Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Standard addition method for rapid, cultivation-independent quantification of Legionella pneumophila cells by qPCR in biotrickling filters†
Gerhard
Schwaiger
a,
Marco
Matt
a,
Philipp
Streich
 a,
Sarah
Bromann
b,
Marcus
Clauß
b,
Martin
Elsner
a,
Sarah
Bromann
b,
Marcus
Clauß
b,
Martin
Elsner
 a and
Michael
Seidel
a and
Michael
Seidel
 *a
*a
aInstitute of Water Chemistry, Chair of Analytical Chemistry and Water Chemistry, Technical University of Munich, Lichtenbergstraße 4, 85748 Garching, Germany. E-mail: michael.seidel@mytum.de
bThuenen-Institute for Agricultural Technology, Bundesallee 47, D-38116 Braunschweig, Germany
First published on 5th April 2024
Abstract
Cultivation-independent molecular biological methods are essential to rapidly quantify pathogens like Legionella pneumophila (L. pneumophila) which is important to control aerosol-generating engineered water systems. A standard addition method was established to quantify L. pneumophila in the very complex matrix of process water and air of exhaust air purification systems in animal husbandry. Therefore, cryopreserved standards of viable L. pneumophila were spiked in air and water samples to calibrate the total bioanalytical process which includes cell lysis, DNA extraction, and qPCR. A standard addition algorithm was employed for qPCR to determine the initial concentration of L. pneumophila. In mineral water, the recovery rate of this approach (73%–134% within the concentration range of 100–5000 Legionella per mL) was in good agreement with numbers obtained from conventional genomic unit (GU) calibration with DNA standards. In air samples of biotrickling filters, in contrast, the conventional DNA standard approach resulted in a significant overestimation of up to 729%, whereas our standard addition gave a more realistic recovery of 131%. With this proof-of-principle study, we were able to show that the molecular biology-based standard addition approach is a suitable method to determine realistic concentrations of L. pneumophila in air and process water samples of biotrickling filter systems. Moreover, this quantification strategy is generally a promising method to quantify pathogens in challenging samples containing a complex microbiota and the classical GU approach used for qPCR leads to unreliable results.
Introduction
Legionella is a group of rod-shaped human pathogens found in various engineered water systems like hot water tanks,1,2 cooling towers3,4 and evaporative cooling systems.5 By law, Legionella prevalence must be monitored in these systems.6 There are around 60 known different species (spp.) of Legionella, while Legionella pneumophila (L. pneumophila) is responsible for over 90% of legionellosis diseases.7L. pneumophila can be divided into 16 serogroups (Sg) based on different lipopolysaccharide (LPS) structures on their membrane surface.8 According to the legislation, cultivation is the only accepted method for quantifying Legionella spp.6 However, the long time between sampling and obtaining results of 9–15 days,9 the potential for overgrowth of other microorganisms, and the presence of viable but non-culturable cells (VBNC)10 have increased the demand for culture-independent methods.11 These methods use either antigens on the cell membrane of bacteria or genomic DNA to quantify rapidly bacteria cells. The serotype-specific LPS of L. pneumophila can be used as an antigen for antibody-based methods like ELISA,12 lateral flow-immunoassays,13 or chemiluminescence sandwich microarray immunoassays.14 Immunomagnetic separation coupled with flow cytometry (IMS-FCM) is a technique that is able to quantify intact bacterial cells of either L. pneumophila Sg 1 or Sg 1–15, depending on the used panel of antibodies.15 The advantage of antibody-based methods is that no additional sample pretreatment is needed. Nevertheless, no information for Legionella spp. is gained with these methods.Molecular biological methods, such as quantitative polymerase chain reaction (qPCR) assays, are widely used and have a high sensitivity and the ability to simultaneously quantify both Legionella spp. and L. pneumophila.15 Several companies provide qPCR assay kits, including Biotecon, Bio-Rad, Promega, Minerva Biolabs, NZYTech, Creative Biogene and Thermo Scientific. The assay kits are all designed and validated according to the DIN ISO 12869:201916 guideline for the quantification of Legionella spp. with qPCR. According to this guideline, the quantification is based on Genomic Units (GU) standards which is stated to work in all types of water, except when the accompanying flora interferes with the determination.16 Environmental samples with a very high microbial load are a challenge in many ways. First, false positive results can be obtained when non-specific amplification of non-target DNA occur.17,18 Additionally, when using silica filters or magnetic beads for DNA extraction, a high total DNA amount can cause overloading, decreasing the efficiency.19,20 Another challenge are biochemical reagents that inhibit the polymerase and decrease the qPCR efficiency.21–24 All these factors, can cause unreliable results and since the cultivation approach achieves often false negative results due overgrowth, the results cannot be verified. In our study we show that agricultural exhaust air purification systems from pig fattening farms are one example where very strong matrices in air and process water occur. Due to their use of an active microbiome to degrade nitrogen and a circulating pH neutral wash water, they have a very high microbial load.
