Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Surface-enhanced Raman scattering (SERS) and tip-enhanced Raman scattering (TERS) in label-free characterization of erythrocyte membranes and extracellular vesicles at the nano-scale and molecular level†
Tetiana
Stepanenko
 abc,
Kamila
Sofińska
abc,
Kamila
Sofińska
 d,
Natalia
Wilkosz
d,
Natalia
Wilkosz
 c,
Jakub
Dybas
c,
Jakub
Dybas
 e,
Ewelina
Wiercigroch
e,
Ewelina
Wiercigroch
 f,
Katarzyna
Bulat
f,
Katarzyna
Bulat
 c,
Ewa
Szczesny-Malysiak
c,
Ewa
Szczesny-Malysiak
 e,
Katarzyna
Skirlińska-Nosek
e,
Katarzyna
Skirlińska-Nosek
 ad,
Sara
Seweryn
ad,
Sara
Seweryn
 ad,
Joanna
Chwiej
ad,
Joanna
Chwiej
 c,
Ewelina
Lipiec
c,
Ewelina
Lipiec
 *d and
Katarzyna M.
Marzec
*d and
Katarzyna M.
Marzec
 *cg
*cg
aJagiellonian University, Doctoral School of Exact and Natural Sciences, Lojasiewicza 11, Krakow, Poland
bJagiellonian University, National Synchrotron Radiation Centre SOLARIS, Czerwone Maki 98 Str., 30-392 Krakow, Poland
cAGH University of Krakow, Faculty of Physics and Applied Computer Science, Al. Mickiewicza 30, 30-059 Krakow, Poland
dJagiellonian University, M. Smoluchowski Institute of Physics, Łojasiewicza 11, 30-348 Kraków, Poland. E-mail: ewelina.lipiec@uj.edu.pl
eJagiellonian University, Jagiellonian Centre for Experimental Therapeutics, Bobrzyńskiego 14 Str., 30-348 Krakow, Poland
fJagiellonian Center of Innovation, Bobrzyńskiego 14 Str., 30-348 Krakow, Poland
gŁukasiewicz Research Network – Krakow Institute of Technology, 73 Zakopiańska Str., 30-418 Krakow, Poland. E-mail: kmarzec@agh.edu.pl; katarzyna.marzec@kit.lukasiewicz.gov.pl
First published on 18th December 2023
Abstract
The manuscript presents the potential of surface-enhanced Raman spectroscopy (SERS) and tip-enhanced Raman spectroscopy (TERS) for label-free characterization of extracellular microvesicles (EVs) and their isolated membranes derived from red blood cells (RBCs) at the nanoscale and at the single-molecule level, providing detection of a few individual amino acids, protein and lipid membrane compartments. The study shows future directions for research, such as investigating the use of the mentioned techniques for the detection and diagnosis of diseases. We demonstrate that SERS and TERS are powerful techniques for identifying the biochemical composition of EVs and their membranes, allowing the detection of small molecules, lipids, and proteins. Furthermore, extracellular vesicles released from red blood cells (REVs) can be broadly classified into exosomes, microvesicles, and apoptotic bodies, based on their size and biogenesis pathways. Our study specifically focuses on microvesicles that range from 100 to 1000 nanometres in diameter, as presented in AFM images. Using SERS and TERS spectra obtained for REVs and their membranes, we were able to characterize the chemical and structural properties of microvesicle membranes with high sensitivity and specificity. This information may help better distinguish and categorize different types of EVs, leading to a better understanding of their functions and potential biomedical applications.
Introduction
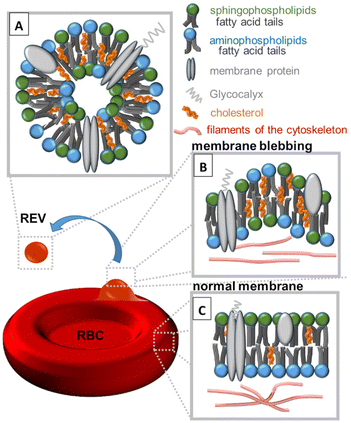
Studies of extracellular microvesicles (EVs) delivered from red blood cells (RBCs) are a rapidly evolving area of research, and scientists are continuously working to understand the full range of functions and implications of these structures. The release of EVs from RBCs (REVs) contributes to a variety of physiological and pathological processes, including inflammation,1 coagulation,2 and immune regulation.3 REVs can be classified into three main categories based on their size. The smallest exosomes (REXs), with a diameter ranging from 30 to 100 nanometres, are formed within the endosomal system and are released during the fusion of multivesicular bodies with the plasma membrane.4 Microvesicles,5–8 also known as RBC microparticles (RMPs), ranging in size from 100 to 1000 nanometres, are formed by membrane blebbing and outward budding of the plasma membrane and are released into the extracellular space.9–11 Apoptotic bodies, ranging in size from 1000 to 5000 nanometres, are formed during programmed cell death and their release is mediated by the plasma membrane breakdown.12Before the formation of REVs (>100 nm), erythrocyte membranes become more rigid and lose their flexibility, and the cytoskeleton becomes disrupted, leading to membrane blebbing followed by EV formation (Fig. 1). During such processes, significant changes in the distribution of membrane lipids occur.13 Normally, aminophospholipids (AP) such as phosphatidylserine (PS), phosphatidylethanolamine (PE) and phosphatidylcholine (PC) are found predominantly in the inner leaflet of the erythrocyte membrane. However, as presented in Fig. 1, during membrane blebbing and REV formation, PS becomes exposed on the outer surface of the blebs and REVs. Externalization of PS is considered to be mediated by the activation of enzymes known as scramblases and floppases that flip PS from the inner leaflet to the outer leaflet of the membrane. Simultaneously, an increase in the levels of PE and PC on the REVs occurs. Sphingophospholipids (SP) such as sphingomyelin (SM) and ceramide (Cer) are also present in the erythrocyte membrane and play an important role in regulating the membrane structure and function. An increase in cholesterol in blebs and REVs is due to the activation of enzymes known as cholesterol esterases, which hydrolyze cholesterol esters to free cholesterol.14
REVs are derived from RBCs, but the composition of their membranes is not identical. According to Yang et al.,15 in REVs, cytoskeletal proteins such as spectrin, actin, and bands 4.1, 4.2, and 4.9 are partially or even completely absent, but glycosylphosphatidylinositol (GPI) connexins and lipid raft markers such as ganglioside M1 or stomata are relatively enriched. REVs carry a variety of proteins, as well as metabolites, cytokines, and growth factors that influence cellular function and signalling.16 Due to their role in cellular communication REVs are highly enriched with tetraspanins (CD9, CD63, CD81, and CD82), which are involved in fusion and cell penetration. The CD47 protein enables REVs to escape macrophage clearance. REVs’ cytosol contains heat shock proteins (HSP70, HSP90) attached to antibodies involved in antigen anchoring, multivesicular body (MVB) formation proteins (Alix, TSG101) engaged in exosomal release15 and specific proteins such as denaturated hemoglobin or enzymes (carbonic anhydrase, acetylcholinesterase16) (Fig. 1A). Similar to RBC membranes, REVs’ membranes contain membrane-associated proteins (stomatin, flotillin), glycoproteins (glycophorin, Gp) and anion transport proteins (Band 3).15 Amino acids (AAs), the building blocks of proteins, are also a crucial component of extracellular vesicles.16
Modern proteomic techniques such as mass spectrometry have been instrumental in identifying many membrane proteins in EVs, shedding light on their molecular composition and function.17 EVs contain transmembrane proteins that span the lipid bilayer one or several times, with both external hydrophilic domains and a hydrophobic domain (typically with an alpha-helix structure) directly interacting with membrane lipids.
