Open Access Article
Open Access ArticleThe arachidonic acid metabolome reveals elevation of prostaglandin E2 biosynthesis in colorectal cancer†
Cuiping
Zhang‡
 a,
Zuojian
Hu‡
a,
Zuojian
Hu‡
 be,
Ziyue
Pan
c,
Zhaodong
Ji
a,
Xinyi
Cao
a,
Hongxiu
Yu
*bd,
Xue
Qin
*e and
Ming
Guan
*a
be,
Ziyue
Pan
c,
Zhaodong
Ji
a,
Xinyi
Cao
a,
Hongxiu
Yu
*bd,
Xue
Qin
*e and
Ming
Guan
*a
aDepartment of Laboratory Medicine, Shanghai Medical College, Huashan Hospital, Fudan University, 200040, Shanghai, China. E-mail: guanming88@yahoo.com
bInstitutes of Biomedical Sciences, Fudan University, Shanghai, 200032, China
cShanghai Tongji Hospital Affiliated to Tongji University, Shanghai, China
dShanghai Stomatological Hospital & School of Stomatology, Fudan University, Shanghai, China
eDepartment of Clinical Laboratory, First Affiliated Hospital of Guangxi Medical University, Nanning 530021, China. E-mail: qinxue@gxmu.edu.cn
First published on 3rd February 2024
Abstract
Arachidonic acid metabolites are a family of bioactive lipids derived from membrane phospholipids. They are involved in cancer progression, but arachidonic acid metabolite profiles and their related biosynthetic pathways remain uncertain in colorectal cancer (CRC). To compare the arachidonic acid metabolite profiles between CRC patients and healthy controls, quantification was performed using a liquid chromatography-mass spectrometry-based analysis of serum and tissue samples. Metabolomics analysis delineated the distinct oxidized lipids in CRC patients and healthy controls. Prostaglandin (PGE2)-derived metabolites were increased, suggesting that the PGE2 biosynthetic pathway was upregulated in CRC. The qRT-PCR and immunohistochemistry analyses showed that the expression level of PGE2 synthases, the key protein of PGE2 biosynthesis, was upregulated in CRC and positively correlated with the CD68+ macrophage density and CRC development. Our study indicates that the PGE2 biosynthetic pathway is associated with macrophage infiltration and progression of CRC tumors.
Introduction
Colorectal cancer (CRC) is the third most prevalent type of cancer and the second leading cause of cancer-related deaths worldwide.1 Abnormal lipid metabolism contributes to different aspects of cancer development, such as cancer cell growth, apoptosis, proliferation, and differentiation.2 Arachidonic acid (AA) metabolites are 20-carbon bioactive lipids derived from the metabolism of polyunsaturated fatty acids (PUFAs). They can modulate various biological processes, including cell adhesion, proliferation, and migration, as well as angiogenesis and inflammatory responses.3 AA metabolism includes three major pathways: cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP) pathways. Prostanoids are the most important and predominant AA metabolites in humans and are involved in many physiological processes, such as the regulation of blood clotting, maintenance of renal homeostasis, modulation of immune responses, inhibition and stimulation of neurotransmitter release, and motility and protection of the gastrointestinal mucosa.4 Prostanoids, which are derived from the COX pathway, are also associated with many pathological conditions, including inflammation, cardiovascular disease, and various cancers.3 Among the prostanoids, prostaglandin E2 (PGE2) is the most well-known member of the PG family and the main mediator of inflammation.5 PGE2 is involved in pathological conditions, including colon, lung, breast, and head and neck cancers and is often associated with poor prognosis.6Several studies have indicated that tumor cells and the immune cells in the tumor microenvironment produce a broad spectrum of AA metabolites.7,8 Microsomal prostaglandin E synthase-1 (mPGES-1), microsomal prostaglandin E synthase-2 (mPGES-2) and cytosolic PGES (cPGES) are three known PGE2 synthases.9 mPGES-2 is encoded by the PTGES2 gene. The biological relevance of AA metabolism and the PGE2 biosynthetic pathway in CRC has not yet been fully established. Drugs aiming to inhibit PGE2 production by suppressing the COX activity, such as nonsteroidal anti-inflammatory drugs (NSAIDs), are commonly used as clinical therapies, but are associated with several adverse effects.10 Therefore, therapies targeting the specific downstream molecules of PGE2 signaling could be considered as a promising alternative approach.
In this study, we aimed to explore the AA metabolism in CRC. We examined the AA metabolite profiles of CRC patients’ serum and tissue samples using the liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) approach. Metabolomics analysis indicated that metabolites derived from the PGE2 synthetic pathway were elevated and associated with a poor prognosis in CRC patients. Furthermore, we used quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and immunohistochemistry (IHC) assays to validate the expression of mPGES-2 in CRC. Our data suggest that mPGES-2 is upregulated in colon adenocarcinoma and positively correlated with both tumor macrophage infiltration and disease progression. These findings may provide new ideas and directions for clinical targets for treating CRC.
Materials and methods
Sample collection
The serum samples of 83 participants (37 patients and 46 controls) were obtained from the First Affiliated Hospital of Guangxi Medical University between September 2020 and May 2021. According to the TNM classification, the number of patients with CRC in each stage was as follows: TNM I n = 8; TNM II n = 12; TNM III n = 10; TNM IV n = 7. The serum of the healthy controls was obtained from abandoned blood samples of apparently healthy individuals which were provided by the health examination center. The tissue samples of 16 patients were obtained from Huashan Hospital of Fudan University. The inclusion criteria of CRC patients are as follows: (1) the patients were diagnosed by colonoscopy and pathological examination, (2) none had received any preoperative cancer treatment, and (3) none had any other systemic disease, including severe metabolic diseases, hematological diseases, or other cancers. CRC stages are based on the Chinese Protocol of Diagnosis and Treatment of Colorectal Cancer (2020 edition) from the National Health Commission of the People's Republic of China.11 The tissue microarray (HColA150CS02) was purchased from Shanghai Outdo Biotech Company. It contains 71 colon cancer tissues and 71 adjacent noncancerous tissues along with each patient's gender, age, lymphatic metastasis status and clinical stage. The use of tissue microarray for research purposes was approved by the Ethics Committee of Shanghai Outdo Biotech Company (SHYJS-CP-1707009). The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (2019-KY-E-160) and the Ethics Committee of the Huashan Hospital of Fudan University approved this study (KY2020-1300). The study conformed to the ethical standards for medical research involving human subjects, as laid out in the 1964 Declaration of Helsinki and its later amendments. All experiments were performed in compliance with relevant laws and institutional guidelines and approved by the ethics committee at First Affiliated Hospital of Guangxi Medical University and Huashan Hospital of Fudan University. Written informed consent was obtained from the human participants of this study.Reagents
High-performance liquid chromatography grade methanol, ethanoic acid, and ethyl acetate were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Formic acid and butyl hydroxytoluene (BHT) were purchased from Sigma-Aldrich (St Louis, MO, USA). Isotope labeled internal standards (IS) were purchased from Cayman Chemical (Ann Arbor, MI, USA).Sample preparation
The metabolite extraction method was performed as previously reported.12,13 Briefly, for serum extraction, 100 μL of serum was mixed with a cocktail of IS in 550 μL of ice-cold methanol containing 0.01 mol L−1 BHT and vortexed for 2 minutes. If necessary, ultrasonic treatment can help full disaggregation. The IS solution contains an equal amount of internal standard (a combination of PGE2-d4, LTB4-d4, 14,15-DHET-d11, 13-HODE-d4, 12-HETE-d8, 11,12-EET-d11, and TXB2-d4). Then, the samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 minutes at 4 °C. The supernatant was mixed with 700 μL of water and 1 mL ethyl acetate and vortexed for 30 s. After centrifugation at 12
000g for 5 minutes at 4 °C. The supernatant was mixed with 700 μL of water and 1 mL ethyl acetate and vortexed for 30 s. After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes at 4 °C, the supernatant was transferred to another tube. The extraction process was repeated on the remaining sample. The supernatants obtained from the two extractions were combined and freeze-dried. The remaining residues were reconstituted in 50 μL methanol before LC-MS/MS analysis. For tissue metabolite extraction, a total of 0.5 g of frozen tissue was macerated and homogenized with a homogenizer, and then the extraction steps were followed as mentioned above.
