Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of lipid-specific proteins with high-density lipid-immobilized beads†
Masayuki
Morito
,
Hiroki
Yasuda
,
Takaaki
Matsufuji
,
Masanao
Kinoshita
and
Nobuaki
Matsumori
 *
*
Department of Chemistry, Graduate School of Science, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan. E-mail: matsmori@chem.kyushu-univ.jp
First published on 3rd June 2024
Abstract
In biological membranes, lipids often interact with membrane proteins (MPs), regulating the localization and activity of MPs in cells. Although elucidating lipid–MP interactions is critical to comprehend the physiological roles of lipids, a systematic and comprehensive identification of lipid-binding proteins has not been adequately established. Therefore, we report the development of lipid-immobilized beads where lipid molecules were covalently immobilized. Owing to the detergent tolerance, these beads enable screening of water-soluble proteins and MPs, the latter of which typically necessitate surfactants for solubilization. Herein, two sphingolipid species—ceramide and sphingomyelin—which are major constituents of lipid rafts, were immobilized on the beads. We first showed that the density of immobilized lipid molecules on the beads was as high as that of biological lipid membranes. Subsequently, we confirmed that these beads enabled the selective pulldown of known sphingomyelin- or ceramide-binding proteins (lysenin, p24, and CERT) from protein mixtures, including cell lysates. In contrast, commercial sphingomyelin beads, on which lipid molecules are sparsely immobilized through biotin–streptavidin linkage, failed to capture lysenin, a well-known protein that recognizes clustered sphingomyelin molecules. This clearly demonstrates the applicability of our beads for obtaining proteins that recognize not only a single lipid molecule but also lipid clusters or lipid membranes. Finally, we demonstrated the screening of lipid-binding proteins from Neuro2a cell lysates using these beads. This method is expected to significantly contribute to the understanding of interactions between lipids and proteins and to unravel the complexities of lipid diversity.
Introduction
Biological membranes, composed of a wide variety of lipids and diverse membrane proteins (MPs), are involved in many important physiological functions, such as membrane transport and cell signaling.1 In biological membranes, lipids often interact with MPs, regulating the localization and activity of MPs in cells.2–6 For instance, the interaction of cholesterol with β2-adrenergicreceptor (β2AR), a type of G-protein-coupled receptor, is necessary for dimerization of the protein.7 This interaction disrupts the interplay between β2AR and G-protein and plays a major role in β2AR signaling. Therefore, elucidation of lipid–MP interactions is critical to understanding the physiological roles of lipids. Although these lipid–MP interactions have often been identified by X-ray crystallography,8–10 novel and powerful methods for identifying MP-binding lipids have been recently developed including native MS11–14 and our self-assembled monolayer-based methods.15,16 Although these methods have advanced the identification of “MP-binding lipids”, they are intrinsically inefficient in the systematic and comprehensive identification of “lipid-binding proteins”.A conventional method for the identification of lipid-binding proteins is liposome coprecipitation.17–20 This method has revealed, for example, that coronin1A, a critical regulator of branched actin network, binds to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2).21 In addition, nakanori was identified as a protein that specifically recognizes and binds sphingomyelin/cholesterol domains.22 However, this method is incompatible with the presence of surfactants, rendering it unsuitable for screening detergent-solubilized MPs.
Besides, affinity beads are used to identify target proteins of small molecules, including pharmaceuticals.23–27 This method is highly valuable for identifying binding proteins because it can be applied to various immobilized ligands and protein sources. This method has been applied to identifying binding proteins of bioactive lipids, including ceramide28–30 and PI(4,5)P2.31,32 In those studies, conventional immobilization methods used for small molecules, where ligands are immobilized sparsely on the bead surface, have been employed for lipid immobilization. In fact, commercially available lipid-coated beads33 are prepared based on the biotin–streptavidin binding, and thus, the density of immobilized lipids is relatively low. This would be suitable for identifying proteins that recognize a single lipid molecule, but identifying proteins that bind to or are embedded in lipid membranes through lipid-MP interactions is challenging.
In this study, to mimic lipid membranes on the bead surface, we developed lipid-immobilized affinity beads, on which lipid molecules were densely immobilized. The lipid-immobilized beads were designed so that the acyl chains of the lipid molecules are covalently bound to the bead surface and the hydrophilic head of the lipid is exposed to the outside. To achieve this, lipid derivatives featuring aminated acyl chain termini were synthesized and immobilized on magnetic nanobeads with high capacity for ligand immobilization. The covalent immobilization of lipid molecules enables screening of not only water-soluble proteins, but also MPs, which typically necessitate the use of surfactants for solubilization. Importantly, the prepared beads mimicked lipid membranes and could selectively capture lipid-specific proteins. We used these beads to capture lipid-binding proteins from cell lysates and identified several lipid-specific protein candidates.
