Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Alterations of the chemical profile of cholesterol in cancer tissue as traced with ToF-SIMS†
Auraya
Manaprasertsak
 a,
Julhash U.
Kazi
a,
Julhash U.
Kazi
 b,
Catharina
Hagerling
b,
Catharina
Hagerling
 a,
Kenneth J.
Pienta
a,
Kenneth J.
Pienta
 c,
Per
Malmberg
c,
Per
Malmberg
 d and
Emma U.
Hammarlund
d and
Emma U.
Hammarlund
 *a
*a
aTissue Development and Evolution (TiDE) Group, Department of Experimental Medical Sciences, Lund University, Lund, Sweden. E-mail: emma.hammarlund at med.lu.se
bDivision of Translational Cancer Research, Department of Laboratory Medicine, Lund University, Lund, Sweden
cThe Cancer Ecology Center, Brady Urological Institute, Johns Hopkins School of Medicine, Baltimore, MD, USA
dDepartment of Chemistry and Chemical Engineering, Chalmers University of Technology, Gothenburg, Sweden
First published on 27th September 2024
Abstract
Cancer has become one of the leading causes of death, with approximately ten million people worldwide dying from cancer each year. In most cases, cancer spreads to remote organs and develops a resistance to therapy. To reduce the deadly impact of cancer, novel targets for markers for early detection are necessary. Given the notable influence of rapid chemical turnover on isotope effects, the heightened turnover rate of cholesterol in cancer offers a promising way for investigation. Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS) offers a valuable tool of tracking cholesterol dynamics. Consequently, we employed ToF-SIMS to assess cholesterol alterations, aiming to uncover potential diagnostic vulnerabilities stemming from heightened cholesterol synthesis. Our study explored the chemical profile of cholesterol influenced by cancer cell metabolism using mammary glands from mice, both with and without cancer. Results revealed a significant increase in the fractional abundance of fragment cholesterol peaks (C27H45+) in cancerous tissues, indicating dysregulated cholesterol metabolism within cancer cells. This suggests potential structural weaknesses or incomplete synthesis. Further investigation into carbon isotope incorporation suggests that the isotopic patterns might be due to the integration of heavier carbon isotopes, although these patterns could be affected by other isotopic influences. Nevertheless, understanding isotope effect of cholesterol profiles have the potential to advance our understanding of cancer biology and improve diagnostic approaches.
Introduction
Markers for early cancer detection are imperative, highlighted by the staggering fact that approximately 20 million people worldwide are diagnosed with cancer annually.1 While the majority of cancers detected at a late stage remain lethal due to metastasis and therapy resistance, cancers detected early are generally curable. Therefore, any novel methods that enable early detection of cancer have the potential to significantly reduce its deadly impact.2 Today, cancer detection typically relies on symptoms, physical examinations, or via screening by radiologic tests or for certain biomarkers. Biomarkers play pivotal roles in early cancer detection and intense research is focused on exploring and utilizing biomolecules that are altered by cancer.Biomolecules used for detecting cancer include DNA, RNA, protein expression levels, and specific antibody expressions.3 Biomarkers indicate molecular changes as the cancer cell population or its impact increases. Biomolecules can also change their chemical properties as they are used or synthesized at a higher rate.4,5 The energy budget of a biochemical process is influenced by the mass differences of the reactants, which leads to varying reaction rates among isotopes of an element. Consequently, in the context of a rapidly growing tumour, certain isotopes might be preferentially utilized due to the specific energy demands of the process. Therefore, it is likely that isotopic variations can affect the bioenergetics of cancer cells, potentially influencing cell proliferation and metabolism. This influence is evidenced by changes in elemental concentrations in blood6,7 and alterations in isotopic balances in tissues, blood or serum.8–11 The balance of isotopes is believed to shift in transformed tissues as a result of increased biosynthesis rates and uptake by cancer cells.9,12 For example, in tumour tissues of breast cancer patients, the proportion of the 64Zn isotope increases compared to the 66Zn isotope.9 Moreover, there are notable isotopic differences between healthy and cancerous tissues and cell lines, with cancerous tissues and cells typically showing 15N depletion and 13C enrichment.12
The isotope effect is a phenomenon in chemistry where the rate of a chemical reaction is altered by the substitution of one isotope for another.13,14 This effect can influence various aspects of chemical reactions, including bond breaking and formation, reaction rates, and reaction pathways.15,16 The effect can manifest in several ways, as isotopic substitutions affect the energy and stability of a molecule.17 A change in stability means that isotopic substitution can lead to a molecule undergoing fragmentation. Such changes in the relative abundance of the fragmented molecules are measurable by mass spectrometry.18 The distribution of the molecule and its fragments is detected with mass spectrometry,19,20 which facilitates detailed analysis and characterization. This technique also allows for tissue imaging providing both spatial and chemical information.21
Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS) provides a robust method for tracking chemical fragmentation processes. One of the benefits of modern cluster ion-based ToF-SIMS is its relatively soft ionization technique, which results in less fragmentation of the sample.22,23 In addition, the focused ion beam enables high-resolution chemical mapping. ToF-SIMS is particularly effective for analysing the surface distribution, identification, and localization of elements, isotopes, and molecules, as well as fragmentations in tissue sections.24 Among the molecules well traced by ToF-SIMS is cholesterol which is known to be poorly ionized by many other techniques. A notable study utilizing ToF-SIMS observed a higher level of cholesterol in the homogeneous tumour region of human glioblastoma multiforme (GBM).25 The sensitivity of the technique allows for the detection of small amounts of cholesterol, while its specificity enables the characterization of individual cholesterol fragments.26
Cholesterol is a membrane lipid that plays a crucial role for all cells including cancer cells.27 During the uncontrolled growth of cancer cells, both cholesterol uptake and synthesis rates typically increase, leading to abnormal metabolism.28–30 Several studies have documented a positive correlation between cancer cell proliferation and cholesterol synthesis rate.31,32 Consequently, increased cholesterol synthesis may compromise the integrity of the cholesterol molecule. Fidelity in this context refers to the structural accuracy and stability of the cholesterol molecule, particularly its resistance to fragmentation during ionization processes. Therefore, we hypothesise that increased cholesterol synthesis during tissue transformation results in altered fidelity of the cholesterol molecule and would also alter the cholesterol fragmentation pattern or isotope profile.
Here, we investigate whether the fidelity of the cholesterol molecule is altered in transformed tissues, subsequently changing its fragmentation pattern. Using ToF-SIMS and desorption electrospray ionization mass spectrometry (DESI-MS), we map the distribution of cholesterol in the mammary glands from a genetically modified mouse model that develop breast cancer (MMTV-PyMT) and wild-type (WT) mice (FVB). Additionally, we attempt to interpret the results in the context of changes to the composition of abundance carbon isotopes.
Materials and methods
Animal and tissue collection: mouse tissue
MMTV-PyMT mice, maintained on FVB/n and WT FVB/n female mice, were purchased from Scanbur, Denmark. All procedures were approved and conducted according to the guidelines from the regional Ethics Committee for Animal Research (Dnr 5.8.18-15383/2019). Mice arrived at the age of 4 weeks. They were kept until mammary tumours in the MMTV-PyMT model approached 1.5 cm, at which point they were 10–15 weeks old. Upon the sacrifice of a tumour-bearing mouse, a WT FVB/n mouse was also sacrificed, and the mammary glands were collected. In total, mammary glands were dissected from 19 WT and 19 MMTV-PyMT mice and snap-frozen in liquid nitrogen and stored in a −80 °C freezer until cryo-sectioning.Mammary gland sectioning and staining
Cryo-sectioning was performed at −25 °C with a cryostat microtome (Leica CM3050S, Germany) without embedding to avoid contamination. Fresh frozen tissues, sectioned to a thickness of 10 μm, were placed on the glass slides and kept at −80 °C until ToF-SIMS analysis. In addition, the tissue sections were stained with haematoxylin and eosin (H&E) for cell and tissue morphology.Sample preparation prior to ToF-SIMS analysis
ToF-SIMS analysis
The samples were investigated using ToF-SIMS V (ION-TOF, Munster, Germany), equipped with a bismuth (Bi3+) liquid metal ion gun as a primary ion source (25 keV). The ToF-SIMS spectra were recorded in positive mode. The current primary ion was approximately 0.2–0.4 pA. The surface spectra analysis was carried out in an area of 500 × 500 μm2. To establish the necessary number of tissue areas, up to 10 randomized areas were compared. The variability within a tissue sample was confidently captured when four randomized regions were analysed (Fig. S1†). Thereafter, four randomized areas were analysed in every tissue sample, and each area was scanned 20 times. All ToF-SIMS spectra were analysed with the Surface Lab software (version 7.0; ION-TOF, GmbH). The mass spectra were internally calibrated to signals of [C]+, [CH]+, [CH2]+, [CH3]+, [C5H15PNO4]+ and [C27H45]+ that are commonly used for calibration in the positive ion mode of tissue analysis. The peaks from cholesterol were selected, and their peak areas were exported with normalization of total ion intensity.The calculation of peak areas
The peak areas of each cholesterol peak were calculated into fractional abundances by dividing the area of each individual peak by the total sum of all cholesterol peak areas. Furthermore, the peak areas were analyzed to calculate several ratios, particularly focusing on the fragment-to-intact cholesterol and various isotope patterns. Normalized ratios were calculated using fold-change, setting the control group to a baseline value of 1.DESI-MS analysis
The mouse tissue sections were investigated using SELECT SERIES MRT for DESI-MS (Desorption Electrospray Ionization Mass Spectrometry). The spectra were recorded in positive mode at high spatial resolution with their identity assigned using high mass accuracy. The DESI source was operated at an ionization voltage of 1.1 kV, a distance of 1–3 mm between the sprayer and the sample surface with a 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol/water mixture with 0.1% formic acid delivered and 50 pg μL−1 leucine enkephalin at a rate of 2.0–1.0 μL min−1. The MRT mass analyser, controlled by Mass Lynx version 4.2. The DESI stage was operated to achieve two-dimensional data with a 50 μm pixel size. All of the experiments were performed using a lock mass, leucine enkephalin, which corresponds to 556.27658 Da as [M + H − 1e]+ as observed during positive mode DESI experiments.