000g for 10 minutes at 4 °C, the supernatant was transferred to another tube. The extraction process was repeated on the remaining sample. The supernatants obtained from the two extractions were combined and freeze-dried. The remaining residues were reconstituted in 50 μL methanol before LC-MS/MS analysis. For tissue metabolite extraction, a total of 0.5 g of frozen tissue was macerated and homogenized with a homogenizer, and then the extraction steps were followed as mentioned above.
LC-MS/MS analysis
LC-MS/MS analyses were performed with the LC-ESI-MS/MS system (the QTRAP 6500+ LC-MS/MS system, SCIEX, Framingham, MA, USA). Separation was performed using a C18 chromatographic column 250 mm × 4.6 mm, 5 μm (Milford Waters, MA, USA) at a temperature of 35 °C. The mobile phase A consisted of 0.05% formic acid in water, and the mobile phase B was acetonitrile. The injection volume was 5 μL. The effluent was connected to an ESI-triple quadrupole-linear ion trap (QTRAP)-MS. The negative ion ESI mode was used and the ESI source operation parameters were as follows: the source temperature was set at 350 °C and the ion spray voltage was −4200 V. Qualitative analysis of the sample metabolites was completed according to special ion transition from secondary spectrum information. Quantitative analysis of the metabolites was performed by normalizing the data from the ratio of the peak area between the endogenous metabolites and the labeled IS by using Skyline. The relative intensity of each metabolite was obtained. The optimized declustering potential (DP) and collision energy (CE) for individual multiple reaction monitoring (MRM) transitions were provided (Table S1†).Immunohistochemistry (IHC)
The formalin-fixed paraffin-embedded samples of CRC patients were sectioned and manufactured as a tissue chip array. The tissue chip was deparaffinized and rehydrated with an ethanol gradient solution. For antigen retrieval, the slides were incubated in 0.01 M citrate buffer (pH 6.0) for 10 minutes in a microwave oven. The endogenous peroxidase activity was blocked using a 3% hydrogen peroxide solution for 10 minutes at room temperature. After rinsing with distilled water and PBS, the slides were incubated overnight at 4 °C with primary antibodies. The primary antibodies and dilutions were as follows: anti-PTGES2/Gbf1 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Abcam, Cambridge, UK, ab300052) and anti-CD68 antibody (1
100, Abcam, Cambridge, UK, ab300052) and anti-CD68 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, DAKO, Carpinteria, CA, USA, M0876). The chip was washed in PBS three times and then incubated with a secondary antibody for 30 minutes at room temperature. Finally, the signal was developed using 3,3′-diaminobenzidine tetrahydrochloride (DAB) at room temperature, and then the slides were counterstained with hematoxylin.
500, DAKO, Carpinteria, CA, USA, M0876). The chip was washed in PBS three times and then incubated with a secondary antibody for 30 minutes at room temperature. Finally, the signal was developed using 3,3′-diaminobenzidine tetrahydrochloride (DAB) at room temperature, and then the slides were counterstained with hematoxylin.
qRT-PCR
Total RNA was isolated from frozen tissue samples using TRIzol (Invitrogen, Thermo Fisher Scientific) according to the manufacturer's instructions. cDNA was prepared with 500 ng of RNA using the PrimeScript RT master mix and SYBR premix Ex Taq (TaKaRa, Otsu, Japan). Real-time PCR was performed using a 7500 real-time PCR system (Applied Biosystems, USA). The expression levels of target genes were normalized to GAPDH and quantified by qRT-PCR. The sequences of the specific primers for GAPDH were 5′-TGATGACATCAAGAAGGTGG-3′ (forward) and 5′-TTACTCCTTGGAGGCCATGT-3′ (reverse). The sequences of the specific primers for PTGES2 were 5′-GAGAAAGCTCGCAACAACTAAAT-3′ (forward) and 5′-TCATGGCTGGGTAGTAGGT-3′ (reverse).Statistical analyses
The specific chromatogram peaks of metabolites were extracted using the Skyline software (MacCoss Lab, UW). Data analyses, including partial least squares discriminant analysis (PLS-DA), ROC analysis, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, correlation analysis, heat map and volcano plot, were performed using MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) and GraphPad software 8.0 version (GraphPad Software Inc, La Jolla, CA, USA). The analyses of mPTGES-2 expression in pan-cancer and CRC patients were performed in GEPIA 2 (gene expression profiling interactive analysis 2) (https://gepia2.cancer-pku.cn/). Data are expressed as mean ± standard deviation (SD). Student's t tests were used for comparisons between two groups. P < 0.05 (two-tailed) was considered statistically significant.Results
Serum arachidonic acid metabolite profiles of participants
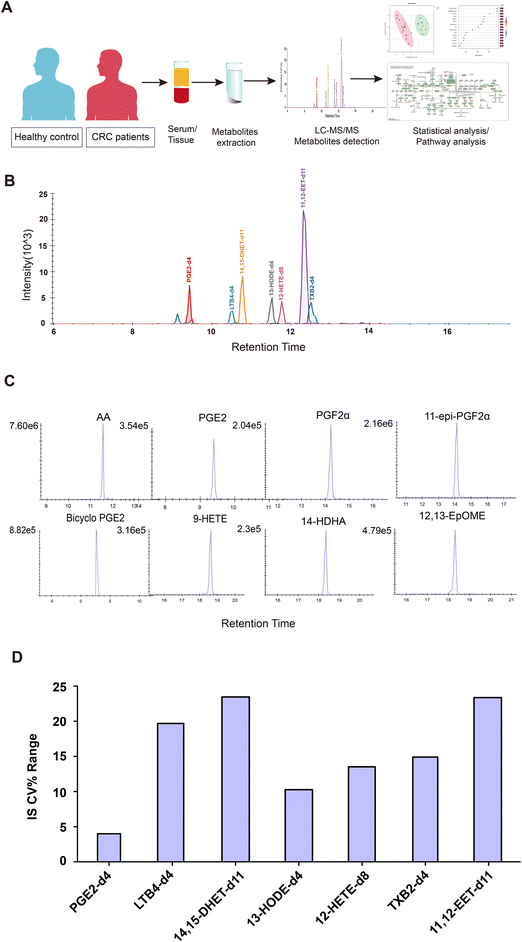
