Open Access Article
Open Access ArticleA polyhydroxyalkanoate synthesised by halophilic archaeon Natrialba swarupiae
Seema Prabhudev
Rodge
 a,
Maruti Jayram
Dhanavade
a,
Maruti Jayram
Dhanavade
 b,
Swapnil Chandrakant
Kajale
c and
Niranjan Prakashrao
Patil
b,
Swapnil Chandrakant
Kajale
c and
Niranjan Prakashrao
Patil
 *a
*a
aDepartment of Microbiology, MES, Abasaheb Garware College, Pune, Maharashtra, India. E-mail: niranjan75@gmail.com
bDepartment of Microbiology, Bharati Vidyapeeth's, Dr Patangrao Kadam Mahavidyalaya, Sangli, Maharashtra, India
cNational Centre for Microbial Resource, National Centre for Cell Science, Pune, Maharashtra, India
First published on 22nd June 2023
Abstract
Monomers and heteropolymers known as biodegradable polyhydroxyalkanoates (PHAs) are promising alternatives to fossil fuel-based polymers. During times of nutrient scarcity, microbes like extremophilic archaea store energy by accumulating PHA in the form of inclusions. However, knowledge of the mechanisms and PHA accumulation capabilities of extremophilic archaea is limited. In this study, an extremophilic haloarcheon, Natrialba swarupiae, isolated from a hypersaline lake in India has been investigated for its ability to produce PHA. The Sudan black B staining method was utilized for an initial assessment of potential PHA synthesizing ability. Nile blue A staining confirmed the presence of PHA granules. PHA concentrations were measured using crotonic acid assays. Nab. swarupiae yielded maxima of 0.27 ± 0.0094 g L−1 biomass and 0.14 ± 0.0017 g L−1 PHB, measuring 54.40 ± 1.386% PHB accumulation. The NMR analysis revealed the presence of a copolymer of PHA containing monomers of 3-hydroxybutyrate (3HB), 3-hydroxyvalerate (3HV), and 3-hydroxyoctanoate (3HO), poly-(3HB-co-3HV-co-3HO). The annotated genome sequence was used to detect the presence of the PHA synthase gene in Nab. swarupiae. The genome-scale metabolic model was built based on the ModelSEED pipeline using the Build Metabolic Model app in KBase. The homology model of PHA synthase was built by using SWISS-MODEL. The molecular docking study was carried out between PHA synthase and 3-hydroxybutyryl-CoA (3HBCA) along with 3-hydroxyoctanoyl-CoA (3HOCA) using PATCH-DOCK. The computational studies revealed good hydrogen bonding interactions between PHA synthase and 3-hydroxybutyryl-CoA. The molecular modeling studies could point out that the amino acid residues of PHA synthase such as TYR76, ASP183, MET152, ASN225, ASP235, THR228, ASP235, GLU238, TYR272 and ASN245 might play a major role in enzyme catalysis during PHA granule production.
Environmental significancePlastic pollution is causing havoc on planet earth. There is an immediate need to take action to find a solution to the pollution caused by plastic. One of the ways to address this problem is by using alternatives to plastic. Polyhydroxyalkanoates (PHA) are an alternative to plastic. Extremophiles are explored for their ability to synthesize PHA, which is produced by their cells in response to stress conditions. To understand the mechanism of PHA biosynthesis, there is a need to study the PHA synthase enzyme responsible for producing PHA. In this study, the PHA synthesizing ability of Natrialba swarupiae was explored, and studies on the structure of PHA synthase and molecular docking were performed to better understand and gain insight into the biochemical process. To the best of our knowledge, this is the first report on PHA synthesis by the Natrialba genus. |
Introduction
It's no secret that plastic trash is one of the biggest issues for our planet right now. More studies are being conducted on plastic pollution, allowing for more precise assessments and facilitating the development of more long-lasting solutions. There is already enough evidence to suggest that immediate actions should be taken rapidly and broadly to stop plastic discharge into the ecosystem and reduce future damage.1 Bans on single-use plastics, taxes on plastic production, and restrictions on the production of products containing microplastics are just a few examples of the measures that governments and non-governmental organizations around the world have taken to address this problem. Storage polymers called PHAs are produced by various microorganisms, including bacteria and archaea. Poly(3-hydroxybutyrate) P(3HB) is the most prevalent PHA and has desired thermoplastic-like characteristics.2 PHAs are biodegradable and non-toxic, making them a great substitute for single-use plastics in consumer products.The unique metabolic capabilities of extremophilic microbes, and especially the haloarchaea group (Archaea domain), have attracted a great deal of scientific interest. Halophilic archaea are unique microorganisms that have adapted to living in places with a lot of salt and the biomolecules they synthesize may have interesting properties.3 Some haloarchaeal species have also demonstrated the capacity to produce bioplastics.4
At the moment, halophiles are applied for the production of bacterioruberin, squalene, bacteriorhodopsin, and diether-/tetraether-lipids, which are the only commercially available products of archaeal cell factories. Additional archaeal products, like carotenoids, biohydrogen, PHAs, and methane, are at different levels of development.5
