Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Native mass spectrometry of complexes formed by molecular glues reveals stoichiometric rearrangement of E3 ligases†
Cara
Jackson
and
Rebecca
Beveridge
 *
*
Department of Pure and Applied Chemistry, University of Strathclyde, Glasgow, UK. E-mail: rebecca.beveridge@strath.ac.uk
First published on 16th April 2024
Abstract
In this application of native mass spectrometry (nMS) to investigate complexes formed by molecular glues (MGs), we have demonstrated its efficiency in delineating stoichiometric rearrangements of E3 ligases that occur during targeted protein degradation (TPD). MGs stabilise interactions between an E3 ligase and a protein of interest (POI) targeted for degradation, and these ternary interactions are challenging to characterise. We have shown that nMS can unambiguously identify complexes formed between the CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 E3 ligase and the POI GSPT1 upon the addition of lenalidomide, pomalidomide or thalidomide. Ternary complex formation was also identified involving the DCAF15
DDB1 E3 ligase and the POI GSPT1 upon the addition of lenalidomide, pomalidomide or thalidomide. Ternary complex formation was also identified involving the DCAF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1
DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 E3 ligase in the presence of MG (E7820 or indisulam) and POI RBM39. Moreover, we uncovered that the DCAF15
DDB1 E3 ligase in the presence of MG (E7820 or indisulam) and POI RBM39. Moreover, we uncovered that the DCAF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1
DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 E3 ligase self-associates into dimers and trimers when analysed alone at low salt concentrations (100 mM ammonium acetate) which dissociate into single copies of the complex at higher salt concentrations (500 mM ammonium acetate), or upon the addition of MG and POI, forming a 1
DDB1 E3 ligase self-associates into dimers and trimers when analysed alone at low salt concentrations (100 mM ammonium acetate) which dissociate into single copies of the complex at higher salt concentrations (500 mM ammonium acetate), or upon the addition of MG and POI, forming a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ternary complex. This work demonstrates the strength of nMS in TPD research, reveals novel binding mechanisms of the DCAF15 E3 ligase, and its self-association into dimers and trimers at reduced salt concentration during structural analysis.
1 ternary complex. This work demonstrates the strength of nMS in TPD research, reveals novel binding mechanisms of the DCAF15 E3 ligase, and its self-association into dimers and trimers at reduced salt concentration during structural analysis.
Introduction
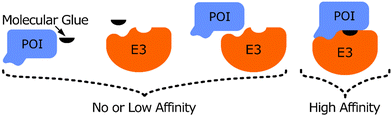
Protein–protein interactions play pivotal roles in many cellular processes and are therefore regarded as promising targets for drug discovery.1 The classic approach of targeting protein–protein interactions has been to inhibit their formation, often with the use of small molecules2,3 or engineered peptides.4 Additionally, the use of protein–protein interaction stabilisers has also garnered significant attention,5 especially in the area of targeted protein degradation (TPD).Under normal physiological conditions, the function of an E3 ligase is to catalyse the ubiquitination of a specific protein, which targets it for proteasomal degradation.6,7 This process requires specific interactions between the E3 ligase and its substrate to achieve regulatory specificity during protein degradation. By artificially creating or strengthening these interactions by using small molecules such as proteolysis-targeting chimeras (PROTACs)8 or Molecular Glues (MGs),9 this function of the E3 ligase can be hijacked and an unwanted protein of interest (POI), such as an oncogenic protein,10 can be targeted for degradation by the cell (Fig. 1). Such molecules draw the POI and the E3 ligase into close spatial proximity, resulting in ubiquitination of the POI by the E3 ligase and subsequent proteasomal degradation of the POI.