Studying the protein content of REVs is an active area of research, as it holds promise for diagnostic and therapeutic applications. Understanding specific proteins and their functions in REVs can provide insights into their roles in intercellular communication and disease progression.
We have previously reported9,18 that atomic force microscopy (AFM) is a powerful imaging technique to analyse the size of REVs formed in situ on the membrane of intact RBCs. The high resolution, non-destructive, in situ imaging capabilities, ability to probe mechanical properties and versatility of AFM make it a great technique for REV characterisation. However, to understand the role of REVs, it is important to analyse their chemical composition. Traditionally, this has been done using labelling techniques such as fluorescent or radioactive tags, which can specifically target certain molecules within the REVs.9,18 However, these methods can be time-consuming and expensive and can alter the natural composition of the REVs. On the other hand, label-free techniques do not require any modification of REVs and can provide information on their natural chemical composition. Finding a suitable technique that could analyze the chemical composition of REVs in a label-free manner is important because it would allow for a more accurate and comprehensive understanding of the roles of REVs in various biological processes. It could also provide a basis for developing diagnostic and therapeutic approaches that can target REVs and their biomolecules. Label-free techniques that provide the possibility of measurements with high sensitivity, molecular specificity, non-destructiveness, spatial resolution and versatility are based on Raman scattering (RS). RS was previously found to be a method that can identify and quantify various biomolecules within animal cells, such as proteins, lipids, carbohydrates and nucleic acids.19 With the application of RS, i.e. with surface-enhanced Raman scattering (SERS) and tip-enhanced Raman scattering (TERS), REVs could be analysed at the nanoscale or even at the single-molecule level. Both SERS and TERS amplify the Raman signal from all types of biomolecules present in the RBC and REV membranes in close proximity to metal, either metal surfaces or nanoscale metal tips.
For instance, REVs contain helical fragments of proteins on their outer membranes. Upon their interaction with gold nanoparticles or a gold tip apex, they result in signal amplification in SERS and TERS. Aromatic amino acids, e.g., tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe), possess an aromatic ring in their side chains. The π-electrons in these aromatic rings can strongly interact with the metal surface, resulting in an enhancement of Raman signals during SERS and TERS experiments.20 Furthermore, all other biomolecules present on the cell membranes (peptides, lipids, carbohydrates) that possess Raman-active bands could potentially be enhanced in SERS and TERS; hence these techniques could be used to analyse the lipid and protein composition of the RBC and REV membranes.
SERS and TERS are powerful spectroscopic techniques used to enhance the Raman signals of the molecules adsorbed on or proximal to metal surfaces. These methods are particularly useful for studying complex biological systems, such as cells, extracellular vesicles or biomolecules, where traditional Raman spectroscopy may not provide sufficient sensitivity.
Although SERS and TERS can be highly effective in enhancing the Raman signals of specific molecules (proteins or lipids), the overlapping of bands from different molecules can complicate the analysis of results. Different molecules may have similar vibrational modes, leading to the overlapping of bands in the Raman spectra. Additionally, TERS provides the distribution of the sample components with nanometer spatial resolution and allows the analysis of spatial correlation between distributions of specific components/particular functional group characteristic of membrane lipids and proteins.
Therefore, the objective of this work was to design the appropriate methodology and define the capabilities of the application of SERS and TERS techniques for RBC membrane and REV analysis, especially for the identification of proteins and lipids. We focused on the RBC membranes and REVs delivered from RBCs obtained from 24 weeks old male C57BL/6J (N = 3) mice.21–24
Experimental
Materials and methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min, respectively). Subsequently, REVs were washed with PBS and centrifuged at 16
000g, 10 min, respectively). Subsequently, REVs were washed with PBS and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600g for 36 min.
600g for 36 min.
CFSE and TER-119 assessment with flow cytometry
To confirm the presence of red blood cell-derived extracellular microvesicles in the obtained material, the samples were stained with 40 μM and 20 μM of CFSE CellTrace Cell Proliferation Kit (Invitrogen, Cat No. 34554) together with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 anti-TER-119 antibody (BioLegend, Cat No. 116228), according to the protocol provided by the manufacturer. Unstained samples served as the control group. Analyses were conducted using a standard flow-cytometry method. Multiparametric flow cytometry acquisition was performed with a BD LSII flow cytometer (Becton Dickinson). Daily instrument quality control was performed to ensure identical operation. Stained samples were diluted in PBS. For each sample, 150
100 anti-TER-119 antibody (BioLegend, Cat No. 116228), according to the protocol provided by the manufacturer. Unstained samples served as the control group. Analyses were conducted using a standard flow-cytometry method. Multiparametric flow cytometry acquisition was performed with a BD LSII flow cytometer (Becton Dickinson). Daily instrument quality control was performed to ensure identical operation. Stained samples were diluted in PBS. For each sample, 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were acquired in log mode for height forward side scatter (FSC-H), area forward scatter (FSC-A) and fluorescent signals.
000 events were acquired in log mode for height forward side scatter (FSC-H), area forward scatter (FSC-A) and fluorescent signals.
Singlets were gated according to the FSC-H to FSC-A ratio. Subsequently, the population of REVs was separated from cellular debris and other microvesicles according to the simultaneous presence of fluorescence signals derived from CFSE and anti-TER-119. Cytometric analysis confirmed the presence of REVs in the material obtained (ESI – Fig. SI1†).
SERS measurement of isolated RBC membranes and RS measurements of standards
Gold nanoparticles (AuNP)27 (Ted Pella, gold colloids, size 60 nm, 2.6 × 1010 particles per ml, Cat No. 15709-20) were added to a suspension of isolated RBC membranes. A drop of the solution was transferred on the CaF2 substrate for SERS measurements. A set of protein (glycophorin, E-cadherin), amino acid (His, Tyr, Trp, Phe) and lipid (SM, PE, PC, CHL) standard compounds were measured in their solid form as a reference.SERS and RS measurements were performed using a WITec CRM alpha 300 confocal Raman microscope (WITec GmbH, Ulm, Germany) with an air-cooled semiconductor laser operating at 633 nm and a CCD detector cooled to −60 °C. The laser was connected to the microscope with a 50 μm diameter optical fibre. The spectrometer monochromator was calibrated with the radiation spectrum of a xenon lamp (WITec UV light). The Zeiss W Plan-Apochromat water-dipping objective (63×/1.0 NA) was used for SERS and the Zeiss EC Epiplan-Neofluar air objective (20×/0.5 NA) for RS measurements. The laser power at the sample position was about 5 mW (2.12 × 106 W cm−2) in case of SERS and 10–20 mW in case of normal RS measurements of standard compounds.