Halophiles have shown promise in overcoming these challenges.6 Because of their unique halophilic properties, some halophiles can grow well in media at elevated pH as well as elevated NaCl levels at higher temperatures. This lets fermentation processes run continuously and without contamination in non-sterile environments.7 Using renewable, low-cost carbon sources, the halophilic archaea, as cell factories, are believed to synthesize PHA, lowering the cost of fermentation in comparison to other extremophiles.8
Several studies have shown that haloarcheon Haloferax mediterranei can utilize a variety of industrial and domestic wastes as carbon sources for making PHA in a very efficient way, e.g., olive oil waste water, crude glycerol phase, starch waste, surplus whey.9–12
Also, a lot of research has shown that the PHA produced by Haloferax mediterranei is a mixture of 3-hydroxybutyrate and 3-hydroxyvalerate, when different carbon sources were used in the growth medium. The most important feature of Hfx. mediterranei is that this strain is able to make PHA copolyesters consisting of 3HB and 3HV even in the absence of a precursor that is structurally similar to 3HV. Instead, the strain just needs simple carbon sources like glycerol or sugars in order to complete the production of the copolyester. In contrast to the vast majority of other strains, which accumulate the P(3HB) homopolyester from such simple carbon, this strain accumulates the copolyester.13 PHBV is a commercially superior and more sustainable polymer than PHB.14 It has been shown that some species of the archaebacterial family Halobacteriaceae accumulate polyhydroxyalkanoates in the form of P(3HB). One of the members of this family, Archaebacterium strain 56, is halophilic in nature and is able to synthesise high levels of P(3HB). “Strain 56” was later classified as Halopiger aswanensis DSM 13151.15,16 PHA synthase catalyzes the polymerization of the hydroxyalkanoate monomer and thus serves as the principal enzyme in PHA biosynthesis. PHA synthase in haloarchaea is classified as class III and consists of two subunits, PhaC and PhaE. PHA synthase in halophilic bacteria, on the other hand, consists solely of the PhaC component and is classified as class I.17 In comparison to their halophilic counterparts, haloarchaeal PHA synthase has been observed to possess a broader substrate range, higher activity, and greater stability when subjected to pH and temperature fluctuations.18
Haloarcula marismortui was able to make up to 21% of its dry weight out of P(3HB) when it was grown in a minimal medium containing glucose as a source of carbon. phaEHm and phaCHm, which possibly code for two subunits of a class III PHA synthase, were reported in this organism.19 The remarkable execution and capacity of PhaC to incorporate both short chain length and medium chain length monomers have attracted considerable interest from the scientific community. The lack of understanding of the three-dimensional structure of PhaC has restricted our comprehension of the polymerization reaction, the factors that regulate chain length and polydispersity, and the molecular basis of the enzyme's specificity. Comprehending the processes by understanding and exploring three-dimensional structures is vital to determining the economical sustainability of these materials.20 The molecular docking approach can be applied to mimic the atomic-level interaction between a small molecule and a protein to characterize the behaviour of molecules in the binding domain of target proteins and reveal crucial biochemical processes.21
The present study focuses on investigating the ability of the haloarchaeal strain Nab. swarupiae to synthesize PHA and explores the mechanism of PHA synthase gene at the molecular level. A homology model of PHA synthase which is a stable structure was validated by different online servers. After PHA synthase model validation the molecular docking studies with 3-hydroxybutyryl-CoA (3HBCA) and 3-hydroxyoctanoyl-CoA (3HOCA) reported proper hydrogen bonding interactions. The amino acid residues of PHA synthase are found to interact with 3HBCA and 3HOCA.
Materials and methods
Procurement of the Nab. swarupiae isolate
The isolate was obtained from the National Centre for Microbial Resource, National Centre for Cell Science, Pune, Maharashtra, India (NCMR), after being initially isolated from a salt pan of the Sambhar Salt Lake in Rajasthan. The isolate was grown in Zobell marine broth with a pH of 9 and a salinity of 25% NaCl, supplemented with 2% glucose, and incubated at 42 °C for 7 days at 120 rpm in a shaking incubator.22Primary screening of the PHA synthesis ability
Secondary screening of the PHA synthesis ability
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz and an acquisition time of 3.27 s. Twenty milligrams of the extracted PHA were mixed with CDCl3 as a solvent for its analysis.27
000 Hz and an acquisition time of 3.27 s. Twenty milligrams of the extracted PHA were mixed with CDCl3 as a solvent for its analysis.27
Import of the genome and gene annotation
The whole genome sequence of Nab. swarupiae available at the public domain was imported at kbase.29 The RAST algorithm RASTtk – v1.073v1.9 was used to annotate the genome of Nab. swarupiae, which contains 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 201