TPD is currently in an era of significant growth, and many innovative technologies are in development to address challenges in the approaches. These challenges include expanding the availability of E3 ligases that can be hijacked in TPD,11 achieving selectivity of a degrader to a specific POI,12 and the availability of analytical methodologies to directly measure the formation of ternary complexes formed between the E3 ligase, the glue, and the POI.13,14
Made infamous as the teratogenic morning sickness medication, the immunomodulatory drug thalidomide has gained newfound therapeutic use in the treatment of multiple myeloma.15 Thalidomide and its derivatives lenalidomide and pomalidomide act as MGs between the E3 ubiquitin ligase consisting of cereblon (CRBN) and damaged DNA binding protein 1 (DDB1) (CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1), and various target proteins. In addition to these MGs, an additional group named the splicing inhibitor sulfonamides are becoming widely used in clinical trials, either as a single agent or in combination with other treatments.16 E7820 and indisulam are the most documented splicing inhibitor sulfonamides, used to recruit RNA binding protein 39 (RBM39) for degradation by the E3 ubiquitin ligase complex herein named the DCAF15 complex. The DCAF15 complex consists of the proteins DDB1 and CUL4 associated factor 15 (DCAF15), DET1 and DDB1 associated protein 1 (DDA1), and damaged DNA binding protein 1 (DDB1). RBM39 and the DCAF15 complex are known to have a low affinity for each other, but molecular glues can greatly enhance the strength of this interaction (Table 1).17
DDB1), and various target proteins. In addition to these MGs, an additional group named the splicing inhibitor sulfonamides are becoming widely used in clinical trials, either as a single agent or in combination with other treatments.16 E7820 and indisulam are the most documented splicing inhibitor sulfonamides, used to recruit RNA binding protein 39 (RBM39) for degradation by the E3 ubiquitin ligase complex herein named the DCAF15 complex. The DCAF15 complex consists of the proteins DDB1 and CUL4 associated factor 15 (DCAF15), DET1 and DDB1 associated protein 1 (DDA1), and damaged DNA binding protein 1 (DDB1). RBM39 and the DCAF15 complex are known to have a low affinity for each other, but molecular glues can greatly enhance the strength of this interaction (Table 1).17
| E3 ligase | Protein of interest | MGs |
|---|---|---|
CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 DDB1 |
GSPT1 | Lenalidomide, pomalidamide, thalidomide |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex 1 complex |
||
| DCAF15 complex | RBM39 | Indisulam, |
(DCAF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1 DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1) 1 DDB1) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex 1 complex |
E7820 |
Native mass spectrometry (nMS) is a means of analysing proteins18,19 and protein complexes in their native state.20,21 Here, large proteins can be transferred from solution into the gas phase as multiply charged ions, in a process known as electrospray ionisation.22 nMS tends to use nanoelectrospray ionisation,23 which produces finer droplets, allowing for native topologies, stoichiometries, and non-covalent interactions to remain while transferring proteins from a nondenaturing solution, commonly ammonium acetate (AmAc),24 into the gas phase.25 nMS is effective in separating individual species that exist in a stoichiometric mixture, capturing transient interactions, and comparing stability of complexes.20,26,27
nMS has previously been used to predict the efficacy of PROTACs,28 to analyse complexes formed between MGs and model peptides,21 and to elucidate the contributions of covalent versus non-covalent binding events that govern aldehyde-based covalent molecular glue activity.29 A recent preprint by our group30 and work published by Huang et al.31 demonstrates the efficacy of nMS to MGs and multimeric E3 complexes. Here, we demonstrate the important and unusual effect of salt concentration on the oligomerisation of the E3 ligase DCAF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1
DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 and its ability to form an MG-induced complex with the substrate protein RBM39.
DDB1 and its ability to form an MG-induced complex with the substrate protein RBM39.
Materials and methods
Proteins were provided by Triana Biomedicines Inc, and details of their expression and purification are given in the ESI.†Preparation of samples for nMS
AmAc solutions were prepared from ultra-pure water (18.2 MΩ cm, Millipore) and analytical grade AmAc solid (Fisher Scientific, Loughborough, Leicestershire, UK). Proteins were dialysed into 200 mM AmAc using 96-well Microdialysis plates (Thermo Fisher Scientific, Waltham, MA USA). The protein concentrations were then measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific Waltham, MA USA) using the A280 method, and subsequently diluted to a protein concentration of 20 μM with 200 mM AmAc. To analyse ternary complex formation, an E3 and POI were then combined 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to give a mixture consisting of 10 μM E3 and 10 μM POI in 200 mM AmAc. 20 mM MG (WuXi AppTec Co., Ltd, Shanghai, China) in 100% DMSO (Sigma-Aldrich, St Louis, MO, USA) was diluted to 200 μM in 1% DMSO using deionised water. MG in 1% DMSO was combined 1
1 to give a mixture consisting of 10 μM E3 and 10 μM POI in 200 mM AmAc. 20 mM MG (WuXi AppTec Co., Ltd, Shanghai, China) in 100% DMSO (Sigma-Aldrich, St Louis, MO, USA) was diluted to 200 μM in 1% DMSO using deionised water. MG in 1% DMSO was combined 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with the 10 μM E3 + 10 μM POI in 200 mM AmAc mixture to give final analytical concentrations of 5 μM E3, 5 μM POI, 100 μM MG in 100 mM AmAc, 0.5% DMSO. Samples were diluted to analytical concentration the day of analysis. For samples involving only one protein, it was added to an equivalent volume of 200 mM AmAc before being added to the MG. For samples involving no MG, 1% DMSO was added to the proteins to ensure equivalent DMSO concentrations. For samples involving lower amounts of MG, the MG was diluted to 2× the working concentration with 1% DMSO prior to being mixed with the protein, also to ensure equivalent DMSO concentrations. For samples in 500 mM and 250 mM AmAc, proteins were initially dialysed into and diluted with 1 M or 500 mM AmAc, respectively, prior to being added to the MG.
