Open Access Article
Open Access ArticleIn vitro dioxygenase activity characterization using headspace stir bar sorptive extraction (HSSE)†
Lucía
Morote
a,
Lourdes
Gómez-Gómez
 ab,
Alberto
López-Jimenez
ac,
Oussama
Ahrazem
ab,
Alberto
López-Jimenez
ac,
Oussama
Ahrazem
 *ac and
Ángela
Rubio-Moraga
*ac and
Ángela
Rubio-Moraga
 *ac
*ac
aInstituto Botánico, Universidad de Castilla-La Mancha, Campus Universitario s/n, Albacete 02071, Spain. E-mail: Angela.Rubio@uclm.es; Oussama.ahrazem@uclm.es
bFacultad de Farmacia, Departamento de Ciencia y Tecnología Agroforestal y Genética, Universidad de Castilla-La Mancha, Dr. José Maria Sánchez Ibañez, s/n, Albacete 02071, Spain
cEscuela Técnica Superior de Ingeniería Agronómica y de Montes y Biotecnología, Departamento de Ciencia y Tecnología Agroforestal y Genética, Universidad de Castilla-La Mancha, Campus Universitario s/n, Albacete 02071, Spain
First published on 2nd August 2024
Abstract
An analytical approach employing headspace sorptive extraction coupled with gas chromatography-mass spectrometry (HSSE-GC-MS) has been successfully developed for the determination of apocarotenoid volatiles arising from the enzymatic activity of carotenoid cleavage enzymes (CCDs) in Escherichia coli. The GjCCD4a enzyme derived from gardenia, known for its cleavage specificity at 7,8 and 7′,8′ double bonds across diverse carotenoid substrates, was utilized as a reference enzyme, using β-carotene as the substrate for the enzymatic activity assays. Optimal headspace conditions for analysis were established following a 5 hours induction period of the recombinant GjCCD4a protein within E. coli cells, engineered to produce β-carotene. The analytical method demonstrated linearity, with correlation coefficient (R2 > 0.95) in calibration, while achieving detection and quantification limits conducive to the accurate determination of β-cyclocitral. Notably, this methodological framework significantly reduced both the handling complexity and sample processing time in comparison to conventional liquid chromatography methods employed for the detection of cleavage products and determination of CCD activities. The proposed HSSE-GC-MS approach not only enhances the efficiency of apocarotenoid analysis but also provides a sensitive means for unraveling the intricate enzymatic processes associated with CCD-mediated carotenoid cleavage in a bacterial model system.
1. Introduction
Carotenoids are isoprenoid compounds produced mainly by plants, some bacteria and fungi, and more recently by certain invertebrates.1 Carotenoids can be defined by a common C40 polyene backbone carrying 3 to 11 conjugated double bonds with different stereo-configurations. Introduction of end-ring structures, hydroxylation, oxygenation, and further modifications give rise to the large structural diversity found among the more than 700 known carotenoids. The polyene structure of carotenoids responsible for their role in photosynthesis, anti-oxidation capacity, and color, makes them vulnerable to oxidation that splits double bonds, forming carbonyl products called apocarotenoids (designated based on the C-atom number at the cleavage site).2 Although not all organisms can biosynthesize carotenoids. Practically, all living organism can catabolize carotenoids by a non-enzymatic and unspecific reaction such as lipoxygenase co-oxidation or photo-oxidation or by an enzymatic oxidative breakdown reaction,3–5 producing apocarotenoids.Apocarotenoids (APOs) are involved in many aspects of plant–environment interactions, such as defense against pathogens and herbivores, serving as chemoattractants or repellents, indicating predation and seed dispersal, serving as allelochemical compounds that give plants a competitive edge, and assisting in the formation of symbiotic relationships. Additionally, they play a crucial role in the development of plants by functioning as phytohormones.6–11
APOs have a huge impact in human health generating health benefits by preventing or managing chronic disease or its symptoms. Within plants, APOs can manifest as either non-volatile or volatile apocarotenoids. Among volatile apocarotenoids, two distinct structural and perceptual classes exist: linear apocarotenoids, exemplified by compounds like 6-methyl-5-hepten-2-one (MHO), geranylacetone, and pseudoionone, and cyclic apocarotenoids, including β-ionone, β-cyclocitral, and safranal.12