201![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486 nucleotides in 99 contigs. The existing gene features were cleared and replaced by standard gene features called Prodigal and Glimmer3. Additionally, scans were conducted for rRNA, tRNA, selenoproteins, pyrrolysoproteins, repeat regions, and crispr. Using Kmers V2, Kmers V1, and protein similarity methods, genomic features were functionally annotated.30–32
486 nucleotides in 99 contigs. The existing gene features were cleared and replaced by standard gene features called Prodigal and Glimmer3. Additionally, scans were conducted for rRNA, tRNA, selenoproteins, pyrrolysoproteins, repeat regions, and crispr. Using Kmers V2, Kmers V1, and protein similarity methods, genomic features were functionally annotated.30–32
Metabolic model
The metabolic pathway map of Nab. swarupiae was generated using the Kbase ModelSEED pipeline using the Build Metabolic Map app, a database for annotating metabolic pathways. The annotated genome information of Nab. swarupiae was loaded into Kbase ModelSEED and the metabolic pathway prediction algorithm was executed, using homology-based inference and genomic information to predict metabolic pathways and reactions.33–35 The generated metabolic pathway map was analyzed to identify the key metabolic pathways of PHA synthesis and was experimentally validated using relevant methods such as Sudan black B and Nile blue A staining and crotonic acid assay.Secondary structural analysis and homology modeling studies of PHA synthase
The gene sequence and amino acid sequence of PHA synthase from Nab. swarupiae were extracted from RASTtk – v1.073 available on the kBase server (https://www.kbase.us/).29 The online server SOPMA program (Self-Optimized Prediction Method with Alignment) was used for secondary structure analysis of the PHA synthase sequence obtained from RASTtk – v1.073.36 This SOPMA analysis gives results that show the role of individual amino acids in building the secondary structure with their positions. Further, the amino acid sequence of PHA synthase from Nab. swarupiae was subjected to a SWISS-MODEL server to obtain its three-dimensional (3D) structure.37–39 Then the predicted 3D structure of PHA synthase was validated by PROCHECK and PROSA to check the model quality.40 After validating the PHA synthase model it was further used for molecular docking studies with 3-hydroxybutyryl-CoA.Molecular docking of PHA synthase with 3HBCA and 3HOCA
Molecular docking studies are now well explored to examine the intramolecular relations between receptors and ligands.41–45 The 2-D model of 3HBCA and 3HOCA was extracted from PubChem: (COMPOUND_CID > 644![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 065) and (COMPOUND_CID > 44
065) and (COMPOUND_CID > 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 441
441![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 888) respectively. Then this 2D structure was converted into a 3D structure with MolView (https://molview.org/). The molecular docking studies were carried out between the predicted model of PHA synthase and 3HOCA by using AutoDock software.46 After molecular docking completion, the docked complex of PHA synthase with 3HBCA and 3HOCA with the lowest energy was further analyzed by using structural visualization software such as CHIMERA.47
888) respectively. Then this 2D structure was converted into a 3D structure with MolView (https://molview.org/). The molecular docking studies were carried out between the predicted model of PHA synthase and 3HOCA by using AutoDock software.46 After molecular docking completion, the docked complex of PHA synthase with 3HBCA and 3HOCA with the lowest energy was further analyzed by using structural visualization software such as CHIMERA.47
Results and discussion
Primary screening for PHA synthesis
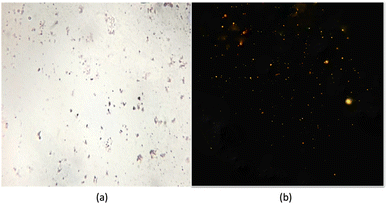
Haloarchaea are a group of archaea that require high salt concentrations for growth and are often found in hypersaline environments such as salt lakes and salt pans. These organisms are acknowledged to synthesise large quantities of lipids as a survival strategy in these extreme environments.48 A haloarchaeal strain, Nab. swarupiae, was recently identified from a salt pan of the Sambhar Salt Lake in Rajasthan, India. Colonies of coccoid, nonmotile, Gram-negative cells turned a pinkish red due to the presence of bacterioruberins which are C50 carotenoid pigments. Similar kinds of pigments have also been reported in Haloterrigena sp. strain SGH1, isolated from the Atacama Desert, Chile.49The optimal temperature range for this strain's aerobic growth is 40 °C, with the NaCl concentration being between 20% and 25% and pH 9, the organism can be considered as polyextremophile.21 Sudan black B staining is a commonly used method to visualize and quantify PHAs in archaea. When haloarchaea Nab. swarupiae were subjected to Sudan black B staining, they showed the presence of intense staining due to the presence of lipid droplets in their cells. The lipid droplets appear as dark inclusions in the cells (Fig. 1a). The presence of PHA granules was further confirmed using the Nile blue A staining method. When observed under a ZEISS fluorescence microscope with a blue filter having a 465 nm excitation wavelength the Nab. swarupiae emitted fluorescence confirming the presence of PHA granules; similar results were reported in case of Halococcus and Halorubrum spp. (Fig. 1b).23,50