1 methanol/water mixture with 0.1% formic acid delivered and 50 pg μL−1 leucine enkephalin at a rate of 2.0–1.0 μL min−1. The MRT mass analyser, controlled by Mass Lynx version 4.2. The DESI stage was operated to achieve two-dimensional data with a 50 μm pixel size. All of the experiments were performed using a lock mass, leucine enkephalin, which corresponds to 556.27658 Da as [M + H − 1e]+ as observed during positive mode DESI experiments.
Statistical analysis and machine learning
The comparison between the means of control (without cancer) and cancer samples was conducted using the Mann–Whitney U test. This test was chosen for its robustness in analysing non-normally distributed data. Statistical significance was determined at a significance level of p < 0.05. Principal Component Analysis (PCA) was applied for dimensionality reduction, using sklearn.decomposition.PCA. In the Partial Least Squares Discriminant Analysis (PLS-DA) workflow, the dataset was divided into training and test sets, comprising 75% and 25% of the data, respectively. The training dataset was used to fit the PLS regression model via sklearn.cross_decomposition.PLSRegression, and the test set was utilized to assess the model's performance. Variable Importance in Projection (VIP) scores were calculated to evaluate feature significance within the PLS-DA model, following the method described.33 This approach helps identify which variables most significantly contribute to the model based on their association with the response variable. For feature selection using a Random Forest approach, a binary classification model was constructed with sklearn.ensemble.RandomForestClassifier. The built-in feature importance attribute of the RandomForestClassifier was leveraged to identify key features that are most predictive of the outcome, aiding in understanding feature relevance and impact on the model predictions.Results and discussion
Transformed mammary gland displayed higher density of cells
In our study, we hypothesized that tissue transformation could influence the structural integrity and metabolic processing of cholesterol molecules. Specifically, we aimed to detect variations in cholesterol fragmentation patterns and assess their potential association with the altered metabolic incorporation of 13C in breast cancer. To explore this hypothesis, we utilized a mouse model of breast cancer and commenced our investigations by examining mammary gland tissues using haematoxylin and eosin (H&E) staining. Mammary glands derived from the FVB mouse, which served as the control group, exhibited a higher density of adipocytes, indicative of normal adipose distribution within the mammary glands (Fig. 1A and B). In contrast, mammary tissue from the MMTV-PyMT mice displayed a significantly increased number of malignantly transformed cells. These cells are characterized by their irregular morphology and increased nuclear-to-cytoplasmic ratio, features that are typical of neoplastic transformation (Fig. 1C and D).ToF-SIMS analysis of cholesterol fragmentation in standard and mammary glands
To assess the cholesterol fragmentation in breast tumours, we first compared the ToF-SIMS spectra of cholesterol in a pure standard (Fig. 2A) with those obtained from tissues of the mammary gland in both WT and breast cancer-bearing (MMTV-PyMT) mice (Fig. 2B and C†). The spectra encompass two groups of cholesterol fragments, within the mass-to-charge (m/z) ranges of 367 to 371 and 383 to 387, respectively. Thus, the presence of cholesterol in the tissue samples is effectively characterized using this method. | ||
| Fig. 2 The overlay spectra of (A) pure standard cholesterol, and mammary glands from (B) WT mouse (FVB) and (C) tumour-bearing mouse (MMTV-PyMT). | ||
Fig. 3 illustrates the differential fragmentation of cholesterol due to ionization. During these measurements, the cholesterol molecule may lose either a hydrogen (H) or a hydroxyl group (–OH). Therefore, the ionization process offers a route to quantify the stability of the cholesterol molecule. Cholesterol molecules that lose the hydroxyl group are represented in the spectra by five distinct peaks, termed the ‘fragment group’ (Fig. 3A), with masses ranging from m/z 367 to 371. Conversely, those losing a hydrogen atom are depicted by five peaks in the ‘intact group’ (Fig. 3B), with masses in the range of m/z 383 to 387.