1 with the 10 μM E3 + 10 μM POI in 200 mM AmAc mixture to give final analytical concentrations of 5 μM E3, 5 μM POI, 100 μM MG in 100 mM AmAc, 0.5% DMSO. Samples were diluted to analytical concentration the day of analysis. For samples involving only one protein, it was added to an equivalent volume of 200 mM AmAc before being added to the MG. For samples involving no MG, 1% DMSO was added to the proteins to ensure equivalent DMSO concentrations. For samples involving lower amounts of MG, the MG was diluted to 2× the working concentration with 1% DMSO prior to being mixed with the protein, also to ensure equivalent DMSO concentrations. For samples in 500 mM and 250 mM AmAc, proteins were initially dialysed into and diluted with 1 M or 500 mM AmAc, respectively, prior to being added to the MG.
nMS
Measurements were carried out using a Waters Synapt G2-Si mass spectrometer (Waters Corporation, Manchester, UK) equipped with a nano-electrospray ionisation source. Nano electrospray tips were pulled in house from thin-walled borosilicate glass capillaries (i.d. 0.78 mm, o.d. 1.0 mm) (Sutter Instrument Co., Novato, CA, USA) using a flaming/brown micropipette puller (Sutter Instrument Co., Novato, CA, USA). A positive potential of 0.9–1.5 kV was applied to the solution using a thin platinum wire (d. 0.125 mm) (Goodfellow, Huntingdon, UK). Other non-default instrument settings include: sampling cone voltage 60–200 V, source offset voltage 80–150 V, collision voltage 2–4 V, trap gas flow 4–5 ml min−1, source temperature 40 °C. Data shown in Fig. 2, 3(A, B) and 4(A, C, D) are representative spectra of experiments that were repeated on a minimum of two separate days. Data shown in Fig. 3C and 4B were one-off experiments. | ||
Fig. 2 (A) GSPT1 + CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 (B) GSPT1 + CRBN DDB1 (B) GSPT1 + CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 + lenalidomide. Protein concentrations are 5 μM and lenalidomide is 100 μM when present. Measured masses of the labelled species are as follows: GSPT1 24 DDB1 + lenalidomide. Protein concentrations are 5 μM and lenalidomide is 100 μM when present. Measured masses of the labelled species are as follows: GSPT1 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 Da. DDB1 93 058 Da. DDB1 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 247 Da. CRBN 247 Da. CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 139 DDB1 139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 775 Da. Ternary complex 164 775 Da. Ternary complex 164![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 090 Da. Masses given to the nearest 1 Da, and further details are given in Table S2.† 090 Da. Masses given to the nearest 1 Da, and further details are given in Table S2.† | ||
 | ||
Fig. 3 (A) RBM39 + DCAF15 complex (DCF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1 DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1). (B) RBM39 + DCAF15 complex + E7820. (C) DCAF15 complex + E7820. Protein concentrations are 5 μM and E7820 is 100 μM when present. Measured masses of the labelled species are as follows: RBM39 12 DDB1). (B) RBM39 + DCAF15 complex + E7820. (C) DCAF15 complex + E7820. Protein concentrations are 5 μM and E7820 is 100 μM when present. Measured masses of the labelled species are as follows: RBM39 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 978.0 Da. DDB1 93 978.0 Da. DDB1 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 247 Da. DDA1 247 Da. DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 104 DDB1 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 948 Da. DCAF15 complex (monomer) 162 948 Da. DCAF15 complex (monomer) 162![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 392 Da. DCAF15 complex (dimer) 324 392 Da. DCAF15 complex (dimer) 324![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 792 Da. DCAF15 complex (trimer) 487 792 Da. DCAF15 complex (trimer) 487![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 208 Da. Ternary complex 175 208 Da. Ternary complex 175![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 725 Da. DCAF15 complex (dimer) + 2xE7820 325 725 Da. DCAF15 complex (dimer) + 2xE7820 325![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 463 Da. DCAF15 complex (trimer) + 3xE7820 488 463 Da. DCAF15 complex (trimer) + 3xE7820 488![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 219 Da. Masses given to the nearest 1 Da, and further details are given in Table S3.† 219 Da. Masses given to the nearest 1 Da, and further details are given in Table S3.† | ||
Size exclusion chromatography
Experiments were carried out using a Waters Alliance HPLC system (Waters Corporation, Manchester, UK) equipped with a Waters 176003596 XBridge BEH200 SEC 3.5 μm 7.8 × 300 column (Waters Corporation, Manchester, UK). Samples were run at a flow rate of 1 mL min−1 using 100 mM AmAc containing 0.5% DMSO (Fig. S8†) or 50 mM HEPES, 0.5 mM TCEP, pH 7.4 containing NaCl at concentrations of 20 mM or 200 mM (Fig. S9†). The column was equilibrated with the corresponding solution prior to each experiment. Concentrations in Fig. S8(A), (C), and (D)† are DCAF15 complex (1 μM), RBM39 (1 μM), and E7820 (20 μM) when present, and in Fig. S8(B),† RBM39 (5 μM). Concentrations were altered due to column loading constraints, and the injection volume was 100 μL. DCAF15 complex concentration in HEPES buffer is 1 μM per 50 μL injection volume. Rationale for peak assignments is given in Fig. S8,† as the SEC was performed as a standalone experiment, separate from nMS.Data processing
Mass spectrometry data was processed using MassLynx (Version 4.2, Waters Corporation, Manchester, UK) and mass spectra were plotted using OriginPro (Version 2022, OriginLab Corporation, Northampton, MA, USA). Figures were created using Inkscape (Version 1.2.1, Inkscape.org) and the molecular glue structures in Table S1† were generated using Chemdraw Professional (Version 20.0.0.41, PerkinElmer Informatics Inc., Waltham, MA, USA).Results
nMS identifies MG-mediated interactions between CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) DDB1 and GSPT1
DDB1 and GSPT1
We first sought to investigate the complexes formed by the MGs lenalidomide, thalidomide and pomalidomide with the CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 E3 ligase and GSPT1. CRBN
