalytic functions upon biomolecules. However, their non-covalent modes of binding render them difficult targets for profiling studies.6,7 AfBPs targeting zinc-metalloproteins have previously been made through combination of a zinc-binding group (ZBG) with a photoactivatable moiety and a peptidic or small molecule recognition motif (Fig. 1b and c).4–9 ZBGs including hydroxamic acids, phosphinic acids, phosphonamides and sulfonamides have been incorporated into AfBPs to target metalloproteins including matrix metalloproteases (MMPs) and carbonic anhydrases (CA).10–15 However, this existing library of AfBPs employ a tiny fraction of all known ZBGs.16,17 Furthermore, only a small subset of these AfBPs have been successfully translated to profiling studies in live cells. Therefore, the development of cell-permeable AfBPs incorporating less explored metal-binding groups has major potential to advance the study of the metalloproteome.
Within the ligandable proteome, there remains a large subset of metalloproteins which have thus far eluded detection. These include metalloproteins with key rolls underlying proteostasis, cell cycle regulation and metabolic function.18–22 We hypothesized that using 8-mercaptoquinole (8MQ) a ZBG which has exhibited distinctive metalloenzyme inhibitory activity would allow us to profile an entirely different set of biologically important metalloproteins.18,23,24 A ZBG library screen by Perez et al. had previously discovered that 8MQ effectively inhibited Rpn11, a zinc-dependent member of the JAB1/MPN/Mov34 metalloenzyme (JAMM) family.18,23 Rpn11 forms part of the 26S proteasome, the multiprotein complex responsible for degradation of damaged and misfolded proteins in eukaryotic cells.25,26 Inhibition of Rpn11 activity stalls the function of the 26S proteasome, leading to accumulation of redundant proteins and induction of apoptosis.23,27 Perez et al. also identified that 8MQ efficiently inhibited the activity of several members of the JAMM metalloprotease family, while more traditional ZBGs poorly inhibited the activity of these enzymes.18,23 The screening hit was optimized towards a more potent and selective cell-permeable inhibitor for Rpn11, Capzimin (Fig. S1, ESI†).23
In 2019, Hameed et al. incorporated 8MQ in a non-covalent probe for detection of JAMM metalloproteases. Ub-8MQ (Fig. S2, ESI†) was capable of enriching several overexpressed JAMM deubiquitinases including Rpn11 from cell lysates.24 The poor cell permeability imparted by the ubiquitin recognition motif prevented profiling studies to be performed in live cells, while the absence of a method for covalent capture impacted the sensitivity of the probe. However, this work still served as an exciting proof of concept and it was recognized that developing a cell-permeable AfBP based upon a minimally functionalized 8MQ scaffold could provide a tool for efficient profiling of several therapeutically significant metalloproteins. We therefore set about designing an AfBP based upon the 8MQ metal-binding motif as a novel profiling tool.
Results and discussion
Probe design and synthesis began with the selection of suitable photoactivatable functional groups. Recent publications describing photoaffinity probes have tended towards the application of diazirines and benzophenones, with alternative moieties such as aryl azides appearing less commonly.2,28–31 Benzophenones are typically popular due to their relatively long wavelengths for activation, reversibility of solvent quenching, ease of synthesis and stability.32 However, they also introduce significant steric bulk and high lipophilicity, potentially decreasing the overall selectivity of the probe.31,33 By contrast, diazirines introduce minimal steric bulk and minimal impact on lipophilicity, but are susceptible to solvent quenching and present a more challenging target for synthesis.34–36 From a design perspective, the diazirine group appeared favourable for application in this AfBP, as it occupies a similar steric footprint to the reported series of Rpn11 inhibitors.18 A benzophenone analogue meanwhile was anticipated to be a less favourable binder due to its bulkier size. Both diazirine and benzophenone probes were prepared to compare their labelling properties. A terminal alkyne was used as a minimal reporter handle to enable downstream functionalization with a biotin affinity tag using copper-catalyzed alkyne–azide cycloaddition (CuAAC). Previous studies had utilized 3-carboxamide analogues of 8MQ to introduce a variety of substituents for analysis of structure–activity relationships.18,23 It was concluded that the photoactivatable group and alkyne reporter handle would also be installed at this position, leaving the remainder of the 8MQ unmodified, thus minimizing the risk of disrupting metal binding.The JAMM deubiquitinase inhibitor 20 was selected for synthesis as a competitive, non-covalent control. Intermediates 8, 11 and 18 were each prepared using modified literature conditions (Fig. 2). Amide coupling partners were combined to generate the target probes as disulfide-bridged dimers. The yields for amide coupling reactions using intermediate 18 were low, but consistent with results reported in the literature under these conditions.18
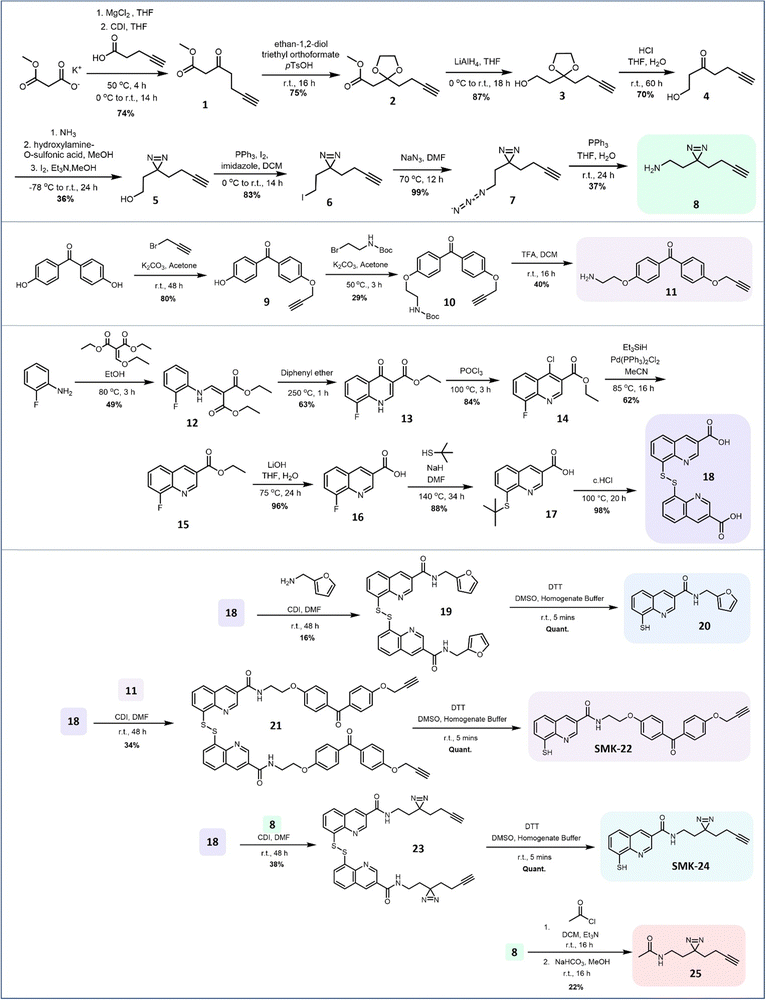 | ||
| Fig. 2 Synthetic route for the preparation of 8MQ AfBPs SMK-22 and SMK-24. Intermediates 8 and 18 enabled preparation of the 8MQ-bearing 20 and the non-zinc-binding diazirine probe 25 as controls. | ||
Disulfides 19, 21 and 23 were treated with the reducing agent dithiothreitol (DTT) immediately prior to use to liberate the corresponding thiols 20, benzophenone SMK-22 and diazirine SMK-24 respectively. Diazirine 25 was also prepared as a non-zinc-binding photoaffinity control probe.