Secondary screening of PHA
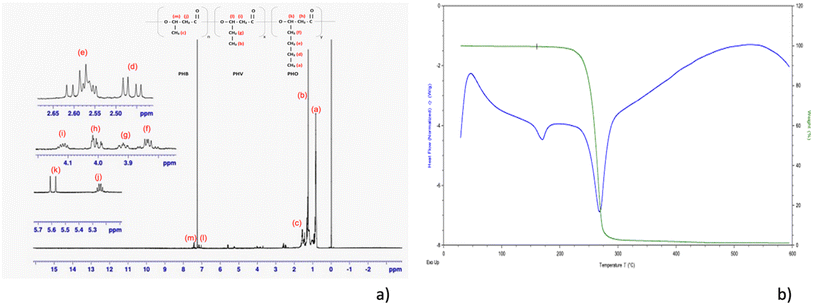
Crotonic acid assay was conducted with the haloarchaeon species Nab. swarupiae. The results showed that the maximum biomass and PHB concentration were 0.27 ± 0.0094 g L−1 and 0.14 ± 0.0017 g L−1, respectively. Additionally, the PHA content as a percentage of dry cell weight was found to be 54.40 ± 1.386%. These results indicate that Nab. swarupiae was able to produce a substantial amount of PHA under the conditions of the experiment, with a high PHA content relative to the biomass produced. This information can be useful for understanding the potential of this species for use in industrial applications, such as bioplastics production. The PHA extracted from the Nab. swarupiae was subjected to NMR analysis. It showed the following results: 1H NMR (500 MHz, CDCl3) δ 7.43 (1H, t), δ 7.15 (1H, q), δ 5.60 (1H, d), δ 5.24 (2H, m), δ 4.10 (2H, m), δ 4.00 (2H, m), δ3.91 (2H, m), δ 3.87 (2H, d), δ 2.57 (2H, m), δ 2.46 (2H, dd), δ 1.56 (3H, m), δ 1.253 (3H, m), δ 0.83 (3H, m). The NMR analysis revealed the presence of a copolymer of PHA containing monomers of 3-hydroxybutyrate (3HB), 3-hydroxyvalerate (3HV), and 3-hydroxyoctanoate (3HO), poly-(3HB-co-3HV-co-3HO) (Fig. 2a). Similar NMR results were observed by Shamala et al. while analysing PHA obtained by R. meliloti and recombinant E. coli.51,52 The TGA of the PHA showed the high purity of the sample. As per the literature survey rapid thermal degradation occurs in the range of 250 to 320 °C for the homopolymer or copolymer of PHA.53 The values of Td10% (246 °C) and Tdmax (290 °C) were found to be comparable to the degradation temperatures reported in the literature (Fig. 2b).28 The thermal behaviour of PHA could be investigated using DSC. Tm was measured to be 149 °C. An exothermic transition at about 50 °C was observed which could be considered as a cold crystallization.54 An endothermic peak was observed at 246 °C depicting the thermal degradation of the molecule. The thermal behaviour of the sample is comparable to that reported in the literature.28 As the organism showed the presence of PHA granules, the organism's ability to synthesize PHA was biochemically confirmed. To understand the PHA synthesis at the molecular level, the presence of the PHA synthase gene was confirmed using gene annotation of the draft genome of Nab. swarupiae.Gene annotation
The input genome of Nab. swarupiae had 4612 CDS and 48 RNA, which includes 45 tRNA and 3 rRNA (5S, 23S and 16S each). Kajale et al. also stated that the genome had 2948 “hypothetical” proteins and 1664 “putatively functional” genes.21 After processing the genome using the RASTtk – v1.073v1.9 application in kbase, 4708 features were called, of which 94 are non-coding and 4614 are coding genes. The output genome has several different types of non-coding features, including non-coding genes (51), non-coding ncRNA (2), non-coding rRNA (3), non-coding repeats (48), non-coding RNA (46), and non-coding tRNA (46), for the identification of new features. The genes in the output genome have 1416 distinct functions, with 1990 genes having SEED annotation ontology across 801 distinct SEED functions. This information is useful in understanding the various roles that the genes play in the genome and the biological processes they are involved in.Metabolic mapping
The metabolic pathway map of Nab. swarupiae was generated using Kbase ModelSEED, a database for annotating metabolic pathways.33 The metabolic model is available at the URL https://doi.org/10.25982/106247.14/1924621 (Fig. 3). The metabolic model built shows the presence of 882 reactions and 968 compounds. Butanoate metabolism can be viewed by following the steps as shown in Fig. 3. Butanoate metabolism consists of 59 reactions and 41 compounds.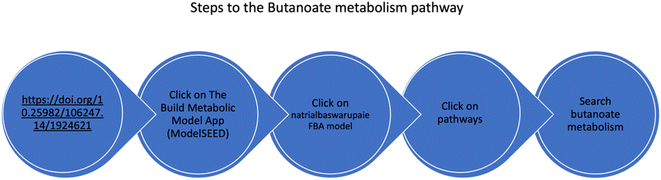 | ||
| Fig. 3 Schematic diagram of the steps to be followed to view the metabolic map of Natrialba swarupiae. | ||
Analysis of PHA synthase gene
Polyhydroxyalkanoic acid synthase gene was found to be located on contig NZ_VTAW01000010.1 on the negative strand from 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 697 to 31
697 to 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182. It is referred to as GCF_008245225.1.CDS.35. The protein PHA synthase has a length of 494 amino acids and its sequence is hydrophobic and has both positive and negative charged regions. The DNA sequence is 1485 base pairs long. It is associated with mRNA and CDS child features. The poly(3-hydroxyalkanoate) polymerase subunit PhaC of Haloferax mediterranei (strain ATCC 33500) was studied and was found to have the presence of 492 amino acids. It showed the presence of 3 active sites at the positions of amino acid numbers 150, 305, and 334.55–57 Meanwhile, the poly(3-hydroxyalkanoate) polymerase subunit PhaC of Haloarcula hispanica strain ATCC 33960 showed the presence of 474 amino acids. It was found to have the presence of 3 active sites at the positions of 162, 317, and 346.58,59