DDB1 E3 ligase and GSPT1. CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 and GSPT1 (5 μM each) were analysed in a mixture from 100 mM AmAc in the absence and presence of the MG lenalidomide (Fig. 2A and B, respectively). AmAc is the most popular solvent in nMS as it is volatile and provides the required pH (6.8) for native protein analysis.24 In the absence of MGs, no interactions are observed between GSPT1 and CRBN
DDB1 and GSPT1 (5 μM each) were analysed in a mixture from 100 mM AmAc in the absence and presence of the MG lenalidomide (Fig. 2A and B, respectively). AmAc is the most popular solvent in nMS as it is volatile and provides the required pH (6.8) for native protein analysis.24 In the absence of MGs, no interactions are observed between GSPT1 and CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 (Fig. 2A). Here, CRBN
DDB1 (Fig. 2A). Here, CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 presents in charge states 17+ to 23+, monomeric DDB1 presents in charge states 14+ to 19+ and the POI GSPT1 presents in three charge states, from 8+ to 10+. GSPT1 is of higher signal intensity than DDB1 or the CRBN
DDB1 presents in charge states 17+ to 23+, monomeric DDB1 presents in charge states 14+ to 19+ and the POI GSPT1 presents in three charge states, from 8+ to 10+. GSPT1 is of higher signal intensity than DDB1 or the CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 complex, which is likely due to a higher ionisation efficiency because of its smaller size. Upon the addition of lenalidomide at 100 μM (Fig. 2B), new peaks corresponding to the E3
DDB1 complex, which is likely due to a higher ionisation efficiency because of its smaller size. Upon the addition of lenalidomide at 100 μM (Fig. 2B), new peaks corresponding to the E3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MG
MG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POI complex can be observed in six charge states from 20+ to 25+. The same is observed upon the addition of additional MGs thalidomide and pomalidomide (Fig. S1†), and all results are in agreement with those obtained by Huang et al.31 Changes in relative signal intensity are due to a reduction in free CRBN
POI complex can be observed in six charge states from 20+ to 25+. The same is observed upon the addition of additional MGs thalidomide and pomalidomide (Fig. S1†), and all results are in agreement with those obtained by Huang et al.31 Changes in relative signal intensity are due to a reduction in free CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 amounts as it becomes incorporated into the ternary complex, which is less apparent for GSPT1 due to the aforementioned higher ionisation efficiency.
DDB1 amounts as it becomes incorporated into the ternary complex, which is less apparent for GSPT1 due to the aforementioned higher ionisation efficiency.
Lenalidomide was introduced to the E3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) POI mixture at lower concentrations of 50 μM and 5 μM, and ternary complex is still observed in both cases, albeit at a low relative intensity (Fig. S2†). No peaks corresponding to binary species POI
POI mixture at lower concentrations of 50 μM and 5 μM, and ternary complex is still observed in both cases, albeit at a low relative intensity (Fig. S2†). No peaks corresponding to binary species POI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MG were observed in the mixture of all three components (Fig. 2, S1 and S2†). The width of the nMS peaks corresponding to CRBN
MG were observed in the mixture of all three components (Fig. 2, S1 and S2†). The width of the nMS peaks corresponding to CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 do not allow to definitively rule out its binding to lenalidomide as a binary complex (Fig. S3†), but it can be assigned that at least some portion of the E3 ligase remains in its unbound form. Immunomodulatory drugs have previously been shown to bind CRBN with weak KDs of 10–65 μM, which we would not expect to observe with nMS.32 The measured vs. expected mass of all proteins used in this study is given in Table S2.†
DDB1 do not allow to definitively rule out its binding to lenalidomide as a binary complex (Fig. S3†), but it can be assigned that at least some portion of the E3 ligase remains in its unbound form. Immunomodulatory drugs have previously been shown to bind CRBN with weak KDs of 10–65 μM, which we would not expect to observe with nMS.32 The measured vs. expected mass of all proteins used in this study is given in Table S2.†
The DCAF15 complex forms dimers and trimers in the absence of MG and POI
We next turned our attention to the DCAF15-targeting MG E7820 (Fig. 3). The DCAF15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDA1
DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 complex was used, lacking the proline-rich, atrophin-homology domain of DCAF15 (amino acids 276–383) which has been used in previous studies, and we refer to here as the DCAF15 complex.17 The POI used is the RRM2 domain of RBM39, which we refer to as RBM39 hereafter. In this case, the control experiment containing RBM39 (4+ to 6+) and the DCAF15 complex in the absence of MG yielded extremely surprising results. Monomeric DCAF15 complex (charge states 19+ to 24+) is only observed to a very low extent, and most of the signal intensity of this species corresponds to dimers and trimers of the DCAF15 complex (Fig. 3A). The dimer of the DCAF15 complex presents in nine charge states from 29+ to 37+ and the trimer also presents in nine charge states, 36+ to 44+. Unbound DDB1 is also present in charge states 14+ to 18+, and the DDA1
DDB1 complex was used, lacking the proline-rich, atrophin-homology domain of DCAF15 (amino acids 276–383) which has been used in previous studies, and we refer to here as the DCAF15 complex.17 The POI used is the RRM2 domain of RBM39, which we refer to as RBM39 hereafter. In this case, the control experiment containing RBM39 (4+ to 6+) and the DCAF15 complex in the absence of MG yielded extremely surprising results. Monomeric DCAF15 complex (charge states 19+ to 24+) is only observed to a very low extent, and most of the signal intensity of this species corresponds to dimers and trimers of the DCAF15 complex (Fig. 3A). The dimer of the DCAF15 complex presents in nine charge states from 29+ to 37+ and the trimer also presents in nine charge states, 36+ to 44+. Unbound DDB1 is also present in charge states 14+ to 18+, and the DDA1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 dimer is present in charge states 15+ to 17+.