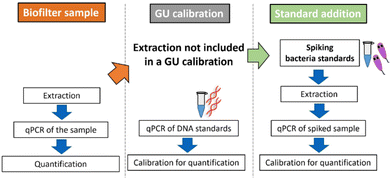
Quantification by qPCR is based on calibration with DNA standards provided by the manufacturers, expressed in genomic units. The cycle threshold (Ct) value obtained for the sample is then calculated, considering the dilution factors and sample volume used, and expressed as GU per mL. Since GU standards do not undergo sample preparation steps such as filtration, cell lysis, and DNA extraction, the quantitative results may lack trueness. Loss of DNA during extraction is inevitable. The extraction efficiency is around 20 to 90% and depends on the method used, sample type, and person who performs it.25–27 Simpler extraction methods can increase the DNA yield and assay sensitivity but also increase matrix effects.28,29 Even, when the extraction efficiency is known in a certain matrix such as mineral water, the transfer to other matrix is not satisfactory and the measured quantity value is not in the agreement of a true quantity value. Furthermore, microbiome-containing matrices of environmental samples are influenced by various factors such as sampling site, temperature, weather, and transportation-(time) until measurement, making the matrix effects never identical.30–32 Additionally, often no blank samples are available since ubiquitous bacteria like Legionella spp. are always present, and removing a single bacterium without influencing other microorganisms is impossible. All these factors lead to the conclusion that a GU calibration or a single calibration in a simulated or even real matrix always entails a certain degree of inaccuracy. Also the addition of DNA33,34 or non-target bacteria35,36 into the sample to estimate the efficiency of the qPCR is a known strategy, but cannot be directly compared with the efficiency of the target quantification. To account for all of the mentioned factors, we conclude that the inclusion of the entire sample preparation procedure can be favorable for calibration in environmental samples, treating the whole process as one combined bioanalytical method (Fig. 1). The addition of the viable target L. pneumophila to calibrate the total bioanalytical workflow is derived from a standard addition approach well known from instrumental analysis such as gas chromatography,37–39 atomic absorption spectroscopy,40 and isotope analysis.41,42 Standard addition is performed by spiking defined levels of the analyte of interest. Using linear regression, analyte concentrations in the original sample can be back-calculated. This requires a linear response behavior of the analytical method and the exact dosability of the analyte. For the quantification of Legionella spp. in environmental samples with qPCR, defined living L. pneumophila cryopreserved standards are added directly into it. The semilogarithmic calibration of qPCR shows linearity between Ct value and bacterial concentration only on a logarithmic scale and therefore adjustments need to be made to obtain quantitative results through the standard addition approach used. Therefore, a standard addition algorithm designed for immunoassays is applied to overcome this limitation.43
In the present work, for the first time to our knowledge, this standard addition approach for qPCR was used to overcome matrix effects in complex air and water samples of a biotrickling filter allowing a more reliable quantification than possible with commonly used GU standards. Since cultivation lead to false negative results, the IMS-FCM was used as complementary cultivation-independent method to confirm the results. While calibration with GU standards is faster and needs less laboratory work, the standard addition approach can be utilized to quantify pathogens in complex water or other environmental matrices, such as those with a high microbial load if quantification with GU standards leads to unreliable results. It also enables the use of simpler DNA extraction methods, thereby enhancing the sensitivity, accuracy and reliability of qPCR.
Experimental
Bacteria cryopreserved standards
Cryopreserved standards of viable L. pneumophila Sg 1 subtype Bellingham were prepared according to standardized method developed by rqmicro15 to ensure a stable and well-defined cell count in the spiking experiments. Two quality controls via flow cytometry, one after growing in media and the second after conditioning in mineral water, were performed. In each quality control, the Total Cell Count (TCC), the Total Legionella Count (TLC), and the Intact Legionella Count (ILC) were measured via flow cytometry on a rqmicro.COUNT device (rqmicro AG, Schlieren, Switzerland). The cryopreserved standards were only used if they were of sufficient purity (TLC/TCC > 90%) and the viability was sufficiently high (ILC/TLC > 60%). The cryo buffer consisted of 2× PBS (5.4 mM KCl, 20 mM Na2HPO4, 3.3 mM KH2PO4, 0.27 M NaCl, pH 7.0), 10 g L−1 BSA (Sigma Aldrich, St. Louis, USA) and 120 g L−1 dextran 40 (Sigma Aldrich, St. Louis, USA). The standards were stored at −80 °C and directly used after thawing at room temperature. Each day, a new cryopreserved standard was used from the same batch. The long-term storage and thawing stability of the cryopreserved standards was shown elsewhere by Streich et al.44 The initial concentration after 8 months was slightly reduced by 7.7 ± 1.2%. The TLC of the used cryopreserved standard was 6.7 × 107 bacteria per mL and the ILC was 3.4 × 107 bacteria per ml.Sample preparation
For calibration in mineral water, a dilution series of cryopreserved bacteria standards were performed in Evian water (1.5 L-bottles, Danone, Paris, France). 1 mL of sample was spiked with 10 μL of diluted standards. For extraction of environmental samples, 1 mL of the sample was directly used and spiked with 10 μL of standard solution. Samples and samples spiked with cryopreserved standards were centrifuged at 8000g for 5 min, followed by DNA extraction of a formed pellet using foodproof® StarPrep Two Kit Procedure B (Biotecon Diagnostics, Potsdam, Germany). Extraction works by a combination of mechanical lysis and heat. After lysis, samples were centrifuged, and supernatants (100 μL) were used for analysis. The DNA extracts were stored at 4 °C and used the next day.Quantitative polymerase amplification reaction