SERS measurements were performed in the line-scan mode. During the experiment, hundreds of individual Raman spectra were obtained from the droplet of the RBCs’ membrane solution. The SERS signal occurred solely in the places where a part of the RBC membrane molecule interacted with the surface of the AuNP. The acquisition time per spectrum was 0.5 s and the spectral resolution was approximately equal to 4 cm−1.
Normal RS measurements of standard compounds were measured in single-point mode. Each standard was measured three times, and the acquired single spectra were subsequently averaged. Each single spectrum was accumulated 20 times with 1s integration time.
Raman measurements and data analysis were performed using WITec Project Plus 2.10 (WITec GmbH), OPUS (Bruker Optics), and OriginPro (OriginLab) software. All SERS spectra were pre-processed (cosmic ray removal, smoothing—13 smoothing points, background removal) and normalized.
AFM measurements of isolated REVs
Isolated REVs were placed on a CaF2 slide and dried in the air at room temperature. AFM measurements were performed on a CRM alpha 300 Raman confocal microscope (WITec, Ulm, Germany) combined with atomic force microscopy. Standard force modulation probes with a nominal spring constant of 2.8 N m−1 (WITec, Ulm, Germany) and AC mode was applied. Samples of isolated REVs were illuminated by a dry Zeiss EC EPIPLAN 20×/0.4 objective. AFM images of 512 × 512 lines from an area of 10 × 10 μm were collected in air. In order to determine the size (diameter and height) of each REV, topographic cross-sections of the obtained AFM images were analysed separately. To define the diameter of REVs, cross sections were made in two perpendicular planes. The average diameter of each REV was calculated as the mean of two values of diameter (presented in figures as diameter 1 and 2) measured in the middle of the height on REV cross-section profile. Data analysis was performed using OriginPro software. The obtained data of the average REV diameters did not have a normal distribution, so they are presented in the figure as a box showing the Q1–Q3 quartiles, median line and min–max whiskers.TERS measurement of isolated REVs
AFM cantilevers (VIT P/TK) were coated with 5 nm of a Ti wetting layer and then with 400 nm of Au using an evaporation system (under the pressure of 10−7 mbar) with a constant deposition rate of 0.05 nm s−1 VIT P/TK (TipsNano) probes coated with Ti/Au were used for EVs imaging in the air prior to TERS spectra acquisition. The spring constant of these cantilevers was 25–95 N m−1, and the resonance frequency was in the range of 200–400 kHz. AFM topographies were collected in AC mode using a SmartSPM microscope (Horiba) at a scan resolution of 256 × 256 pixels, scan rates of 0.1–0.4 Hz and scan sizes of ca. 500 nm.Based on the acquired AFM topographies and phase images, areas of interest for TERS spectra and map collection were selected manually. TERS measurements were performed using a SmartSPM microscope (Horiba) integrated via OmegaScope with a Labram HR Raman spectrometer (Horiba) equipped with a CCD camera cooled to −79 °C. A HeNe laser operating at 633 nm was applied with a laser power of 12.5 mW (1.3 × 106 W cm−2). TERS spectra were acquired in the spectral range of 650–1800 cm−1 with a spectral resolution of 2 cm−1. The acquisition time of spectra was set depending on the signal-to-noise ratio (SNR). We also carefully monitored the TERS signal in terms of features of amorphous carbon as a result of the thermal decomposition of the sample.28 Based on an investigation into spectral features (the presence of the Phe and amide bands), we continuously diagnosed the TERS signal and adjusted the beam power to prevent sample thermal decomposition, as demonstrated elsewhere.29 Calculation of Pearson correlation coefficients was conducted in the MATLAB (RRID:SCR_001622) environment from MathWorks (Natick, USA) using built-in functions, assuming that the relationship is approximately linear.
A knife-edge method was applied to calculate the spatial resolution of TERS mapping. Details of the procedure were previously described.30 According to calculations, the best-estimated spatial resolution was ca. 86 ± 44 nm as presented in Fig. SI4.† The relatively high value of uncertainty results from the Shannon–Nyquist theorem.
Results and discussion
Deciphering the chemical composition of isolated red blood cell membranes via SERS analysis
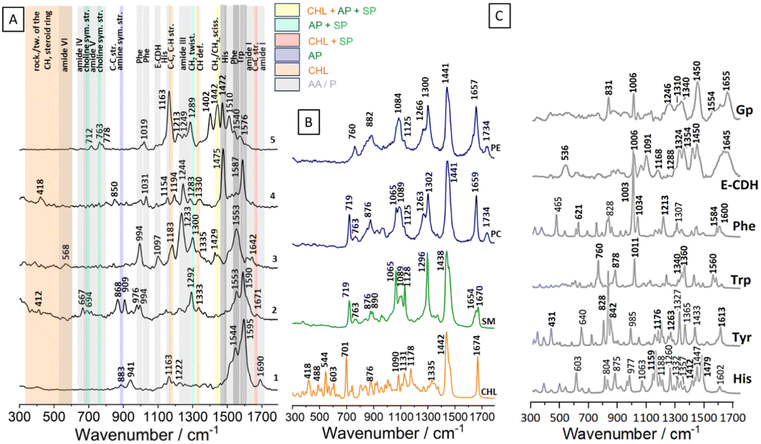
The first step of the analysis was focused on deciphering the chemical composition of isolated RBC membranes using SERS [Fig. 2A]. This step was necessary to disentangle the source of information obtained via TERS analysis from the microvesicles released by the RBCs.The exemplary SERS spectra of RBC membranes, collected at a wavelength of 633 nm in the presence of gold nanoparticles, are presented in Fig. 2A. The spectra contain bands originating mainly from proteins and membrane lipids (sphingolipids: ceramide42,43 and SM, as well as aminophospholipids: PS,44 PE, PC).
