Open Access Article
Open Access ArticleA microfluidic approach to study variations in Chlamydomonas reinhardtii alkaline phosphatase activity in response to phosphate availability†
Alireza
Rahnama‡
a,
Manibarathi
Vaithiyanathan‡
a,
Luis
Briceno-Mena
a,
Travis M.
Dugas
a,
Kelly L.
Yates
a,
Jose A.
Romagnoli
a and
Adam T.
Melvin
 *ab
*ab
aCain Department of Chemical Engineering, Louisiana State University, Baton Rouge, Louisiana 70803, USA. E-mail: melvina@clemson.edu
bDepartment of Chemical and Biomolecular Engineering, Clemson University, Clemson, South Carolina 29634, USA
First published on 12th June 2024
Abstract
Algal growth depends strongly on phosphorus (P) as a key nutrient, underscoring the significance of monitoring P levels. Algal species display a sensitive response to fluctuations in P availability, notably through the expression of alkaline phosphatase (AP) when challenged with P-depletion. As such, alkaline phosphatase activity (APA) serves as a valuable metric for P availability, offering insights into how algae utilize and fix available P resources. However, current APA quantification methods lack single cell resolution, while also being time- and reagent consuming. Microfluidics offers a promising cost-effective solution to these limitations, providing a platform for precise single-cell analysis. In this study, a trap-based microfluidic device was integrated with a commercially available AP live stain to study the single cell APA response of a model algae strain, Chlamydomonas reinhardtii, when exposed to different exogenous P levels. A three-step culture-starve-spike process was used to induce APA in cells cultured under two different basal P levels (1 and 21 mM). When challenged with different spiked P levels (ranging from 0.1–41 mM), C. reinhardtii cells demonstrated a highly heterogeneous APA response. Two-way ANOVA confirmed that this response is influenced by both spiked and basal P levels. Utilizing an unsupervised machine learning approach (HDBSCAN), distinct subpopulations of C. reinhardtii cells were identified exhibiting varying levels of APA at the single-cell level. These subpopulations encompass significant groups of individual cells with either notably high or low APA, contributing to the overall behavior of the cohorts. Considerable intrapopulation differences in APA were observed across cohorts with similar average behavior. For instance, while some cohorts exhibited a concentrated distribution around the overall average APA, others displayed subpopulations dispersed across a wider range of APA levels. This underscores the potential bias introduced by analyzing a small number of cells in bulk, which may skew results by overrepresenting extreme behavioral subpopulations. The findings if this study highlight the need for analytical approaches that account for single cell heterogeneity in APA and demonstrate the utility of microfluidics as a well-suited means for such investigations. This study illuminates the complexities of APA regulation at the single cell level, providing crucial insights that advance our understanding of algal phosphorus metabolism and environmental responses.
Introduction
Phosphorous (P) is one of the primary essential nutrients that support the growth of thousands of different algae species in both fresh and salt bodies of water. Compared to other nutrients required for algal growth, P has been shown to be a limiting element in most marine ecosystems.1,2 As such, close monitoring and documentation of P availability is essential when tracking the growth of algal species as it can be directly correlated with abnormal behavior including enhanced or diminished growth rates.3,4 One such example is eutrophication, or the excessive growth of algae resulting from an abrupt increase in one or more of the limiting nutrients, which has been linked to the abnormal changes in growth of microorganisms dating back to the late 1960's.5,6 Eutrophication can lead to the proliferation of algal blooms, a phenomenon observed in coastal regions worldwide. The extent of these blooms varies based on nutrient levels, temperature fluctuations, tidal patterns, and allelochemical signaling interactions between different algal species.7–9 In some cases, the microorganisms increase so abundantly that their pigments discolor inside the water, which is commonly referred to as “red-tide”.10 Such algal blooms can lead to harmful consequences (referred to as Harmful Algal Blooms) such as biotic turbidity, increased bacterial decomposition that promote hypoxia or anoxia events, or the production of toxic secondary metabolites such as hepatotoxin, microcystin, and allelopathic chemical that could in turn pose a threat to both marine and human lives.11–13 HABs in fresh and marine bodies of water can impact human health through the consumption of contaminated seafood or through inhalation of air- and waterborne toxins.14 Although there are studies that have shown no uniform trend in the frequency and the distribution of HABs, neither globally nor specifically in the U.S., it is established that the problem is expanding into new regions.15,16 The highlighted significance of algal blooms (including but not limited to the harmful incidents), calls for a better understanding of the phenomenon and how different factors can contribute to it. Therefore, it is essential to investigate how changes in nutrient levels can affect algal growth. Specifically, phosphorus (P) is recognized as a key contributor to eutrophication.17,18P is available in water in either particulate (PP) or dissolved (DP) form, out of which dissolved inorganic P (DIP) is the most available from and the only form which can be directly consumed by algal species.19 Several phytoplankton species have evolved the ability to adapt through a variety of metabolic process to ensure survival under oligotrophic conditions where there is limited, or insufficient, DIP sources.20 Most significantly, algal species require alkaline phosphatase (AP) for hydrolysis of dissolved organic phosphorus (DOP) to DIP for their consumption. AP is an inducible enzyme that is expressed on the exterior of the cellular membrane when needed to catalyze hydrolysis of DOP.21 As such, a negative correlation exists between AP activity (APA) and DIP-deficiency, where higher APA is indicative of sparsity of DIP availability for algal consumption and subsequent DOP-utilization.22,23 Understating this correlation is essential for assessments involving dynamics of nutrient availability and can be leveraged for improving aquatic health. Several studies have suggested that APA is highly variable with organism-dependent kinetics that are sensitive to P availability which has suggested that APA has potential as a metric in measuring P availability especially in the study of marine species.24,25 When it comes to marine species, the most common approach in investigating APA is the bulk measurement of APA using soluble substrates or enzyme-labeled fluorescence (ELF).26,27 Traditionally, this requires the implementation of multiple incubation, washing, and centrifugation steps to isolate the cells and add the reagents which oftentimes leads to substantially high sample loss and a relatively low number of cells to study (∼20–50). Recently, others have aimed to improve upon these methods by developing new approaches to quantify APA. Xie et al. investigated the effect of different types of P resources on algal growth as well as APA using similar bulk approach.28 Recently, Lv et al. developed a ratiometric fluorescent approach using nitrogen-doped carbon quantum dots in a purified enzyme assay providing an alternative means to quantifying APA.29 A significant limitation of this bulk-view approach lies in their tendency to quantify APA across an entire population, thereby reporting an average value that overlooks individual cell behavior. However, delving into single cell responsiveness offers a unique opportunity to uncover subpopulations with distinct behaviors, providing deeper insights into cellular dynamics and potential regulatory mechanisms. Examination at the single cell level allows for the identification of rare or outlier cells that may be masked in bulk population analyses. Additionally, understanding the variability in single cell responses can shed light on the underlying factors driving cellular function and the regulatory networks governing complex biological processes. González-Gil et al. conducted one of the pioneering studies utilizing a combination of ELF and flow cytometry. Their research aimed to determine the measurability of APA levels and their source within individual algal cells.30 While flow cytometry can offer significantly high throughput in single cell analysis, it is a method that is heavily reliant on highly sophisticated data collection and analysis systems and can exhibit sensitivity issues depending on the choice of fluorescent stain or protein marker.31 Alternatively, microfluidics presents a cost-effective and accessible alternative to flow cytometry and established bench-scale methods, offering advantages such as reduced reagent costs, ease of use, reproducibility, and compatibility with various fluorescent microscopy techniques. This approach is particularly suitable for applications requiring a lower cell count and seamless mixing of reagents with samples.32–34 In a study recently by Girault et al., the application of combined ELF labeling with a droplet microfluidic system to analyze the dynamic kinetics of APA in Tetraselmis sp. was demonstrated.35 Their work focused primarily on the development and integration of a novel microfluidic device with fluorescence microscopy which they used to show time-dependent changes in APA and single cell variability. While this was an innovative new approach to study APA in algae at the single cell level; the scope of that study did not investigate the role of altered exogenous P levels on APA kinetics. As such, it is still unclear how single algal cells leverage altered APA in response to nutrient variation.