The qPCR Kit microproof® Legionella Quantification LyoKit (Biotecon, Potsdam, Germany) was used for all experiments. Measurements were performed on the qTOWER3G (Analytik Jena, Jena, Germany) instrumentation. 25 μL of the DNA extract was added to the qPCR reaction mix according to the manual delivered by Biotecon. A negative control (PCR-H2O) was added for every run. Thermal cycling conditions were chosen according to the given manual. First, a pre-incubation was performed (1 cycle) at 37 °C for 4 minutes and 95 °C for 5 minutes. Followed by 50 amplification cycles containing 3 steps. Step 1: 95 °C for 5 s, step 2: 60 °C for 60 s and step 3: 72 °C for 60 s. The fluorescence measurement was performed in step 2. Four different fluorescence channels were measured, Cy5 (internal control), HEX (Legionella spp.), FAM (L. pneumophila Sg 1–15), and ROX (L. pneumophila Sg 1). If one identical DNA extract was measured in replicates, it is indicated as a technical replicate. If, for each measurement, a different replicate of a DNA extract was measured, it is stated as a biological replicate. A negative control (PCR-grade H20) was always added to a qPCR run.Data evaluation
The evaluation sheet, based on the ISO 12869:2019, included in the qPCR Kit microproof® Legionella Quantification LyoKit (Biotecon Diagnostics, Potsdam, Germany) was used for GU calibration. According to this standard a limit of quantification of 130 cells per mL was achieved for Legionella spp., L. pneumophila and L. pneumophila Sg 1. A limit of detection of 15 cells per mL for Legionella spp. and 25 cells per mL for L. pneumophila and L. pneumophila Sg 1 was obtained. The published Excel-Solver sheet from Pang et al.43 developed for standard addition approaches for immunoassays was used to quantify L. pneumophila by qPCR. For adaption to qPCR, instead of signal values, medium cycle threshold (Ct) values of each spiking concentration were inserted in the sheet and an estimated concentration of the unspiked concentration was inserted as a starting point for the algorithmic-based calculation. Afterwards, the Excel-Solver algorithm was able to calculate the exact value.Measurements with IMS-FCM
L. pneumophila were counted by combining immunomagnetic separation with flow cytometry (IMS-FCM). All measurements were performed on the device rqmicro.COUNT (rqmicro, Schlieren, Switzerland). Intact Legionella counts (ILC) were measured by the Legionella kit L.p. SG1–15 DETECT Kit (31102) (rqmicro, Schlieren, Switzerland). The assay was performed as described by the manufacturer with the following changes. Instead of the enrichment of samples with filtration of 100 mL sample, 3 mL of the clean gas samples were filtrated through a prefilter (5 μm diameter pore size, 39002, rqmicro, Schlieren, Switzerland) and then caught on a sterile filter (0.22 μm diameter pore size, 39001, rqmicro, Schlieren, Switzerland). Only a prefiltration (5 μm diameter pore size) was performed for the process water samples. Without any further enrichment, the analysis was proceeded as described in the kit. To measure the total microbial load, the total cell count (TCC)-Kit (30010) (rqmicro, Schlieren, Switzerland) was used and performed according to the kit manual.Sampling at the biotrickling filter
At a fattening pig house with 2000 animals, the clean gas, and process water of an installed biotrickling filter were sampled. The cleaned gas was sampled at 3 different timepoints directly at the outlet of the unit. To protect the bioaerosol collector from wind, a tube (1 m deep, 1 m in diameter) was installed for the sampling. A Coriolis® μ device (Bertin Technologies SAS, Montigny-le-Bretonneux, France) was used as collection system. The collection vessels were filled with 15 ml sterile 0.9% NaCl solution. The sampling duration consisted of 3 rounds, each lasting 10 minutes, with a flow rate of 300 L min−1. Three individual samples were combined into one composite sample, corresponding to a total air volume of 9 m3 per sample. The collected total volume was measured and divided by the initial volume of 45 mL to get the evaporation factor of liquid during measurement. This factor was used when calculating the raw data from the measurements to the unit cells per m3. Two process water samples at 2 different timepoints were taken from the middle of the basin using a 500 mL PE bottle. All samples were transported in sealed bottles, kept in the dark, without active cooling, to the laboratory of the Thünen Institute. They were then directly sent via express delivery (arriving around 9 a.m. the following day) to our institute at TUM, where the measurements were conducted.Results and discussion
Spiking strategy for standard addition calibration in qPCR
For standard addition, samples with unknown concentrations of Legionella spp. or L. pneumophila must be spiked with a defined concentration of intact bacteria cells. This necessitates the use of a reliable bacterial standard with a well-defined concentration, requiring quality control measures. Therefore, a cryopreserved standard of L. pneumophila Sg 1 subtype Bellingham was applied for the standard addition approach shown in Fig. 2.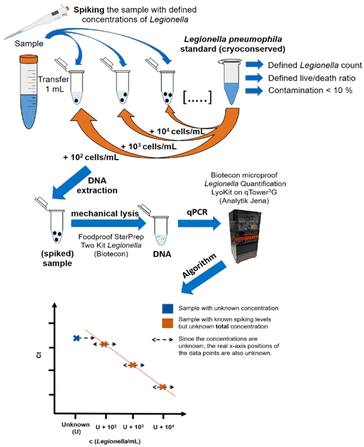 | ||
| Fig. 2 Workflow of the standard addition approach. 1 mL of the sample is spiked with different concentrations of a Legionella pneumophila Sg 1 subtype Bellingham bacteria standard to adjust the spiking concentrations. Afterwards, unspiked sample plus the spiked dilution series are extracted and measured identically according to the kit manual. For the quantification, an algorithm43 is used that is alternating the unknown concentration U until the residual sum of squares is minimal, resulting in the maximum linearity of the data points. | ||
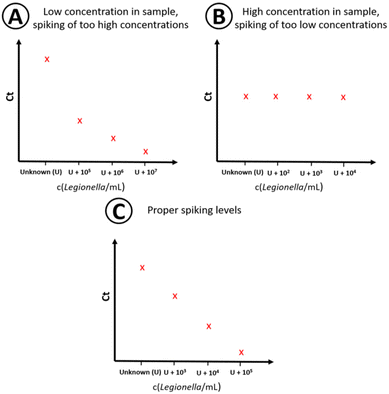
Since the concentration in the sample is unknown (U), the corresponding position on the x-axis of the plotted data points in the regression plot is unknown, too. The used algorithm by Pang et al.43 for the quantitative analysis utilizes a linear behavior on a logarithmic scale within the linear working range of the qPCR assay when U is close to the true value. U is changed by a numeric algorithm until the highest linearity is found. This means the residual variation from the calculated values of the regression line and the observed values is the smallest, R2 is the closest to 1. At least four different spiking concentrations in the sample are needed as data points for standard addition.43 To minimize sample volume, time and costs, the amount of data points should be limited as much as possible while still generating quantitative results. For that, the spiking levels must be chosen so that the total concentration (sample concentration + spiking concentration) is within the linear range of the method to generate a proper concentration dependency. Both too high (Fig. 3A) and too low (Fig. 3B) spike concentrations need to be avoided, since either the signal of the actual concentration in the sample is dwarfed by the standard addition, or no concentration dependency is visible due to the high initial concentration in the non-spiked sample (Fig. 3B). This is achieved by properly estimating the sample concentration by preliminary measurements and the knowledge of the linear working range of the assay (see Fig. 3C).
Proof-of-principle study
The qPCR with standard addition was initially tested using a sterile Evian mineral water matrix. Drinking water does not show significant influences on qPCR applications. It is a well-defined water matrix ideal for quantitative calibration studies of pathogens. In this experiment, Evian water was spiked with L. pneumophila Sg 1 subtype Bellingham at varying concentrations. Biological triplicates were conducted, where each was measured as a triplicate using qPCR. The spiked concentrations were calculated by standard addition and in comparison, a GU calibration was conducted according to the manual of the qPCR kit. For GU calibration, a calibration curve was constructed using four different levels of GUs, ranging from 25 to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 GU per reaction. The Ct values of the measurements were fitted for GU calibration using an evaluation Excel sheet based on the ISO 12869:2019 provided by the company Biotecon. 102–106 cells per mL were spiked into Evian water to perform the standard addition approach. The algorithm described in the Excel-Solver program by Pang et al.43 was applied to determine the unknown concentration of L. pneumophila. The calibration results are shown in Fig. 4. The summary of the data is shown in Table 1. For the standard addition, a linear behavior within the logarithmic scale is needed. In all 3 fluorescence channels, a high linearity of the data points was observed. For the Legionella spp. channel, a R2 of 0.998 was achieved. For the L. pneumophila and L. pneumophila Sg 1 channel a R2 of 0.999 was determined. Blank measurements (sterile mineral water with no added L. pneumophila Sg 1) showed no Ct values for the L. pneumophila primer set and very high Ct values for Legionella spp. and L. pneumophila Sg 1 primer set, indicating a minor carryover of DNA during DNA extraction or qPCR runs in this experiment. On average, both quantification principles slightly overestimated the true value with a mean of 139% (GU approach) and 135% (standard addition approach).
000 GU per reaction. The Ct values of the measurements were fitted for GU calibration using an evaluation Excel sheet based on the ISO 12869:2019 provided by the company Biotecon. 102–106 cells per mL were spiked into Evian water to perform the standard addition approach. The algorithm described in the Excel-Solver program by Pang et al.43 was applied to determine the unknown concentration of L. pneumophila. The calibration results are shown in Fig. 4. The summary of the data is shown in Table 1. For the standard addition, a linear behavior within the logarithmic scale is needed. In all 3 fluorescence channels, a high linearity of the data points was observed. For the Legionella spp. channel, a R2 of 0.998 was achieved. For the L. pneumophila and L. pneumophila Sg 1 channel a R2 of 0.999 was determined. Blank measurements (sterile mineral water with no added L. pneumophila Sg 1) showed no Ct values for the L. pneumophila primer set and very high Ct values for Legionella spp. and L. pneumophila Sg 1 primer set, indicating a minor carryover of DNA during DNA extraction or qPCR runs in this experiment. On average, both quantification principles slightly overestimated the true value with a mean of 139% (GU approach) and 135% (standard addition approach).
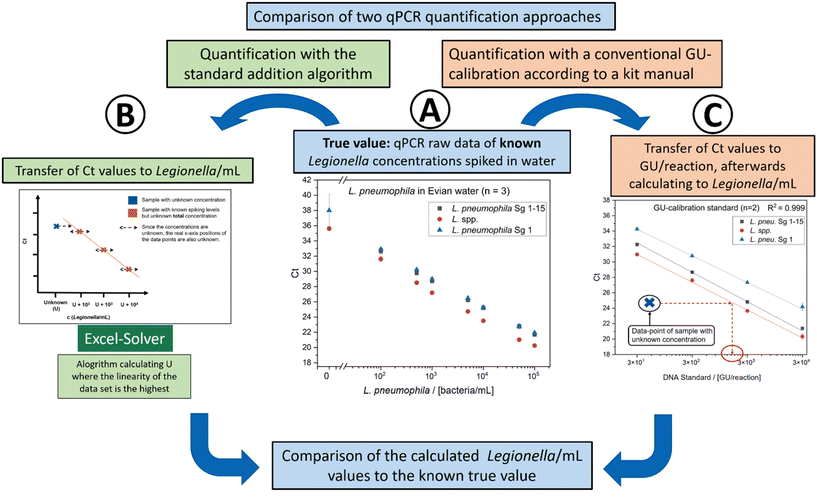 | ||
| Fig. 4 Calibration with different concentrations of L. pneumophila Sg 1 subtype Bellingham in Evian water. Three primer sets are included, detecting Legionella spp., L. pneumophila Sg 1–15 and L. pneumophila Sg 1 (number of measurements m = 8, number of biological replicates n = 3). (A) qPCR raw data. Ct values corresponding to the spiked L. pneumophila concentrations. This represents the known “true value” of bacteria in the Evian water. The known values are treated as unknown and were calculated with the standard addition approach (B) and the kit GU-calibration (C). (B) Quantification of the data points of (A) using the standard addition algorithm and the Excel-Solver program from Pang et al.43 As described previously, the algorithm is used to calculate U, resulting in the highest linearity of the plot, which gives the value of U. (C) Calibration of the Biotecon GU standard as described in the kit manual (m = 4, n = 2). The data points of A are fitted in the calibration to get the GU number of the data points and afterward transferred in cells per mL. | ||
| Spiked value in L. pneumophila Sg 1 cells per mL | Calculated value with standard addition in Legionella spp./mL (recovery) | Calculated value with GU-calibration in Legionella spp. cells per mL (recovery) |
|---|---|---|
| Legionella spp. | ||
| 0 | 18 | <15 |
| 100 | 106 (105%) | <130 |
| 500 | 567 (113%) | 630 (126%) |
| 1000 | 1222 (122%) | 1500 (150%) |
| 5000 | 3659 (73%) | 7100 (142%) |
| Spiked value in L. pneumophila Sg 1 cells per mL | Calculated value with standard addition in L. pneumophila Sg 1–15 cells per mL (recovery) | Calculated value with GU-calibration in L. pneumophila Sg 1–15 cells per mL (recovery) |
|---|---|---|
| L. pneumophila Sg 1–15 | ||
| 0 | ||
| 100 | 128 (128%) | <130 |
| 500 | 737 (147%) | 620 (124%) |
| 1000 | 1343 (134%) | 1200 (120%) |
| 5000 | 6603 (132%) | 5700 (114%) |
| Spiked value in L. pneumophila Sg 1 cells per mL | Calculated value with standard addition in L. pneumophila Sg 1 cells per mL (recovery) | Calculated value with GU-calibration in L. pneumophila Sg 1 cells per mL (recovery) |
|---|---|---|
| L. pneumophila Sg 1 | ||
| 0 | 11 | <15 |
| 100 | 138 (138%) | <130 |
| 500 | 606 (121%) | 620 (124%) |
| 1000 | 1198 (120%) | 1200 (120%) |
| 5000 | 3913 (78%) | 5700 (114%) |
Based on the data, this suggests that the standard addition approach yields comparable results in the drinking water matrix, where the GU calibration is considered as the gold standard. The approach demonstrates a recovery rate of 73%–134% within a concentration range of 100–5000 Legionella per mL (Table 1). All recovery values are within the acceptable range of −0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 unit and +0.3
log10 unit and +0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 unit defined in the ISO12869:2019,16 calculated by the decimal logarithm of the measured value divided by the true value. Consequently, it can be concluded that the standard addition method has been effectively adapted from the immunoassay approach proposed by Pang et al.43 to a qPCR method.
log10 unit defined in the ISO12869:2019,16 calculated by the decimal logarithm of the measured value divided by the true value. Consequently, it can be concluded that the standard addition method has been effectively adapted from the immunoassay approach proposed by Pang et al.43 to a qPCR method.