The most common bands enhanced in our SERS measurements and yet absent in the reference lipid spectra were the amide vibrations, commonly observed in proteins.36 Amide I appeared at 1690 and 1642 cm−1 in spectra 1 and 3, respectively. The amide I band being present at 1690 cm−1 is typically associated with a protein having a high proportion of β-sheet secondary structure. The Raman band at 1642 cm−1 is indicative of the presence of random coils and turns in the protein's secondary structure; the position of this band is close to the amide I band on the E-CDH spectrum36 [Fig. 2C]. Amide II bands, typically present in the range of 1480–1575 cm−1, are very weak in the Raman spectra.36 Strong bands present in the SERS spectra 1–5 at 1587 and 1553 cm−1 may originate from the protein backbone. The prominent band at 1587 cm−1 could be assigned to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C out-of-phase stretching of the phenylalanine's ring. The band at 1553 cm−1 originates from the indole ring stretching vibrations of tryptophan. Strong bands at 1475 and 1472 cm−1 in spectra 4 and 5, respectively, could come from histidine. The band at 1402 cm−1, derived from C–H and CH2 bending in proteins, was enhanced in spectrum 5.32,36 Amide III bands are observed at the wavenumbers of 1292, 1283, 1249, 1244, 1233, and 1222 cm−1. In the presented SERS spectra, bands from hydrophobic amino acids inside chains were recognised. The signals from glycine or proline, such as the 1031 cm−1 band, may arise from C–N stretching, while the 941 cm−1 band could be attributed to proline and valine within the 924–943 cm−1 range.36 The presence of a strong band at 994 cm−1 in spectrum 3 and a weak band at 1031 cm−1 in spectrum 4 suggests the enhancement of the phenylalanine signal from the proteins. Besides the amide I and amide III regions, the spectra show marker bands, in particular for the aromatic residues such as tryptophan (at around 763, 883 and 1553 cm−1) and tyrosine (at around 868 cm−1).36
C out-of-phase stretching of the phenylalanine's ring. The band at 1553 cm−1 originates from the indole ring stretching vibrations of tryptophan. Strong bands at 1475 and 1472 cm−1 in spectra 4 and 5, respectively, could come from histidine. The band at 1402 cm−1, derived from C–H and CH2 bending in proteins, was enhanced in spectrum 5.32,36 Amide III bands are observed at the wavenumbers of 1292, 1283, 1249, 1244, 1233, and 1222 cm−1. In the presented SERS spectra, bands from hydrophobic amino acids inside chains were recognised. The signals from glycine or proline, such as the 1031 cm−1 band, may arise from C–N stretching, while the 941 cm−1 band could be attributed to proline and valine within the 924–943 cm−1 range.36 The presence of a strong band at 994 cm−1 in spectrum 3 and a weak band at 1031 cm−1 in spectrum 4 suggests the enhancement of the phenylalanine signal from the proteins. Besides the amide I and amide III regions, the spectra show marker bands, in particular for the aromatic residues such as tryptophan (at around 763, 883 and 1553 cm−1) and tyrosine (at around 868 cm−1).36
Most bands in the fingerprint region that could be assigned to membrane lipids exhibited SERS enhancement [Fig. 2A]. Their positions were comparable to those observed in all reference membrane lipid spectra (Fig. 2B, Table 1). The position of the band at 1671 cm−1 is close to the position of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching bands of cholesterol, SM and ceramide.42,43 In spectrum 5 (Fig. 2A.5.), the band at 1442 cm−1 is characteristic of CH2 scissoring in lipids and could come from PC, PE, SM as well as from CHL. Cholesterol-associated bands from C–C and C–H stretching were present at 1330, 1333, and 1183 cm−1. Ethanolamine from PE appeared in the SERS spectrum at 883 cm−1 (Fig. 2A.1.). C–C ring breathing of tyrosine is present at 850 cm−1.36 The positions of the bands at 712 and 763 cm−1 in the SERS spectra suggest that choline groups from PC and SM were enhanced. Upon comparing the spectra of the Raman standards with the SERS spectra, it becomes evident that the bands up to about 600 cm−1 are attributed to the CH groups and steroid ring of cholesterol.
C stretching bands of cholesterol, SM and ceramide.42,43 In spectrum 5 (Fig. 2A.5.), the band at 1442 cm−1 is characteristic of CH2 scissoring in lipids and could come from PC, PE, SM as well as from CHL. Cholesterol-associated bands from C–C and C–H stretching were present at 1330, 1333, and 1183 cm−1. Ethanolamine from PE appeared in the SERS spectrum at 883 cm−1 (Fig. 2A.1.). C–C ring breathing of tyrosine is present at 850 cm−1.36 The positions of the bands at 712 and 763 cm−1 in the SERS spectra suggest that choline groups from PC and SM were enhanced. Upon comparing the spectra of the Raman standards with the SERS spectra, it becomes evident that the bands up to about 600 cm−1 are attributed to the CH groups and steroid ring of cholesterol.
| AP | SP | CHL | AA | Proteins | Assignment | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PE | PC | SM | CHL | His | Tyr | Trp | Phe | E-CHD | Gp | |
| 1736 | 1736 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching31,32 O stretching31,32 |
||||||||
| 1671 | 1602 | 1613 | 1600 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C in-phase stretching of the ring32–35 C in-phase stretching of the ring32–35 |
||||||
| 1668 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching (E isomer)31,32 C stretching (E isomer)31,32 |
|||||||||
| 1658 | 1653 | 1642 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching (Z isomer)31,32 C stretching (Z isomer)31,32 |
|||||||
| 1655 | Amide I, a-helix31,36,37 | |||||||||
| 1645 | Amide I, random coil31,36,37 | |||||||||
| 1584 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C out of phase stretching of the ring34 C out of phase stretching of the ring34 |
|||||||||
| 1554 | Amide II, weak31,36,37 | |||||||||
| 1550 | Indole ring vibration37,38 | |||||||||
| 1479 | N–H imidazole ring39 | |||||||||
| 1450 | 1450 | CH deformation36 | ||||||||
| 1447 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and N–C stretching imidazole ring39 C and N–C stretching imidazole ring39 |
|||||||||
| 1436 | 1441 | 1439 | (CH2/CH3) scissoring32,40,41 | |||||||
| 1439 | CH2 scissoring32 | |||||||||
| 1410 | N–C stretching imidazole ring39 | |||||||||
| 1360 | 1354 | 1340 | Fermi resonance (Trp)36–38 | |||||||
| 1340 | 1324 | 1310 | ||||||||
| 1332 | CH deformation31,34 | |||||||||
| 1357 | N–H band in the imidazole ring35 | |||||||||
| 1332 | N–C stretching and N–H bending39 | |||||||||
| 1288 | Amide III, a-helix31,36,37 | |||||||||
| 1296 | 1299 | 1293 | CH2 in-phase twisting32 | |||||||
| 1262 | Phenolic C–O stretching37 | |||||||||
| 1260 | C–H, NH3 bending in the im. ring35,39 | |||||||||
| 1263 | 1260 | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H in plane deformation32 C–H in plane deformation32 |
||||||||
| 1213 | C–C stretching34 | |||||||||
| 1188 | C–N stretching and NH3 bending in the imidazole ring35,39 | |||||||||
| 1176 | C–C, C–H stretching in the steroid ring32 | |||||||||
| 1176 | ||||||||||
| 1159 | C–H, N–H bending in the im. ring35,39 | |||||||||
| 1120 | 1120 | 1125 | 1132 | C–C stretching32,33,40 | ||||||
| 1083 | 1083 | 1086 | 1086 | C–C stretching32 | ||||||
| 1060 | C–H, NH3 mixed vibrations39 | |||||||||
| 1066 | 1060 | C–C stretching32 | ||||||||
| 1034 | C–C in phase stretching and C–C–H in phase deformation34 | |||||||||
| 1011 | Benzene ring breathing37,38 | |||||||||
| 1003 | C–C–C deformation of the ring34 | |||||||||
| 977 | NH3 rocking, C–H bending39 | |||||||||
| 889 | C–C–N+ symmetric stretching40 | |||||||||
| 881 | –C–NH2 asymmetric stretching32 | |||||||||
| 878 | N–H stretching of the indole ring37,38 | |||||||||
| 875 | C–H, NH3 mixed vibrations39 | |||||||||
| 869 | 875 | N+(CH3)3 asymmetric stretching32,33 | ||||||||
| 842 | Fermi resonance (ring breathing)37 | |||||||||
| 828 | 831 | Fermi resonance (ring breathing)37 | ||||||||
| 804 | Im. ring. def, CH, NH3 bend.39 | |||||||||
| 760 | 766 | N+(CH3)3 symmetric stretching32,33 | ||||||||
| 760 | in-phase symmetric breathing vibration of the benzene and pyrrole ring37,38 | |||||||||
| 754 | –C–NH2 symmetric stretching32,33 | |||||||||
| 700 | In plane deformation of the ring31–33 | |||||||||
| 715 | 712 | N+(CH3)3 symmetric stretching32,33 | ||||||||
| Amide IV32,36 | ||||||||||
| Amide V32,36 | ||||||||||
| Amide VI32,36 | ||||||||||
| 621 | Phenyl ring breathing vibrations34 | |||||||||
| 603 | Ring def., C–C–N bend39 | |||||||||
| 536 | S–S36 | |||||||||
| 603 | Rocking/twisting of the CH groups, steroid ring32,33 | |||||||||
| 542 | ||||||||||
| 490 | ||||||||||
| 422 | ||||||||||
SERS spectra obtained in this study exhibit a high level of compatibility with data obtained from conventional Raman spectroscopy, indicating the effectiveness and sensitivity of this technique for erythrocyte membrane studies. The presence of gold nanoparticles enhances the detection range, thus making it suitable for measuring low concentrated samples. SERS spectra from 1 to 5, acquired from the erythrocyte membranes, reveal bands originating from the plasmon-enhanced resonance of various membrane fragments. The high compatibility with the existing literature supports the use of these data in the analysis of microvesicle membranes derived from the erythrocyte membranes. The part of our work presenting TERS research shows correlations with the single-point spectra, facilitating the assessment of the similarities in chemical composition between the erythrocyte membranes and the microvesicles.