To identify the AA metabolites in CRC patient samples, we performed a targeted metabolomics analysis with LC-MS/MS. As depicted in the workflow (Fig. 1A), serum or tissue samples from patients with CRC and healthy controls were collected. Isotope labeled IS were added to the samples before metabolite extraction. Then, the metabolites in the samples were analyzed with an MRM-based LC-MS/MS method. With our MRM method, 98 oxidized lipids can be detected, including AA metabolites, as well as those of linoleic acid (LA) metabolism, unsaturated fatty acid biosynthesis, and glycerolipid metabolism. Seven isotope labeled IS can be completely separated using the LC conditions. The total ion chromatogram (TIC) and retention times for each IS are shown in Fig. 1B. The extraction ion chromatograms (XICs) of the selected AA metabolites in the measured samples are shown in Fig. 1C. We normalized the data from the ratio of the peak area between endogenous metabolites and labeled IS using Skyline, obtaining the relative intensity of each metabolite. Furthermore, we investigated the reproducibility of this method. The overall coefficient of variation (CV%) of each labeled IS in the six QC samples was less than 25% (Fig. 1D), demonstrating that this method had high reproducibility.Table 1 shows the clinicopathological features of the patients and healthy controls. There was no statistically significant difference in age or sex distribution between the CRC group and healthy controls. The CRC group consisted of 37 patients, while the healthy control group had 46 individuals. According to the TNM classification, the number of patients with CRC in each stage was as follows: TNM I n = 8; TNM II n = 12; TNM III, n = 10; TNM IV n = 7. According to pathological staging, the numbers were as follows: six cases with high differentiation, 28 cases with medium differentiation and three cases with low differentiation. We performed a targeted metabolomics analysis aimed at oxidized lipids using high-resolution MS. After excluding peaks with inconsistent retention times or low intensities (<3 signal/noise), a total of 48 oxidized lipids were detected in serum, as shown in the heatmap (Fig. 2A). To explore any differences in these oxidized lipids between CRC patients and healthy controls, we chose a fold change (FC) ≥1.5 or ≤0.67 and P values ≤0.05 as the screening criteria similar to previous studies.14–16 As shown in the volcano plot (Fig. 2B), the CRC group had 20 upregulated metabolites, including PGE2, prostaglandin F2α (PGF2α), leukotriene C4 (LTC4), leukotriene B4 (LTB4), and more, derived from AA metabolism and four downregulated metabolites, including 11-hydroxy-5Z,8Z,11E,14Z-eicosatetraenoic acid (11-HETE), resolvin D1, thromboxane B3 (TXB3) and 13,14-dihydro-15-keto PGF2a (PGFM). Next, a partial least squares discriminant analysis (PLS-DA) was performed to maximize variations between groups in oxidized lipid levels. The specific metabolites that contributed significantly to the variations were selected according to variable important in projection (VIP) scores (Fig. 2C and D). Metabolites were well separated between the healthy control and CRC groups, suggesting that oxidized lipids may be helpful for differentiating the case group from the control group (Fig. 2C). Among the top 15 metabolites (VIP scores >1) that contributed significantly to the variations, Maresin 1,9-hydroxy-10,12-octadecadienoic acid (9-HODE), (12R,13S)-(9Z)-12,13-epoxyoctadecenoic acid (12,13-EpOME), (10E,12Z)-9-oxooctadeca-10,12-dienoic acid (9-oxoODE), 9,10-epoxy-12-octadecenoic acid (9,10-EpOME), and PGE2 were upregulated significantly in the case group relative to the healthy control group, demonstrating that these metabolites could be considered as potential CRC biomarkers (Fig. 2D). These metabolite profiles delineate the differences in oxidized lipids in serum samples of CRC patients and healthy controls, suggesting that changes in AA metabolites may be related to CRC development.
| Characteristics | CRC patients | Healthy controls | P value |
|---|---|---|---|
| Total | 37 | 46 | |
| Sex | 0.25 | ||
| Male | 22 | 33 | |
| Female | 15 | 13 | |
| Age (years) | 0.36 | ||
| Mean | 58.86 | 56.80 | |
| Range | 29–75 | 45–90 | |
| TNM stage | |||
| I | 8 | ||
| II | 12 | ||
| III | 10 | ||
| IV | 7 | ||
| Pathological differentiation | |||
| L | 6 | ||
| M | 28 | ||
| H | 3 | ||
The AA metabolite profiles show that the PGE2 biosynthetic pathway is upregulated in CRC
Furthermore, to investigate the changes in oxidized lipid levels in CRC, we performed a targeted metabolomics analysis in CRC tissues (n = 16) and healthy tissues (n = 10) using the same workflow as shown in Fig. 1A. Our data show that nine metabolites were upregulated and seven metabolites were downregulated in the CRC tissues (Fig. 3A). Subsequently, we compared the changes in oxidized lipid profiles between CRC serum and CRC tissue samples, finding that the changes of five oxidized lipids were overlapping (Fig. 3B). Compared with the findings in healthy controls, PGE2, PGF2α and AA metabolites were upregulated and 11-HETE and resolvin D1 were downregulated in both CRC serum and tissue samples (Fig. 3C). Then, we examined these metabolites in serum with diagnostic efficacy analysis to assess their ability to predict CRC occurrence (Fig. 3D). The receiver operating characteristic (ROC) curves showed that PGE2 and resolvin D1 had the highest sensitivity values, 91% and 83%, respectively, for distinguishing CRC patients from healthy controls (area under curve, AUC = 0.767 and 0.839, respectively). In addition, 11-HETE and PGF2α differentiated patients with CRC from healthy controls with high specificity, 89% and 81%, respectively (AUC = 0.740 and 0.771, respectively). Based on previous studies17,18 and the KEGG database (https://www.kegg.jp), we performed the pathway analysis and drew a scheme of AA metabolism with the differential metabolite profiling in CRC (Fig. 3E). In the map of AA metabolism, the red words represent up-regulated metabolites in CRC, and the blue words represent down-regulated metabolites in CRC. The grey boxes represent that the metabolites can be detected by our method, while the metabolites in the outlined boxes were not measured with our current assay. The levels of PGE2-derived metabolites, like PGF2α, PGA2 (prostaglandin A2), and 15-keto-PGE2 (15-keto-prostaglandin E2), were significantly increased in CRC. These results collectively indicate that the PGE2 biosynthetic pathway was upregulated in the CRC samples.mPGES-2 is upregulated in colon adenocarcinoma