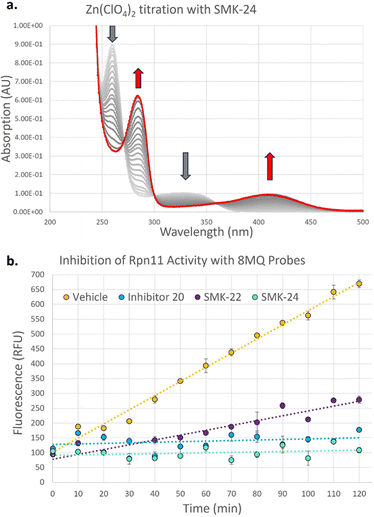
To investigate the binding kinetics of the 8MQ motif with zinc(II), a UV-vis titration experiment was performed with diazirine probe SMK-24 (Fig. 3a). Upon gradual addition of Zn(ClO4)2 to a solution of SMK-24, a significant bathochromic shift was observed for absorption maxima at 260 nm and 328 nm to 282 nm and 411 nm respectively. Binding kinetics were modelled in ReactLab and found to be consistent with formation of a Zn(SMK-24)2 species with a binding constant of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 = 15.2. This result supported the hypothesis that 8MQ acts as a strongly binding bidentate ligand to zinc(II), most similar to hydroxamic acids (log
k2 = 15.2. This result supported the hypothesis that 8MQ acts as a strongly binding bidentate ligand to zinc(II), most similar to hydroxamic acids (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 = 17.5)37 and sulfonamides (log
k2 = 17.5)37 and sulfonamides (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 = 14.7).38 By contrast, phosphinic acids (log
k2 = 14.7).38 By contrast, phosphinic acids (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 = 6.4)39 and sulfonyl ureas (log
k2 = 6.4)39 and sulfonyl ureas (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) k2 = 3.9)40,41 exhibit lower association constants as metal chelators, indicating that AfBPs bearing these groups rely upon additional interactions with the protein target to achieve strong binding.
k2 = 3.9)40,41 exhibit lower association constants as metal chelators, indicating that AfBPs bearing these groups rely upon additional interactions with the protein target to achieve strong binding.
In order to test the synthesized probes on a zinc-metalloenzyme, expression of a recombinant Rpn11/Rpn8 hetereodimer was carried out in BL21 competent E. coli cells. Purified Rpn11/Rpn8 was incubated with a fluorogenic substrate, Ub-AMC (Fig. S3, ESI†), and an increase in fluorescence over time indicated that the purified Rpn11 enzyme exhibited functional deubiquitinase activity. Treatment of Rpn11/Rpn8 with literature inhibitor 20 resulted in concentration dependent inhibition of enzymatic activity (93% inhibition at 20 μM) (Fig. 3b). Comparable inhibition could be observed with diazirine SMK-24 (97% inhibition at 20 μM), however benzophenone SMK-22 exhibited poorer enzyme inhibition (66% inhibition at 20 μM). This indicated that while the incorporation of the aliphatic diazirine and terminal alkyne of SMK-24 had not significantly perturbed binding to Rpn11 relative to literature inhibitor 20, the adjustment to a bulkier benzophenone group of SMK-22 had a detrimental impact on binding.