The strongest bands in the SERS spectra are located in the spectra region of around 1525–1590 cm−1 (Fig. 2A.1–4, Fig. SI2† Supplement: average spectrum from 15 SERS spectra) and they are originated from the aromatic ring's vibrations (Trp, Tyr, Phe). In some spectra, the most intense bands could be related to the presence of histidine (Fig. 2A.5). Aromatic rings contain a system of alternating single and double bonds, creating a conjugated π-electron system. This allows for an efficient charge transfer and delocalization of electrons within the ring structure. When these aromatic molecules are adsorbed onto the AuNp, the π-electron system of the aromatic ring can couple with the localized surface plasmons of metal nanoparticles. This coupling leads to several effects that contribute to the strong enhancement of Raman signals from aromatic rings in SERS.45
Common Raman bands coming from other biomolecules are also present in the SERS spectra, but their signals are lower than those of the aromatic rings of Trp, Tyr, and Phe (Fig. 2A).
In order to identify lipid bands in the SERS spectra, exemplary spectra were obtained with a 633 nm laser for typical membrane lipids: SM, PE, PC, and CHL and are presented in Fig. 2B and their assignment is given in Table 1. The positioning of bands in the Raman spectra of lipids was consistent with the literature data for measurements of these compounds using a 532 nm laser.32
Exemplary spectra of amino acids and erythrocyte proteins were measured with 633 nm laser and presented in Fig. 2C to enable the identification of the protein bands on the SERS spectra. The side chain of His contains a five-membered imidazole ring with two nitrogen atoms within its structure, manifested as distinctive Raman bands: 1602, 1479, 1447, 1357, 1332 1260, 1188, 1159, 804, and 603 cm−1.35,39 The bands at 1060, 977, 875, 804 and 603 cm−1 in the His spectrum may be assigned to the different vibrations of the C–H and NH3.39 Aromatic amino acid back chains’ bands could be visible on the proteins’ spectra. Bands’ characteristic of three aromatic amino acids: Tyr, Trp and Phe are shown in bold in Fig. 2C. These bands are associated with aromatic ring vibrations: 1613, 1176, 842, 828 cm−1 (Tyr), 1550, 1360, 1340, 1011, 878, 760 cm−1 (Trp), 1600, 1584, 1034, 1003, and 621 cm−1 (Phe). Moreover, phenolic C–O stretching at 1262 cm−1 is characteristic for Phe. These bands do not overlap with other bands in the proteins’ spectra and could be used in the SERS spectra analysis.34 Additional bands on the aromatic amino acids’ spectra are also assigned and presented in Table 1. Two membrane proteins: E-cadherin (E-CDH) and glycophorin (Gp), are measured and presented in Fig. 2C and assigned in Table 1. Their spectra contain bands from amide bands, disulfide bonds, C–C stretching and, interestingly, aromatic amino acid chains: Trp Fermi resonance at 1340/1310 cm−1 (Gp) and 1354/1324 cm−1 (E-CDH), the Phe band at 1006 cm−1 E-CDH, Gp, Tyr Fermi resonance at 831 cm−1 (Gp), respectively.
In our studies, we were able to determine the presence of AP and SP in extracellular vesicles, which reflects the complex lipid composition of these vesicles and suggests that they may play a key role in the biological activities and functions of REVs. The specific lipid composition of extracellular vesicles can affect their stability, interactions with target cells and overall biological effects. Unfortunately, using spectroscopic techniques, we were unable to distinguish bands coming from a single group of lipids. However, it was possible to distinguish the lipid moiety from protein-derived chemical group modes. Moreover, differentiation between different amino acids and even specific proteins was accomplished.
The similarity between the chemical composition of erythrocyte membranes and EVs can indeed pose challenges when trying to differentiate them using certain analytical techniques. Both erythrocyte membranes and EVs are composed of phospholipid bilayers and proteins, which are major components in their structure; however as presented in Fig. 1, some chemical groups are present in the majority only on the REVs’ surface. On the other hand, the similarity in composition can lead to the overlapping of bands in Raman spectra, making it difficult to distinguish between the two objects based solely on their chemical signatures.
AFM experiment
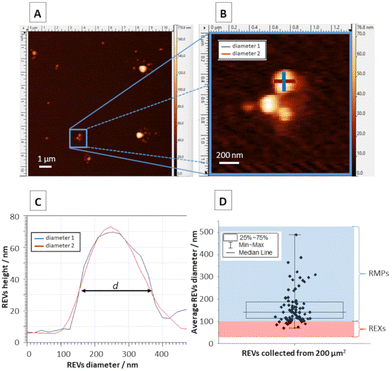
Obtained topographic AFM images (Fig. 3A) allowed the analysis of REVs’ diameter (Fig. 3B) by determining this parameter in each analysed object in two perpendicular planes at the half-height on the topographic cross-section and calculating the average (Fig. 3C). The performed analysis enabled the determination of diameter distribution and the average diameter of REVs (Fig. 3D). AFM analyses show that most of the REVs can be characterized as RMPs (89% of measured REVs) their sizes spanning between 100 and 500 nm. Only a small portion of REXs with a diameter below 100 nm was observed (11% of analysed REVs). For further TERS measurements, REVs with a diameter of around 200 nm were selected, in order to analyze only those microparticles that definitely belong to the RMP group and were created due to the process of membrane blebbing and outward budding of the plasma membrane.TERS experiment
Fig. 4 shows the representative results of the TERS experiment on REVs (Fig. 4A and B). More TERS spectra are shown in the supplement in Fig. SI3.† In all single-point TERS spectra presented in Fig. 4C, the amide II bands indicating the N–H bending, C–N, and C–C stretching vibrations are well pronounced. The position of bands at 1579 and 1572 cm−1 may suggest the presence of Trp (indole ring vibration36,46) or His46 in the peptide chain of proteins composing microvesicles. However, this spectral position is also characteristic of the β-sheet secondary structure or unordered regions of proteins.47 Thus, proteins present in the structure of REVs may contain a relatively high load of these secondary structures. Moreover, amide II bands present at 1543 cm−1, and 1500 cm−1 in the third TERS spectrum (brown) indicate the local contribution of proteins abundant in α-helix secondary structure, and His, respectively.46,48 However, the unambiguous interpretation of the bands in the amide II region is challenging. As mentioned above, band characteristic of protein secondary structure may overlap with bands attributed to aromatic amino acid residues (Trp, Tyr, Phe, His). Here, it is worth noting that the observed low intensity or the absence of the amide I band in the TER spectra is to some extent related to the photo-dissociation of the peptide backbone in the plasmonic hot-spot upon interaction with hot carriers (electrons) from the metalized probe apex.49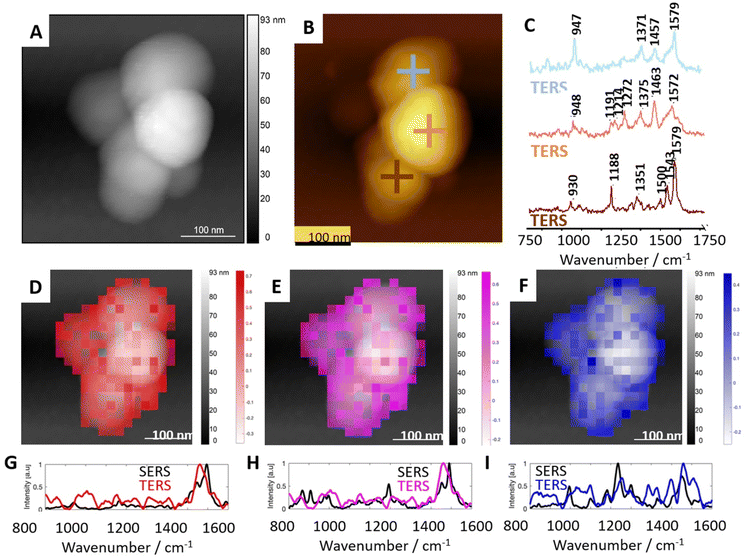 | ||
| Fig. 4 Results of TERS measurements of selected microvesicles in comparison with average SERS results. (A) The topography image of a microvesicle selected for TERS mapping. (B) The topographical image with exemplary spots of TERS spectra acquisition. (C) The single point TERS spectra collected at locations marked in (B). (D–F) Distribution of correlation coefficient calculated from TERS maps imposed on AFM topographies and three marker SERS spectra (presented in Fig. 2). (G–I) Pearson correlation of TERS spectra most similar to the marker SERS spectra. | ||
Bands at 1457 cm−1 and 1463 cm−1, pronounced in TERS spectra 1 and 2, respectively, represent CH, CH2, and CH3 deformational vibrations in proteins.46,47 However, methyl and methylene motions in this spectral region are also characteristic of CHL, SP, and AP. Therefore, a clear-cut determination of the origin of these vibrations in the TERS spectra is challenging. In all single-point TERS spectra presented in Fig. 4C(1–3), the band present in the spectral range of 1380–1350 cm−1 may correspond to the Cα–H/N–H bending mode characteristic for β-sheets in proteins44 or CH3 symmetric bending in CHL. A distinct band visible at 1272 cm−1 in TERS spectrum 2 (orange) is specific for the symmetric stretching of the PO2− group characteristic of aminophospholipids like PE and PC. This spectrum also displays bands at 1214 cm−1 and 1191 cm−1, which may suggest the contribution of β-sheet secondary structure and side residues of amino acids composing proteins (e.g., Tyr), respectively.47 The position of the band at 1188 cm−1 in TERS spectrum 3 (brown) corresponds to the C–C stretching in SM or aminophospholipids (PS, PE). The general spectral characteristics of this TERS spectrum corresponds well to the SERS spectrum 1. These spectra may reflect a local presence of a SM mixture with proteins in the RBC membranes (SERS) and EVs (TERS). Bands at 947 and 948 cm−1 present in TERS spectra 1 and 2, respectively, are characteristic of the CH banging vibration of CHL. The spectral positions pronounced in TERS spectrum 1 (blue) correspond well to bands enhanced in SERS spectrum 2 in Fig. 2. The general spectral shape of these bands suggests that TERS spectrum 1 and SERS spectrum 2 were collected at spots abundant in CHL and proteins. TERS spectrum 2 shows similarities in the spectral shape of SERS spectra 3–5 (also shown in Fig. 2). The positions of bands suggest the local spectral characteristics of AP with protein inclusions.
Pearson correlation is a useful method for examining patterns and making presumptions about the relationship between them. A statistical measure obtained during calculation is the correlation coefficient. It ranges between −1 and 1, where 1 indicates a perfect positive linear relationship (when one variable increases, the other tends to increase proportionally), −1 indicates a perfect negative linear relationship (when one variable increases, the other tends to decrease proportionally), and 0 indicates no linear relationship between the variables. However, it is important to note that correlation does not imply causation, meaning that a strong correlation between two variables does not necessarily indicate that changes in one variable cause changes in the other. The distribution of correlation coefficients was calculated from TERS maps and the three SERS spectra (spectra 1, 2, and 3 presented in Fig. 2) representing mixtures of peptides/proteins with cholesterol and also proteins with AP and sphingolipids presented in Fig. 4D–F respectively. The distribution of each of these three spectra on the EVs’ surface was found to be heterogeneous. Such heterogeneity is likely related to the formation of lipid domains triggered by the membrane proteins. Due to high spatial resolution, TERS enabled nanoscale label-free observation of protein/cholesterol-rich and lipid-rich domains, similarly to the results obtained for the cellular membrane by Zenobi and co-workers.50 However, the presence of the domains was never demonstrated for such small objects as EVs in a label-free and non-destructive manner.
Conclusions
This article highlights the use of SERS and TERS in the analysis of extracellular vesicles, particularly red blood cell-derived microvesicles and their membranes.The SERS spectra revealed prominent bands corresponding to amide vibrations commonly observed in proteins. These bands were attributed to the presence of specific secondary structures in the proteins, such as β-sheets and random coils/turns. Numerous bands in the SERS spectra were attributed to membrane lipids, including sphingolipids (sphingomyelin) and aminophospholipids (phosphatidylserine, phosphatidylethanolamine, and phosphatidylcholine). The positions of these bands matched those observed in the reference membrane lipid spectra. Specific bands corresponding to cholesterol were observed in the SERS spectra, providing information about the presence and structure of cholesterol in the RBC membranes. The SERS spectra obtained in the study were highly compatible with the data from conventional Raman spectroscopy, demonstrating the effectiveness and sensitivity of the SERS technique for detecting erythrocyte membranes.
In order to characterize the surface of EVs, the TERS technique was applied. We collected TERS spectra comprising distinct bands related to various molecular vibrations derived from the microvesicle membrane components, such as proteins, cholesterol, sphingomyelin, and aminophospholipids. The TERS spectra obtained from different spots on the REV surface exhibited heterogeneity, suggesting variations in the local molecular composition (protein secondary structures, e.g., β-sheets and α-helixes, as well as lipids, such as cholesterol or aminophospholipids). This heterogeneity may be related to the presence of lipid domains formed by membrane proteins. TERS provides high spatial resolution, enabling nanoscale observation of protein-/cholesterol-rich and lipid-rich domains on the surface of EVs. This level of detail was previously demonstrated for cellular membranes, but not for EVs in a label-free and non-destructive manner.
Raman spectroscopy, the basis for both SERS and TERS, provides specific information on the molecular composition of microvesicles. This enables the identification of biomolecules such as proteins and lipids within the microvesicle membrane. In SERS, vibrations within the membrane components can be enhanced, and the technique is highly sensitive, although it provides information from random parts of the sample, depending on the nanoparticle's position.
On the other hand, TERS offers information from the local parts of the membrane's outer surface, providing an analysis of the membrane surface and demonstrating high heterogeneity in the calculated TERS correlation maps. Both, TERS and SERS are powerful techniques for the surface analysis of REVs, offering high sensitivity, molecular specificity, non-destructiveness, high spatial resolution, and versatility. They are expected to significantly contribute to our understanding of these biologically important entities at the single-molecule level.
Author contributions
Conceptualization and visualization: T. S., E. L., K. S. and K. M. M.; methodology: T. S., K. S., J. D., E. W., K. B., E. S.-M., and S. S.; formal analysis: T. S., K. S., N. W., J. D., K. B., E. S.-M., K. S.-N., E. L., and K. M. M.; investigation: T. S., K. S., J. D., E. W., K. B., E. S.-M., and S. S.; data curation, K. M. M.; writing – original draft preparation: T. S., K. S., N. W., J. D., K. B., and K. M.; writing – review and editing: J. D., E. S.-M., J. C., E. L., K. S., and K. M. M.; supervision, E. L. and K. M. M.; project administration, K. M. M.; funding acquisition, K. M. M. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Polish National Science Centre, No. UMO-2020/38/E/ST4/00197.References
- F. Gaignier, C. Legrand-Frossi, E. Stragier, J. Mathiot, J. L. Merlin, C. Cohen-Salmon, L. Lanfumey and J. P. Frippiat, Front. Physiol., 2018, 9, 514, DOI:10.3389/fphys.2018.00514.
- N. Maugeri, L. Campana, M. Gavina, C. Covino, M. De Metrio, C. Panciroli, L. Maiuri, A. Maseri, A. D'Angelo, M. E. Bianchi, P. Rovere-Querini and A. A. Manfredi, J. Thromb. Haemostasis, 2014, 12, 2074–2088, DOI:10.1111/jth.12710.
- K. W. Witwer and C. Théry, J. Extracell. Vesicles, 2019, 8(1), 1648167, DOI:10.1080/20013078.2019.1648167.
- C. Sun, H. Wang, S. Mao, J. Liu, S. Li and J. Wang, Immunol. Lett., 2015, 164(2), 65–71, DOI:10.1016/j.imlet.2015.02.007.
- M. Schmelz, Front. Med., 2019, 6, 167, DOI:10.3389/fmed.2019.00167.
- L. Gámez-Díaz, E. C. Sigmund, V. Reiser, W. Vach, S. Jung and B. Grimbacher, Front. Immunol., 2018, 9, 720, DOI:10.3389/fimmu.2018.00720.
- I. I. Høiland, R. A. Liang, K. Hindberg, N. Latysheva, O. L. Brekke, T. E. Mollnes and J. B. Hansen, Thromb. Res., 2018, 169, 50–56, DOI:10.1016/j.thromres.2018.06.019.
- Y. Hisada and N. Mackman, Curr. Opin. Hematol., 2019, 26(5), 349–356, DOI:10.1097/moh.0000000000000521.
- M. Kaczmarska, M. Grosicki, K. Bulat, M. Mardyla, E. Szczesny-Malysiak, A. Blat, J. Dybas, T. Sacha and K. M. Marzec, Nanomedicine, 2020, 28, 102221, DOI:10.1016/j.nano.2020.102221.
- E. Pariset, S. Malkani, E. Cekanaviciute and S. V. Costes, Int. J. Radiat. Biol., 2021, 97, 132–150, DOI:10.1080/09553002.2020.1820598.
- H. Devarbhavi, D. Ganga, M. Menon, K. Kothari and R. Singh, J. Gastroenterol. Hepatol., 2020, 35, 1223–1228, DOI:10.1111/jgh.15213.
- M. Sala, M. Ros and F. Saltel, Front. Oncol., 2020, 10, 1620, DOI:10.1080/09553002.2020.1820598.
- M. Zarà, G. F. Guidetti, M. Camera, I. Canobbio, P. Amadio, M. Torti, E. Tremoli and S. S. Barbieri, Int. J. Mol. Sci., 2019, 20(11), 2840, DOI:10.3390/ijms20112840.
- K. Thangaraju, S. N. Neerukonda, U. Katneni and P. W. Buehler, Int. J. Mol. Sci., 2020, 22, 153, DOI:10.3390/IJMS22010153.
- L. Yang, S. Huang, Z. Zhang, Z. Liu and L. Zhang, Int. J. Mol. Sci., 2022, 23, 5927, DOI:10.3390/ijms23115927.
- W. Chiangjong, P. Netsirisawan, S. Hongeng and S. Chutipongtanate, Front. Med., 2021, 8, 761362, DOI:10.3389/fmed.2021.761362.
- L. J. Bruce, R. Beckmann, M. L. Ribeiro, L. L. Peters, J. A. Chasis, J. Delaunay, N. Mohandas, D. J. Anstee and M. J. A. Tanner, Blood, 2003, 101(10), 4180–4188, DOI:10.1182/blood-2002-09-2824.
- E. Szczesny-Malysiak, T. Mohaissen, K. Bulat, M. Kaczmarska, A. Wajda and K. M. Marzec, Haematologica, 2021, 106, 2779–2782, DOI:10.3324/HAEMATOL.2021.278895.
- C. R. F. Cafezeiro, M. P. Lopes, C. T. Silva, M. S. Ávila, L. F. B. C. Seguro, S. Mangini, I. W. Campos, F. A. Gaiotto, F. G. Marcondes-Braga and F. Bacal, Am. J. Transplant., 2020, 20(5), 1451–1453, DOI:10.1111/ajt.15744.
- X. X. Han, R. S. Rodriguez, C. L. Haynes, Y. Ozaki and B. Zhao, Nat. Rev. Methods Primers, 2021, 1, 87, DOI:10.1038/s43586-021-00083-6.
- E. Szczesny-Malysiak, J. Dybas, A. Blat, K. Bulat, K. Kus, M. Kaczmarska, A. Wajda, K. Malek, S. Chlopicki and K. M. Marzec, Biochim. Biophys. Acta, Mol. Cell Res., 2020, 1867(11), 118803, DOI:10.1016/j.bbamcr.2020.118803.
- T. Stepanenko, G. Zając, A. Czajkowski, W. Rutkowska, A. Górecki, K. M. Marzec and J. Dybas, Biochim. Biophys. Acta, Mol. Cell Res., 2023, 1870(1), 119378, DOI:10.1016/j.bbamcr.2022.119378.
- A. Blat, T. Stepanenko, K. Bulat, A. Wajda, J. Dybas, T. Mohaissen, F. C. Alcicek, E. Szczesny-Malysiak, K. Malek, A. Fedorowicz and K. M. Marzec, Int. J. Mol. Sci., 2021, 22, 1–15, DOI:10.3390/ijms22052660.
- K. Bulat, J. Dybas, M. Kaczmarska, A. Rygula, A. Jasztal, E. Szczesny-Malysiak, M. Baranska, B. R. Wood and K. M. Marzec, Analyst, 2020, 5(145), 1749–1758, 10.1039/c9an01707k.
- J. Dybas, K. Bulat, A. Blat, T. Mohaissen, A. Wajda, M. Mardyla, M. Kaczmarska, M. Franczyk-Zarow, K. Malek, S. Chlopicki and K. M. Marzec, Biochim. Biophys. Acta, Mol. Basis Dis., 2022, 146, 116481, DOI:10.1016/j.bbadis.2020.165972.
- A. Blat, J. Dybas, M. Kaczmarska, K. Chrabaszcz, K. Bulat, R. B. Kostogrys, A. Cernescu, K. Malek and K. M. Marzec, Anal. Chem., 2019, 91(15), 9867–9874, DOI:10.1021/acs.analchem.9b01536.
- A. L. Chang, Y. Kim, A. P. Seitz, R. M. Schuster, A. B. Lentsch and T. A. Pritts, Shock, 2017, 47, 632–637, DOI:10.1097/SHK.0000000000000780.
- K. F. Domke, D. Zhang and B. Pettinger, J. Phys. Chem. C, 2007, 111, 8611–8616, DOI:10.1021/jp071519l.
- E. Lipiec, A. Japaridze, J. Szczerbiński, G. Dietler and R. Zenobi, Small, 2016, 12, 4821–4829, DOI:10.1002/SMLL.201601711.
- E. Lipiec, D. Perez-Guaita, J. Kaderli, B. R. Wood and R. Zenobi, Angew. Chem., Int. Ed., 2018, 57, 8519–8524, DOI:10.1002/ANIE.201803234.
- K. Malek, Vibrational Spectroscopy: From Theory to Applications, PWN, 2016, ISBN: 978-83-01-18893-1 Search PubMed.
- K. Czamara, K. Majzner, M. Z. Pacia, K. Kochan, A. Kaczor and M. Baranska, J. Raman Spectrosc., 2015, 46(1), 4–20, DOI:10.1002/jrs.4607.
- C. Krafft, L. Neudert, T. Simat and R. Salzer, Spectrochim. Acta, Part A, 2005, 61(7), 1529–1535, DOI:10.1016/j.saa.2004.11.017.
- B. Hernández, F. Pflüger, S. G. Kruglik and M. Ghomi, J. Raman Spectrosc., 2013, 44(6), 827–833, DOI:10.1002/jrs.4290.
- H. Takeuchi, Biopolymers, 2003, 72(5), 305–317, DOI:10.1002/bip.10440.
- A. Rygula, K. Majzner, K. M. Marzec, A. Kaczor, M. Pilarczyk and M. Baranska, J. Raman Spectrosc., 2013, 44, 1061–1076, DOI:10.1002/JRS.4335.
- C. H. Chuang and Y. T. Chen, J. Raman Spectrosc., 2009, 40(2), 150–156, DOI:10.1002/jrs.2097.
- C. W. Lai, M. Schwab, S. C. Hill, J. Santarpia and Y.-L. Pan, Opt. Express, 2016, 24(11), 11654–11667, DOI:10.1364/oe.24.011654.
- S. Kumar, A. K. Rai, S. B. Rai and D. K. Rai, Indian J. Phys., 2010, 84, 563–573, DOI:10.1007/s12648-010-0039-6.
- H. Akutsu and Y. Kyogoku, Chem. Phys. Lipids, 1975, 14(2), 113–122, DOI:10.1016/0009-3084(75)90053-5.
- M. Gniadecka, H. C. Wulf, N. N. Mortensen, O. F. Nielsen and D. H. Christensen, J. Raman Spectrosc., 1997, 28, 125–129, DOI:10.1002/(SICI)1097-4555(199702)28:2/3<125::AID-JRS65>3.0.CO;2-%23.
- M. Wegener, R. Neubert, W. Rettig and S. Wartewig, Int. J. Pharm., 1996, 126(1–2), 203–213, DOI:10.1016/0378-5173(95)04309-8.
- M. Janssens, G. S. Gooris and J. A. Bouwstra, Biochim. Biophys. Acta, Biomembr., 2009, 1788(3), 732–742, DOI:10.1016/j.bbamem.2009.01.003.
- J. F. Hsu, P. Y. Hsieh, H. Y. Hsu and S. Shigeto, Sci. Rep., 2015, 5, 1–8, DOI:10.1038/srep17541.
- C. Gao, L. Yu, L. Ma, X. Lu, S. Wu, P. Song and L. Xia, Chem. Phys. Lett., 2020, 745, 137273, DOI:10.1016/J.CPLETT.2020.137273.
- G. Zhu, X. Zhu, Q. Fan and X. Wan, Spectrochim. Acta, Part A, 2011, 78, 1187–1195, DOI:10.1016/j.saa.2010.12.079.
- E. Lipiec, J. Kaderli, J. Kobierski, R. Riek, K. Skirlińska-Nosek, K. Sofińska, M. Szymoński and R. Zenobi, Angew. Chem., Int. Ed., 2021, 60, 4545–4550, DOI:10.1002/ANIE.202010331.
- V. A. Shashilov, V. Sikirzhytski, L. A. Popova and I. K. Lednev, Methods, 2010, 52(1), 23–37, DOI:10.1016/j.ymeth.2010.05.004.
- J. Szczerbiński, J. B. Metternich, G. Goubert and R. Zenobi, Small, 2020, 16(4), 1–9, DOI:10.1002/smll.201905197.
- D. Mrđenović, W. Ge, N. Kumar and R. Zenobi, Angew. Chem., Int. Ed., 2022, 61(43), 1–8, DOI:10.1002/ANIE.202210288.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01658g |
| This journal is © The Royal Society of Chemistry 2024 |