Quantification in complex matrices: a proof-of-principle study on a biotrickling filter
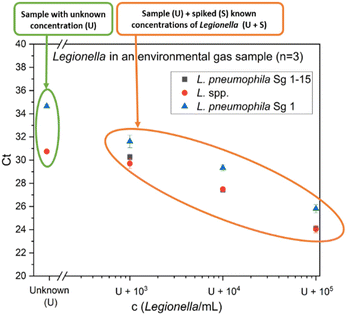
As an example for a challenging real matrix, environmental samples from agricultural biotrickling filters were used. Clean gas samples were collected with a cyclone sampler and the measurement with flow cytometry resulted in a high microbial load of 3.1 × 107 total cells per m3 (Fig. S1A†). For the analysis of Legionella using the qPCR standard addition method, spiking levels of 103–105 cells per mL L. pneumophila Sg 1 were chosen since a concentration in this range was expected in the sample. Fig. 5 presents the qPCR results of one gas sample (sample 3). 3 samples were analyzed and the results are summarized in Table 2. For all samples, L. species concentrations determined by the GU calibration are 292–380% higher than the results of the standard addition approach. Since no blank samples were available and the true concentration of Legionella spp. in the sample was not known, an orthogonal reference method was needed. Therefore, as a comparison to the qPCR results, the concentration of L. pneumophila was also determined by IMS-FCM (Fig. S1B†). Combining immunomagnetic separation with subsequent flow cytometry allows counting L. pneumophila labeled by a panel of antibodies specific for Sg 1–15. This system generates reliable results on drinking water45 and was shown to work also well on gas samples, sampled with the Coreolis μ15 as well as for process water of cooling towers.44 Prefiltration was conducted to remove larger particles and prevent clogging of the flow paths in the applied cartridge. Regarding our samples, the loss during prefiltration is expected to be minimal in clean gas samples and minor for process water samples, making it as a suitable reference method. The standard addition methods overestimate the FCM-IMS value (823 L. pneumophila cells per m3) for L. pneumophila Sg 1 by 131%. In contrast, the conventional GU calibration resulted in an overestimation of 729%. The standard addition approach is closer to the results of the IMS-FCM. The GU calibration has a relative overestimation when compared to both the standard addition method (557%) and IMS-FCM (729%). One plausible explanation is that the fluorescence channel, which measures the internal standard, is used as a correction factor for other fluorescence channels that detect the amplicons of different primer sets. If inhibitory effects exist in the internal standard channel, the measured value is corrected, leading to potential overestimation if the inhibition of the internal standard primer set differs from the analyte primer set. Another reason can be an increase in DNA extraction efficiency. While efficiency generally decreases with higher microbial load,46 a larger and stronger bacterial pellet forms during the initial centrifugation step when there is a higher amount of bacteria. The presence of substantial amounts of other cells and DNA may lead to a higher recovery of Legionella spp. DNA in biotrickling filter samples compared to water matrices, where the system is calibrated by the provider. This difference can result in overestimation. Furthermore, the GU-calibration initially provides the value per reaction, which then needs to be calculated based on the volume of DNA extract used in the qPCR reaction relative to the total DNA extract volume and the sample volume before extraction. All these mathematical corrections increase the likelihood of error propagation. In contrast, our alternative approach – the described qPCR with standard addition – takes into account the whole sample preparation process for calibration. No presumed correction factor is required for DNA extraction and importantly, the unit can be expressed in bacteria per mL, making it more comparable to other methods, such as cultivation, immunoassays and flow cytometric approaches.| Biotrickling filter clean gas sample | Calculated concentration according to standard addition | Calculated concentration according to GU-calibration |
|---|---|---|
| Sample 1 c(Legionella spp. cells per m3) | 2010 | 5875 |
| Sample 2 c(Legionella spp. cells per m3) | 1305 | 4000 |
| Sample 3 c(Legionella spp. cells per m3) | 9710 | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 850 |
| Sample 3 c(L. pneumophila cells per m3) | 1150 | 6410 |
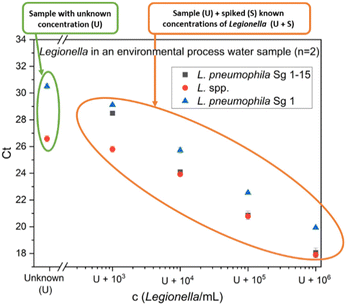
For a more in-depth analysis of the prevalence of Legionella spp. in biotrickling filters, process water samples were analyzed, posing an even more significant challenge. The process water, although initially fresh water, continuously accumulates deposited dust, ammonia, and organic substances during the operation of the system. This results in a cloudy (139 ± 9 NTU (Nephelometric Turbidity Unit)) yellow-brownish sample with a very high microbial load (TCC = 2.2 × 107 cells per mL, Fig. S2†). Due to the anticipated high concentrations of Legionella spp. in process water, samples were diluted at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 before extraction. Moreover, higher concentrations were used for spiking levels to prevent saturation effects. The qPCR results of one process water sample (sample 2) are presented in Fig. 6.
100 before extraction. Moreover, higher concentrations were used for spiking levels to prevent saturation effects. The qPCR results of one process water sample (sample 2) are presented in Fig. 6.