The goal of this study was to investigate how APA in single Chlamydomonas reinhardtii cells is altered due to variations in the P concentrations using a microfluidic approach. C. reinhardtii, is a unicellular flagellate species commonly used in laboratory scale as a model algae cell line. Algal species with characteristics similar to those of C. reinhardtii are significant in nature as they play key roles in the biogeochemistry of oceans by fixing CO2 and being impactful on the deep sea carbon influx.36 Microfluidics allows for minimized assay steps, incubation time, and reagent volumes, facilitating an all-in-one device to investigate population-based heterogeneity by quantifying single cell APA. To gain a better understanding of how APA is altered by C. reinhardtii in response to variations in P availability, a three-step culture-starve-spike process was implemented to induce APA in response to changes in spiked exogeneous P levels in cells cultured under two different basal P levels. Following that, APA was quantified at the single cell level using a commercially available stain. Extensive analysis of single cell data in this study highlights the substantial impact of both the basal P levels to which cells are acclimated and the spiked P level they encounter during abrupt changes in nutrient availability. These findings underscore the significance of single cell investigation in this subject and highlight the wealth of data and potentially impactful interpretations that are often overlooked in bulk analysis.
Materials and methods
Cell culture and reagents
C. reinhardtii (D66 strain, a kind gift from Dr Naohiro Kato, Department of Biological Sciences, Louisiana State University) were cultured in 100 mL of nitrogen tris-acetate-phosphate (NTAP) media at two different P concentrations in a 250 mL flask stoppered with a sponge plug under a 12 h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h (light
12 h (light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dark, 45 W LED grow light) cycle at 25 °C on a continuous rotary shaker (110 rpm). NTA (NTAP excluding the phosphate contents) was prepared as previously described37 without adding the phosphate solution. A stock phosphate solution (3.31 M of K2HPO4, 2.12 M of KH2PO4, 0.40 M Tris, and 0.32 M glacial acetic acid in 50 mL of deionized water) was used to supplement the NTA to achieve either 1 or 21 mM final P concentrations. All the mentioned salts were purchased from Sigma Aldrich (St Louis, MO, USA) and chemicals were purchased from VWR. Throughout the rest of this article, DIP as the primary form of P for algae consumption will be the focus and herein referred to as “P”.
dark, 45 W LED grow light) cycle at 25 °C on a continuous rotary shaker (110 rpm). NTA (NTAP excluding the phosphate contents) was prepared as previously described37 without adding the phosphate solution. A stock phosphate solution (3.31 M of K2HPO4, 2.12 M of KH2PO4, 0.40 M Tris, and 0.32 M glacial acetic acid in 50 mL of deionized water) was used to supplement the NTA to achieve either 1 or 21 mM final P concentrations. All the mentioned salts were purchased from Sigma Aldrich (St Louis, MO, USA) and chemicals were purchased from VWR. Throughout the rest of this article, DIP as the primary form of P for algae consumption will be the focus and herein referred to as “P”.
Microfluidic device design and fabrication
The microfluidic device was fabricated by a combination of soft lithography and polydimethylsiloxane (PDMS) replication. The geometry was designed in AutoCAD (Autodesk, USA) and printed onto an iron oxide/chrome photo mask (Front Range) prior to fabrication. A silicon master was fabricated using a negative photoresist polymer, SU-8 2025 (MicroChem). The SU-8 was deposited on a clean 3′′ silicon wafer and baked at 65 °C for 15 min followed by a second baking step at 95 °C for 30 min. After cooling down, the wafer was exposed to UV light with 1.2 mW cm−2 power intensity for 40 s using an iron oxide/chrome photo mask to create the fluidic channels. Following the UV exposure, the wafer was baked again at 65 °C for 15 min and then at 95 °C for 30 min. The silicon wafer was developed with a SU-8 developer solution (Microchem) to remove the non-crosslinked SU-8 and yield the microfluidic patterns. Following development, the wafer was hard baked once more 150 °C for 30 min to increase wafer durability. PDMS replicas (Sylgard 184, Ellsworth Adhesives) were generated by mixing the base- and curing agents in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio followed by degassing in a vacuum chamber to create a bubble-free mixture. This PDMS was poured on the silicon master and was cured for a minimum of 8 h at 65 °C. Once cured, the PDMS was removed from the wafer and the inlet and outlet ports were punched using a blunted 18-gauge needle. The PDMS replicas were permanently bonded to 25 × 75 mm glass slides (Corning) using an O2 Harrick Plasma PDC-32G basic plasma cleaner through a 30 s exposure to plasma. The devices were left overnight to ensure proper bonding between the PDMS and the glass before being used for experiment.