Results
Preparation of lipid-immobilized beads
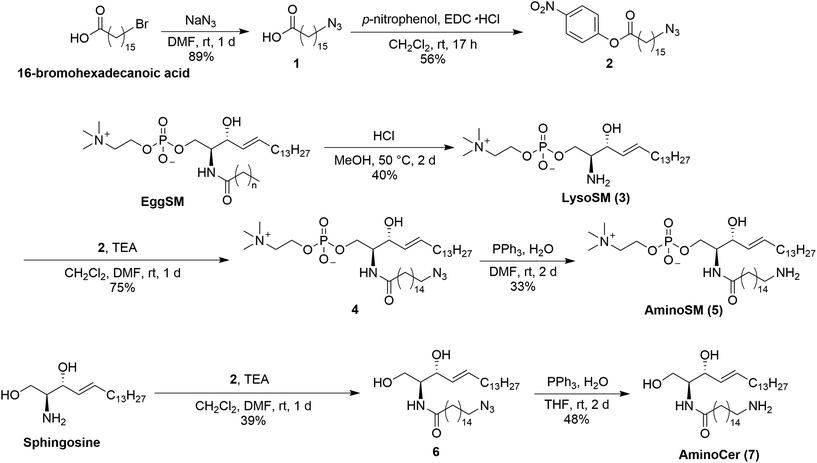
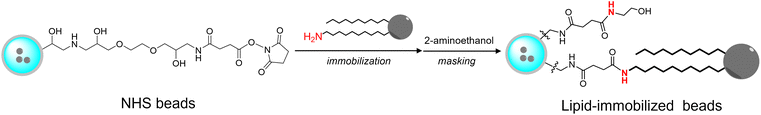
Because lipid rafts—functional domains of biological membranes enriched in sphingolipids and cholesterol—are proposed to be platforms for the accumulation and function of MPs,34–37 sphingomyelin (SM) and ceramide (Cer), which are major players in lipid rafts, were chosen for immobilization.38 Because proteins localized in lipid rafts potentially recognize not only the rigidity but the constituent lipid molecules of the rafts, these beads are expected to identify raft-related proteins. First, derivatives of SM and Cer were synthesized by introducing amino groups at the acyl chain terminals of lipid molecules (Scheme 1). Synthesis of aminoSM 5 commenced with deacylation of eggSM in methanolic HCl to yield lysosphingomyelin 3. Subsequent introduction of the azido acyl chain to 3, followed by reduction of the azido group through the Staudinger reaction yielded the desired product 5. AminoCer 7 was synthesized according to a similar scheme utilizing sphingosine as a substrate instead of lysosphingomyelin. The synthesized lipid derivatives were immobilized on FG beads (Scheme 2). Because FG beads (nanomagnetic beads) have a high density of functional groups on their surface,39 they would be well-suited for mimicking lipid membranes. The aminated lipids were immobilized by mixing with NHS-activated FG beads, where carboxy groups were activated as N-hydroxysuccinimide (NHS) ester groups. Subsequently, unreacted NHS ester groups were masked with aminoethanol. In addition to these lipid-immobilized beads, negative control beads were prepared by reacting with dodecylamine.Lipids were densely immobilized on the beads
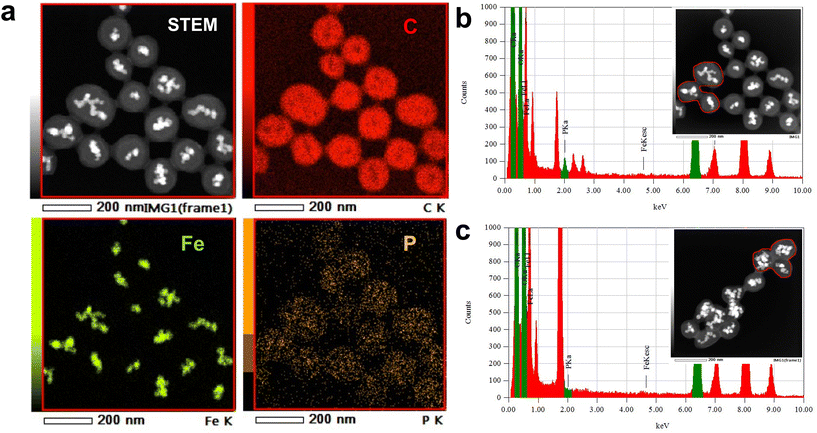
To confirm the dense immobilization of lipid molecules on the bead surface, phosphorus atoms present in the head of SM were used to quantify immobilized lipid. Because phosphorus atoms are not contained in either the bead carrier or linker, the distribution and quantification of lipid molecules on the beads was simultaneously estimated through phosphorus detection.Initially, scanning transmission electron microscopy (STEM) and energy-dispersive X-ray spectroscopy (EDS) analyses were conducted to confirm secure immobilization of SM molecules. In addition to carbon and Fe atoms, which were present in FG beads, phosphorus atoms were detected and distributed on SM beads (Fig. 1a). In the case of SM beads, a 2.0 keV peak, derived from phosphorus atoms, was detected in the EDS spectrum (Fig. 1b), whereas no phosphorus atoms were detected in control beads (Fig. 1c).
Next, the amount of immobilized SM was determined through phosphorus quantification. The beads were decomposed with sulfuric acid and hydrogen peroxide. The phosphoric acid formed was quantified by the molybdenum blue method.40 The amount of immobilized SM molecules per milligram of beads was determined as 73.2 ± 3.0 nmol mg−1, indicating an occupation area of 51.4 ± 2.1 Å2 per SM molecule on the bead surface (Table S1†). This value is comparable to that of an authentic lipid bilayer (40–60 Å2),41,42 suggesting that a lipid membrane structure is reproduced on the bead surface.
Taken together, these results confirmed uniform immobilization of lipid molecules at high density on the bead surface mimicking a lipid membrane structure.
