Open Access Article
Open Access ArticleSurface-enhanced Raman scattering enhancement using a hybrid gold nanoparticles@carbon nanodot substrate for herbicide detection†
Naghmeh
Aboualigaledari
a,
Anitha
Jayapalan
a,
Panesun
Tukur
a,
Mengxin
Liu
a,
Frank
Tukur
a,
Yanling
Zhang
c,
Gerald
Ducatte
c,
Madan
Verma
c,
Janet
Tarus
c,
Simona E.
Hunyadi Murph
 *b and
Jianjun
Wei
*b and
Jianjun
Wei
 *a
*a
aDepartment of Nanoscience, Joint School of Nanoscience and Nanoengineering, University of North Carolina at Greensboro, Greensboro, NC 27401, USA. E-mail: j_wei@uncg.edu
bEnvironmental and Legacy Management Department, Savannah River National Laboratory (SRNL), Aiken, SC 29808, USA. E-mail: Simona.Murph@srnl.doe.gov
cSyngenta Crop Protection Inc., Greensboro, NC 27409, USA
First published on 29th August 2024
Abstract
The widespread distribution of herbicides in the environment poses a significant risk to human health and wildlife. Surface-enhanced Raman scattering (SERS) has emerged as a powerful technique for detecting and analyzing herbicides. However, developing a low-cost, highly sensitive, reproducible, stable, and Raman-active nanostructured substrate for herbicide detection remains a particular challenge. In this research, a nanohybrid substrate consisting of gold nanoparticles@carbon nanodots (AuNPs@CNDs) was synthesized by reducing HAuCl4 in the presence of CNDs at 100 °C. The optical, chemical, and physical properties of CNDs, AuNPs, and the hybrid AuNPs@CND substrates were thoroughly investigated using various techniques including UV-vis spectrometry, Raman spectroscopy, X-ray photoelectron spectroscopy (XPS), scanning electron microscopy (SEM), transmission electron microscopy (TEM), and CytoViva darkfield and hyperspectral imaging. The SERS effect of the substrates was evaluated using rhodamine 6G (Rh6G), a Raman-active probe, and two groups of herbicides containing mesotrione or S-metolachlor. The results demonstrated a significant signal amplification in the SERS spectra of Rh6G and herbicide molecule detection using the AuNPs@CND substrate compared to bare CNDs and AuNPs alone. This suggests that the nanohybrid AuNPs@CND SERS substrate holds promise for the detection of herbicides and other organic compounds in environmental applications.
1. Introduction
The widespread distribution of herbicides in the environment poses a significant risk to human health and wildlife.1 Accurate, quantitative monitoring of herbicide residues, especially in water environments, is critical for their management, removal or treatment. Traditionally, a broad range of analytical methods, such as gas chromatography (GC),2 high performance liquid chromatography (HPLC), GC-mass spectrometry (GC-MS), electrochemical and spectrophotometric techniques, have been used for the extraction and determination of their concentration.3–6 Advancements in nanomaterials and nanotechnology have provided alternative advantageous sensor technologies, such as nano-electrochemical sensors,7 molecularly imprinted polymers,8 and nanoplasmonic surface enhanced Raman spectroscopy (SERS),9 which may enable easy operation, improved sensitivity, portability and real-time and on-site applications.The surface-enhanced Raman scattering (SERS) technique relies on plasmonic metallic nanostructures to concentrate electromagnetic energy, thereby enhancing the molecular Raman signal. SERS spectroscopy is a powerful technique with diverse applications in medical science and analytical chemistry.10 Tailored nanostructures enable fast, sensitive, nondestructive, and highly efficient detection of probe molecules, even facilitating single molecule analysis.11–13 In SERS, the amplification of the Raman signals of probe molecules results from the excitation of localized surface plasmon resonance (LSPR) at the metal–dielectric interface of the substrate. Therefore, the magnitude of SERS enhancement critically depends on the SERS substrate.14
Gold and silver nanostructures are typical metals used as SERS substrates due to their LSPR bands covering the visible and near-infrared (NIR) wavelength regimes necessary to excite Raman modes.15–18 While silver nanoparticles (AgNPs) offer higher plasmon enhancement, they are susceptible to oxidation, leading to poor stability.19 In contrast, gold exhibits superior chemical stability.20,21 Recent SERS studies have focused on developing and optimizing substrates with promising enhancement effects.22–25 For instance, a gold nanorod array was developed and optimized for SERS detection of the herbicide atrazine.26 With advancements in nanotechnology, SERS substrates have evolved from single-composition nanomaterials to multi-component nanomaterials.27 This is due to their ability to serve as multi-modal platforms for enhanced sensing of analytes, sorbent capability, and magnetic recyclability, among others.28 Hybrid nanomaterials and complex nanostructures comprising carbon nanomaterials and metal NPs, such as AuNPs, have demonstrated exceptional SERS enhancement capabilities.29,30
Carbon dots (CDs) with diameters usually less than 10 nm are classified as carbon quantum dots (CQDs), carbon nanodots (CNDs), and graphene QDs (GQDs).31 Compared to conventional semiconductor quantum dots and semiconductors, CNDs offer advantages such as low toxicity, excellent biocompatibility, multifunctionality, and straightforward synthesis.32–34 CNDs represent a novel class of carbon nanomaterials characterized by a spherical morphology and typically an amorphous lattice structure. The functional groups present in CNDs play a crucial role in enhancing surface-enhanced Raman scattering (SERS). These functional groups are typically introduced during hydrothermal synthesis through doping heteroatoms like nitrogen (N), sulfur (S), boron (B), phosphorus (P), and others.35–38
Recent findings indicate that N-doped CNDs contribute to the improvement of SERS signals, likely because N-doped CNDs facilitate excited state charge transfer to the target molecule through π–π interactions.39–41 By incorporating CNDs as a shell component into AuNPs with optimized thickness, the chemical enhancement can be improved. This enhancement occurs through increased adsorption of analytes and the prevention of signal interference from probe molecules caused by direct contact with gold surfaces.42
The utilization of AuNPs achieves high SERS enhancement due to their appropriate size, shape, and matching of the laser line with the AuNPs’ LSPR, which leads to an increase in the surface area and local electromagnetic (EM) field enhancement.42 Previous studies have demonstrated the effectiveness of AuNPs or silver NP-based SERS substrates for detecting herbicides, including atrazine, diquat, flumetsulam, glyphosate, paraquat, and prometryn.26,43–52 SERS detection of relatively new types of herbicides, such as mesotrione and S-metolachlor, has been rarely reported. In this study, microwave-synthesized hybrid AuNP@CND core–shell composite NPs are optimized for Raman signal amplification as SERS probes for the sensitive detection of mesotrione and S-metolachlor molecules, which are notably the active components in the formulation complex. This approach capitalizes on the advantageous synergistic effects of both chemical and EM field enhancement from the CND and AuNP components,29 thereby improving SERS sensing capabilities.
