Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploiting Locusta migratoria as a source of bioactive peptides with anti-fibrosis properties using an in silico approach†
Carla S. S. Teixeira a,
Rita Biltesa,
Caterina Villaa,
Sérgio F. Sousa
a,
Rita Biltesa,
Caterina Villaa,
Sérgio F. Sousa bc,
Joana Costa
bc,
Joana Costa a,
Isabel M. P. L. V. O. Ferreira
a,
Isabel M. P. L. V. O. Ferreira a and
Isabel Mafra
a and
Isabel Mafra *a
*a
aREQUIMTE-LAQV, Faculdade de Farmácia, Universidade do Porto, Rua de Jorge Viterbo Ferreira, 228, 4050-313 Porto, Portugal. E-mail: isabel.mafra@ff.up.pt
bAssociate Laboratory i4HB – Institute for Health and Bioeconomy, Faculty of Medicine, University of Porto, 4200-319 Porto, Portugal
cUCIBIO – Applied Molecular Biosciences Unit, BioSIM – Department of Biomedicine, Faculty of Medicine, University of Porto, 4200-319 Porto, Portugal
First published on 1st December 2023
Abstract
Edible insects have been proposed as an environmentally and economically sustainable source of protein, and are considered as an alternative food, especially to meat. The migratory locust, Locusta migratoria, is an edible species authorised by the European Union as a novel food. In addition to their nutritional value, edible insects are also sources of bioactive compounds. This study used an in silico approach to simulate the gastrointestinal digestion of selected L. migratoria proteins and posteriorly identify peptides capable of selectively inhibiting the N-subunit of the somatic angiotensin-I converting enzyme (sACE). The application of the molecular docking protocol enabled the identification of three peptides, namely TCDSL, IDCSR and EAEEGQF, which were predicted to act as potential selective inhibitors of the sACE N-domain and, therefore, possess bioactivity against cardiac and pulmonary fibrosis.
1. Introduction
Edible insects have been proposed as an environmentally and economically sustainable source of protein, and are considered as an alternative food, especially to meat.1 Although their nutrient composition is affected by several factors (e.g., species, sex, developmental stage, habitat, diet, type of processing and preparation), when compared to common meats, edible insects generally contain higher percentages of highly digestible proteins2–4 containing all essential amino acids in adequate proportions for human needs.1 Insects can be obtained from nature or they can be reared. Insect production requires less water and soil than that of other animals such as poultry, pork or cattle. Moreover, their facilities do not require expensive technologies and they have the possibility of being fed with industrial and agricultural waste streams. They also have a high feed conversion efficiency, an elevated rate of reproduction5 and a high percentage of edible mass (up to 80%), emitting lower greenhouse gases6 than animals. The edible insect sector contributes directly to 7 of the 17 United Nations Sustainable Development Goals (SDG) that are intended to “stimulate action over the next 15 years in areas of critical importance for humanity and the planet”, namely SDG 2, 6, 9, 12, 13, 15 and 16.7The migratory locust, Locusta migratoria (order Orthoptera, family Acrididae) is a very unpopular species around the world due to its ability to cause extensive damage to crops. However, the dry adult insect has a rich nutritional composition (50.4 ± 2.0% crude protein, 19.6 ± 0.8% crude fat, 4.8 ± 0.7% carbohydrates, 15.6 ± 1.7% crude fibre, 6.2 ± 0.5% ash and 3.8 ± 0.2% moisture), providing 490.4 ± 4.0 calories per 100 g of the dry product8 and is consumed as a food delicacy in several regions of the world for centuries.9 In the European Union, it was classified as a novel food according to Regulation (EU) 2015/2283 on novel foods10 and recently considered safe for human consumption.11 The European Union legislation allows it to be placed on the EU market as frozen adult insects without legs and wings, dried without legs and wings and ground with legs and wings.11
Besides the nutritional value of insects, the exploitation of nutraceutical properties can be a stimulus for increased interest in their intake in Western diets. In a particular case of migratory locust, Ochiai et al. demonstrated that the dietary administration of insect powder to rats improved fat metabolism and promoted therapeutic/ameliorating effects against dyslipidemia.12
Most studies concerning the bioactive properties of edible insects focused on the identification of peptides with antihypertensive, antidiabetic and antioxidant properties.13 In a previous work, we reported that insects can also be a source of bioactive peptides with antifibrosis properties. An in silico protocol that simulated gastrointestinal (GI) digestion of the house cricket (Acheta domesticus) identified several peptides capable of selectively inhibiting the N-subunit of the somatic angiotensin-I converting enzyme (sACE; EC 3.4.15.1).14 To our knowledge, that was the first study associating the consumption of insects with potential antifibrosis effects.14
The sACE is a 150–180 kDa dipeptidyl carboxypeptidase that also displays endopeptidase activity and can be found in epithelial, endothelial and neuroepithelial cells.15 It is a promiscuous enzyme that has several substrates (e.g., angiotensin I (Ang I), N-acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP), substance P, enkephalins, gonadotropin-releasing hormone (GnRH), N-formylmethionine-leucyl-phenylalanine luteinizing hormone-releasing hormone (LHRH), neurotensin, kinins and amyloid-beta), being implicated in several physiological processes, including blood pressure control, fibrosis, erythropoiesis, haematopoiesis, myelopoiesis immunomodulation, renal development and function, cell signalling or amyloid-beta clearance.16
sACE is composed of two homologous catalytic domains, known as the N- and C-domains, each harbouring a catalytic zinc ion at the active site. Overall, the two domains share 65% sequence homology with each other,17 though for their catalytic pocket residues, the homology can reach 89%. The detailed analysis of their active sites showed that although most key residues involved in inhibitor-binding are common among both active sites, there are some domain specific structural differences that are responsible for their different kinetic profiles.18 The C-domain is primarily responsible for catalysing the last step in the production of the mitogenic and hypertensive octapeptide, angiotensin II (Ang II), through the cleavage of the His-Leu dipeptide from the C-terminus of angiotensin I (Ang I). Both C and N domains inactivate the vasodilator nonapeptide bradykinin (BK) with the same efficiency through sequential cleavage of Phe-Arg and Ser-Pro dipeptides.19 Since C-domain inhibition is an effective way to reduce blood pressure, sACE inhibitors are extensively used for the treatment of hypertension and cardiovascular disease. The current problem is that most of the available sACE inhibitors are not domain-specific, resulting in elevated levels of bradykinin that can trigger side effects, such as cough and angioedema.20
The N-domain is exclusively responsible for the hydrolysis of the tetrapeptide Ac-SDKP (N-acetyl-Ser–Asp–Lys–Pro). AcSDKP is a natural inhibitor of hematopoietic stem cell proliferation21 and also prevents the proliferation of fibroblasts in the myocardium, aorta and kidneys, leading to the reduction of collagen deposition in hematopoietic stem cells of cardiac and pulmonary tissues.15,22,23 Since excess collagen deposition in these tissues has been identified as a mechanism for the pathogenesis of fibrosis, the selective inhibition of the N-domain of sACE has been proposed as a possible treatment for cardiac and pulmonary fibrosis, without interfering with the C-domain function in blood pressure and water and salt homeostasis.18
The aim of this work is to apply a molecular docking protocol to identify peptides originating from the simulated GI digestion of L. Migratoria proteins, capable of selectively inhibiting the N domain sACE and, therefore, with potential antifibrosis effects.