1 ratio followed by degassing in a vacuum chamber to create a bubble-free mixture. This PDMS was poured on the silicon master and was cured for a minimum of 8 h at 65 °C. Once cured, the PDMS was removed from the wafer and the inlet and outlet ports were punched using a blunted 18-gauge needle. The PDMS replicas were permanently bonded to 25 × 75 mm glass slides (Corning) using an O2 Harrick Plasma PDC-32G basic plasma cleaner through a 30 s exposure to plasma. The devices were left overnight to ensure proper bonding between the PDMS and the glass before being used for experiment.
Single cell analysis of APA via the microfluidic device
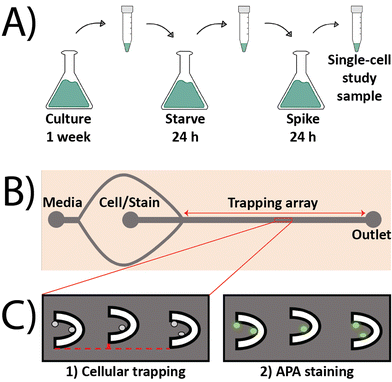
To induce the expression of AP in algae cells and investigate the impact of change in P-levels on APA, a three-step process of culture, starve, and spike was followed. Cells were first cultured in NTAP with 1 mM P for a week to sensitize them to this baseline P concentration. Following the week-long culture, 10 mL from the cell culture suspension was collected and centrifuged at 4000 rcf for 3.5 min at room temperature. Following the removal of the supernatant, the resulting cell pellet was re-suspended in 10 mL of NTA media (0 mM P content) and left for 24 h to starve the cells of P. After this starvation period, cells were once again isolated using the same centrifuge step as described above, but this time the pellet was re-suspended in NTA supplemented with increasing P concentrations of 0.1, 0.5, 1, 11, 21, 31, and 41 mM and incubated for another 24 h, making up the spike period. The selection of these spike concentrations was to investigate the impact of sudden decrease (0.1 and 0.5 mM) or increase (11, 21, 31, and 41 mM) in P-levels on APA. Following the 24 h spike period, cells were isolated as described above and re-suspended in 1 mL of the corresponding spiked NTAP. Prior to loading the algae cells inside the microfluidic device, both inlet ports on the device (media and cells), each had a syringe filled with 5 mL of spiked NTAP connected to them using Tygon tubing (Cole Palmer). A flow rate of 5 μL min−1 was induced on both inlets using syringe pumps (Harvard apparatus) for approximately 10 min to ensure that the device was filled with fluid and there were no air bubbles trapped in the array. The media flow was then diminished to 2 μL min−1 on the media inlet, and the syringe connected to the cell inlet was replaced with a new 1 mL syringe containing the cell suspension to load the algae into the device at a flowrate of 0.5 μL min−1. Upon successful trapping of the algae cells, the algae syringe at the cell inlet was swapped with another syringe containing 2.5 mL of spiked NTAP supplemented with 5 μL of AP live stain (ThermoFisher Scientific). Flow from the syringe connected to the media inlet was halted and the AP stain solution was infused into the device at 0.5 μL min−1 for 60 minutes in the dark. A DMi8 inverted fluorescent microscope (Leica microsystems) was used to collect images across the entire trapping array using brightfield and FITC (λEx: 440–520 nm and λEm: 497–557 nm) filter sets (Chroma Tech. Corp, Bellows Falls, VT, USA). A Hamamatsu-Flash 4-CL-880338 camera (Hamamatsu Photonics K.K., Japan) was used to collect images using a digitization of 16 bits and quality mode of 209.8 MHz in all experiments. The fluorescence intensities of individual cells were quantified in gray values using thresholding in the Leica Application Suite X (LAS X) software by defining regions of interest (ROIs) around each individual cell. The normalized fluorescence intensities (AU) of single algae cells were calculated using the following equation: | (1) |
Results and discussion
Single cell analysis of APA in intact algal cells by integrating the AP live stain and a trap-based microfluidic device
Most methods to quantify APA rely on performing bulk measurements across a population of cells or in enzyme-only activation/inhibition studies, both of which cannot account for the substantial heterogeneity associated with the single cell response. This can ultimately mask sub-populational responsiveness of algal species which can lead to potential incorrect extrapolation on the behavior of larger populations of algae in an HAB when only the average value of certain metrics is used to represent the sizeable environment. Moreover, most bulk measurement methods are time- and reagent-consuming and oftentimes result in substantial loss of sample due to the numerous wash and centrifugation steps. Conversely, a microfluidic approach offers a time- and cost-effective alternative by minimizing the volume of expensive reagents (e.g., commercially available stains) and allowing for facile and rapid trapping of single algal cells. The commercially available AP live stain was selected to stain algal cells to quantify APA based on its prior success as a stem cell imaging product that allows users to differentially stain pluripotent stem cells (PSCs).38 The AP live stain offers an easy method in quantifying APA; however, it has not been used with algal cells and is mostly reported in studies of mammalian cells. Prior to its inclusion in the microfluidic device, it was important to first verify that the AP live stain could be used to quantify APA in algae. This was accomplished by first testing it in a bulk population of C. reinhardtii cells outside of the microfluidic device. To induce APA, C. reinhardtii cells were cultured in NTAP with 1 mM P for one week followed by a 24 h P starvation period in NTA followed by a 24 h P spike period in NTAP with 1 mM P (Fig. 1A). This protocol was designed to induce APA in the algal cells by depriving them of exogeneous P followed by exposing them to it like a serum starvation step in mammalian cells. After the spike period, the cells were incubated with the AP live stain and then imaged using fluorescence microscopy. As expected, the AP live stain worked in algal cells and was able to generate a green fluorescence signal in cells (Fig. S1C†). It was confirmed that the observed signal was not due to autofluorescence as cells imaged in the absence of the AP live stain did not exhibit any fluorescence signal (Fig. S1D†).The trap-based microfluidic device was fabricated to isolate and trap single algal cells while allowing for the facile incorporation of the AP live stain. The microfluidic approach was designed based on prior devices by first by Tran et al. and then by Landwehr et al. who used a similar geometry to isolate and trap cancer cells.39,40 The trap-based device in this study consisted of a main channel that was 250 μm wide and 8000 μm long, containing a 110-member array of U-shaped traps to isolate the algae. The traps were spaced 300 μm apart and vertically offset by 10 μm in an alternating pattern. The device had two inlets for the media and cells (Fig. 1B). The media inlet splits into two flows that converge past the cell inlet to allow for the formation of a sheath flow surrounding the cells to focus them into the trapping array located in the center line of the main channel. This focusing of the cell flow in a central streamline was demonstrated by Tran et al. and Landwehr et al. to achieve higher trapping efficiency. The media inlet was also useful during assay incorporation as it allowed for the facile incorporation of the AP live stain while avoiding potential bubble nucleation that is associated with the syringe swaps between experimental steps. The device offered an average trapping efficiency of ∼72% attained from >60 experiments where 50% of the traps collected two or more cells and 22% of the traps contained one cell.