According to the oxidized lipidomic analysis, the AA-derived metabolites, including PGE2, PGF2α, PGA2, and 15-keto-PGE2, were increased in CRC. To further understand the PGE2 biosynthetic pathway in CRC, we investigated mPGES-2 expression in colon adenocarcinoma. Firstly, the pan-cancer analysis with GEPIA revealed that mPGES-2 expression levels were upregulated in CRC patient datasets (Fig. 4A). Additionally, the analysis of data from 275 CRC and 349 normal control samples from TCGA (the cancer genome atlas) and GTEx (genotype-tissue expression) databases showed that mPGES-2 levels were elevated in colon adenocarcinoma tissues compared with those in normal colon tissues (Fig. 4B). Results from the GEO (gene expression omnibus) database also showed that mPGES-2 expression levels were higher in CRC tissues than in the paired normal tissues (Fig. 4C). Then, we collected clinical CRC and adjacent normal tissue samples and performed qPCR and IHC analyses for validation. The results indicated that the mRNA levels of mPGES-2 were significantly higher in tumor tissues than in normal tissues (Fig. 4D). Regarding the IHC analysis of the tissue microarray, Table S2† shows the clinicopathological features of the patients. At the protein level, mPGES-2 expression was observed in the cytoplasm and cell membrane by IHC staining (Fig. 4E). Furthermore, the pathological score of mPGES-2 was significantly higher in the cytoplasm of the tumor cells than in the adjacent normal cells (Fig. 4F). These results revealed that mPGES-2, the key protein of the PGE2 synthetic pathway, is upregulated in CRC.Elevation of the PGE2 biosynthetic pathway is associated with macrophage infiltration and CRC progression
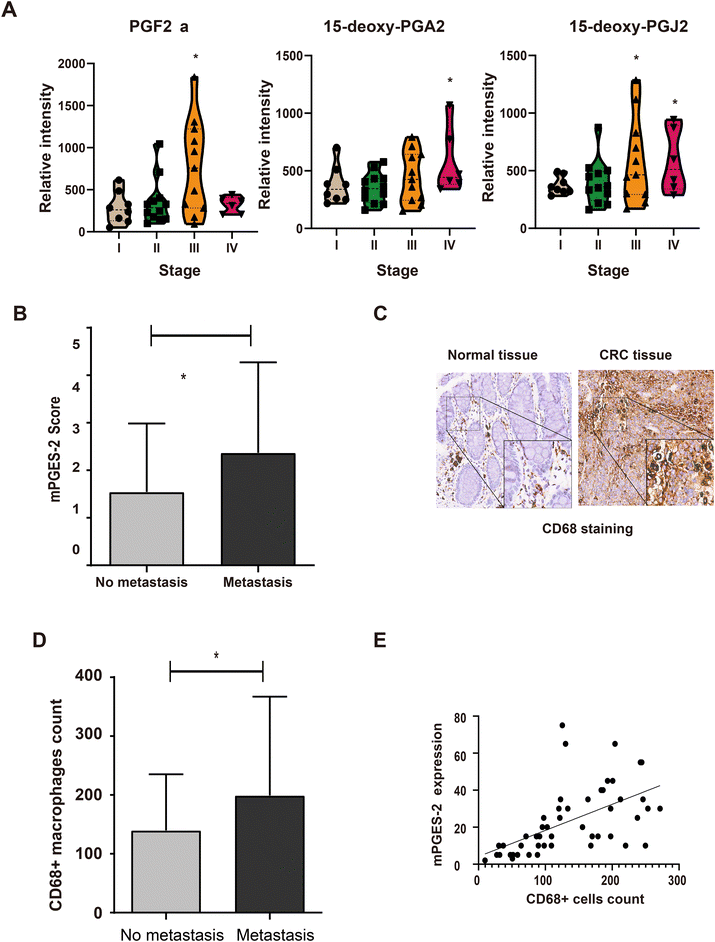
To explore the relationships between the PGE2 biosynthetic pathway and CRC progression, the PGE2-derived metabolites from patients at differential clinical stages were examined by LC-MS/MS. CRC patients with higher TNM stages (III and/or IV) had significantly higher levels of PGE2 biosynthetic pathway-associated metabolites (Fig. 5A). Moreover, higher mPGES-2 expression levels were observed in the CRC patients with metastasis than those without metastasis (Fig. 5B). Additionally, the CD68 protein expression was examined by IHC staining for the macrophage density. CD68+ macrophages were observed mostly in the tumor stroma of CRC tissue specimens (Fig. 5C). Interestingly, the CRC patients with metastasis had an elevated macrophage density in the tumor tissues compared with those without metastasis (Fig. 5D). Notably, in tumor stroma with high macrophage infiltration, mPGES-2 expression was elevated. A positive correlation was observed between the macrophage density and the mPGES-2 protein expression (linear regression r = 0.71, P < 0.001; Fig. 5E). These results indicated that elevation of the PGE2 biosynthetic pathway is associated with macrophage infiltration and CRC progression.Discussion
CRC progression is often accompanied by lipid metabolism reprogramming, such as the metabolism of fatty acids, phospholipids, and sphingolipids.2,12 The AA metabolic pathway involves a large group of oxidized lipids that participate in inflammatory and neuropsychiatric processes.19 In addition to being part of lipogenesis and decomposition processes, some oxidized lipids can also act as second messengers in the signaling pathway.20 In recent years, researchers have begun to focus on AA metabolism in tumor biological functions and immune regulation.21,22 In this study, we established a targeted metabolomics method for detecting AA metabolites using LC-MS/MS. We investigated the AA metabolite profiles of serum and tissue samples from CRC patients and healthy individuals, finding that the PGE2 biosynthetic pathway was upregulated in CRC and was positively associated with macrophage infiltration.Numerous assays have been used by other studies to quantitate AA metabolites. Immunoassays, including enzyme-linked immunosorbent assays (ELISAs) and radio immune assays (RIAs), have also been used for AA metabolite detection, but the specific antibodies required to quantitate each eicosanoid make them expensive and inefficient. These approaches also lack the ability to examine multiple metabolites simultaneously.23,24 GC-MS (gas chromatography-mass spectrometry) methods can largely overcome these deficiencies, but they have limitations. The volatility and thermal stability may not be the same for all analytes, which can affect the efficiency of chemical derivatization in the GC-MS methods.25 Over the last decade, LC-MS/MS for proteomics and metabolomics research has rapidly grown, with the associated higher throughput and sensitivity. In our study, an LC-MS/MS method was developed to profile 98 oxidized lipids within a 20-minute analysis time. We added an equal amount of seven isotopically labeled internal standards into each sample. Because it is difficult to obtain the isotope internal standard corresponding to each metabolite, we selected the 1–3 isotope internal standards corresponding to metabolites from each metabolic pathway of arachidonic acid for data normalization.18 For example, PGE2-d4 corresponds to COX pathway metabolites, 14,15-DHET-d11 and 11,12-EET-d11 correspond to CYP pathway metabolites, 13-HODE-d4, 12-HETE-d8, and TXB2-d4 correspond to LOX pathway metabolites, and LTB4-d4 corresponds to ALOX5 pathway metabolites.
