Open Access Article
Open Access ArticleHelix-based screening with structure prediction using artificial intelligence has potential for the rapid development of peptide inhibitors targeting class I viral fusion†
Satoshi
Suzuki
 a,
Mio
Kuroda
b,
Keisuke
Aoki
bc,
Kumi
Kawaji
d,
Yoshiki
Hiramatsu
a,
Mina
Sasano
d,
Akie
Nishiyama
a,
Kazutaka
Murayama
a,
Mio
Kuroda
b,
Keisuke
Aoki
bc,
Kumi
Kawaji
d,
Yoshiki
Hiramatsu
a,
Mina
Sasano
d,
Akie
Nishiyama
a,
Kazutaka
Murayama
 e,
Eiichi N.
Kodama
adf,
Shinya
Oishi
e,
Eiichi N.
Kodama
adf,
Shinya
Oishi
 bc and
Hironori
Hayashi
bc and
Hironori
Hayashi
 *d
*d
aDepartment of Infectious Diseases, Tohoku University Graduate School of Medicine, 2-1, Seiryo-machi, Aoba-ku, Sendai, Miyagi 980-8575, Japan
bLaboratory of Medicinal Chemistry, Kyoto Pharmaceutical University, 1, Misasagi-Shichono-cho, Yamashina-ku, Kyoto 607-8412, Japan
cGraduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan
dDivision of Infectious Diseases, International Research Institute of Disaster Science, Tohoku University, 2-1, Seiryo-machi, Aoba-ku, Sendai, Miyagi 980-8575, Japan. E-mail: hironori.hayashi.b1@tohoku.ac.jp
eDivision of Biomedical Measurements and Diagnostics, Graduate School of Biomedical Engineering, Tohoku University, 2-1, Seiryo-machi, Aoba-ku, Sendai, Miyagi 980-8575, Japan
fDepartment of Infectious Disease, Graduate School of Medicine and Tohoku Medical Megabank Organization, Tohoku University, 2-1, Seiryo-machi, Aoba-ku, Sendai, Miyagi 980-8575, Japan
First published on 7th November 2023
Abstract
The rapid development of drugs against emerging and re-emerging viruses is required to prevent future pandemics. However, inhibitors usually take a long time to optimize. Here, to improve the optimization step, we used two heptad repeats (HR) in the spike protein (S protein) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a model and established a screening system for peptide-based inhibitors containing an α-helix region (SPICA). SPICA can be used to identify critical amino acid regions and evaluate the inhibitory effects of peptides as decoys. We further employed an artificial intelligence structure-prediction system (AlphaFold2) for the rapid analysis of structure–activity relationships. Here, we identified that critical amino acid regions, DVDLGD (amino acids 1163–1168 in the S protein), IQKEIDRLNE (1179–1188), and NLNESLIDL (1192–1200), played a pivotal role in SARS-CoV-2 fusion. Peptides containing these critical amino acid regions efficiently blocked viral replication. We also demonstrated that AlphaFold2 could successfully predict structures similar to the reported crystal and cryo-electron microscopy structures of the post-fusion form of the SARS-CoV-2 S protein. Notably, the predicted structures of the HR1 region and the peptide-based fusion inhibitors corresponded well with the antiviral effects of each fusion inhibitor. Thus, the combination of SPICA and AlphaFold2 is a powerful tool to design viral fusion inhibitors using only the amino-acid sequence of the fusion protein.
Introduction
In the current global era, the occurrence of pandemics of respiratory viral diseases is difficult to prevent. Since coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported,1 at least six million patients worldwide have died from COVID-19 as of mid 2023.2 This high death toll was because of the weak therapeutic efficacy and limited clinical use of antiviral drugs and the emergence of vaccine-escaping SARS-CoV-2 variants.3–5 In preparation for future unpredictable viral outbreaks, novel strategies for rapid drug development are required to generate treatments for infected individuals and to control the viral transmission.4Certain enveloped viruses, e.g., SARS-CoV-2, human immunodeficiency virus (HIV), Ebola virus, respiratory syncytial virus, and measles virus (MeV), express class I fusion proteins (FP) on the surface of the viral envelop.6–12 The class I FP induces viral membrane fusion by regulated structural transitions after binding of the virus to its receptor on the host cell surface (Fig. 1 and Fig. S1, ESI†). Briefly, the N-terminal hydrophobic domains of FP, which are termed the fusion peptides, are inserted into the host cell membrane. Following the insertion, the C-terminal α-helix region, heptad repeat 2 (HR2), interacts with the trimeric N-terminal α-helix region of the FP, heptad repeat 1 (HR1). The helix–helix interaction (HHI) between HR1 and HR2 causes intramolecular folding and forms a six-helix bundle (6HB), which drives membrane fusion resulting in viral entry.6–12
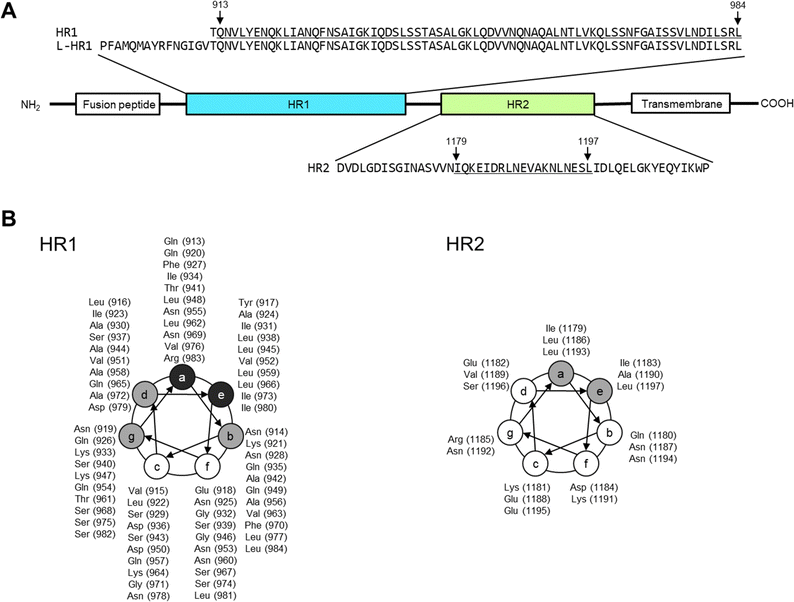 | ||
| Fig. 1 Amino-acid sequences of viral fusion proteins of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), heptad repeat 1 and 2 (HR1 and 2, respectively). The amino acid numbers in the parentheses are derived from the entire Wuhan strain spike protein (S protein), EPI_ISL_408667 (GISAID).10,48 (A) The positions of fusion peptide, HR1, HR2, and transmembrane domain are shown. According to 6LXT10 in the protein data bank, Gln913 to Leu984 of HR1 and Ile1179 to Leu1197 of HR2 form an α-helix structure (underlined regions). (B) Helical wheel depicted with amino acids and sequence numbers. Amino acids indicated in gray mainly form the interface between HR1 and 2, and those in black are involved in HR1 trimer formation. | ||
We have developed a rational strategy for peptide design targeting the HR1–HR2 interaction and have identified fusion inhibitors against HIV-1,13–16 MeV,17 and SARS-CoV.18 During the COVID-19 pandemic, an mRNA vaccine was rapidly developed using only the nucleotide sequence information.19 Our strategy, which is based on only the amino-acid or nucleotide sequence information, is expected to enable the design of peptide fusion inhibitors against the HR1–HR2 interaction in a short time (Fig. S1, ESI†). Fusion inhibitors against various viruses, including SARS-CoV-2, have been previously reported.10,20–22 However, further optimization of the peptide design to enable a quick response to unpredictable viral outbreaks requires efficient methods to rapidly (i) identify critical amino acid regions for inhibition of the HR1–HR2 interaction and evaluate the inhibitory effects and (ii) determine the three-dimensional (3D) structures of HR1–HR2 peptide complexes using only nucleotide sequence information. Such methods will facilitate the rapid development of effective drugs with less effort, even for viruses that are challenging to propagate in vitro or study because of biological safety limitations.