Single algal cells exhibit heterogeneous APA when exposed to varying exogeneous P concentrations
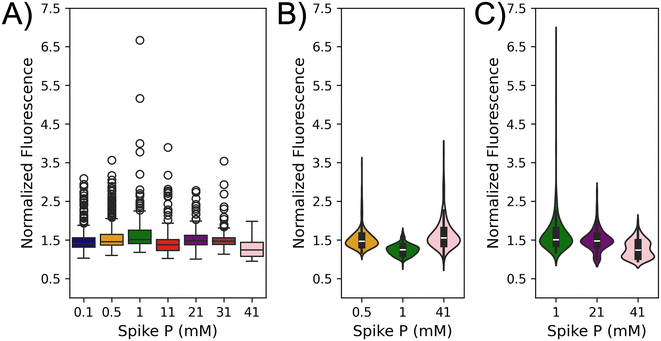
Algal cells in both fresh water and marine environments are commonly exposed to a range of exogeneous P concentrations due to numerous factors. Based on this, it was hypothesized that single algal cells could exhibit a range of APA when challenged with different P concentrations after being acclimated to a 1 mM basal P level. The microfluidic approach was deployed using the P starvation and spike methodology to investigate this (Fig. 1). After the 24 h starvation period, seven cohorts of C. reinhardtii cells were incubated for 24 h in NTAP spiked with increasing P concentrations (0.1, 0.5, 1, 11, 21, 31, or 41 mM). It was found that single algal cells exhibited a high variability at both inter- and intra-cohort levels with regards to measured fluorescence signals. Representative fluorescence images of algae from the 0.5 mM spiked P cohort (Fig. 2A and B) show cells with either enhanced or diminished APA. While this small sample size illustrates significant heterogeneity within a single cohort (brighter vs. dimmer signal), it underscores the importance of analyzing larger populations to obtain a more comprehensive understanding of single cell APA responses and their impact on cohort behavior. To further investigate this difference in behavior, single cell APA was quantified across all seven cohorts of cells exposed to increasing spiked exogeneous P concentrations (Fig. 2C) using a minimum of 183 cells and an average of 260 cells quantified for each P concentration (Table S1†). Statistical analyses including Fisher's test and analysis of variance (ANOVA) were performed across all seven cohorts (Table S2†). Analysis of the seven different cohorts led to interesting observations at both the single cell and population level. The cells that were spiked with the basal P concentration (1 mM) demonstrated a mostly homogenous response as evidenced by the lowest overall mean fluorescence signal and standard deviation among all cohorts (1.245 ± 0.144, green). This suggests that these cells do not exhibit an adaptive response when challenged with the same P levels as what they were grown in. On the other hand, a greater degree of heterogeneity was observed from the individual cells challenged to either diluted (0.1, and 0.5 mM) or enhanced (11, 21, 31, and 41 mM) P concentrations. The highest mean fluorescence signal was observed in the 41 mM cohort (1.646, pink) while the second highest was observed in the 0.5 mM cohort (1.531, orange) suggesting that the magnitude of the spiked P concentration does not directly govern APA as cells exposed to both diluted and enhanced P levels exhibit enhanced APA. Looking at the single cell APA variability among different cohorts, the highest observed heterogeneity was found in the 21 mM cohort with a standard deviation of 0.404, a 1.8-fold higher value compared to the standard deviation from the 1 mM cohort. This highlights the substantial variability in APA responses across different P concentrations. When looking across the subpopulation of cells from the diluted cohorts (0.1 and 0.5 mM), it was found that greater than 75% of the cells from the diluted cohorts demonstrated a higher APA than the third quadrant of the basal cohort. Moreover, it was seen that alterations in APA are dependent on the magnitude of change (increase/decrease) in P levels. Decreasing P from 1 mM to 0.1 mM and 0.5 mM resulted in significant increases in mean APA of these cohorts by 18% and 23%, respectively. Conversely, increasing P to 11, 21, and 31 mM yielded moderate increases in mean APA, with increments of 10%, 14%, and 11%, respectively. A notable deviation occurred at 41 mM P, where a substantial escalation in mean APA was observed (32%). In the pairwise comparison of individual cohorts, a statistically significant difference in means is evident across nearly all cohorts (18/21 combinations), with the exception of three: 0.1 vs. 21 mM, 11 vs. 21 mM, and 21 vs. 31 mM (Table S2†). While there are noticeable variations in the data distribution among these three cohort pairs (Fig. 2C), the statistical similarity in their means may stem from the presence of subpopulations with notably higher or lower APA levels, influencing the overall cohort averages. These findings underscore the sensitivity of APA to changes in exogenous P concentration with the magnitude of change exerting differential effects on enzyme activity. The non-linear relationship between P concentration and APA suggests a complex interplay, potentially indicative of threshold effects at higher phosphorus concentrations. It is interesting to determine if this non-linear relationship is observed across an algal population that has been acclimated to a different basal P level.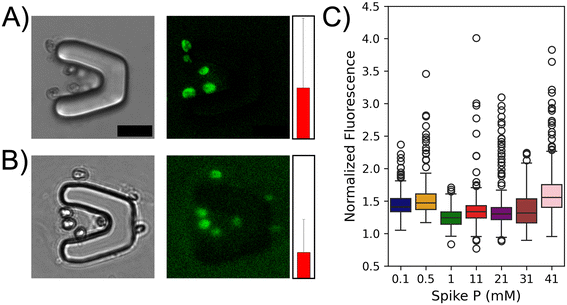 | ||
| Fig. 2 C. reinhardtii cells acclimated to 1 mM P in their culture media, demonstrate variable levels of heterogeneity in their APA when exposed to different spike P concentrations. Representative fluorescence microscopy images (brightfield and FITC for AP live stain) of cells cultured in NTAP spiked with 0.5 mM P show diminished (1.75 ± 0.88, A) or enhanced APA (3.29 ± 0.16, B). The red bar next to each image indicates the mean normalized fluorescence value corresponding to APA from the cells in the respective image. Scale bar is 20 μm. (C) Box plots of normalized fluorescence for all seven cohorts of spike P concentration (mM) acclimated to a basal P of 1 mM in NTAP. Cells from each cohort demonstrate different mean and standard deviation values (Table S1†). Statistical analyses were performed using Origin and the numerical values are presented in Table S2.† | ||
Basal phosphorus levels significantly shape the single cell APA response in C. reinhardtii