We identified 48 oxidized lipids in CRC patient serum samples using the LC-MS/MS technology. The results showed that there were differences in the profiles of AA metabolites between the CRC group and the healthy control group, indicating that the AA metabolic process changed during the development. Among the oxidized lipids upregulated in the CRC patient serum, PGE2, PGF2α, and PGA2 were metabolized from AA through the COX pathway, while LTC4 and LTB4 were metabolized from the LOX pathway. These metabolites are all known as potential markers of lipid peroxidation and associated with metabolic diseases,26 inflammation, and tumors.27–29 11-HETE is an AA-derived oxidized lipid and is a marker of lipid peroxidation, but its biological mechanism is not clear yet. A previous study suggested that 11-HETE as an anti-inflammatory mediator and a peroxisome proliferator-activated receptor (PPAR) ligand could be implicated in tumor suppression.30–32 We also found that the resolvin D1 levels were significantly lower compared with those in the healthy controls. Resolvin D1 is a member of the specialized pro-resolving mediators and has been found to possess significant anti-inflammatory, anti-allergic, pro-hemolytic, pro-healing, and pain-relieving properties.33 Interestingly, the metabolites of PGE2, PGF2α and AA were upregulated, while 11-HETE and resolvin D1 were downregulated both in the CRC serum and tissue samples. It is known that AA can be converted to various lipid mediators through the enzyme-dependent pathway. Therefore, a link between the protein expression from tissue and the concentration of lipids from serum might exist. This could be affected by different clearance rates from blood for the various compounds and variations of clearance of these compounds among individuals.17 In our study, LA metabolites, including 13-hydroxyoctadecadienoic acid (13-HODE), 9-OxoODE, 9,10-EpOME, and 12,13-EpOME, were elevated in the CRC group. The EpOME metabolites may mediate inflammation and contribute to immunosuppression.34 A previous study observed upregulation of 12,13-EpOME in CRC, which is consistent with our findings. Moreover, the accumulation of 12,13-EpOME can promote the epithelial–mesenchymal transition and metastasis in CRC.35,36 Among the oxidized lipids downregulated in the CRC patient serum samples, TXB3 belongs to the ω-3 class of PUFAs, which reportedly have anti-CRC properties.37 We then performed ROC analyses of the screened metabolites in serum to determine their ability to predict the CRC occurrence. The AUC and diagnostic sensitivity values of PGE2 and PGF2α for distinguishing CRC patients from healthy controls were higher than those of the carbohydrate antigen 19-9 (CA19-9), which is a conventional tumor marker for clinical diagnosis of CRC.38,39 Previous studies have reported that PGE2 signaling can support an inflammatory microenvironment, which is required for early-stage tumorigenesis and can promote tumor invasion and metastasis.40–44 In summary, our metabolomics analysis and pathway analysis mapping from KEGG showed that PGE2-derived metabolites were significantly increased in the CRC samples, suggesting that the PGE2 biosynthetic pathway is upregulated in CRC.
Among inflammatory mediators, PGE2 plays an important role in the pathogenesis of a number of diseases, including kidney disease, rheumatoid arthritis, diabetes, and various cancers.45–48 mPGES-2, mPGES-1 and cPGES are three known PGE2 synthases.9 Expression of mPGES-1 is activated by proinflammatory stimulation. In contrast to the inducibility of mPGES-1, mPGES-2 is relatively constitutively expressed in cells and is not appreciably induced by inflammation or cell damage.47 Several previous studies have demonstrated that mPGES-2 was expressed specifically in human colorectal adenocarcinoma cell lines and tissues, with low levels of expression in other cells or tissues (brain, heart, liver, lungs).47–49 Consistent with these results, we found that mPGES-2 expression levels were upregulated in publicly available CRC patient datasets, as well as in our qPCR and IHC validation experiments. Because the mature mPGES-2 protein exists in an N-terminally truncated cytosolic form, mPGES-2 can function as a cytosolic enzyme rather than a membrane-bound enzyme.9 In our study, elevated mPGES-2 protein expression was observed in the cytoplasm of the CRC tumor cells compared with the findings in the adjacent normal cells. PGE2 synthases are highly upregulated in inflammatory diseases and tumors.50 Notably, levels of PGE2 and its metabolites are markedly increased in many types of human cancers, including colon, lung, breast, and bladder cancers. They are often associated with a poor prognosis.51–54 However, the possibility of tissue-specific or pathological roles of mPGES-2 in CRC remains unknown. In our study, we explored associations between the PGE2 biosynthetic pathway and CRC progression. The results suggested that the CRC patients with higher TNM stages (III and/or IV stages) had significantly higher levels of PGE2 metabolites. Moreover, higher mPGES-2 expression levels were observed in the CRC patients with metastasis than those without metastasis. Our results suggest that the elevated mPGES-2 expression and PGE2 metabolite levels are associated with a poor prognosis in CRC patients. Tumor-associated macrophages (TAMs) play a pivotal role in the connection between inflammation and cancer, including in tumor progression, suppression of anti-tumor immunity, and tumor cell dissemination.55 Several studies have reported that activation of the PGE2 biosynthetic pathway has a major impact on tumoral stroma inflammatory cells and can promote the development of an immunosuppressive microenvironment.41,42,56 However, the biological role of mPGES-2 in immunoregulation in CRC remains unclear. Consistent with the results of previous studies,40 our data demonstrated higher macrophage infiltration in CRC tumor stroma that is significantly correlated with CRC progression. In addition, we found that macrophage infiltration in CRC patient tissues was significantly associated with elevated mPGES-2 expression levels. Overall, these results suggest that increased mPGES-2 expression is associated with macrophage infiltration and CRC progression.