2. Materials and methods
2.1. Chemicals and reagents
Hydrogen tetrachloroaurate (HAuCl4) solution (from Fisher Scientific, Au 44% w/w), sodium citrate (from Fisher Scientific, ≥99%), rhodamine 6G (from ACROS Organics, 99%), citric acid (from ACROS Organics, 99%), and ethylenediamine (EDA, from Fisher Scientific, 99%) were utilized in this study without further purification.The herbicides used in the research, including mesotrione (2-(4-methylsulfonyl-2-nitrobenzoyl)cyclohexane-1,3-dione, ZA1296E), A12738A formulation, and S-metolachlor ((S)-2-chloro-N-(2-ethyl-6-methyl-phenyi)-N-(2-methoxy-1-methyl-ethyl)-acetamide, CGA77102), as well as its formulation LUMAX EZ (A19414A), were obtained from Syngenta Crop Protection, LLC.
2.2. Materials characterization
The morphology of both the AuNPs and AuNPs@CNDs was examined using field-emission scanning electron microscopy (FE-SEM) conducted with a Carl Zeiss Auriga-BU FIB FESEM Microscope. The measurements were performed at an accelerating voltage of 5.0 kV. Additionally, transmission electron microscopy (TEM) was employed using a Carl Zeiss Libra 120 PLUS instrument to further investigate the morphology.The elemental composition of the samples was determined using X-ray photoelectron spectroscopy (XPS) utilizing a Thermo Scientific ESCALAB Xi+ instrument. Raman spectroscopy was conducted using a Horiba XploRA One Raman Confocal Microscope system, with a 532 nm laser serving as the excitation source.
To assess the optical properties of the CNDs, AuNPs, and AuNPs@CNDs, ultraviolet-visible spectrophotometry was performed using a Varian Cary 6000i spectrophotometer. Fluorescence spectrophotometry was carried out using a Varian Cary Eclipse instrument. A Varian Agilent 710 inductively coupled plasma optical emission spectrometer (ICP-OES) with trace elemental analysis capability for wavelengths from 177 to 785 nm was used to determine the concentration of Au in AuNPs@CND and AuNP suspensions.
2.3. Nanoparticle synthesis
To record the SERS spectra of 4 herbicide samples, 10 μL of each herbicide sample were added to 50 μL of AuNPs@CND aqueous suspension and sonicated for 30 min. The mixture of samples was dried on cover glasses for 3 h in a fume hood. SERS spectra were obtained on dried samples using 532 nm excitation, a 50× objective, a 1200 gr per nm grating, an accumulation time of 5 s, and an acquisition time of 35 s.
The first group of herbicides included CGA77102 (S-metolachlor, 86.6% by weight) and A19414A formulation (S-metolachlor, 27.1% by weight). The sample solutions containing the desired concentrations of CGA77102 and A19414A formulation ranging from 10−12–10−3 M were analysed with respect to the effective S-metolachlor component without extraction. The same procedure was carried out for the second group of herbicides, ZA1296E (mesotrione, 60.0 by wt%) and A12738A formulation (mesotrione, 40.0 wt%).
3. Results and discussion
3.1. Characterization of CNDs, AuNPs, and AuNPs@CNDs
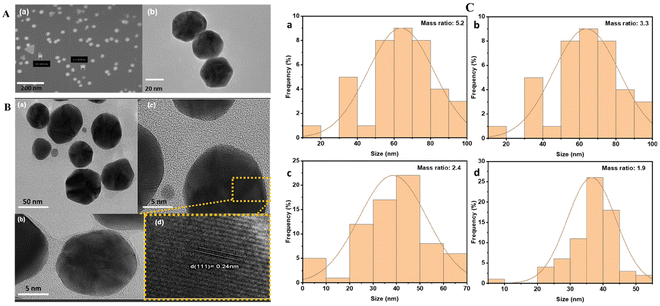
The size and morphology of the NPs were characterized using SEM and TEM. The SEM and TEM images of AuNPs (Fig. 1A) reveal the spherical shape of AuNPs with a size of approximately 50 nm, consistent with the 540 nm absorption of the UV-vis spectra of AuNPs (Fig. 2A). The TEM images of hybrid AuNPs@CNDs (Fig. S1A†) at different HAuCl4/CND mass ratios (5.2, 3.3, 2.4, and 1.9) show that the AuNPs@CNDs obtained with the mass ratio of 2.4 have a spherical shape and the NPs are not agglomerated and more uniformly distributed compared to the other mass ratios. To confirm the hybrid structure of AuNPs@CNDs, SEM and energy dispersive X-ray spectroscopy (EDS) were applied to map the elemental distribution. Fig. S1B† shows the carbon-capped core–shell configuration. Additionally, the HR-TEM images of 2.4-ratio AuNPs@CNDs (Fig. 1B) suggest successful wrapping of AuNP cores by CND structures. Fig. 1B(d) confirms a 0.24 nm lattice fringe of its crystalline structure corresponding to the (111) planes of AuNPs.55 The size distribution of AuNPs@CNDs as a function of the HAuCl4/CND mass ratio is shown in Fig. 1C and AuNPs@CNDs with a mass ratio of 2.4 have a size range between 40 and 50 nm, similar to a previous report.53 It is known that carboxylate and nitrogen-containing groups including amine and pyridine have a strong binding affinity to gold,56,57 which allows the CNDs to act as a capping agent that wraps around the surface of AuNPs to form a core–shell structure.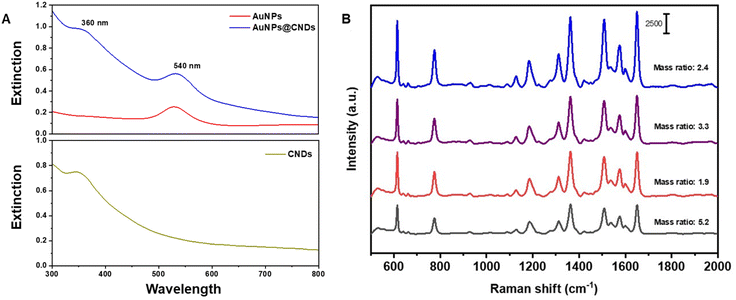 | ||
| Fig. 2 (A) UV-Vis absorption spectra of AuNPs, CNDs and hybrid AuNPs@CNDs and (B) SERS spectra of Rh6G with the hybrid AuNPs@CNDs generated with different mass ratios. | ||
To further confirm the formation of hybrid AuNPs@CNDs, UV-visible absorption spectra were recorded and compared with those of three samples, i.e., the AuNPs, CNDs and the hybrid. In Fig. 2A, the spectrum of AuNPs shows a SPR peak at 540 nm, which indicates the formation of AuNPs with a size of around 50 ± 7 nm.42 The absorption spectrum of CNDs displays a typical peak at 360 nm, which is attributed to the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O.58 The absorption spectrum of AuNPs@CNDs presents both the plasmon absorption peak at 540 nm of the AuNP core53 and the 360 nm peak for CNDs, corroborating the production of the hybrid NPs. Comparing the UV-vis absorption from different mass ratio products (Fig. S2†), the 2.4 mass ratio AuNPs@CNDs have the highest plasmonic peak around 540 nm along with a distinct peak at 360 nm for the CNDs, implying potential for Raman signal enhancement.
O.58 The absorption spectrum of AuNPs@CNDs presents both the plasmon absorption peak at 540 nm of the AuNP core53 and the 360 nm peak for CNDs, corroborating the production of the hybrid NPs. Comparing the UV-vis absorption from different mass ratio products (Fig. S2†), the 2.4 mass ratio AuNPs@CNDs have the highest plasmonic peak around 540 nm along with a distinct peak at 360 nm for the CNDs, implying potential for Raman signal enhancement.
The SERS performance of the hybrid AuNPs@CND substrates obtained from different mass ratios was evaluated using Rh6G. Fig. 2B presents the SERS spectra of Rh6G at a concentration of 48 × 10−8 M using AuNPs@CND substrates produced from different HAuCl4/CND mass ratios. It is apparent that the 2.4 ratio hybrid substrate provides the best SERS signal enhancement of Rh6G, plausibly due to the size range of the generated AuNPs and optimum surface coverage. The results further confirm the plasmonic properties observed in the UV-vis spectra. Therefore, the 2.4 mass ratio AuNPs@CND hybrid substrate is selected for further investigation and applied in herbicide sensing studies.
The structural, composition and spectroscopic properties of the synthesized NPs were further performed and analyzed. The Raman spectra of both CNDs and AuNPs@CNDs (Fig. S3a†) show two distinct peaks at 1340 cm−1 and 1590 cm−1, which correspond to the D and G bands of the carbon component, respectively. The D band corresponds to structural defects in graphitic sp3-hybridized carbon, whereas the G band is related to the vibration of sp2 carbon atoms in a 2D hexagonal lattice.59,60 The relative intensity of D and G bands (D/G ratio) in the Raman spectra of AuNPs@CNDs is 0.780, whereas this ratio is 0.741 for CNDs. The D and G band intensities of AuNPs@CNDs are much higher than those of CNDs, suggesting the enhanced defect effect of AuNPs.61,62 The photoluminescence (PL) spectra of CNDs and AuNPs@CNDs (Fig. S3b†) show that the PL intensity of AuNPs@CNDs is decreased compared to that of CNDs when excited with 360 nm light, which indicates that the PL of CNDs is quenched in the AuNPs@CND system. This phenomenon is attributed to a photo-induced electron transfer process between the CNDs and AuNPS.53,61
The full XPS scan spectra of CNDs and AuNPs@CNDs exhibit three peaks at 285.5, 399.0 and 531.0 eV, which are attributed to C 1s, N 1s, and O 1s, respectively.63 Compared to the XPS spectrum of CNDs, the XPS spectrum of AuNPs@CNDs presents a peak of the Au element at 335 eV (Fig. S4a and b†). The C 1s XPS spectra of CNDs and AuNPs@CNDs (Fig. S4d and e†) show peaks at 284.3, 285.4, and 287.5 eV, which are attributed to C–C, C–O–C, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively. The N 1s XPS spectra shown in Fig. S4c† feature two peaks corresponding to pyridinic nitrogen (398 eV) and pyridone nitrogen (399.6 eV).64 The O 1s band (Fig. S4f†) can be deconvoluted into two peaks located at 531.4 and 531.9 eV, which represent the C–O and C
O groups, respectively. The N 1s XPS spectra shown in Fig. S4c† feature two peaks corresponding to pyridinic nitrogen (398 eV) and pyridone nitrogen (399.6 eV).64 The O 1s band (Fig. S4f†) can be deconvoluted into two peaks located at 531.4 and 531.9 eV, which represent the C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively.
O groups, respectively.
A dark-field CytoViva hyperspectral imaging system was used to measure light scattering from AuNPs and AuNPs@CNDs distributed on a glass substrate. A CytoViva hyperspectral imaging system enables both optical and hyperspectral imaging of samples, which can be used for spectral characterization and spectral mapping of nanoscale samples.58,65 In experimental measurement, unpolarized plain light (broad band) from a halogen lamp was used as incident light to illuminate the nanoparticle samples from the top. The reflecting light signal captured by a 60× objective lens is then collected using a darkfield CytoViva hyperspectral imaging system (Fig. 3). The peak at around 550 nm corresponds to the SPR of AuNPs, which is in accordance with the UV-vis spectra and confirms that the SPR of AuNPs occurs at around 540 nm. It is noteworthy that the peak at around 580 nm can be attributed to the SPR of the aggregation of AuNPs.66 The intensities of the SPR peaks of AuNPs@CNDs are much higher than those of AuNPs, suggesting the enhancement effect of AuNPs@CNDs. A new resonant mode located at 615 nm appeared after AuNPs are covered by CNDs, and this is probably because of coupling between AuNPs and CNDs.55
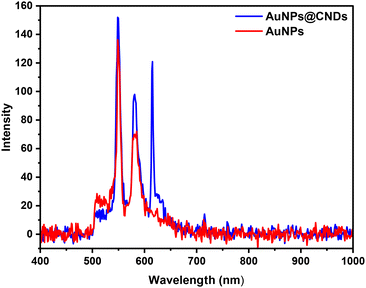 | ||
| Fig. 3 The extracted reflection spectra obtained from the CytoViva hyperspectral imaging measurements. | ||
3.2. SERS properties of NPs
Rh6G was used as a Raman reporter to perform the SERS evaluation using three NPs, i.e., AuNPs@CNDs, CNDs, and AuNPs, prior to the application in herbicide detection. Fig. 4A displays the SERS spectra of 48 × 10−8 M Rh6G after mixing with AuNPs@CNDs, AuNPs, CNDs, respectively, and Rh6G only for comparison. An obviously increased SERS signal was observed from the AuNPs@CND hybrid substrate. The great signal amplification (>10×) of the main signature peaks of Rh6G obtained using AuNPs@CNDs was obtained compared to that obtained using the AuNPs for Rh6G. The band at 614 cm−1 was assigned to the C–C–C ring in-plane in the xanthene ring, whereas those around 771, 1127, and 1183 cm−1 were attributed to the C–H ring in the xanthene or phenyl ring.67 The prominent peaks in the range of 1311, 1362, 1507, 1573, and 1650 cm−1 were due to the symmetric modes of in-plane C–C stretching vibrations in the xanthene or phenyl ring, which are the characteristic Raman scattering features of Rh6G.20,68 These results confirm that the hybrid AuNPs@CND system can significantly improve the SERS performance in terms of signal enhancement, implying enhanced sensitivity and accuracy for sensing applications.Next, we collected the SERS responses of Rh6G at different concentrations ranging from 48 × 10−12 to 48 × 10−8 M using the same AuNPs@CND substrate. Fig. 4B shows that there is an increase in the Raman signal intensity of the characteristic Rh6G peaks upon increasing the concentration of Rh6G. By plotting log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I1650 (logarithm of SERS peak intensity at 1650 cm−1) versus log
I1650 (logarithm of SERS peak intensity at 1650 cm−1) versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (logarithm of Rh6G concentration), a linear calibration curve was obtained, where the square coefficient of determination (R2) is 0.991, demonstrating a linear relationship between the Raman intensity and the concentration.
C (logarithm of Rh6G concentration), a linear calibration curve was obtained, where the square coefficient of determination (R2) is 0.991, demonstrating a linear relationship between the Raman intensity and the concentration.
The reproducibility and stability of the SERS substrate for sensing are important for the detection of analytes of interest, especially when deployed in the field.9 The SERS responses collected at 18 random points on the same sample using AuNPs@CNDs and 48 × 10−8 M Rh6G display the uniformity of the sample (Fig. 5A) based on a standard deviation of 0.9% from the peak intensity at 1650 cm−1. The uniformity in SERS signals recorded confirms the good reproducibility and homogeneity of using the hybrid AuNPs@CNDs in sample preparation and detection. Furthermore, to confirm the batch-to-batch reproducibility, 6 different batches of AuNPs@CND substrates were used for SERS measurement of Rh6G and the SERS intensity at the 1650 cm−1 peak was recorded at 3 random points of each substrate. Fig. 5B demonstrates the averaged Raman intensity for the six samples, showing excellent reproducibility. The UV-visible spectra of AuNPs@CNDs and SERS spectra of 48 × 10−8 M Rh6G with AuNPs@CNDs in Fig. 5C and D show their stability during storage for up to 60 days, respectively. There are no significant changes in both spectra, which confirms the high stability of the hybrid AuNPs@CND substrate.
3.3. SERS detection of herbicide molecules
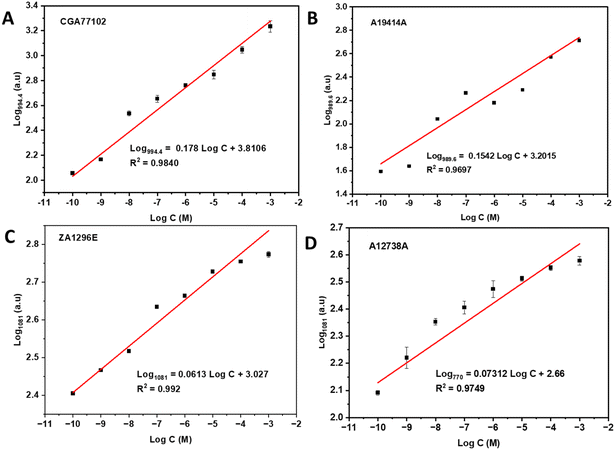
The 2.4-ratio AuNPs@CND substrate in solution was further applied for the detection of two different groups of herbicides. As described in the Experimental section, the first group of herbicides includes CGA77102 (S-metolachlor, 86.6 wt%) and A19414A formulation (S-metolachlor, 27.1 wt%). S-metolachlor is a member of the chloroacetanilide family of herbicides, which is used for grass and broadleaf controls in corn, soybean, etc. The signal amplification of the main Raman peaks at 994.4 and 989.6 cm−1 obtained using AuNPs@CNDs compared to that of bare AuNPs is calculated to be 10.8 and 7.3-fold for CGA77102 and A19414A, respectively (Fig. S5a and b†).The second group of herbicides includes ZA1296E (mesotrione, 60 wt%) and A12738A formulation (mesotrione, 40 wt%) with the main effective component of mesotrione. Mesotrione has an aromatic ketone and is a toxic synthetic herbicide in the agriculture industry for the selective contact and residual control of broadleaf weeds in field corn. According to Fig. S5c and d,† the SERS peaks at 1526, 1552, and 1608 cm−1 are attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching in carotenoids, the central 16-membered-ring vibrations and the C–C stretching of the pyrrole ring in chlorophyll, and the ν phenyl ring in phenolic compounds, respectively.69 Similarly, the hybrid AuNPs@CNDs provide greater Raman signal enhancement than the AuNPs.
C stretching in carotenoids, the central 16-membered-ring vibrations and the C–C stretching of the pyrrole ring in chlorophyll, and the ν phenyl ring in phenolic compounds, respectively.69 Similarly, the hybrid AuNPs@CNDs provide greater Raman signal enhancement than the AuNPs.
Fig. 6 shows the SERS spectra of the four herbicide samples using the hybrid AuNPs@CNDs as Raman enhancement probes. The spectra of CGA77102 and A19414A with different concentrations indicate a decline in Raman signal intensity along with decreasing concentration. Similarly, the SERS spectra of ZA1296E and A12738A at different concentrations show a decline in Raman intensity with decreasing concentration. Linear calibration curves as a function of the herbicide concentration ranging from 10−10 to 10−3 M are achieved by plotting log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I994.4 and log
I994.4 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I989.6versus log
I989.6versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C for CGA77102 and A19414A (Fig. 7A and B), which demonstrate a linear detection response with R2 values of 0.984 and 0.970, respectively. The lowest detection limit (LOD) is estimated to be 0.1 nM using a signal/noise ratio of 3.32 A small shift of the characteristic peak position between CGA77102 and A19414A (994.4 vs. 989.6 cm−1) could be attributed to the different components in the original samples or interaction between the substrates and analytes. The logarithm of the SERS intensity of the 1081 cm−1 peak versus the logarithm of the concentration of ZA1296E and A12738A in the range of 10−10–10−3 M was plotted with good linearity with R2 = 0.992 and 0.9749, respectively, as shown in Fig. 7C and D.
C for CGA77102 and A19414A (Fig. 7A and B), which demonstrate a linear detection response with R2 values of 0.984 and 0.970, respectively. The lowest detection limit (LOD) is estimated to be 0.1 nM using a signal/noise ratio of 3.32 A small shift of the characteristic peak position between CGA77102 and A19414A (994.4 vs. 989.6 cm−1) could be attributed to the different components in the original samples or interaction between the substrates and analytes. The logarithm of the SERS intensity of the 1081 cm−1 peak versus the logarithm of the concentration of ZA1296E and A12738A in the range of 10−10–10−3 M was plotted with good linearity with R2 = 0.992 and 0.9749, respectively, as shown in Fig. 7C and D.
3.4 Detection of herbicides in tap water and recovery experiments
To test the capability of using the hybrid AuNPs@CNDs as a Raman probe for herbicide detection, we spiked tap water with herbicide samples and obtained the SERS spectra to evaluate the recovery rate, i.e., the percentage of the measured concentration relative to the spiked concentration. In these experiments, three distinct concentration levels at low (10−10 M), medium (10−6 M), and high (10−3 M) concentrations were prepared, and the SERS spectra were obtained (Fig. S6†). Based on the Raman signal intensity at the marker peak of each sample, we obtained the measured concentrations according to the calibration curves shown in Fig. 7. The recovery rate of known amounts of spiked herbicides in tap water was calculated with respect to the spiked concentration and is reported in Table 1. The results indicate that the recovery rate at medium and high levels is >90%, higher than the lower level in the range of 87–88%. These results demonstrate the effectiveness of using AuNPs@CNDs for the detection of herbicides in tap water.| Herbicide | Spiked herbicide (M) | Measured (M, averaged) | Recovery rate (%) |
|---|---|---|---|
| CGA77102 | 10−3 | 0.95 × 10−3 | 95 ± 2 |
| 10−6 | 9.66 × 10−7 | 96 ± 1.8 | |
| 10−10 | 6.73 × 10−11 | 87 ± 1.5 | |
| A19414A | 10−3 | 0.95 × 10−3 | 95 ± 2.1 |
| 10−6 | 9.82 × 10−7 | 98 ± 1.2 | |
| 10−10 | 8.71 × 10−11 | 87 ± 1.9 | |
| ZA1296E | 10−3 | 0.87 × 10−3 | 87 ± 4.8 |
| 10−6 | 9.91 × 10−7 | 89 ± 3.0 | |
| 10−10 | 7.69 × 10−11 | 87 ± 4.0 | |
| A12738A | 10−3 | 0.88 × 10−3 | 88 ± 1.0 |
| 10−6 | 9.61 × 10−7 | 96 ± 1.1 | |
| 10−10 | 8.71 × 10−11 | 87 ± 3.5 |
3.5 A comparison of NP-based SERS detection of herbicides
To the best of our knowledge, SERS detection of herbicides mesotrione and S-metolachlor has been rarely reported in the literature. AuNPs or AgNP-based SERS substrates were used for the detection of a few other herbicides. Table 2 summarizes the representative work on herbicide detection, including the SERS substrates used, synthesis methods, detection ranges and sensitivities (LOD), for comparison with this work. This work, to the best of our knowledge, for the first time uses hybrid AuNPs@CNDs as SERS substrates for the detection of two herbicides and demonstrates superior sensitivity and a broad dynamic range of concentrations.| Herbicides | SERS substrate | Synthesis method | Detection range | LOD (sensitivity) | Ref. |
|---|---|---|---|---|---|
| Paraquat | AgNPs@Si | In situ growth | 1–1000 ppm | 1 ppm | 43 |
| Atrazine | CNDs@AgNPs | Hydrothermal | 10–1000 nM | 10 nM (2 ppm) | 44 |
| Atrazine | Au nanorod array | Seed growth | 0.01–10 mM | 0.1 mM | 26 |
| Paraquat | AuNPs-3D structure | Laser engraving | 2.7 ppb–27 ppm | 2.7 ppb | 45 |
| Flumetsulam | AuNPs-MgSO4 | HAuCl4 reduction | 0.005–1 ppm | 10 ppb | 46 |
| Glyphosate | Si-AgNPs | 0.1–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM 000 nM |
0.01 nM | 47 | |
| Diquat | Au nanobipyramids (AuBPs) MoS2@tinfoil box | Layer-by-layer | 1 pM–0.1 mM | 1 pM (in urine) | 48 |
| Paraquat | Au nanostars | Seed growth | 0.2–50 ppm | 0.2 ppm | 49 |
| Prometryn | AgNPs | Colloidal synthesis | 4.0–800 μM | 28 ppb (pH 11) | 50 |
| 8.0–800 μM | 128 ppb (pH 7) | ||||
| Atrazine | AgNPs@electrode | Colloidal synthesis | 1 nm–1 mM | — | 51 |
| Paraquat | AuNPs-aptamer | Deposition on chip | 0.25–2.5 μM | 0.1 μM | 52 |
| Mesotrione | AuNPs@CNDs | Microwave-assisted | 0.1 nM–1 mM | 0.1 nM | This work |
| S-metolachlor | 0.1 nM–1 mM | 0.1 nM |
4. Conclusion
A hybrid AuNPs@CND SERS substrate was synthesized through a simple approach using CNDs as reducing and stabilizing agents. SERS substrates with different nominal mass ratios of CNDs to HAuCl4 were evaluated using Rh6G and it turned out that a ratio of 2.4 provides the highest Raman enhancement response. The enhancement is believed to arise from electronic coupling at the interface of gold and carbon structures with a specific size and composition for an optimized localized electromagnetic field in SERS. The hybrid AuNPs@CND substrates produce a significant enhanced Raman signal with excellent response and almost an order of magnitude signal amplification in Rh6G measurements when compared to AuNPs or CNDs alone used as the SERS substrate. The SERS measurements of four different herbicide samples containing an active component, mesotrione or S-metolachlor, respectively, coupled to the hybrid subststrate illustrate excellent repeatability, and reproducibility. Additionally, the long term stability demonstrated in this study for sensitive determination of the herbicides spiked in tap water promises for use in remote environmental areas.Author contributions
Conceptualization, Jianjun Wei; data curation, Naghmeh Aboualigaledari, Panesun Tukur, Mengxin Liu and Frank Tukur; funding acquisition, Simona Murph and Jianjun Wei; investigation, Naghmeh Aboualigaledari, Anitha Jayapalan, Panesun Tukur, Mengxin Liu and Frank Tukur; methodology, Naghmeh Aboualigaledari, Anitha Jayapalan, Yanling Zhang and Jianjun Wei; project administration, Jianjun Wei; resources, Gerald Ducatte, Madan Verma, Janet Tarus and Simona Murph; supervision, Simona Murph and Jianjun Wei; writing – original draft, Naghmeh Aboualigaledari; and writing – review & editing, Jianjun Wei.Data availability
The datasets supporting this article have been uploaded as part of the ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Department of Energy Minority Serving Institution Partnership Program (MSIPP) managed by the Savannah River National Laboratory under BSRA contract TOA # 0961 (G-SOW-A-02375) and by a Syngenta Inc. (Greensboro, NC, USA) grant via a Cooperative Agreement (PO# 1400105841).This work was performed at the JSNN, a member of the Southeastern Nanotechnology Infrastructure Corridor (SENIC) and the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (ECCS-1542174).
References
- V. M. Pathak, V. K. Verma, B. S. Rawat, B. Kaur, N. Babu, A. Sharma, S. Dewali, M. Yadav, R. Kumari, S. Singh, A. Mohapatra, V. Pandey, N. Rana and J. M. Cunill, Front. Microbiol., 2022, 13, 962619 CrossRef PubMed.
- J. Kvalvåg, Analyst, 1974, 99, 666–669 RSC.
- E.-C. Zhao, W.-L. Shan, S.-R. Jiang, Y. Liu and Z.-Q. Zhou, Microchem. J., 2006, 83, 105–110 CrossRef CAS.
- Y. Wang, L. Shen, Z. Gong, J. Pan, X. Zheng and J. Xue, Water Environ. Res., 2019, 91, 1009–1024 CrossRef CAS PubMed.
- W. L. Budde, Mass Spectrom. Rev., 2004, 23, 1–24 CrossRef CAS PubMed.
- S. G. Han and T. G. Nam, Appl. Biol. Chem., 2024, 67, 12 CrossRef CAS.
- H. Du, Y. Xie and J. Wang, TrAC, Trends Anal. Chem., 2021, 135, 116178 CrossRef CAS.
- R. D. Ayivi, S. O. Obare and J. Wei, TrAC, Trends Anal. Chem., 2023, 167, 117231 CrossRef CAS.
- R. D. Ayivi, B. O. Adesanmi, E. S. McLamore, J. Wei and S. O. Obare, Chemosensors, 2023, 11, 203 CrossRef CAS.
- A. I. Pérez-Jiménez, D. Lyu, Z. Lu, G. Liu and B. Ren, Chem. Sci., 2020, 11, 4563–4577 RSC.
- Á. I. López-Lorente, Anal. Chim. Acta, 2021, 1168, 338474 CrossRef PubMed.
- Z. Zeng, Y. Liu and J. Wei, TrAC, Trends Anal. Chem., 2016, 75, 162–173 CrossRef CAS.
- S. H. Murph, G. K. Larsen and K. J. Coopersmith, Anisotropic and Shape-Selective Nanomaterials: Structure-Property Relationships, SpringerCham, 1st edn, 2017 Search PubMed.
- Shinki and S. Sarkar, in Encyclopedia of Materials: Electronics, ed. A. S. M. A. Haseeb, Academic Press, Oxford, 2023, pp. 442–458, DOI:10.1016/B978-0-12-819728-8.00051-6.
- W. Wu and M. Pauly, Mater. Adv., 2022, 3, 186–215 RSC.
- Y. Guo, Z. Xu, A. G. Curto, Y.-J. Zeng and D. Van Thourhout, Prog. Mater. Sci., 2023, 138, 101158 CrossRef CAS.
- H. Yu, W. Wu and Z.-Y. Li, in Encyclopedia of Nanomaterials, ed. Y. Yin, Y. Lu and Y. Xia, Elsevier, Oxford, 1st edn, 2023, pp. 496–510, DOI:10.1016/B978-0-12-822425-0.00045-2.
- K. Bhardwaj and A. Jaiswal, Analyst, 2023, 148, 562–572 RSC.
- N. V. Doroshina, O. A. Streletskiy, I. A. Zavidovskiy, M. K. Tatmyshevskiy, G. I. Tselikov, O. O. Kapitanova, A. V. Syuy, R. Romanov, P. Mishra, V. Bobrovs, A. M. Markeev, D. I. Yakubovsky, I. A. Veselova, A. V. Arsenin, V. S. Volkov and S. M. Novikov, Heliyon, 2024, 10, e27538 CrossRef CAS PubMed.
- K. Sridhar, B. S. Inbaraj and B.-H. Chen, Chemosphere, 2022, 301, 134702 CrossRef CAS PubMed.
- F. D. Cortes Vega, P. G. Martinez Torres, J. Pichardo-Molina, N. M. Gomez Ortiz, V. G. Hadjiev, J. Zarate Medina and F. C. Robles Hernandez, J. Mater. Chem. C, 2017, 5, 4959–4966 RSC.
- J. Son, G.-H. Kim, Y. Lee, C. Lee, S. Cha and J.-M. Nam, J. Am. Chem. Soc., 2022, 144, 22337–22351 CrossRef CAS PubMed.
- H. Chen, Z. Cheng, X. Zhou, R. Wang and F. Yu, Anal. Chem., 2022, 94, 143–164 CrossRef CAS PubMed.
- D. García-Lojo, S. Núñez-Sánchez, S. Gómez-Graña, M. Grzelczak, I. Pastoriza-Santos, J. Pérez-Juste and L. M. Liz-Marzán, Acc. Chem. Res., 2019, 52, 1855–1864 CrossRef PubMed.
- J. Langer and D. Jimenez de Aberasturi, et al. , ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- N. Albarghouthi, P. MacMillan and C. L. Brosseau, Analyst, 2021, 146, 2037–2047 RSC.
- J.-F. Li, Y.-J. Zhang, S.-Y. Ding, R. Panneerselvam and Z.-Q. Tian, Chem. Rev., 2017, 117, 5002–5069 CrossRef CAS PubMed.
- G. K. Larsen, W. Farr and S. E. H. Murph, J. Phys. Chem. C, 2016, 120, 15162–15172 CrossRef CAS.
- E. G. d. L. Oliveira, H. P. de Oliveira and A. S. L. Gomes, SN Appl. Sci., 2020, 2, 1491 CrossRef CAS.
- T. Wang, S. Wang, Z. Cheng, J. Wei, L. Yang, Z. Zhong, H. Hu, Y. Wang, B. Zhou and P. Li, Chem. Eng. J., 2021, 424, 130323 CrossRef CAS.
- J. Liu, R. Li and B. Yang, ACS Cent. Sci., 2020, 6, 2179–2195 CrossRef CAS PubMed.
- D. M. Arvapalli, A. T. Sheardy, K. C. Alapati and J. Wei, Talanta, 2020, 209, 120538 CrossRef CAS PubMed.
- Z. Ji, A. Sheardy, Z. Zeng, W. Zhang, H. Chevva, K. Allado, Z. Yin and J. Wei, Molecules, 2019, 24, 152 CrossRef PubMed.
- Z. Ji, Z. Yin, Z. Jia and J. Wei, Langmuir, 2020, 36, 8632–8640 CrossRef CAS PubMed.
- H. Safardoust-Hojaghan, O. Amiri, M. Hassanpour, M. Panahi-Kalamuei, H. Moayedi and M. Salavati-Niasari, Food Chem., 2019, 295, 530–536 CrossRef CAS PubMed.
- S. Khan and A. K. Narula, J. Mater. Sci., 2023, 34, 1955 CAS.
- N. Far'ain Md Noor, M. A. Saiful Badri, M. M. Salleh and A. A. Umar, Opt. Mater., 2018, 83, 306–314 CrossRef.
- M. Azami, J. Wei, M. Valizadehderakhshan, A. Jayapalan, O. O. Ayodele and K. Nowlin, J. Phys. Chem. C, 2023, 127, 7360–7370 CrossRef CAS PubMed.
- R. Das, S. Parveen, A. Bora and P. K. Giri, Carbon, 2020, 160, 273–286 CrossRef CAS.
- L. Wang, C. Li, Y. Luo and Z. Jiang, Materials, 2018, 11, 1655 CrossRef PubMed.
- Z. Zeng, W. Zhang, D. M. Arvapalli, B. Bloom, A. Sheardy, T. Mabe, Y. Liu, Z. Ji, H. Chevva, D. H. Waldeck and J. Wei, Phys. Chem. Chem. Phys., 2017, 19, 20101–20109 RSC.
- M. Berge, A. Dowek, P. Prognon, F.-X. Legrand, A. Tfayli, L. Minh Mai Lê and E. Caudron, Spectrochim. Acta, Part A, 2022, 268, 120628 CrossRef CAS PubMed.
- T. C. Dao, T. Q. N. Luong, T. A. Cao, N. H. Nguyen, N. M. Kieu, T. T. Luong and V. V. Le, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6, 035012 Search PubMed.
- J. Tang, W. Chen and H. Ju, Talanta, 2019, 201, 46–51 CrossRef CAS PubMed.
- R. Botta, P. Eiamchai, M. Horprathum, S. Limwichean, C. Chananonnawathorn, V. Patthanasettakul, R. Maezono, A. Jomphoak and N. Nuntawong, Sens. Actuators, B, 2020, 304, 127327 CrossRef CAS.
- M. Han, H. Lu and Z. Zhang, Molecules, 2020, 25, 4662 CrossRef CAS PubMed.
- K. A. López-Castaños, L. A. Ortiz-Frade, E. Méndez, E. Quiroga-González, M. A. González-Fuentes and A. Méndez-Albores, Front. Chem., 2020, 8, 612076 CrossRef PubMed.
- Q.-Y. Jiang, D. Li, Y. Liu, Z.-S. Mao, Y. Yu, P. Zhu, Q. Xu, Y. Sun, L. Hu, J. Wang, J. Chen, F. Chen and Y. Cao, Sens. Actuators, B, 2021, 344, 130290 CrossRef CAS.
- M.-H. Lin, L. Sun, F. Kong and M. Lin, Food Control, 2021, 130, 108280 CrossRef CAS.
- R. J. G. Rubira, L. N. Furini, C. J. L. Constantino and S. Sanchez-Cortes, Vib. Spectrosc., 2021, 114, 103245 CrossRef CAS.
- N. Albarghouthi, M. M. Eisnor, C. C. Pye and C. L. Brosseau, J. Phys. Chem. C, 2022, 126, 9836–9842 CrossRef CAS.
- N. Kamkrua, T. Ngernsutivorakul, S. Limwichean, P. Eiamchai, C. Chananonnawathorn, V. Pattanasetthakul, R. Ricco, K. Choowongkomon, M. Horprathum, N. Nuntawong, T. Bora and R. Botta, ACS Appl. Nano Mater., 2023, 6, 1072–1082 CrossRef CAS.
- P. Luo, C. Li and G. Shi, Phys. Chem. Chem. Phys., 2012, 14, 7360–7366 RSC.
- G. Frens, Nat. Phys. Sci., 1973, 241, 20–22 CrossRef CAS.
- X. Miao, S. Wen, Y. Su, J. Fu, X. Luo, P. Wu, C. Cai, R. Jelinek, L.-P. Jiang and J.-J. Zhu, Anal. Chem., 2019, 91, 7295–7303 CrossRef CAS PubMed.
- H. Al-Johani, E. Abou-Hamad, A. Jedidi, C. M. Widdifield, J. Viger-Gravel, S. S. Sangaru, D. Gajan, D. H. Anjum, S. Ould-Chikh, M. N. Hedhili, A. Gurinov, M. J. Kelly, M. El Eter, L. Cavallo, L. Emsley and J.-M. Basset, Nat. Chem., 2017, 9, 890–895 CrossRef CAS PubMed.
- Y. Lyu, L. M. Becerril, M. Vanzan, S. Corni, M. Cattelan, G. Granozzi, M. Frasconi, P. Rajak, P. Banerjee, R. Ciancio, F. Mancin and P. Scrimin, Adv. Mater., 2024, 36, 2211624 CrossRef CAS PubMed.
- B. Bagra, W. Zhang, Z. Zeng, T. Mabe and J. Wei, Langmuir, 2019, 35, 8903–8909 CrossRef CAS PubMed.
- B. D. Mansuriya and Z. Altintas, Nanomaterials, 2021, 11, 2525 CrossRef CAS PubMed.
- N. Naskar, M. Wagner, H. J. Räder, H. Qi, U. Kaiser, T. Weil and S. Chakrabortty, Adv. Photonics Res., 2022, 3, 2100092 CrossRef CAS.
- R. Liu, H. Huang, H. Li, Y. Liu, J. Zhong, Y. Li, S. Zhang and Z. Kang, ACS Catal., 2014, 4, 328–336 CrossRef CAS.
- X. Fei, Z. Liu, Y. Li, G. Yang, C. Su, H. Zhong, Z. Zhuang and Z. Guo, J. Alloys Compd., 2017, 725, 1084–1090 CrossRef CAS.
- H. Qiu, M. Wang, M. Cao, L. Zhang, S. Ji, J. Dou, Y. Ji, S. Kou, J. Guo and Z. Yang, Appl. Surf. Sci., 2019, 489, 1010–1018 CrossRef CAS.
- M. Ayiania, M. Smith, A. J. R. Hensley, L. Scudiero, J.-S. McEwen and M. Garcia-Perez, Carbon, 2020, 162, 528–544 CrossRef CAS.
- Z. Zeng, T. Mabe, W. Zhang, B. Bagra, Z. Ji, Z. Yin, K. Allado and J. Wei, ACS Appl. Bio Mater., 2018, 1, 802–807 CrossRef CAS PubMed.
- J. Waitkus, Y. Chang, L. Liu, S. V. Puttaswamy, T. Chung, A. M. Molina Vargas, S. J. Dollery, M. R. O'Connell, H. Cai, G. J. Tobin, N. Bhalla and K. Du, Adv. Mater. Interfaces, 2023, 10, 2201261 CrossRef CAS PubMed.
- X. N. He, Y. Gao, M. Mahjouri-Samani, P. N. Black, J. Allen, M. Mitchell, W. Xiong, Y. S. Zhou, L. Jiang and Y. F. Lu, Nanotechnology, 2012, 23, 205702 CrossRef CAS PubMed.
- C. Kavitha, K. Bramhaiah, N. S. John and B. E. Ramachandran, Chem. Phys. Lett., 2015, 629, 81–86 CrossRef CAS.
- P. Vítek, K. Novotná, P. Hodaňová, B. Rapantová and K. Klem, Spectrochim. Acta, Part A, 2017, 170, 234–241 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: UV-vis and SERS spectra for synthesis optimization, additional TEM, XPS spectra, SERS spectra, and calibration curves. See DOI: https://doi.org/10.1039/d4an00649f |
| This journal is © The Royal Society of Chemistry 2024 |