The lowest mean fluorescence signal (correlating to APA) and standard deviation were observed in the cohort challenged with the basal P concentration (1 mM), which motivated a second study to explore the impact of basal P concentration on APA in response to spiked concentrations. Basal P concentration is not universally constant in nature with variations reported in different regions and under different circumstances such as HABs.41–44 A similar three-step (culture-starve-spike) approach was employed here as described in Fig. 1A, except that the cells were conditioned to NTAP with a basal P concentration of 21 mM for a week prior to the starvation/spike period. The 21 mM P value was selected because if fell roughly in the middle of the range of spiked concentrations studied in the prior section. Cells acclimated to the 21 mM basal P concentration exhibited the same morphology as those acclimated to the 1 mM basal P concentration (data not shown); however, these cells exhibited a significantly higher growth rate when compared to the 1 mM basal P (Fig. S2†). This can be explained by the fact that higher exogeneous P levels have been linked to enhanced growth rates which has been well documented in the literature.45 APA was quantified across the same seven cohorts of spiked P concentrations (0.1–41 mM) using a minimum of 135 and average of 289 cells per cohort (Table S3†). Statistical analyses including Fisher's test and analysis of variance (ANOVA) were performed across all seven cohorts which showed a statistically significant difference between nearly all the cohorts (17/21 combinations) excluding four: 0.5 vs. 1 mM, 0.1 vs. 21 mM, 0.1 vs. 31 mM, and 21 vs. 31 mM (Table S4†). Similar to the discussion in the previous section, differences in data distribution among these cohort pairs (Fig. 3A) suggest potential subpopulations with notably higher or lower APA levels distributed differently across cohorts. These variations may counterbalance the distribution effect, leading to statistically similar cohort means. Interestingly, the APA of the cells acclimated to 21 mM P levels exhibited vastly different trends compared to the basal 1 mM P population. The cells challenged with 1 mM P yielded the highest mean APA (1.659 ± 0.519) followed by the 0.5 and 0.1 mM P cohorts (Fig. 3A and Table S3†). The 21 and 31 mM cohorts exhibited intermediate APA while the 41 mM cohort exhibited the lowest mean APA. This contrasts with the behavior of cells challenged with 41 mM P in the 1 mM basal P cohort which exhibited the highest mean APA. This supports the findings that basal P levels are also a driving factor for APA. The driving force behind these different population averages could, in part, be described by significant heterogeneity at the single cell level. For example, the cohorts challenged with 1, 0.5, and 0.1 mM P exhibited the highest variances across all seven cohorts whereas the cells challenged with 41 mM P exhibited the lowest variance (Table S3†) suggesting that the higher (or lower) mean fluorescence values are driven by distinct subpopulations of high versus low APA.A comparative analysis was performed given the substantially different behavior across the cohorts from the 1 and 21 mM basal P concentration. For the sake of clarity, the naming scheme for the cohorts will be listed as two dashed numbers where first number is the basal P concentration and the second number is the spiked P concentration (e.g., the cohort challenged with 1 mM P from the 21 mM basal P concentration is 21–1). Interestingly, the 21–21 cohort exhibited a completely different response to the 1–1 cohort with the 1–1 cohort exhibiting the lowest overall APA whereas the 21–21 cohort exhibited intermediate APA (Tables S1, S3 and Fig. S3†). In looking at the mean and variance of these four cohorts, it was discovered that the 1–1 cohort (1.245 ± 0.144) exhibited a similar mean to the 1–21 cohort mean (1.415 ± 0.404) but a vastly difference variance (Fig. S3†). A different trend was observed between the 21–1 cohort (1.659 ± 0.519) and 21–21 cohort (1.502 ± 0.288) where the 21–1 cohort exhibited the highest mean and standard deviation. Moreover, looking at pairwise combinations of these four cohorts, Fisher's test shows a statistically significant difference across all four cohorts (Table S5†) which suggests a significant impact from both basal and spiked concentrations. This motivated performing a two-factor ANOVA looking at both basal and spiked P concentration which found a significant difference for both factors in addition to an interaction parameter suggested that both basal and spiked concentration influence APA (Table S6†). These findings highlight the intricate interplay between basal and spiked P concentrations and the adaptive mechanisms of cells to varying P levels. To further highlight this, the violin plots were created looking at the 1–0.5, 1–1, and 1–41 cohorts (Fig. 3B) and the 21–1, 21–21, and 21–41 cohorts (Fig. 3C) to visualize the different distributions across the cohorts. A comparison between the 1–1 and 21–1 cohorts shows a similar shape of the violin plots with a similar width near the mean values for both cohorts suggesting a similar frequency in APA between most of the cells; however, the higher peak in the 21–1 cohort supports the presence of a subpopulation of cells with a very high fluorescence signal driving the observed increase in mean APA. Conversely, the shape of the 1–41 and 21–41 cohorts differs with the former exhibiting a smoother distribution of the data while the latter appears to have two large subpopulations in the upper and lower quartiles. This, combined with the higher peak in the 1–41 cohort compared to the 21–41, suggests distinct subpopulations with high versus low APA driving the observed difference in mean APA. Moreover, a notable observation is the more uniform and concentrated distribution of data in the 1–1 cohort compared to the 21–21 mM cohort. The shape of the 21–21 cohort suggests that most cells exhibit the mean fluorescence signal; however, the higher and lower peaks along with the width of the lower peak further supports the existence of distinct subpopulations with varying APA driving difference in the mean fluorescence values. This observation suggests that cells acclimated to lower basal P concentrations exhibit a tighter regulation of the APA response resulting in a more homogeneous distribution of APA activity levels. In contrast, cells acclimated to higher basal phosphate concentrations display a broader range of APA activity levels, indicating a potentially less tightly regulated response. All these findings support the need to perform additional analysis to identify subpopulations of cells across all cohorts that could be driving the difference in the population averages.