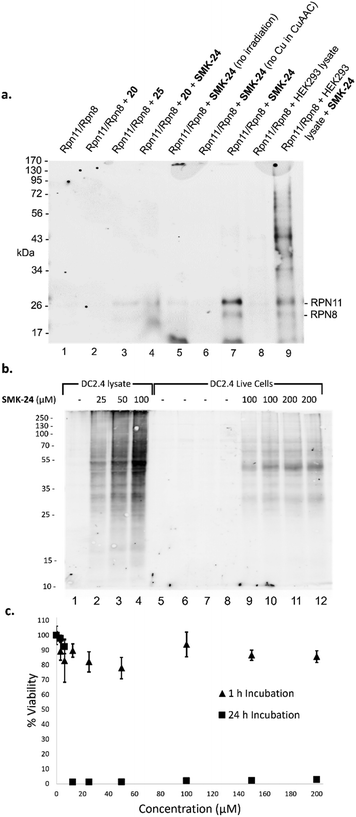
Next, the capacity for covalent capture of Rpn11 was tested. Recombinant Rpn11/Rpn8 was incubated with the 8MQ AfBPs and subject to UV irradiation. Labelled proteins were conjugated to biotin-PEG4-azide using CuAAC. Proteins were resolved by gel electrophoresis and visualized by western blotting (Fig. 4a) and silver staining (Fig. S4, ESI†). Diazirine probe SMK-24 was found to successfully label Rpn11 (Fig. 4a, lane 7). Labelling was competitively diminished when an excess of 8MQ-bearing 20 was used (Fig. 4a, lane 4), while minimal labelling could be seen in the absence of UV-irradiation (Fig. 4a, lane 5) and no labelling could be visualized if CuSO4 was not included in the CuAAC reaction mixture (Fig. 4a, lane 6). Incubation with an equivalent concentration of control diazirine 25 showed only trace levels of non-specific labelling (Fig. 4a, lane 3). When Rpn11/Rpn8 was spiked into the protein extract of HEK293 lysate, labelling of the recombinant enzyme could still be observed (Fig. 4a, lane 9). Significantly, diazirine probe SMK-24 was found to specifically label several other proteins in this complex cell extract mixture, indicating that the probe may have also labelled endogenous cellular metalloproteins. A comparable experiment was performed with benzophenone SMK-22, though poorer labelling of Rpn11 was observed and lower specificity labelling was found in HEK293 lysate (Fig. S5, ESI†).
The potential for translation of AfBPs SMK-22 and SMK-24 to live cell labelling experiments was now considered. As the 8MQ probes were based on a series on cell-permeable JAMM inhibitors, it was anticipated that they should also permeate live mammalian cells. A parallel artificial membrane permeability assay (PAMPA) was performed to test this hypothesis (Fig. S6, ESI†). Diazirine SMK-24 effectively permeated the artificial membrane (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pe = −5.47) to a comparable degree with the cell active inhibitor 20 (log
Pe = −5.47) to a comparable degree with the cell active inhibitor 20 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pe = −5.23) and positive permeation control compound carbamazepine (log
Pe = −5.23) and positive permeation control compound carbamazepine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pe = −5.16). However, benzophenone SMK-22 exhibited poor permeability in this assay (log
Pe = −5.16). However, benzophenone SMK-22 exhibited poor permeability in this assay (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pe = −6.32), comparable to the low permeation control compound furosemide (log
Pe = −6.32), comparable to the low permeation control compound furosemide (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pe = −6.76). SMK-22 was also observed to exhibit very poor solubility in aqueous media. Based on its superior performance in experiments to this stage, diazirine SMK-24 was selected for further study and no further investigations were carried out using benzophenone SMK-22.
Pe = −6.76). SMK-22 was also observed to exhibit very poor solubility in aqueous media. Based on its superior performance in experiments to this stage, diazirine SMK-24 was selected for further study and no further investigations were carried out using benzophenone SMK-22.
Anticipating the translatability of SMK-24 to profiling Rpn11 in live cells, immortalized murine dendritic cells (DC2.4) were selected for initial study. As a well-characterized immune cell line, DC2.4 cells offered the opportunity to study the impact of SMK-24 on the activity of both the 26S proteasome and immunoproteasome. To examine the permeation of diazirine probe SMK-24, a labelling experiment was performed in parallel using both intact and lysed DC2.4 cells. SMK-24 was incubated with the cell lysate or live DC2.4 cells prior to irradiation. The protein content of labelled live cells was thereafter harvested. All samples were then subject to CuAAC conjugation with biotin-PEG4-azide and proteins were resolved by SDS-PAGE. Western blotting revealed that the major protein bands which were labelled in the lysate samples were also observable in the live cell samples (Fig. 4b). This supported the viability of SMK-24 to permeate live cells and to label endogenously expressed proteins without the need to disrupt cell structure.
To assess the effect upon DC2.4 cells of incubation with probe SMK-24, an MTT cell viability assay was performed. A concentration range of 0–200 μM SMK-24 was incubated in DC2.4 cells, with most cells remaining viable even at high probe concentrations following incubation for 1 hour (Fig. 4c). However, when cells were incubated for a longer period of 24 hours, viability dropped close to zero in samples containing >10 μM SMK-24. The apparent cytotoxicity of diazirine probe SMK-24 at concentrations >10 μM with prolonged incubation times guided future protein profiling experiments in live cells to be conducted with an incubation time of just 1 hour.
With diazirine probe SMK-24 established as a cell-permeable and efficient zinc-binding probe capable of competitively labelling a model zinc metalloenzyme, focus moved to translating this probe towards application in proteomic profiling experiments. Having previously observed that SMK-24 was capable of labelling discrete protein bands in HEK293 lysate (Fig. 4a), DC2.4 lysate (Fig. 4b) and live cells follow-up studies were performed: proteins labelled with SMK-24 in HEK293 lysate, DC2.4 lysate and finally in live cell experiments were enriched on avidin beads and subjected to tryptic digest. The resulting peptides were analyzed by LC-MS/MS. Identification, assignment and analysis of constituent peptides was performed using MaxQuant and Perseus.