There are still several limitations of this study that need to be addressed. The application of target metabolomic analysis can improve specificity and selectivity, but some other potentially relevant metabolites are not identified because of the absence of MRM transitions. Although we used standard substances for normalization, the metabolomics data involved relative quantification rather than absolute quantification. Thus, it is necessary to establish a method with isotope labeled IS and absolute quantification that can cover additional metabolites. This can be used to validate the concentration of any differential metabolites. Another limitation of the study is the limited sample size. The metabolic profiles of oxidized lipids vary greatly among individuals within the population. However, only a small number of samples from a single center were included in our research. Therefore, the clinical application of oxidized lipid metabolic profiles requires larger sample sizes from multiple centers in further work. Our data of AA metabolite profiling and mPGES-2 expression patterns still indicate that the PGE2 biosynthetic pathway is upregulated in CRC. Further research is needed to confirm the specific molecular mechanisms involved in macrophage infiltration and immunoregulation.
In conclusion, our study explored AA metabolite profile differences between CRC patients and healthy controls and then further investigated several metabolites derived from the PGE2 biosynthetic pathway. These PGE2-derived metabolites were elevated in the CRC group and associated with a poor prognosis. Furthermore, we demonstrated that the expression of mPGES-2, a PGE2 synthase, was upregulated in CRC and was positively correlated with macrophage infiltration in tumors and CRC progression.
Abbreviations
| 11-HETE | 11-Hydroxy-5Z,8Z,11E,14Z-eicosatetraenoic acid |
| 12,13-EpOME | (12R,13S)-(9Z)-12,13-Epoxyoctadecenoic acid |
| 9,10-EpOME | 9,10-Epoxy-12-octadecenoic acid |
| 9-HODE | 9-Hydroxy-10,12-octadecadienoic acid |
| 9-oxoODE | (10E,12Z)-9-Oxooctadeca-10,12-dienoic acid |
| AA | Arachidonic acid |
| BHT | Formic acid and butyl hydroxytoluene |
| CRC | Colorectal cancer |
| CYP | Cytochrome P450 |
| DAB | 3,3′-Diaminobenzidine tetrahydrochloride |
| LA | Linoleic acid |
| LOX | Lipoxygenase |
| LTB4 | Leukotriene B4 |
| LTC4 | Leukotriene C4 |
| TIC | Total ion chromatogram |
| TXB3 | Thromboxane B3 |
| VIP | Variable important in projection |
Author contributions
Ming Guan, Xue Qin and Hongxiu Yu designed the experiments. Cuiping Zhang and Zuojian Hu performed the sample collection. Ziyue Pan, Zhaodong Ji, and Xinyi Cao performed the experiments and analyzed the data. Cuiping Zhang and Xue Qin wrote the paper. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Foundation of Shanghai Municipal Health Commission (20204Y0426) and the National Natural Science Foundation of China (no. 8200081939).References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed.
- A. Pakiet, J. Kobiela, P. Stepnowski, T. Sledzinski and A. Mika, Changes in lipids composition and metabolism in colorectal cancer: a review, Lipids Health Dis., 2019, 18(1), 29 CrossRef PubMed.
- R. N. Gomes, S. Felipe da Costa and A. Colquhoun, Eicosanoids and cancer, Clinics, 2018, 73(suppl 1), e530s CrossRef PubMed.
- A. M. Johnson, E. K. Kleczko and R. A. Nemenoff, Eicosanoids in Cancer: New Roles in Immunoregulation, Front. Pharmacol., 2020, 11, 595498 CrossRef CAS PubMed.
- D. Wang and R. N. Dubois, Prostaglandins and cancer, Gut, 2006, 55(1), 115–122 CrossRef CAS PubMed.
- A. Greenhough, H. J. Smartt, A. E. Moore, H. R. Roberts, A. C. Williams, C. Paraskeva and A. Kaidi, The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment, Carcinogenesis, 2009, 30(3), 377–386 CrossRef CAS PubMed.
- G. Y. Moore and G. P. Pidgeon, Cross-Talk between Cancer Cells and the Tumour Microenvironment: The Role of the 5-Lipoxygenase Pathway, Int. J. Mol. Sci., 2017, 18(2), 236 CrossRef PubMed.
- J. M. Poczobutt, M. Gijon, J. Amin, D. Hanson, H. Li, D. Walker, M. Weiser-Evans, X. Lu, R. C. Murphy and R. A. Nemenoff, Eicosanoid profiling in an orthotopic model of lung cancer progression by mass spectrometry demonstrates selective production of leukotrienes by inflammatory cells of the microenvironment, PLoS One, 2013, 8(11), e79633 CrossRef CAS PubMed.
- S. Hara, D. Kamei, Y. Sasaki, A. Tanemoto, Y. Nakatani and M. Murakami, Prostaglandin E synthases: Understanding their pathophysiological roles through mouse genetic models, Biochimie, 2010, 92(6), 651–659 CrossRef CAS PubMed.
- D. Zhong, J. Cai, C. Hu, J. Chen, R. Zhang, C. Fan, S. Li, H. Zhang, Z. Xu, Z. Jia, D. Guo and Y. Sun, Inhibition of mPGES-2 ameliorates NASH by activating NR1D1 via heme, Hepatology, 2023, 78(2), 547–561 CrossRef PubMed.
- National Health Commission of the People's Republic of China, Chinese Protocol of Diagnosis and Treatment of Colorectal Cancer (2020 edition), Zhonghua Waike Zazhi, 2020, 58(8), 561–585 Search PubMed.
- Z. Pan, Z. Hu, L. Guan, L. Zhang, X. Gao, L. Yang, T. Gong, Y. Hu, Y. Zhao and H. Yu, Diagnostic value of serum sphingolipids in patients with colorectal cancer, Analyst, 2022, 147(10), 2189–2197 RSC.
- D. Sacks, B. Baxter, B. C. V. Campbell, J. S. Carpenter, C. Cognard, D. Dippel, M. Eesa, U. Fischer, K. Hausegger, J. A. Hirsch, M. Shazam Hussain, O. Jansen, M. V. Jayaraman, A. A. Khalessi, B. W. Kluck, S. Lavine, P. M. Meyers, S. Ramee, D. A. Rufenacht, C. M. Schirmer and D. Vorwerk, Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke, Int. J. Stroke, 2018, 13(6), 612–632 Search PubMed.
- H. Ruan, Z. Wang, K. Chen, X. Tang, T. Ding, C. Zhang and M. Guan, The characteristics of cerebrospinal fluid anaplastic large cells in a patient with primary leptomeningeal anaplastic large cell lymphoma, Clin. Chim. Acta, 2022, 537, 46–50 CrossRef CAS PubMed.
- H. Zha, Y. Cai, Y. Yin, Z. Wang, K. Li and Z. J. Zhu, SWATHtoMRM: Development of High-Coverage Targeted Metabolomics Method Using SWATH Technology for Biomarker Discovery, Anal. Chem., 2018, 90(6), 4062–4070 CrossRef CAS PubMed.