The concentration values of two different samples are summarized in Table 3. Once again, the qPCR results indicate higher quantities of L. pneumophila than to IMS-FCM (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 L. pneumophila cells per mL). When employing the standard addition approach, a smaller overestimation is observed in contrast to the GU calibration method. It is worth noting that alternative quantification method IMS-FCM may slightly underestimate the presence of L. pneumophila in process water due to prefiltration using a 5 μm pore-sized filter. This filtration step is implemented to prevent cartridge blockage and has been observed in previous experiments to retain some Legionella spp. that are bound to bigger particles or due to clump formation of the bacteria, resulting in a slight reduction in concentration within the filtered sample. Furthermore, also clump formation due a high bacteria concentration inside the cartridge during measurement can result in an underestimation. One bacteria aggregate containing more than one L. pneumophila can result in only one count. Nevertheless, the tendency that was shown in the gas samples, where the IMS-FCM is in better agreement with the SA-approach is also observed in process water. The GU calibration results in a factor of 3.9 to 11.9 higher Legionella spp. concentration than the SA approach. For L. pneumophila concentration, the GU calibration is 3.8 times higher than the SA approach.
740 L. pneumophila cells per mL). When employing the standard addition approach, a smaller overestimation is observed in contrast to the GU calibration method. It is worth noting that alternative quantification method IMS-FCM may slightly underestimate the presence of L. pneumophila in process water due to prefiltration using a 5 μm pore-sized filter. This filtration step is implemented to prevent cartridge blockage and has been observed in previous experiments to retain some Legionella spp. that are bound to bigger particles or due to clump formation of the bacteria, resulting in a slight reduction in concentration within the filtered sample. Furthermore, also clump formation due a high bacteria concentration inside the cartridge during measurement can result in an underestimation. One bacteria aggregate containing more than one L. pneumophila can result in only one count. Nevertheless, the tendency that was shown in the gas samples, where the IMS-FCM is in better agreement with the SA-approach is also observed in process water. The GU calibration results in a factor of 3.9 to 11.9 higher Legionella spp. concentration than the SA approach. For L. pneumophila concentration, the GU calibration is 3.8 times higher than the SA approach.
| Biotrickling filter process water | Calculated concentration according to standard addition | Calculated concentration according to GU-calibration |
|---|---|---|
| Sample 1 c(Legionella spp. cells per mL) | 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Sample 2 c(Legionella spp. cells per mL) | 257![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| Sample 2 c(L. pneumophila cells per mL) | 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
In summary, the standard addition approach proved to generate more reliable results in the biotrickling filter process water than the GU calibration, thus enhancing the analysis of these complex samples.
Conclusion
In this study a novel standard addition approach for qPCR was implemented for the first time, encompassing the entire sample preparation process. Cryopreserved L. pneumophila standards of known concentrations were used for spiking, enabling the generation of quantitative results in mineral water comparable to those obtained with a GU calibration. In contrast, when applied to biotrickling filter samples with high microbial loads, the conventional GU calibration yielded unreliable results. Here, the standard addition approach proved to be more reliable in generating accurate results. It could be demonstrated, that biotrickling filters are a potential source of Legionella spp. and the developed method is a valuable tool for further investigation of these systems. Furthermore, the qPCR approach enables the quantification of both, Legionella spp. and L. pneumophila, allowing for a more comprehensive analysis compared to immunoassays. It is important to acknowledge that this method entails higher costs and increased workload, making it less suitable for routine measurements with a large number of samples to analyze on a daily basis. However, it exhibits significant potential for analyzing complex and poorly characterized samples where the impact of the matrix on traditional calibration methods is unknown. Particularly in cases where the sample composition is unstable and subject to change, the standard addition approach can outperform classical calibration methods, providing more dependable results. This reliable qPCR method becomes particularly crucial when cultivating the target bacteria, as observed in biotrickling filters, is not feasible. Quantitative results are then unattainable through culture-based techniques and subsequent analyses such as MALDI-TOF are not possible. Other methods like digital PCR (dPCR) using absolute quantification is due to the separation in small reaction chambers less affected to matrix effects. This results in a more robust assay in comparison to classical qPCR approaches.47 Nevertheless, the availability of qPCR devices in most laboratories and accessibility of different kits on the market is still bigger than for dPCR. Additionally, the DNA extraction in dPCR has the same challenges as qPCR. The efficiency can highly depend on the matrix. While the quantification is different between qPCR and dPCR, our work can be adapted and be applied in the future for dPCR approaches to overcome matrix influences during DNA extraction by spiking living bacteria standards. For qPCR the SA addition approach can also enable the use of a simpler DNA extraction method resulting in higher yields and better sensitivity of the assay since it is less affected by matrix effects. Furthermore, if the cryopreserved standards are added directly after sampling, not only DNA extraction but also the transportation can be covered in the calibration. The strength of the standard addition approach lies in its versatility, allowing its application to a wide range of environmental analytical inquiries where the detection of microorganisms and consideration of matrix effects are essential factors.Author contributions
The manuscript was written through contributions of all authors. Gerhard Schwaiger planed and performed the experiments of the quantification with the help of Marco Matt. Gerhard Schwaiger evaluated the data and wrote the manuscript with the input from the co-authors. Philipp Streich characterized the cryopreserved bacteria standards. Sarah Bromann and Marcus Clauß were taking the samples. M. Elsner and M. Seidel supervised the project and were responsible for funding acquisition and resources. All authors have given approval to the final version of the manuscript.Notes
The AI tool “Grammarly” was used to improve the English written language.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the funding of the project by the BMEL. The authors would like to thank Analytik Jena for deployment of the qPCR device and rqmicro for their flow cytometry device rqmicro.COUNT.References
- F. Martinelli, A. Caruso, L. Moschini, A. Turano, C. Scarcella and F. Speziani, A comparison of Legionella pneumophila occurrence in hot water tanks and instantaneous devices in domestic, nosocomial, and community environments, Curr. Microbiol., 2000, 41(5), 374–376 CrossRef CAS PubMed.
- S. F. Dufresne, M. C. Locas, A. Duchesne, C. Restieri, J. Ismaïl, B. Lefebvre, A. C. Labbé, R. Dion, M. Plante and M. Laverdière, Sporadic Legionnaires’ Disease: The role of domestic electric hot-water tanks, Epidemiol. Infect., 2012, 140(1), 172–181 CrossRef CAS PubMed.