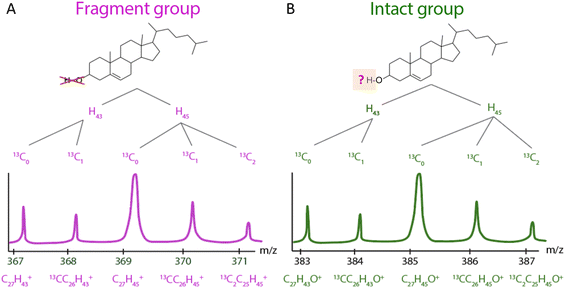 | ||
| Fig. 3 Schematic diagram of the difference between each peak of cholesterol. (A) fragment group and (B) intact group. | ||
Within each group, the analysis method further distinguishes cholesterol molecules that contain one or two 13C isotopes. In the fragment group, cholesterol with one 13C is identified at m/z 368 and 370, while in the intact group, it appears at m/z 384 and 386. Additionally, distinctions are made based on the loss of two additional hydrogen atoms. Within both groups, the molecules can contain either 43 or 45 hydrogens. For instance, the m/z range of 367 to 371 in the fragment group reflects the loss of hydroxyl group, resulting in molecular ions C27H43+ and C27H45+. Similarly, in the intact group, the range of m/z 383 to 387 demonstrates the loss of a hydrogen atom, leading to molecular ions of C27H43O+ or C27H45O+. Each set of peaks further differentiates by the numbers of the heavier 13C isotope.
ToF-SIMS analysis of cholesterol from mammary glands
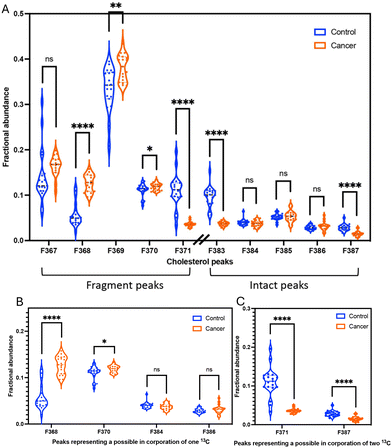
We calculated the fractional abundance of ten peaks from cholesterol from the mammary gland tissues of 19 WT mice and 19 tumour-bearing mice (MMTV-PyMT) (Fig. 4A).The fractional abundances of fragment (C27H45+) and intact (C27H45O+) of cholesterol in mammary glands
To investigate differential fragmentation, we measured both fragmented and intact cholesterol in mammary gland tissues from 19 WT mice and 19 MMTV-PyMT mice. We observed that the fractional abundances of fragmented cholesterol (m/z 369: C27H45+) is statistically significant higher in transformed tissues compared to control tissues (Fig. 4A). In contrast, the fractional abundances of intact cholesterol when H45 (m/z 385: C27H45O+) showed no difference between control and cancer samples (Fig. 4A). However, the fractional abundances of intact cholesterol when H43 (C27H43O+: m/z 383) were significantly lower in transformed tissues (Fig. 4A).Additionally, we performed a calculation to determine the ratio between the fragment peak and the intact peak of cholesterol within the same sample. This approach confirmed the trend observed in the fractional abundance analysis. Specifically, the ratio of fragmented cholesterol (C27H45+: m/z 369) over intact cholesterol (C27H45O+: m/z 385) exhibited a higher proportion in cancer samples (Fig. S2†). This notable distinction implies the potential to distinguish a difference in the fidelity of the cholesterol molecule via ToF-SIMS.
To assess whether the observed alterations in cholesterol structure are consistent across different ionization techniques, we expanded our analysis to include Desorption Electrospray Ionization Mass Spectrometry (DESI-MS). This method was applied to mammary gland samples from both control mice and mice with breast cancer. We observed a significant increase in the proportion of fragmented cholesterol (m/z 369: C27H45+) in the mammary gland samples from mice with breast cancer (Fig. S3†). However, the trend of the m/z 385 peak differing from that observed in the ToF-SIMS data. Therefore, while these findings suggest a consistent fragmentation pattern of cholesterol at m/z 369 across different ionization methods, the data also indicate some variability, which could be influenced by the specific analytical techniques used.