- C. Jiao and Z. Gu, iTRAQ-based proteomic analysis reveals changes in response to sodium nitroprusside treatment in soybean sprouts, Food Chem., 2019, 292, 372–376 CrossRef CAS PubMed.
- G. Rodriguez-Blanco, P. C. Burgers, L. J. Dekker, J. J. Ijzermans, M. F. Wildhagen, E. A. Schenk-Braat, C. H. Bangma, G. Jenster and T. M. Luider, Serum levels of arachidonic acid metabolites change during prostate cancer progression, Prostate, 2014, 74(6), 618–627 CrossRef CAS PubMed.
- C. Zhang, Z. Hu, K. Wang, L. Yang, Y. Li, H. Schluter, P. Yang, J. Hong and H. Yu, Lipidomic profiling of virus infection identifies mediators that resolve herpes simplex virus-induced corneal inflammatory lesions, Analyst, 2020, 145(11), 3967–3976 RSC.
- Y. S. Chhonker, V. Bala and D. J. Murry, Quantification of eicosanoids and their metabolites in biological matrices: a review, Bioanalysis, 2018, 10(24), 2027–2046 CrossRef CAS PubMed.
- T. Shimizu, Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation, Annu. Rev. Pharmacol. Toxicol., 2009, 49, 123–150 CrossRef CAS PubMed.
- K. Cao, Y. Lyu, J. Chen, C. He, X. Lyu, Y. Zhang, L. Chen, Y. Jiang, J. Xiang, B. Liu and C. Wu, Prognostic Implication of Plasma Metabolites in Gastric Cancer, Int. J. Mol. Sci., 2023, 24(16), 12774 CrossRef CAS PubMed.
- E. Vassiliou and R. Farias-Pereira, Impact of Lipid Metabolism on Macrophage Polarization: Implications for Inflammation and Tumor Immunity, Int. J. Mol. Sci., 2023, 24(15), 12032 CrossRef CAS PubMed.
- B. Gomolka, E. Siegert, K. Blossey, W. H. Schunck, M. Rothe and K. H. Weylandt, Analysis of omega-3 and omega-6 fatty acid-derived lipid metabolite formation in human and mouse blood samples, Prostaglandins Other Lipid Mediators, 2011, 94(3–4), 81–87 CrossRef CAS PubMed.
- B. Rago and C. Fu, Development of a high-throughput ultra performance liquid chromatography-mass spectrometry assay to profile 18 eicosanoids as exploratory biomarkers for atherosclerotic diseases, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2013, 936, 25–32 CrossRef CAS PubMed.
- S. Ogawa, K. Tomaru, N. Matsumoto, S. Watanabe and T. Higashi, LC/ESI-MS/MS method for determination of salivary eicosapentaenoic acid concentration to arachidonic acid concentration ratio, Biomed. Chromatogr., 2016, 30(1), 29–34 CrossRef CAS PubMed.
- L. Zu, G. Guo, B. Zhou and W. Gao, Relationship between metabolites of arachidonic acid and prognosis in patients with acute coronary syndrome, Thromb. Res., 2016, 144, 192–201 CrossRef CAS PubMed.
- N. Wang, X. Zhao, J. Huai, Y. Li, C. Cheng, K. Bi and R. Dai, Arachidonic acid metabonomics study for understanding therapeutic mechanism of Huo Luo Xiao Ling Dan on rat model of rheumatoid arthritis, J. Ethnopharmacol., 2018, 217, 205–211 CrossRef CAS PubMed.
- N. Wang, X. Zhao, W. Wang, Y. Peng, K. Bi and R. Dai, Targeted profiling of arachidonic acid and eicosanoids in rat tissue by UFLC-MS/MS: Application to identify potential markers for rheumatoid arthritis, Talanta, 2017, 162, 479–487 CrossRef CAS PubMed.
- P. C. Calder, Eicosanoids, Essays Biochem., 2020, 64(3), 423–441 CrossRef CAS PubMed.
- J. Zhang, Q. Yang, J. Li, Y. Zhong, L. Zhang, Q. Huang, B. Chen, M. Mo, S. Shen, Q. Zhong, H. Liu and C. Cai, Distinct differences in serum eicosanoids in healthy, enteritis and colorectal cancer individuals, Metabolomics, 2017, 14(1), 4 CrossRef PubMed.
- M. L. Edin, F. B. Lih, B. D. Hammock, S. Thomson, D. C. Zeldin and D. Bishop-Bailey, Vascular Lipidomic Profiling of Potential Endogenous Fatty Acid PPAR Ligands Reveals the Coronary Artery as Major Producer of CYP450-Derived Epoxy Fatty Acids, Cells, 2020, 9(5), 1096 CrossRef CAS PubMed.
- M. Hada, M. L. Edin, P. Hartge, F. B. Lih, N. Wentzensen, D. C. Zeldin and B. Trabert, Prediagnostic Serum Levels of Fatty Acid Metabolites and Risk of Ovarian Cancer in the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Cancer Epidemiol. Biomarkers Prev., 2019, 28(1), 189–197 CrossRef PubMed.
- C. A. M. Silva, B. G. Graham, K. Webb, M. N. Islam, M. Harton, M. A. de Mello Marques, F. Marques de Carvalho, R. O. Pinheiro, J. Spencer, E. N. Sarno, G. M. Batista Pereira, M. C. Vidal Pessolani, C. Santos de Macedo and J. T. Belisle, Polyunsaturated Fatty Acid-Derived Lipid Mediators as Potential Biomarkers for Leprosy Among Individuals with Asymptomatic Mycobacterium leprae Infection, ACS Infect. Dis., 2023, 9(8), 1458–1469 CrossRef CAS PubMed.
- K. Hildreth, S. D. Kodani, B. D. Hammock and L. Zhao, Cytochrome P450-derived linoleic acid metabolites EpOMEs and DiHOMEs: a review of recent studies, J. Nutr. Biochem., 2020, 86, 108484 CrossRef CAS PubMed.
- F. da Costa Souza, A. C. G. Grodzki, R. K. Morgan, Z. Zhang, A. Y. Taha and P. J. Lein, Oxidized linoleic acid metabolites regulate neuronal morphogenesis in vitro, Neurochem. Int., 2023, 164, 105506 CrossRef CAS PubMed.
- C. Kong, X. Yan, Y. Zhu, H. Zhu, Y. Luo, P. Liu, S. Ferrandon, M. F. Kalady, R. Gao, J. He, F. Yin, X. Qu, J. Zheng, Y. Gao, Q. Wei, Y. Ma, J. Y. Liu and H. Qin, Fusobacterium Nucleatum Promotes the Development of Colorectal Cancer by Activating a Cytochrome P450/Epoxyoctadecenoic Acid Axis via TLR4/Keap1/NRF2 Signaling, Cancer Res., 2021, 81(17), 4485–4498 CrossRef CAS PubMed.