Lipid-binding proteins interacted with the beads
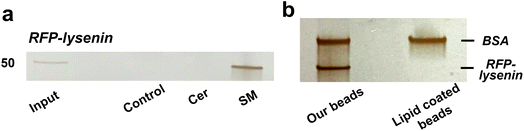
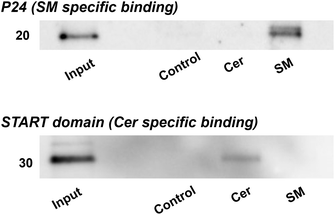
To validate the efficacy of lipid-immobilized beads in obtaining lipid-specific binding proteins, pulldown experiments were conducted using known lipid-binding proteins. Initially, a pulldown assay was executed using lysenin, which is a toxic protein sourced from earthworms and specifically recognizes and binds SM clusters.43–45 SM-, Cer-, and control beads were incubated with a solution containing red-fluorescent protein (RFP)-labelled lysenin and bovine serum albumin (BSA) and washed. Fluorescence microscopy showed that RFP-lysenin bound specifically to SM beads and not the other beads (Fig. S1†). Bead-bound proteins were recovered through heat treatment and visualized using silver-stained SDS-PAGE. Lysenin exhibited specific binding to SM beads (Fig. 2a). Then, these lipid-immobilized beads were compared with commercially available lipid-coated beads (Echelon Biosciences), on which lipids are sparsely immobilized through biotin–streptavidin linkage. Our SM beads and commercial SM beads were incubated with a mixture of lysenin and BSA, and subjected to pulldown experiments. Consequently, our beads had a markedly higher binding capacity for lysenin than the commercial beads (Fig. 2b).To further demonstrate the capacity of lipid-immobilized beads, we examined whether known lipid-binding proteins could be pulled down from complex protein mixtures such as cell lysates. The cell lysate of Neuro2a was incubated with the beads and the recovered proteins were identified through western blotting with antibodies targeting p24,46 an SM-specific binding protein, and START domain of CERT,47,48 a Cer-specific binding domain. The results revealed the specific binding of p24 to SM beads (Fig. 3, top) and the START domain to Cer beads (Fig. 3, bottom). Thus, we confirmed the applicability of lipid-immobilized beads in isolating lipid-binding proteins from complex protein mixtures and identifying lipid-specific proteins.
Screening lipid-specific binding proteins from Neuro2a cells
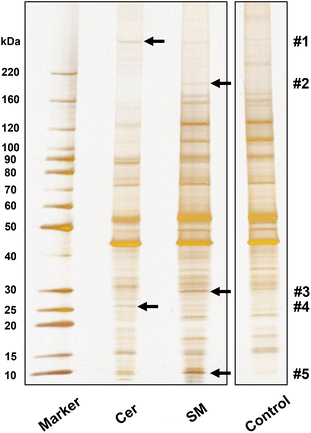
To demonstrate screening of lipid-binding proteins, proteins recovered from lipid-immobilized beads incubated with Neuro2a cell lysates were separated with SDS-PAGE, to obtain several distinctive bands (indicated by arrows, Fig. 4). The proteins in these bands were identified by proteomic analysis (Table S2†). For each band, proteins with top three Mascot scores are presented in Table 1, and proteins from control beads were excluded as nonspecific bindings. We focused on tumor protein D54 (TPD54), an SM-binding protein, identified in band #3. Pulldown experiments with authentic TPD54 were performed, confirming the specific binding of TPD54 to SM beads (Fig. 5).| Band | Lipid | Protein name | Mascot score | Mass | Number of peptides |
|---|---|---|---|---|---|
| #1 | Cer | Keratin, type I cytoskeletal 42 | 123 | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 444 |
4 |
| Bone marrow stromal antigen 2 | 91 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 311 311 |
3 | ||
| Nesprin-2 | 87 | 787![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 997 997 |
5 | ||
| #2 | SM | Proline-, glutamic acid- and leucine-rich protein 1 | 99 | 119![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 306 306 |
4 |
| YLP motif-containing protein 1 | 80 | 155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 146 146 |
4 | ||
| GRB10-interacting GYF protein 2 | 79 | 149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 387 387 |
3 | ||
| #3 | SM | Tumor protein D54 | 805 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 085 085 |
32 |
| MICOS complex subunit Mic19 | 229 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 546 546 |
6 | ||
| Vimentin | 222 | 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 712 |
11 | ||
| #4 | Cer | Protein FAM3C | 200 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 022 022 |
8 |
| Protein SCO2 homolog, mitochondrial | 183 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 097 097 |
7 | ||
| Bone marrow stromal antigen 2 | 167 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 311 311 |
4 | ||
| #5 | SM | ATP synthase subunit g | 273 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417 417 |
13 |
| Dynein light chain 1 | 160 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 530 |
4 | ||
| ATP synthase subunit e | 144 | 8230 | 6 |
Discussion
In this study, lipid-immobilized beads were developed to identify lipid-binding proteins. To mimic lipid membranes, sphingolipid derivatives with aminated acyl chain terminals were synthesized from lysosphingolipids (Scheme 1). These derivatives were subsequently amide-linked to the beads such that their hydrophilic heads were directed outward. EDS analysis and phosphorus quantification not only confirmed secure immobilization, but also revealed a high density of immobilized lipids equivalent to authentic lipid membranes, suggesting that the bead surface imitates lipid membranes (Fig. 1). Thus, we succeeded in preparing of detergent-tolerant and membrane-mimeticlipid-immobilized beads for screening membrane-associated proteins.Pulldown of a known SM-binding protein, lysenin, showed specific binding to SM beads (Fig. 2a). Comparison of the performance between our beads and commercially available beads further demonstrated the superiority of our SM beads in recognizing lysenin (Fig. 2b). Lysenin recognizes and binds SM domains rather than a single SM molecule.45 Thus, the commercial beads with sparse SM immobilization likely failed to capture lysenin. This result further supports the formation of a lipid membrane-like structure on the surface of our beads. Additionally, p24 and START domain of CERT in Neuro2a cell lysates exhibited specific binding to SM and Cer beads, respectively (Fig. 3), demonstrating that endogenous lipid-binding proteins can be detected from complex protein mixtures. The START domain is thought to be produced by proteolysis of CERT. These findings underscore the utility of this approach in obtaining lipid-specific binding proteins. Although lipid-like control beads were used in this study along with SM and Cer beads, it is hard to completely rule out the possibility that the proteins examined in this study cross-react with other lipids. To address this concern, we are currently preparing beads immobilized with various lipids, including phosphatidylcholine and glycolipids, and will conduct pulldown experiments using this series of lipid-immobilized beads.