The observed prevalence of fragmented cholesterol in transformed tissues indicate a vulnerability in the molecular structure. This vulnerability may manifest as increased susceptibility to fragmentation under the ionizing condition of ToF-SIMS. To explore this possibility, we compared the 369/385 ratio between pure cholesterol, control tissue, and cancer tissue. The ratios were 447%, 620%, and 948%, respectively (Fig. S4†). These results suggest that the extent of cholesterol fragmentation is significantly higher in cancer tissue, potentially supporting the hypothesis that incomplete synthesis or other biochemical alterations in cancerous tissues contribute to the increased fragmentation. It is noted that the synthesis of cholesterol intermediates squalene and lanosterol are deregulated in transformed tissue.30,34–36 Although our analysis does not specifically target these intermediates, the possibility that incomplete synthesis contributes to the observed fragmentation of cholesterol remains a plausible explanation. This could potentially lead to an accumulation of biochemically unstable precursors that are more prone to fragmentation.
Moreover, another possibility is the matrix effect in ToF-SIMS. The matrix effect refers to the influence that the chemical environment or matrix of a sample can have on the accuracy and reliability of analytical measurements. In mass spectrometry techniques, the matrix effect can lead to changes in the signal intensity or response of the analyte of interest.37,38 To mitigate the matrix effect, we examined the distribution of peaks from the phosphatidylcholine head group (C5H12N+: m/z 86 and C5H15PNO4+: m/z 184) and salts (Na+: m/z 23 and K+: m/z 39). The mammary glands from wild-type mice and mice with breast cancer exhibited similar distribution patterns (Fig. S5†) and peak areas (Fig. S6†). This suggests that the matrix composition of both tissues is quite similar. Additionally, we performed the ToF-SIMS analysis of the enriched 13C and 13C2 cholesterol standard (Fig. 5) to see if it showed a similar effect to the non-enriched cholesterol. The results revealed no significant difference in the ratios of 3 cholesterol standards (Fig. S7†). The spectra from all standards exhibit similar peak patterns and intensities, with minor differences attributable to the presence of 13C isotopes in the enriched standard, likely due to oxidation effects in the standard samples (Fig. S8†). Since no variations are observed in these pure standard samples, any variations seen in the cancer tissue are likely attributable solely to the tissue samples.
 | ||
| Fig. 5 The overlay spectra of (A) non-enriched cholesterol standard (C27H46O), (B) enriched 13C cholesterol standard (13CC26H46O) and (C) enriched 13C2 cholesterol standard (13C2C25H46O). | ||
In this study, we investigated whether this change in molecular fidelity is related to the rate of lipid synthesis, which may necessitate the incorporation of more of the heavier carbon isotope, 13C. Due to their differing mass, isotopes can affect the stability and fragmentation pathways of cholesterol molecules during ionization and subsequent analysis. Therefore, we proceeded to examine the ratios of various isotopic patterns of cholesterol in both mammary glands with and without cancer to observe the potential incorporation of 13C.
The fractional abundances of the isotopic patterns of cholesterol from the mammary glands
Due to their differing mass, isotopes can affect the stability and fragmentation, the different isotopic patterns of cholesterol were also investigated. The different isotopic patterns of cholesterol are 13CC26H43+ (368), 13CC26H45+ (370), 13C2C25H45+ (371), 13CC26H43O+ (384), 13CC26H45O+ (386), and 13C2C25H45O+ (387). The significant difference in the fractional abundances of isotopic patterns between control and cancer could be explained by the potential incorporation of one 13C in transformed tissue (13CC26H43+ (368) and 13CC26H45+ (370)). The molecules with one 13C are overrepresented in transformed tissues (Fig. 4B).However, it appears that the molecules with two 13C are overrepresented in normal tissue for both 13C2C25H45+ (371) and 13C2C25H45O+ (387) (Fig. 4C). These trends are also evident in an alternative method used to calculate peak area ratios of the isotopic patterns from cholesterol (Fig. S9†). This opposite and significant trend is counterintuitive. Therefore, we first asked if the data accurately captured peaks of carbon isotope. The variation could be due to other factors, such as protonation effects in ToF-SIMS,39 or interference from 2H and 18O. The limitations of ToF-SIMS means that weights for e.g., a cholesterol molecule with one 13C appears to have the same weight as a molecule with one hydrogen atom. This process is called protonation. Protonation is arguably the most important means of ionization in the positive mode of soft ionization mass spectrometry methods.40,41 Protonation may contribute to our quantification and therefore overestimate some of our isotope peaks. However, for this overestimation to be responsible for our opposing trends, protonation must contribute in a discriminatory way whether the tissue is normal and transformed. With our current knowledge, it appears inexplicable how protonation would contribute unevenly between tissues. Additional influence on the cholesterol fidelity within transformed tissues may be isotopic effects stemming from variations in hydrogen or oxygen isotopes. The presence of isotopes, notably deuterium (2H) or oxygen isotopes (18O), can significantly modify the observed fragmentation patterns of cholesterol. The natural abundances of deuterium (2H) and oxygen-18 (18O) are approximately 0.0156% and 0.204%, respectively. Given that a cholesterol molecule contains 27 hydrogen atoms, the probability of 2H substitution is consequently increased. The substitution of 2H and 18O can induce changes in bond strengths, thereby impacting the likelihood of specific fragmentation pathways.15 This phenomenon is often referred to as the isotopic effect or isotopic substitution effect.13,14 Hydrogen (1H vs.2H) and oxygen (16O vs.18O) isotopes are typically influence due to their higher natural abundances and large mass differences. However, the impact of oxygen isotopes on fragmentation may be less pronounced compared to hydrogen isotopes due to the fewer number of oxygen atom in cholesterol molecule.