182. It is referred to as GCF_008245225.1.CDS.35. The protein PHA synthase has a length of 494 amino acids and its sequence is hydrophobic and has both positive and negative charged regions. The DNA sequence is 1485 base pairs long. It is associated with mRNA and CDS child features. The poly(3-hydroxyalkanoate) polymerase subunit PhaC of Haloferax mediterranei (strain ATCC 33500) was studied and was found to have the presence of 492 amino acids. It showed the presence of 3 active sites at the positions of amino acid numbers 150, 305, and 334.55–57 Meanwhile, the poly(3-hydroxyalkanoate) polymerase subunit PhaC of Haloarcula hispanica strain ATCC 33960 showed the presence of 474 amino acids. It was found to have the presence of 3 active sites at the positions of 162, 317, and 346.58,59
Secondary structure analysis
The results obtained from SOPMA analysis for PHA synthase revealed that about 47.77% of the residues form an alpha helix, 11.74% of the residues produce an extended strand region, and 35.63% the residues form random coils. Meanwhile, only 4.86% of the amino acid residues form a beta-turn (Fig. 4).36 The overall SOPMA result states that there is about 35.63% part of the protein occupied by a random coil but still there are a good number of amino acids that reside in the regions of the alpha helix and extended strand. Considering the SOPMA result, it is confirmed that the total percentage of the random coil and beta-turn forming regions is very low which indicates that the good quality of the model can be predicted using the PHA synthase sequence (Fig. 5).Structural validation of PHA synthase
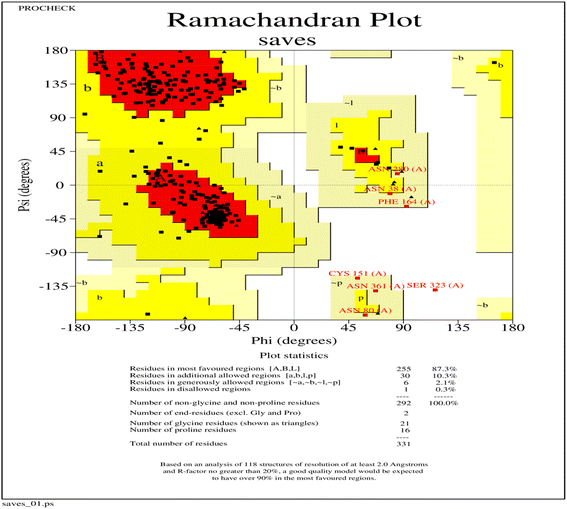
The three-dimensional structure of PHA synthase from Nab. swarupiae was built by using SWISS-MODEL and shows good quality.37 The predicted model of PHA synthase when analyzed using PROCHECK shows that about 87.3% of the residues fall in the most favoured regions, 10.3% of the residues are present in additional allowed regions and 2.1% of the residues belong to generously allowed regions (Fig. 6). A total of 99.7% of the amino acid residues are present in the favoured regions whereas only 0.3% of the residues are in the outer region which indicates good quality of the predicted model (Fig. 6). The model quality was evaluated by another validation tool PROSA which gives a Z score. The PROSA analysis PHA synthase model was given a Z score of −5.8. This Z score is within the acceptable range of X-ray and NMR studies (Fig. 7a). The PROSA II test showed a maximum number of residues having negative interaction energy hence it confirms that the PHA synthase model is of good quality (Fig. 7b).60 After validating the PHA synthase model using all these validation servers it was confirmed that the predicted model of PHA synthase is of good quality and suitable to dock with 3HBCA and 3HOCA.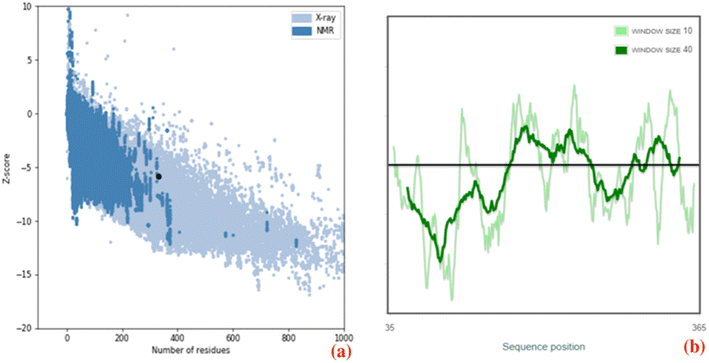 | ||
| Fig. 7 (a) ProSA-web Z-score plot of the PHA synthase of Natrialba swarupiae (b) ProSA-web plot of residue scores of a native protein structure of the PHA synthase of Natrialba swarupiae. | ||
Analysis of a docked complex of PHA synthase with 3HBCA and 3HOCA
The docked complex of PHA synthase with 3HBCA and 3HOCA was analyzed to check the molecular interactions as well as its energy level. The docked complex had lower energy, hence it was stable (Fig. 8). The hydrogen bonding interactions of the PHA synthase with 3HBCA and 3HOCA could be playing the main role in structural stability as shown in Table 1. The PHA synthase-like amino acid residues such as TYR76, ASP183, MET152, ASN225, ASP235, THR228, ASP235, GLU238, TYR272 and ASN245 showed strong hydrogen bonding interactions with 3HBCA (Fig. 9 and Table 1). Hence these amino acid residues could play a vital role in the activity of the PHA synthase enzyme. Molecular docking studies have provided insights into the molecular interactions of PHA synthase with 3HBCA and 3HOCA. This information can help researchers understand the structural and functional features of the target protein.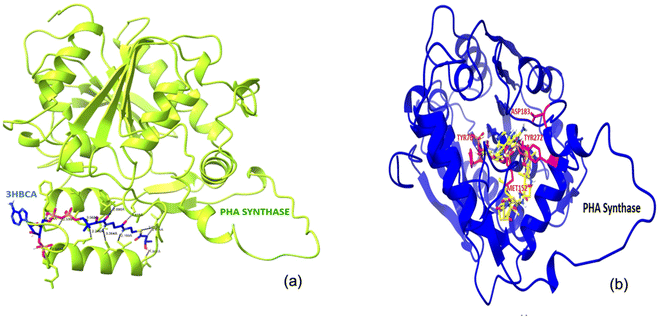 | ||
| Fig. 8 (a)The docked complex of PHA synthase with 3HBCA; (b) the docked complex of PHA synthase with 3HOCA. | ||
| Sr. no. | Hydrogen bonding interactions between the amino acid residues of PHA synthase and 3-hydroxybutyryl-CoA | Distance in Å |
|---|---|---|
| 1 | ASN 245 CA⋯3HBCA 1 H33: | 1.474 |
| 2 | 3HBCA 1 H7⋯ASN 225 OD1: | 1.801 |
| 3 | THR 228 CG2⋯3HBCA 1 H12: | 2.199 |
| 4 | ASP 235 OD2⋯3HBCA 1 C22: | 2.246 |
| 5 | 3HBCA 1 C3⋯LEU 213 CD1: | 2.414 |
| 6 | 3HBCA 1 O65⋯VAL 215 CG2: | 2.418 |
| 7 | ASN 245 OD1⋯3HBCA 1 H28: | 2.539 |
| 8 | GLU 238 C⋯3HBCA 1 O57: | 2.543 |
| Sr. no. | Hydrogen bonding interactions between the amino acid residues of PHA synthase and 3-hydroxyoctyryl-CoA | Distance in Å |
|---|---|---|
| 1 | TYR 272 OH⋯3HOCA 1 H5: | 1.075 |
| 2 | TYR 76 HH⋯3HOCA 1 O10: | 2.244 |
| 3 | ASP 183 OD1⋯3HOCA 1 H5: | 2.252 |
| 4 | ASP 183 OD2⋯3HOCA 1 H6: | 2.299 |
| 5 | MET 152 CE⋯3HOCA 1 C27: | 2.595 |
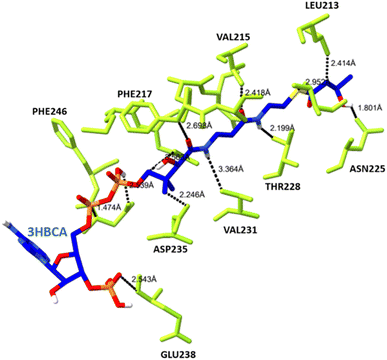 | ||
| Fig. 9 The hydrogen bonding interactions between the amino acid residues of PHA synthase (yellow) and 3HBCA (blue). | ||
Han et al. extensively studied polyhydroxyalkanoate synthase in halophilic archaea. In their study they have mentioned four Natrialba species, Nab. chahannaoensis CGMCC 1.1977, Nab. hulunbeirensis CGMCC 1.1986, Nab. magadii CGMCC 1.1966, and Natrialba sp. CGMCC 1.1968, out of which 3 Natrialba spp. showed the presence of PhaC gene, but none of them showed PHA accumulation.61 To the best of our knowledge this is the first report on Nab. swarupaie showing PHA accumulation.