The specific enzymes catalyzing the oxidative cleavage of double bonds in the carotenoid polyene chain generating apocarotenoid precursors are known as carotenoid cleavage dioxygenases (CCDs), which also recognize and cleavage apocarotenoids. Interestingly, CCDs enzymes are present in almost all living organisms, so that all are capable of synthesizing apocarotenoid compounds.1
Several methodologies have been developed to characterize CCDs activities,13 among them the most used, for its simplicity, is by in vivo analyses in carotenoid-producing bacteria. However, this approach presents several limitations as the number of substrates available for the analyses, and the fact that some apocarotenoids are degraded or modified by the bacteria. In order to detect and quantify the products of CCDs activities, liquid chromatography (LC) and gas chromatography (GC) analyses have been developed.14,15
Liquid chromatography (LC) serves as an analytical tool for determining the nonvolatile byproducts resulting from the enzymatic cleavage of carotenoids. However, inherent limitations underscore the method's applicability. Firstly, the necessity of extracting the nonvolatile product from the bacterial pellet constitutes a time-intensive process, requiring approximately 2 hours utilizing a speed vacuum system prior to injection into the LC. Moreover, a more critical limitation arises from the metabolic activity of the bacteria themselves, leading to the inherent degradation of the target product. In contrast, volatile apocarotenoids present an alternative way for analysis, particularly through Gas Chromatography (GC). Among the diverse methodologies employed for capturing volatile apocarotenoids generated through Carotenoid Cleavage Dioxygenase (CCD) activities, Headspace Solid-Phase Microextraction (HS-SPME) stands out as a widely utilized technique.16–18 However, the use of Stir Bar Sorptive Extraction or SBSE methodology followed by gas chromatography coupled to mass spectrometry (GC-MS) analysis,19 has not been used to capture apocarotenoids volatiles generated by CCDs.
In this study, an optimize method has been developed to assess Carotenoid Cleavage Dioxygenase (CCD) activities in vivo, utilizing the headspace Stir Bar Sorptive Extraction (HSSE) technique, followed by subsequent analysis through Gas Chromatography coupled with Mass Spectrometry (GC-MS). This innovative approach serves as a significant advancement in the determination of CCD activities within living systems, offering enhanced precision and sensitivity in the measurement of apocarotenoid products.
2. Material and methods
2.1. Standard
A standard solution of β-cyclocitral purchased form Supelco (purity ≥99.0%) was prepared at a final concentration of 1000 ppb (1000 μg L−1) in MeOH. For calibration purposes, five concentration levels were prepared using the standard solution, in the range of 25 to 1000 μg L−1. All these solutions were stored at 4 °C until their use.2.2. Vector construction
Gene sequence of GjCCD4a20 (accession number ARU08109.1) was synthesized de novo by NZYTech Gene Synthesis service (https://www.nzytech.com/products-services/category/molecular-services/gene-synthesis) and amplified by PCR with specific primers (pThio-G-CCD4a-NYTF: cgcccttggcgaattcATGGATGCTTTTTCTTCAAGTTTCTTGTCAC and pThio-G-CCD4a-NYTR: taccctcgaggaattcTTATAGTTTATTCAATTCAGACTCACGCACAAAGAG). The obtained product was purified with the Wizard® SV Gel and PCR Clean-Up System (Promega) and was cloned into the EcoRI site of pTHIO-Dan1 vector using an In-Fusion® HD Cloning Plus CE kit (Clontech, https://www.clontech.com).The resulting expression plasmid, named pThio-GjCCD4a was sequenced to confirm the correct assembly and the gene sequence. The vector was then transformed into E. coli BL21 strain engineered with a plasmid for the production of β-carotene.21 pAC-BETA was a gift from Francis X Cunningham Jr (Addgene plasmid # 53272; https://n2t.net/addgene:53272; RRID:Addgene_53272). The empty pTHIO-Dan1 vector was also used to transform E. coli BL21 strain engineered with a plasmid to produce β-carotene (pAC-Beta from Addgene org (https://www.addgene.org)) as a negative control.
2.3. In vitro analysis
Positive colonies were selected from the Luria–Bertani solid medium containing ampicillin (50 μg mL−1) and chloramphenicol (30 μg mL−1) to initiate a pre-culture in a volume of 3 mL LB containing the appropriate antibiotic, and grow overnight at 37 °C. The cultured cells were transferred then to 50 mL 2× yeast extract tryptone medium supplemented with ampicillin (25 μg mL−1) and chloramphenicol (15 μg mL−1) and further cultured at 30 °C with shaking until an OD600 of 0.7 was reached. Cells were then induced with 0.2% L-arabinose as described before.222.4. Calibration curve
Quantification was based on calibration curves of the respective standard by adding 80 μL of β-cyclocitral standard dilution into 8 mL of E. Coli BL21 engineered with a plasmid capable of producing β-carotene and induced with a concentration of 0.2% L-arabinose as described before. Five concentrations were prepared using the standard solutions in the range of 0.125 to 1.0 μg L−1. The extraction bars were introduced in the adapter holders, the vials were capped and stirred at 150 rpm with an extraction temperature of 30 °C for 24 h.2.5. Volatile APOS sampling
After arabinose induction, 8 mL of the cultures were transferred to a 20 mL vial designed for headspace analysis, supplied by Gerstel (Mulheim an der Ruhr, Germany). The extraction bars, commercially known as Twister (Gerstel), were introduced then in adapter holders with a small opening in its bottom to keep it in the upper part of the vial. Then the vials were tightly capped for extraction with an aluminum cap and a PTFE/silicone septum. The volatile extraction procedures were performed during 5 h, 24 h and 48 h after arabinose induction by using triplicates for each incubation. The vials were stirred at 150 rpm with an extraction temperature of 30 °C. The identical culture under the same circumstances, free of arabinose, served as the negative control.2.6. HSSE desorption and GC-MS analysis
After the incubation period was completed, the vials were unsealed and the sorptive extraction devices (twisters, PDMS; 10 mm length; 0.5 mm film thickness) were gently retrieved from the headspace. Following that, the twisters were cleaned with distilled water before being dried with a cellulose tissue. The purified twisters were then placed in thermal desorption tubes and GC/MS analysis was performed. The volatile analysis was performed using an automated thermal desorption unit (TDU, Gerstel, Mülheim and der Ruhr, Germany) mounted on an Agilent gas chromatograph system (GC) coupled to a quadrupole electron ionization mass spectrometric detector (Agilent Technologies, Palo Alto, CA, USA) equipped with a fused silica capillary column (BP21 stationary phase, 30 m length, 0.25 mm i.d., and 0.25 μm film thickness) (SGE, Ringwood, Australia). The carrier gas was helium with a constant column pressure of 20.75 psi.The stir bars were thermally desorbed in a stream of helium carrier gas at a flow rate of 75 mL min−1 with the TDU programmed from 40 to 295 °C (held 5 min) at a rate of 60 °C min−1 at splitless desorption mode. The analytes were focused on a programmed temperature vaporizing injector (CIS-4, Gerstel), containing a packed liner (20 mg tenax TA), held at −10 °C with cryo cooling prior to injection. After desorption and focusing, the CIS-4 was programmed from −10 °C to 260 °C (held for 5 min) at 12 °C min−1 to transfer the trapped volatiles onto the analytical column. The GC oven temperature was programmed to 40 °C (held for 2 min), raised to 80 °C (5 °C min−1, held for 2 min), raised to 130 °C (10 °C min−1, held for 5 min), raised to 150 °C (5 °C min−1, held for 5 min), and then raised to 230 °C (10 °C min−1, held for 5 min). The MS was operated in scan acquisition (m/z 27–300) with an ionization energy of 70 eV. The temperature of the MS transfer line was maintained at 230 °C. MS data acquisition was carried out in positive scan mode, although to avoid matrix interferences, the MS quantification was performed in the single ion-monitoring mode using their characteristic m/z values. The identification and quantification of the β-cyclocitral were performed using the NIST library and confirmed by comparison with the mass spectra and retention time of pure standard (Sigma-Aldrich, Steinheim, Germany).23
2.7. Non-volatile APOS and carotenoids sampling
50 mL culture of E. coli transformed as previously described, induced with L-arabinose, were harvested by centrifugation after 5 h, 24 h and 48 h. All extraction were performed in triplicate. The pellets were extracted sequentially by the addition of 4 mL of chloroform and methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). After that, samples were sonicated for 10 minutes mili Q, vortex and centrifuged at 8000 rpm × 10 min. After centrifugation two phases were visible. The colored one was taken and transferred to a new clean 2 mL microcentrifuge tube and dried in a vacuum centrifuge. Dried pigment residues from the E. coli culture were stored at −80 °C until analysis by HPLC where they were resuspended in 500 μL tert-butyl methyl ether and centrifuged at 12
1). After that, samples were sonicated for 10 minutes mili Q, vortex and centrifuged at 8000 rpm × 10 min. After centrifugation two phases were visible. The colored one was taken and transferred to a new clean 2 mL microcentrifuge tube and dried in a vacuum centrifuge. Dried pigment residues from the E. coli culture were stored at −80 °C until analysis by HPLC where they were resuspended in 500 μL tert-butyl methyl ether and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm × 10 min. After that, the tert-butyl methyl ether supernatants were transferred to a 1 mL sample vials and analyzed by HPLC.24
000 rpm × 10 min. After that, the tert-butyl methyl ether supernatants were transferred to a 1 mL sample vials and analyzed by HPLC.24
2.8. HPLC-DAD analysis
High-performance liquid chromatography (HPLC) analysis was performed in an Agilent liquid chromatography 1100 system equipped with a diode-array detector, which was set to scan from 200 to 600 nm. A YMC Carotenoid C30 column (5 μm column, 250 × 4.6 mm) kept at 28 °C was used as stationary phase. The injection volume was set at 20 μL. The flow rate was pumped at 1 mL min−1. The solvents and the chromatography method was modified from the described before:25 solvent A was 98% methanol; solvent B was 95% methanol and solvent C was 100% tetramethyl butyl ether. The elution conditions were as follows: from 80% A and 20% C to 60% A and 40% C in 3 min; from for 60% A and 40% C to 60% B and 40% C in 1 min; from 60% B and 40% C to 100% C in 8 min; from 100% C to 80% A and 20% C in 1 min. This original mixture was maintained in isocratic conditions for 3 min. The crocetin dial and β-carotene was characterized by its absorption spectra and retention time using the standard purchased by Supelco and CaroteNautre, respectively.3. Results and discussion
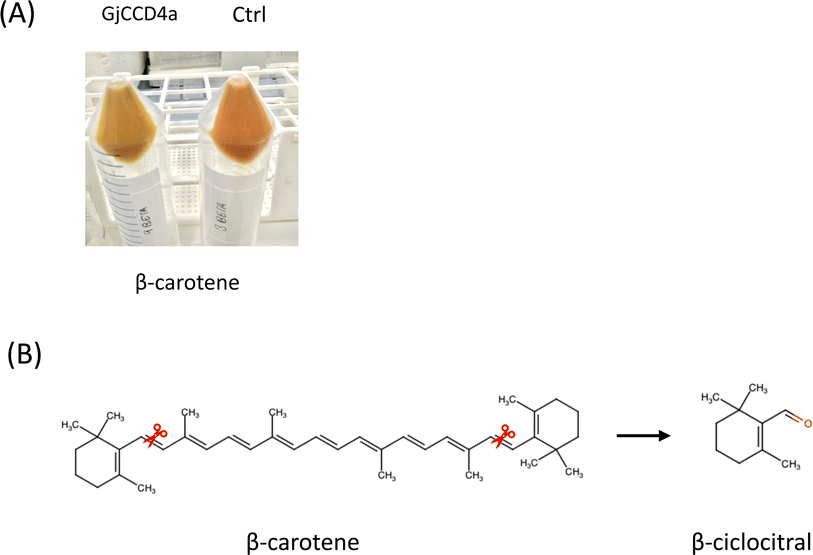
To evaluate the activity of CCD enzymes, two basic approaches are currently used: in vitro and/or in vivo method. CCDs from several plant species were used in these studies.26 CCDs were expressed in carotenoid-accumulating E. coli strains containing plasmids that allowed the accumulation of different carotenoids substrates. The in vitro method allows for the testing of many more substrates, including apocarotenoids. Thin-layer chromatography (TLC), HPLC, LC-MS, or GC-MS are used to detect and identify the products generated in both cases.An analytical protocol combining headspace sorptive extraction and gas chromatography-mass spectrometry (HSSE-GC-MS) has been developed in this study to accurately determine apocarotenoid volatiles that result from the enzymatic catalysis of carotenoid cleavage enzymes (CCDs) in E. coli. We chose headspace sorptive extraction combined with gas chromatography-mass spectrometry (HSSE-GC-MS) since it minimizes interference from non-volatile matrix components, simplifies the sample preparation process and offers high sensitivity for identifying volatile compounds when working with samples that have low quantities of the target analytes. To demonstrate that HSSE-GC-MS is a powerful and adaptable analytical method with advantages in terms of simplicity, broad applicability, selectivity, and sensitivity, making it an excellent choice for the analysis of volatile apocarotenoids, we determined the activity of the gardenia enzyme GjCCD4a, which was assembled using in Fusion strategy (ESI, Fig. 1†). The GjCCD4a cleaves β-carotene at 7,8 and 7′,8′ double bonds and generates crocetin and the volatile β-cyclocitral generating a loss of color in the precipitate of the bacteria by cleavage of β-carotene (Fig. 1). For the calibration curve, the resulting concentrations were as follows: 0.125, 0.25, 0.5, 0.75 and 1 μg L−1. Calibration was performed in this study by directly introducing different concentration of standard solution into the vials containing the E. Coli BL21 β-carotene producers after the induction with 0.2% of L-arabinose, maintaining the same experimental conditions in this way. The data obtained allowed the linearity with a R2 = 0.95.
3.1. Optimization of the extraction conditions
Headspace sorptive extraction (HSSE) has been found to be influenced by a number of variables, including sample amount, extraction time, agitation speed, and salt addition. Although the extraction temperature is a parameter that could influence the process, in the present study we did not focus on it because it is necessary to maintain an induction temperature below 37 °C in order to prevent the formation of inclusion bodies produced by bacteria. More specifically, with regard to the extraction procedure, the sample volume was considered negligible in the context of HSSE. For each replica in our experimental design, 8 mL of E. coli culture was used; this amount was selected for handling convenience and to reduce the possibility of contact problems with the sorptive extraction equipment (twister) during the incubator's agitation process. In any case, when contrasted with alternative E. coli cultures that employed organic solvent extraction for the detection of apocarotenoid volatiles (APOs) influenced by additional carotenoid cleavage enzymes (CCDs) through gas chromatography-mass spectrometry (GC-MS) methods,27,28 the sample volume utilized in our approach was notably reduced. Given the primary objective of retrieving the enzyme's cleavage product in vivo while ensuring minimal impact on bacterial growth and maintaining simplicity, the addition of salt was not deemed necessary. Notably, the alternative headspace approach utilized to identify CCDs' cleavage products in vitro, distinct from our methodology, did not involve the use of salt or tenax desorption tubes, as observed in solid-phase microextraction (SPME) fibers29 or conventional tubes.30 This strategic deviation underscores the unique considerations and methodological choices made in our experimental design to achieve precise insights into CCD-mediated carotenoid cleavage within the E. coli system.HSSE-GC-MS combines Stir Bar Sorptive Extraction (HSSE) and Gas Chromatography-Mass Spectrometry. HSSE uses a stir bar coated with a sorptive substance capable to adsorb and concentrate the analytes of interest from a sample matrix. The stir bar can be agitated into the sample or suspended in the HS as was done during this assay, increasing the extraction efficiency of the desired analytes that demonstrate the effectivity of the enzyme (Fig. 2A). GC-MS then separates these molecules based on their chemical characteristics before identifying and quantifying them using their mass-to-charge ratio. This combination provides great specificity and sensitivity in identifying and measuring volatile chemicals. HSSE-GC-MS has various strengths. This method works incredibly well for removing apocarotenoids at trace quantities from complex biological materials such as a bacterial culture. Apocarotenoids can be transferred from the aqueous phase to the sorptive coating by suspending the stir bar in the HS volume. The extraction's efficiency can be maximized by optimizing the retention of the analytes of interest in the stir bar mainly by controlling time and avoiding sample contamination. Following the specified extraction duration, the stir bar is taken out, dried, and prepared for analysis. Moreover, gas chromatography and mass spectrometry are combined in GC-MS to identify and measure the compounds present in a sample. Accurate and repeatable results are obtained by lowering detection limits and eliminating sample matrix effects through the use of an integrated strategy. The combination of GC-MS yields great sensitivity and specificity for volatile and semi-volatile chemicals, allowing for exact identification and quantification. HSSE efficiently concentrates tiny quantities of chemicals from large sample volumes, increasing detection limits and frequently requiring minimal sample preparation while removing the requirement for organic solvents. However, it is best suited for volatile and semi-volatile molecules, limiting its usefulness for non-volatile substances. Furthermore, the combination of HSSE and GC-MS necessitates specific equipment and expertise, making it more complicated and perhaps more expensive than other approaches. On the other hand, HPLC is incredibly adaptable and can analyze a large variety of chemicals, including polar, non-volatile, and thermally unstable ones. It offers extremely accurate quantitative analysis and is standard calibrated for accurate measurement, however it frequently calls for significant amounts of organic solvents, which can be expensive and harmful to the environment. Furthermore, HPLC is typically less sensitive for volatile compounds than GC-MS, which makes GC-MS the most desirable option for the detection of these compounds.