DDB1 dimer is present in charge states 15+ to 17+.
To identify whether this multimerization is concentration dependent, the DCAF15 complex was analysed alone at concentrations of 5 μM and 2.5 μM (Fig. S4†) which yielded very similar complex distributions, suggesting that the formation of the dimers and trimers is not heavily dependent on concentration in this range that is suitable for nMS analysis. Upon close observation, peaks can also be identified corresponding to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex between the DCAF15 complex and RBM39, as indicated by asterisks in Fig. 3, S5 and S6.† This is in agreement with the literature, as RBM39 is known to be a native substrate of the DCAF15 complex, and a weak interaction of 4–6 μM has previously been measured between the two species.17
1 complex between the DCAF15 complex and RBM39, as indicated by asterisks in Fig. 3, S5 and S6.† This is in agreement with the literature, as RBM39 is known to be a native substrate of the DCAF15 complex, and a weak interaction of 4–6 μM has previously been measured between the two species.17
Upon addition of the MG E7820 (Fig. 3B) to the DCAF15 complex and the RBM39, a stoichiometric rearrangement of the DCAF15 complex occurs and the main signal intensity now corresponds to a single copy of the complex bound to E7820 and RBM39 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (20+ to 25+). Low amounts of dimeric DCAF15 complex remain present bound to two molecules of E7820 (Fig. 3B), and the trimeric complex is almost completely eradicated. The relative signal intensity of the ternary complex is higher than that of the unbound DCAF15 species, as the signal is now more monodisperse and spread over fewer separate species. To investigate whether this stoichiometric rearrangement of the DCAF15 complex is due to the MG or the POI, the DCAF15 complex and E7820 were sprayed together in the absence of RBM39. In this case, the DCAF15 complex is mainly observed as dimers bound to E7820 in a 2
1 stoichiometry (20+ to 25+). Low amounts of dimeric DCAF15 complex remain present bound to two molecules of E7820 (Fig. 3B), and the trimeric complex is almost completely eradicated. The relative signal intensity of the ternary complex is higher than that of the unbound DCAF15 species, as the signal is now more monodisperse and spread over fewer separate species. To investigate whether this stoichiometric rearrangement of the DCAF15 complex is due to the MG or the POI, the DCAF15 complex and E7820 were sprayed together in the absence of RBM39. In this case, the DCAF15 complex is mainly observed as dimers bound to E7820 in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry (Fig. 3C and S7†). Very low signal for the trimer of the DCAF15 complex is observed bound to three E7820 molecules (3
2 stoichiometry (Fig. 3C and S7†). Very low signal for the trimer of the DCAF15 complex is observed bound to three E7820 molecules (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 complex), but the intensity of the trimer is much lower than for the DCAF15 complex in the absence of MG (Fig. 3A and S4†). We therefore hypothesise either that the MG destabilises the DCAF15 complex trimer and causes preference for the dimer, or that the MG binds to the monomer and prevents DCAF15 complex self-assembly into trimers. No interaction was seen between the monomeric DCAF15 complex and E7820, which has been previously measured to have a KD > 50 μM.33 This is a weak interaction that would not be expected to be observed with nMS, but the fact that the 2
3 complex), but the intensity of the trimer is much lower than for the DCAF15 complex in the absence of MG (Fig. 3A and S4†). We therefore hypothesise either that the MG destabilises the DCAF15 complex trimer and causes preference for the dimer, or that the MG binds to the monomer and prevents DCAF15 complex self-assembly into trimers. No interaction was seen between the monomeric DCAF15 complex and E7820, which has been previously measured to have a KD > 50 μM.33 This is a weak interaction that would not be expected to be observed with nMS, but the fact that the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex is seen suggests that the MGs have a stronger interaction with the dimer than with the monomer (Fig. S7†). Equivalent data for the indisulam MG is shown in Fig. S5.†
2 complex is seen suggests that the MGs have a stronger interaction with the dimer than with the monomer (Fig. S7†). Equivalent data for the indisulam MG is shown in Fig. S5.†
As a complementary approach to nMS and to consolidate these findings, size exclusion chromatography (SEC) was employed to compare the interactions of the DCAF15 complex, RBM39 and E7820 (Fig. S8†). SEC was used as a standalone method, separate from the mass spectrometry experiments. SEC confirmed the presence of higher order oligomers of the DCAF15 complex when analysed alone or in the presence of RBM39 (without MG), as well as the formation of the ternary complex in the presence of both E7820 and RBM39.