Furthermore, we conducted a screening from cell lysates as a demonstration experiment, and identified interesting proteins as candidate lipid-binding proteins (Fig. 4 and Table 1). ATP synthase subunit g was identified as an SM-specific protein at band #5. Given that ATP synthase was reported to localize in lipid rafts,49 SM recognition of ATP synthase may be attributed to its preference for lipid rafts. Protein FAM3C, identified as a Cer-binding protein in this study, is a biomarker for Alzheimer's disease and known to reduce the production of amyloid β (Aβ) protein.50 Besides, Cer is involved in the elevation of Aβ.51,52 Therefore, the interaction between Cer and FAM3C might control the production of Aβ.
TPD54 was confirmed as SM-specific by the pulldown experiments (Fig. 5). TPD54 is one of the most abundantly expressed proteins in cancer cells, and quantitative proteomics of HeLa cells revealed that TPD54 is the 180th most abundant of 8804 identified proteins.53 Recently, the presence of a membrane transport pathway facilitated by intracellular nanovesicles (diameter: ∼30 nm) was revealed. TPD54 was reported to bind to the surface of the nanovesicles by inserting an amphipathic lipid-packing sensor (ALPS) motif.54,55 Because the ALPS motif senses lipid packing,56 TPD54 likely recognizes and binds densely-packed SM membranes, and not individual SM molecules, through the ALPS motif. This again indicates the applicability of our beads for obtaining proteins that recognize lipid membranes, which is otherwise difficult with conventional approaches.
Conclusions
Thus far, sparsely immobilized lipid-coated beads have been employed for screening lipid-binding proteins. However, these approaches face serious challenges when screening under conditions reflective of biological membranes, as evidenced by the pulldown of lysenin (Fig. 2b). In this context, our membrane-mimetic lipid-immobilized beads have opened a new avenue for identifying lipid-specific MPs that recognize not only a single lipid molecule but also lipid clusters or lipid membranes. Additionally, magnetic beads enable efficient pulldown through magnetic force, facilitating high-throughput analysis and potential automation. The synthesis of aminated derivatives allows for the immobilization of diverse lipid species and development of a variety of lipid-immobilized beads. With an expanded repertoire of immobilized lipid species, high-throughput analysis of various lipid–protein interactions becomes feasible. The comprehensive analysis of lipid-binding proteins can significantly contribute to understanding lipid–protein interactions and the complexities of lipid diversity.Experimental
General
NHS-activated FG beads were purchased from Tamagawa Seiki Co., Ltd (Nagano, Japan). SM beads were purchased from Echelon Biosciences (Salt Lake City, UT, USA). Antibodies were purchased from Proteintech Group, Inc. (Rosemont, IL, USA). All other reagents were purchased from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan), Tokyo Chemical Inc. (Tokyo, Japan), Nacalai Tesque Inc. (Kyoto, Japan), or Sigma-Aldrich (St Louis, MO, USA). For details of the synthesis of compounds 1–7, see ESI.†Preparation of lipid-immobilized beads
NHS-activated beads (1.0 mg) were washed three times with 200 μL of DMF and incubated with 10 mM triethylamine and 1 mM aminated lipids in 200 μL of DMF overnight at room temperature. The beads were washed three times with 200 μL of DMF, and unreacted NHS ester groups were masked with 1.0 M ethanolamine in DMF. After washing three times with 50% MeOH, these beads were resuspended in 50 μL of 50% MeOH, and stored at 4 °C. Negative control beads were prepared by reacting with 1 mM dodecylamine in DMF, instead of aminated lipids.Phosphorus quantification
Next, 25% H2SO4 (225 μL) was added to 0.2 mg of SM and control beads, respectively. Each suspension was heated at 200 °C for 25 min, cooled for 5 min, mixed with 75 μL of H2O2, and heated at 200 °C for 30 min. After cooling for 5 min, Milli-Q water (1950 μL), 10% ascorbic acid (250 μL), and 2.5% hexaammonium heptamolybdate (250 μL) were sequentially added to the suspension and the mixture was heated at 100 °C for 7 min. Absorbance of the supernatant was measured at 820 nm using a JASCO V-730BIO (JASCO Corporation, Tokyo, Japan).STEM and EDS analysis
SM and control beads were washed with water twice and resuspended in 50 μL of Milli-Q water. An aliquot of 7 μL suspension was placed on a carbon-coated copper grid (Okenshoji, Tokyo, Japan), excess water was removed, and the sample was dried in vacuo overnight. STEM images were observed using a JEM-ARM200F instrument equipped with a JED-2300T (JEOL, Tokyo, Japan) at an acceleration voltage of 200 kV. The JEOL Analysis Station software was used to control STEM-EDS mapping.Neuro-2a cell lysate preparation
Neuro-2a cells were cultured in E-MEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin at 37 °C with 5% CO2. Cells were suspended in lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, and 1% Nonidet P-40) containing protease inhibitor cocktail (Nacalai Tesque Inc., Kyoto, Japan), incubated on ice for 1 h, and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, for 20 min at 4 °C. The supernatant was collected to prepare the lysate. The protein content in the lysate was quantified with BCA Protein Assay Kit (Takara Bio Inc., Shiga, Japan), and the lysate was diluted to a concentration of 1 mg mL−1.