Here, we established a screening system for peptide-based inhibitors containing an α-helix region (SPICA) using SARS-CoV-2 as a model. Using SPICA, critical amino acid regions for the HR1–HR2 interaction in the SARS-CoV-2 spike protein (S protein) were rapidly identified without requiring peptide synthesis. In addition, the inhibitory effect of the peptides on the HR1–HR2 interaction was examined (Fig. 2(A)). In SPICA, HR1-fused maltose binding protein (MBP) and HR2-fused alkaline phosphatase (ALP) were used (Fig. S2, ESI†), the same as for the F-ELISA13,18 that we have reported previously. We focused on HR2 in the present study because the HR2 sequences are conserved23 and HR1 has a largely unstructured conformation in solution.12,24–26 Additionally, we employed artificial intelligence to predict the 3D structures of peptides containing critical amino acid regions that could interact with the HR1 of the S protein.
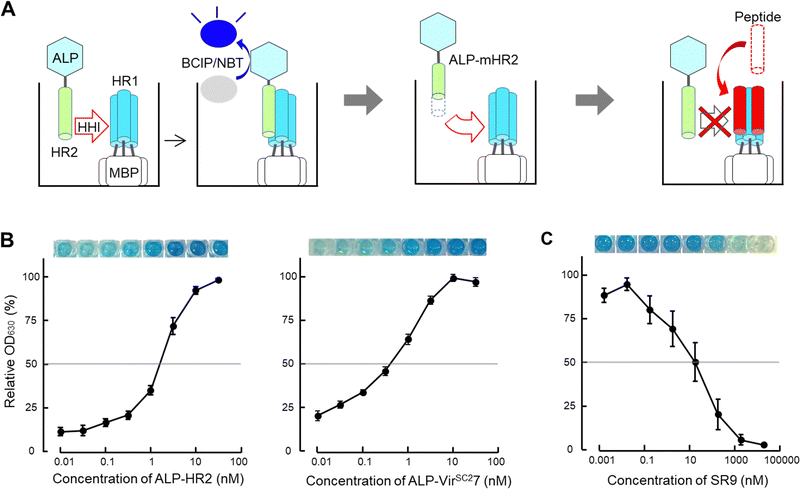 | ||
| Fig. 2 Scheme of the screening system for peptide-based inhibitors containing an α-helix region (SPICA). (A) The helix–helix interaction (HHI) of alkaline phosphatase (ALP) fused with genetically generated heptad repeat 2 (HR2) with plate-coated maltose binding protein (MBP) fused HR1 was examined on an ELISA plate. The HR1–HR2 interactions were detected from the ALP activity (ALP-HR2; see also Fig. S2, ESI†) using the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) color reaction (left). SPICA was used to detect the HR1–HR2 interaction using HR2 or deleted and/or alanine substituted HR2 (modified HR2, mHR2) (middle). SPICA was used to examine the inhibitory effects of peptides toward the HR1–HR2 interaction (right). (B) Interaction profiles of HR1 and HR2 (left) and HR1 and VIRSC27 (right) are shown with the relative OD630 values. The OD630 values dose dependently increased up to 100 nM. The HR1 and HR2 regions of the Wuhan strain of SARS-CoV-2 were used (the sequence is shown in Table 1). (C) The inhibitory profile of SR9 (the sequence of SR9 is shown in Table 2) used in SPICA. | ||
Results and discussion
Identification of critical amino acid regions in HR2 for the interaction with HR1
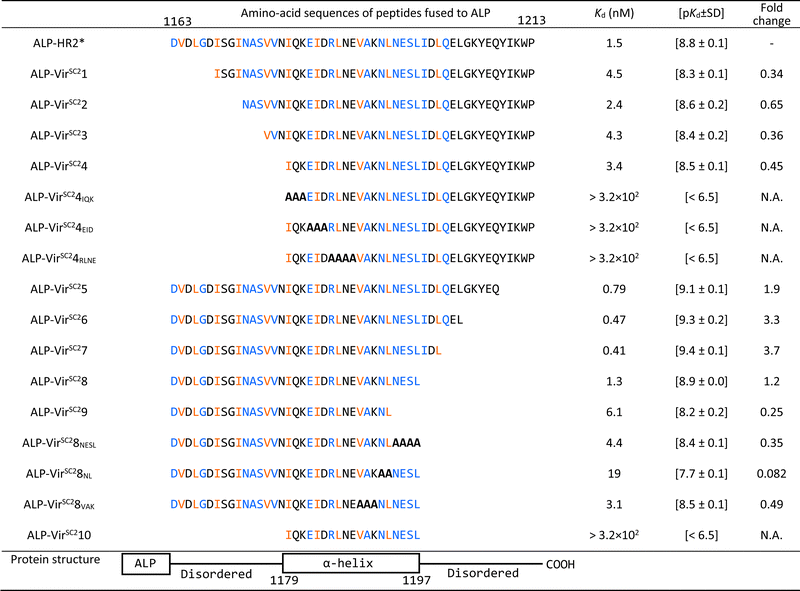
First, we optimized the pH value of the interaction buffer for SPICA (Fig. S3, ESI†). The optimal pH value was determined to be 6.4–7.0. Therefore, SPICA could be used to investigate antiviral effects at near neutral pH values. Then, we determined that non-specific binding of ALP did not appreciably affect the interactions under investigation. The ALP activity was observed to be dependent on the HR1–HR2 interaction (Fig. S4, ESI†).To identify the critical amino acid regions, we introduced amino acid deletions and/or alanine substitutions27 in the HR2 region and the log-transformed dissociation constants (pKd values) of each ALP-fused modified HR2 region (ALP-mHR2) with MBP-fused HR1 (MBP-HR1) were determined (Fig. 2(B) and Table 1). A comparison of the pKd values of ALP-N-terminal amino acid deleted HR2 fusion peptides (ALP-VirSC21 to ALP-VirSC24) with ALP-HR2 (1163–1213, 51 amino acids) indicated that ALP-VirSC21 showed a weak interaction. However, ALP-VirSC22, ALP-VirSC23, and ALP-VirSC24 had interactions comparable with ALP-HR2. The alanine-substituted ALP-VirSC24IQK, ALP-VirSC24EID, and ALP-VirSC24RLNE showed poor interactions. These data indicated that the N-terminal regions DVDLGD (1163–1168) and IQKEIDRLNE (1179–1188) in HR2 were important for the interaction with HR1.