Interactions of the DCAF15 complex are strongly regulated by salt concentration
To further investigate this unexpected oligomerisation of the DCAF15 complex, it was analysed from solutions of increased AmAc concentration to examine the effect of ionic strength on the interactions (Fig. 4). When ionised from a solution of 100 mM AmAc (Fig. 4A), the DCAF15 complex is present in a mixture of monomers, dimers and trimers, with the signal corresponding to the dimer being the most intense. Upon increasing the AmAc concentration of the starting solution to 250 mM (Fig. 4B), the majority of the signal corresponds to monomeric DCAF15, with a low amount of signal corresponding to the dimeric form (∼35% intensity relative to the monomer) and very little signal remaining for the trimer (marginally above baseline). Upon analysis from 500 mM AmAc, the signal corresponding to the dimeric DCAF15 complex is lower still (∼10% intensity relative to the monomer), and the signal corresponding to the trimer is abolished. This means that higher AmAc concentrations, and therefore higher ionic strength, shifts the DCAF15 complex from dimers and trimers towards the monomeric form. To determine whether the DCAF15 is still able to form a ternary complex in these conditions it was analysed in the presence of E7820 and RBM39 (Fig. 4D) which resulted in the formation of ternary complex. However, in these high salt conditions, there remains a portion of unbound DCAF15 complex, which is not present when analysed from the low salt solution (Fig. 3B). This implies that the higher ionic strength solution is disrupting interactions between the DCAF15 complexes, as well as with the RBM39 POI. The dependence on salt concentration suggests that the DCAF15 interactions to form homodimers and those required for ternary complex formation with E7820 and RBM39 are mainly electrostatic in nature. Indeed, the crystal structure presented by Du et al.17 confirms that specific interactions involved in the ternary complex are mainly hydrogen bonds and salt bridges. We therefore expect that the same types of interactions lead to multimerisation of the DCAF15 complex.To validate whether the oligomerisation of the DCAF15 complex at low salt concentration also occurs in common biological buffers, we performed SEC using HEPES buffer with low (20 mM) and high (200 mM) concentrations of NaCl (Fig. S9†). At high NaCl concentration, the major peak elutes at ∼13 mL, corresponding to a monomer of the DCAF15 complex. Some earlier eluting species are also present, indicating low amounts of self-association. Upon reduction of the NaCl concentration to 20 mM the main peak shifts to elute at ∼7 mL indicating no remaining monomeric form of the DCAF15 complex. These results therefore follow the same trend as changing the AmAc concentration, with the DCAF15 complex eluting earlier in the low salt buffer, indicating its multimerization.
This dissociation of DCAF15 multimers in response to high salt concentration is not a feature displayed across all proteins. In a study by Gavriilidou et al.,34 it was shown that increasing AmAc concentrations up to 500 mM promoted tetramer formation of Concanavalin A over the dimeric form. In this same study it was shown that protein–ligand interactions are affected differently by high AmAc concentration, with interactions increasing in affinity between lysozyme–NaG3 (tri-N-acetylchitotriose) and trypsin–pefabloc, and interactions decreasing in affinity between carbonic anhydrase II-chlorothiazide and β-lactoglobulin–lauric acid. Despite the importance of AmAc solutions in native mass spectrometry to preserve protein structure there remains little evidence in the literature regarding its optimum concentration. However, this significant change in the ability of the DCAF15 complex to multimerise and form ternary complexes may be a feature of the protein that contributes to its activity and regulation, perhaps in response to osmotic stress. For example, an increase in intracellular salt concentration as a result of dehydration could activate DCAF15, causing increased protein ubiquitination. The plausibility of this hypothesis may be uncovered during future research.