000 rpm, for 20 min at 4 °C. The supernatant was collected to prepare the lysate. The protein content in the lysate was quantified with BCA Protein Assay Kit (Takara Bio Inc., Shiga, Japan), and the lysate was diluted to a concentration of 1 mg mL−1.
Pulldown with lipid-immobilized beads
Each type of bead (0.5 mg) was washed with 200 μL of lysis buffer before use, followed by dispersion in 50 μL of protein solution. After incubation at 4 °C for 4 h, the supernatant was removed through magnetic separation. The beads were washed twice with 200 μL of lysis buffer and then boiled in SDS sample buffer (240 mM Tris-HCl [pH6.8], 8% SDS, 40% glycerol, 0.1% bromophenol blue, and 20% 2-mercaptoethanol). The eluted proteins were applied to 4%–20% SDS-PAGE. Proteins were detected by silver staining or western blotting.Lysenin pulldown with lipid-immobilized beads and commercial beads
SM-immobilized beads (0.2 mg) were incubated with a protein mixture (10 μg mL−1 RFP-lysenin and 1 mg mL−1 BSA in lysis buffer, 50 μL). The subsequent procedure was the same as above. By contrast, commercial SM beads (Echelon Bioscience, 40 μL) were washed with 200 μL of lysis buffer before use, followed by dispersion in 50 μL of the same protein mixture. After incubation at 4 °C for 4 h, the supernatant was removed by centrifugation at 800g for 4 min. The beads were washed twice with 200 μL of lysis buffer, boiled in SDS sample buffer, and centrifuged at 800g for 4 min. The supernatant was applied to a 4%–20% SDS-PAGE gel, followed by silver staining.Western blotting
Proteins were electrophoretically transferred from SDS-PAGE gels to PVDF membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked with 2% BSA in PBS containing 0.05% Tween-20 (PBS-T) for 1 h at room temperature, incubated with primary antibodies (anti-p24 antibody and anti-CERT antibody) in PBS-T containing 2% BSA overnight at 4 °C, washed three times with PBS-T, incubated with HRP-conjugated secondary antibodies for 1 h, and washed three times with PBS-T. Signals were detected with an imaging system (ImageQuant LAS 4000 mini, Cytiva, Marlborough, MA, US).Proteomic analysis
Each selected band was cut into pieces and subjected to LC-MS/MS analysis. The gel fragments were destained and digested with mass spectrometry-grade trypsin (PROMEGA, Madison, WI, USA) in 25 mM ammonium bicarbonate. nLC-MS/MS analysis was conducted using Advance UHPLC (Bruker, Billerica, MA, USA) and Orbitrap Velos Pro (Thermo Fisher Scientific, Waltham, MA, USA). Trypsin-digested samples were separated by SilicaTip (0.100 mm i.d. × 15 cm, Nikkyo Technos, Japan) packed with a 3 μm C18 L-column (Chemical Evaluation and Research Institute, Japan) using a linear gradient (30 min, acetonitrile/0.1% formic acid) at a flow rate of 320 nL min−1. The resulting MS and MS/MS data were searched against the Swiss-Prot database using the Mascot search engine software (ver. 2.8.0.1, Matrix Science, Boston, MA, USA). Significance threshold was set to p < 0.05. The proteins with at least three peptide matches were listed. Nonspecific proteins identified in control samples were excluded from the list of identified proteins.Author contributions
M. Morito: investigation, methodology, visualization, and writing original draft; H. Yasuda: investigation; T. Matsufuji: investigation; M. Konoshita: investigation; N. Matsumori: writing original draft, funding acquisition, supervision, conceptualization, and project administration. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge Prof. Junichi Ikenouchi, Kyushu University, for providing RFP-lysenin. We also thank Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University, for technical support of proteomic analysis. The electron microscopic observations were conducted at Ultramicroscopy Research Center (Kyusyu University, Fukuoka, Japan). This work was supported by the Japan Society for the Promotion of Science KAKENHI (JP15H03121, JP16H00773, JP20H00405, JP22K19115, and JP23H04881 to N. M.; JP20K06590 and JP23K05719 to M. K.), and JST SPRING (JPMJSP2136 to M. M.).References
- G. van Meer and A. I. de Kroon, Lipid map of the mammalian cell, J. Cell Sci., 2011, 124, 5–8 CrossRef CAS PubMed.
- A. G. Lee, Lipid-protein interactions in biological membranes: a structural perspective, Biochim. Biophys. Acta, 2003, 1612, 1–40 CrossRef CAS PubMed.
- P. A. Janmey and P. K. Kinnunen, Biophysical properties of lipids and dynamic membranes, Trends Cell Biol., 2006, 16, 538–546 CrossRef CAS PubMed.
- C. Hunte and S. Richers, Lipids and membrane protein structures, Curr. Opin. Struct. Biol., 2008, 18, 406–411 CrossRef CAS PubMed.
- R. Phillips, T. Ursell, P. Wiggins and P. Sens, Emerging roles for lipids in shaping membrane-protein function, Nature, 2009, 459, 379–385 CrossRef CAS PubMed.
- P. L. Yeagle, Non-covalent binding of membrane lipids to membrane proteins, Biochim. Biophys. Acta, 2014, 1838, 1548–1559 CrossRef CAS PubMed.
- V. Cherezov, D. M. Rosenbaum, M. A. Hanson, S. G. Rasmussen, F. S. Thian, T. S. Kobilka, H. J. Choi, P. Kuhn, W. I. Weis, B. K. Kobilka and R. C. Stevens, High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor, Science, 2007, 318, 1258–1265 CrossRef CAS PubMed.
- S. B. Long, X. Tao, E. B. Campbell and R. MacKinnon, Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment, Nature, 2007, 450, 376–382 CrossRef CAS PubMed.
- G. Gimpl, Interaction of G protein coupled receptors and cholesterol, Chem. Phys. Lipids, 2016, 199, 61–73 CrossRef CAS PubMed.
- V. Corradi, B. I. Sejdiu, H. Mesa-Galloso, H. Abdizadeh, S. Y. Noskov, S. J. Marrink and D. P. Tieleman, Emerging Diversity in Lipid-Protein Interactions, Chem. Rev., 2019, 119, 5775–5848 CrossRef CAS PubMed.
- J. Gault, I. Liko, M. Landreh, D. Shutin, J. R. Bolla, D. Jefferies, M. Agasid, H. Y. Yen, M. Ladds, D. P. Lane, S. Khalid, C. Mullen, P. M. Remes, R. Huguet, G. McAlister, M. Goodwin, R. Viner, J. E. P. Syka and C. V. Robinson, Combining native and ‘omics’ mass spectrometry to identify endogenous ligands bound to membrane proteins, Nat. Methods, 2020, 17, 505–508 CrossRef CAS PubMed.
- X. Cong, Y. Liu, W. Liu, X. Liang and A. Laganowsky, Allosteric modulation of protein-protein interactions by individual lipid binding events, Nat. Commun., 2017, 8, 2203 CrossRef PubMed.
- C. E. Norris, J. E. Keener, S. Perera, N. Weerasinghe, S. D. E. Fried, W. C. Resager, J. G. Rohrbough, M. F. Brown and M. T. Marty, Native Mass Spectrometry Reveals the Simultaneous Binding of Lipids and Zinc to Rhodopsin, Int. J. Mass Spectrom., 2021, 460, 116477 CrossRef CAS PubMed.
- K. Gupta, J. Li, I. Liko, J. Gault, C. Bechara, D. Wu, J. T. S. Hopper, K. Giles, J. L. P. Benesch and C. V. Robinson, Identifying key membrane protein lipid interactions using mass spectrometry, Nat. Protoc., 2018, 13, 1106–1120 CrossRef CAS PubMed.
- M. Inada, M. Kinoshita, A. Sumino, S. Oiki and N. Matsumori, A concise method for quantitative analysis of interactions between lipids and membrane proteins, Anal. Chim. Acta, 2019, 1059, 103–112 CrossRef CAS PubMed.
- S. Wangamnuayporn, M. Kinoshita, T. Kawai and N. Matsumori, Gold nanoparticle-powered screening of membrane protein-specific lipids from complex lipid mixtures, Anal. Biochem., 2023, 687, 115447 CrossRef PubMed.
- A. L. Capriotti, G. Caracciolo, G. Caruso, C. Cavaliere, D. Pozzi, R. Samperi and A. Lagana, Analysis of plasma protein adsorption onto DC-Chol-DOPE cationic liposomes by HPLC-CHIP coupled to a Q-TOF mass spectrometer, Anal. Bioanal. Chem., 2010, 398, 2895–2903 CrossRef CAS PubMed.
- R. Pattipeiluhu, S. Crielaard, I. Klein-Schiphorst, B. I. Florea, A. Kros and F. Campbell, Unbiased Identification of the Liposome Protein Corona using Photoaffinity-based Chemoproteomics, ACS Cent. Sci., 2020, 6, 535–545 CrossRef CAS PubMed.
- M. Frick, C. Schwieger and C. Schmidt, Liposomes as Carriers of Membrane-Associated Proteins and Peptides for Mass Spectrometric Analysis, Angew. Chem., Int. Ed., 2021, 60, 11523–11530 CrossRef CAS PubMed.
- M. Tanaka, A. Arakaki and T. Matsunaga, Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria, Mol. Microbiol., 2010, 76, 480–488 CrossRef CAS PubMed.
- K. Tsujita, T. Itoh, A. Kondo, M. Oyama, H. Kozuka-Hata, Y. Irino, J. Hasegawa and T. Takenawa, Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides, J. Biol. Chem., 2010, 285, 6781–6789 CrossRef CAS PubMed.
