Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sodium fluoride preserves blood metabolite integrity for biomarker discovery in large-scale, multi-site metabolomics investigations†
Wenzheng
Xiong
 ab,
Daniel C.
Anthony
b,
Suzie
Anthony
c,
Thi Bao Tien
Ho
ab,
Daniel C.
Anthony
b,
Suzie
Anthony
c,
Thi Bao Tien
Ho
 b,
Edouard
Louis
d,
Jack
Satsangi
e and
Daniel E.
Radford-Smith
b,
Edouard
Louis
d,
Jack
Satsangi
e and
Daniel E.
Radford-Smith
 *ab
*ab
aDepartment of Chemistry, University of Oxford, Oxford, UK. E-mail: daniel.radford-smith@some.ox.ac.uk
bDepartment of Pharmacology, Medical Sciences Division, University of Oxford, Oxford, UK
cDepartment of Radiology, Oxford University Hospitals NHS Foundation Trust, Oxford, UK
dDepartment of Gastroenterology, University Hospital CHU of Liège, Liège, Belgium
eTranslational Gastroenterology Unit, Nuffield Department of Experimental Medicine, University of Oxford, Oxford, UK
First published on 12th January 2024
Abstract
Background: Metabolite profiling of blood by nuclear magnetic resonance (NMR) is invaluable to clinical biomarker discovery. To ensure robustness, biomarkers require validation in large cohorts and across multiple centres. However, collection procedures are known to impact on the stability of biofluids that may, in turn, degrade biomarker signals. We trialled three blood collection tubes with the aim of solving technical challenges due to preanalytical variation in blood metabolite levels that are common in cohort studies. Methods: We first investigated global NMR-based metabolite variability between biobanks, including the large-scale UK Biobank and TwinsUK biobank of the general UK population, and more targeted biobanks derived from multicentre clinical trials relating to inflammatory bowel disease. We then compared the blood metabolome of 12 healthy adult volunteers when collected into either sodium fluoride/potassium oxalate, lithium heparin, or serum blood tubes using different pre-processing parameters. Results: Preanalytical variation in the method of blood collection strongly influences metabolite composition within and between biobanks. This variability can largely be attributed to glucose and lactate. In the healthy control cohort, the fluoride oxalate collection tube prevented fluctuation in glucose and lactate levels for 24 hours at either 4 °C or room temperature (20 °C). Conclusions: Blood collection into a fluoride oxalate collection tube appears to preserve the blood metabolome with delayed processing up to 24 hours at 4 °C. This method may be considered as an alternative when rapid processing is not feasible.
Introduction
Metabolite profiling of serum and plasma is emerging as an invaluable tool spanning multiple domains of medical research.1 Nuclear magnetic resonance (NMR)-based metabolomics techniques involve the simultaneous profiling of many metabolites (small molecular intermediates) in a biofluid or other medium.2 Inexpensive, high-throughput, and highly reproducible, global metabolite quantification by NMR is now being employed at the frontier of medicine, ranging from population-wide health profiling3,4 to predicting diagnosis,5 progression,6 and response to treatment7 in a spectrum of diseases.Clinical biomarkers require validation in large cohorts across multiple centres.8 NMR-based metabolomics requires minimal sample preparation compared to other metabolomics techniques (i.e. mass spectrometry methods).9 Nevertheless, centrifugation speed and time,10 incubation temperature,11–13 storage time at −20 °C and −80 °C,13,14 the type of collection tube,15,16 and the number of freeze–thaw cycles11,14,17,18 are just some of the pre-analytical variables known to strongly influence metabolite composition in blood. While many of these variables may be able to be well-controlled within a given study, the time between blood collection and erythrocyte separation by centrifugation and the time to sample storage at −80 °C present unique challenges due to the pragmatic requirements of large-scale, multi-centre clinical trials from which clinical biomarkers may be robustly identified.13,19,20 These two factors have been shown to drastically influence the relative abundance of metabolites involved in glycolysis.10–12,14,18,21–26 To date, it has not been investigated whether the type of blood collection tube used affects metabolite integrity due to a delay in centrifugation and/or sample measurement. Moreover, conflicting evidence exists as to whether other metabolites not directly involved in glycolysis, such as lipoproteins, are affected by time to centrifugation.14,21 In this study we aimed to systematically evaluate and compare the relative efficacy of sodium fluoride/potassium oxalate, lithium heparin, and additive-free serum blood tubes in preserving metabolite integrity for up to 24-hours before centrifugation, and up to 48-hours before sample measurement after NMR sample preparation. We expected that sodium fluoride/potassium oxalate blood tubes, which inhibit glycolysis, preserve the metabolic state at the time of blood collection more effectively than the widely used lithium heparin or serum blood vacutainers.
Ideally, standard operating procedures (SOPs) are, prospectively, put in place in advance of longitudinal, multicentre research projects to reduce pre-analytical variability in metabolite levels within and between biobanks. In practice, this may be difficult to achieve depending on the scale and resources of individual collection sites. Previous studies have demonstrated conflicting efficacy of keeping blood tubes refrigerated at 4 °C prior to centrifugation, aliquoting, and freezing on the variability in metabolite levels.13,18,26 Moreover, a secure cold chain may not be feasible in all sites, and the level of compliance to SOPs is unclear and is often left unreported. Most importantly, all previous studies addressing these pre-analytical variables treated all samples identically when varying time to erythrocyte separation and time at room temperature (RT). Therefore, it is largely unsurprising that, in at least one analytical dimension, inter-individual differences are maintained. In practice, this identical treatment of individual differences does not occur; samples may be processed at any time between, for example, 0 and upwards of 24 hours.
Guidelines on how to prepare samples and observations on the effects of delayed erythrocyte separation are insufficient to advance methods of reducing biomarker variability for NMR. Similarly, z-scoring of metabolite concentrations to facilitate comparisons across cohorts3,27 impedes the discovery of healthy and disease-associated metabolite ranges that may be clinically relevant. Practical solutions to stably preserve blood metabolite levels are required, and clear reporting guidelines are required to explain potential non-biological between-sample variability. In this study, we first investigate global NMR-based metabolite variability between biobanks, including the large-scale UK Biobank and TwinsUK biobank of the general UK population, and more targeted biobanks derived from multicentre clinical trials relating to IBD. We then compared the blood metabolome of 12 healthy adult volunteers when collected into either sodium fluoride/potassium oxalate, lithium heparin, or serum blood tubes using different pre-processing parameters, with the aim of determining the optimal tube type for preserving an individual's blood metabolomic fingerprint with significantly delayed sample processing of up to 24 hours.
Methods
Cohorts
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411 individuals of middle and old age (mean 56.5 years, range 37–73 years). Written, informed consent was provided by all participants. The UK Biobank has generic ethical approval from the Northwest Multi-centre Research Ethics Committee (ref. 11/NW/03820). The current study is registered under the approved research ID 95409. At the time of this study, NMR metabolomic data was available on ∼120
411 individuals of middle and old age (mean 56.5 years, range 37–73 years). Written, informed consent was provided by all participants. The UK Biobank has generic ethical approval from the Northwest Multi-centre Research Ethics Committee (ref. 11/NW/03820). The current study is registered under the approved research ID 95409. At the time of this study, NMR metabolomic data was available on ∼120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 participants. Individuals were typically non-fasting prior to blood sampling (median fasting time 3 hours, standard deviation 2.4 hours). Whole blood was collected into spray-coated K2EDTA tubes and immediately centrifuged at 2500g for 10 minutes at 4 °C. Plasma was not aliquoted. Rather, tubes were transported to a central storage facility at 4 °C for aliquoting and archiving at −80 °C (mean time from venepuncture to freezing 24 hours, standard deviation 2.5 hours).28 Acquisition of NMR metabolite data was performed by Nightingale Health Ltd and is described in detail elsewhere.4
000 participants. Individuals were typically non-fasting prior to blood sampling (median fasting time 3 hours, standard deviation 2.4 hours). Whole blood was collected into spray-coated K2EDTA tubes and immediately centrifuged at 2500g for 10 minutes at 4 °C. Plasma was not aliquoted. Rather, tubes were transported to a central storage facility at 4 °C for aliquoting and archiving at −80 °C (mean time from venepuncture to freezing 24 hours, standard deviation 2.5 hours).28 Acquisition of NMR metabolite data was performed by Nightingale Health Ltd and is described in detail elsewhere.4
In both STORI and SPARE, biomarkers associated with relapse was a pre-defined secondary endpoint. The standard operating procedure for serum collection and processing is provided in the ESI.† Briefly, blood was collected into BD Vacutainer SST II Advance Tubes (BD 367958), allowed to clot, and centrifuged at room temperature for 10 minutes at 3000g. The target for clotting time at room temperature was between 0.5 and 2 h, and the target time-to-freezing of serum aliquots was within 4 hours of the blood draw. Serum aliquots were then stored at −80 °C. All patients provided informed consent and ethical approval was obtained. All patients had been treated with a combination therapy of infliximab (IFX) and anti-metabolite >8 months and had been in sustained steroid-free remission >6 months. Patients were not fasted prior to blood collection.
Blood was collected into BD Vacutainer blood tubes® under the conditions described in Table 1. Vacutainer tubes were centrifuged initially at 300g for 10 minutes at 4 °C. After the supernatant (either serum or plasma) was carefully collected into a new tube, the sample was again centrifuged at 5000g for 10 minutes at 4 °C and the supernatant collected into a new tube and stored at −80 °C prior to sample preparation.
| Tube | Time to centrifugation (hours) | Delay between sample preparation and measurement (hours) |
|---|---|---|
| Serum (Red, BD 367837) | 0.5 RT | 0 (RT), 12 (4 °C), 24 (4 °C), 48 (4 °C) |
| 24 RT | 0 (RT) | |
| 24 4 °C | 0 (RT) | |
| Lithium Heparin Plasma (Green, BD 367885) | 0.5 RT | 0 (RT), 12 (4 °C), 24 (4 °C), 48 (4 °C) |
| 24 RT | 0 (RT) | |
| 24 4 °C | 0 (RT) | |
| Fluoride/Oxalate Plasma (Grey, BD 368921) | 0.5 RT | 0 (RT), 12 (4 °C), 24 (4 °C), 48 (4 °C) |
| 24 RT | 0 (RT) | |
| 24 4 °C | 0 (RT) |
NMR spectroscopy
NMR spectroscopy and global metabolite quantification from serum and plasma was performed in-house for the SPARE, STORI, and healthy volunteer cohorts. On the day of NMR spectroscopy, serum or plasma samples were thawed at room temperature, mixed by pipetting, and combined with 75 mM sodium phosphate buffer prepared in D2O (pH 7.4) as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio of serum to buffer (total volume 600 μL) in a 5 mm borosilicate glass NMR tube (Norell 502-7).
5 ratio of serum to buffer (total volume 600 μL) in a 5 mm borosilicate glass NMR tube (Norell 502-7).
1H NMR spectra were acquired at 298 K using a 700 MHz Bruker AVIII spectrometer operating at 16.4 T equipped with a 1H [13C/15N] TCI cryoprobe (Department of Chemistry, University of Oxford). 1D NOESY and CPMG experiments were conducted on each sample, and CPMG spectra were used for downstream analysis. Pooled plasma samples were spread throughout the run of the SPARE and STORI samples to monitor technical variation.
NMR Spectra were phased, baseline corrected (using a fifth-degree polynomial), and referenced to the lactate-CH3 doublet resonance at δ = 1.33 ppm in Topspin 4.1.4 (Bruker). Spectra were exported to ACD/Labs Spectrus Processor Academic Edition 12.01 (Advanced Chemistry Development, Inc.) for data-preprocessing. The NMR spectra were segmented into 100 integral regions using a manual binning approach, with exclusion of noise and water signals. The integration of metabolite peaks was performed judiciously to minimize overlapping and aligned with literature assignments. Resonances were assigned to metabolites using a combination of literature values and 2D spectra of human plasma and serum, leading to the confident identification of 39 metabolites. Spectral regions with overlapped metabolites were annotated in the assignment using a ‘/’ and ‘[ ]’ notation. The absolute integral values were subject to the main statistical analysis unless stated otherwise. Additional statistical analyses using probabilistic quotient normalisation (PQN)33 and sum normalisation were included in the ESI.† In the healthy control cohort, lactate and glucose levels were also calculated based on the absolute mmol concentration using a standard curve.
Statistical analysis
Statistical analysis was performed in R 4.1.2 and GraphPad Prism 9. After pareto scaling of the absolute integral values, multivariate principal component analysis (PCA) was performed using the ropls package to visualise and compare individual metabolite profiles in an unsupervised fashion.34Post-hoc analysis of TwinsUK, STORI, and SPARE NMR data was limited to univariate t-testing of glucose and lactate metabolite levels, based on inspection of the corresponding PCA loadings plots. UK Biobank NMR metabolite data were further correlated against time of day of blood collection, fasting time, and delay in sample measurement using Pearson's correlation coefficient. In the healthy control cohort, individual metabolites (39 identified metabolites) were subject to repeated measures two-way ANOVA with Šidák post hoc test,35 and Benjamini–Hochberg method was used for p value adjustment to control the false discovery rate across all metabolites. Adjusted two-tailed p values (q values) ≤ 0.05 were considered statistically significant. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Results
Lactate and glucose drive metabolite variation between and within biobanks
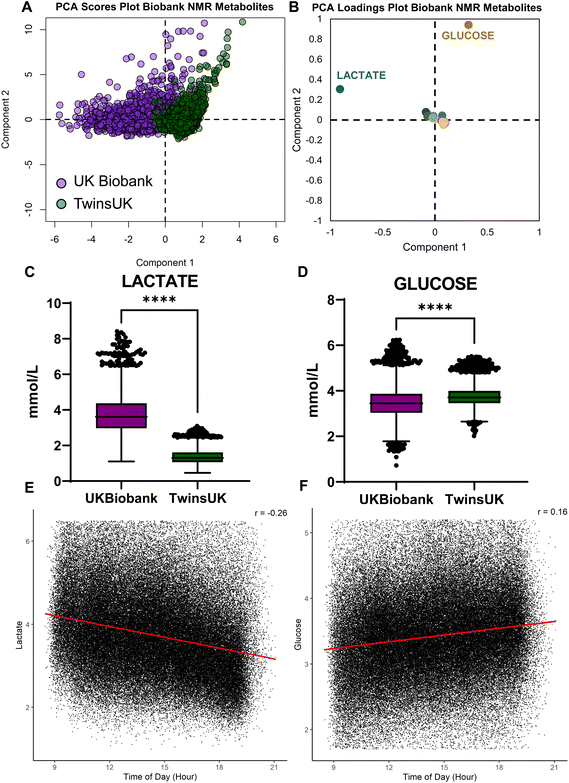
NMR metabolite data was collated between the UK Biobank and TwinsUK cohorts. As the TwinsUK cohort is predominately female sex, a female cohort from the UK Biobank (n = 4830) was randomly sampled for comparison. Principal component analysis revealed greater variation in the metabolome within the UK Biobank cohort compared to the TwinsUK cohort along the first principal component, as well as systematic variation between the two biobanks along the same component (Fig. 1A). Inspection of the corresponding loadings plot revealed the key role of lactate in driving this variation along the first principal component, and glucose in driving within-biobank variation along the second principal component (Fig. 1B).Lactate levels were increased in the UK Biobank cohort (mean 3.7) relative to the TwinsUK cohort (mean 1.37; mean difference 2.36 [Welch's t-test 95%CI 2.33–2.40], Fig. 1C). Glucose levels were decreased in the UK Biobank cohort (mean 3.48) relative to the TwinsUK cohort (mean 3.76; mean difference 0.29 [Welch's t-test 95%CI 0.26–0.31], Fig. 1D). Using available metadata pertaining to the UK Biobank, we identified systematic, linear variation in plasma lactate (Pearson's r = −0.26, n = 118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021, Fig. 1E) and glucose (Pearson's r = 0.16, n = 118
021, Fig. 1E) and glucose (Pearson's r = 0.16, n = 118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 021, Fig. 1F) levels according to the time of sample collection. This may correspond to a systematic delay between sample collection and centrifugation according to queued collected samples.