Conclusions
In summary, nMS has been demonstrated as an asset for the determination of MG ternary complex formation, successfully and clearly showing the presence of two ternary complexes between disease relevant proteins. It is a sensitive, fast, label-free technique requiring low sample consumption. This work has shown that nMS can show these complexes with intact proteins, in addition to the model peptides which have previously been used.21 nMS was able to directly show the existence of the MG ternary complexes, which will be beneficial in the screening of small molecule libraries for further glues that modulate this interaction.Our results are largely in agreement with a very recent paper by Huang et al.31 who interrogated the ability of nMS and mass photometry to determine ternary complex formation with MGs. The authors similarly reported that interactions between CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 and GSPT1 are stabilised by immunomodulatory drug MGs, and describe an elegant multiplexed experiment in which multiple POIs are pooled together and their recruitment to CRBN
DDB1 and GSPT1 are stabilised by immunomodulatory drug MGs, and describe an elegant multiplexed experiment in which multiple POIs are pooled together and their recruitment to CRBN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DDB1 by pomalidomide is simultaneously measured. The authors also report on the multimerization of the DCAF15 complex in 100 mM AmAc with both nMS and mass photometry, and nMS revealed that a single copy of the DCAF15 complex is incorporated into the MG ternary complex. Our work provides the additional important interrogation into the effect of salt concentration on DCAF15 self-association, and the effect that the salt concentration has on the propensity of the DCAF15 complex to form ternary complexes with MG and GSPT1.
DDB1 by pomalidomide is simultaneously measured. The authors also report on the multimerization of the DCAF15 complex in 100 mM AmAc with both nMS and mass photometry, and nMS revealed that a single copy of the DCAF15 complex is incorporated into the MG ternary complex. Our work provides the additional important interrogation into the effect of salt concentration on DCAF15 self-association, and the effect that the salt concentration has on the propensity of the DCAF15 complex to form ternary complexes with MG and GSPT1.
This ability of the DCAF15 complex to self-associate into dimers and trimers, which are disrupted either in the presence of (i) E7820/indisulam and RBM39 or (ii) higher salt concentrations, is an unexpected and important outcome of this study. As the intracellular concentrations of Na+ and K+ are ∼12 mM and ∼150 mM, respectively,35,36 it can be predicted that the DCAF15 complex is present as an equilibrium of monomers and multimers in physiological conditions. Despite the DCAF15 complex being extensively studied in structural biology due to its potential role in TPD, salt-dependent oligomerisation has not previously been reported, to our knowledge. Such findings are paramount in understanding E3 ligases for their manipulation in TPD, and we expect that analysis of complexes via nMS will eventually be routine in guiding drug design.
Author contributions
CJ collected and analysed the data and wrote the first draft of the manuscript. RB acquired the research funding, conceptualised and supervised the project and reviewed and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by Triana Biomedicines Inc, Waltham, MA, USA. RB acknowledges support of a UKRI Future Leaders Fellowship (Grant Reference MR/T020970/1) and the University of Strathclyde for a Chancellor's Fellowship (2020–2022). CJ is supported by an EPSRC studentship. AstraZeneca is thanked for providing CJ with a CASE top-up. The authors acknowledge the MVLS Structural Biology and Biophysical Characterisation Facility, University of Glasgow, for the SEC analysis.References
- D. E. Scott, et al., Small molecules, big targets: drug discovery faces the protein–protein interaction challenge, Nat. Rev. Drug Discovery, 2016, 15(8), 533–550 CrossRef CAS PubMed.
- M. J. Gorczynski, et al., Allosteric Inhibition of the Protein-Protein Interaction between the Leukemia-Associated Proteins Runx1 and CBFβ, Chem. Biol., 2007, 14(10), 1186–1197 CrossRef CAS PubMed.
- X. Yin, et al., Low molecular weight inhibitors of Myc–Max interaction and function, Oncogene, 2003, 22(40), 6151–6159 CrossRef CAS PubMed.
- L. Dietrich, et al., Cell Permeable Stapled Peptide Inhibitor of Wnt Signaling that Targets β-Catenin Protein-Protein Interactions, Cell Chem. Biol., 2017, 24(8), 958–968 CrossRef CAS PubMed.
- P. Thiel, M. Kaiser and C. Ottmann, Small-Molecule Stabilization of Protein–Protein Interactions: An Underestimated Concept in Drug Discovery?, Angew. Chem., Int. Ed., 2012, 51(9), 2012–2018 CrossRef CAS PubMed.
- Q. Yang, et al., E3 ubiquitin ligases: styles, structures and functions, Mol. Biomed., 2021, 2(1), 23 CrossRef PubMed.
- L. Buetow and D. T. Huang, Structural insights into the catalysis and regulation of E3 ubiquitin ligases, Nat. Rev. Mol. Cell Biol., 2016, 17(10), 626–642 CrossRef CAS PubMed.
- K. M. Sakamoto, et al., Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(15), 8554–8559 CrossRef CAS PubMed.
- M. Słabicki, et al., The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K, Nature, 2020, 585(7824), 293–297 CrossRef PubMed.
- J. Salami, et al., Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance, Commun. Biol., 2018, 1(1), 100 CrossRef PubMed.
- L. T. Kramer and X. Zhang, Expanding the landscape of E3 ligases for targeted protein degradation, Curr. Res. Chem. Biol., 2022, 2, 100020 CrossRef CAS.
- B. E. Smith, et al., Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase, Nat. Commun., 2019, 10(1), 131 CrossRef PubMed.
- M. S. Gadd, et al., Structural basis of PROTAC cooperative recognition for selective protein degradation, Nat. Chem. Biol., 2017, 13(5), 514–521 CrossRef CAS PubMed.