- A. Makino, M. Abe, R. Ishitsuka, M. Murate, T. Kishimoto, S. Sakai, F. Hullin-Matsuda, Y. Shimada, T. Inaba, H. Miyatake, H. Tanaka, A. Kurahashi, C. G. Pack, R. S. Kasai, S. Kubo, N. L. Schieber, N. Dohmae, N. Tochio, K. Hagiwara, Y. Sasaki, Y. Aida, F. Fujimori, T. Kigawa, K. Nishibori, R. G. Parton, A. Kusumi, Y. Sako, G. Anderluh, M. Yamashita, T. Kobayashi, P. Greimel and T. Kobayashi, A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C, FASEB J., 2017, 31, 1301–1322 CrossRef CAS PubMed.
- C. Kuramori, M. Azuma, K. Kume, Y. Kaneko, A. Inoue, Y. Yamaguchi, Y. Kabe, T. Hosoya, M. Kizaki, M. Suematsu and H. Handa, Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus, Biochem. Biophys. Res. Commun., 2009, 379, 519–525 CrossRef CAS PubMed.
- T. Hirota, J. W. Lee, P. C. St John, M. Sawa, K. Iwaisako, T. Noguchi, P. Y. Pongsawakul, T. Sonntag, D. K. Welsh, D. A. Brenner, F. J. Doyle, P. G. Schultz and S. A. Kay, Identification of Small Molecule Activators of Cryptochrome, Science, 2012, 337, 1094–1097 CrossRef CAS PubMed.
- G. R. Rosania, Y.-T. Chang, O. Perez, D. Sutherlin, H. Dong, D. J. Lockhart and P. G. Schultz, Myoseverin, a microtubule-binding molecule with novel cellular effects, Nat. Biotechnol., 2000, 18, 304–308 CrossRef CAS PubMed.
- S. M. Khersonsky, D.-W. Jung, T.-W. Kang, D. P. Walsh, H.-S. Moon, H. Jo, E. M. Jacobson, V. Shetty, T. A. Neubert and Y.-T. Chang, Facilitated Forward Chemical Genetics Using a Tagged Triazine Library and Zebrafish Embryo Screening, J. Am. Chem. Soc., 2003, 125, 11804–11805 CrossRef CAS PubMed.
- T. Ito, H. Ando, T. Suzuki, T. Ogura, K. Hotta, Y. Imamura, Y. Yamaguchi and H. Handa, Identification of a Primary Target of Thalidomide Teratogenicity, Science, 2010, 327, 1345–1350 CrossRef CAS PubMed.
- V. Kota, Z. M. Szulc and H. Hama, Identification of C6-ceramide-interacting proteins in D6P2T Schwannoma cells, Proteomics, 2012, 12, 2179–2184 CrossRef CAS PubMed.
- Z. Zhu, J. Chen, G. Wang, A. Elsherbini, L. Zhong, X. Jiang, H. Qin, P. Tripathi, W. Zhi, S. D. Spassieva, A. J. Morris and E. Bieberich, Ceramide regulates interaction of Hsd17b4 with Pex5 and function of peroxisomes, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2019, 1864, 1514–1524 CrossRef CAS PubMed.
- J. N. Kong, Z. Zhu, Y. Itokazu, G. Wang, M. B. Dinkins, L. Zhong, H. P. Lin, A. Elsherbini, S. Leanhart, X. Jiang, H. Qin, W. Zhi, S. D. Spassieva and E. Bieberich, Novel function of ceramide for regulation of mitochondrial ATP release in astrocytes, J. Lipid Res., 2018, 59, 488–506 CrossRef CAS PubMed.
- A. E. Lewis, L. Sommer, M. O. Arntzen, Y. Strahm, N. A. Morrice, N. Divecha and C. S. D’Santos, Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction, Mol. Cell. Proteomics, 2011, 10, M110.003376 CrossRef PubMed.
- S. K. Kim, H. Kim, Y. R. Yang, P. G. Suh and J. S. Chang, Phosphatidylinositol phosphates directly bind to neurofilament light chain (NF-L) for the regulation of NF-L self assembly, Exp. Mol. Med., 2011, 43, 153–160 CrossRef CAS PubMed.
- V. R. Rao, M. N. Corradetti, J. Chen, J. Peng, J. Yuan, G. D. Prestwich and J. S. Brugge, Expression cloning of protein targets for 3-phosphorylated phosphoinositides, J. Biol. Chem., 1999, 274, 37893–37900 CrossRef CAS PubMed.
- L. J. Pike, The challenge of lipid rafts, J. Lipid Res., 2009, 50(Suppl), S323–S328 CrossRef PubMed.
- D. Lingwood and K. Simons, Lipid rafts as a membrane-organizing principle, Science, 2010, 327, 46–50 CrossRef CAS PubMed.
- I. Levental, K. R. Levental and F. A. Heberle, Lipid Rafts: Controversies Resolved, Mysteries Remain, Trends Cell Biol., 2020, 30, 341–353 CrossRef CAS PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, The mystery of membrane organization: composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS PubMed.
- G. van Meer and S. Hoetzl, Sphingolipid topology and the dynamic organization and function of membrane proteins, FEBS Lett., 2010, 584, 1800–1805 CrossRef CAS PubMed.
- K. Nishio, Y. Masaike, M. Ikeda, H. Narimatsu, N. Gokon, S. Tsubouchi, M. Hatakeyama, S. Sakamoto, N. Hanyu, A. Sandhu, H. Kawaguchi, M. Abe and H. Handa, Development of novel magnetic nano-carriers for high-performance affinity purification, Colloids Surf., B, 2008, 64, 162–169 CrossRef CAS PubMed.