- J. Aldoori, A. J. Cockbain, G. J. Toogood and M. A. Hull, Omega-3 polyunsaturated fatty acids: moving towards precision use for prevention and treatment of colorectal cancer, Gut, 2022, 71(4), 822–837 CrossRef CAS PubMed.
- H. Rao, H. Wu, Q. Huang, Z. Yu and Z. Zhong, Clinical Value of Serum CEA, CA24–2 and CA19–9 in Patients with Colorectal Cancer, Clin. Lab., 2021, 67(4), 1079–1089 CAS.
- S. Hou, J. Jing, Y. Wang, L. Du, B. Tian, X. Xu, T. Sun and Y. Shi, Evaluation of Clinical Diagnostic and Prognostic Value of Preoperative Serum Carcinoembryonic Antigen, CA19–9, and CA24–2 for Colorectal Cancer, Altern. Ther. Health Med., 2023, 29(6), 192–197 Search PubMed.
- D. Che, S. Zhang, Z. Jing, L. Shang, S. Jin, F. Liu, J. Shen, Y. Li, J. Hu, Q. Meng and Y. Yu, Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE(2)/beta-catenin signalling pathway, Mol. Immunol., 2017, 90, 197–210 CrossRef CAS PubMed.
- F. Gomez-Valenzuela, E. Escobar, R. Perez-Tomas and V. P. Montecinos, The Inflammatory Profile of the Tumor Microenvironment, Orchestrated by Cyclooxygenase-2, Promotes Epithelial-Mesenchymal Transition, Front. Oncol., 2021, 11, 686792 CrossRef CAS PubMed.
- T. Wang, B. Jing, D. Xu, Y. Liao, H. Song, B. Sun, W. Guo, J. Xu, K. Li, M. Hu, S. Liu, J. Ling, Y. Kuang, T. Zhang, S. Zhang, F. Yao, B. P. Zhou and J. Deng, PTGES/PGE(2) signaling links immunosuppression and lung metastasis in Gprc5a-knockout mouse model, Oncogene, 2020, 39(15), 3179–3194 CrossRef CAS PubMed.
- K. Echizen, H. Oshima, M. Nakayama and M. Oshima, The inflammatory microenvironment that promotes gastrointestinal cancer development and invasion, Adv. Biol. Regul., 2018, 68, 39–45 CrossRef CAS PubMed.
- G. Yan, H. Zhao, Q. Zhang, Y. Zhou, L. Wu, J. Lei, X. Wang, J. Zhang, X. Zhang, L. Zheng, G. Du, W. Xiao, B. Tang, H. Miao and Y. Li, A RIPK3-PGE(2) Circuit Mediates Myeloid-Derived Suppressor Cell-Potentiated Colorectal Carcinogenesis, Cancer Res., 2018, 78(19), 5586–5599 CrossRef CAS PubMed.
- R. Nasrallah, R. Hassouneh and R. L. Hebert, PGE2, Kidney Disease, and Cardiovascular Risk: Beyond Hypertension and Diabetes, J. Am. Soc. Nephrol., 2016, 27(3), 666–676 CrossRef CAS PubMed.
- D. Wang and R. N. DuBois, Role of prostanoids in gastrointestinal cancer, J. Clin. Invest., 2018, 128(7), 2732–2742 CrossRef PubMed.
- M. Murakami, K. Nakashima, D. Kamei, S. Masuda, Y. Ishikawa, T. Ishii, Y. Ohmiya, K. Watanabe and I. Kudo, Cellular prostaglandin E2 production by membrane-bound prostaglandin E synthase-2 via both cyclooxygenases-1 and -2, J. Biol. Chem., 2003, 278(39), 37937–37947 CrossRef CAS PubMed.
- D. Zhong, Z. Wan, J. Cai, L. Quan, R. Zhang, T. Teng, H. Gao, C. Fan, M. Wang, D. Guo, H. Zhang, Z. Jia and Y. Sun, mPGES-2 blockade antagonizes beta-cell senescence to ameliorate diabetes by acting on NR4A1, Nat. Metab., 2022, 4(2), 269–283 CrossRef CAS PubMed.
- L. A. Jania, S. Chandrasekharan, M. G. Backlund, N. A. Foley, J. Snouwaert, I. M. Wang, P. Clark, L. P. Audoly and B. H. Koller, Microsomal prostaglandin E synthase-2 is not essential for in vivo prostaglandin E2 biosynthesis, Prostaglandins Other Lipid Mediators, 2009, 88(3–4), 73–81 CrossRef CAS PubMed.
- M. Isono, T. Suzuki, K. Hosono, I. Hayashi, H. Sakagami, S. Uematsu, S. Akira, Y. A. DeClerck, H. Okamoto and M. Majima, Microsomal prostaglandin E synthase-1 enhances bone cancer growth and bone cancer-related pain behaviors in mice, Life Sci., 2011, 88(15–16), 693–700 CrossRef CAS PubMed.
- Y. Schumacher, T. Aparicio, S. Ourabah, F. Baraille, A. Martin, P. Wind, R. Dentin, C. Postic and S. Guilmeau, Dysregulated CRTC1 activity is a novel component of PGE2 signaling that contributes to colon cancer growth, Oncogene, 2016, 35(20), 2602–2614 CrossRef CAS PubMed.
- T. Wang, B. Jing, B. Sun, Y. Liao, H. Song, D. Xu, W. Guo, K. Li, M. Hu, S. Liu, J. Ling, Y. Kuang, Y. Feng, B. P. Zhou and J. Deng, Stabilization of PTGES by deubiquitinase USP9X promotes metastatic features of lung cancer via PGE(2) signaling, Am. J. Cancer Res., 2019, 9(6), 1145–1160 CAS.
- A. V. Kurtova, J. Xiao, Q. Mo, S. Pazhanisamy, R. Krasnow, S. P. Lerner, F. Chen, T. T. Roh, E. Lay, P. L. Ho and K. S. Chan, Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance, Nature, 2015, 517(7533), 209–213 CrossRef CAS PubMed.
- P. Nandi, G. V. Girish, M. Majumder, X. Xin, E. Tutunea-Fatan and P. K. Lala, PGE2 promotes breast cancer-associated lymphangiogenesis by activation of EP4 receptor on lymphatic endothelial cells, BMC Cancer, 2017, 17(1), 11 CrossRef PubMed.
- G. Hamilton, B. Rath, L. Klameth and M. J. Hochmair, Small cell lung cancer: Recruitment of macrophages by circulating tumor cells, OncoImmunology, 2016, 5(3), e1093277 CrossRef PubMed.
- Y. Han, W. Guo, T. Ren, Y. Huang, S. Wang, K. Liu, B. Zheng, K. Yang, H. Zhang and X. Liang, Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis, Cancer Lett., 2019, 440–441, 116–125 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01723k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |