Open Access Article
Open Access ArticleLung-selective nucleic acid vectors generated by in vivo lung-targeting-protein decoration of polyplexes†
Xu
Pu
a,
Zejuan
Li
a,
Ran
Chen
a,
Junqiu
Shi
a,
Jinlong
Qin
 *b,
Yunqing
Zhu
*b,
Yunqing
Zhu
 *a and
Jianzhong
Du
*a and
Jianzhong
Du
 *ab
*ab
aDepartment of Polymeric Materials, School of Materials Science and Engineering, Tongji University, 4800 Caoan Road, Shanghai 201804, China. E-mail: 1019zhuyq@tongji.edu.cn
bDepartment of Gynecology and Obstetrics, Shanghai Key Laboratory of Anesthesiology and Brain Functional Modulation, Clinical Research Center for Anesthesiology and Perioperative Medicine, Translational Research Institute of Brain and Brain-Like Intelligence, Shanghai Fourth People's Hospital, School of Medicine, Tongji University, Shanghai 200434, China. E-mail: jzdu@tongji.edu.cn
First published on 22nd May 2024
Abstract
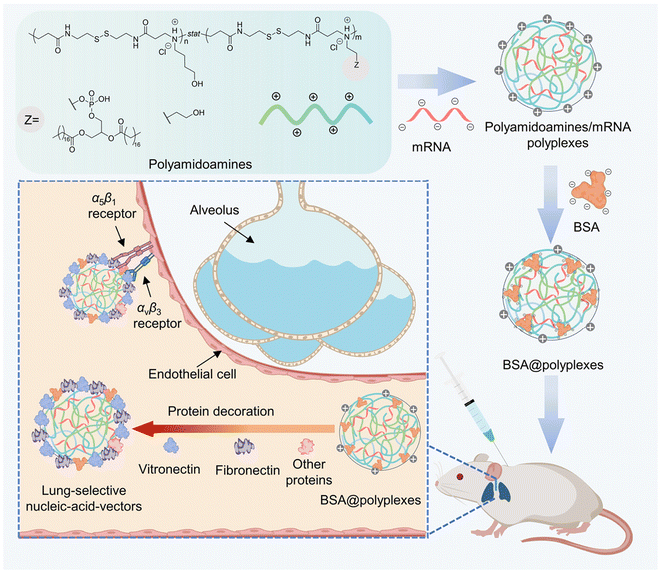
Nucleic acid drugs show immense therapeutic potential, but achieving selective organ targeting (SORT) for pulmonary disease therapy remains a formidable challenge due to the high mortality rate caused by pulmonary embolism via intravenous administration or the mucus barrier in the respiratory tract via nebulized delivery. To meet this important challenge, we propose a new strategy to prepare lung-selective nucleic-acid vectors generated by in vivo decoration of lung-targeting proteins on bioreducible polyplexes. First, we synthesized polyamidoamines, named pabol and polylipo, to encapsulate and protect nucleic acids, forming polyamidoamines/mRNA polyplexes. Second, bovine serum albumin (BSA) was coated on the surface of these polyplexes, called BSA@polyplexes, including BSA@pabol polyplexes and BSA@polylipo polyplexes, to neutralize excess positive charge, thereby enhancing biosafety. Finally, after subcutaneous injection, proteins, especially vitronectin and fibronectins, attached to the polyplexes, resulting in the formation of lung-selective nucleic-acid vectors that achieve efficient lung targeting.
Introduction
Nucleic acids are delivered into cells by vectors and work by either increasing or correcting gene expression or silencing/blocking gene expression.1 In recent years, an increasing number of nucleic acid drugs have been approved for clinical applications.2,3 However, nucleic acids alone face significant challenges in entering cells and coming into effect. For example, the existence of nucleases will lead to the degradation of nucleic acids. Besides, it is difficult for negatively charged nucleic acids to enter cells, as the membrane also displays a negative charge.1,4 To protect nucleic acids and promote cellular uptake, many delivery systems have been developed, including lipid nanoparticles (LNPs), polymeric delivery vectors and cell-penetrating peptides (CPPs).5 With the development of polymer chemistry and the emergence of new functional monomers, more and more polymers with good biosafety have been utilized in the field of nucleic acid delivery,6 such as poly(β-amino ester)s (PBAEs)7,8 and polyamidoamines.9–11 Polyamidoamines can be facilely synthesized via the aza-Michael polyaddition of amines and bis(acrylamide)s under mild conditions.12 This allows the convenient synthesis of polyamidoamines with specific properties by simply changing the structure and composition of the monomers.At present, the safe and effective delivery of nucleic acid drugs to specific organs remains an important challenge. Nucleic acid drugs are usually accumulated in the liver after systemic administration, largely due to mononuclear phagocytosis (MPS) and the liver's vascular permeability.13 It may cause acute hepatic cell toxicity, leading to tissue damage and inflammation.14 In addition, the drugs may be cleared by MPS, jeopardizing the drug delivery targeting other organs.15 Therefore, achieving selective organ targeting (SORT) remains a key challenge in the field of nucleic acid delivery.
The lung, a crucial organ facilitating gas exchange between the human body and the external environment, distinguishes itself by its direct connection to the outside world through the respiratory tract. This unique feature renders the lung susceptible to invasion and infection by external pollutants, pathogens, and harmful substances.16 Such vulnerability underscores the potential for lung damage, posing a significant threat to human health and well-being. Promising avenues for the treatment of various respiratory diseases, such as cystic fibrosis,17 lung cancer,18,19 and asthma,20–22 involve the delivery of nucleic acids to the lungs. Therefore, the development of vectors capable of specifically delivering nucleic acid drugs to lung cells and tissues becomes paramount. This precise delivery ensures that therapeutic interventions achieve their intended goals while minimizing off-target effects.