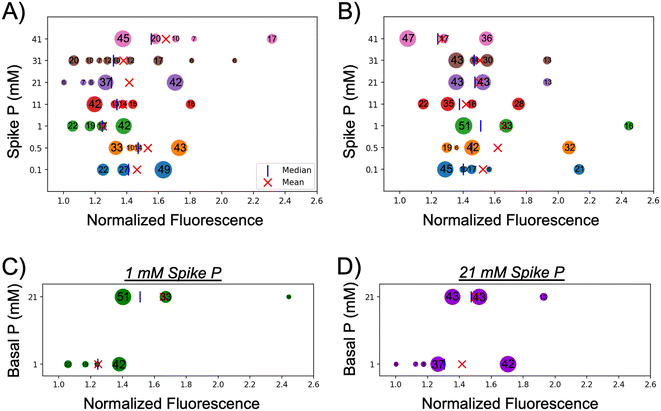
Unsupervised machine learning identifies distinct subpopulations of C. reinhardtii cells with enhanced or diminished APA due to adaptive behavior
To attain a greater understanding on the individual cell response under different basal and spiked conditions, and to be able to identify potential subpopulations of distinct characteristics, an unsupervised machine learning approach was implemented to cluster the complete dataset (ESI† methods and Fig. 4). This analysis identified a range of 4–9 clusters of various sizes based on the average normalized fluorescence where the number inside each bubble represents the percent of the population that falls within that cluster. The population mean and median values for each cohort are marked on the plot as well. As was demonstrated in the box plots and violin plots (Fig. 2 and 3), the distribution of cells based on APA was highly varied for each cohort in cells acclimated to either 1 mM basal P (Fig. 4A) or 21 mM basal P (Fig. 4B). Looking at cells acclimated to 1 mM basal P (Fig. 4A) it is seen that cohorts spiked with P levels equivalent to, or lower than the basal level consists of a fewer number of clusters of larger size when compared to cohorts with spiked P levels higher than basal levels. All four clusters from the 1–0.5 cohort (orange) and two of the three clusters (68% of total population) from the 1–0.1 (navy) cohort exhibit an APA response that is above the population average of the 1 mM cohort (green). The population increase in APA seen in the 1–0.1 and 1–0.5 cohorts observed in Fig. 2, coupled with the largest subpopulations showing above average APA, suggest these cells attempt to satisfy their nutrient requirements through up-regulation of AP after being exposed to spiked concentrations lower than they were acclimated to. Increasing the spiked P concentrations (11–41 mM) yielded an increasing number of clusters of decreasing size showing a more pronounced heterogeneity in cellular response to variations in P availability. These four cohorts all show a higher population average APA than that of the 1 mM; however, three of the cohorts (1–11, 1–21, and 1–31) also contain clusters with diminished APA compared to the 1 mM population average. All these spiked cohorts contain subpopulations with extremely high APA that are driving the higher population average for each cohort. Interestingly, the size and number of these subpopulations varies across the spiked P cohorts with the 1–0.1, 1–0.5, and 1–21 cohorts showing a single large subpopulation with enhanced APA whereas the 1–11, 1–31, and 1–41 cohorts yields multiple, small subpopulations with APA. These findings suggest that altered exogenous P level, either by increasing or decreasing, results in a heterogeneous cellular response driving either enhanced or diminished APA in select subpopulations of cells.The cohorts of cells acclimated to the higher 21 mM basal P levels (Fig. 4B) exhibit a slightly different behavior. All the cohorts were found to contain a similar number of clusters (3–5); however, the distribution of these clusters varied depending on the spike P levels (Fig. 4B). Three cohorts exposed to lower spiked P levels (21–1, 21–0.5, and 21–0.1) had a large cluster below the population average (51%, 42%, and 45%, respectively); however, they also contained a cluster of significant size (16%, 32%, and 21%, respectively) with APA in the higher ends of the normalized range. A similar trend was observed in the 21–21 and 21–31 cohorts; however, the size of the subpopulation with enhanced APA was not as prominent. The 21–41 cohort exhibited a distribution; however, the normalized fluorescence signal of the enhanced subpopulation (1.545, 36% of the population) is offset by the lowest signal for the diminished subpopulation (1.051, 47% of the population) which drives the lowest population average of the 21 mM basal P cohorts. The 21–11 cohort exhibited a similar behavior to the 21–41 cohort lacking a subpopulation with enhanced APA compared to the other cohorts which could potentially explain the lower population average for these cohorts. This suggests the upregulation of AP in a select number of cells as their primary adaptation strategy to altered basal P levels; however, most of the cells continue to rely on their internal sources of P that they are accumulated to. A comparison between the cohorts acclimated to different basal P levels yields some similar behaviors. Cells acclimated to 1 mM basal P levels yielded a similar distribution of clusters when spiked with either 1 or 21 mM P (Fig. 4C and D). The 1–1 and 1–21 cohorts both contained one subpopulation (42% of the population) with enhanced APA and 3–4 subpopulations with diminished APA (Fig. 4C and D, bottom row). Conversely, the 21–1 and 21–21 cohorts both contained three subpopulations, one with diminished APA, one with intermediate APA, and one with enhanced APA all equally similar size (Fig. 4C and D, top row). These similar distributions suggest that basal P levels potentially drive a difference in adaptability with a subset of cells acclimated to higher basal P levels being predisposed to exhibit extremely enhanced APA compared those acclimated to lower basal P levels.
Together, these findings suggest a somewhat basic understanding of the highly adaptive behavior among individual C. reinhardtii cells in response to deviations in nutrient availability. This adaptive behavior among algal populations has been studied in a variety of strains and under different nutrient limitations and/or enrichments, demonstrating a strong dependence on the species- and strain-specific physiological characteristics. Šupraha et al. studied two batch cultures of Helicosphaera carteri from contrasting nutrient regimes originated in different regions under phosphorus-replete conditions and showed how differently the two batches adapt to the new condition in terms of overall growth rates and inorganic carbon production.46 In another study, Yang et al. investigated how co-limitation of P and iron (Fe) simultaneously would induce a responsive behavior in C. watsonii observing a similar response employed by co-nutrient-depleted cells to that of single-nutrient-limited behavior, such as smaller cell size.47 In the study presented here, it is also demonstrated that the algal populations are highly adaptive in response to changes in key nutrient availability, but they also exhibit the adaptive behavior of individual cells is non-uniform across different populations of variable basal and spiked P concentrations. These observations also highlight the flaw in only looking at the population average value from a sample of cells suggesting that findings can be heavily biased depending on which subpopulation they belong to, and the total number of cells studied. Typical methods to quantify APA in algal cells collected from HABs only look at a small number of cells due to the size of the sample the substantial loss of cells using established approaches. As such, it is possible that some observations on APA in cells are incorrect due to only looking at cells from these subpopulations with enhanced or diminished APA.