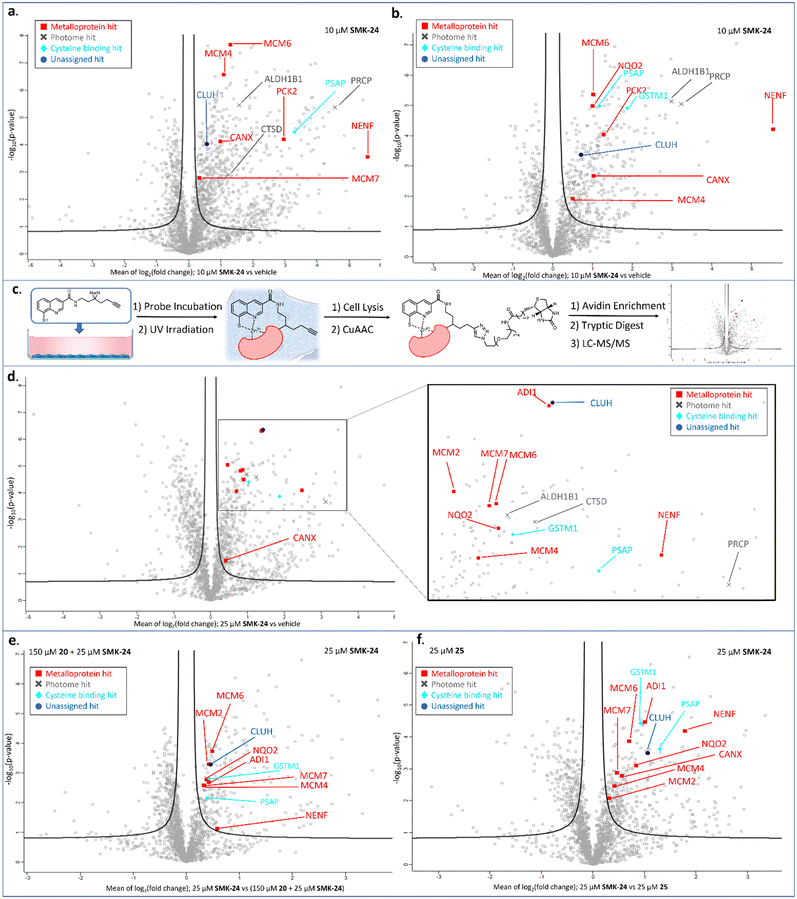
Proteomics results for diazirine probe SMK-24 identified several metalloproteins enriched from cell lysate labelling experiments (Fig. 5a and b). Labelling and enrichment experiments were then translated to live DC2.4 cells using an adapted labelling workflow (Fig. 5c) and were subject to proteomic analysis (Fig. 5d). A comparison was made between the cell lysate and live cell labelling experiments and a table of consistently enriched protein hits was compiled (Fig. S7, ESI†). The robustness of metalloprotein hits were examined through further control experiments. Samples treated with both SMK-24 and 8MQ-bearing 20 in competition were contrasted with samples containing SMK-24 alone (Fig. 5e). Significantly diminished enrichment of protein hits in competition samples indicated protein binding occurred through engagement of the 8MQ motif. Control experiments with the non-zinc-binding diazirine 25 helped identify proteins enriched through non-specific labelling (Fig. 5f). Those hits which passed these thresholds and could be detected in repeat experiments were considered robust.
The validated metalloprotein hits included N-ribosyldihydronicotinamide quinone reductase 2 (NQO2), neudesin neurotrophic factor (NENF), phosphoenolpyruvate carboxykinase 2 (PCK2), calnexin (CANX), acireductone dioxygenase 1 (ADI1) and minichromosome maintenance complex component 6 (MCM6). NQO2, a thioredoxin enzyme dependent upon a zinc co-factor for catalytic activity, plays an important role in stabilizing the tumor suppressor gene p53.42,43 NENF is an iron-binding protein which promotes cell proliferation in undifferentiated neural progenitor cells and is a prognostic marker in renal cancer cells.44,45 PCK2 is a mitochondrial kinase dependent upon the binding of a manganese cofactor to enable phosphorylation of oxaloacetate in the citric acid cycle and it's activity has been implicated in chemotherapeutic drug resistance.21,46 CANX is a calcium and zinc-binding protein which acts as a chaperone in the formation of glycoproteins at the endoplasmic reticulum and is a prognostic marker in colorectal cancer.47,48 ADI1 is an iron and nickel binding enzyme which functions as a methionine scavenger and exerts tumor suppressor effects in prostate cancer.49 Excitingly, SMK-24 was found to enrich a significantly different subset of the metalloproteome than can be profiled using the existing library of metal-binding AfBPs (Fig. 6).4–8,10–15,50–53 An interesting outcome of the metalloprotein hits discussed above show that 8MQ may also chelate metalloproteins bearing non-zinc metal ion cofactors.
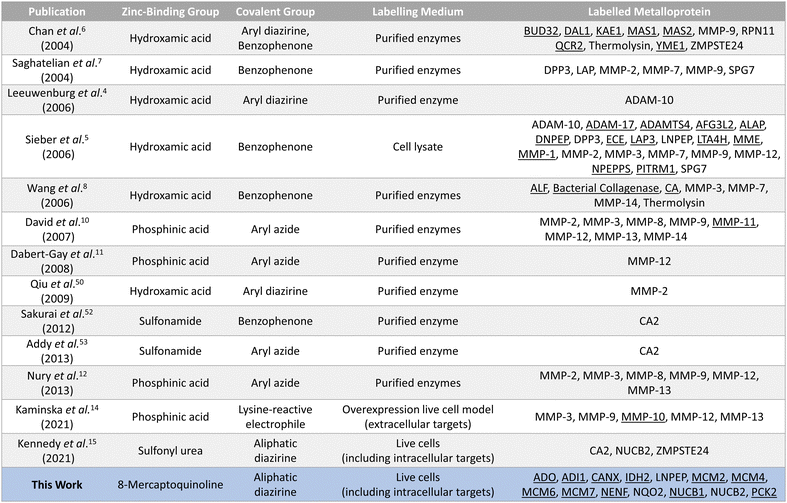 | ||
| Fig. 6 Summary of the reported AfBPs for profiling members of the metalloproteome. Key structural motifs and the corresponding metalloprotein(s) labelled are provided. Unique entries are underlined. | ||
In addition to metal-binding hits, several other commonly enriched proteins were observed in our data sets. A subset of these hits including cathepsin D (CTSD), prolylcarboxypeptidase (PRCP) and aldehyde dehydrogenase 1 family member B1 (ALDH1B1) are reported members of the ‘photome’, a selection of proteins described by Kleiner et al. which are readily and non-specifically labelled by diazirine-containing AfBPs.31 As enrichment of these photome hits was unlikely to be indicative of 8MQ binding, these hits were disregarded. A consistently enriched subset of hits was also found to include cysteine-rich or cysteine-binding proteins such as prosaposin (PSAP), cysteine and glycine rich protein 1 (CSRP1) and glutathione S-transferase mu 1 (GSTM1). Detection of these proteins indicates that SMK-24 is capable of undergoing in situ oxidation to form protein-probe disulfide complexes prior to photoactivation and labelling. Finally, a subset of consistently enriched proteins were identified with no easily assignable binding mode. One such example is clustered mitochondria protein homologue (CLUH), a cytosolic mRNA binding protein, which was consistently and competitively enriched in cell lysate and live cell experiments with diazirine probe SMK-24 (Fig. 5d–f and Fig. S8, S9, ESI†).
To our knowledge, there has been no report of CLUH binding metal ion co-factors. However, our results suggest that CLUH may be capable of binding a metal ion co-factor or associating with a secondary metalloprotein in the cellular environment.
Despite labelling several metalloproteins, we were surprised to discover that SMK-24 had not enriched endogenously expressed Rpn11 from the extracts of either HEK293 or DC2.4 cells. Increasing the loading of SMK-24 to high concentrations (≤200 μM) was not found to result in Rpn11 enrichment. A possible explanation for this finding is that SMK-24 may have bound effectively to the purified recombinant Rpn11/Rpn8 heterodimer, but not to endogenous Rpn11 in the greater structure of the 26S proteasome. Alternatively, the selection of the diazirine photoactivatable group in this probe may have been suboptimal for Rpn11 labelling.
The metalloprotein hit most consistently enriched by diazirine probe SMK-24 in lysate and live cell labelling experiments was minichromosome maintenance complex component 6 (MCM6). MCM6 is a nucleosomal protein which forms part of a hexameric DNA helicase complex in once per cell-cycle DNA replication.54,55 The complex is formed through interaction of MCM components 2–7 (MCM2–7) and is recruited to DNA replication origins by CDT1 and CDC6.56,57 In addition to MCM6 enrichment, MCM4 and MCM7 were also labelled in lysate experiments (Fig. 5a and b). The MCM components share several common structural motifs, including an N-terminal zinc finger domain which is understood to play a key role in hexamer formation and binding of DNA for helicase activity.57,58 Given that diazirine probe SMK-24 enriched MCM6 in a competitive manner with 8MQ-bearing 20, and was not enriched by non-zinc-binding control 25, it was rationalized that SMK-24 was binding MCM6 via the zinc-finger domain. MCM6 overexpression has been found to be an adverse prognostic marker in several cancer types, including lung cancer, breast cancer, glioma, lymphoma and oesophageal squamous cell carcinoma.55,59–61 MCM4 and MCM7 overexpression has been found to be adversely prognostic in cases of oesophageal carcinoma and squamous cell carcinoma.61–63 The overexpression of MCM2 has been associated with oral squamous cell carcinoma and cervical carcinoma, while MCM3 and MCM5 have been found as biomarkers of dysplastic oral lesions and renal cell carcinoma respectively.19,20,64 An AfBP for detection of MCM components could therefore prove to be a valuable tool in diagnostics and research. As a result, focus moved towards establishing the viability of SMK-24 as a tool for detection of MCM components.
We were delighted to discover that several MCM components, including MCM2, MCM4, MCM6 and MCM7 could be successfully enriched through application of diazirine probe SMK-24 in live DC2.4 cell labelling experiments (Fig. 5d). Labelling of these components was found to be competitive with 8MQ-bearing 20 (Fig. 5e), while enrichment was observed above the non-zinc-binding diazirine 25 (Fig. 5f and Fig. S9, ESI†). A follow-up live cell labelling experiment was performed in primary bone marrow derived dendritic cells (BMDC) (Fig. S11, ESI†), where MCM6 was enriched as the only detectable MCM component.
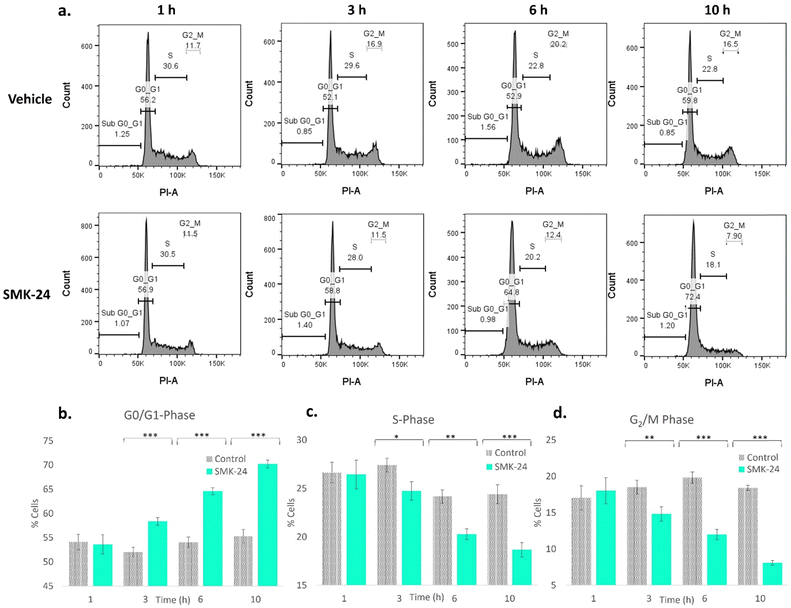
It was recognized that in binding MCM components via a functionally crucial zinc-finger domain, diazirine probe SMK-24 may disrupt the formation or activity of the MCM helicase complex. This would affect the cell's ability to duplicate DNA, hence stalling its transition from the growth (G1) phase to the DNA synthesis (S) phase of the cell cycle. To test whether treatment with SMK-24 caused cells to stall in the quiescent (G0) or G1 phase, a study was performed where HEK293 cells were incubated with SMK-24 for 1–10 h, after which cells were fixed and treated with the DNA stain propidium iodide.65 The DNA content of the cells was analyzed using flow cytometry (Fig. 7a and Fig. S12, ESI†).
Analysis of cells treated with diazirine probe SMK-24 revealed significant changes in cell cycle population over the duration of the experiment. The relative population of cells in G0/G1-phase increased over time (Fig. 7b), while the population of cells in S-phase (Fig. 7c) and growth 2 (G2) phase and mitosis (M) (Fig. 7d) decreased. This result is consistent with disruption or inactivation of the MCM helicase complex, resulting in greater numbers of cells stalling in G0/G1 phase. This effect may be augmented by cellular stresses resulting from off-target engagement of SMK-24, as well as zinc scavenging effects which may disrupt cellular signalling and cause a greater proportion of cells to enter the G0 quiescence state. No significant change was observed in the population of apoptotic cells in the sub-G0/G1 population over the course of this experiment, indicating that population changes were not significantly driven by apoptotic cell death.
The MCM components are perhaps the most significant metalloproteins to be detected using probe SMK-24. While 8MQ-containing compounds are known to exhibit off-target activity, the identification of several binding partners implicated in key DNA replication machinery is potentially significant. These results would justify further investigations to detect whether other 8MQ-containing compounds exert similar effects on the cell cycle. This may reveal an exciting opportunity for future research into a diagnostically significant class of metalloproteins.
Conclusions
We have developed a photoactivatable affinity-based probe for profiling a range of metalloproteins in live cell experiments. Diazirine probe SMK-24 was demonstrated to successfully labelled Rpn11 in a simple model system, but proved not to be translatable to detection of the endogenously expressed native enzyme. Instead, several therapeutically significant metalloproteins were successfully enriched from lysate and live cell labelling experiments and identified using proteomics. We discovered several components of the minichromosome maintenance complex as novel metalloprotein binding partners of SMK-24. In particular, we recognized MCM6, an adverse prognostic biomarker in several cancer variants, to be consistently and sensitively enriched with SMK-24 in the mammalian cell lines tested. Finally, we demonstrated that treatment of SMK-24 in HEK293 cells resulted in significant changes in cell cycle population, consistent with inactivation of the MCM helicase complex. We anticipate SMK-24 to be an important tool in the study of several therapeutically significant metalloproteins.Author contributions
Probe design was performed by S. M. M. and J. F. M. Experiment design was performed by S. M. M., J. F. M., S. I. V. K., B. I. F. and D. M. Z. All practical chemistry and biology was performed by S. M. M. LC-MS/MS data acquisition was performed by B. I. F. Data processing and analysis was performed by S. M. M. The manuscript was written by S. M. M. and J. F. M.Data availability
Characterisation data for all compounds is available in the ESI† as are further supporting experimental data referenced in the manuscript. Chemical and biochemical experimental methods are also included as are NMR spectra of the probe disulfide precursor compounds. Proteomics data is available through the PRIDE partner repository using the identifier: PXD052663.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This publication was supported by the Synthesis and Solid State Pharmaceutical Centre (SSPC), funded by Science Foundation Ireland (SFI) Grant no. 12/RC/2275_P2 (J. F. M.), Trinity College Dublin (J. F. M.), European Molecular Biology Organisation, Scientific Exchange Grant no. 9879 (S. M. M.) and ERC CoG KineTic grant number 865175 (S. I. V. K.). We wish to thank Professor Andreas Martin of UC Berkley for generously permitting use of the plasmid for Rpn11/Rpn8 expression and Dr Dharjath Hameed of Leiden University Medical Center, Leiden University for supplying the plasmid and providing valuable guidance on expression and purification. We also wish to thank Dr John O’Brien and Dr Gary Hessman in Trinity College Dublin School of Chemistry for support in compound characterization and to Dr Barry Moran in Trinity College Dublin School of Biochemistry and Immunology for support in flow cytometry. Finally, we wish to thank Dr Amber Barendrecht, Constant Tellinga, Dr Nina Ligthart, Thijmen Mostert and Dr Tyrza van Leeuwen of Leiden Institute of Chemistry, Leiden University for kindly donating cells for lysate labelling experiments. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD052663.66Notes and references
- B. F. Cravatt, A. T. Wright and J. W. Kozarich, Annu. Rev. Biochem., 2008, 77, 383–414 CrossRef CAS PubMed.
- S. M. McKenna, E. M. Fay and J. F. McGouran, ACS Chem. Biol., 2021, 16, 2719–2730 CrossRef CAS PubMed.
- H. Fang, B. Peng, S. Y. Ong, Q. Wu, L. Li and S. Q. Yao, Chem. Sci., 2021, 12, 8288–8310 RSC.
- M. A. Leeuwenburgh, P. P. Geurink, T. Klein, H. F. Kauffman, G. A. Van Der Marel, R. Bischoff and H. S. Overkleeft, Org. Lett., 2006, 8, 1705–1708 CrossRef CAS PubMed.
- S. A. Sieber, S. Niessen, H. S. Hoover and B. F. Cravatt, Nat. Chem. Biol., 2006, 2, 274–281 CrossRef CAS PubMed.
- E. W. S. Chan, S. Chattopadhaya, R. C. Panicker, X. Huang and S. Q. Yao, J. Am. Chem. Soc., 2004, 126, 14435–14446 CrossRef CAS PubMed.
- A. Saghatelian, N. Jessani, A. Joseph, M. Humphrey and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10000–10005 CrossRef CAS PubMed.
- J. Wang, M. Uttamchandani, J. Li, M. Hu and S. Q. Yao, Chem. Commun., 2006, 3783–3785 RSC.
- R. Nakai, C. M. Salisbury, H. Rosen and B. F. Cravatt, Bioorg. Med. Chem., 2009, 17, 1101–1108 CrossRef CAS PubMed.
- A. David, D. Steer, S. Bregant, L. Devel, A. Makaritis, F. Beau, A. Yiotakis and V. Dive, Angew. Chem., Int. Ed., 2007, 46, 3275–3277 CrossRef CAS PubMed.
- A. S. Dabert-Gay, B. Czarny, L. Devel, F. Beau, E. Lajeunesse, S. Bregant, R. Thai, A. Yiotakis and V. Dive, J. Biol. Chem., 2008, 283, 31058–31067 CrossRef CAS PubMed.
- C. Nury, B. Czarny, E. Cassar-Lajeunesse, D. Georgiadis, S. Bregant and V. Dive, ChemBioChem, 2013, 14, 107–114 CrossRef CAS PubMed.
- K. Teruya, K. F. Tonissen and S. A. Poulsen, MedChemComm, 2016, 7, 2045–2062 RSC.
- M. Kaminska, P. Bruyat, C. Malgorn, M. Doladilhe, E. Cassar-Lajeunesse, C. Fruchart Gaillard, M. De Souza, F. Beau, R. Thai, I. Correia, A. Galat, D. Georgiadis, O. Lequin, V. Dive, S. Bregant and L. Devel, Angew. Chem., 2021, 133, 18420–18427 CrossRef.
- C. R. Kennedy, A. Goya Grocin, T. Kovačič, R. Singh, J. A. Ward, A. R. Shenoy and E. W. Tate, ACS Chem. Biol., 2021, 16, 982–990 CrossRef CAS PubMed.
- A. Agrawal, S. L. Johnson, J. A. Jacobsen, M. T. Miller, L. H. Chen, M. Pellecchia and S. M. Cohen, ChemMedChem, 2010, 5, 195–199 CrossRef CAS PubMed.
- J. A. Jacobsen, J. L. Fullagar, M. T. Miller and S. M. Cohen, J. Med. Chem., 2011, 54, 591–602 CrossRef CAS PubMed.
- C. Perez, J. Li, F. Parlati, M. Rouffet, M. Yuyong, A. L. Mackinnon, T. F. Chou, R. J. Deshaies and S. M. Cohen, J. Med. Chem., 2017, 60, 1343–1361 CrossRef CAS PubMed.
- H. Zhong, B. Chen, H. Neves, J. Xing, Y. Ye, Y. Lin, G. Zhuang, S. D. Zhang, J. Huang and H. F. Kwok, Cancer Manag. Res., 2017, 9, 637–647 CrossRef CAS PubMed.
- Y. Li, J. Zou, Q. Zhang, F. Quan, L. Cao, X. Zhang, J. Liu and D. Wu, Front. Oncol., 2021, 11, 1–17 CAS.
- Z. Jing, J. Gao, J. Li, F. Niu, L. Tian, P. Nan, Y. Sun, X. Xie, Y. Zhu, Y. Zhao, F. Liu, L. Zhou, Y. Sun and X. Zhao, Cancer Lett., 2021, 519, 46–62 CrossRef CAS PubMed.
- Y. De Chu, H. Y. Lai, L. M. Pai, Y. H. Huang, Y. H. Lin, K. H. Liang and C. T. Yeh, Cell Death Dis., 2019, 10, 240 CrossRef PubMed.
- J. Li, T. Yakushi, F. Parlati, A. L. MacKinnon, C. Perez, Y. Ma, K. P. Carter, S. Colayco, G. Magnuson, B. Brown, K. Nguyen, S. Vasile, E. Suyama, L. H. Smith, E. Sergienko, A. B. Pinkerton, T. D. Y. Chung, A. E. Palmer, I. Pass, S. Hess, S. M. Cohen and R. J. Deshaies, Nat. Chem. Biol., 2017, 13, 486–493 CrossRef CAS PubMed.
- D. S. Hameed, A. Sapmaz, L. Burggraaff, A. Amore, C. J. Slingerland, G. J. P. van Westen and H. Ovaa, Angew. Chem., Int. Ed., 2019, 58, 14477–14482 CrossRef CAS PubMed.
- N. C. Taylor and J. F. McGouran, Front. Chem., 2020, 7, 914 CrossRef PubMed.
- T. E. T. Mevissen and D. Komander, Annu. Rev. Biochem., 2017, 86, 159–192 CrossRef CAS PubMed.
- H. A. Bustamante, N. Albornoz, E. Morselli, A. Soza and P. V. Burgos, Cell. Signal., 2023, 101, 1–7 CrossRef PubMed.
- J. Wang, Q. Chen, Y. Shan, X. Pan and J. Zhang, Trends Anal. Chem., 2019, 115, 110–120 CrossRef CAS.
- D. P. Murale, S. C. Hong, M. M. Haque and J. S. Lee, Proteome Sci., 2017, 15, 1–34 Search PubMed.
- J. R. Hill and A. A. B. Robertson, J. Med. Chem., 2018, 61, 6945–6963 CrossRef CAS PubMed.
- P. Kleiner, W. Heydenreuter, M. Stahl, V. S. Korotkov and S. A. Sieber, Angew. Chem., Int. Ed., 2017, 56, 1396–1401 CrossRef CAS PubMed.
- J. P. Holland, M. Gut, S. Klingler, R. Fay and A. Guillou, Chem. - Eur. J., 2020, 26, 33–48 CrossRef CAS PubMed.
- L. L. Punzalan, L. Jiang, D. Mao, A. Das Mahapatra, S. Sato, Y. Takemoto, M. Tsujimura, K. Kusamori, M. Nishikawa, L. Zhou and M. Uesugi, Cell Chem. Biol., 2020, 27, 708–718 CrossRef CAS PubMed.
- L. Dubinsky, B. P. Krom and M. M. Meijler, Bioorg. Med. Chem., 2012, 20, 554–570 CrossRef CAS PubMed.
- A. V. West, G. Muncipinto, H. Y. Wu, A. C. Huang, M. T. Labenski, L. H. Jones and C. M. Woo, J. Am. Chem. Soc., 2021, 143, 6691–6700 CrossRef CAS PubMed.
- H. Park, J. Y. Koo, Y. V. V. Srikanth, S. Oh, J. Lee, J. Park and S. B. Park, Chem. Commun., 2016, 52, 5828–5831 RSC.
- P. Buglyó, E. M. Nagy, E. Farkas, I. Sóvágó, D. Sanna and G. Micera, Polyhedron, 2007, 26, 1625–1633 CrossRef.
- T. Koike, E. Kimura, I. Nakamura, Y. Hashimoto and M. Shiro, J. Am. Chem. Soc., 1992, 114, 7338–7345 CrossRef CAS.
- M. Lukáš, M. Kývala, P. Hermann, I. Lukeš, D. Sanna and G. Micera, J. Chem. Soc. Dalt. Trans., 2001, 2850–2857 RSC.
- B. Y. Guo, W. L. Wei and J. M. Lin, J. Chromatogr. Sci., 2009, 47, 116–120 CAS.
- P. A. Shah, J. V. Shah, M. Sanyal and P. S. Shrivastav, Int. J. Pharm. Pharm. Sci., 2015, 7, 105–111 CAS.
- K. K. K. Leung and B. H. Shilton, J. Biol. Chem., 2013, 288, 11242–11251 CrossRef CAS PubMed.
- Z. Lin, X. Wang, K. A. Bustin, K. Shishikura, N. R. McKnight, L. He, R. M. Suciu, K. Hu, X. Han, M. Ahmadi, E. J. Olson, W. H. Parsons and M. L. Matthews, ACS Cent. Sci., 2021, 7, 1524–1534 CrossRef CAS PubMed.
- H. Ohta, I. Kimura, M. Konishi and N. Itoh, Front. Mol. Biosci., 2015, 2, 1–5 Search PubMed.
- I. Kimura, Y. Nakayama, Y. Zhao, M. Konishi and N. Itoh, Front. Neurosci., 2013, 7, 1–5 Search PubMed.
- M. Escós, P. Latorre, J. Hidalgo, R. Hurtado-Guerrero, J. A. Carrodeguas and P. López-Buesa, Biochem. Biophys. Rep., 2016, 7, 124–129 Search PubMed.
- D. B. Williams, J. Cell Sci., 2006, 119, 615–623 CrossRef CAS PubMed.
- D. Ryan, S. Carberry, Á. C. Murphy, A. U. Lindner, J. Fay, S. Hector, N. McCawley, O. Bacon, C. G. Concannon, E. W. Kay, D. A. McNamara and J. H. M. Prehn, J. Transl. Med., 2016, 14, 1–10 CrossRef PubMed.
- S. W. Oram, J. Ai, G. M. Pagani, M. R. Hitchens, J. A. Stern, S. Eggener, M. Pins, W. Xiao, X. Cai, R. Haleem, F. Jiang, T. C. Pochapsky, L. Hedstrom and Z. Wang, Neoplasia, 2007, 9, 643–651 CrossRef CAS PubMed.
- W. Qiu, J. Xu, X. Li, L. Zhong, J. Li, J. Li and F. Nan, Chin. J. Chem., 2009, 27, 825–833 CrossRef CAS.
- P. P. Geurink, T. Klein, L. Prèly, K. Paal, M. A. Leeuwenburgh, G. A. Van Der Marel, H. F. Kauffman, H. S. Overkleeft and R. Bischoff, Eur. J. Org. Chem., 2010, 2100–2112 CrossRef CAS.
- K. Sakurai, M. Tawa, A. Okada, R. Yamada, N. Sato, M. Inahara and M. Inoue, Chem. – Asian J., 2012, 7, 1567–1571 CrossRef CAS PubMed.
- P. S. Addy, B. Saha, N. D. P. Singh, A. K. Das, J. T. Bush, C. Lejeune, C. J. Schofield and A. Basak, Chem. Commun., 2013, 49, 1930–1932 RSC.
- N. Tuteja and R. Tuteja, Eur. J. Biochem., 2004, 271, 1835–1848 CrossRef CAS PubMed.
- L. Cheng, Z. Tan, Z. Huang, Y. Pan, W. Zhang and J. Wang, Med. Sci. Monit., 2020, 26, 1–15 Search PubMed.
- Z. Wei, C. Liu, X. Wu, N. Xu, B. Zhou, C. Liang and G. Zhu, J. Biol. Chem., 2010, 285, 12469–12473 CrossRef CAS PubMed.
- Y. Zhai, E. Cheng, H. Wu, N. Li, P. Y. K. Yung, N. Gao and B. K. Tye, Nat. Struct. Mol. Biol., 2017, 24, 300–308 CrossRef CAS PubMed.
- Z. You, Y. Ishimi, H. Masai and F. Hanaoka, J. Biol. Chem., 2002, 277, 42471–42479 CrossRef CAS PubMed.
- T. Zeng, Y. Guan, Y. Kun Li, Q. Wu, X. Jun Tang, X. Zeng, H. Ling and J. Zou, Clin. Chim. Acta, 2021, 517, 92–98 CrossRef CAS PubMed.
- Y. Gu, X. Hu, X. Liu, C. Cheng, K. Chen, Y. Wu and Z. Wu, BMC Cancer, 2021, 21, 1–14 CrossRef PubMed.
- T. Cao, S. J. Yi, L. X. Wang, J. X. Zhao, J. Xiao, N. Xie, Z. Zeng, Q. Han, H. O. Tang, Y. K. Li, J. Zou and Q. Wu, BioMed, 2020, 3574261 CAS.
- C. J. Feng, H. J. Li, J. N. Li, Y. J. Lu and G. Q. Liao, Anticancer Res., 2008, 28, 3763–3769 CAS.
- S. Yang, Y. Yuan, W. Ren, H. Wang, Z. Zhao, H. Zhao, Q. Zhao, X. Chen, X. Jiang and L. Zhang, Front. Oncol., 2022, 12, 1–23 Search PubMed.
- A. G. Lameira, F. S. C. Pontes, D. M. Guimarães, A. C. G. Alves, A. S. de Jesus, H. A. R. Pontes, D. dos and S. Pinto, J. Oral Pathol. Med., 2014, 43, 427–434 CrossRef CAS PubMed.
- M. J. Meegan, S. Nathwani, B. Twamley, D. M. Zisterer and N. M. O’Boyle, Eur. J. Med. Chem., 2017, 125, 453–463 CrossRef CAS PubMed.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. Garcia-Seisdedos, S. Hewapathirana, S. Kamatchinathan, D. J. Kundu, A. Prakash, A. Frericks-Zipper, M. Eisenacher, M. Walzer, S. Wang, A. Brazma and J. A. Vizcaíno, Nucleic Acids Res., 2021, 50, D543–D552 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00053f |
| This journal is © The Royal Society of Chemistry 2024 |