2. Materials and methods
This study applied an in silico protocol previously adopted by our14 and other research groups.24–26 It begins with the selection of the target L. migratoria proteins, and their simulated GI digestion followed by the application of a docking protocol to evaluate the binding interactions between the selected peptides and protein drug targets (N- and C-domains of sACE). Finally, the allergenic potential of the potentially bioactive peptides was evaluated. The procedure is detailed in the next subsections.2.1 Protein selection
The proteins selected for this work were obtained from the analysis of all the available L. migratoria proteins in the UniProtKB database (https://www.uniprot.org/; accessed on March 2023) and the National Center for Biotechnology Information (NCBI) protein database (https://www.ncbi.nlm.nih.gov/; accessed on March 2023).To raise the probability of obtaining new peptides, characteristic and/or specific L. migratoria species with unknown bioactivity, the protein selection criterion was the lowest percentage of sequence identity (percentage of residues identical between two proteins divided by the length of the shortest sequence)27 compared to proteins from other non-target species when a protein BLAST was performed on NCBI. However, since the proteome of L. migratoria is not complete, a certain degree of sequence identity was accepted for this analysis, depending on the available sequences and the database from where they were collected.
2.2 Simulated GI digestion
The in silico digestion of the proteins selected in the previous section was performed using the “enzyme(s) action” tool available in the BIOPEP-UWM server (https://www.uwm.edu.pl/biochemia/index.php/en/biopep).28 Each protein sequence was in silico digested through the sequential application of endoproteases: pepsin (EC 3.4.23.1; pH 1.3), trypsin (EC 3.4.21.4) and chymotrypsin A (EC 3.4.21.1) involved in the in vivo GI digestion.2.3 Evaluation of the potential biological activity
All peptides obtained in the previous section were submitted to the “Profiles of potential biological activity” tool of the BIOPEP-UWM server (https://www.uwm.edu.pl/biochemia/index.php/en/biopep)28 to evaluate if they have any previously identified biological activity. Only the peptides with unknown bioactivity were selected for the next steps.Additionally, the peptides selected for the last steps were also submitted to the Food-derived bioactive peptides (DFBP)29 database (https://www.cqudfbp.net/) and a literature check was performed to confirm that they do not have any previously identified biological activity.
2.4 Target-peptide docking
When preparing the structures for protein–ligand docking, water molecules and additional ligands were removed. The ligands Ang-II and BJ2 were saved for later use to optimise the docking protocol for each target. This was accomplished through redocking and comparing the predicted poses with the experimental ones by root mean square deviation (RMSD). Preparation of the structure of the targets and addition of the hydrogen atoms was done using the GOLD software31 according to the recommended protocol, as in previous studies involving other proteins.32–35
Docking was performed using GOLD software using the PLP scoring function.38 The co-crystallised ligands, BJ2 and Ang-II were used as the reference to evaluate and optimise the accuracy of the docking protocol for each target. The conformation of each ligand was randomised, and the ligands were redocked against their initial target. Different settings were considered to ensure an accurate reproduction of the pose of the reference ligands. The optimised parameters included the position of the centre and radius of the docking region, and a number of independent genetic algorithm (GA) runs.
The final optimised protocol for each target was applied to dock all 245 peptides against each target structure. The most stable predicted conformations for each peptide were recorded and analysed. The PLP binding score for each peptide to each target protein was also registered and compared with the binding scores of the reference molecules Ang-II and BJ2. The GOLD's PLP scoring values are non-dimensional, with higher values indicating a stronger association.
2.5 Evaluation of potential allergenicity
Sequences of the selected L. migratoria proteins were used to predict their linear B-cell epitopes. For this purpose, the Bepipred Linear Epitope Prediction 2.0 tool from the IEDB analysis resource (https://tools.iedb.org/bcell/) (Jespersen, Peters, Nielsen, & Marcatili, 2017) was used. The selected potentially bioactive peptides were then compared with the obtained sequences of the predicted epitopes to search for common regions. Peptide allergenicity prediction was carried out using the AllerTOP v.2.0 bioinformatic tool (https://www.ddg-pharmfac.net/AllerTOP/index.html) (Dimitrov, Bangov, Flower, & Doytchinova, 2014).3. Results and discussion
3.1 Protein selection
The search in the UniProtKB and NCBI databases for L. migratoria proteins resulted in 93 (reviewed by Swiss-Prot) and 3598 protein sequences, respectively. To restrict the number of target sequences and raise the probability of obtaining peptides with unidentified biological activities, a selection criterion was defined based on the percentage of identity compared to proteins from other species when a protein BLAST was performed on NCBI.As the main objective of this study was to target new and highly specific peptides of L. migratoria, a selection criterion of sequence identity below 81% was used to analyse the 93 identified proteins, reviewed by Swiss-Prot, available at the UniProtKB database. Similarly, for the 3598 protein sequences available in NCBI database, a criterion of less than 55% sequence identity was established for protein selection. As the number of proteins obtained from the two databases was significantly different, the selection criteria were adapted to each one to restrict the number of proteins selected for the next steps.
The adopted procedure resulted in a total of 9 protein sequences, 3 obtained from UniProtKB database and 6 obtained from the NCBI protein database. Their NCBI/UniProtKB accession numbers and relevant features are described in Table 1.
| Database | Accession number | Protein name | Length | Sequence | Species with higher sequence identity | Percentage of sequence identity | Number of fragments obtained after simulated GI digestion | Number of peptides (>1aa) with known biological activity | Number of peptides (>1aa) with unknown biological activity |
|---|---|---|---|---|---|---|---|---|---|
| NA – no sequence identity found. | |||||||||
| NCBI | ALD51386.1 | Odorant receptor 126 | 428 | MEFESMMGPGLPLMRLTGLWQMGRQGGGVSRGLRLATIVLSVLLVVAGSTLHLVFDTPDQFEDITLCGFNIDIVSLDLLKGVLFVVQGAPLRELVQLLCDARAGFTFADINHAIRGRYEAVADRMRILLQATVVLPLVGWLSAPLMSRLAAGAGGSRAPRQLPVPAWLPVDIHATPTYELLYALQAFGCTAAGAFSICVDAFFIRLMLLISAEIEVLCENISAIGVPHPAQGSGGCICRCQPNAADLACTCKGCVKAFTSSPEEASDEMYQLLVKAVRHHQTIIRMVALLQQTMDALVFIVLFANMANLCCSLFATAILLQRGGSLTKTLKGLSAVPVVLYQTSLYCLFGHIVTDQSEKLYNAAISCGWVNCDARFKRSLLIFMVEAMKPLEITVGKFCKLSRQMLLQVFHSSYALMNLLYYYHYNTE | Ceracris kiangsu | 55% | 167 | 21 | 62 |
| AKN21235.1 | Cchamide-2 precursor | 133 | MSAKQHTAVALLGDAAPSAVHAARRIRRRPVRRAADRGGVPVRAAPSAGHAARRRRQTWVHGVRALVLRRPRQARRPGAGGGGAGRGGGGSGGAAGRPGGAGGGGGAGGGGTAIPPVAVPAAVAAACLPATVS | NA | NA | 34 | 7 | 14 | |
| AMO66175.1 | Defensin 4 | 69 | MKNSTVFFLVGLLTTAGIAFCSAAPAQSVQDDRQAHLTCDSLSALGVPCAAVRCVKGAYCQHGVCHCRV | Schistocerca cancellata | 52% | 21 | 1 | 12 | |
| API80737.1 | Autophagy-related protein 2 | 220 | MRGMSLLLLSNEHSFPCIAEIVSTVRIEILLTHSYFNSLSNNRKPGIKLLFHQKLYFMFVSEAMFSNCRFLQLSVFNMLLVFNTAIRNSMNGLTYVIDIINPETETVLYSAVHLLYILRRTEDWSHTLSTQDSSCFAGYMVQKGRNTIGDSSSECSIFSDFPFLEMLPLHHMNHRHFFRRKVVNFRLQLVLTRNSSRSVYQCVQNGSLLRCMMETLCIEI | NA | NA | 109 | 22 | 24 | |
| AQY60265.1 | CYP3117C1 | 499 | MSVWLVFGAALATVCACALAVASWLWATRLQTAAPGPPTWPLLGNVQHFLKQPVLLEHAADLYKQYGDVFRFYVGPKLVVVVTKPEDVKRVLVTTKWQERDPYFLGTLRKVTGNGLLINSGEVWQRHRKALEPTFHYTALHRYLDTFNKEVCLLSERLAAMGGQESDVLPLMCLSSLRITMCALGGMEYDVVEPDQYQQQQLASEFIGFLKVFQATMFRPWKAINSLMWMSEDGRKLKKIIGMAKDVTNRYLAALRVYNTKLEITSHFSSLLLEEKPEMQEMDDKISDEVVTVAVTATETMAGALAYALSALGLYPEWQVKAQQQLDEVFGEGGDFLRPATLEDIGKLTVIDAIVKETLRLFTVVPFLPRIIDEDIPLAGGRYVAPRGCCVAVASFLTHRDPDLYPEPDKFDPGRFLPGGSATSRKPFSYIPFGAGSRVCLGSSFATLEMKVTLATLLRQFTVVSGSTRKDLEHTLFSITAHPLKGFRLSFRARKEQSL | Schistocerca cancellata | 55% | 197 | 23 | 81 | |
| DAA64589.1 | Serine protease-like protein 1 | 276 | MIKEAVLVLALAACVSAAVLPVRRIAHSGPLRKTGLKQGRIVGGKEAEEGQFPYSVSIQWQLSGVSSHFCGGALVKDDVVVTAGQCAHVVTYGLTTVVAGRVYMDESVYGQSTLWEISHPEYKVVNNHAINDIAVFTLTVGFDLSDKINVIGLPSQDQKPYAQAATLSGWGSTSNSMLPETSDTLHYAEVTVIPTVNCYALMTDDSTFNNNNICSGPVTGKISSCVGDIGSPLVQYGNLIGVVSWNTVPCGTFGMPIVYTRVSAYSDWIKEYMDTK | Schistocerca nitens | 51% | 83 | 13 | 45 | |
| Uniprot | P14570 | ATP synthase protein 8 | 52 | MPQMSPMMWFSLFIMFSMTMMLFNQLNFFSYKPNKIMSSNNKIKKKNINWMW | Oedaleus asiaticus | 80.77% | 36 | 8 | 4 |
| P19872 | Adipokinetic prohormone type 3 | 77 | MQVRAVLVLAVVALVAVATSRAQLNFTPWWGKRALGAPAAGDCVSASPQALLSILNAAQAEVQKLIDCSRFTSEANS | Schistocerca gregaria | 71.43% | 24 | 5 | 9 | |
| P80059 | Pars intercerebralis major peptide D1 | 54 | SCTEKTCPGTETCCTTPQGEEGCCPYKEGVCCLDGIHCCPSGTVCDEDHRRCIQ | Schistocerca cancellata | 71.15% | 9 | 0 | 6 | |
3.2 Peptides from simulated GI digestion
The simulated in silico GI digestion of the 9 selected proteins generated a total of 680 fragments ranging between 1 and 31 residues in size (Table S1, ESI†). After removing the single amino acid fragments and duplicates, 314 peptides were obtained. From those, 73 have been described to possess bioactive properties in the BIOPEP-UWM server and 241 (Table S2, ESI†) are peptides with unknown bioactivity. The latter were selected for molecular docking studies.3.3 Molecular docking analysis
To evaluate the potential of each of the 241 peptides as selective inhibitors of the N-domain of sACE and, thus, possess bioactivity against cardiac and pulmonary fibrosis, a molecular docking simulation was performed.To assess the differences in the affinities between the two sACE domains, two crystallographic structures were used:
(1) the N-domain complexed with BJ2 (PDB ID: 6EN5), a potent inhibitor that is 84-fold more selective towards the N-domain than towards the C-domain (Fig. 1a);
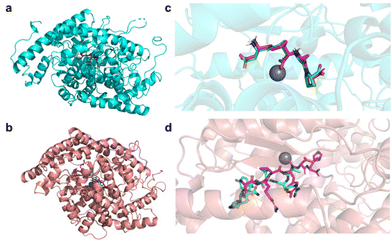 | ||
| Fig. 1 Pymol representation of the crystallographic structures (represented in the cartoon) of (a) N-domain of sACE complexed with BJ2 (PDB ID: 6EN5) and (b) C-domain of sACE complexed with Ang-II (PDB ID: 4APH). Pymol representation of the superimposition of the binding poses of the reference inhibitors BJ2 and Ang-II in the complex crystallographic structure (represented in licorice and coloured in magenta) and after the re-docking protocol (represented in licorice and coloured in green) in (c) the N-domain of sACE and (d) the C-domain of sACE. Zn2+ is represented in grey. | ||
(2) the C-domain complexed with the reaction product and competitive inhibitor Ang-II (PDB ID: 4APH) (Fig. 1b).
To validate the docking algorithm, the co-crystallised inhibitors (reference inhibitors Ref1 (BJ2) and Ref2 (Ang-II)) were removed from the crystallographic structure of the protein, randomised and re-docked into their active site. The crystallographic and re-docked poses were superimposed and represented in Fig. 1c and d.
For the N-domain both inhibitor structures are well aligned inside the enzyme's active site (Fig. 1c), suggesting that the adopted molecular docking protocol can reproduce the crystallographic pose very well and, therefore, it can be applied to predict the binding poses of the studied peptides. For the C-domain, the docking algorithm was not so effective in reproducing the crystallographic pose (Fig. 1d). This result was not surprising since the crystallographic structure was reported to present some difficulties in defining the exact coordinates for all residues of the ligand, Ang-II, suggesting that it was able to bind to the C-domain active site in two different ‘sliding’ conformations.30 The docking scores obtained for the 241 peptides and the reference inhibitors (Ref1 and Ref2) against the two targets (PDB ID: 4APH and 6EN5) are reported in Table S2 of the ESI.†
Since the aim of this work is to identify peptides that are potential selective inhibitors of the N-domain of sACE, three selection criteria (SC1–3) were defined and sequentially applied to the docking scores obtained for the 6EN5 structure.
(SC1) Peptides with docking score (ds) values higher than that obtained for the reference inhibitor (Ref1, ds = 119.68) means that the peptides may have a higher affinity for the active site than the crystallographic inhibitor.
(SC2) Peptides in which the difference between the docking scores for the N- and C-domains (ds N-domain minus ds C-domain) is higher than 20 indicates that the peptides have higher affinity for the N-domain than for the C-domain.
(SC3) Peptides with a ratio between the docking score and the number of non-H atoms higher than 2 eliminate the peptides whose docking score was positively influenced by the size of the peptide avoiding the bias of the result.
After applying the first criterion (SC1), 49 peptides were selected. The application of the second criterion (SC2) restricted the selection to 15 peptides which were reduced to 14 after removing peptide #118 which did not comply with the third criterion (SC3) (Table 2). Therefore, a total of 14 peptides with potential selectivity for the N-domain of sACE were obtained and followed for further analysis. The absence of known biological activities associated with the selected peptides was confirmed using the DFBP database.
| SC1 | SC2 | SC3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Peptide ID | Peptide code | ds 6EN5 (>119.68) | ds 4APH | ds 6EN5–ds 4APH (>20) | Non-H atoms | ds/non-H atoms (>2) | Potential IgE binding | Potential cross-reactivity |
| #17 | VIDIIN | 129.46 | 90.24 | 39.22 | 48 | 2.70 | No | Per a 1, Periplaneta americana |
| #31 | TTAGIAF | 123.93 | 103.49 | 20.44 | 48 | 2.58 | No | No |
| #40 | TCDSL | 127.47 | 94.89 | 32.58 | 36 | 3.54 | Yes | No |
| #45 | SVSIQW | 131.06 | 102.03 | 29.03 | 51 | 2.57 | No | No |
| #77 | QTIIR | 135.05 | 98.24 | 36.81 | 44 | 3.07 | No | Sar s 1, Sarcoptes scabiei |
| #81 | QQQQL | 122.3 | 97.04 | 25.26 | 45 | 2.72 | Yes | Allergenic protein, Oryza sativa |
| #85 | QGGGVSR | 130.16 | 108.32 | 21.84 | 46 | 2.83 | No | Asp f 9, Aspergillus fumigatus |
| #107 | PEPDK | 122.26 | 96.28 | 25.98 | 41 | 2.98 | Yes | No |
| #109 | PEDVK | 121.45 | 97.46 | 23.99 | 41 | 2.96 | No | No |
| #130 | IDCSR | 129.11 | 102.2 | 26.91 | 40 | 3.23 | Yes | No |
| #145 | GGQESDVL | 133.44 | 113.31 | 20.13 | 56 | 2.38 | No | Asp FII, Aspergillus fumigatus |
| #164 | EITVGK | 129.72 | 104.97 | 24.75 | 45 | 2.88 | No | No |
| #173 | EAEEGQF | 138.65 | 116.28 | 22.37 | 57 | 2.43 | No | No |
| #186 | DESVY | 130.1 | 103.07 | 27.03 | 43 | 3.03 | Yes | No |
3.4 Molecular interaction analysis
To assess whether the 14 selected peptides have a common molecular feature that differentiates them and justifies their distinctive docking scores, their intermolecular interactions with the active site residues of both enzymatic targets were examined using the PLIP server and reported in Table S3 of the ESI.†Most of the identified interactions are established between the peptides and residues that are conserved among both active sites. Some of those conserved N-domain/C-domain residues are: Gln259/281, Lys489/511, Tyr498/520, His331/353, His491/513, Tyr501/523, Ala334/356,18 His361/383, His365/387, Phe435/457, Phe505/52740 and Asp336/358.41
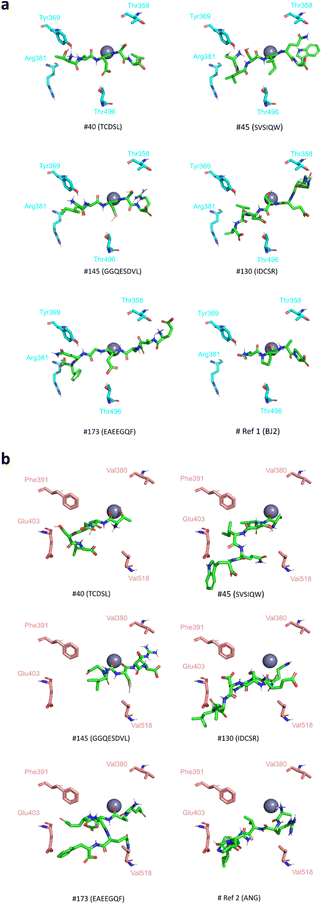
According to the literature, the molecular basis beyond the selectivity of the N- and C-domains relies on the replacement of some residues of the binding pocket with others possessing different chemical properties.18 The literature refers to five main N-domain/C-domain substitutions, namely the replacement of a positively charged arginine (Arg381) in the N-domain with a negatively charged glutamate (Glu403) in the C-domain and the replacement of four hydrophilic residues in the N-domain (Tyr369, Ser357, Thr358 and Thr496) with four hydrophobic residues in the C-domain (Phe391, Val379, Val380, Val518). These substitutions affect the sACE binding site moieties influencing the type of interactions established between the ligands and the active site. The aforementioned residues have already been used as part of a successful targeting approach towards the design of the N-domain selective inhibitor BJ2, which was co-crystallised with the sACE structure selected for the present docking studies.18 Considering that the BJ2 inhibitor showed potent inhibition and was 84-fold more selective toward the N-domain, the potential selectivity of the 14 peptides selected in this work was determined by evaluating their capability to establish intermolecular interactions with the Arg381, Tyr369, Ser357, Thr358 and Thr496 residues of the N-domain active site. To discard the possibility of the interaction of peptides with the corresponding C-domain residues, their capability to bind to residues Glu403, Phe391, Val379, Val380, and Val518 of the C-domain was also evaluated. The result of the analysis of the intermolecular interactions between the domain specific residues and the selected peptides are presented in Table 3, which shows that:
(1) all peptides establish at least one interaction with one of the four N-domain specific residues Arg381, Tyr369, Thr358 and Thr496;
(2) for 12 (#17, #31, #40, #77, #81, #85, #107, #109, #130, #164, #173 and #186) out of the 14 peptides, the redocked (Ref1) and crystallographic BJ2 poses establish a hydrogen bond with Tyr369;
(3) peptide #145, excluded from point (2) establishes a hydrophobic interaction with Tyr369;
(4) 5 peptides only interact with N-domain specific residues and not with any C-domain specific residues (#40, #45, #130, #145, and #173).
| Enzyme target | Type of interaction | Peptide ID | 17 | 31 | 40 | 45 | 77 | 81 | 85 | 107 | 109 | 130 | 145 | 164 | 173 | 186 | Ref1 (BJ2) | Ref2 (Ang ii) | Crystallographic pose | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residue | ||||||||||||||||||||
| x, molecular interaction between the peptide and enzyme residue. −, no molecular interaction. | ||||||||||||||||||||
| N-domain | Hydrogen bonds | 358 | THR | − | − | − | x | − | − | − | − | − | − | − | − | x | − | − | − | − |
| 369 | TYR | x | x | x | − | x | x | x | x | x | x | − | x | x | x | x | − | x | ||
| 496 | THR | x | − | x | − | − | x | − | − | x | − | x | x | x | − | − | − | − | ||
| Hydrophobic interactions | 358 | THR | x | − | − | − | x | x | − | − | − | − | x | − | − | − | − | x | − | |
| 369 | TYR | − | − | − | − | − | − | − | − | − | − | x | − | − | − | − | − | − | ||
| 381 | ARG | − | − | − | − | − | − | − | − | − | − | x | − | x | − | − | − | − | ||
| 496 | THR | x | − | − | x | x | − | − | − | − | − | − | x | − | − | − | − | x | ||
| C-domain | Hydrogen bonds | 403 | GLU | − | − | − | − | − | − | − | − | x | − | − | − | − | − | − | − | − |
| Hydrophobic interactions | 391 | PHE | x | x | − | − | x | − | − | x | x | − | − | − | − | − | − | − | x | |
| 403 | GLU | − | − | − | − | − | − | x | − | − | − | − | − | − | − | − | x | − | ||
| 518 | VAL | − | − | − | − | − | x | − | x | − | − | − | x | − | x | − | x | x | ||
| Salt bridges | 403 | GLU | − | − | − | − | x | − | − | − | − | − | − | − | − | − | − | − | − | |
There are several conditions for the last group of 5 peptides to be considered as potential N-domain selective inhibitors: they establish a hydrogen bond or/and a hydrophobic interaction with at least one specific residue of the active site of the N-domain and they do not interact with any specific residue of the active site of the C-domain (Fig. 2). Considering that the hydrogen bond with Tyr369 may positively influence the selectivity of the molecule, this final selection may be restricted to 3 peptides: TCDSL (#40), IDCSR (#130) and EAEEGQF (#173).
This was the first study that reported the potential bioactivity of these 3 peptides and the second that demonstrated that GI digestion of edible insect proteins can produce peptides with antifibrosis bioactivity. Information in the literature on specific inhibitors of the N-domain of sACE is still very scarce and, although requiring in vitro/in vivo validation, these results are a valuable contribution to the rational design of specific inhibitors of the sACE N- and C-domains. It also demonstrates that including insects in diets can bring unexplored health benefits.
3.5 Allergenicity prediction
As a novel food, L. migratoria can be a source of allergens that might lead to inadvertently allergic reactions in sensitised individuals. Therefore, the evaluation of the potential allergenicity of the selected 14 peptides is essential to avoid health risks in allergic individuals after their ingestion, contact or inhalation. Presently, there are no allergenic proteins from L. migratoria identified as allergens by the WHO/IUIS Allergen Nomenclature Sub-Committee (https://allergen.org/). However, L. migratoria, as well as most of the edible insect species, share homologous proteins with arthropods, such as crustaceans and dust mites, which can lead to IgE cross-reactivity in allergic patients.42,43 Firstly, the prediction of linear B-cell epitopes for each of the selected L. migratoria proteins was performed by IEDB analysis (Bepipred Linear Epitope Prediction 2.0 tool44) and the results are summarised in Table 1. This analysis demonstrated homology between the predicted epitopes obtained by the IEDB and five of the 14 selected peptides TCDSL (#40), QQQQL (#81), PEPDK (#107), DESVY (#186) and IDCSR (#130), indicating that these peptides could have the potential to induce IgE-binding in sensitised individuals. The AllerTop v.2.0 tool45 was then used to assess the potential allergenicity of each peptide and the results displayed five peptides as probable allergens, namely QTIIR (#77), GGQESDVL (#145), QGGGVSR (#85), VIDIIN (#17) and QQQQL (#81). The identified sequences corresponded to known allergens of itch mite, house dust mite, cockroach (Periplaneta americana), and fungus (Aspergillus fumigatus), among others. These peptides could lead to potential IgE cross-reactivity in patients with pre-existent allergies. However, their actual allergenic risk needs to be tested in vitro/in vivo, since there are no reports confirming this potential cross-reactivity.4. Conclusions
The present work applied an in silico approach, for the first time, to determine the capability of obtaining peptides that selectively inhibit the N-domain of sACE, after the GI digestion of L. migratoria. Due to the large number of available protein sequences from this species, the in silico protocol was only applied to a restricted number of proteins that share a maximum of sequence identity of 81% with other protein sequences. This approach successfully met the objective of this work, because out of 314 peptides obtained from simulated GI digestion of the 9 selected proteins, 241 have not been previously related to any bioactive properties.The selection criteria applied allowed us to filter the initial pool of peptides and propose a group of 14 peptides for an in-depth evaluation. After a careful assessment of the intermolecular interactions between each peptide, the reference inhibitor and the residues lining the active sites of the two sACE domains, a group of 14 peptides was restricted to 3 peptides: TCDSL (#40), IDCSR (#130) and EAEEGQF (#173). The allergenicity prediction tools suggested that although peptides #40 and #130 could have the potential to induce IgE-binding in sensitised individuals, apparently none of the three peptides leads to IgE cross-reactivity in patients with pre-existent allergies. However, these results should be confirmed experimentally.
In conclusion, peptides TCDSL (#40), IDCSR (#130) and EAEEGQF (#173) are potential selective inhibitors of the sACE N-domain and their bioactivity against fibrosis should be evaluated through in vitro and in vivo studies.
Author contributions
Carla S. S. Teixeira: methodology; formal analysis; investigation; conceptualization; writing – original draft; and writing – review & editing. Rita Biltes: methodology; formal analysis; investigation; conceptualization; writing – original draft; and writing – review & editing. Caterina Villa: methodology; formal analysis; investigation; conceptualization; writing – original draft; and writing – review & editing. Sérgio F. Sousa: methodology; formal analysis; investigation; conceptualization; supervision; writing – original draft; and writing – review & editing. Joana Costa: methodology; formal analysis; investigation; conceptualization; supervision; and writing – review & editing. Isabel M. P. L. V. O. Ferreira: conceptualization; investigation; funding acquisition; project administration; supervision; and writing – review & editing. Isabel Mafra: conceptualization; investigation; funding acquisition; project administration; supervision; and writing – review & editing.Conflicts of interest
There are no conflicts to declare.References
- A. Orkusz, Edible Insects versus Meat—Nutritional Comparison: Knowledge of Their Composition Is the Key to Good Health, Nutrients, 2021, 13(4), 1207 CrossRef CAS PubMed.
- T. A. Churchward-Venne, P. J. M. Pinckaers, J. J. A. van Loon and L. J. C. van Loon, Consideration of insects as a source of dietary protein for human consumption, Nutr. Rev., 2017, 75, 1035–1045 CrossRef PubMed.
- M. Rodríguez-Rodríguez, F. G. Barroso, D. Fabrikov and M. J. Sánchez-Muros, In Vitro Crude Protein Digestibility of Insects: A Review, Insects, 2022, 13(8), 682 CrossRef PubMed.
- L. Hammer, D. Moretti, L. Abbühl-Eng, P. Kandiah, N. Hilaj, R. Portmann and L. Egger, Mealworm larvae (Tenebrio molitor) and crickets (Acheta domesticus) show high total protein in vitro digestibility and can provide good-to-excellent protein quality as determined by in vitro DIAAS, Front. Nutr., 2023, 10, 1150581 CrossRef PubMed.
- J. M. Wilkinson, Re-defining efficiency of feed use by livestock, Animal, 2011, 5, 1014–1022 CrossRef CAS PubMed.
- I. A. Hansen, D. G. A. B. Oonincx, J. van Itterbeeck, M. J. W. Heetkamp, H. van den Brand, J. J. A. van Loon and A. van Huis, An Exploration on Greenhouse Gas and Ammonia Production by Insect Species Suitable for Animal or Human Consumption, PLoS One, 2010, 5(12), e14445 CrossRef PubMed.
- R. Moruzzo, S. Mancini and A. Guidi, Edible Insects and Sustainable Development Goals, Insects, 2021, 12(6), 557 CrossRef PubMed.
- E. H. Mohamed, Determination of nutritive value of the edible migratory locust Locusta migratoria, Linnaeus, 1758 (Orthoptera: Acrididae), Int. J. Adv. Pharm., Biol. Chem., 2015, 144–148 CAS.
- F. Lourenço, R. Calado, I. Medina and O. M. C. C. Ameixa, The Potential Impacts by the Invasion of Insects Reared to Feed Livestock and Pet Animals in Europe and Other Regions: A Critical Review, Sustainability, 2022, 14(10), 6361 CrossRef.
- Regulation (EU) 2015/2283, Regulation (EU) 2015/2283 of the European Parliament and of the Council of 25 November 2015 on novel foods, amending Regulation (EU) No 1169/2011 of the European Parliament and of the Council and repealing Regulation (EC) No 258/97 of the European Parliament and of the Council and Commission Regulation (EC) No 1852/2001, Official Journal of the European Union, 2015, L327, 1–22 Search PubMed.
- Regulation (EU) 2021/1975, Commission Implementing Regulation (EU) 2021/1975 of 12 November 2021 authorising the placing on the market of frozen, dried and powder forms of Locusta migratoria as a novel food under Regulation (EU) 2015/2283 of the European Parliament and of the Council and amending Commission Implementing Regulation (EU) 2017/2470, Official Journal of the European Union, 2021, L402, 10–16 Search PubMed.
- M. Ochiai, K. Tezuka, H. Yoshida, T. Akazawa, Y. Komiya, H. Ogasawara, Y. Adachi and M. Nakada, Edible insect Locusta migratoria shows intestinal protein digestibility and improves plasma and hepatic lipid metabolism in male rats, Food Chem., 2022, 396, 133701 CrossRef CAS.
- C. S. S. Teixeira, C. Villa, J. Costa, I. M. P. L. V. O. Ferreira and I. Mafra, Edible Insects as a Novel Source of Bioactive Peptides: A Systematic Review, Foods, 2023, 12(10), 2026 CrossRef CAS PubMed.
- C. S. S. Teixeira, C. Villa, S. F. Sousa, J. Costa, I. M. P. L. V. O. Ferreira and I. Mafra, An in silico approach to unveil peptides from Acheta domesticus with potential bioactivity against hypertension, diabetes, cardiac and pulmonary fibrosis, Food Res. Int., 2023, 169, 112847 CrossRef CAS PubMed.
- A. Rousseau, A. Michaud, M.-T. Chauvet, M. Lenfant and P. Corvol, The Hemoregulatory Peptide N-Acetyl-Ser-Asp-Lys-Pro Is a Natural and Specific Substrate of the N-terminal Active Site of Human Angiotensin-converting Enzyme, J. Biol. Chem., 1995, 270, 3656–3661 CrossRef CAS.
- G. E. Cozier, E. C. Newby, S. L. U. Schwager, R. E. Isaac, E. D. Sturrock and K. R. Acharya, Structural basis for the inhibition of human angiotensin–1 converting enzyme by fosinoprilat, FEBS J., 2022, 289, 6659–6671 CrossRef CAS PubMed.
- F. Soubrier, F. Alhenc-Gelas, C. Hubert, J. Allegrini, M. John, G. Tregear and P. Corvol, Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 9386–9390 CrossRef CAS PubMed.
- S. Fienberg, G. E. Cozier, K. R. Acharya, K. Chibale and E. D. Sturrock, The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme, J. Med. Chem., 2017, 61, 344–359 CrossRef PubMed.
- A. Kuoppala, K. A. Lindstedt, J. Saarinen, P. T. Kovanen and J. O. Kokkonen, Inactivation of bradykinin by angiotensin-converting enzyme and by carboxypeptidase N in human plasma, Am. J. Physiol.: Heart Circ. Physiol., 2000, 278, H1069–H1074 CrossRef CAS PubMed.
- Z. H. Israili and W. D. Hall, Cough and Angioneurotic Edema Associated with Angiotensin-Converting Enzyme Inhibitor Therapy, Ann. Intern. Med., 1992, 117, 234–242 CrossRef CAS PubMed.
- M. Lenfant, J. Wdzieczak-Bakala, E. Guittet, J. C. Prome, D. Sotty and E. Frindel, Inhibitor of hematopoietic pluripotent stem cell proliferation: purification and determination of its structure, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 779–782 CrossRef CAS PubMed.
- G. Masuyer, R. G. Douglas, E. D. Sturrock and K. R. Acharya, Structural basis of Ac-SDKP hydrolysis by Angiotensin-I converting enzyme, Sci. Rep., 2015, 5, 13742 CrossRef CAS PubMed.
- R. G. Douglas, M. R. Ehlers and E. D. Sturrock, Antifibrotic peptideN-acetyl-Ser-Asp-Lys-Pro (Ac-SDKP): Opportunities for angiotensin-converting enzyme inhibitor design, Clin. Exp. Pharmacol. Physiol., 2013, 40, 535–541 CrossRef CAS PubMed.
- J. A. Koh, J. H. Ong, F. Abd Manan, K. Y. Ee, F. C. Wong and T. T. Chai, Discovery of Bifunctional Anti-DPP-IV and Anti-ACE Peptides from Housefly Larval Proteins After In silico Gastrointestinal Digestion, Biointerface Res. Appl. Chem., 2022, 12, 4929–4944 CAS.
- J. H. Ong, J. A. Koh, Y. Q. Siew, F. A. Manan, F. C. Wong and T. T. Chai, In silico discovery of multifunctional bioactive peptides from silkworm cocoon proteins following proteolysis, Curr. Top. Pept. Protein Res., 2021, 22, 47–57 CAS.
- J. H. Ong, C. E. Liang, W. L. Wong, F. C. Wong and T. T. Chai, Multi-target anti-sars-cov-2 peptides from mealworm proteins: An in silico study, Malays. J. Biochem. Mol. Biol., 2021, 24, 83–91 Search PubMed.
- D. Kanduc, Homology, similarity, and identity in peptide epitope immunodefinition, J. Pept. Sci., 2012, 18, 487–494 CrossRef CAS PubMed.
- P. Minkiewicz, A. Iwaniak and M. Darewicz, BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities, Int. J. Mol. Sci., 2019, 20(23), 5978 CrossRef CAS PubMed.
- D. Qin, W. Bo, X. Zheng, Y. Hao, B. Li, J. Zheng, G. Liang and Z. Lu, DFBP: a comprehensive database of food-derived bioactive peptides for peptidomics research, Bioinformatics, 2022, 38, 3275–3280 CrossRef CAS PubMed.
- G. Masuyer, S. L. U. Schwager, E. D. Sturrock, R. E. Isaac and K. R. Acharya, Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides, Sci. Rep., 2012, 2, 717 CrossRef PubMed.
- G. Jones, P. Willett, R. C. Glen, A. R. Leach and R. Taylor, Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen, J. Mol. Biol., 1997, 267, 727–748 CrossRef CAS PubMed.
- D. Lapaillerie, C. Charlier, V. Guyonnet-Dupérat, E. Murigneux, H. S. Fernandes, F. G. Martins, R. P. Magalhães, T. F. Vieira, C. Richetta, F. Subra, S. Lebourgeois, C. Charpentier, D. Descamps, B. Visseaux, P. Weigel, A. Favereaux, C. Beauvineau, F. Buron, M.-P. Teulade-Fichou, S. Routier, S. Gallois-Montbrun, L. Meertens, O. Delelis, S. F. Sousa and V. Parissi, Selection of Bis-Indolyl Pyridines and Triphenylamines as New Inhibitors of SARS-CoV-2 Cellular Entry by Modulating the Spike Protein/ACE2 Interfaces, Antimicrob. Agents Chemother., 2022, 66, e00083–22 CrossRef PubMed.
- C. M. M. Coelho, R. B. Pereira, T. F. Vieira, C. M. Teixeira, M. J. G. Fernandes, A. R. O. Rodrigues, D. M. Pereira, S. F. Sousa, A. Gil Fortes, E. M. S. Castanheira and M. S. T. Gonçalves, Synthesis, computational and nanoencapsulation studies on eugenol-derived insecticides, New J. Chem., 2022, 46, 14375–14387 RSC.
- S. Silva, J. Marto, L. M. Gonçalves, H. S. Fernandes, S. F. Sousa, A. J. Almeida and N. Vale, Development of Neuropeptide Y and Cell-Penetrating Peptide MAP Adsorbed onto Lipid Nanoparticle Surface, Molecules, 2022, 27(9), 2734 CrossRef CAS PubMed.
- R. P. Magalhães, T. F. Vieira, A. Melo and S. F. Sousa, Identification of novel candidates for inhibition of LasR, a quorum-sensing receptor of multidrug resistant Pseudomonas aeruginosa, through a specialized multi-level in silico approach, Mol. Syst. Des. Eng., 2022, 7, 434–446 RSC.
- T. Sander, J. Freyss, M. von Korff and C. Rufener, DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis, J. Chem. Inf. Model., 2015, 55, 460–473 CrossRef CAS PubMed.
- N. M. O’Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, Open Babel: An open chemical toolbox, J. Cheminf., 2011, 3, 33 Search PubMed.
- O. Korb, T. Stützle and T. E. Exner, Empirical Scoring Functions for Advanced Protein–Ligand Docking with PLANTS, J. Chem. Inf. Model., 2009, 49, 84–96 CrossRef CAS PubMed.
- S. Salentin, S. Schreiber, V. J. Haupt, M. F. Adasme and M. Schroeder, PLIP: fully automated protein–ligand interaction profiler, Nucleic Acids Res., 2015, 43, W443–W447 CrossRef CAS PubMed.
- G. E. Cozier, S. L. Schwager, R. K. Sharma, K. Chibale, E. D. Sturrock and K. R. Acharya, Crystal structures of sampatrilat and sampatrilat–Asp in complex with human ACE – a molecular basis for domain selectivity, FEBS J., 2018, 285, 1477–1490 CrossRef CAS PubMed.
- G. J. Kramer, A. Mohd, S. L. U. Schwager, G. Masuyer, K. R. Acharya, E. D. Sturrock and B. O. Bachmann, Interkingdom Pharmacology of Angiotensin-I Converting Enzyme Inhibitor Phosphonates Produced by Actinomycetes, ACS Med. Chem. Lett., 2014, 5, 346–351 CrossRef CAS PubMed.
- L. De Marchi, A. Wangorsch and G. Zoccatelli, Allergens from Edible Insects: Cross-reactivity and Effects of Processing, Curr. Allergy Asthma Rep., 2021, 21, 35 CrossRef CAS PubMed.
- I. Pali-Schöll, P. Meinlschmidt, D. Larenas-Linnemann, B. Purschke, G. Hofstetter, F. A. Rodríguez-Monroy, L. Einhorn, N. Mothes-Luksch, E. Jensen-Jarolim and H. Jäger, Edible insects: Cross-recognition of IgE from crustacean- and house dust mite allergic patients, and reduction of allergenicity by food processing, World Allergy Organ. J., 2019, 12(1), 100006 CrossRef PubMed.
- M. C. Jespersen, B. Peters, M. Nielsen and P. Marcatili, BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes, Nucleic Acids Res., 2017, 45, W24–W29 CrossRef CAS PubMed.
- I. Dimitrov, I. Bangov, D. R. Flower and I. Doytchinova, AllerTOP v.2—a server for in silico prediction of allergens, J. Mol. Model., 2014, 20, 2278 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3fo04246d |
| This journal is © The Royal Society of Chemistry 2024 |