- V. A. Mouchtouri, G. Goutziana, J. Kremastinou and C. Hadjichristodoulou, Legionella species colonization in cooling towers: risk factors and assessment of control measures, Am. J. Infect. Control, 2010, 38(1), 50–55 CrossRef PubMed.
- K. Paranjape, É Bédard, L. G. Whyte, J. Ronholm, M. Prévost and S. P. Faucher, Presence of Legionella spp. in cooling towers: The role of microbial diversity, Pseudomonas, and continuous chlorine application, Water Res., 2020, 169, 115252 CrossRef CAS PubMed.
- B. Crook, L. Willerton, D. Smith, L. Wilson, V. Poran, J. Helps and P. McDermott, Legionella risk in evaporative cooling systems and underlying causes of associated breaches in health and safety compliance, Int. J. Hyg. Environ. Health, 2020, 224, 113425 CrossRef PubMed.
- Zweiundvierzigste Verordnung zur Durchführung des Bundes-Immissionsschutzgesetzes (42. BImSchV), BGBI. I p. 2379; 2018 I p. 202.
- T. Kuroki, Y. Watanabe, H. Teranishi, S. Izumiyama, J. Amemura-Maekawa and F. Kura, Legionella Prevalence and Risk of Legionellosis in Japanese Households, Epidemiol. Infect., 2017, 145(7), 1398–1408 CrossRef CAS PubMed.
- A. Katsiaflaka, S. Pournaras, I. Kristo, V. A. Mouchtouri, M. Kyritsi, E. Velonakis, A. C. Vatopoulos and C. Hadjichristodoulou, Epidemiological investigation of Legionella pneumophila serogroup 2 to 14 isolates from water samples by amplified fragment length polymorphism and sequence-based typing and detection of birulence traits, Appl. Environ. Microbiol., 2016, 82(20), 6102–6108 CrossRef CAS PubMed.
- Water Quality - Enumeration of Legionella (ISO 11731:2017); 2017.
- A. K. T. Kirschner, Determination of viable legionellae in engineered water systems: do we find what we are looking for?, Water Res., 2016, 93, 276–288 CrossRef CAS PubMed.
- K. Pancer and H. Stypułkowska-Misiurewicz, Pontiac fever - non-pneumonic legionellosis, Przegl. Epidemiol., 2003, 57(4), 607–612 Search PubMed.
- P. L. Elverdal, C. S. Jørgensen, K. A. Krogfelt and S. A. Uldum, Two years’ performance of an in-house ELISA for diagnosis of legionnaires’ disease: detection of specific IgM and IgG antibodies against Legionella pneumophila serogroup 1, 3 and 6 in human serum, J. Microbiol. Methods, 2013, 94(2), 94–97 CrossRef CAS PubMed.
- A. Y. W. Wong, A. T. A. Johnsson, A. Iversen, S. Athlin and V. Özenci, Evaluation of four lateral flow assays for the detection of legionella urinary antigen, Microorganisms, 2021, 9(3), 493 CrossRef PubMed.
- A. Wunderlich, C. Torggler, D. Elsässer, C. Lück, R. Niessner and M. Seidel, Rapid Quantification method for Legionella pneumophila in surface water, Anal. Bioanal. Chem., 2016, 408(9), 2203–2213 CrossRef CAS PubMed.
- L. Heining, L. Welp, A. Hugo, M. Lsner and M. Seidel, Immunomagnetic separation coupled with flow cytometry for the analysis of Legionella pneumophila in aerosols, Anal. Bioanal. Chem., 2023, 415, 5139–5149 CrossRef CAS PubMed.
- Water Quality—Detection and quantification of Legionella spp. and/or Legionella pneumophila by concentration and genic amplification by quantitative polymerase chain reaction (qPCR) (ISO/TS 12869:2019), 2019.
- J. J. Lahoz–Monfort, G. Guillera–Arroita and R. Tingley, Statistical approaches to account for false–positive errors in environmental DNA samples, Mol. Ecol. Resour., 2016, 16(3), 673–685 CrossRef PubMed.
- M. Sugimoto, J.-Y. Wu, S. Abudayyeh, J. Hoffman, H. Brahem, K. Al-Khatib, Y. Yamaoka and D. Y. Graham, Unreliability of results of PCR detection of Helicobacter Pylori in clinical or environmental samples, J. Clin. Microbiol., 2009, 47(3), 738–742 CrossRef PubMed.
- S. P. Van Tongeren, J. E. Degener and H. J. M. Harmsen, Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR, Eur. J. Clin. Microbiol. Infect. Dis., 2011, 30(9), 1053–1061 CrossRef CAS PubMed.
- K. Rudi, M. Kroken, O. J. Dahlberg, A. Deggerdal, K. S. Jakobsen and F. Larsen, Rapid, Universal Method to Isolate PCR-Ready DNA Using Magnetic Beads, BioTechniques, 1997, 22(3), 506–511 CrossRef CAS PubMed.
- R. F. Lance and X. Guan, Variation in inhibitor effects on qPCR assays and implications for eDNA surveys, Can. J. Fish. Aquat. Sci., 2020, 77(1), 23–33 CrossRef CAS.
- M. Sidstedt, P. Rådström and J. Hedman, PCR inhibition in qPCR, dPCR and MPS—mechanisms and solutions, Anal. Bioanal. Chem., 2020, 412(9), 2009–2023 CrossRef CAS PubMed.
- A. M. McKee, S. F. Spear and T. W. Pierson, The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples, Biol. Conserv., 2015, 183, 70–76 CrossRef.
- K. Uchii, H. Doi, T. Okahashi, I. Katano, H. Yamanaka, M. K. Sakata and T. Minamoto, Comparison of inhibition resistance among PCR reagents for detection and quantification of environmental DNA, Environ. DNA, 2019, 1(4), 359–367 CrossRef.
- B. M. Kemp, M. Winters, C. Monroe and J. L. Barta, How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16® system using the Qiagen MinElute purification kit, Hum. Biol., 2014, 86(4), 313–329 Search PubMed.
- L. I. Moreno, The effect of sample and sample matrix on DNA processing: mechanisms for the detection and management of inhibition in forensic samples, Doctor of Philosophy Chemistry, Florida International University, 2015 Search PubMed.