- M. J. Roy, et al., SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate, ACS Chem. Biol., 2019, 14(3), 361–368 CrossRef CAS PubMed.
- J. Yamamoto, et al., Discovery of CRBN as a target of thalidomide: a breakthrough for progress in the development of protein degraders, Chem. Soc. Rev., 2022, 51(15), 6234–6250 RSC.
- B. Milojkovic Kerklaan, et al., A phase I, dose escalation, pharmacodynamic, pharmacokinetic, and food-effect study of α2 integrin inhibitor E7820 in patients with advanced solid tumors, Invest. New Drugs, 2016, 34(3), 329–337 CrossRef CAS PubMed.
- X. Du, et al., Structural Basis and Kinetic Pathway of RBM39 Recruitment to DCAF15 by a Sulfonamide Molecular Glue E7820, Structure, 2019, 27(11), 1625–1633 CrossRef CAS PubMed.
- M. Karas, U. Bahr and T. Dülcks, Nano-electrospray ionization mass spectrometry: addressing analytical problems beyond routine, Fresenius’ J. Anal. Chem., 2000, 366(6–7), 669–676 CrossRef CAS PubMed.
- A. C. Leney and A. J. R. Heck, Native Mass Spectrometry: What is in the Name?, J. Am. Soc. Mass Spectrom., 2017, 28(1), 5–13 CrossRef CAS PubMed.
- R. Beveridge, et al., Mass spectrometry locates local and allosteric conformational changes that occur on cofactor binding, Nat. Commun., 2016, 7(1), 12163 CrossRef CAS PubMed.
- J. Bellamy-Carter, et al., Discovering protein–protein interaction stabilisers by native mass spectrometry, Chem. Sci., 2021, 12(32), 10724–10731 RSC.
- J. B. Fenn, et al., Electrospray Ionization for Mass Spectrometry of Large Biomolecules, Science, 1989, 246(4926), 64–71 CrossRef CAS PubMed.
- M. Wilm and M. Mann, Analytical Properties of the Nanoelectrospray Ion Source, Anal. Chem., 1996, 68(1), 1–8 CrossRef CAS PubMed.
- L. Konermann, Addressing a Common Misconception: Ammonium Acetate as Neutral pH “Buffer” for Native Electrospray Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2017, 28(9), 1827–1835 CrossRef CAS PubMed.
- M. Sharon and C. V. Robinson, The role of mass spectrometry in structure elucidation of dynamic protein complexes, Annu. Rev. Biochem., 2007, 76(1), 167–193 CrossRef CAS PubMed.
- M. Sugiyama, et al., Structural characterization of the circadian clock protein complex composed of KaiB and KaiC by inverse contrast-matching small-angle neutron scattering, Sci. Rep., 2016, 6(1), 35567 CrossRef CAS PubMed.
- J. Cveticanin, et al., Insight into the Autosomal-Dominant Inheritance Pattern of SOD1-Associated ALS from Native Mass Spectrometry, J. Mol. Biol., 2020, 432(23), 5995–6002 CrossRef CAS PubMed.
- R. Beveridge, et al., Native Mass Spectrometry Can Effectively Predict PROTAC Efficacy, ACS Cent. Sci., 2020, 6(7), 1223–1230 CrossRef CAS PubMed.
- C. J. A. Verhoef, et al., Tracking the mechanism of covalent molecular glue stabilization using native mass spectrometry, Chem. Sci., 2023, 14(24), 6756–6762 RSC.
- C. Jackson and R. Beveridge, Native Mass Spectrometry of Complexes Formed by Molecular Glues Revealed Stoichiometric Rearrangement of E3 Ligases, bioRxiv, 2023 DOI:10.1101/2023.02.03.526954.
- X. Huang, et al., Oligomeric Remodeling by Molecular Glues Revealed Using Native Mass Spectrometry and Mass Photometry, J. Am. Chem. Soc., 2023, 145(27), 14716–14726 CrossRef CAS PubMed.
- A. A. Akuffo, et al., Ligand-mediated protein degradation reveals functional conservation among sequence variants of the CUL4-type E3 ligase substrate receptor cereblon, J. Biol. Chem., 2018, 293(16), 6187–6200 CrossRef CAS PubMed.
- D. E. Bussiere, et al., Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex, Nat. Chem. Biol., 2020, 16(1), 15–23 CrossRef CAS PubMed.
- A. F. M. Gavriilidou, B. Gülbakan and R. Zenobi, Influence of Ammonium Acetate Concentration on Receptor–Ligand Binding Affinities Measured by Native Nano ESI-MS: A Systematic Study, Anal. Chem., 2015, 87(20), 10378–10384 CrossRef CAS PubMed.
- G. Madelin, et al., A method for estimating intracellular sodium concentration and extracellular volume fraction in brain in vivo using sodium magnetic resonance imaging, Sci. Rep., 2014, 4(1), 4763 CrossRef PubMed.
- M. Zacchia, et al., Potassium: From Physiology to Clinical Implications, Kidney Dis., 2016, 2(2), 72–79 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an00110a |
| This journal is © The Royal Society of Chemistry 2024 |