Resolving what ion contributes to the mass balance alteration can be resolved by either another method to quantify the isotope contribution at higher resolution (like GC-HR-MS), or with the magnetic resonance mass spectrometry with higher mass resolution (like MRMS). With the current techniques used here, it the observations are consistent with the predicted isotope pattern change that cholesterol synthesis in transformed tissue incorporates additional 13C, but only up until one molecule.
Thus, we conclude that the isotopic patterns may be explained by incorporation of heavy carbon isotopes but obscured by other isotopic effects. Although the resolution fails to shed light on the exact ion that contributes to the mass difference, the fidelity of the cholesterol molecule appears altered during tissue transformation.
Machine learning of cholesterol from mammary glands
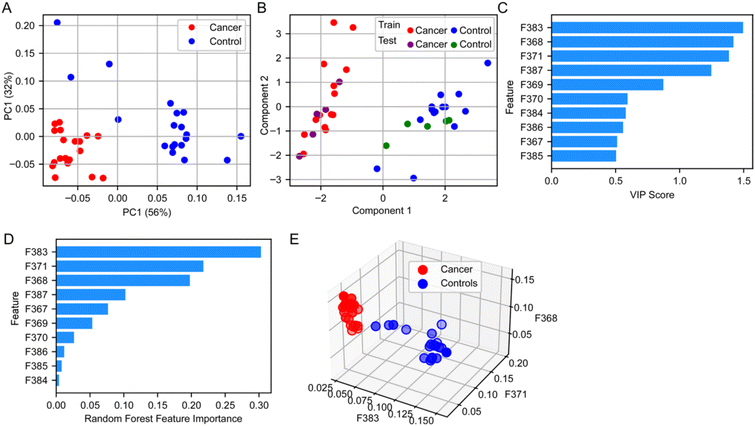
To investigate the discriminative power of the fractional abundances of 10 cholesterol fragments in distinguishing between cancer and control samples, we initially employed PCA. This analysis enabled us to visually assess the separation between the two groups. As anticipated, the projection onto the first two principal components resulted in a clear distinction between all cancer and control samples (Fig. 6A). Building on PCA analysis, we evaluated the classification efficacy of these fractional abundances using PLS-DA. We allocated 75% of the dataset for training the PLS-DA model and reserved the remaining 25% for testing. The PLS-DA model proved to be highly effective, correctly classifying all samples in the test set (Fig. 6B). To further elucidate which of the six ratios contributed most significantly to the model's predictive performance, we calculated the VIP scores within the PLS-DA framework. This analysis revealed that three specific fractional abundances, for m/z 383, 368, and 371 were particularly influential in the model (Fig. 6C). In parallel, we utilized a Random Forest Classifier to identify the most relevant features. This method corroborated the findings from the PLS-DA analysis, indicating a similar set of important features (Fig. 6D). Lastly, to visually confirm the discriminative capability of these three key fractional abundances, we generated a three-dimensional scatter plot of their raw values. This plot demonstrated a complete separation of the cancer and control samples, reinforcing the effectiveness of these fractional abundances in distinguishing between the two groups (Fig. 6E).Conclusion
We investigated whether the increased rate of cholesterol synthesis during tissue transformation leaves a detectable chemical signature in the cholesterol molecule. Our study revealed a significant increase in the fragmentation of cholesterol in transformed tissues, measured as a higher fractional abundance of fragmented (ionized) cholesterol. This increase in fragmentation suggests a possible alteration in the molecular integrity of cholesterol, potentially linked to the metabolic changes associated with tissue transformation. Furthermore, the observation of an altered structural fidelity would be consistent with increased incorporation of 13C, if also obscured by other mass alterations. Future research should aim to determine whether these changes in cholesterol structure persist throughout tumor progression, whether they can be detected in blood samples, and whether they involve specific isotopic mechanisms.Author contributions