Conclusion
This study investigated the ability of Nab. swarupiae, an extremophilic haloarcheon isolated from a hypersaline lake in India, to produce biodegradable PHAs. The Sudan black B staining method and Nile blue A staining showed potential PHA production. The highest concentration of PHA (0.14 g L−1) was achieved with 54.40% PHA/DCW. The genome sequence revealed a PHA synthase gene, and a genome-scale metabolic model was built. A molecular docking study was carried out, which showed good hydrogen bonding interactions between PHA synthase and 3-hydroxybutyryl-CoA and also 3-hydroxyoctanoyl-CoA. The molecular docking studies between PHA synthase and 3HBCA revealed that there are strong hydrogen bonding interactions between them. The amino acid residues of PHA synthase such as TYR76, ASP183, MET152, ASN225, ASP235, THR228, ASP235, GLU238, TYR272 and ASN245 could play a major role in enzyme catalysis. This molecular modelling study supports the view that PHA synthase could efficiently interact with 3HBCA and 3HOCA to produce PHA granules. This molecular docking study has provided valuable insights into the molecular interactions between PHA synthase and its substrate, and can aid in understanding molecular mechanisms and designing protein engineering experiments.Data availability
The metabolic pathway map of Natrialba swarupiae was generated using Kbase ModelSEED, a database for annotating metabolic pathways (Henry et al. 2010). The metabolic model is available at the URL https://doi.org/10.25982/106247.14/1924621 (Fig. 3).Author contributions
Seema Rodge: conceptualization, writing – original draft, writing – review and editing, methodology, visualisation and investigation. Maruti Dhanavade: software, formal analysis, writing – original draft, investigation, validation and visualization. Swapnil Kajale: resources, data curation, investigation. Niranjan Patil: conceptualization, investigation, writing – review and editing, and supervision.Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript. The authors have no financial or proprietary interests in any material discussed in this article.Acknowledgements
The authors declare that no funds, grants, or other support was received during the preparation of this manuscript and research. We are grateful to Dr Avinash Sharma, Scientist, NCMR, Pune, India for providing us with the Natrialba swarupiae isolate. We are thankful to the Principal, Abasaheb Garware College, Pune and Head, Department of Microbiology, Abasaheb Garware College, Pune, India.References
- A. A. Horton, Plastic pollution: When do we know enough?, J. Hazard. Mater., 2022, 422, 126885, DOI:10.1016/j.jhazmat.2021.126885.
- D. Nagarajan, et al., Microbial cell factories for the production of polyhydroxyalkanoates, Essays Biochem., 2021, 65, 337–353, DOI:10.1042/EBC20200142.
- A. Singh and A. K. Singh, Haloarchaea: worth exploring for their biotechnological potential, Biotechnol. Lett., 2017, 39, 1793–1800, DOI:10.1007/s10529-017-2434-y.
- M. Koller and S. K. Rittmann, Haloarchaea as Emerging Big Players in Future Polyhydroxyalkanoate Bioproduction: Review of Trends and Perspectives, Curr. Res. Biotechnol., 2022, 4, 377–391, DOI:10.1016/j.crbiot.2022.09.002.
- K. Pfeifer, et al., Archaea Biotechnology, Biotechnol. Adv., 2021, 47, 107668, DOI:10.1016/j.biotechadv.2020.107668.
- L. Simó-Cabrera, et al., Haloarchaea as Cell Factories to Produce Bioplastics, Mar. Drugs, 2021, 19, 159, DOI:10.3390/md19030159.
- J. Yin, et al., Halophiles, coming stars for industrial biotechnology, Biotechnol. Adv., 2015, 33, 1433–1442, DOI:10.1016/j.biotechadv.2014.10.008.
- F. Karray, et al., Production of Poly(3-Hydroxybutyrate) by Haloarcula, Halorubrum, and Natrinema Haloarchaeal Genera Using Starch as a Carbon Source, Archaea, 2021, 2021, 8888712, DOI:10.1155/2021/8888712.
- D. Alsafadi and O. Al-Mashaqbeh, A one-stage cultivation process for the production of poly-3-(hydroxybutyrate-co-hydroxyvalerate) from olive mill wastewater by Haloferax mediterranei, N. Biotechnol., 2017, 34, 47–53, DOI:10.1016/j.nbt.2016.05.003.