Conclusions
This study aimed to investigate the single cell APA response of C. reinhardtii when challenged with fluctuations in P availability under both basal and spiked conditions. This was accomplished by leveraging a trap-based microfluidic device integrated with the commercially available AP live stain, as an all-in-one platform for trapping, staining, and imaging of single algal cells. A three-step culture-starve-spike cycle was used to induce APA in C. reinhardtii cells. Initial results from cells acclimated to a basal P level of 1 mM and challenged with 0.1–41 mM spiked P, demonstrated a highly heterogenous, nonlinear single cell APA response. This motivated a second study to investigate the potential significance of basal P levels by acclimating cells to elevated P levels (21 mM) in a week-long culture and challenged with similar spiked concentrations from the first study. Results demonstrated similar heterogeneity of single cell APA behavior yet with different trends from what was observed with the 1 mM basal P cohorts. For instance, when cells were exposed to spiked P similar to their basal level, the 1 mM spiked cohort (acclimated to 1 mM P) showed both the lowest mean and standard deviation in APA across all cohorts acclimated to 1 mM P. In contrast, the 21 mM spiked cohort (acclimated to 21 mM P) had the third lowest of both mean and standard deviation in APA (among cohorts acclimated to 21 mM P). Moreover, in cells acclimated to 1 mM basal P, the 0.1 mM spike cohort (lowest spike concentration) showed higher mean and standard deviation compared to the 1 mM spiked cohort, whereas in the 21 mM basal group, the mean APA of the 0.1 mM spiked cohort was statistically similar to that of the 21 mM spiked cohort. Conversely, the 41 mM spiked P (highest spike concentration) induced the highest mean APA across the 1 mM basal cohorts, and the lowest among the 21 mM basal cohorts. These differences highlighted the significance of both basal and spike P levels, which was confirmed by two-way ANOVA. To obtain a deeper understanding of the intrapopulation APA response of C. reinhardtii, an unsupervised machine learning (HDBSCAN) was implemented to cluster the data. This analysis identified subpopulations of cells with distinct behaviors that included diminished, average, or enhanced APA. Notably, distinct subpopulations were observed across cohorts of similar average/bulk behavior. For example, in the 1 mM basal cohorts, the mean APA of the 21 and 31 mM spiked P cohorts are statistically similar; however, it was seen that the 21 mM cohort consisted of fewer, larger subpopulations located mostly near the mean, whereas the 31 mM cohort consisted of more, smaller subpopulation more dispersed along a wider range, with one subpopulation of highly enhanced APA. The presence of these subpopulations suggest that individual cells utilize unique adaptation mechanisms when faced with sudden fluctuations in environmental phosphorus (P) levels and that this depends upon both the basal P levels they are acclimated to in addition to acute spiked concentration. Further investigations are warranted to explore the intracellular mechanisms underlying the distinct behaviors exhibited by individual cells. In conclusion, this study provides valuable insights into the nuanced single cell response of C. reinhardtii to phosphorus availability, highlighting the multifaceted interplay between basal and spiked P concentrations. By uncovering distinct APA behaviors at the single cell level, this study sets the stage for further investigations into the intricate mechanisms underlying algal nutrient utilization. Ultimately, this knowledge can inform strategies for managing nutrient dynamics in aquatic ecosystems, contributing to scientists understanding of environmental sustainability.Author contributions
Conceptualization, A.T.M., M.V., K.L.Y.; data curation, A.R., M.V., L.B.M.; formal analysis, A.R., M.V., L.B.M., J.A.R.; investigation, M.V., T.M.D., K.L.Y.; methodology, M.V., T.M.D., K.L.Y.; project administration, A.T.M.; resources, A.T.M.; software, L.B.M.; supervision, A.T.M.; validation: A.R., M.V.; visualization: A.R., writing of original draft: A.R., L.B.M.; writing-review and editing: A.T.M. All authors have read and agreed to the published version of the manuscript.Data availability
All raw and processed data presented in this manuscript has been uploaded and stored to the Clemson University OneDrive where it is securely maintained under the direction of the corresponding author. Back-ups of all the data are also maintained on external hard drives located in the office of the corresponding author. All data, raw and processed, can and will be made available to any interested parties. They are encouraged to directly contact the corresponding author (Dr Adam Melvin, melvina@clemson.edu) to request access to the data.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The work was funded by National Science Foundation (CBET1509713, ATM). The authors would like to thank David Englehardt, Sharif Rahman, and Devin Manning (LSU Chemical Engineering) for their contribution to initial data collection and help in the early stages of the project. The authors would also like to thank Drs Naohiro Kato and Gabela Nelson (LSU Biological Sciences) for providing the algal cells and their assistance with the algal cell culture conditions.References
- J. Ahlgren, L. Tranvik, A. Gogoll, M. Waldebäck, K. Markides and E. Rydin, Environ. Sci. Technol., 2005, 39, 867–872 CrossRef CAS.
- D. Correll, Poult. Sci., 1999, 78, 674–682 CrossRef CAS.
- R. Quinlan, A. Filazzola, O. Mahdiyan, A. Shuvo, K. Blagrave, C. Ewins, L. Moslenko, D. K. Gray, C. M. O'Reilly and S. Sharma, Limnol. Oceanogr., 2021, 66, 392–404 CrossRef CAS.
- W. Feng, F. Yang, C. Zhang, J. Liu, F. Song, H. Chen, Y. Zhu, S. Liu and J. P. Giesy, Environ. Pollut., 2020, 265, 114838 CrossRef CAS.
- D. W. Schindler, Limnol. Oceanogr., 2006, 51, 356–363 CrossRef.
- R. A. Vollenweider, Scientific fundamentals of the eutrophication of lakes and flowing waters, with particular reference to nitrogen and phosphorus as factors in eutrophication, Parisa, France, 1968 Search PubMed.
- E. M. Rodgers, Biol. Lett., 2021, 17, 20210442 CrossRef CAS.
- S. A. Reynolds and D. C. Aldridge, Front. Environ. Sci., 2021, 8 DOI:10.3389/fenvs.2020.551803.
- P. Zhang, W. Luo, M. Fu, J. Zhang, M. Cheng and J. Xie, Front. Mar. Sci., 2022, 9 DOI:10.3389/fmars.2022.961560.
- D. M. Anderson, Ocean Coast. Manag., 2009, 52, 342–347 CrossRef.
- A. R. Brown, M. Lilley, J. Shutler, C. Lowe, Y. Artioli, R. Torres, E. Berdalet and C. R. Tyler, Rev. Aquacult., 2020, 12, 1663–1688 CrossRef.