- C. Katevatis, A. Fan and C. M. Klapperich, Low concentration DNA extraction and recovery using a silica solid phase, PLoS One, 2017, 12(5), e0176848 CrossRef PubMed.
- L. M. Schiebelhut, S. S. Abboud, L. E. Gómez Daglio, H. F. Swift and M. N. Dawson, A comparison of DNA extraction methods for high–throughput DNA analyses, Mol. Ecol. Resour., 2017, 17(4), 721–729 CrossRef CAS PubMed.
- T. Särkinen, M. Staats, J. E. Richardson, R. S. Cowan and F. T. Bakker, How to ppen the treasure chest? optimising DNA extraction from herbarium specimens, PLoS One, 2012, 7(8), e43808 CrossRef PubMed.
- C. A. Evans, P. J. Coombes and R. H. Dunstan, Wind, rain and bacteria: The effect of weather on the microbial composition of roof-harvested rainwater, Water Res., 2006, 40(1), 37–44 CrossRef CAS PubMed.
- J. F. Griffith, K. C. Schiff, G. S. Lyon and J. A. Fuhrman, Microbiological Water Quality at Non-Human Influenced Reference Beaches in Southern California during Wet Weather, Mar. Pollut. Bull., 2010, 60(4), 500–508 CrossRef CAS PubMed.
- M. Zaninotto, A. Tasinato, A. Padoan, G. Vecchiato, A. Pinato, L. Sciacovelli and M. Plebani, Effects of sample transportation on commonly requested laboratory tests, Clin. Chem. Lab. Med., 2012, 50(10), 1755–1760 CAS.
- Y. Huang, X. Yin, C. Zhu, W. Wang, D. Grierson, C. Xu and K. Chen, Standard addition quantitative real-time PCR (SAQPCR): A novel approach for determination of transgene copy number avoiding PCR efficiency estimation, PLoS One, 2013, 8(1), e53489 CrossRef CAS PubMed.
- S. L. R. Ellison, K. R. Emslie and Z. Kassir, A standard additions method reduces inhibitor-induced bias in quantitative real-time PCR, Anal. Bioanal. Chem., 2011, 401(10), 3221–3227 CrossRef CAS PubMed.
- D. M. Stoeckel, E. A. Stelzer and L. K. Dick, Evaluation of two spike-and-recovery controls for assessment of extraction efficiency in microbial source tracking studies, Water Res., 2009, 43(19), 4820–4827 CrossRef CAS PubMed.
- E. Scarsella, A. Zecconi, M. Cintio and B. Stefanon, Characterization of microbiome on feces, blood and milk in dairy cows with different milk leucocyte pattern, Animals, 2021, 11(5), 1463 CrossRef PubMed.
- A. G. Garrido, J. L. M. Vidal, J. L. F. Moreno and R. Romero-González, Compensation for matrix effects in gas chromatography–tandem mass spectrometry using a single point standard addition, J. Chromatogr. A, 2009, 1216(23), 4798–4808 CrossRef PubMed.
- Y. Zhu, G. Li, Y. Duan, S. Chen, C. Zhang and Y. Li, Application of the standard addition method for the determination of acrylamide in heat-processed starchy foods by gas chromatography with electron capture detector, Food Chem., 2008, 109(4), 899–908 CrossRef CAS PubMed.
- S. Asthana, K. Karsauliya, S. Dixit, A. Tripathi, A. Kumar, S. P. Singh and M. Das, Development and validation of the ultra performance liquid chromatography-tandem mass spectrometer method for quantification of methylenecyclopropylglycine in litchi fruits using the standard addition method, Food Anal. Methods, 2019, 12(9), 2086–2093 CrossRef.
- F. M. Fortunato, M. A. Bechlin, J. A. G. Neto, G. L. Donati and B. T. Jones, Internal standard addition calibration: determination of calcium and magnesium by atomic absorption spectrometry, Microchem. J., 2015, 122, 63–69 CrossRef CAS.
- Z. Lu, J.-M. Zhu, D. Tan, T. M. Johnson and X. Wang, Double spike-standard addition technique and its application in measuring isotopes, Anal. Chem., 2023, 95(4), 2253–2259 CrossRef CAS PubMed.
- E. T. Tipper, P. Louvat, F. Capmas, A. Galy and J. Gaillardet, Accuracy of stable Mg and Ca isotope data obtained by MC-ICP-MS using the standard addition method, Chem. Geol., 2008, 257(1–2), 65–75 CrossRef CAS.
- S. Pang and S. Cowen, A generic standard additions based method to determine endogenous analyte concentrations by immunoassays to overcome complex biological matrix interference, Sci. Rep., 2017, 7(1), 17542 CrossRef PubMed.
- P. Streich, J. Redwitz, S. Walser-Reichenbach, C. Herr, M. Elsner and M. Seidel, Culture-independent quantification of Legionella pneumophila in evaporative cooling systems using immunomagnetic separation coupled with flow cytometry, Appl. Microbiol., 2024, 4, 284–296 CrossRef.
- H. P. Füchslin, S. Kötzsch, H.-A. Keserue and T. Egli, Rapid and quantitative detection of Legionella pneumophila applying immunomagnetic separation and flow cytometry, Cytometry, Part A, 2010, 77(3), 264–274 CrossRef PubMed.
- A. W. Luk, S. Beckmann and M. Manefield, Dependency of DNA extraction efficiency on cell concentration confounds molecular quantification of microorganisms in groundwater, FEMS Microbiol. Ecol., 2018, 94(10), fiy146 CrossRef CAS PubMed.
- L. Falzone, G. Gattuso, C. Lombardo, G. Lupo, C. Grillo, D. Spandidos, M. Libra and M. Salmeri, Droplet Digital PCR for the Detection and Monitoring of Legionella pneumophila, Int. J. Mol. Med., 2020, 46(5), 1777–1782 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: IMS-FCM and FCM data for the biofilter samples. Fig. S1 shows TLC and ILC dot plots of a clean gas sample. Fig. S2 shows TCC and ILC dot plots of a process water sample. See DOI: https://doi.org/10.1039/d3an02207b |
| This journal is © The Royal Society of Chemistry 2024 |