- C. Hermann-Krauss, et al., Archaeal production of polyhydroxyalkanoate (PHA) co-and terpolyesters from biodiesel industry-derived by-products, Archaea, 2013, 2013, 129268, DOI:10.1155/2013/129268.
- C. W. Chen, et al., Enzymatic extruded starch as a carbon source for the production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by Haloferax mediterranei, Process Biochem., 2006, 41, 2289–2296, DOI:10.1016/j.procbio.2006.05.026.
- M. Koller, et al., Biosynthesis of high quality polyhydroxyalkanoate co-and terpolyesters for potential medical application by the archaeon Haloferax mediterranei, Macromol. Symp., 2007, 253, 33–39 CrossRef CAS.
- T. M. Don, C. W. Chen and T. H. Chan, Preparation and characterization of poly (hydroxyalkanoate) from the fermentation of Haloferax mediterranei, J. Biomater. Sci., Polym. Ed., 2006, 17, 1425–1438 CrossRef CAS PubMed.
- J. Han, et al., Biosynthesis, characterization, and hemostasis potential of tailor-made poly(3-hydroxybutyrate-co-3-hydroxyvalerate) produced by Haloferax mediterranei, Biomacromol, 2015, 16, 578–588, DOI:10.1021/bm5016267.
- F. F. Hezayen, A. Steinbüchel and B. H. Rehm, Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56, Arch. Biochem. Biophys., 2002, 403, 284–291, DOI:10.1016/s0003-9861(02)00234-5.
- F. F. Hezayen, et al., Halopiger aswanensis sp. nov., a polymer-producing and extremely halophilic archaeon isolated from hypersaline soil, Int. J. Syst. Evol. Microbiol., 2010, 60, 633–637, DOI:10.1099/ijs.0.013078-0.
- R. Mitra, et al., Current developments on polyhydroxyalkanoates synthesis by using halophiles as a promising cell factory, Microb. Cell Fact., 2020, 19, 86, DOI:10.1186/s12934-020-01342-z.
- J. Han, et al., Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui, Appl. Environ. Microbiol., 2007, 73, 6058–6065, DOI:10.1128/AEM.00953-07.
- M. F. Chek, et al., Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics, Sci. Rep., 2017, 7, 5312, DOI:10.1038/s41598-017-05509-4.
- B. J. McConkey, V. Sobolev and M. Edelman, The performance of current methods in ligand-protein docking, Curr. Sci., 2002, 83, 845–855 CAS.
- S. Kajale, et al., Natrialba swarupiae sp. nov., a halophilic archaeon isolated from a hypersaline lake in India, Int. J. Syst. Evol. Microbiol., 2020, 70, 1876–1881, DOI:10.1099/ijsem.0.003986.
- K. L. Burdon, Fatty Material in Bacteria and Fungi Revealed by Staining Dried, Fixed Slide Preparations, J. Bacteriol., 1946, 52, 665–678, DOI:10.1128/jb.52.6.665-678.1946.
- A. G. Ostle and J. G. Holt, Nile blue A as a fluorescent stain for poly-beta-hydroxybutyrate, Appl. Environ. Microbiol., 1982, 44, 238–241, DOI:10.1128/aem.44.1.238-241.1982.
- J. H. Law and R. A. Slepecky, Assay of poly-beta-hydroxybutyric acid, J. Bacteriol., 1961, 82, 33–36, DOI:10.1128/jb.82.1.33-36.1961.
- S. Palmieri, et al., Effects of different pre-treatments on the properties of polyhydroxyalkanoates extracted from sidestreams of a municipal wastewater treatment plant, Sci. Total Environ., 2021, 801, 149633, DOI:10.1016/j.scitotenv.2021.149633.
- D. Heinrich, M. H. Madkour, M. A. Al-Ghamdi, Shabbaj II and A. Steinbüchel, Large scale extraction of poly(3-hydroxybutyrate) from Ralstonia eutropha H16 using sodium hypochlorite, AMB Express, 2012, 2(1), 59, DOI:10.1186/2191-0855-2-59.
- Y. Doi, et al., Nuclear Magnetic Resonance Studies of Poly(3-Hydroxybutyrate) and Polyphosphate Metabolism in Alcaligenes eutrophus, Appl. Environ. Microbiol., 1989, 55, 2932–2938, DOI:10.1128/aem.55.11.2932-2938.1989.
- L. Lorini, et al., Characterization of Polyhydroxyalkanoates Produced at Pilot Scale From Different Organic Wastes, Front. Bioeng. Biotechnol., 2021, 9, 628719, DOI:10.3389/fbioe.2021.628719.
- A. Arkin, et al., KBase: The United States Department of Energy Systems Biology Knowledgebase, Nat. Biotechnol., 2018, 36, 566–569, DOI:10.1038/nbt.4163.
- R. K. Aziz, et al., The RAST Server: rapid annotations using subsystems technology, BMC Genomics, 2008, 9, 75, DOI:10.1186/1471-2164-9-75.
- R. Overbeek, et al., The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST), Nucleic Acids Res., 2014, 42, D206–D214, DOI:10.1093/nar/gkt1226.
- T. Brettin, et al., RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes, Sci. Rep., 2015, 5, 8365, DOI:10.1038/srep0836.
- C. S. Henry, et al., High-throughput generation, optimization and analysis of genome-scale metabolic models, Nat. Biotechnol., 2010, 28, 977–982, DOI:10.1038/nbt.1672.
- M. Latendresse, Efficiently gap-filling reaction networks, BMC Bioinf., 2014, 15, 225, DOI:10.1186/1471-2105-15-225.