To maximize bacterial growth, three trials were carried out, each in triplicate, using a regulated agitation speed of 150 rpm. The durations of the extraction were adjusted to be 5 hours, 24 hours, and 48 hours after the introduction of arabinose in E. coli culture. With a particular focus on the “total chromatographic area” linked to the direct product of β-cyclocitral's cleavage of β-carotene, the purpose of varying the extraction durations was to ascertain their influence on the experimental results (Fig. 2B). We aimed to investigate the complex dynamics of bacterial growth and its impact on the targeted enzymatic cleavage process by examining these different temporal intervals after induction. The experimental outcomes were correlated with the principal ion of this volatile, specifically at 137 atomic mass units (amu) (ESI, Fig. 2†). The length of the extraction time is found to be a crucial factor that has a significant and favorable impact. Increasing the extraction time has been shown to have a noticeable and significant effect, improving the extraction yield or efficiency of the desired compounds, however in our case, it is interesting to note that of β-cyclocitral production during five hours of incubation generated an output that was roughly seven times greater than what was produced after twenty-four hours. This finding highlights the β-cyclocitral generation's temporal sensitivity and shows that a 5 hours incubation period is significantly more favourable for optimum and increased production levels. The significant variation in β-cyclocitral concentrations between the three incubation times highlights the significance of careful temporal optimization in order to maximize the capture of the enzymatic cleavage product. A same behaviour has been observed when volatiles from south African wine were analysed authors reported that lower alcohols and esters start to decrease as the time increases, pointing out that probably is due to them being released from the PDMS layer to the headspace.31 The depletion of volatiles from the adsorbent could arise from the equilibrium dynamics inherent in the adsorption process. This phenomenon may result in the release or desorption of the volatiles that have been collected under particular equilibrium conditions due to the complex interplay of parameters influencing the interaction between the adsorbent material and the volatile molecules. Nevertheless, the cleavage product was consistently detectable during each of the assessed incubation periods. As a result, the finding suggests that it may be possible to evaluate the enzymatic activity using this methodology with shorter incubation times, like the five-hours period. But it's important to remember that using an “overnight” incubation period is also a workable and useful option that can be applied in some experimental scenarios or workflow situations.
This approach has been used widely and thoroughly evaluated for the detection and measurement of volatile compounds in a variety of sample matrices. Its application in the analysis of volatile profiles in beer,32 aromatic and medicinal plant33 are noteworthy examples of its successful implementation together with honey,34 and wine.31,35 Although the HSSE-GC-MS method provides significant advances in the qualitative analysis of CCD activities in E. coli cultures, offering important information on the activity and variety of enzyme products, its current form has limitations in quantitative applications. The intrinsic unpredictability of biological systems, such as E. coli cultures, as well as the basal induction exhibited by some plasmids, makes it difficult to obtain reliable and consistent quantitative data, making it difficult to quantify enzyme characteristics, such as affinities and rates of substrate of reaction. This is why an attempt has been made to optimize the extraction time since it is a crucial and controllable factor in the method. Future studies may focus on improving the quantitative capabilities of the HSSE-GC-MS method in enzymatic studies performed in cell cultures.
3.2. Comparative study against HPLC
To thoroughly test whether the optimized HSSE methodology was providing best result to HPLC methodology, the same induced E. coli transformed cultures were used in the same conditions and incubation times. By using HSSE-GC-ME analytical method, detection of the volatile product was possible at all incubation times while using HPLC the detection of the inner compound resulting from the enzymatic activity of GjCCD4 was not successful. Upon GjCCD4a expression, color intensity of E. coli pellets was significantly reduced compared with the negative control without GjCCD4a expression, suggesting efficient cleavage of the substrate β-carotene by GjCCD4a (Fig. 1A). The decrease in the area of the substrate β-carotene can also be perfectly appreciated in the chromatogram, which underlines the assertion of enzyme activity (Fig. 3). Despite all the aforementioned, the detection of the inner product from the enzyme cleavage 7,8 was impossible to determine, this disappearance of the internal cleavage product may be due to the metabolization by the bacteria as it is a carbon source. Taking into account this uncertainty of not always detecting the reaction product, the HSSE-GC-MS method is presented as a suitable option to evaluate de CCDs activity overcoming all the previously described limitations of the HPLC methodology.In addition, the extraction for the consecutive analysis by HPLC has many drawbacks compared to HSSE-GC-MS, namely the sample volume needed for analysis is much larger than the used for HSSE and the extraction procedure carried out from the pellet is a time-consuming action that requires the use of organic solvents. In contrast, the twister method allows for continuous sample extraction without requiring the removal and processing of the cell culture, which is a significant advantage over the SPME method. This methodology provides a distinct advantage by allowing operation at a lower temperature of 30 °C, thereby not only increasing overall efficiency but also significantly streamlining the sampling process.
The HSSE-GC-MS technique stands out for its ability to analyze volatile products obtained through CCDs activity in vitro and continuously, without the need to stop the cell culture. This capability addresses a major gap in the detection of volatile compounds, which are often difficult to measure accurately using traditional methods. One of the important advantages of this method is its ability to provide accurate results using large and small sample volumes. The efficiency of the HSSE-GC-MS method reduces the time required for sample processing and analysis. This not only accelerates the pace of research but also allows for the high-throughput analysis of multiple samples, thereby increasing the overall productivity of research projects. Furthermore, the elimination of organic solvents in the HSSE-GC-MS method addresses both environmental and health concerns associated with solvent use. This makes the method more sustainable and safer for researchers, aligning with green chemistry principles.
4. Conclusions
HSSE is an appropriate method to determine the volatile apocarotenoids product in a in vivo continuous process. The results that have been obtained are similar to those achieved by other more widely recognized techniques, such as HS-SPME. However, HSSE presents some advantages, such as a significant reduction of the volume of the samples and the time required for the processing of those samples. This technique eliminates the need to remove and process the cell culture and enables continuous sample extraction. Furthermore, the times of the experimentation are considerably reduced by using HSSE-GC-MS, being proved in this article how the best detection values are achieved already 5 hours after 0.2% L-arabinose induction.Data availability
The data supporting our findings are available in the manuscript file or from the corresponding author upon request.Author contributions
LGG, and ARM conceived and designed the research. LM, ALJ and ARM conducted the experiments. LGG, OA, LM and ARM analyzed the data. LGG, ARM, LM and OA drafted the manuscript. All authors reviewed and contributed to the final manuscript which was approved by all.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by grants BIO2016-77000-R, PIB2020-114761RB-I00 and FJC2021-046632-I (to M. E.) from the Ministerio de Ciencia e Innovación (MCIN), SBPLY/17/180501/000234, and SBPLY/21/180501/000012 from the Junta de Comunidades de Castilla-La Mancha (co-financed European Union FEDER funds) to LGG and OA. AZA and RSG for technical support and writing assistance for material and methods.References
- T. Maoka, J. Nat. Med., 2020, 74, 1–16 CrossRef CAS PubMed.
- O. Ahrazem, L. Gómez-Gómez, M. J. Rodrigo, J. Avalos and M. C. Limón, Int. J. Mol. Sci., 2016, 17, 1781 CrossRef PubMed.
- M. D. White and E. Flashman, Curr. Opin. Chem. Biol., 2016, 31, 126–135 CrossRef CAS PubMed.
- M. Havaux, Plant J. Cell Mol. Biol., 2014, 79, 597–606 CrossRef CAS PubMed.
- X. Sui, P. D. Kiser, J. von Lintig and K. Palczewski, Arch. Biochem. Biophys., 2013, 539, 203–213 CrossRef CAS PubMed.
- M. H. Walter, D. S. Floss and D. Strack, Planta, 2010, 232, 1–17 CrossRef CAS PubMed.
- L. A. Cáceres, S. Lakshminarayan, K. K.-C. Yeung, B. D. McGarvey, A. Hannoufa, M. W. Sumarah, X. Benitez and I. M. Scott, J. Chem. Ecol., 2016, 42, 107–117 CrossRef PubMed.
- A. Rodríguez, B. Alquézar and L. Peña, New Phytol., 2013, 197, 36–48 CrossRef PubMed.
- F. A. Macías, A. López, R. M. Varela, A. Torres and J. M. G. Molinillo, Phytochemistry, 2004, 65, 3057–3063 CrossRef PubMed.
- M. DellaGreca, C. Di Marino, A. Zarrelli and B. D'Abrosca, J. Nat. Prod., 2004, 67, 1492–1495 CrossRef CAS PubMed.
- K. Akiyama, K. Matsuzaki and H. Hayashi, Nature, 2005, 435, 824–827 CrossRef CAS PubMed.
- J. T. Vogel, D. M. Tieman, C. A. Sims, A. Z. Odabasi, D. G. Clark and H. J. Klee, J. Sci. Food Agric., 2010, 90, 2233–2240 CrossRef CAS PubMed.
- L. Gómez-Gómez, G. Diretto, O. Ahrazem and S. Al-Babili, Methods Mol. Biol., 2020, 2083, 63–74 CrossRef PubMed.
- J. Mi, K.-P. Jia, J. Y. Wang and S. Al-Babili, Anal. Chim. Acta, 2018, 1035, 87–95 CrossRef CAS PubMed.
- J. Leroux, T. T. Truong, B. J. Pogson and R. P. McQuinn, Methods Enzymol., 2022, 670, 311–368 CAS.
- F.-C. Huang, P. Molnár and W. Schwab, J. Exp. Bot., 2009, 60, 3011–3022 CrossRef CAS PubMed.
- J. Wang, N. Zhang, M. Zhao, T. Jing, J. Jin, B. Wu, X. Wan, W. Schwab and C. Song, J. Agric. Food Chem., 2020, 68, 1684–1690 CrossRef CAS PubMed.
- A. Ilg, P. Beyer and S. Al-Babili, FEBS J., 2009, 276, 736–747 CrossRef CAS PubMed.
- O. Zuloaga, N. Etxebarria, B. González-Gaya, M. Olivares, A. Prieto and A. Usobiaga, in Solid-Phase Extraction, ed. C. F. Poole, Elsevier, 2020, pp. 493–530 Search PubMed.
- Z. Xu, X. Pu, R. Gao, O. C. Demurtas, S. J. Fleck, M. Richter, C. He, A. Ji, W. Sun, J. Kong, K. Hu, F. Ren, J. Song, Z. Wang, T. Gao, C. Xiong, H. Yu, T. Xin, V. A. Albert, G. Giuliano, S. Chen and J. Song, BMC Biol., 2020, 18, 63 CrossRef CAS PubMed.
- F. X. Cunningham, B. Pogson, Z. Sun, K. A. McDonald, D. DellaPenna and E. Gantt, Plant Cell, 1996, 8, 1613–1626 CAS.
- L. Gómez-Gómez, G. Diretto, O. Ahrazem and S. Al-Babili, in Plant and Food Carotenoids: Methods and Protocols, ed. M. Rodríguez-Concepción and R. Welsch, Springer US, New York, NY, 2020, pp. 63–74 Search PubMed.
- R. Sánchez-Gómez, A. Zalacain, G. L. Alonso and M. R. Salinas, Food Chem., 2016, 204, 499–505 CrossRef PubMed.
- L. Morote, M. Lobato-Gómez, O. Ahrazem, J. Argandoña, B. Olmedilla-Alonso, A. J. López-Jiménez, G. Diretto, R. Cuciniello, P. Bergamo, S. Frusciante, E. Niza, Á. Rubio-Moraga, S. Crispi, A. Granell and L. Gómez-Gómez, J. Funct. Foods, 2023, 101, 105432 CrossRef CAS.
- L. Morote, Á. Rubio-Moraga, A. J. López-Jiménez, J. Argandoña, E. Niza, O. Ahrazem and L. Gómez-Gómez, Plant Sci. Int. J. Exp. Plant Biol., 2023, 329, 111609 CAS.
- L. Gómez-Gómez, G. Diretto, O. Ahrazem and S. Al-Babili, Methods Mol. Biol., 2020, 2083, 63–74 CrossRef PubMed.
- Z. Sun, J. Hans, M. H. Walter, R. Matusova, J. Beekwilder, F. W. A. Verstappen, Z. Ming, E. van Echtelt, D. Strack, T. Bisseling and H. J. Bouwmeester, Planta, 2008, 228, 789–801 CrossRef CAS PubMed.
- A. J. Simkin, B. A. Underwood, M. Auldridge, H. M. Loucas, K. Shibuya, E. Schmelz, D. G. Clark and H. J. Klee, Plant Physiol., 2004, 136, 3504–3514 CrossRef CAS PubMed.
- F.-C. Huang, P. Molnár and W. Schwab, J. Exp. Bot., 2009, 60, 3011–3022 CrossRef CAS PubMed.
- A. Rubio, J. L. Rambla, M. Santaella, M. D. Gómez, D. Orzaez, A. Granell and L. Gómez-Gómez, J. Biol. Chem., 2008, 283, 24816–24825 CrossRef CAS PubMed.
- B. T. Weldegergis, A. G. J. Tredoux and A. M. Crouch, J. Agric. Food Chem., 2007, 55, 8696–8702 CrossRef CAS PubMed.
- J. E. Ruvalcaba, E. Durán-Guerrero, C. G. Barroso and R. Castro, Foods, 2020, 9, 255 CrossRef CAS PubMed.
- C. Bicchi, C. Cordero, C. Iori, P. Rubiolo and P. Sandra, J. High Resolut. Chromatogr., 2000, 23, 539–546 CrossRef CAS.
- J. I. Cacho, N. Campillo, P. Viñas and M. Hernández-Córdoba, J. Chromatogr. A, 2015, 1399, 18–24 CrossRef CAS PubMed.
- M. Pérez-Jiménez and M. Á. Pozo-Bayón, Food Res. Int., 2019, 121, 97–107 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay00827h |
| This journal is © The Royal Society of Chemistry 2024 |