| The interactions of HR1 with HR2 derivatives are shown. The amino acid numbers are the sequence numbers of the entire viral S protein.10 The HR2* sequence used in this study was reported previously.10 Alanine substitutions are indicated with bold letters. The blue and orange-colored amino acids have hydrogen bonds and hydrophobic interactions with HR1, respectively, as determined from 6LXT10 and 6XRA.35 |
|---|
|
For ALP-mHR2 with amino acid deleted at the C-terminus of HR2 (ALP-VirSC25 to ALP-VirSC29), ALP-VirSC25, ALP-VirSC26, and ALP-VirSC27 showed comparable interactions with HR1, compared with ALP-HR2, while ALP-VirSC28 had a lower pKd value than ALP-VirSC27. ALP-VirSC29 had a weak interaction compared with ALP-VirSC28. For other alanine substitutions, ALP-VirSC28NESL had a lower pKd value compared with ALP-VirSC28, indicating the importance of the NESL (1194–1197) region. Moreover, the introduction of alanine substitutions for N and L (1192 and 1193) (ALP-VirSC28NL) reduced the strength of the interaction by one log or more compared with ALP-VirSC28. These data indicated that the C-terminal region NLNESLIDL (1192–1200) of HR2 was important for the interaction with HR1. Finally, SPICA indicated three critical regions, DVDLGD (amino acids 1163–1168 in the S protein), IQKEIDRLNE (1179–1188), and NLNESLIDL (1192–1200) that formed hydrogen bonds and hydrophobic interactions to HR1 trimer (Fig. S5–S7, ESI†).
These results demonstrated that SPICA could be used to determine the Kd values of the HR1–HR2 interaction without the need to use infectious viruses, and to identify the critical amino acid regions of HR2. Of note, previously reported fusion inhibitory peptides against SARS-CoV-2 also contain this critical amino acid region (Table S1, ESI†).28–34
Analysis of structure–activity relationships using crystal and cryo-electron microscopy structures
To investigate the utility of SPICA, the structure–activity relationships were evaluated using a previously reported crystal structure of the SARS-CoV-2 HR1–HR2 complex (PDB ID: 6LXT)10 and a cryo-electron microscopy (cryo-EM) structure of the S protein (PDB ID: 6XRA).35 The ALP-HR2 peptides with N-terminal amino acid deletions, ALP-VirSC21 to ALP-VirSC24, exhibited weaker interactions, by 0.5 log or less, compared with ALP-HR2, in SPICA. Especially, the Kd value of ALP-HR2 was decreased three-fold by the deletion of the DVDLGD region. However, the deletion of N-terminal amino acids (DISGINASVVN) in HR2 (ALP-VirSC22 to ALP-VirSC24) had little effect on the Kd values. In the crystal and cryo-EM structures (Table S2, ESI†), the DVDLG region showed similar interactions, including hydrogen bonds (H-bonds) and hydrophobic interactions, but the DISGINASVVN region formed different or undetected structures, indicating that the DVDLG and DISGINASVVN regions formed strong and weak interactions, respectively, with HR1 (Fig. 3(A), (B) and (D)). Previously reported fusion inhibitors against human parainfluenza also have helical and disordered regions.36 Previous studies of SARS-CoV-2 have reported the importance of the N-terminal amino acids of HR2.37,38 The results of SPICA also indicated that, for fusion inhibitors against SARS-CoV-2, a disordered region and the stability of the α-helical structure were important to exhibit an antiviral effect (Tables 1 and 2). The importance of the disordered regions was reinforced by the crystal structure (Fig. 3 and Table S2, ESI†). Comparing the crystal and cryo-EM structures showed that the N-terminal disordered region included an unstable region (near the DISGINASVVN region) between two stable interaction sites (Fig. 3(B) and (D)). The optimization of this unstable region to induce an interaction with HR1 could lead to the development of more effective fusion inhibitors.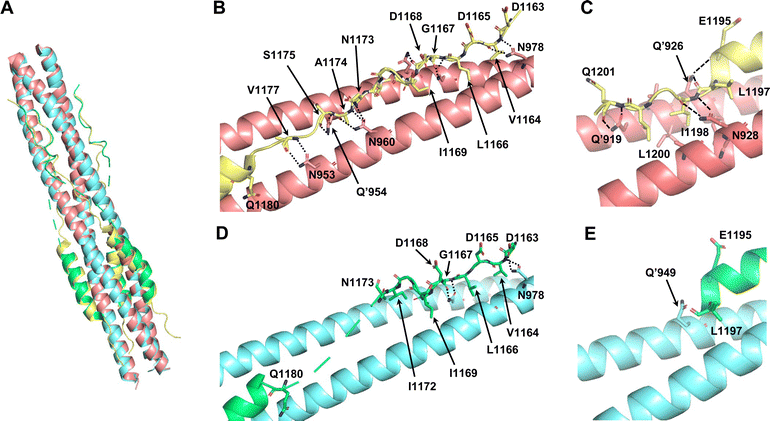 | ||
| Fig. 3 Comparison of the reported crystal structure and cryo-electron microscopy (cryo-EM) structure of 6HB from the SARS-CoV-2 S protein. The light pink and light yellow chains are HR1 and HR2 in 6HB (6LXT),10 respectively. The light blue and light green chains are HR1 and HR2 in the S protein (6XRA),35 respectively. (A) Superimposition of the crystal structure and cryo-EM structure (6LXT and 6XRA, respectively). (B) Structure of the disordered region flanking the N-terminal helical region of HR2 in 6LXT. (C) Structure of the C-terminal helical region of HR2 in 6LXT. (D) Structure of the disordered region flanking the N-terminal helical region of HR2 in 6XRA. The amino acids located from Ala1174 to Ile1179 are not shown. (E) Structure around the C-terminal helical region of HR2 in 6XRA. The amino acids located after Leu1197 are not shown. | ||
| Peptide or compound | Sequences of compounds | IC50 (μM) [pIC50 ± SD] | ||
|---|---|---|---|---|
| HR1 and HR2 in SPICA were the full-length peptide from the Wuhan strain of SARS-CoV-2 as depicted in Fig. 1. EK1 has been reported as a fusion inhibitor of SARS-CoV-2. VP-14637, C45, and T112 have been reported as fusion inhibitors of RSV, and M1 has been reported as a fusion inhibitor of MeV. longHR2_42 contains all the critical amino acid region determined by SPICA. | ||||
| longHR2_42 | AcHN– | PDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL | –CONH2 | 0.0070 [8.2 ± 0.2] |
| EK1 | H2N– | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL | –COOH | 0.0010 [9.0 ± 0.3] |
| SR9 | AcHN– | ISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL | –CONH2 | 0.023 [7.6 ± 0.7] |
| VP-14637 | C25H22N10O3 | 26 [4.6 ± 0.2] | ||
| C45 | H2N– | NFYDPLVFPSDEFDASISQVNEKINQSLAFIRKSDELLHNVNAGK | –COOH | >20 [<4.7] |
| T112 | AcHN– | VFPSDEFDASISQVNEKINQSLAFIRKSDELLHNV | –CONH2 | >20 [<4.7] |
| M1 | AcHN– | ISLERLDVGTNLGNAIAKLEDAKELLESSDQILRS | –CONH2 | >10 [<5.0] |
Regarding the ALP-C-terminal amino acid deleted HR2 fusion peptides (ALP-VirSC25 to ALP-VirSC28), ALP-VirSC26 and ALP-VirSC27 clearly exhibited strong interactions compared with ALP-HR2 and ALP-VirSC28 as indicated by the difference in the pKd values of 0.5 or more by SPICA. ALP-VirSC26 and ALP-VirSC27 have additional sequences, IDLQEL and IDL, respectively, compared with ALP-VirSC28. These amino acids formed H-bonds and hydrophobic interactions in the structure of the HR1–HR2 complex (PDB ID: 6LXT) (Fig. 3(C)). Compared with ALP-VirSC26 and ALP-VirSC27, the weak interaction of ALP-VirSC25 with MBP-HR1 was caused by additional unstable or weak interactions of the amino-acid region (GKYEQ). The cryo-EM structure of the peptide failed to reveal the C-terminal structure containing IDLQELGKYEQ (PDB ID: 6XRA) (Fig. 3(E)).
Structure–activity relationship analysis using AlphaFold2
For additional analysis of the effects of deletions and substitutions, we employed structure prediction using AlphaFold2.39 The predicted structure of the 6HB constructed from HR1 with an additional 15 N-terminal amino acids (L-HR1 shown in Fig. 1(A)) and HR2 was similar (root mean square deviation; RMSD = 0.6897 for 331 common Cα atoms) to the crystal structure of the HR1 and HR2 complex (Fig. 4(A)). Furthermore, the C-terminal amino acid regions, IDL and IDLQEL, in the AlphaFold2 predictions of the HR1-VirSC26 and ALP-VirSC27 complexes were also similar to those in the crystal structures (Fig. 3(C) and 4(C), (D)). Regarding the HR1 and VirSC25 complex, AlphaFold2 predicted that the addition of C-terminal amino acids could affect the C-terminal structure of the peptide (Fig. 4(B)). The structures of VirSC24, VirSC28, and derivatives interacting with HR1 were predicted to investigate the effect of introducing alanine substitutions. All the predicted structures, except for the VirSC28NL complex, were similar to the crystal structures (Fig. S8 and S9, ESI†) but some H-bonds and/or hydrophobic interactions had disappeared. As in the case for VirSC28NL that showed a weak interaction affinity with HR1 in the SPICA results, AlphaFold2 predicted that alanine substitutions would destabilize the helical structure (Fig. S9, ESI†).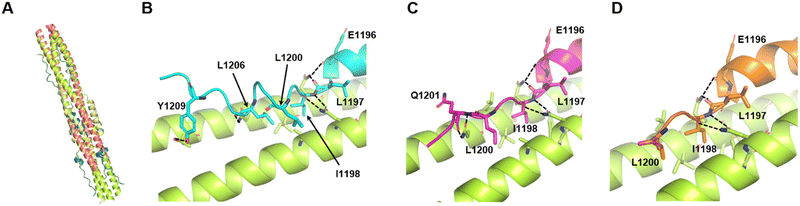 | ||
| Fig. 4 Predicted structures of HR2, VirSC25, VirSC26, and VirSC27 with the L-HR1 trimer in AlphaFold2. (A) Predicted structure of the 6HB constructed from HR2 and HR1 with an additional 15 N-terminal amino acids (L-HR1) superimposed with the previously reported crystal structure (PDB code: 6LXT).10 These two structures were very similar (root mean square deviation; RMSD: 0.6897). (B) to (D) C-terminal amino acids in the predicted structures of VirSC25 (cyan; B), VirSC26 (magenta; C), and VirSC27 (orange; D) complexes with trimeric L-HR1 (light green). Dotted lines indicate predicted H-bonds. | ||
Rapid drug development is required to suppress pandemics by generating treatments against unpredictable emerging and re-emerging viruses. The present study suggested that, in the near future, AlphaFold2 will be used to successfully predict 6HB structures from the amino-acid sequences of HR1 and HR2 (Fig. 4 and Fig. S8, S9, ESI†) without crystallization. Thus, the combination of SPICA and AlphaFold2 can be a useful tool to rapidly design, not only viral fusion inhibitors, but also peptide-based inhibitors targeting intra- and inter-molecular HHI or protein–protein interactions. Furthermore, because fusion inhibitors act in the extracellular space, the effect of such compounds should not be appreciably influenced by the ability for cell penetration. Therefore, antiviral effects predicted from SPICA and AlphaFold2 can be directly correlated with the in vitro effect. Thus, SPICA and AlphaFold2 would be useful in the development of treatments for fatal and highly infectious viruses with class I FP. However, the structures predicted using the current version of AlphaFold2 may still be unsatisfactory for various viral 6HBs and need further optimization for the detailed analysis of protein–protein interactions, and currently, analysis of crystal structures is still required.
Inhibitory effect of HR2-derived peptides and other inhibitors in SPICA
The inhibition of the HR1–HR2 interaction by previously reported fusion inhibitors was investigated using SPICA. As shown in Fig. 2(C) and Table 2, longHR2_42,37 which contains all the critical amino acid regions determined by SPICA, efficiently blocked the HR1–HR2 interaction with an IC50 value in the order of 10−9 M (pIC50 = 8.2 ± 0.2). The IC50 value of SR918 that has an N-terminal amino acid sequence deleted from longHR2_42 was in the order of 10−8 M (pIC50 = 7.6 ± 0.7). EK1, a SARS-CoV-2 fusion inhibitor based on the HR2 of another corona virus strain, HCoV-OC43, with an EK motif,10,22,40,41 also inhibited the interaction between HR1 and HR2 of SARS-CoV-2 (pIC50 = 9.0 ± 0.3). As expected, fusion inhibitors for respiratory syncytial virus and MeV (VP-14637,42 C45,43 T112,25 and M117,25), as negative controls, failed to block the HR1–HR2 interaction of SARS-CoV-2. A viral RNA polymerase inhibitor, remdesivir44,45 also showed no inhibitory effect using SPICA. SPICA demonstrated the ability to selectively identify peptide inhibitors.Antiviral effect in cells
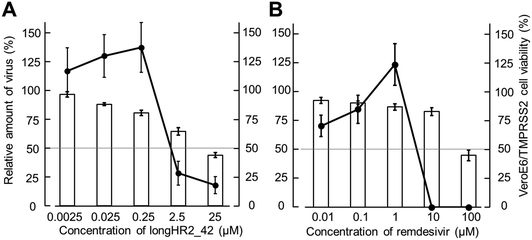
The antiviral effects of longHR2_42 and remdesivir were determined against SARS-CoV-2 in VeroE6/TMPRSS2 cells46 (Fig. 5(A) and (B)). Quantitative RT-PCR analysis indicated that remdesivir and longHR2_42 inhibited SARS-CoV-2 replication with pEC50 values of 5.4 ± 0.2 and 5.8 ± 0.4, respectively. The cytotoxicity of longHR2_42 was relatively high as shown in Fig. 5.Even though the peptides exhibited a strong inhibitory effect (pIC50 = 8.2 ± 0.2) in SPICA, longHR2_42 showed only a moderate antiviral effect and incompletely suppressed approximately 80% of viral replication. The inhibitory effects obtained from SPICA predicted relatively higher values than those from the antiviral assay. SARS-CoV-2 has multiple entry pathways, such as endocytosis.8,11,47 Certain pathways, which are less dependent on fusion by the S protein, might cause escape from the suppression by peptide inhibitors.
Conclusions
SPICA is a useful tool to identify critical amino acid regions, which is required to develop viral entry inhibitors targeting the HR1–HR2 interaction without requiring assays with infectious viruses and/or peptide synthesis. SPICA and structural analysis showed that the combination of α-helical and disordered regions, or the α-helix stability, are important for the inhibition of the HR1–HR2 interaction. Moreover, structures predicted by AlphaFold2 may support the rapid development of viral class I fusion inhibitors. We have demonstrated that SPICA is useful for an initial screening strategy for future pandemics without the need for peptide synthesis or viral assays. Thus, these results will contribute to the development of novel entry inhibitors to rapidly and broadly suppress future pandemics.Experimental
Virus and cells
The SARS-CoV-2 strain JPN/TY/WK-521 (GISAID: EPI_ISL_408667)48 was kindly provided by the National Institute of Infectious Diseases (Tokyo, Japan). The virus was propagated in VeroE6/transmembrane protease serine 2 (TMPRSS2) cells46 derived from African green monkey kidney cells (VeroE6) with TMPRSS2 transfection. The cells were obtained from the Japanese Cancer Research Resources Bank (JCRB) cell bank (Sennan, Japan) and maintained in Dulbecco's modified eagle media (DMEM) (Merck, Darmstadt, Germany) with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 1 mg mL−1 G-418 (Fujifilm Wako Chemicals, Osaka, Japan).Plasmid construction
The DNA fragment of the ALP coding region (amino acids 22–471) without the secretory signal sequence was amplified by PCR from Escherichia coli, JM109 genome (K12 strain, JM109, NIPPON GENE, Tokyo, Japan). The ALP cDNA49,50 was fused with cDNA coding a four glycine linker (Gly4) at the 3′-terminus of the ALP cDNA by PCR using an appropriate primer set (Table S3, ESI†). The resultant ALP-Gly4 cDNA was cloned into a pET47b vector (pET47b-ALP) (Novagen, Billerica, MA, USA) via BamHI/HindIII sites. The various lengths of DNA fragments coding the HR2 region (modified HR2, mHR2) were amplified by RT-PCR from SARS-CoV-2 infected VeroE6/TMPRSS2 cells. The sequences of the primers are listed in Table S3 (ESI†). The amplified fragments were inserted into the pET47b-ALP vector (pET47b-ALP-peptides) via HindIII/XhoI sites. In the vectors, the ALP-HR2 derivatives were designed to be expressed with a hexa-histidine tag fused to the N-terminus of ALP. The HR1 region was amplified in the same way as HR2. The amplified fragments were inserted via SalI/BamHI sites into a pMAL-c6T vector (New England Biolabs, Ipswich, MA, USA), resulting in MBP being fused to the N-terminus of HR1 (Fig. S2, ESI†).Protein expression and purification
The pET47b-ALP-peptide and pMALc6T-HR1 were verified by DNA sequencing and then each transformed into E. coli BL21-Codon Plus (DE3)-RIL (Agilent Technologies, Santa Clara, CA, USA) for bacterial expression. Both cultures were grown in a shake flask containing Luria broth with kanamycin and chloramphenicol for pET47b-ALP-mHR2, or with ampicillin and chloramphenicol for pMALc6T-HR1 at 37 °C. The cultures were grown in the flasks to an optical density of 0.6 at 600 nm at 37 °C. The expression of each protein was induced by addition of isopropyl β-D-thiogalactopyranoside (0.3 and 1.0 mM for ALP-mHR2 and MBP-HR1, respectively). Then, expression of ALP-mHR2 and MBP-HR1 were induced for 16 h at 25 °C,51 and at 4 h at 37 °C, respectively. After incubation, the E. coli were spun down for pellet collection. For purification of ALP-mHR2 and MBP-HR1, each pellet was resuspended in 10 mM Tris–HCl pH 8.0 and sonicated. The cell lysates were centrifuged and the supernatants were collected. The expressed ALP-mHR2 and MBP-HR1 were purified by Ni-NTA agarose (Qiagen, Venlo, Netherlands) and amylose resin (New England Biolabs), respectively. The purified proteins were analyzed using sodium dodecyl sulfate-poly-acrylamide gel electrophoresis (SDS-PAGE) to determine the molecular sizes and purity of the proteins. The concentrations of each protein were determined from the quantities of the protein bands of SDS-PAGE using photos (Chemi Doc MP imaging system, Bio-Rad Hercules, CA, USA) and ImageJ.52 Bovine serum albumin (BSA; Thermo Fisher Scientific) was used as a standard to construct the calibration curve.Detection of the critical amino acid region in ALP-mHR2
MBP-HR1 was diluted to 50 nM with 50 mM sodium carbonate buffer (pH 9.4) and coated on a 96-well ELISA plate (Costar 1 × 8 Stripwell™ high binding EIA/RIA plate; Corning, NY, USA) at 4 °C overnight. After the plate was washed three times with PBS containing 0.025% Tween 20 (T-PBS) (pH 7.4), the plate was subjected to blocking with 0.1% BSA in T-PBS at 4 °C for 2.5 h, and then washed three times with T-PBS. ALP-mHR2 in 100 mM 2-(N-morpholino) ethanesulfonic acid (MES) buffer (pH 6.4) with 100 mM NaCl and 2 mM MgCl2 was added to the plate and diluted to the desired concentrations. The plate was incubated at 37 °C for 1.5 h and then washed three times with 100 mM MES buffer (pH 6.4) containing 0.025% Tween 20. Phosphatase substrate, 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT, BluePhos Microwell Phosphatase Substrate; SeraCare Life Sciences, Milford, MA, USA) containing 1 mM Zn(CH3COO)2 and 2 mM MgCl2 was added and the plate was incubated at 37 °C for 0.5 h. The optical density at 630 nm (OD630) was measured using a microplate reader (BioTek 800 TS, Agilent Technologies). The relative OD630 values were calculated as a ratio of the OD630 value at each concentration and the highest OD630 value in the dilution. Using the two ALP-mHR2 concentrations flanking the concentration at which the relative OD630 value was 50%, the log-transformed dissociation constants (pKd) were calculated using the following equation:| pKd = −{log(A/B) × (50 − C)/(D − C) + log(B)} |
Structural model calculations
Three-dimensional structures of 6HB containing HR1 and various HR2 regions were predicted by AlphaFold ver. 2.0.39 The calculations were conducted using the “multimer” mode. Superimposition was conducted between the 6HB model structure by AlphaFold2 and the crystal structure of SARS-CoV-2 (PDB-ID: 6LXT) by using Superpose in CCP4i program package.53Peptide synthesis
All peptides were synthesized by standard 9-fluorenylmethoxy carbonyl (Fmoc)-based solid-phase techniques.17,41 The peptides for biological tests were purified by high performance liquid chromatography on a preparative Cosmosil 5C18 AR-II column (Nacalai Tesque Inc., Kyoto, Japan) using a linear gradient of H2O/acetonitrile containing 0.05% trifluoroacetic acid.17Inhibition of HR1–HR2 interaction in SPICA
MBP-HR1 was diluted to 20 nM with 50 mM sodium carbonate buffer and coated overnight following the method described above. The plates were washed and blocked as described above. The drug candidates were added to the plate and serially diluted to give concentrations in the range of 0.0001–50 μM using 100 mM MES buffer (pH 6.4) containing 100 mM NaCl and 2 mM MgCl2 in the plate. After serial dilution, the plate was incubated at 37 °C for 1 h. After the incubation, 5 nM (final concentration) ALP-HR2 in the same buffer was added and the plate was incubated at 37 °C for 1 h. The plate was processed the same as described above, incubated at 37 °C for 1 h, and the OD630 values were measured as above. The relative OD630 value was calculated as a ratio of the OD630 value at each drug concentration and the drug-free OD630 value. The pIC50 value was calculated the same as for the pKd value.Quantification of SARS-CoV-2
Quantitative RT-PCR was performed as described previously.54 Briefly, the designated concentrations of drug candidates were prepared by 10-fold serial dilutions in flat bottom 96-well microtiter plates (Thermo Fisher Scientific) as above. SARS-CoV-2 strain JPN/TY/WK-521 (GISAID: EPI_ISL_408667), which is a variant of Wuhan strain isolated in Japan, and VeroE6/TMPRSS2 cells (2.0 × 104 cells per well) were added to the plates and the plates were incubated for 3 days. After incubation, viral RNA copies in culture supernatant from each well were quantified according to the company's recommended protocols using the SARS-CoV-2 detection kit, N1 set (Toyobo, Ltd, Biotech Support Department, Osaka, Japan).55,56 PCR was run for 45 cycles using a Takara Thermal Cycler Dice Real Time System Lite Model TP700 and analyzed using Takara Dice Real Time software (Takara Bio Inc., Shiga, Japan) using the ΔΔCt method. The relative amounts of SARS-CoV-2 were calculated as the ratios of the viral RNA copies with and without drugs. The log-transformed 50% effective concentration (pEC50) was defined as the concentration of drug at which the relative copy number of SARS-CoV-2 was 50%.Cytotoxicity
MTT assay was performed as described previously.57 The drug candidates in DMEM were added to flat bottom 96-well microtiter plates (Thermo Fisher Scientific) and serially diluted as described above. Then, VeroE6/TMPRSS2 cells (2.0 × 104 cells per well) were seeded. After 3 days, 20 μL of 0.5% MTT (Dojindo Laboratories, Kumamoto, Japan) in PBS was added to each well and incubated for 3 h at 37 °C. After incubation, 170 μL of medium was removed. To solubilize the formazan crystals, 170 μL of isopropanol with 10% octylphenoxypolyethoxyethanol (Fujifilm Wako Chemicals) and 0.4% HCl was added. The formazan crystals were completely solubilized by pipetting. The optical density at 560 nm (OD560) was measured using the microplate reader 800TS. The relative OD560 values were calculated as the ratio of the OD560 value at each drug concentration and the drug-free OD560 value of cells without virus.Author contributions
S. S.: investigation. M. K, K. A., and S. O.: resources. K. K.: investigation and validation. Y. H., M. S., and A. N.: validation. K. M.: formal analysis. E. N. K. and H. H.: project administration and funding acquisition.Conflicts of interest
Satoshi Suzuki received a fellowship from Takeda Science Foundation (Osaka, Japan).Acknowledgements
We gratefully acknowledge all data contributors for EPI_ISL_408667, National Institute of Infectious Diseases for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based. We are grateful to the kind support of Emiko Usui and Kazushige Hirata. We acknowledge support from the Co-creation Center for Disaster Resilience, IRIDeS, Tohoku University and the Biomedical Research Core of the Tohoku University Graduate School of Medicine. This work was partly supported by research grants from the Japan Society of the Promotion for Science (JSPS no. JP16H05346, JP18H02555, JP21K19366) and AMED under grant numbers JP20ak0101140, and JP22gm1610007 (AMED-CREST). This work was also supported in part by a research grant from the AEON MALL Co. Ltd. Victoria Muir, PhD, from Edanz (https://jp.edanz.com/ac) edited a draft of this manuscript. K. A is grateful for “The Graduate Program for Medical Innovation (MIP)” and for the Research Fellowships for Young Scientists from JSPS, Japan.References
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS.
- World Health Organization (WHO), https://www.who.int/emergencies/diseases/novel-coronavirus-2019, accessed August 2023.
- L. D. Saravolatz, S. Depcinski and M. Sharma, Clin. Infect. Dis., 2023, 76, 165–171 CrossRef.
- A. M. Edwards, R. S. Baric, E. O. Saphire and J. B. Ulmer, Science, 2022, 375, 1133–1139 CrossRef CAS.
- N. Higashi-Kuwata, K. Tsuji, H. Hayashi, H. Bulut, M. Kiso, M. Imai, H. Ogata-Aoki, T. Ishii, T. Kobayakawa, K. Nakano, N. Takamune, N. Kishimoto, S. ichiro Hattori, D. Das, Y. Uemura, Y. Shimizu, M. Aoki, K. Hasegawa, S. Suzuki, A. Nishiyama, J. Saruwatari, Y. Shimizu, Y. Sukenaga, Y. Takamatsu, K. Tsuchiya, K. Maeda, K. Yoshimura, S. Iida, S. Ozono, T. Suzuki, T. Okamura, S. Misumi, Y. Kawaoka, H. Tamamura and H. Mitsuya, Nat. Commun., 2023, 14, 1076 CrossRef CAS.
- B. J. Bosch, R. van der Zee, C. A. M. de Haan and P. J. M. Rottier, J. Virol., 2003, 77, 8801–8811 CrossRef CAS.
- J. Bosch, B. E. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. R. Heck, R. De Groot, A. D. M. E. Osterhaus and P. J. M. Rottier, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8455–8460 CrossRef PubMed.
- R. Watanabe, S. Matsuyama, K. Shirato, M. Maejima, S. Fukushi, S. Morikawa and F. Taguchi, J. Virol., 2008, 82, 11985–11991 CrossRef CAS PubMed.
- R. J. G. Hulswit, C. A. M. de Haan and B. J. Bosch, Adv. Virus Res., 2016, 96, 29–57 CAS.
- S. Xia, M. Liu, C. Wang, W. Xu, Q. Lan, S. Feng, F. Qi, L. Bao, L. Du, S. Liu, C. Qin, F. Sun, Z. Shi, Y. Zhu, S. Jiang and L. Lu, Cell Res., 2020, 30, 343–355 CrossRef CAS PubMed.
- C. B. Jackson, M. Farzan, B. Chen and H. Choe, Nat. Rev. Mol. Cell Biol., 2022, 23, 3–20 CrossRef CAS PubMed.
- P. M. Colman and M. C. Lawrence, Nat. Rev. Mol. Cell Biol., 2003, 4, 309–319 CrossRef CAS.
- H. Nishikawa, E. Kodama, A. Sakakibara, A. Fukudome, K. Izumi, S. Oishi, N. Fujii and M. Matsuoka, Antiviral Res., 2008, 80, 71–76 CrossRef CAS PubMed.
- K. Izumi, K. Kawaji, F. Miyamoto, K. Shimane, K. Shimura, Y. Sakagami, T. Hattori, K. Watanabe, S. Oishi, N. Fujii, M. Matsuoka, M. Kaku, S. G. Sarafianos and E. N. Kodama, Int. J. Biochem. Cell Biol., 2013, 45, 908–915 CrossRef CAS.
- H. Nishikawa, S. Nakamura, E. Kodama, S. Ito, K. Kajiwara, K. Izumi, Y. Sakagami, S. Oishi, T. Ohkubo, Y. Kobayashi, A. Otaka, N. Fujii and M. Matsuoka, Int. J. Biochem. Cell Biol., 2009, 41, 891–899 CrossRef CAS PubMed.
- T. Naito, K. Izumi, E. Kodama, H. Nishikawa, K. Watanabe, Y. Sakagami, K. Kajiwara, S. G. Sarafianos, S. Oishi, N. Fujii and M. Matsuoka, Antimicrob. Agents Chemother., 2009, 53, 1013–1018 CrossRef CAS PubMed.
- M. Watanabe, K. Hashimoto, Y. Abe, E. N. Kodama, R. Nabika, S. Oishi, S. Ohara, M. Sato, Y. Kawasaki, N. Fujii and M. Hosoya, PLoS One, 2016, 11, e0162823 CrossRef PubMed.
- M. Ujike, H. Nishikawa, A. Otaka, N. Yamamoto, N. Yamamoto, M. Matsuoka, E. Kodama, N. Fujii and F. Taguchi, J. Virol., 2008, 82, 588–592 CrossRef CAS.
- N. Pardi, M. J. Hogan and D. Weissman, Curr. Opin. Immunol., 2020, 65, 14–20 CrossRef CAS PubMed.
- J. L. McKimm-Breschkin, S. Jiang, D. S. Hui, J. H. Beigel, E. A. Govorkova and N. Lee, Antiviral Res., 2018, 149, 118–142 CrossRef CAS PubMed.
- E. M. Contreras, I. A. Monreal, M. Ruvalcaba, V. Ortega and H. C. Aguilar, Curr. Opin. Virol., 2021, 51, 34–47 CrossRef CAS PubMed.
- S. Xia, L. Yan, W. Xu, A. S. Agrawal, A. Algaissi, C.-T. K. Tseng, Q. Wang, L. Du, W. Tan, I. A. Wilson, S. Jiang, B. Yang and L. Lu, Sci. Adv., 2019, 5, eaav4580 CrossRef CAS PubMed.
- L. Agrawal, T. Poullikkas, S. Eisenhower, C. Monsanto, R. K. Bakku, M. H. Chen and R. S. Kalra, Antibodies, 2021, 10, 3 CrossRef CAS.
- C. T. Wild, D. C. Shugars, T. K. Greenwell, C. B. McDanal and T. J. Matthews, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 9770–9774 CrossRef CAS.
- D. M. Lambert, S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka and S. R. Petteway Jr, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 2186–2191 CrossRef CAS.
- M. Lu, S. C. Blacklow and P. S. Kim, Nat. Struct. Mol. Biol., 1995, 2, 1075–1082 CrossRef CAS.
- B. C. Cunningham and J. A. Wells, Science, 1989, 244, 1081–1085 CrossRef CAS PubMed.
- S. Xia, Y. Zhu, M. Liu, Q. Lan, W. Xu, Y. Wu, T. Ying, S. Liu, Z. Shi, S. Jiang and L. Lu, Cell. Mol. Immunol., 2020, 17, 765–767 CrossRef CAS PubMed.
- Y. Zhu, D. Yu, H. Yan, H. Chong and Y. He, J. Virol., 2020, 94, e00635 CAS.
- M. Kandeel, M. Yamamoto, H. Tani, A. Kobayashi, J. Gohda, Y. Kawaguchi, B. K. Park, H. J. Kwon, J. I. Inoue and A. Alkattan, Biomol. Ther., 2021, 29, 282–289 CrossRef CAS.
- D. Yu, Y. Zhu, T. Jiao, T. Wu, X. Xiao, B. Qin, H. Chong, X. Lei, L. Ren, S. Cui, J. Wang and Y. He, Emerging Microbes Infect., 2021, 10, 1227–1240 CrossRef CAS.
- J. Zhou, W. Xu, Z. Liu, C. Wang, S. Xia, Q. Lan, Y. Cai, S. Su, J. Pu, L. Xing, Y. Xie, L. Lu, S. Jiang and Q. Wang, Acta Pharm. Sin. B, 2022, 12, 1652–1661 CrossRef CAS.
- Y. Behzadipour and S. Hemmati, Int. J. Pept. Res. Ther., 2022, 28, 42 CrossRef CAS PubMed.
- K. Tsuji, K. Baffour-Awuah Owusu, Y. Miura, T. Ishii, K. Shinohara, T. Kobayakawa, A. Emi, T. Nakano, Y. Suzuki and H. Tamamura, RSC Adv., 2023, 13, 8779–8793 RSC.
- Y. Cai, J. Zhang, T. Xiao, H. Peng, S. M. Sterling, R. M. Walsh Jr, S. Rawson, S. Rits-Volloch and B. Chen, Science, 2020, 369, 1586–1592 CrossRef CAS.
- V. K. Outlaw, R. W. Cheloha, E. M. Jurgens, F. T. Bovier, Y. Zhu, D. F. Kreitler, O. Harder, S. Niewiesk, M. Porotto, S. H. Gellman and A. Moscona, J. Am. Chem. Soc., 2021, 143, 5958–5966 CrossRef CAS.
- K. Yang, C. Wang, A. J. B. Kreutzberger, K. I. White, R. A. Pfuetzner, L. Esquivies, T. Kirchhausen and A. T. Brunger, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2300360120 CrossRef CAS PubMed.
- Y. Hu, Y. Zhu, Y. Yu, N. Liu, X. Ju, Q. Ding and Y. He, Antiviral Res., 2023, 212, 105571 CrossRef CAS.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS.
- S. Marqusee and R. L. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 8898–8902 CrossRef CAS.
- A. Otaka, M. Nakamura, D. Nameki, E. Kodama, S. Uchiyama, S. Nakamura, H. Nakano, H. Tamamura, Y. Kobayashi, M. Matsuoka and N. Fujii, Angew. Chem., Int. Ed., 2002, 41, 2937–2940 CrossRef.
- J. L. Douglas, M. L. Panis, E. Ho, K.-Y. Lin, S. H. Krawczyk, D. M. Grant, R. Cai, S. Swaminathan and T. Cihlar, J. Virol., 2003, 77, 5054–5064 CrossRef CAS PubMed.
- X. Zhao, M. Singh, V. N. Malashkevich and P. S. Kim, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14172–14177 CrossRef CAS.
- T. K. Warren, R. Jordan, M. K. Lo, A. S. Ray, R. L. Mackman, V. Soloveva, D. Siegel, M. Perron, R. Bannister, H. C. Hui, N. Larson, R. Strickley, J. Wells, K. S. Stuthman, S. A. Van Tongeren, N. L. Garza, G. Donnelly, A. C. Shurtleff, C. J. Retterer, D. Gharaibeh, R. Zamani, T. Kenny, B. P. Eaton, E. Grimes, L. S. Welch, L. Gomba, C. L. Wilhelmsen, D. K. Nichols, J. E. Nuss, E. R. Nagle, J. R. Kugelman, G. Palacios, E. Doerffler, S. Neville, E. Carra, M. O. Clarke, L. Zhang, W. Lew, B. Ross, Q. Wang, K. Chun, L. Wolfe, D. Babusis, Y. Park, K. M. Stray, I. Trancheva, J. Y. Feng, O. Barauskas, Y. Xu, P. Wong, M. R. Braun, M. Flint, L. K. McMullan, S. S. Chen, R. Fearns, S. Swaminathan, D. L. Mayers, C. F. Spiropoulou, W. A. Lee, S. T. Nichol, T. Cihlar and S. Bavari, Nature, 2016, 531, 381–385 CrossRef CAS PubMed.
- M. Wang, R. Cao, L. Zhang, X. Yang, J. Liu, M. Xu, Z. Shi, Z. Hu, W. Zhong and G. Xiao, Cell Res., 2020, 30, 269–271 CrossRef CAS.
- S. Matsuyama, N. Nao, K. Shirato, M. Kawase, S. Saito, I. Takayama, N. Nagata, T. Sekizuka, H. Katoh, F. Kato, M. Sakata, M. Tahara, S. Kutsuna, N. Ohmagari, M. Kuroda, T. Suzuki, T. Kageyama and M. Takeda, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 7001–7003 CrossRef CAS.
- N. Khan, X. Chen and J. D. Geiger, Front. Pharmacol., 2020, 11, 595888 CrossRef CAS PubMed.
- S. Khare, C. Gurry, L. Freitas, M. B. Schultz, G. Bach, A. Diallo, N. Akite, J. Ho, R. T. C. Lee, W. Yeo, G. C. C. Team and S. Maurer-Stroh, China CDC Wkly, 2021, 3, 1049–1051 Search PubMed.
- Y. Kikuchi, K. Yoda, M. Yamasaki and G. Tamura, Nucleic Acids Res., 1981, 9, 5671–5678 CrossRef CAS.
- J. Dodt, T. Schmitz, T. Schäfer and C. Bergmann, FEBS Lett., 1986, 202, 373–377 CrossRef CAS PubMed.
- A. I. Derman and J. Beckwith, J. Bacteriol., 1995, 177, 3764–3770 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer, D. K. Chu, T. Bleicker, S. Brünink, J. Schneider, M. L. Schmidt, D. G. Mulders, B. L. Haagmans, B. van der Veer, S. van den Brink, L. Wijsman, G. Goderski, J. L. Romette, J. Ellis, M. Zambon, M. Peiris, H. Goossens, C. Reusken, M. P. Koopmans and C. Drosten, Eurosurveillance, 2020, 25, 2000045 Search PubMed.
- R. Takeda, H. Sawa, M. Sasaki, Y. Orba, N. Maishi, T. Tsumita, N. Ushijima, Y. Hida, H. Sano, Y. Kitagawa and K. Hida., Sci. Rep., 2022, 12, 14050 CrossRef CAS PubMed.
- J. Yoo, M. Sasaki, S. X. Cho, Y. Kasuga, B. Zhu, R. Ouda, Y. Orba, P. de Figueiredo, H. Sawa and K. S. Kobayashi., SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis, Nat. Commun., 2021, 12, 6602 CrossRef CAS.
- W. Watanabe, K. Konno, K. Ijichi, H. Inoue, T. Yokota and S. Shigeta, J. Virol. Methods, 1994, 48, 257–265 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cb00166k |
| This journal is © The Royal Society of Chemistry 2024 |