A. M., E. U. H. and P. M. conceived the project, designed the experiments. C. H. designed and provided the access to murine models experiment. A. M. carried out the research. E. U. H., P. M., J. U. K, and K. J. P were involved in the supervision and discussion of data interpretation. All authors were involved in editing the manuscript and have given approval to the final version of the manuscript.Data availability
The raw data generated and analyzed during this study are openly available from Open Science Framework at https://osf.io/f8phj/?view_only=10c745b793584f00be550156912f1aa0.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Sjöberg Foundation for funding to explore ‘The Elemental Fingerprint of Early Cancer’. We thank Chalmers Materials Analysis Laboratory (CMAL) at Chalmers University of Technology for the support with ToF-SIMS experiments and to Waters Corporation, UK for conducting the DESI-MS experiments. We also thank to the BMC animal facilities staff, the EMV, Dr. Sofie Mohlin, and TiDE members for their valuable assistance and discussions. Additionally, we are grateful to the Development and Promotion of Science and Technology Talents Project (DPST), Thailand for the funding support.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, CA-Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- D. Crosby, S. Bhatia, K. M. Brindle, L. M. Coussens, C. Dive, M. Emberton, S. Esener, R. C. Fitzgerald, S. S. Gambhir and P. Kuhn, Science, 2022, 375, eaay9040 CrossRef CAS PubMed.
- D. Liu, Biomarker Res., 2019, 7, 25 CrossRef PubMed.
- N. Koundouros and G. Poulogiannis, Br. J. Cancer, 2020, 122, 4–22 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- R. Radišauskas, I. Kuzmickienė, E. Milinavičienė and R. Everatt, Medicina, 2016, 52, 89–98 CrossRef PubMed.
- M. V. Liberti and J. W. Locasale, Trends Biochem. Sci., 2016, 41, 211–218 CrossRef CAS PubMed.
- F. Larner, Anal. Bioanal. Chem., 2016, 408, 345–349 CrossRef CAS PubMed.
- F. Larner, L. N. Woodley, S. Shousha, A. Moyes, E. Humphreys-Williams, S. Strekopytov, A. N. Halliday, M. Rehkämper and R. C. Coombes, Metallomics, 2015, 7, 112–117 CrossRef CAS PubMed.
- K. Schilling, F. Larner, A. Saad, R. Roberts, H. M. Kocher, O. Blyuss, A. N. Halliday and T. Crnogorac-Jurcevic, Metallomics, 2020, 12, 752–757 CrossRef CAS PubMed.
- P. Télouk, A. Puisieux, T. Fujii, V. Balter, V. P. Bondanese, A. P. Morel, G. Clapisson, A. Lamboux and F. Albarede, Metallomics, 2015, 7, 299–308 CrossRef PubMed.
- I. Tea, E. Martineau, I. Antheaume, J. Lalande, C. Mauve, F. Gilard, S. Barillé-Nion, A. C. Blackburn and G. Tcherkez, Sci. Rep., 2016, 6, 34251 CrossRef CAS PubMed.
- P. J. Derrick, Mass Spectrom. Rev., 1983, 2, 285–298 CrossRef CAS.
- D. Zakett, R. Flynn and R. Cooks, J. Phys. Chem., 1978, 82, 2359–2362 CrossRef CAS.
- P. E. Hansen, Prog. Nucl. Magn. Reson. Spectrosc., 2020, 120–121, 109–117 CrossRef CAS PubMed.
- D. A. Singleton and Z. Wang, Tetrahedron Lett., 2005, 46, 819–822 CrossRef CAS.
- J. A. Linscott, K. Kapilashrami, Z. Wang, C. Senevirathne, I. R. Bothwell, G. Blum and M. Luo, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E8369–E8378 CrossRef CAS PubMed.
- K. Kantnerová, N. Kuhlbusch, D. Juchelka, A. Hilkert, S. Kopf and C. Neubauer, Nat. Protoc., 2024, 19, 2435–2466 CrossRef PubMed.
- A. Van Langenhove, J. Clin. Pharmacol., 1986, 26, 383–389 CrossRef CAS PubMed.
- L. Bluck and D. A. Volmer, Spectroscopy, 2009, 23, 36 Search PubMed.
- D. R. Klein, E. S. Rivera, R. M. Caprioli and J. M. Spraggins, Anal. Chem., 2024, 96, 5065–5070 CrossRef CAS PubMed.
- J. G. Son, S. Yoon, H. K. Shon, J. H. Moon, S. Joh and T. G. Lee, Biointerphases, 2020, 15, 021011 CrossRef PubMed.
- G. E. Johnson, M. Lysonski and J. Laskin, Anal. Chem., 2010, 82, 5718–5727 CrossRef CAS PubMed.
- H. Nygren, P. Malmberg, C. Kriegeskotte and H. F. Arlinghaus, FEBS Lett., 2004, 566, 291–293 CAS.
- M. H. Philipsen, E. Hansson, A. Manaprasertsak, S. Lange, E. Jennische, H. Carén, K. Gatzinsky, A. Jakola, E. U. Hammarlund and P. Malmberg, ACS Chem. Neurosci., 2023, 14, 1602–1609 CrossRef CAS PubMed.
- S. G. Ostrowski, M. E. Kurczy, T. P. Roddy, N. Winograd and A. G. Ewing, Anal. Chem., 2007, 79, 3554–3560 CrossRef CAS PubMed.
- J. Zhang, Q. Li, Y. Wu, D. Wang, L. Xu, Y. Zhang, S. Wang, T. Wang, F. Liu, M. Y. Zaky, S. Hou, S. Liu, K. Zou, H. Lei, L. Zou, Y. Zhang and H. Liu, Cell Commun. Signaling, 2019, 17, 15 CrossRef PubMed.
- I. Giacomini, F. Gianfanti, M. A. Desbats, G. Orso, M. Berretta, T. Prayer-Galetti, E. Ragazzi and V. Cocetta, Front. Oncol., 2021, 11, 682911 CrossRef CAS PubMed.
- M. Xiao, J. Xu, W. Wang, B. Zhang, J. Liu, J. Li, H. Xu, Y. Zhao, X. Yu and S. Shi, Exp. Mol. Med., 2023, 55, 1982–1995 CrossRef CAS PubMed.
- P. Kucharzewska, H. C. Christianson and M. Belting, PLoS One, 2015, 10, e0116740 CrossRef PubMed.
- S. Silvente-Poirot and M. Poirot, Curr. Opin. Pharmacol., 2012, 6, 673–676 CrossRef PubMed.
- B. Huang, B.-l. Song and C. Xu, Nat. Metab., 2020, 2, 132–141 CrossRef PubMed.
- R. Gosselin, D. Rodrigue and C. Duchesne, Chemom. Intell. Lab. Syst., 2010, 100, 12–21 CrossRef CAS.
- A. Gorin, L. Gabitova and I. Astsaturov, Curr. Opin. Pharmacol., 2012, 12, 710–716 CrossRef CAS PubMed.
- A. D. Nguyen, J. G. McDonald, R. K. Bruick and R. A. DeBose-Boyd, J. Biol. Chem., 2007, 282, 27436–27446 CrossRef CAS PubMed.
- B.-L. Song, N. B. Javitt and R. A. DeBose-Boyd, Cell Metab., 2005, 1, 179–189 CrossRef CAS PubMed.
- F. Fack, S. Tardito, G. Hochart, A. Oudin, L. Zheng, S. Fritah, A. Golebiewska, P. V. Nazarov, A. Bernard, A. C. Hau, O. Keunen, W. Leenders, M. Lund-Johansen, J. Stauber, E. Gottlieb, R. Bjerkvig and S. P. Niclou, EMBO Mol. Med., 2017, 9, 1681–1695 CrossRef CAS PubMed.
- A. F. M. Altelaar, J. van Minnen, R. M. A. Heeren and S. R. Piersma, Appl. Surf. Sci., 2006, 252, 6702–6705 CrossRef CAS.
- N. J. Popczun, L. Breuer, A. Wucher and N. Winograd, J. Phys. Chem. C, 2017, 121, 8931–8937 CrossRef CAS.
- J. B. Fenn, M. Mann, C. K. Meng, S. F. Wong and C. M. Whitehouse, Science, 1989, 246, 64–71 CrossRef CAS PubMed.
- S. Zhou and K. D. Cook, J. Am. Soc. Mass Spectrom., 2000, 11, 961–966 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an01050g |
| This journal is © The Royal Society of Chemistry 2024 |