021, Fig. 1F) levels according to the time of sample collection. This may correspond to a systematic delay between sample collection and centrifugation according to queued collected samples.
As a second example, and on a smaller scale, lactate and glucose were also found to drive serum metabolome variation between clinical biobank data pertaining to the SPARE and STORI cohorts (Fig. 2). Here, data acquisition and relative metabolite quantification were performed in-house. The PCA scores plot (Fig. 2A) and corresponding loadings plot (Fig. 2B) illustrate systematic variation in lactate and glucose levels along the second principal component. Glucose levels were decreased in the STORI cohort (mean 0.0058) relative to the SPARE cohort (mean 0.011; mean difference 0.0055 [Welch's t-test 95%CI 0.0046–0.63]). Lactate levels were increased in the STORI cohort (mean 0.13) relative to the SPARE cohort (mean 0.053; mean difference 0.073 [Welch's t-test 95%CI 0.064–0.082]). These differences are visualised in the median NMR spectrum of each cohort (Fig. 2E).
Effects of delayed sample measurement on absolute plasma and serum metabolite concentrations
UK Biobank metabolomic metadata was used to determine the effect of time between sample preparation and measurement on plasma metabolite levels. Approximately 25% of samples were run within 12 hours of sample preparation, 63% within 24 hours, and 97% within 48 hours (Fig. 3A). The top six metabolites had a Pearson correlation coefficient >0.1 (p < 0.0001) and are shown in Fig. 3B–G. Plasma histidine concentration showed a strong negative correlation with time to sample preparation (Pearson's r = −0.36, Fig. 3B). Glycine (r = −0.14), glutamine (r = −0.13), phenylalanine (r = −0.09), citrate (r = −0.10) were also negative correlated with time to sample preparation, while free cholesterol in intermediate density lipoprotein (r = 0.10) was positive correlated (Fig. 3C–G, respectively).In the healthy control cohort (n = 12 per condition), we found changes in the metabolome as a result of delayed sample measurement (Fig. 4). Consistent with data from the UK Biobank, the effect of delayed sample measurement was small relative to interindividual variation (Fig. 4A, B, and S1†). Most samples shifted in the second component in the PCA scores plot, driving by decreased levels of glucose, high-density lipoprotein (HDL) and glycerophosocholine (Fig. 4C and D). The stability of the serum/plasma metabolome was maintained even after a 12-hour delay in NMR measurement, as demonstrated by minimal changes in metabolite levels (Fig. 4C). However, when LH Plasma samples were subjected to a 24-hour delay in measurement, a significant decrease was observed in 31 out of 39 metabolites. Notably, it was observed that LH Plasma samples started forming precipitates after 12 hours, and all samples exhibited precipitates after 24 hours. In contrast, serum and FX plasma demonstrated better stability than LH Plasma after a 24-hour and even 48-hour delay in measurement as indicated by changes in percentages of most metabolites below 10% and significant alterations observed in only a few metabolites (Table S1†). The majority of metabolites showed a decreasing trend resulting from the delay in measurement, except for glutamate, which tended to increase over time, particularly in FX plasma (Fig. 4C). Albumin, phosphocholine, glycerophosphocholine and glucose levels significantly decrease after 24 hours in at least two types of plasma/serum samples. Aligning with our observations in the UK Biobank dataset, histidine and glutamine levels tend to decrease over time, especially after 24 hours in LH Plasma (Fig. 4E and F, and Table S1†).
Metabolite differences by tube type using optimal processing parameters in the healthy control cohort
We next investigated how abundances of metabolites differed between BD Vacutainers under optimal processing conditions. Absolute metabolite concentrations of LH plasma (green) and serum (red) tubes were comparable (Fig. S2, and Table S2†). In contrast, the total integral of the spectra (spectral intensity) from FX plasma (grey) was reduced in both the NOESY and CPMG experiments due to the decline in albumin and lipoprotein levels in FX plasma (Fig. S2†). When using a PQN or sum-normalised approach to adjust for the total integral between samples and make FX plasma comparable, few differences remained between tube types (Table S2†). Branched chain amino acids (BCAAs, including leucine, isoleucine, valine) were increased in FX tubes relative to LH plasma and serum. And 3-hydroxybutyrate levels increased in FX plasma compared to LH plasma and serum, potentially influenced by variations in the lipoprotein line shape (Fig. S2†).Evaluation of a fluoride/oxalate additive to preserve inter-individual metabolite variation using realistic timepoints
Lastly, we investigated whether the combination of sodium fluoride and potassium oxalate in a BD Vacutainer could mitigate against preanalytical variation in the individual blood metabolome. As expected, 24-hours at either RT or 4 °C resulted in an increase in lactate and a decrease in glucose when stored in either LH plasma or serum tubes (Fig. 5A–D, and Fig. S3†). When LH plasma samples were centrifuged after a 24-hour delay at 4 °C, the lactate levels increased 34% and the glucose levels decreased 18% (Table S2†). The alterations were similar for the serum samples, and further exacerbated when the samples were left at RT prior to centrifugation. Conversely, FX tubes effectively prevented anaerobic glycolysis for at least 24-hours at RT. Only 7% decrease was observed in glucose levels when FX plasma samples were centrifuged after a 24-hour delay at 4 °C. It should be noted that as demonstrated previously, glucose levels decreased 6% in FX plasma (q = 0.096, two-way ANOVA, sidak post hoc, FDR correction across metabolites, Table S1†) after 24 hours delay in NMR measurement at 4 °C. Therefore, the decrease of glucose in FX tubes was likely due to general degradation not due to anaerobic glycolysis.Moreover, the combination of using FX tubes and refrigerating the blood tube during the pre-processing interval effectively prevented metabolite variation between an optimal, 30-minute pre-centrifugation delay and the 24-hour delay (Fig. 5A and E). Without refrigeration, glutamate and glutamine levels changed dramatically after 24 hours at RT (Table S2†).
Discussion
Blood is the main biofluid utilised in epidemiological studies utilising metabolomics to predict disease onset, severity, and treatment response. We hypothesised that fluoride oxalate vacutainer blood collection tubes would thus be a more appropriate blood tube for metabolome studies. To test this hypothesis, we systematically investigated the effect of preanalytical variation in blood sample handling in lithium heparin, fluoride oxalate, and serum blood tubes. To our knowledge, we demonstrate for the first time that the use of FX plasma tubes in combination with refrigeration at 4 °C negates metabolome variation due to preanalytical factors. This is important, as lactate and glucose are established biomarkers for the early diagnosis of cancer,36,37 and may prove important to the diagnosis and prediction of other diseases when integrated into multivariate blood metabolite algorithms.3 Our findings suggest that fluoride oxalate tubes could be beneficial in metabolomic studies where there is variability in blood processing times, pending further validation with a broader range of metabolites and larger sample sizes.Our initial investigations revealed that much of the variation in metabolite levels within and between blood biobank cohorts could be attributed to variations in glycolysis metabolites. This variation is attributed to residual anaerobic glycolysis occurring when erythrocyte separation from serum or plasma is delayed. Delayed sample processing due to a high volume of samples being processed (UK Biobank) or collection methods where the rapid processing of bloods was not prioritised (STORI) led to increased lactate and decreased glucose resonances compared to cohorts where rapid blood processing was prioritised (TwinsUK and SPARE cohorts). As normal lactate levels are <2 mmol,38 the absolute concentrations reported in the UK Biobank are not suitable for clinical translation.
Sodium fluoride and potassium oxalate act in concert to inhibit glycolysis in blood samples.39 Specifically, sodium fluoride inhibits the enzyme enolase, which catalyses the conversion of 2-phosphoglycerate (2-PG) into phosphoenolpyruvate (PEP), the penultimate step in glycolysis. Potassium oxalate precipitates calcium ions to inhibit the clotting cascade. Fluoride oxalate tubes, as expected, showed baseline differences in amino acid levels and glycolytic metabolites compared to serum.15 Consistent with other studies, the metabolite profile of serum and lithium heparin plasma were comparable,15 and behaved similarly in response to delayed erythrocyte separation.21
Previous studies have documented the effects of delayed erythrocyte separation on metabolite levels as quantified by NMR-based metabolomics techniques.10,12–14,17,19,21–24,40 Consistent with our own study, lactate and glucose are typically most affected. However, in contrast to other reports,18 we also clearly show in both the UK Biobank and healthy volunteer data that residual glycolytic metabolism does occur (in the absence of an inhibitor of glycolysis) even when blood is stored at 4 °C prior to centrifugation.
After separation of serum or plasma from erythrocytes, metabolite variation is minimal. In the UK Biobank cohort, only histidine showed a significant effect of delay in sample measurement (r = −0.39) compared to interindividual biological variation. Other metabolites, while significantly correlated with measurement delay, showed smaller effect sizes between 0.01 and 0.21 which are less relevant relative to the observed interindividual variation. In our healthy volunteer study, we also investigated a delay in sample measurement over a similar timeframe, for up to 48 hours at 4 °C. Again, we found minimal variation in metabolite levels, consistent with the UK Biobank data and early UK Biobank pilot studies that tested delays of up to 24 hours at 4 °C.41 Such variation is suitable to be corrected by previously described statistical techniques.42
We also provide suggestions for reporting preanalytical variation to aid in the synergy of future metabolomic studies. The reporting guideline consists of key information relating to the individual, the pre-processing variables, and the post-processing variables that may differ between samples in a single study (Table 2). The guidelines do not include variables that should be controlled for within a study and ideally between studies, for example, long-term storage temperature of frozen samples, and NMR spectra acquisition parameters. We have shown that fasting time can strongly influence the concentration of certain metabolites, primarily ketone bodies (Fig. S4†). Recording the time of blood collection, centrifugation, and storage temperatures allows studies to record SOP compliance, for which data is sorely lacking in biomarker research, and may also help identify mistreated samples. The UK Biobank has implemented pipelines that allow for automated reporting of key information. For example, barcoded blood tubes were used that, when scanned, allowed for automated logging of sample collection and processing.43
| Variable | Reported by |
|---|---|
| Time since last meal (hours) | Participant |
| Time of blood collection | Phlebotomist |
| Storage temperature pre-centrifugation | |
| Time centrifuged | Blood processing technician |
| Temperature during centrifugation | |
| Time frozen | |
| Time of NMR sample preparation | NMR technician |
| Storage temperature pre-data acquisition | |
| Time of data acquisition |
We recognise the limitations of this study. When comparing the UK Biobank and TwinsUK metabolomics datasets, we limited the comparison to non-lipid data (amino acids and energy metabolites) due to non-overlapping metabolite data and uncertainty in how these values were derived between the two cohorts. While our study provided insights using NMR-based metabolite profiling, which focused on metabolites commonly assayed by this technique, we acknowledge that the scope was limited with respect to the range of metabolites analysed. Future research could extend these findings by incorporating a broader spectrum of metabolomic analyses, including mass spectrometry and advanced lipoprotein assays, as well as labile metabolites detectable by NMR such as coenzymes and antioxidants,44 to provide a more comprehensive understanding of preanalytical variability across different platforms.
In the healthy control cohort, blood storage in fluoride oxalate tubes resulted in a slight negative bias in glucose levels under optimal processing conditions, which is consistent with previous reports.45 This bias did not show variation over time. As we suggest that the fluoride oxalate tube type should be implemented between and across studies, this small, systematic difference will not affect metabolome variability. However, the presence of this negative bias may affect the reported prevalence of fasting hyperglycaemia, which may be relevant for the screening of diabetes mellitus in the general population. Studies have also shown that both sodium fluoride46 and potassium oxalate47 have the potential to cause haemolysis after prolonged standing time at room temperature. We considered other, more effective methods of inhibiting glycolysis, such as citrate-buffered tubes,46 which are also less prone to haemolysing samples over time. However, these additives are known to interfere with the NMR spectrum and the concentrations of neighbouring metabolites.15 In our study, haemolysis was detected only in fluoride oxalate tubes left at room temperature, and not at 4 °C. Nevertheless, we encourage the discovery of novel additives that inhibit major metabolic pathways in blood under anaerobic conditions and in the absence of haemolysis.
Conclusions
Pre-analytical variation in blood sample collection cause variation within and between biobank biomarker studies. We trialled three blood collection tubes with the aim of solving technical challenges due to preanalytical variation in blood metabolite levels that are common in cohort studies. We found that a combination of refrigeration at 4 °C and the use of a fluoride/oxalate additive prevented changes in metabolite levels, as compared to optimal processing parameters, for at least 24 hours prior to erythrocyte separation. We also showed that the effect of delayed sample measurement on the stability of quantified metabolites was small, but dependent on the type of blood collection tube used at the pre-processing stage. Using this information, we devised a short list of key reporting guidelines that could be implemented in future cohort studies to improve the reproducibility of blood metabolite signatures as biomarkers of health and disease.Data availability
Metabolomics data have been deposited to the EMBL-EBI MetaboLights database48 (https://doi.org/10.1093/nar/gkz1019, with the identifier MTBLS8861).Author contributions
Conceptualisation – DCA & DRS; data curation – WX & DRS; formal analysis – WX & DRS; investigation – JS, EL, SA, & DRS; methodology – DRS, SA, WX, & HTBT; supervision – DCA & DRS; writing draft – WX & DRS; writing review and editing – WX, DCA, SA, HTBT, EL, JS, & DRS.Conflicts of interest
There are no conflicts to declare.References
- D. S. Wishart, Emerging applications of metabolomics in drug discovery and precision medicine, Nat. Rev. Drug Discovery, 2016, 15(7), 473–484 CrossRef CAS PubMed.
- O. Beckonert, H. C. Keun and T. M. D. Ebbels, et al., Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts, Nat. Protoc., 2007, 2(11), 2692–2703 CrossRef CAS PubMed.
- T. Buergel, J. Steinfeldt and G. Ruyoga, et al., Metabolomic profiles predict individual multidisease outcomes, Nat. Med., 2022, 28(11), 2309–2320 CrossRef CAS PubMed.
- H. Julkunen, A. Cichońska and M. Tiainen, et al., Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank, Nat. Commun., 2023, 14(1), 604 CrossRef CAS.
- D. E. Radford-Smith, E. A. Selvaraj and R. Peters, et al., A novel serum metabolomic panel distinguishes IgG4-related sclerosing cholangitis from primary sclerosing cholangitis, Liver Int., 2022, 42(6), 1344–1354 CrossRef CAS PubMed.
- F. Probert, A. Walsh and M. Jagielowicz, et al., Plasma Nuclear Magnetic Resonance Metabolomics Discriminates Between High and Low Endoscopic Activity and Predicts Progression in a Prospective Cohort of Patients With Ulcerative Colitis, J. Crohns Colitis, 2018, 12(11), 1326–1337 CrossRef.
- G. Caspani, G. Turecki and R. W. Lam, et al., Metabolomic signatures associated with depression and predictors of antidepressant response in humans: A CAN-BIND-1 report, Commun. Biol., 2021, 4(1), 903 CrossRef CAS PubMed.
- D. Vuckovic, Current trends and challenges in sample preparation for global metabolomics using liquid chromatography–mass spectrometry, Anal. Bioanal. Chem., 2012, 403(6), 1523–1548 CrossRef CAS PubMed.
- M. Jacob, A. L. Lopata and M. Dasouki, Abdel Rahman AM. Metabolomics toward personalized medicine, Mass Spectrom. Rev., 2019, 38(3), 221–238 CrossRef CAS PubMed.
- E. Jobard, O. Trédan and D. Postoly, et al., A Systematic Evaluation of Blood Serum and Plasma Pre-Analytics for Metabolomics Cohort Studies, Int. J. Mol. Sci., 2016, 17(12), 2035 CrossRef PubMed.
- P. Yin, A. Peter and H. Franken, et al., Preanalytical aspects and sample quality assessment in metabolomics studies of human blood, Clin. Chem., 2013, 59(5), 833–845 CrossRef CAS.
- D. L. Santos Ferreira, H. J. Maple and M. Goodwin, et al., The Effect of Pre-Analytical Conditions on Blood Metabolomics in Epidemiological Studies, Metabolites, 2019, 9(4), 64 CrossRef PubMed.
- J. L. Alexander, N. J. Wyatt and S. Camuzeaux, et al., Considerations for peripheral blood transport and storage during large-scale multicentre metabolome research, Gut, 2024, 73, 379–383 Search PubMed , [cited 2023 May 23]; available from: https://gut.bmj.com/content/early/2023/02/07/gutjnl-2022-329297.
- J. Pinto, M. R. M. Domingues and E. Galhano, et al., Human plasma stability during handling and storage: impact on NMR metabolomics, Analyst, 2014, 139(5), 1168–1177 RSC.
- J. Sotelo-Orozco, S.-Y. Chen, I. Hertz-Picciotto and C. M. Slupsky, A Comparison of Serum and Plasma Blood Collection Tubes for the Integration of Epidemiological and Metabolomics Data, Front. Mol. Biosci., 2021, 8, 682134 CrossRef CAS PubMed , [cited 2023 May 24]. Available from: https://www.frontiersin.org/articles/10.3389/fmolb.2021.682134.
- A. Vignoli, L. Tenori, C. Morsiani, P. Turano, M. Capri and C. Luchinat, Serum or Plasma (and Which Plasma), That Is the Question, J. Proteome Res., 2022, 21(4), 1061–1072 CrossRef CAS PubMed.
- I. H. Muti, M. Gonzalez Sanchez-Dahl and A. B. Zhong, et al., Designing a quality assurance process for quality control of nuclear magnetic resonance metabolomics studies of human blood, NMR Biomed., 2023, 36(4), e4868 CrossRef CAS PubMed.
- O. Fliniaux, G. Gaillard, A. Lion, D. Cailleu, F. Mesnard and F. Betsou, Influence of common preanalytical variations on the metabolic profile of serum samples in biobanks, J. Biomol. NMR., 2011, 51(4), 457–465 CrossRef CAS PubMed.
- C. Ellervik and J. Vaught, Preanalytical variables affecting the integrity of human biospecimens in biobanking, Clin. Chem., 2015, 61(7), 914–934 CrossRef CAS PubMed.
- G. Anton, R. Wilson and Z. Yu, et al., Pre-Analytical Sample Quality: Metabolite Ratios as an Intrinsic Marker for Prolonged Room Temperature Exposure of Serum Samples, PLoS One, 2015, 10(3), e0121495 CrossRef PubMed.
- J. Debik, S. H. Isaksen and M. Strømmen, et al., Effect of Delayed Centrifugation on the Levels of NMR-Measured Lipoproteins and Metabolites in Plasma and Serum Samples, Anal. Chem., 2022, 94(49), 17003–17010 CrossRef CAS PubMed.
- M. V. Fomenko, L. V. Yanshole and Y. P. Tsentalovich, Stability of Metabolomic Content during Sample Preparation: Blood and Brain Tissues, Metabolites, 2022, 12(9), 811 CrossRef CAS PubMed.
- P. Bernini, I. Bertini, C. Luchinat, P. Nincheri, S. Staderini and P. Turano, Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks, J. Biomol. NMR., 2011, 49(3–4), 231–243 CrossRef CAS PubMed.
- C. Brunius, A. Pedersen and D. Malmodin, et al., Prediction and modeling of pre-analytical sampling errors as a strategy to improve plasma NMR metabolomics data, Bioinformatics, 2017, 33(22), 3567–3574 CrossRef CAS PubMed.
- K. M. McClain, S. C. Moore and J. N. Sampson, et al., Preanalytical Sample Handling Conditions and Their Effects on the Human Serum Metabolome in Epidemiologic Studies, Am. J. Epidemiol., 2021, 190(3), 459–467 CrossRef PubMed.
- T. C. Peakman and P. Elliott, The UK Biobank sample handling and storage validation studies, Int. J. Epidemiol., 2008, 37(suppl_1), i2–i6 CrossRef PubMed.
- C. Barrios, J. Zierer and P. Würtz, et al., Circulating metabolic biomarkers of renal function in diabetic and non-diabetic populations, Sci. Rep., 2018, 8(1), 15249 CrossRef PubMed.
- N. E. Allen, M. Arnold and S. Parish, et al., Approaches to minimising the epidemiological impact of sources of systematic and random variation that may affect biochemistry assay data in UK Biobank, Wellcome Open Res., 2021, 5, 222 Search PubMed.
- A. Moayyeri, C. J. Hammond, D. J. Hart and T. D. Spector, The UK Adult Twin Registry (TwinsUK Resource), in Twin Research and Human Genetics, Cambridge University Press, 2013, vol. 16(1), pp. 144–149 Search PubMed.
- Z. Yu, G. Zhai and P. Singmann, et al., Human serum metabolic profiles are age dependent, Aging Cell, 2012, 11(6), 960–967 CrossRef CAS PubMed.
- E. Louis, J.-Y. Mary and G. Vernier-Massouille, et al., Maintenance of remission among patients with Crohn's disease on antimetabolite therapy after infliximab therapy is stopped, Gastroenterology, 2012, 142(1), 63–70 CrossRef CAS PubMed . e5; quiz e31.
- E. Louis, M. Resche-Rigon and D. Laharie, et al., Withdrawal of infliximab or concomitant immunosuppressant therapy in patients with Crohn's disease on combination therapy (SPARE): a multicentre, open-label, randomised controlled trial, Lancet Gastroenterol. Hepatol., 2023, 8(3), 215–227 CrossRef PubMed.
- S. Cacciatore, L. Tenori, C. Luchinat, P. R. Bennett and D. A. MacIntyre, KODAMA: an R package for knowledge discovery and data mining, Bioinformatics, 2017, 33(4), 621–623 CrossRef CAS PubMed.
- E. A. Thévenot, A. Roux, Y. Xu, E. Ezan and C. Junot, Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses, J. Proteome Res., 2015, 14(8), 3322–3335 CrossRef.
- H. Singmann, B. Bolker, J. Westfall, F. Aust and M. S. Ben-Shachar, afex: Analysis of factorial experiments. R package version 013–145. 2015.
- J. R. Larkin, S. Anthony and V. A. Johanssen, et al., Metabolomic Biomarkers in Blood Samples Identify Cancers in a Mixed Population of Patients with Nonspecific Symptoms, Clin. Cancer Res., 2022, 28(8), 1651–1661 CrossRef PubMed.
- E. Goldberg, S. Ievari-Shariati and B. Kidane, et al., Comparative metabolomics studies of blood collected in streck and heparin tubes from lung cancer patients, PLoS One, 2021, 16(4), e0249648 CrossRef CAS PubMed.
- C. D. Foucher and R. E. TubbenLactic Acidosis, in Lactic Acidosis [Updated 2022 Jul 18], StatPearls Publishing, Treasure Island (FL), Available from: https://www.ncbi.nlm.nih.gov/books/NBK470202/ Search PubMed.
- R. Gambino, Sodium fluoride: an ineffective inhibitor of glycolysis, Ann. Clin. Biochem., 2013, 50(Pt 1), 3–5 CrossRef CAS PubMed.
- C. Cruickshank-Quinn, L. K. Zheng, K. Quinn, R. Bowler, R. Reisdorph and N. Reisdorph, Impact of Blood Collection Tubes and Sample Handling Time on Serum and Plasma Metabolome and Lipidome, Metabolites, 2018, 8(4), 88 CrossRef PubMed.
- R. H. Barton, J. K. Nicholson, P. Elliott and E. Holmes, High-throughput 1H NMR-based metabolic analysis of human serum and urine for large-scale epidemiological studies: validation study, Int. J. Epidemiol., 2008, 37(suppl_1), i31–i40 CrossRef PubMed.
- S. C. Ritchie, P. Surendran and S. Karthikeyan, et al., Quality control and removal of technical variation of NMR metabolic biomarker data in ∼120,000 UK Biobank participants, Sci. Data, 2023, 10(1), 64 CrossRef CAS PubMed.
- P. Downey and T. C. Peakman, Design and implementation of a high-throughput biological sample processing facility using modern manufacturing principles, Int. J. Epidemiol., 2008, 37(suppl_1), i46–i50 CrossRef.
- G. A. Nagana Gowda, V. Pascua and D. Raftery, Anomalous Dynamics of Labile Metabolites in Cold Human Blood Detected Using 1H NMR Spectroscopy, Anal. Chem., 2023, 95(34), 12923–12930 CrossRef CAS PubMed.
- W. S. Waring, L. E. Evans and C. T. Kirkpatrick, Glycolysis inhibitors negatively bias blood glucose measurements: potential impact on the reported prevalence of diabetes mellitus, J. Clin. Pathol., 2007, 60(7), 820–823 CrossRef CAS.
- G. Lippi, M. Nybo, J. Cadamuro, J. T. Guimaraes, E. van Dongen-Lases and A.-M. Simundic, Chapter Four - Blood Glucose Determination: Effect of Tube Additives, in Advances in Clinical Chemistry, ed. G. S. Makowski, Elsevier, 2018. pp. 101–123 [cited 2023 May 24]. Available from: https://www.sciencedirect.com/science/article/pii/S0065242317300689 Search PubMed.
- J. G. Elferink, The mechanism of calcium oxalate crystal-induced haemolysis of human erythrocytes, Br. J. Exp. Pathol., 1987, 68(4), 551–557 CAS.
- K. Haug, K. Cochrane and V. C. Nainala, et al., MetaboLights: a resource evolving in response to the needs of its scientific community, Nucleic Acids Res., 2020, 48(D1), D440–D444 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01359f |
| This journal is © The Royal Society of Chemistry 2024 |