- J. M. Dreyfuss, et al., Reconstruction and validation of a genome-scale metabolic model for the filamentous fungus Neurospora crassa using FARM, PLoS Comput. Biol., 2013, 9, e1003126, DOI:10.1371/journal.pcbi.1003126.
- C. Geourjon and G. Deléage, SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments, Comput. Appl. Biosci., 1995, 11, 681–684, DOI:10.1093/bioinformatics/11.6.681.
- T. Schwede, et al., SWISS-MODEL: An automated protein homology-modeling server, Nucleic Acids Res., 2003, 31, 3381–3385, DOI:10.1093/nar/gkg520.
- A. Waterhouse, et al., SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res., 2018, 46, W296–W303, DOI:10.1093/nar/gky427.
- S. Bienert, et al., The SWISS-MODEL Repository-new features and functionality, Nucleic Acids Res., 2017, 45, D313–D319, DOI:10.1093/nar/gkw1132.
- R. A. Laskowaski, et al., PROCHECK a program to check sterio-chemical quality of a protein structures, J. Appl. Crystallogr., 1993, 26, 283–291, DOI:10.1107/S0021889892009944.
- K. D. Sonawane and M. J. Dhanavade, Molecular Docking Technique to Understand Enzyme-Ligand Interactions, Methods and Algorithms for Molecular Docking-Based Drug Design and Discovery, IGI Global, 2015, pp. 245–264 Search PubMed.
- K. D. Sonawane and M. J. Dhanavade, Computational Approaches to Understand Cleavage Mechanism of Amyloid Beta (Aβ) Peptide. Computational Modeling of Drugs against Alzheimer's Disease, Springer, Neuromethods, 2017, pp. 263–282 Search PubMed.
- M. J. Dhanavade and K. D. Sonawane, Insights into the molecular interactions between aminopeptidase and amyloid beta peptide using molecular modeling techniques, Amino Acids, 2014, 46, 1853–1866, DOI:10.1007/s00726-014-1740-0.
- M. J. Dhanavade, et al., Homology modeling, molecular docking and MD simulation studies to investigate role of cysteine protease from Xanthomonas campestris in degradation of Aβ peptide, Comput. Biol. Med., 2013, 43, 2063–2070, DOI:10.1016/j.compbiomed.2013.09.021.
- M. J. Dhanavade, et al., Molecular modeling approach to explore the role of cathepsin B from Hordeum vulgare in the degradation of Aβ peptides, Mol. BioSyst., 2016, 12, 162–168, 10.1039/c5mb00718f.
- G. M. Morris, et al., AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30, 2785–2791, DOI:10.1002/jcc.21256.
- E. F. Pettersen, et al., UCSF Chimera a visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25, 1605–1612, DOI:10.1002/jcc.20084.
- M. Kates, Biology of halophilic bacteria, Part II. Membrane lipids of extreme halophiles: biosynthesis, function and evolutionary significance, Experientia., 1993, 49, 1027–1036, DOI:10.1007/BF01929909.
- N. Flores, et al., Haloterrigena sp. Strain SGH1, a Bacterioruberin-Rich, Perchlorate-Tolerant Halophilic Archaeon Isolated From Halite Microbial Communities, Atacama Desert, Chile, Front. Microbiol., 2020, 11, 324, DOI:10.3389/fmicb.2020.00324.
- A. Legat, et al., Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species, Appl. Microbiol. Biotechnol., 2010, 87, 1119–1127, DOI:10.1007/s00253-010-2611-6.
- T. R. Shamala, et al., Production and characterization of bacterial polyhydroxyalkanoate copolymers and evaluation of their blends by Fourier transform infrared spectroscopy and scanning electron microscopy, Indian J. Microbiol., 2009, 49, 251–258, DOI:10.1007/s12088-009-0031-z.
- G.-Y. A. Tan, et al., Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review, Polymers, 2014, 6, 706–754, DOI:10.3390/polym6030706.
- S. K. Hahn and Y. K. Chang, A thermogravimetric analysis for poly(3-Hydroxybutyrate) quantification, Biotechnol. Tech., 1995, 9, 873–878, DOI:10.1007/BF00158539.
- R. M. R. Wellen, et al., Melting and crystallization of poly(3-Hydroxybutyrate): effect of heating/cooling rates on phase transformation, Polim., 2015, 25, 296–304, DOI:10.1590/0104-1428.1961.
- S. DasSarma, et al., Methylomes of Two Extremely Halophilic Archaea Species, Haloarcula marismortui and Haloferax mediterranei, Microbiol. Resour. Announce., 2019, 8, 00577719, DOI:10.1128/MRA.00577-19.
- Q. Lu, et al., Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei, J. Bacteriol., 2008, 190, 4173–4180, DOI:10.1128/JB.00134-08.
- J. Han, et al., Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer, J. Bacteriol., 2012, 194, 4463–4464, DOI:10.1128/JB.00880-12.
- J. Han, et al., Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui, Appl. Environ. Microbiol., 2007, 73, 6058–6065, DOI:10.1128/AEM.00953-07.
- H. Liu, et al., Complete genome sequence of Haloarcula hispanica, a Model Haloarchaeon for studying genetics, metabolism, and virus-host interaction, J. Bacteriol., 2011, 193, 6086–6087, DOI:10.1128/JB.05953-11.
- M. Wiederstein and M. J. Sippl, ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins, Nucleic Acids Res., 2007, 35, W407–W410, DOI:10.1093/nar/gkm290.
- J. Han, et al., Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases, Appl. Environ. Microbiol., 2010, 76, 7811–7819, DOI:10.1128/AEM.01117-10.
| This journal is © The Royal Society of Chemistry 2023 |