- J. Murphy and J. P. Riley, A modified single solution method for the determination of phosphate in natural waters, Anal. Chim. Acta, 1962, 27, 31–36 CrossRef CAS.
- P. S. Niemela, S. Ollila, M. T. Hyvonen, M. Karttunen and I. Vattulainen, Assessing the nature of lipid raft membranes, PLoS Comput. Biol., 2007, 3, e34 CrossRef PubMed.
- A. Leftin, T. R. Molugu, C. Job, K. Beyer and M. F. Brown, Area per Lipid and Cholesterol Interactions in Membranes from Separated Local-Field 13 C NMR Spectroscopy, Biophys. J., 2014, 107, 2274–2286 CrossRef CAS PubMed.
- Y. Sekizawa, K. Hagiwara, T. Nakajima and H. Kobayashi, A novel protein, lysenin, that causes contraction of the isolated rat aorta: its purification from the coelomic fluid of the earthworm, Eisenia foetida, Biomed. Res., 1996, 17, 197–203 CrossRef CAS.
- L. De Colibus, A. F. Sonnen, K. J. Morris, C. A. Siebert, P. Abrusci, J. Plitzko, V. Hodnik, M. Leippe, E. Volpi, G. Anderluh and R. J. Gilbert, Structures of lysenin reveal a shared evolutionary origin for pore-forming proteins and its mode of sphingomyelin recognition, Structure, 2012, 20, 1498–1507 CrossRef CAS PubMed.
- R. Ishitsuka, A. Yamaji-Hasegawa, A. Makino, Y. Hirabayashi and T. Kobayashi, A lipid-specific toxin reveals heterogeneity of sphingomyelin-containing membranes, Biophys. J., 2004, 86, 296–307 CrossRef CAS PubMed.
- F. X. Contreras, A. M. Ernst, P. Haberkant, P. Bjorkholm, E. Lindahl, B. Gonen, C. Tischer, A. Elofsson, G. von Heijne, C. Thiele, R. Pepperkok, F. Wieland and B. Brugger, Molecular recognition of a single sphingolipid species by a protein's transmembrane domain, Nature, 2012, 481, 525–529 CrossRef CAS PubMed.
- K. Hanada, K. Kumagai, N. Tomishige and T. Yamaji, CERT-mediated trafficking of ceramide, Biochim. Biophys. Acta, 2009, 1791, 684–691 CrossRef CAS PubMed.
- K. Kumagai and K. Hanada, Structure, functions and regulation of CERT, a lipid-transfer protein for the delivery of ceramide at the ER-Golgi membrane contact sites, FEBS Lett., 2019, 593, 2366–2377 CrossRef CAS PubMed.
- T. J. Bae, M. S. Kim, J. W. Kim, B. W. Kim, H. J. Choo, J. W. Lee, K. B. Kim, C. S. Lee, J. H. Kim, S. Y. Chang, C. Y. Kang, S. W. Lee and Y. G. Ko, Lipid raft proteome reveals ATP synthase complex in the cell surface, Proteomics, 2004, 4, 3536–3548 CrossRef CAS PubMed.
- Y. Zhu, Z. Pu, G. Wang, Y. Li, Y. Wang, N. Li and F. Peng, FAM3C: an emerging biomarker and potential therapeutic target for cancer, Biomarkers Med., 2021, 15, 373–384 CrossRef CAS PubMed.
- M. Jazvinšćak Jembrek, P. R. Hof and G. Šimić, Ceramides in Alzheimer’s Disease: Key Mediators of Neuronal Apoptosis Induced by Oxidative Stress and Abeta Accumulation, Oxid. Med. Cell. Longevity, 2015, 2015, 346783 Search PubMed.
- M. R. Chowdhury, H. K. Jin and J. S. Bae, Diverse Roles of Ceramide in the Progression and Pathogenesis of Alzheimer’s Disease, Biomedicines, 2022, 10, 1956 CrossRef CAS PubMed.
- M. Y. Hein, N. C. Hubner, I. Poser, J. Cox, N. Nagaraj, Y. Toyoda, I. A. Gak, I. Weisswange, J. Mansfeld, F. Buchholz, A. A. Hyman and M. Mann, A human interactome in three quantitative dimensions organized by stoichiometries and abundances, Cell, 2015, 163, 712–723 CrossRef CAS PubMed.
- G. Larocque, J. L.-B. Penelope, N. I. Clarke, N. J. Carter and S. J. Royle, Tumor protein D54 defines a new class of intracellular transport vesicles, J. Cell Biol., 2020, 219, e201812044 CrossRef PubMed.
- A. Reynaud, M. Magdeleine, A. Patel, A. S. Gay, D. Debayle, S. Abelanet and B. Antonny, Tumor protein D54 binds intracellular nanovesicles via an extended amphipathic region, J. Biol. Chem., 2022, 298, 102136 CrossRef CAS PubMed.
- S. Vanni, L. Vamparys, R. Gautier, G. Drin, C. Etchebest, P. F. Fuchs and B. Antonny, Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues, Biophys. J., 2013, 104, 575–584 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fluorescence images of RFP-lysenin bound to each type of bead, amount of immobilized SM molecules per milligram of bead, identified lipid-binding proteins from Neuro2a cells, and synthesis of aminoSM and amino-Cer. See DOI: https://doi.org/10.1039/d4an00579a |
| This journal is © The Royal Society of Chemistry 2024 |