In the realm of lung-targeted delivery, the selection of a delivery route plays a pivotal role in determining the biological distribution and therapeutic efficacy of nucleic acid drugs. Currently, two primary delivery methods are commonly employed for lung targeting: intravenous injection and inhalation. Intravenous injection facilitates lung targeting by leveraging the fact that the entire cardiac output passes through the pulmonary circulation in systemic circulation.23 However, this approach has inherent drawbacks. Intravenous administration can lead to the accumulation of nanoparticles in organs beyond the lungs, thereby diminishing efficacy.24 More critically, it poses risks of severe systemic toxicity25 and pulmonary embolism, which can be fatal.26 On the other hand, inhalation, particularly via nebulized delivery, allows for the direct delivery of drugs to the lungs, reducing systemic toxicity. However, achieving lung-targeted delivery through inhalation presents its own challenges. Nanoparticles must penetrate the mucus barrier in the respiratory tract to reach the lungs,27 often requiring larger drug doses to achieve the desired therapeutic effect.28
The chemical structure of vectors plays a vital role in lung targeting, which has a huge impact on the formation of protein corona.29–32 For example, the lipid tail structure of lipid nanoparticles (LNPs) significantly influences their organ selectivity. Lipid tails with ester bonds primarily target the liver,33 whereas those tails with amide bonds tend to accumulate in the lungs32 owing to the adsorption of different proteins. Encouraged by these insightful discoveries, our endeavour focuses on synthesizing a novel class of polymers with specialized lung-targeting capabilities and strategically optimizing the delivery method to facilitate safe and effective delivery to the lungs.
Herein, we utilized the aza-Michael polyaddition method to synthesize two kinds of polyamidoamine polymers to form polyamidoamines/mRNA polyplexes. Furthermore, bovine serum albumin (BSA) was coated on the surface of the polyplexes to significantly reduce the cytotoxicity and improve the biocompatibility of the drugs. The affinity for proteins contributes to the absorption of proteins especially the lung-targeting-related proteins vitronectin and fibronectin, leading to their accumulation and protein expression in the lungs (Scheme 1).
Experimental
Preparation of BSA@polyplexes
After preparing the polyamidoamines/mRNA polyplexes, bovine serum albumin (BSA) was coated onto the surface of the polyplexes, forming BSA@polyplexes, named BSA@pabol polyplexes and BSA@polylipo polyplexes, respectively. BSA was dissolved in HEPES buffer (20 mM HEPES, 5 wt% glucose in water, pH 7.0) in advance and filtered through a 0.22 μm cellulose acetate filter (Corning, NY). Next, the BSA solution was added dropwise to the polyplex solution (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v = 1
v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) while stirring at 1000 rpm. The solution was allowed to equilibrate for 30 min at room temperature before further operation.
2) while stirring at 1000 rpm. The solution was allowed to equilibrate for 30 min at room temperature before further operation.
Hemolysis
To prepare rabbit red blood cells (RBCs) for hemolysis testing, rabbit red blood was first washed with 1× PBS and centrifuged at 4000 rpm for 5 min several times until the supernatant was no longer red. Next, the RBCs were diluted in PBS at pH 7 to a 4% (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) RBC suspension. Then, a polyplex solution (400 μL) containing 4 μg of mRNA was incubated with 400 μL of the RBC solution at pH 7 and at 37 °C for 1 h. Positive and negative controls were prepared using 1% Triton-X and PBS, respectively. After incubation, the mixture was centrifuged at 4000 rpm for 5 min to obtain the supernatant. The supernatant was then plated in a 96-well plate and measured at 540 nm using a Multiskan FC microplate reader (ThermoFisher Scientific, Shanghai, China). The hemolysis rate was calculated using the following equation:
v) RBC suspension. Then, a polyplex solution (400 μL) containing 4 μg of mRNA was incubated with 400 μL of the RBC solution at pH 7 and at 37 °C for 1 h. Positive and negative controls were prepared using 1% Triton-X and PBS, respectively. After incubation, the mixture was centrifuged at 4000 rpm for 5 min to obtain the supernatant. The supernatant was then plated in a 96-well plate and measured at 540 nm using a Multiskan FC microplate reader (ThermoFisher Scientific, Shanghai, China). The hemolysis rate was calculated using the following equation:where Qs represents the absorbance of the sample, Qn represents the absorbance of the negative control, and Qp represents the absorbance of the positive control.
Encapsulation efficiency
To prepare 1× TE buffer, the 20× TE buffer was diluted using DNase/RNase free water. For the RiboGreen assay, a working solution of the RiboGreen reagent was prepared using 1× TE buffer. Samples (100 μL) with 1 μg of an mRNA and RiboGreen working solution were plated in 96-well plates and incubated for 5 min at room temperature. The samples were measured using a fluorescence microplate reader with excitation of 480 nm and emission of 520 nm. mRNA encapsulation efficiency can be calculated according to the following equation:Agarose gel retardation assay
A pre-prepared polyplex solution with a concentration of 100 μg mL−1 was used for the experiment. The polyplex solution (10 μL) was mixed with 2 μL of loading buffer. The mixture was then loaded onto a 2% agarose gel and electrophoresed at 180 V for 45 min. After the electrophoresis process, the gel was stained with ethidium bromide.In vitro colloidal stability tests
Pabol/mRNA and polylipo/mRNA polyplexes with an mRNA concentration of 10 μg mL−1 in HEPES buffer (20 mM HEPES, 5 wt% glucose in water, pH 7.0) were stored at 4 °C and 25 °C, respectively. The hydrodynamic diameters (Dh) and polydispersity (PD) were measured by dynamic light scattering (DLS) once a week. The serum stability was evaluated by turbidity measurements. Briefly, fetal bovine serum (FBS) (10% v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) was added to the polyplex solution with 90 μL of mRNA (10 μg mL−1) in 96-well plates. The plate was then incubated at 37 °C, and the absorbance of each well at 660 nm was measured at various time intervals.
v) was added to the polyplex solution with 90 μL of mRNA (10 μg mL−1) in 96-well plates. The plate was then incubated at 37 °C, and the absorbance of each well at 660 nm was measured at various time intervals.
GSH reduction responsiveness
The polyplex solution (180 μL) with 1.8 μg of mRNA was mixed with a GSH stock solution (100 mM) at 25 °C. The changes in Dh and the derived count rate were measured by DLS within 6 h.Albumin binding effect
For the polyacrylamide gel electrophoresis, the polyplex solution (8 μL) containing mRNA at a concentration of 10 μg mL−1 and 12 μL of the BSA solution (0.4 mg mL−1) were mixed and then incubated at 25 °C for 20 min. The zeta potential changes of nanoparticles mixed with albumin were measured by ZetaSizer Nano series instrument. Subsequently, loading buffer (5 μL) for native-PAGE was added to the mixture before it was loaded onto the gel. The system was run in Tris-Gly buffer at 180 V for 1 h. Finally, the sample was stained with Coomassie Brilliant Blue overnight and washed several times.For the fluorescence quenching assay, the PEI, jetPEI, pabol, and polylipo particle samples (100 μL) containing 1 μg of mRNA were incubated with 400 μg mL−1 BSA at 37 °C for 30 min. Tryptophan quenching was measured at λex of 280 nm.
Circular dichroism (CD) was used to characterize the denaturation of albumin after binding to nanoparticles. The polyplex solution containing mRNA at a concentration of 10 μg mL−1 was incubated with 100 μg mL−1 BSA at 37 °C for 1 h. Denatured BSA heated at 90 °C for 10 min served as the control group.
Cell viability
To assess the cytotoxicity of polymers and polyplexes to HEK cells, a CCK-8 assay was conducted. Nanoparticles with an mRNA concentration of 50 μg mL−1 were prepared in advance. HEK 293T cells were seeded in 96-well plates at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well with 100 μL of complete medium. The cells were then incubated for 24 h at 37 °C in 5% CO2. Afterward, the medium was replaced with 90 μL of fresh medium mixed with 10 μL of nanoparticle solutions. After 4 h, the medium was further replaced with fresh complete medium. After 24 h of incubation, the samples were further incubated with 100 μL of fresh medium and 10 μL of a CCK-8 solution. The absorbance of each well at 660 nm was measured after approximately 2 to 4 h.
000 cells per well with 100 μL of complete medium. The cells were then incubated for 24 h at 37 °C in 5% CO2. Afterward, the medium was replaced with 90 μL of fresh medium mixed with 10 μL of nanoparticle solutions. After 4 h, the medium was further replaced with fresh complete medium. After 24 h of incubation, the samples were further incubated with 100 μL of fresh medium and 10 μL of a CCK-8 solution. The absorbance of each well at 660 nm was measured after approximately 2 to 4 h.
In vivo biodistribution
Six-eight-week-old BALB/C nude mice were used as experimental models. The mice were subcutaneously injected with a polyplex solution containing 5 μg of firefly luciferase mRNA. After 24 h, the mice were intraperitoneally injected with 100 μL of a D-luciferin solution (30 mg mL−1) before being sacrificed. Subsequently, the organs including the heart, liver, spleen, lungs, and kidneys were harvested to assess the biodistribution of the protein using an in vivo imaging system.Proteomic assay
The polyplex solution (50 μg mL−1) was mixed with plasma in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio and incubated at 37 °C for 1 h. Following the incubation period, the mixture was centrifuged for 30 min at 15
1 volume ratio and incubated at 37 °C for 1 h. Following the incubation period, the mixture was centrifuged for 30 min at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, and the temperature was maintained at 4 °C. The supernatant and plasma were then subjected to analysis by electrospray liquid chromatography mass spectrometry (LC-MS/MS) to determine the protein adsorption of the nanoparticles. The results were normalized using Origin software.
000g, and the temperature was maintained at 4 °C. The supernatant and plasma were then subjected to analysis by electrospray liquid chromatography mass spectrometry (LC-MS/MS) to determine the protein adsorption of the nanoparticles. The results were normalized using Origin software.
Results and discussion
Design and synthesis of polyamidoamines
Polyamidoamines, including poly((N,N′-bis(acryloyl)cystamine-co-4-amino-1-butanol)-co-1,2-distearoyl-sn-glycero-3-phosphoethanolamine) (polylipo), and poly(N,N′-bis(acryloyl)cystamine-co-4-amino-1-butanol) (pabol), were synthesized via aza-Michael polyaddition (Fig. S1†). The existence of protonated tertiary amines allows polyamidoamines to bind to nucleic acids. Additionally, nucleic acids could be rapidly released in the presence of glutathione (GSH) in cells due to the disulfide bond. The syntheses of polymers were evaluated by 1H NMR and size exclusion chromatography (Fig. S2–S5†). The size exclusion chromatography results revealed that the molecular weight of pabol and polylipo was 10.3 kDa and 7.2 kDa, respectively, confirming the successful preparation of these two polymers.Complexation of polyamidoamines with nucleic acids
The size and zeta potential of nanoparticles play crucial roles in organ-targeting and transfection efficiency. Nanoparticles with sizes ranging from approximately 50 to 500 nm and a positive charge within 10 mV have shown potential for organ targeting.29,34–36 Therefore, precise control over particle size and zeta potential is necessary for achieving effective organ targeting.37To obtain lung-targeting nanoparticles with optimal size and zeta potential, the weight ratios of the polymers to nucleic acids were investigated by forming polyamidoamines/mRNA polyplexes, including polylipo/mRNA and pabol/mRNA polyplexes. As shown in Fig. S6†, the polylipo/mRNA polyplexes have a hydrodynamic diameter of 164 nm and zeta potential of +14.8 mV at a polylipo/mRNA weight ratio of 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the pabol/mRNA polyplexes have a hydrodynamic diameter of 142 nm and zeta potential of +23 mV.
1, while the pabol/mRNA polyplexes have a hydrodynamic diameter of 142 nm and zeta potential of +23 mV.
To confirm the effective interaction between the polymers and nucleic acids, the agarose gel retardation assay was performed. The clear, distinct band in the well indicated that the mRNA was efficiently bound to the polymers and prevented from migrating through the gel, confirming the successful and complete encapsulation of mRNA by the polymers (Fig. S7†). As potential nucleic acid delivery vectors, polyamidoamines should be able to encapsulate various nucleic acids efficiently. Therefore, encapsulation of other kinds of nucleic acids by polylipo and pabol was also evaluated. Through optimizing the pH and the weight ratio, a few different nucleic acids were successfully encapsulated by polylipo and pabol with ideal size and zeta potential (Fig. S8 and S9†). In addition, the RiboGreen assay was applied to determine the encapsulation efficiency. As illustrated in Fig. 1d, the encapsulation efficiency of polylipo and pabol reached more than 80%, suggesting the efficient encapsulation of nucleic acids. The successful transfection in HEK cells also validated the effectiveness of polylipo and pabol as nucleic acid delivery vectors (Fig. S10†).
Colloidal stability and redox-responsiveness of the polyamidoamines/mRNA polyplexes
The colloidal stability of nanoparticles is a crucial factor influencing their therapeutic efficacy in medical applications.2,38 Therefore, to assess the colloidal stability of the different polyplexes, DLS studies were performed in HEPES buffer (20 mM HEPES, 5 wt% glucose in water, pH 7.0) at 4 °C and 25 °C over a period of 14 days. The data supported the finding that the pabol/mRNA polyplexes showed better colloidal stability than the polylipo/mRNA polyplexes. When stored at 4 °C, the hydrodynamic diameter change of the pabol/mRNA polyplexes was about 35% after 14 days, while the particle size of the polylipo/mRNA polyplexes increased from approximately 145 nm to 269 nm, with a change of 85%. The polydispersity (PD) of the pabol/mRNA polyplexes and polylipo/mRNA polyplexes was maintained below 0.3 for 14 days (Fig. 2a). In addition, the polylipo/mRNA polyplexes exhibited poor dispersibility at 25 °C and the particle size reached 319 nm after 14 days (Fig. 2b). This difference in the colloidal stability might be due to the introduction of phospholipid side chains that reduce the surface charge of polylipo and increase the tendency of hydrophobic interaction between the particles, leading to aggregation.Having various proteins in the blood may lead to the agglomeration and precipitation of the nanoparticles. Therefore, the serum stability of the polyplexes was also investigated using turbidity measurements. As shown in Fig. 2c, the absorbance of the pabol/mRNA and polylipo/mRNA polyplexes at 660 nm remained unchanged, indicating that both pabol/mRNA and polylipo/mRNA polyplexes remained stable in the presence of serum for at least 2.5 h. Additionally, it was observed that the absorbance of the polyamidoamines/mRNA polyplexes was higher than that of the PEI/mRNA and jetPEI/mRNA polyplexes, indicating that the polyamidoamines/mRNA polyplexes might be able to adsorb more serum proteins after being incubated with serum, which would be of great significance for exploiting protein corona to achieve organ targeting.
Glutathione (GSH) is known to cleave disulfide backbones.39 To demonstrate the bioreducibility of the polyplexes in the presence of GSH, the change in the hydrodynamic diameter of the polylipo/mRNA polyplexes within 6 h was monitored. As shown in Fig. 2d, the presence of GSH resulted in the cleavage of the disulfide backbone, causing the polyplexes to disassemble and release nearly all the mRNA within 6 h.
Albumin adsorption to polyplexes and preparation of BSA@polyplexes
With respect to severe cytotoxicity caused by the high surface charge of polymeric delivery vectors,40 it is necessary to reduce their surface charge and enhance biosafety. Bovine serum albumin (BSA) is an ideal material to address this concern. As a natural biomaterial, BSA possesses biocompatibility, non-immunogenicity, and biodegradability, and it is negatively charged at physiological pH.41 Therefore, it is feasible to directly coat BSA on the surface of the polyamidoamines/mRNA polyplexes to neutralize the excess positive charge and improve biosafety.42In order to verify the feasibility of BSA coating, the affinity of the polyamidoamines/mRNA polyplexes for BSA was assessed. It was observed that there was more obvious gel retardation phenomenon in both pabol/mRNA and polylipo/mRNA polyplexes, compared to other polyplexes based on polymer vectors such as PEI/mRNA, jetPEI/mRNA polyplexes, or cationic liposomes, such as Lipofectamine 2000 nanoparticles (Fig. S11a†). The quantification (Fig. S11b†) showed that there was little change in the Lipofectamine 2000 group, while the relative integrated intensity of the jetPEI/mRNA and PEI/mRNA polyplexes was about 95%, and the relative integrated intensity of the pabol/mRNA and polylipo/mRNA polyplexes decreased to around 85%.
Furthermore, we examined the zeta potential changes of the nanoparticles after incubation with BSA. As shown in Fig. S12,† the zeta potential of the polyplexes changed from positive to negative before and after the incubation with BSA, indicating that the polyplexes were indeed coated with BSA. However, Lipofectamine 2000 was negatively charged, and there was no retardation in the gel spectrum. This indicates that the surface charge of nanocarriers plays an important role in protein adsorption. Nanoparticles can only combine with BSA if the surface charge is positive. As for the difference in the affinity for BSA between the polyamidoamines/mRNA and other polymer/mRNA polyplexes, apart from hydrogen bonds, the presence of an amide bond enhances the interaction between the nanoparticles and BSA, resulting in a better adsorption effect. Tryptophan is usually used as a fluorophore to study interactions between BSA and drugs.43 Co-incubation with pabol/mRNA and polylipo/mRNA polyplexes resulted in noticeable fluorescence quenching, also confirming the superior affinity of polyamidoamines/mRNA polyplexes for BSA (Fig. 3a).
Additionally, to address concerns about the potential endocytosis of denatured proteins by macrophages,44 adsorption-mediated changes in the three-dimensional structure of albumin were characterized using circular dichroism (CD) spectroscopy. It was found that while the pabol/mRNA and polylipo/mRNA polyplexes were able to combine with BSA, they caused negligible secondary structure changes in BSA (Fig. 3b). In other words, BSA could maintain its function even after being combined with polyplexes. Therefore, it is reasonable to conclude that the polyamidoamines/mRNA polyplexes exhibited a strong affinity for BSA, facilitating the coating of the polyplexes with BSA.
The absorption of BSA resulted in significant concentration-dependent reductions in surface charge as it is negatively charged at pH 7.0. Meanwhile, after coating with BSA, the size of the polyplexes also increased. To obtain BSA@polyplexes with optimal size and lower zeta potential for lung targeting, BSA@polyplexes with different weight ratios of BSA and mRNA were prepared, named BSA@pabol polyplexes and BSA@polylipo polyplexes, respectively. Fig. 3d and f demonstrates that the BSA/polylipo/mRNA ratio of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was enough to neutralize the positive surface charge of the polylipo/mRNA polyplexes, whereas the pabol/mRNA polyplexes required a higher amount of BSA (BSA/pabol/RNA, 0.2
1 was enough to neutralize the positive surface charge of the polylipo/mRNA polyplexes, whereas the pabol/mRNA polyplexes required a higher amount of BSA (BSA/pabol/RNA, 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to reduce the positive charge to a lower level. This suggested that the incorporation of a phospholipid moiety might enhance the interaction between the polylipo/mRNA polyplexes and BSA.
1) to reduce the positive charge to a lower level. This suggested that the incorporation of a phospholipid moiety might enhance the interaction between the polylipo/mRNA polyplexes and BSA.
Taking the suitable particle size and zeta potential into consideration, BSA@pabol (203 nm, +14.1 mV) and BSA@polylipo (323 nm, +1.9 mV) polyplexes were used for subsequent experiments. Transmission electron microscopy (TEM) was applied to observe the morphology of BSA@polyplexes (Fig. S13†). TEM images showed that the sizes of the BSA@pabol and BSA@pabol polyplexes were about 267 nm and 379 nm, respectively, which were larger than the sizes obtained by DLS. This difference could be due to the collapse of the polyplexes during sample preparation.45 In addition, BSA@polyplexes were stable for at least 7 d at 4 °C (Fig. S14†). As shown in Fig. S15,† the green fluorescence demonstrated that BSA@polyplexes successfully transfected mouse lung primary microvascular endothelial cells, enabling effective lung targeting.
Cytotoxicity of the polymers and the polyplexes
The cytotoxicity of the polyplexes was evaluated at a concentration of 5 μg mL−1 mRNA, the same as the concentration usually applied in vivo (Fig. 4a).10,46 The polyamidoamines/mRNA polyplexes displayed a relative cell viability of over 50%, while the commercial transfection reagents resulted in significant cell death. For polymeric carriers, the high surface charge was a significant factor contributing to its high cytotoxicity. Therefore, the introduction of BSA coating significantly reduced the cytotoxicity of the nanoparticles and improved their biosafety, resulting in the increase of relative cell viability from 88% to 94% and from 70% to 91% for the BSA@polylipo and BSA@pabol polyplexes, respectively. Similarly, cytotoxicity on mouse lung primary microvascular endothelial cells revealed higher relative cell viability in BSA@pabol and BSA@polylipo (Fig. S16†). Likewise, the hemolysis rates of the polyamidoamines/mRNA and BSA@polyplexes were less than 1%, which are lower than those of commercial transfection reagents (Fig. 4b), indicating that both polyamidoamines/mRNA polyplexes and BSA@polyplexes were hemocompatible.Targeting mechanism
As the presence of certain proteins in the protein corona may lead to the accumulation of nanoparticles in specific organs, the protein corona was considered to be an important factor for in vivo targeting. To identify the proteins that are enriched on the surface of the polyamidoamines/mRNA polyplexes and BSA@polyplexes, a comparative analysis of the protein compositions of the supernatant and plasma was conducted to assess the affinity of the polyplexes for proteins associated with lung targeting.As shown in Fig. 5, after the polyplexes were incubated with plasma, a notable difference was observed in the vitronectin content between the obtained supernatant and the plasma. Specifically, the vitronectin content in pure plasma was three times higher than that in the supernatant of both pabol/mRNA and BSA@pabol polyplexes, and approximately two times higher than that in the polylipo/mRNA and BSA@polylipo polyplexes. In the case of the polylipo/mRNA polyplexes, the adsorption of other proteins could be attributed to the presence of phospholipids, although this phenomenon could be mitigated after albumin coating. Furthermore, significant differences between pure plasma and the supernatant were also observed in coagulation-related proteins, especially fibronectin. It is reported that vitronectin is able to bind αvβ3 integrin, which is highly expressed by lung endothelial cells.47 As for fibronectin it was found that, apart from αvβ3, fibronectin can also bind α5β1, resulting in enhanced endothelial cell adhesion.48–50 Therefore, the presence of these proteins indicates the potential for lung targeting upon the entry of nanoparticles into the body.
In vivo targeting efficiency
After subcutaneous injection, proteins were absorbed on the surface of the polyplexes, forming lung-selective nucleic-acid vectors, including the polylipo, BSA@polylipo, pabol, and BSA@pabol vectors, finally resulting in lung targeting. Encouraged by the proteomics results, firefly luciferase was employed as a reporter protein to assess the in vivo targeting efficiency of the lung-selective nucleic-acid vectors using an in vivo imaging system. Each mouse received a subcutaneous injection of polyamidoamines/mRNA polyplexes or BSA@polyplexes, containing 5.0 μg of mRNA, and was sacrificed after 24 h. The organs were then harvested for luminescent imaging.The results of luciferase expression confirmed the viability of our lung-targeting strategy, with the lung exhibiting the highest accumulation compared with most of the other organs (Fig. 6). Quantitative analysis further confirmed higher levels of luciferase protein expression in the lungs with average radiance an order of magnitude higher than that of the other organs (heart, liver, and spleen; Fig. S17†), suggesting that the vectors were effectively targeted to the lungs.
The quantitative results also demonstrated that the lung-targeting effect of the pabol-based vectors was better than that of the polylipo-based vectors. This could be due to the non-specific adsorption of other proteins. Although the pre-coating of BSA could address this issue to some extent, the targeting efficiency was still inferior to that of the pabol-based vectors. To assess the potential toxicity of the lung-selective nucleic-acid vectors, the main organs such as the heart, liver, spleen, lungs, and kidneys were subjected to H&E staining, which confirmed that the vectors did not induce serious toxic side effects 24 h after injection (Fig. 7). The liver and kidneys play crucial roles in drug metabolism and excretion, respectively. The alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels are indicators of liver function, while blood urea nitrogen (BUN) and creatinine (CREA) levels reflect kidney function. Assessing these markers helps evaluate drug-induced toxicity or damage to these organs. As shown in Fig. S18,† there were no significant differences between the vector and PBS groups, suggesting that the vectors possessed good biosafety.
Conclusions
In conclusion, new nucleic acid delivery vectors with protein coronas were successfully developed. Firstly, we synthesized polyamidoamines, including pabol and polylipo, which complex with nucleic acids to form polyamidoamines/mRNA polyplexes. The encapsulation efficiency of polyamidoamines/mRNA polyplexes reached more than 80%. Second, to neutralize the excess positive charges and improve biosafety, BSA was coated on the surface of these polyplexes, creating BSA@polyplexes. The albumin coating on the polyamidoamines/mRNA polyplexes not only reduced surface charge but also cytotoxicity. Therefore, the relative cell viability for BSA@polylipo and BSA@pabol polyplexes increased from 88% to 94% and from 70% to 91%, respectively. After subcutaneous injection, the interaction between polyplexes and plasma proteins led to the formation of protein corona enriched by vitronectin and fibronectin, forming lung-selective nucleic-acid vectors. Using firefly luciferase mRNA as the reporter, these vectors demonstrated highly efficient lung-targeting capability, with the average radiance an order of magnitude higher in the lungs compared to other organs, including the heart, the liver, and the spleen. This suggests that these vectors, by leveraging protein affinity for effective delivery, hold promise for lung-targeted therapeutics.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Tongji University and approved by the Animal Ethics Committee of Shanghai Tenth People's Hospital affiliated to Tongji University [SHDSYY-2024-1231].Author contributions
Xu Pu: conceptualization, investigation, methodology, formal analysis, writing – original draft, writing – review & editing. Zejuan Li: methodology. Ran Chen: methodology. Junqiu Shi: methodology. Jinlong Qin: conceptualization, methodology, and funding acquisition. Yunqing Zhu: conceptualization, methodology, writing – review & editing, and funding acquisition. Jianzhong Du: conceptualization, methodology, and writing – review & editing, funding acquisition.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This project is supported by the National Key R&D Program of China (2022YFC2402900), the National Natural Science Foundation of China (22335005, 51903190, 22175131 and 21925505), the Innovation Program of Shanghai Municipal Education Commission (2023ZKZD28), and the Shanghai International Scientific Collaboration Fund (21520710100). J.D. is the recipient of the National Science Fund for Distinguished Young Scholars.References
- A. I. S. van den Berg, C. O. Yun, R. M. Schiffelers and W. E. Hennink, J. Controlled Release, 2021, 331, 121–141 CrossRef CAS PubMed.
- N. Chaudhary, D. Weissman and K. A. Whitehead, Nat. Rev. Drug Discovery, 2021, 20, 817–838 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- H. J. Vaughan, J. J. Green and S. Y. Tzeng, Adv. Mater., 2020, 32, 1901081 CrossRef CAS.
- P. S. Kowalski, A. Rudra, L. Miao and D. G. Anderson, Mol. Ther., 2019, 27, 710–728 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- T. T. Smith, S. B. Stephan, H. F. Moffett, L. E. McKnight, W. Ji, D. Reiman, E. Bonagofski, M. E. Wohlfahrt, S. P. S. Pillai and M. T. Stephan, Nat. Nanotechnol., 2017, 12, 813–820 CrossRef CAS PubMed.
- N. N. Parayath, S. B. Stephan, A. L. Koehne, P. S. Nelson and M. T. Stephan, Nat. Commun., 2020, 11, 6080 CrossRef CAS PubMed.
- J. Reyes-Reveles, R. Sedaghat-Herati, D. R. Gilley, A. M. Schaeffer, K. C. Ghosh, T. D. Greene, H. E. Gann, W. A. Dowler, S. Kramer, J. M. Dean and R. K. Delong, Biomacromolecules, 2013, 14, 4108–4115 CrossRef CAS PubMed.
- A. K. Blakney, Y. Zhu, P. F. McKay, C. R. Bouton, J. Yeow, J. Tang, K. Hu, K. Samnuan, C. L. Grigsby, R. J. Shattock and M. M. Stevens, ACS Nano, 2020, 14, 5711–5727 CrossRef CAS PubMed.
- X. Zhang, K. Hong, Q. Sun, Y. Zhu and J. Du, Biomater. Sci., 2021, 9, 5275–5292 RSC.
- W. Cheng, D. Wu and Y. Liu, Biomacromolecules, 2016, 17, 3115–3126 CrossRef CAS PubMed.
- E. Rohner, R. Yang, K. S. Foo, A. Goedel and K. R. Chien, Nat. Biotechnol., 2022, 40, 1586–1600 CrossRef CAS PubMed.
- Y.-N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Ángel Lázaro, C. Castells-Sala, O. Meca-Cortes, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2018, 7, 1800335 CrossRef.
- Z. Tang, X. You, Y. Xiao, W. Chen, Y. Li, X. Huang, H. Liu, F. Xiao, C. Liu, S. Koo, N. Kong and W. Tao, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2304966120–e2304966120 CrossRef CAS PubMed.
- L. Allen, L. Allen, S. B. Carr, G. Davies, D. Downey, M. Egan, J. T. Forton, R. Gray, C. Haworth, A. Horsley, A. R. Smyth, K. W. Southern and J. C. Davies, Nat. Commun., 2023, 14, 693 CrossRef CAS PubMed.
- G. L. Zhao, W. L. Ho, J. X. Chu, X. J. Xiong, B. Hu, K. O. Boakye-Yiadom, X. Y. Xu and X. Q. Zhang, ACS Appl. Mater. Interfaces, 2023, 15, 31273–31284 CrossRef CAS PubMed.
- J. F. Han, Q. Wang, Z. R. Zhang, T. Gong and X. Sun, Small, 2014, 10, 524–535 CrossRef CAS PubMed.
- M. Zhang, H. Jiang, L. Wu, H. Lu, H. Bera, X. Zhao, X. Guo, X. Liu, D. Cun and M. Yang, J. Controlled Release, 2022, 352, 422–437 CrossRef CAS PubMed.
- K. Kurrikoff, K. Freimann, K. L. Veiman, E. M. Peets, A. Piirsoo and Ü. Langel, Sci. Rep., 2019, 9, 19926 CrossRef CAS PubMed.
- Y. R. Xie, N. H. Kim, V. Nadithe, D. Schalk, A. Thakur, A. Kiliç, L. G. Lum, D. J. P. Bassett and O. M. Merkel, J. Controlled Release, 2016, 229, 120–129 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- X. Ke, L. Shelton, Y. Hu, Y. Zhu, E. Chow, H. Tang, J. L. Santos and H. Q. Mao, ACS Appl. Mater. Interfaces, 2020, 12, 35835–35844 CrossRef CAS.
- L. Miao, Y. Zhang and L. Huang, Mol. Cancer, 2021, 20, 41 CrossRef CAS PubMed.
- J. C. Kaczmarek, A. K. Patel, K. J. Kauffman, O. S. Fenton, M. J. Webber, M. W. Heartlein, F. DeRosa and D. G. Anderson, Angew. Chem., Int. Ed., 2016, 55, 13808–13812 CrossRef CAS PubMed.
- P. Mastorakos, A. L. da Silva, J. Chisholm, E. Song, W. K. Choi, M. P. Boyle, M. M. Morales, J. Hanes and J. S. Suk, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 8720–8725 CrossRef CAS PubMed.
- A. K. Patel, J. C. Kaczmarek, S. Bose, K. J. Kauffman, F. Mir, M. W. Heartlein, F. DeRosa, R. Langer and D. G. Anderson, Adv. Mater., 2019, 31, 1805116 CrossRef PubMed.
- Y. Cao, Z. X. He, Q. M. X. Chen, X. Y. He, L. L. Su, W. X. Yu, M. M. Zhang, H. Y. Yang, X. X. Huang and J. F. Li, Nano Lett., 2022, 22, 6580–6589 CrossRef CAS PubMed.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2109256118 CrossRef CAS PubMed.
- S. A. Dilliard, Y. H. Sun, M. O. Brown, Y. C. Sung, S. Chatterjee, L. Farbiak, A. Vaidya, X. Z. Lian, X. Wang, A. Lemoff and D. J. Siegwart, J. Controlled Release, 2023, 361, 361–372 CrossRef CAS PubMed.
- M. Qiu, Y. Tang, J. J. Chen, R. Muriph, Z. F. Ye, C. F. Huang, J. Evans, E. P. Henske and Q. B. Xu, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2116271119 CrossRef CAS PubMed.
- M. Qiu, Z. Glass, J. J. Chen, M. Haas, X. Jin, X. W. Zhao, X. H. Rui, Z. F. Ye, Y. M. Li, F. Zhang and Q. B. Xu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020401118 CrossRef CAS.
- W. Yu, R. Liu, Y. Zhou and H. Gao, ACS Cent. Sci., 2020, 6, 100–116 CrossRef CAS.
- L. M. Kranz, M. Diken, H. Haas, S. Kreiter, C. Loquai, K. C. Reuter, M. Meng, D. Fritz, F. Vascotto, H. Hefesha, C. Grunwitz, M. Vormehr, Y. Huesemann, A. Selmi, A. N. Kuhn, J. Buck, E. Derhovanessian, R. Rae, S. Attig, J. Diekmann, R. A. Jabulowsky, S. Heesch, J. Hassel, P. Langguth, S. Grabbe, C. Huber, O. Tuereci and U. Sahin, Nature, 2016, 534, 396–401 CrossRef PubMed.
- P. S. Kowalski, U. C. Palmiero, Y. Huang, A. Rudra, R. Langer and D. G. Anderson, Adv. Mater., 2018, 30, 1801151 CrossRef.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- K. D. Popowski, A. Moatti, G. Scull, D. Silkstone, H. Lutz, B. L. D. Abad, A. George, E. Belcher, D. S. Zhu, X. Mei, X. Cheng, M. Cislo, A. Ghodsi, Y. H. Cai, K. Huang, J. L. Li, A. C. Brown, A. Greenbaum, P. U. C. Dinh and K. Cheng, Matter, 2022, 5, 2960–2974 CrossRef CAS PubMed.
- C. Lin, Z. Y. Zhong, M. C. Lok, X. L. Jiang, W. E. Hennink, J. Feijen and J. F. J. Engbersen, Bioconjugate Chem., 2007, 18, 138–145 CrossRef CAS PubMed.
- H. Wang, H. Chang, Q. Zhang and Y. Cheng, Top. Curr. Chem., 2017, 375, 62 CrossRef PubMed.
- C. Lei, X. R. Liu, Q. B. Chen, Y. Li, J. L. Zhou, L. Y. Zhou and T. Zou, J. Controlled Release, 2021, 331, 416–433 CrossRef CAS.
- L. Piao, H. Li, L. Teng, B. C. Yung, Y. Sugimoto, R. W. Brueggemeier and R. J. Lee, Nanomed. Nanotechnol., 2013, 9, 122–129 CrossRef CAS PubMed.
- N. Wangoo, C. R. Suri and G. Shekhawat, Appl. Phys. Lett., 2008, 92, 133104 CrossRef.
- M. P. Vincent, S. Bobbala, N. B. Karabin, M. Frey, Y. G. Liu, J. O. Navidzadeh, T. Stack and E. A. Scott, Nat. Commun., 2021, 12, 648 CrossRef CAS PubMed.
- P. Wei, E. J. Cornel and J. Du, Macromolecules, 2021, 54, 7603–7611 CrossRef CAS.
- S. Liu, X. Wang, X. L. Yu, Q. Cheng, L. T. Johnson, S. Chatterjee, D. Zhang, S. M. Lee, Y. H. Sun, T. C. Lin, J. L. Liu and D. J. Siegwart, J. Am. Chem. Soc., 2021, 143, 21321–21330 CrossRef CAS PubMed.
- B. Singh, C. Z. Fu and J. Bhattacharya, Am. J. Physiol.: Lung Cell., 2000, 278, L217–L226 CAS.
- K. Suehiro, J. Gailit and E. F. Plow, J. Biol. Chem., 1997, 272, 5360–5366 CrossRef CAS.
- J. Koo, D. Galanakis, Y. Liu, A. Ramek, A. Fields, X. Ba, M. Simon and M. H. Rafailovich, Biomacromolecules, 2012, 13, 1259–1268 CrossRef CAS.
- J. Koo, M. H. Rafailovich, L. Medved, G. Tsurupa, B. J. Kudryk, Y. Liu and D. K. Galanakis, J. Thromb. Haemostasis, 2010, 8, 2727–2735 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and experimental, synthetic route, 1H NMR spectra of polymers, SEC curves, hydrodynamic diameter and zeta potential of polyplexes, etc. See DOI: https://doi.org/10.1039/d4bm00502c |
| This journal is © The Royal Society of Chemistry 2024 |