- T. Yan, X.-D. Li, Z.-J. Tan, R.-C. Yu and J.-Z. Zou, Harmful Algae, 2022, 111, 102148 CrossRef CAS.
- R. B. Wallace and C. J. Gobler, Mar. Pollut. Bull., 2021, 172, 112908 CrossRef CAS.
- D. M. Anderson, A. D. Cembella and G. M. Hallegraeff, Ann. Rev. Mar. Sci., 2012, 4, 143–176 CrossRef.
- G. M. Hallegraeff, D. M. Anderson, C. Belin, M.-Y. D. Bottein, E. Bresnan, M. Chinain, H. Enevoldsen, M. Iwataki, B. Karlson, C. H. McKenzie, I. Sunesen, G. C. Pitcher, P. Provoost, A. Richardson, L. Schweibold, P. A. Tester, V. L. Trainer, A. T. Yñiguez and A. Zingone, Commun. Earth Environ., 2021, 2, 1–10 CrossRef.
- D. M. Anderson, E. Fensin, C. J. Gobler, A. E. Hoeglund, K. A. Hubbard, D. M. Kulis, J. H. Landsberg, K. A. Lefebvre, P. Provoost, M. L. Richlen, J. L. Smith, A. R. Solow and V. L. Trainer, Harmful Algae, 2021, 102, 101975 CrossRef CAS.
- Q. Zhang, Y. Chen, M. Wang, J. Zhang, Q. Chen and D. Liu, Water Res., 2021, 196, 117048 CrossRef CAS.
- C. L. Schelske, Science, 2009, 324, 722–722 CrossRef CAS.
- S. Huang, W. Kong, Z. Yang, H. Yu and F. Li, Ecotoxicol. Environ. Saf., 2019, 167, 146–160 CrossRef CAS.
- D. M. Karl, Ann. Rev. Mar. Sci., 2014, 6, 279–337 CrossRef.
- S. T. Dyhrman, J. W. Ammerman and B. A. S. Van Mooy, Oceanography, 2007, 20, 110–116 CrossRef.
- M. Jansson, H. Olsson and K. Pettersson, Hydrobiologia, 1988, 170, 157–175 CrossRef CAS.
- S. Duhamel, S. T. Dyhrman and D. M. Karl, Limnol. Oceanogr., 2010, 55, 1414–1425 CrossRef CAS.
- C. Labry, D. Delmas and A. Herbland, J. Exp. Mar. Biol. Ecol., 2005, 318, 213–225 CrossRef CAS.
- K. Rengefors, K. Pettersson, T. Blenckner and D. M. Anderson, J. Plankton Res., 2001, 23, 435–443 CrossRef CAS.
- D. Diaz-de-Quijano, C. N. Stratman and S. A. Berger, Heliyon, 2020, 6, e05582 CrossRef CAS.
- L. Ou, X. Qin, X. Shi, Q. Feng, S. Zhang, S. Lu and Y. Qi, Microb. Ecol., 2020, 79, 459–471 CrossRef CAS.
- E. Xie, Y. Su, S. Deng, M. Kontopyrgou and D. Zhang, Environ. Pollut., 2021, 268, 115807 CrossRef CAS.
- Y. Lv, J. Chen, X. Zhou and X. Su, Anal. Bioanal. Chem., 2022, 414, 7989–7998 CrossRef CAS.
- S. González-Gil, B. Keafer, R. Jovine, A. Aguilera, S. Lu and D. Anderson, Mar. Ecol.: Prog. Ser., 1998, 164, 21–35 CrossRef.
- Y. Jiao, L. Gao, Y. Ji and W. Liu, TrAC, Trends Anal. Chem., 2022, 157, 116796 CrossRef CAS.
- F. C. Jammes and S. J. Maerkl, Microsyst. Nanoeng., 2020, 6, 1–14 CrossRef.
- M. L. Kovarik, D. M. Ornoff, A. T. Melvin, N. C. Dobes, Y. Wang, A. J. Dickinson, P. C. Gach, P. K. Shah and N. L. Allbritton, Anal. Chem., 2013, 85, 451–472 CrossRef CAS.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS.
- M. Girault, T. Beneyton, D. Pekin, L. Buisson, S. Bichon, C. Charbonnier, Y. del Amo and J.-C. Baret, Anal. Chem., 2018, 90, 4174–4181 CrossRef CAS.
- A. R. Longhurst, Limnol. Oceanogr., 1991, 36, 1507–1526 CrossRef CAS.
- T. Ide, S. Mochiji, N. Ueki, K. Yamaguchi, S. Shigenobu, M. Hirono and K. Wakabayashi, Biochem. Biophys. Rep., 2016, 7, 379–385 Search PubMed.
- U. Singh, R. H. Quintanilla, S. Grecian, K. R. Gee, M. S. Rao and U. Lakshmipathy, Stem Cell Rev. Rep., 2012, 8, 1021–1029 CrossRef.
- Q. D. Tran, T. F. Kong, D. Hu, M. Lam and R. H. W. Lam, Lab Chip, 2016, 16, 2813–2819 RSC.
- G. M. Landwehr, A. J. Kristof, S. M. Rahman, J. H. Pettigrew, R. Coates, J. B. Balhoff, U. L. Triantafillu, Y. Kim and A. T. Melvin, Biomicrofluidics, 2018, 12, 054109 CrossRef.
- D. M. Karl and G. Tien, Mar. Chem., 1997, 56, 77–96 CrossRef CAS.
- K. C. Ruttenberg and S. T. Dyhrman, J. Geophys. Res.: Oceans, 2005, 110, C10 CrossRef.
- S. Gu, G. Gruau, R. Dupas, C. Rumpel, A. Crème, O. Fovet, C. Gascuel-Odoux, L. Jeanneau, G. Humbert and P. Petitjean, Sci. Total Environ., 2017, 598, 421–431 CrossRef CAS.
- X. Bai, J. Sun, Y. Zhou, L. Gu, H. Zhao and J. Wang, Chemosphere, 2017, 169, 577–585 CrossRef CAS.
- W. R. Hill, S. E. Fanta and B. J. Roberts, Limnol. Oceanogr., 2009, 54, 368–380 CrossRef CAS.
- L. Šupraha, A. C. Gerecht, I. Probert and J. Henderiks, Sci. Rep., 2015, 5, 16499 CrossRef.
- N. Yang, Y.-A. Lin, C. A. Merkel, M. A. DeMers, P.-P. Qu, E. A. Webb, F.-X. Fu and D. A. Hutchins, ISME J., 2022, 16, 2702–2711 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00619d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |