Open Access Article
Open Access ArticleBacterial transformation of per- and poly-fluoroalkyl substances: a review for the field of bioremediation
Jessica A.
LaFond
 a,
Paul B.
Hatzinger
a,
Paul B.
Hatzinger
 b,
Jennifer L.
Guelfo
b,
Jennifer L.
Guelfo
 a,
Kayleigh
Millerick
a and
W. Andrew
Jackson
*a
a,
Kayleigh
Millerick
a and
W. Andrew
Jackson
*a
aDepartment of Civil, Environmental & Construction Engineering, Texas Tech University, Lubbock, TX 79409, USA. E-mail: andrew.jackson@ttu.edu
bAptim Federal Services, LLC, 17 Princess Road, Lawrenceville, New Jersey, USA
First published on 26th June 2023
Abstract
Per- and polyfluoroalkyl substances (PFAS) have received growing attention as more research on their potential risk and widespread extent of contamination has become available. Remediation schemes are currently being developed to help mitigate exposure to PFAS, especially in aquifers underlying sites where extensive application of aqueous film forming foam (AFFF) for fire supression and/or fire training has resulted in widepread PFAS contamination. To-date, microbial remediation (bioremediation) has not been considered for PFAS treatment due to the recalcitrant nature of these compounds, and the lack of identified organisms that can completely degrade PFAS to non-toxic by-products. However, based on recent literature, the role of microorganisms may be important to our understanding of processes occurring at contaminated sites where transformation of PFAS precursors to terminal end-products has been observed. This paper reviews the current state of knowledge on the biotransformation of PFAS by microorganisms and includes topics of particular interest to bioremediation schemes. First, we review studies linking the exposure of PFAS to microbial toxicity at the cellular level and then expand the viewpoint to observed changes at the microbial community level. Second, we review the different classes of PFAS observed to biotransform and provide and overview of reactions that are likely to occur under different environmental conditions. Third we evaluate the role of co-metabolism and specific enzyme classes in the observed biotransformation of larger PFAS to terminal perfluoroalkyl acids (PFAAs). Lastly, we identify knowledge gaps in the understanding of PFAS biotransformation and provide suggestions for future research that may result in new strategies that include bioremediation as a solution for PFAS in environmental matrices.
1. Introduction
The past two decades have seen a growing awareness and interest in remediation schemes for compounds classified as per- and polyfluoroalkyl substances, collectively known as PFAS. PFAS are anthropogenic compounds synthesized and mass produced for their unique and advantageous properties including hydro- and lipo-phobicity, extreme heat resistance, and high surface-activity.1 Their basic structure consists of a functional group that is typically charged (“head”) attached to a fluoroalkyl backbone (“tail”), and this combination of the “head” and “tail” structure yields molecules that are surface active (i.e. surfactant), particularly at the air–water interface. Surfactant PFAS lower the surface tension of the media which allows ready formation of foams, leading to extensive use of PFAS in fire-fighting foams.2 In addition to foams, PFAS have been used in a range of industrial applications and consumer products. For example, they have been used as components in food packaging liners, stain resistant textiles and water-resistant goods (since PFAS repel both water and oil), as well as in cables, wires, gaskets and hoses, due to their stability under high temperatures. They are also used in engines as lubricants and coatings to protect parts from oxidation.3PFAS are structurally diverse. For example, the head can be a range of functional groups including carboxylates, sulfonates, sulfonamides, phosphonates, and other larger moieties consisting of combinations of the previously listed groups and compounds such as hydrocarbons and amines, and the tail varies by length and degree of fluorination.4 It is also possible to have tails that include both fluoroalkyl and ether moieties, and these PFAS are commonly referred to as per- and polyfluoroalkyl ether acids (PFEAs; e.g., GenX).5 Due to the extreme stability of the carbon–fluorine bond, many PFAS have been shown to be recalcitrant in the environment even if transformation of the head group is observed.6 Widespread use and transport via the ocean and atmosphere has led to global PFAS contamination.7–9 This, along with rising health concerns has led to incorporation of some PFAS in the Stockholm Convention list for Persistent Organic Pollutants and to voluntary phaseouts by major manufacturers.10 Recently, the United States Environmental Protection Agency (EPA) proposed stringent drinking water maximum contaminant levels (MCLs) for perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS) (4 ng L−1) and a composite of perfluorononanoic acid (PFNA), perfluorohexanesulfonic acid (PFHxS), perfluorobutanesulfonic acid (PFBS) and GenX calculated based on their relative hazard indices.11 There is a pressing need for viable methods of treatment and remediation as there are documented PFAS impacts to thousands of water supplies across the US and abroad.11
To-date there has not been an all-encompassing solution for PFAS remediation, particularly with regards to legacy PFAS releases, such as those used for fire-training with aqueous film forming foam (AFFF), where decades of AFFF use have led to extensive soil and groundwater contamination. However, in recent years there have been promising advances in destructive technologies, such as photocatalytic systems with boron nitride and titanium dioxide, showing mineralization capabilities and more recently a non-photocatalytic system using dimethyl sulfoxide (DMSO) proving capable of mineralizing PFOA and GenX at near room temperature through decarboxylation reactions.12–15 Other destructive processes such as ball milling, super critical water oxidation, plasma, and thermal decomposition as well as non-destructive technologies such as adsorption onto activated carbon and removal via ion exchange resin have also shown promising results with a potential to be applied ex situ, and in a few cases in situ (e.g., adsorption to injected colloidal activated carbon or clay media).16–20 In addition to physical and chemical removal and destruction technologies, recent studies suggest that biological processes may be just as important to consider for the overall remediation strategy of impacted sites and therefore are the focus of this review.
Bioremediation has been applied for treatment of a wide variety of compounds and is often the primary in situ remedial approach.21 However, the unique chemistry of PFAS makes enzymatic transformation to non-toxic end-products difficult. That being said, detoxification may only be one component of an overall remediation strategy. In many cases, PFAS must first be extracted from the subsurface before any remedial technique can be effectively applied. Increasing extraction efficiency of specific PFAS through transformation of these compounds, often called “perfluoroalkyl acid (PFAA) precursors” to more mobile, and sometimes smaller, anionic PFAAs could be a more suitable role for bioremediation in PFAS remediation schemes. This is similar to PFAS flushing approaches using oxidative approaches that have been studied at the lab scale.22 Understanding the role of biodegradation and bioremediation in tackling the PFAS problem is critical to effectively remediate sites in the future; therefore, understanding the current literature and knowledge gaps related to PFAS biotransformation is necessary. There have been several literature reviews on microbial transformation of PFAS23–26 and one on microbial mechanisms involved in transformation;27 however, this review will uniquely focus on topics applicable to bioremediation schemes including:
(1) The impact of PFAS on bacteria (i.e. observed toxicity and community shifts).
(2) Currently established bacterial degradation pathways for different classes of PFAS.
(3) Discussion on the role of co-metabolism in observed degradation including prominent enzymes thought to be responsible.
(4) Future work based on identified knowledge gaps.
The goal of this review is to provide a concise update on the status of bacteria–PFAS interactions as it pertains to bioremediation and therefore aid in the dissemination of studies that show potential usefulness for bacteria in remediation schemes.
PFAS are a large class of compounds currently encompassing 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different fluorinated molecules in the EPA CompTox master list at the time of writing. The large number of PFAS and the tendency of naming conventions to vary among different chemical companies and research groups has prompted us to provide a shortened list of PFAS relevant to discussions within the body of this review (Table 1), for acronyms from specific studies mentioned in tables, the reader is referred to the original text.
000 different fluorinated molecules in the EPA CompTox master list at the time of writing. The large number of PFAS and the tendency of naming conventions to vary among different chemical companies and research groups has prompted us to provide a shortened list of PFAS relevant to discussions within the body of this review (Table 1), for acronyms from specific studies mentioned in tables, the reader is referred to the original text.
2. Effects of PFAS on microorganisms
When applying in situ bioremediation approaches, it is important to develop a conceptual site model. For example, understanding redox profiles, available electron donors (i.e. organic material in the soil, hydrocarbons from petroleum products, methane, inorganic donors such as ammonia), and relevant electron acceptors at a given site (such as oxygen, nitrate, sulfate, iron, and carbon dioxide) can help define the dominant bacterial communities and the degradative processes that these communities can mediate. Understanding toxicity impacts may play an important role for some sites where the targeted enzyme(s) or microbial process (e.g., nitrification, reductive dehalogenation) may be inhibited by PFAS contamination. The diversity of both microorganisms and PFAS make it impossible to study exact processes for every situation. However, observations on toxicity mechanisms and overall shifts in community structures due to PFAS exposure can help improve conceptual models of impacted systems. First PFAS toxicity effects on the cellular level will be discussed followed by more generally observed community shifts.2.1 Cellular effects
The effects of PFAS on individual cells are relevant from a practical standpoint for understanding potential impacts on pollutant biodegradation (e.g., on reductive dehalogenation of chlorinated solvents) and from a theoretical standpoint for understanding which cellular characteristics contribute to PFAS toxicity or lack thereof. There have been several recent studies investigating the effects of PFAS on bacterial cells (Table 2). These effects include changes in cell morphology, surface charge, and membrane fluidity.| Source | PFAS | Bacteria | Toxicity effect | Method of observation |
|---|---|---|---|---|
| a TEM, Transmission Electron Microscopy; QCM-D, Quartz Crystal Microbalance with Dissipation Monitoring; qRT-PCR, Quantitative Reverse Transcription Polymerase Chain Reaction; RT-PCR, Reverse Transcription Polymerase Char Reaction; FTIR, Fourier transform infrared spectroscopy; SOD, Superoxide Dismutase; EPS, Extracellular Polymeric Substances; ROS, Reactive Oxygen Species. | ||||
| 28 | PFOA, PFNA, POS, PFHxS (0.01–50 mg L−1) | Alcanivorax borkumenis, synthetic mono-layer and bi-layer phospholipids | Increased membrane fluidity | Compressibility measurements |
| 29 | PFHxS (0.02 mg L−1) | Pseudonomas PS27 and PDMF10 | Morphology changes (rod to round) | TEM |
| 30 | PFBA, PFOA, PFNA, PFBS, PFHxS, PFOS (0.1–50 mg L−1) | Aliivibrio fischeri | Quorum sensing | Luminescence measurements |
| Increased cell membrane permeability | ||||
| 31 | PFBA, PFOA, PFNA, PFBS, PFHxS, PFOS (0.01–50 mg L−1) | Staphylococcus epidermidis, Aliivibrio fischeri DC43 | Partitioning into membrane | QCM-D, zeta potential |
| 32 | PFOA, PFOS (0.1–500 mg L−1) | Escherichia coli | PFOA-DNA oxidative stress | TEM, FTIR, viability assays, RT-PCR, SOD assay, zeta potential |
| PFOS-membrane disruption | ||||
| 33 | PFBA, PFPeA, PFHxA. PFHpA, PFOA, PFNA, PFDA, PFUnA, PFBS, PFHxS, PFOS (0.1, 0.2, 0.5, 1, 2, 5, 10 mg L−1) | Rhodococcus jostii RHA1 | Increased EPS production, increase in 2 stress-related genes (sigF3, pfmA) | qRT-PCR, optical density EPS assay, light microscopy |
| 34 | PFOS (0.1, 1, 5 mg L−1) | Thermophilic anaerobic sludge | Increased EPS production, increased ROS levels, increased membrane permeability | EPS assay, ROS assay, luminescence measurements |
The current studies look only at a small subset of known PFAS and do not include any zwitterionic, fluorotelomer, or sulfonamide compounds, which greatly limits their field applicability given the prevalence of these groups of compounds in historic AFFF used at impacted sites.35 However, they do provide preliminary insight into mechanisms of toxicity, with the most commonly observed mechanism being membrane disruption. Membrane disruption has been observed through visual methods such as transmission electron microscopy (TEM) as well as using luminescence to detect the transfer of dye through the membrane in cell cultures.30,32 Naumann et al. took this a step further and specifically looked at the membrane interactions using isolated membranes from Alcanivorax borkumenis and model phospholipid compounds of both mono- and bi-layer membranes.28 They performed compressibility tests and found that while PFAS did cause lipid condensation, (i.e. caused the phospholipids to become more tightly packed), it still increased membrane fluidity in all cases. This finding is interesting as it implies that intramolecular forces may be disrupted by PFAS with some areas of the membrane being condensed while the others have increased fluidity. They also found a chain length and functional group dependent relationship where longer fluorinated chains were more likely to insert themselves and maintain association with the membrane as were sulfonates.
PFAS have also been observed to trigger stress responses in cells such as the production of extracellular polymeric substances (EPS).33 EPS production is the key component in the development of biofilms and is thought to protect the cells from harmful conditions.36,37 Weathers et al., noted stress induced changes in Rhodococcus RHA1 when exposed to a mixture of eleven PFAAs, including a doubling of cell length, likely due to either EPS production causing cells to remain together after division or incomplete cell division, as well as enhanced biofilm production.38 The specific mechanism of toxicity was studied more in-depth by Liu et al. who differentiated between the effects of PFOA and PFOS on Escherichia coli cells.32 Exposure to PFOS increased membrane fluidity to a greater extent than PFOA which is consistent with the above discussion of membrane affects in which longer chain sulfonates were linked to greater membrane disruption.32 PFOA's main route of toxicity appears to be oxidative damage, especially to DNA, and overall PFOA had a higher observed toxicity than PFOS for E. coli.32 In addition to toxicity effects, PFOS and PFOA may alter the hydrophobicity of the cells themselves: at the lowest dose tested (10 mg L−1) these PFAS appear to stimulate production of porins and lipopolysaccharides, making cell surfaces more hydrophobic.32 It is worth noting here that the lowest doses tested in this study are orders of magnitude higher than typical environmental concentrations in groundwater which average around 100 ng L−1 with high values typically below 10 μg L−1.39 At higher doses (PFOS: 500 mg L; PFOA 15 mg L−1), PFAS can bind to the membrane's hydrophobic sites, leaving the hydrophilic head to interact with the surroundings, and potentially changing the cell charge.32 Significant zeta potential changes were not observed until PFAS exceeded 50 mg L−1, which is consistent with Fitzgerald et al. who found no zeta potential differences in live cells (of both gram-positive and gram-negative bacteria) at concentrations up to 50 mg L−1.31,32 However, Fitzgerald et al. did observe differences in zeta potential with their phospholipid model membrane (separate from actual cells) and theorized that the cell wall prevented the surface charge changes since the PFAS associated with the membrane and the cell wall remained unchanged.31 Exploring this idea of cell wall and PFAS interactions, a preliminary study on the difference between gram-negative (lacking an outer layer of peptidoglycan) and gram-positive bacteria (having an outer layer of peptidoglycan) in relation to sorption of PFOA and PFOS indicated that the gram-positive bacteria sorbed more PFAS.40 This leads to some interesting questions on the role of the cell wall and the presence of peptidoglycan in interactions with PFAS. More research is needed to better understand these interactions.
While these studies cannot be directly applied to all types of cells and PFAS that may be present at a contaminated site, they do provide a good framework for understanding the effect of some PFAS on specific strains of bacteria. These studies also demonstrate that toxicity varies by compound, and it is likely to also vary by microorganism, potentially leading to appreciable shifts in microbial community structure as discussed in the following section.
2.2 Microbial community effects
As PFAS contamination is widespread and may affect natural systems with diverse microbial communities, it is of interest to know how microbial ecology changes with PFAS exposure. These changes may reflect toxicity of specific PFAS (i.e., with some species, and those that depend on them, declining in density), but they also may lead to a better understanding and identification of strains with the ability to biotransform PFAS. These PFAS-transforming organisms may increase in the affected communities, particularly if they gain carbon, energy or nutrients during PFAS biotransformation. To understand community shifts, several studies have performed 16S rRNA surveys on samples from a variety of different sites (Table 3). These studies have shown several consistent trends in community shifts. This section first discusses bacteria observed to increase in numbers after PFAS exposure and then those observed to decline with PFAS exposure.| Source | System | PFAS | Dominated | Decreased |
|---|---|---|---|---|
| a 1. Dose–response, 2. Exposure-response, 3. Existing contaminated system; a. phylum, b. class, c. order, d. genus; SBR sequencing batch reactor (wastewater treatment), AS activated sludge, WS wetland sediments, AD anaerobic digester, WAS waste activated sludge, FC filter cake, AW Antarctic waters, FWP freshwater pond, RS river sediment, LL landfill leachate, GS grassland soil. N.D. no data reported. | ||||
| (41)1 | SBR | PFOS | aProteobacteria, Bacteroidetes | Chloroflexi, Actinobacteria |
| (41)1 | SBR | PFOS | bGammaproteobacteria, Bacteroidia, Alphaproteobacteria | Anaerolineae, Actinobacteria, and Chloroflexia |
| (42)3 | AS | PFOA (20 mg L−1) | aBactroidetes, Proteobacteria, Acidobacteria | Planctomycetes |
| (43)2 | WS | 6:2 FTS | aProteobacteria, Acidobaceria, Gammaproteobacteria | Planctomycetes, bacteroidetes |
| (43)2 | WS | 6:2 FTS | d(a)Methylocaldum (Gammaproteobacteria) | SM1A02 (Planctomycetes), Ohtaekwangia, Taibaiella (Bacteroidetes) |
| (44)2 | AD | AFFF (DCP + PFOS) | N.D. | Decrease in methanogenic activity |
| (45)2 | AD | PFOS (500 mg L−1) | N.D. | No decrease in methanogenic activity |
| (46)2 | WAS, AS, FC | PFOA (10 mg L−1) | bBurkholderiaceae | Hyphomicrobium |
| (46)2 | WAS, AS, FC | PFOA (10 mg L−1) | Acidimicrobium sp. Strain A6 | N.D. |
| (46)2 | WAS, AS, FC | PFOA (10 mg L−1) | dDechloromonas, Desulfobulbus, Desulfosporosinus, Desulfovibrio | N.D. |
| (47)1 | AW | PFOS/PFOA (2–600 ng L−1) | aBacteriodetes, Chloroflexi | N.D. |
| (47)1 | AW | PFOS/PFOA (2–600 ng L−1) | bGammaproteobacteria, Flavobacteriia | N.D. |
| (47)1 | AW | PFOS/PFOA (2–600 ng L−1) | cRhodobacterales | N.D. |
| (48)1 | FWP | PFOA (0.45–5 mg L−1) | aProteobacteria and Chloroflexi | Actinobacteria, baceroidetes, Verrucomicrobia |
| (48)1 | FWP | PFOA (0.45–5 mg L−1) | bBetaproteobacteria | N.D. |
| (49)3 | RS | PFOA (8–465 ng L−1) | dThiobacillus, Sulfurimonas | N.D. |
| (50)1 | LL | 6:2 FTS (650 μg L−1) | aAcidobacteria, actinobacteria | N.D. |
| (51)1 | AS | PFOS (0.01–1 mg L−1) | bBacteroidetes, Verrucomicrobia | Betaproteobacteria |
| (51)1 | AS | PFOS (0.01–1 mg L−1) | dComamonas | Zoogloea, Pseudomonas, |
| (52)1 | GS | OBS (1–100 mg kg−1) | Ammonia oxidizing archaea | Ammonia oxidizing bacteria |
| (53)1 | WS | PFBA, PFPA, PFHxA, PFHpA, PFOA, PFHxS, PFBS, PFOS (0.2–4.2 μg/L) | aProteobacteria, Verrucomicrobia | Actinobacteria |
| Cyanobacteria | ||||
It is important to put these studies into context of environmental sites and recognize that although laboratory studies have shown impacts on microbial communities, PFAS may be significantly less important than general environmental parameters such as pH and oxidation–reduction potential (ORP) at sites with long term exposure.57 Further analysis of the metadata generated by these studies is needed before any firm conclusions are possible on the effect of PFAS on community structure. It is also important to recognize that 16S rRNA gene surveys have inherent bias that selects for microorganisms that are already characterized both in the sample preparation steps and in the bioinformatics analysis. So, although PFAS does appear to induce community structure change, especially at a lab-scale, more in depth studies linking community composition, PFAS concentration, and other environmental factors are needed.
3. Microbial transformation of PFAS
The next section of this review focuses on observed biotransformation of PFAS by different bacterial communities and individual strains. Identifying degradation pathways can help inform our understanding of what occurs once PFAS is released into the environment from both a forensic and predictive standpoint. Our understanding of microbial transformation of PFAS has evolved over the past twenty years, as is often the case for emerging contaminants.58 Initially there were conflicting reports on whether or not PFAS biotransformed at all. In the early 2000s, studies showed the expected environmental half-life varying from 10 to 1000 years for the same fluoroacrylate polymer.59,60 One study indicated that PFOA was biologically inert, while others showed significant transformation of the functional head groups of perfluorinated compounds produced by electrochemical fluorination (ECF) and polyfluorinated compounds produced from the fluorotelomerization process (FT) leading to the idea that only transformation of the functional group, and not the fluorinated tail, was likely.61,62 In more recent studies, instances of defluorination for PFOA and the fluorotelomer alcohols (FTOHs) and transformation of FT-precursors has been observed as well as increased reports of a number of ECF-precursor functional groups undergoing bacterial transformation. Further discussions of FT-PFAS degradation pathways, ECF-PFAS degradation pathways, and terminal PFAA defluorination follow and are based on the studies cited in Table 4.| Category | Source | Starting compound | Organism | Bio-transformation products | Transformation efficiency | Experiment length/temp (°C) | Half-life reported | Starting concentration |
|---|---|---|---|---|---|---|---|---|
| FT precursors | 63 | FTU and HMU | Aerobic soils (both agricultural land and forest) | PFOA, PFHpA, and PFHxA, 7:2 s FTOH, 8:2 FTOH | Forest soil (24–27%) | 180d (22) | FTU: 126–148d, HMU: 478–667d | 100 μg g−1 |
| 64 | 8:2 FTAC and FTMAC | Aerobic soils | PFOA, FTOH | 10.3% mol production of PFOA | 105d (R) | 8:2 FTAC <5d and FTMAC 15 d | 75–100 μg g−1 | |
| 65 | 4:2 6:2 and 8:2 FtTAoS, | Aerobic soil from AFFF site | 4:2, 6:2, 8:2 FtS, 6:2 FtUCA, 5:3 FtCA, C4–C8 PFCAs. Newly characterized: 6:2 and 8:2 FtSOAoS and FtSO2AoS (4 total) | >95% yield to intermediates | 60d (30) | n.d. | 26 μM 6:2 FTTAoS, 0.04 μM 8:2 FtTAoS and 0.005 μM 4:2 FtTAoS | |
| 66 | 6:2 FTAA and 6:2 FTAB | Activated sludge | 6:2 FTOH, 6:2 FTCA, 6:2 FTUCA, 5:3 FTCA, PFHxA, PFPeA, PFBA | 16 and 6 mol% quantifiable products | 109d (R) | 419d FTAA and 1817d FTAB | 600 μg/400 mL | |
| 55 | 6:2 FTAB, 6:2 FTSA | Gordonia sp. strain NB4–1Y | *degradation was not tracked, gene expression instead was measured | NR (30) | 60 μM | |||
| 67 | 6:2 FTOH, 4:2 FTOH, TFMAA | Mixed landfill soil culture (aerobic) | 5:3 FTCA, PFHxA, PFPeA, PFBA, TFA, | ∼100% for FTOHs and >7% for TFMAA | 32d (R) | 1.5d 6:2 FTOH | 10 ppb for FTOHs and 300 ppb for TFMAA | |
| 43 | 6:2 FTS | Aerobic and anoxic wetland sediments | 5:3 acid, PFHxA, PFPeA | Aerobic 8.9%. anaerobic, 0.7 mole% | 120d (30) | n.d. | 160 μg/20 mL for aerobic and 100 μg/20 mL for anoxic | |
| 50 | 6:2 FTS, 8:2 FTOH | Landfill leachate | PFBA, PFPeA, PFHxA, PFHpA, PFOA | 60d (20) | 650 μg L−1 | |||
| 68 | 6:2 FTSA | Rhodococcus jostii RHA1 | 6:2 FTCA, 6:2 FTUCA, α-OH 5:3 FTCA | >3 log reduction | 6d (30) | n.d. | 40 μM | |
| 69 | 6:2 FTSA | Sediments (aerobic and anaerobic) | 5:3 acid, PFPeA, PFHxA | 98.9% aerobic, 0% anaerobic | 100d (R) | <5d aerobic | 1.3 mg L−1 | |
| 70 | 6:2 FTSA and 6:2 FTAB | Gordonia sp. strain NB4–1Y | PFBA, PFPeA, PFHxA, 4:3 FTCA, 5:3 FTCA, 6:2 FTCA, 6:2 FTUA, 5:2 sFTOH, 5:2 FT ketone and 6:2 FTOH | 99.9% FTSA and 70.4% FTAB | 7d (30) | n.d. | 60 μM FTSA and 60 μM FTAB | |
| 71 | 6:2 FtTAoS | Anaerobic AFFF impacted soil | 6:2 FtTP | 75% | 270d (30) | n.d. | 20 μM | |
| 72 | 6:2 FtTAoS | Aerobic soil culture | 6:2 FTS, PFHxA, PFPeA, PFBA | 100% | 20d (R) | 3.81d to 6.65d | 12–16 μM | |
| 45 | PFOS. 6:2 FTSA, PFBS, TFA, PFPrA, PFPeA, 3 H-PFPrA, trifluoropropanoic acid, 2,2-bis(trifluoromethyl)propanoic acid, 5H-PFPeA, trifluoropentanoic acid | Aerobic and anaerobic wastewater sludge | n.d. | n.d. | 1241d (R) | n.d. | 200–500 ppm | |
| 73 | 6:2 PAPs | Pseudomonas oleovorans, Pseudomonas butanovora, and Pseudomonas fluorescens DSM 8341 | 6:2 FTOH, 6:2 FTCA, 6:2 FTUCA, 5:2 ketone, 5:2 sFTOH, PFHxA, 5:3 acid, 5:3 uacid | 8.6 mol% per gram of cells | 30d (30) | n.d. | 6.9 ppm of 6:2 FTOH equivalents | |
| 74 | AFFF, 6:2 FTAB | Soil from site contaminated from AFFF and from a crude oil spill | PFCs, PFHxA, PFHpA, PFPeA, PFBA, 5:3 FTCA, 6:2 FTUA, 5:2 ketone, 6:2 FTSA | Reduction in total measured PFAS: 24% | 60d (R) | 31d | PFOS: <2100 ng g−1, PFOA: <850 ng g−1, 6:2 FTAB: 10 μg g−1 | |
| 75 | 6:2 FtTAoS | Aerobic and anoxic soils | 6:2 FtS, PFHxA | 84% removal | 282d (30) | n.d. | 35 μM | |
| ECF precursors | 76 | Cationic quaternary ammonium compounds (QACs): PFOAAmS and PFOSAmS | Non-impacted area close to AFFF site | PFOA | 30 mol% yield to PFOA. | 180d (22) | PFOAAmS: 142d, PFOSAmS: n.d. | 1 μg g−1 |
| 77 | EtFOSA | Non-AFFF-impacted soil | PFOS, FOSA, FOSAA, EtFOSAA | 4% yield to PFOS | 182d (22) | 13.9 ± 2.1 d | 3.12 nmol g−1 | |
| 78 | EtFOSA | Carrot microcosm | FOSAA, FOSA, PFOS | 81% depletion EtFOSA | 80d (25) | 35.8 ± 3.7d to 40 ± 7.8d | 3.8 mg kg−1 | |
| 79 | EtFOSE | Aerobic soils (acidic, alkaline) | EtFOSAA, EtFOSA, FOSAA, FOSA, PFOS | (1.06, 5.49) mol% PFOS, | (180, 210)d (22) | (26, 31)d | 3.7 μg g−1 | |
| 80 | EtFOSE | St. Bernard soil | EtFOSA, EtFOSAA, FOSA, PFOS | >99% | 105d (22) | 8.7d branched, 9.6d linear | 4.72 nmol g−1 of EtFOSE, sum of branched and linear. | |
| 81 | MeFBSE and EtFOSE | Anaerobic digester communities | MeFBSAA and PFBSI; ETFOSAA and PFOS | 75% depletion MeFBSAA and 3% depletion EtFOSE | 108d (20–26) | 35.8d MeFBSAA 1860d EtFOSE | 1 ppm | |
| 82 | PFOANO and PFOSNO | Aerobic soils | PFOA and PFOS | PFOA 15–21 mol%, PFOS: 2 mol% | 90d (25) | PFOANO: 3-7d, PFOSNO: 15d | PFOANO: 800 ng g−1, PFOSNO: 2000 ng g−1 | |
| 83 | PFOANO, PFOAAmS, PFOAB, PFOSNO, PFOSAmS, PFOSB | Aerobic soils | FOSAA, FOSA, PFOS, PFOA | 0.06–32.6% | 90 d-150d (22) | 14–675d | ∼2 μg g−1 | |
| 84 | AmPr-FHxSA | Aerobic soil enrichments | FHxSA-PrA, FHxSA, PFHxS | 30% | 70d (30) | n.d. | 0.2 μM | |
| PFAAs transformation in complex systems | 85 | PFOA | Activated sludge- enriched, photocatalytic, synergistic system | n.d. | 79.7% removal | n.d. (25) | 4d | 500 μg L−1 |
| 47 | PFOS | Antarctic ocean | n.d. | 50% | 6d (R) | 43h for PFOS | 2–600 ng L−1 | |
| 86 | PFOS and PFOA | Chemoorgano-heterotrophic bacteria and total yeast and molds | n.d. | 46–69% PFOS depletion and 16–36% PFOA depletion | 28d (28) | 20.3d PFOS and 38.9d PFOA | 1 μg mL−1 for total PFAAs | |
| 87 | PFOS and PFOA | Activated sludge | n.d. | 15–42% | 1d (22) | 1.2d | 100, 250, 500 ng L−1 | |
| Complex precursor mixes | 88 | PFAS released from carpet | Methanogenic community | Unable to conclude. 13.7 times higher PFAS measured in biotic than abiotic treatments | n.d. | 552d (37) | n.d. | n.d. |
| 57 | AFFF | Soil samples taken from impacted site for community analysis | observed decrease of 8:2 FTS with increase of PFOA, PFHpA, | n.d. | n.d. (n.d.) | n.d. | n.d. | |
| 89 | AFFF impacted soil | Aerobic and anaerobic soil columns | PFAAs | n.d. | 1.5d (R) | n.d. | n.d. | |
| Terminal end-product transformation | 90 | PFOS | Pseudomonas plecoglossicida 2.4-D | n.d. | 75% | 6d (28) | 3d PFOS pure culture, 3 months in soil | 0.1 w/v% |
| 91 | PFOA and PFOS | Acidimicrobium sp. strain A6 | PFBA, PFPeA, PFHxA, PFHpA | 60% | 100d (30) | n.d. | 0.24 mM PFOA and 0.2 mM PFOS | |
| 46 | WWTP biosolids (with PFOA) | Acidimicrobium sp. strain A6 | PFBA, PFPeA, PFHxA, PFHpA | 68% | 150d (25) | n.d. | 0.2–10 mg L−1 PFOA | |
| 92 | PFOA | Pseudomonas parafulva YAB1 | n.d. | 48.1% | 2.5d (30) | n.d. | 500 mg L−1 | |
| 93 | PFOA | Pseudomonas parafulva YAB1 – gene shuffled | n.d. | 58.6% | 4d (30) | n.d. | 500 mg L−1 | |
| 94 | PFCAs | Dehalococcoides, KB-1, mixed culture | Defluorinated PFCAs | ∼100% | 150d (R) | n.d. | 75 μM | |
| 95 | PFMeUPA, FTMeUPA | Dehalococcoides, KB-1, mixed culture | Hydrogenated and defluorinated transition products | >90% | n.d. (34) | 130d | 75 μM | |
| 96 | PFOS and PFOA | Ensifer adhaerens strain M1 | n.d. | 100% | 6d (26–28) | 3d for PFOS and 2d PFOA | 1 g L−1 | |
| 90 | PFOS | Pseudomonas plecoglossicida 2.4-D | n.d. | 75% | 6d (28) | 3d PFOS pure culture, 3 months in soil | 0.1 w/v% | |
3.1 FTOH biotransformation
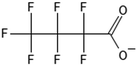
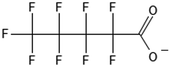
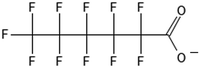
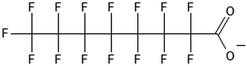
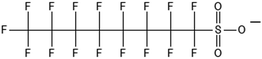
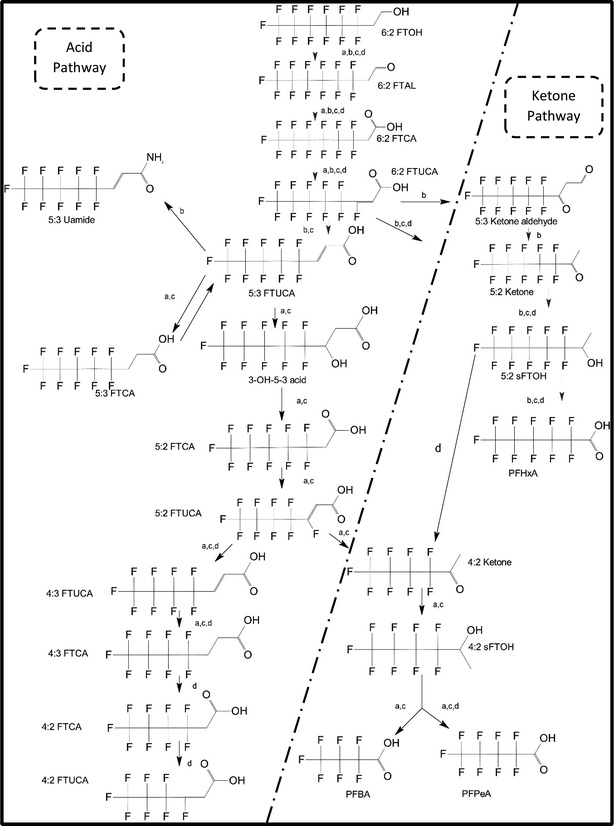
One of the first classes of PFAS found to biotransform, and therefore a commonly studied one, is the N:2 FTOHs, specifically 6:2 and 8:2 FTOH, which have partially fluorinated chains with a CH2 group at the end of the fluorinated tail. N:2 FTOHs, such as 6:2 FTOH (Fig. 1), generally transform through a series of intermediates into either PFOA, shorter chain carboxylates (PFBA, PFPeA, PFHxA) or saturated acids, through what was termed the “one carbon removal pathway”.97–101 During N:2 FTOH biotransformation, the unsaturated carboxylic acid (N:2 FTUCA) appears to be a key intermediate from which different pathways branch.97 From N:2 FTUCA, one pathway produces acids and the other produces ketones.100,102 The acid pathway releases two CF2 while the ketone pathway only releases one.100,102 It has been noted that the pathway used can be influenced by the addition of specific co-substrates (e.g., lactate), opening possibilities for influencing the type of end-products that are produced from N:2 FTOH degradation and possibly allowing for selection of acid products instead of ketones.100,102 There remains some ambiguity concerning the proposed metabolic pathway from 6:2 FTOH to PFBA, which may reflect differences in bacterial culture; but overall the acid pathway continues to produce acids until it branches off into a ketone pathway, which produces terminal end-products.70,103 It is also interesting to note that the soil culture used in Liu et al. appeared to add an amide group to 5:2 FTUCA producing 5:3 Uamide, a reaction that currently appears unique to that study.1043.2 FTOH precursor degradation
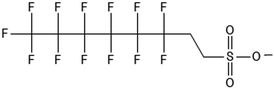
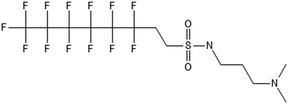
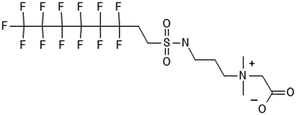
There are many PFAS identified that have a FT structure (i.e. partially fluorinated carbon chain) and larger complex functional groups. These “FT-precursors”, especially those with sulfonamide functional groups, can undergo cleavage of the ester bond and transform to FTOHs (Fig. 2), and then proceed along the established degradation pathway for FTOHs into the terminal short chain perfluorinated carboxylic acids (PFCAs) and other ketone and acid intermediates discussed above.43,69,70 These precursor studies have included soils (unsaturated), sediments (saturated), activated sludge, and pure cultures. Each of these microbial systems will be discussed in more detail below.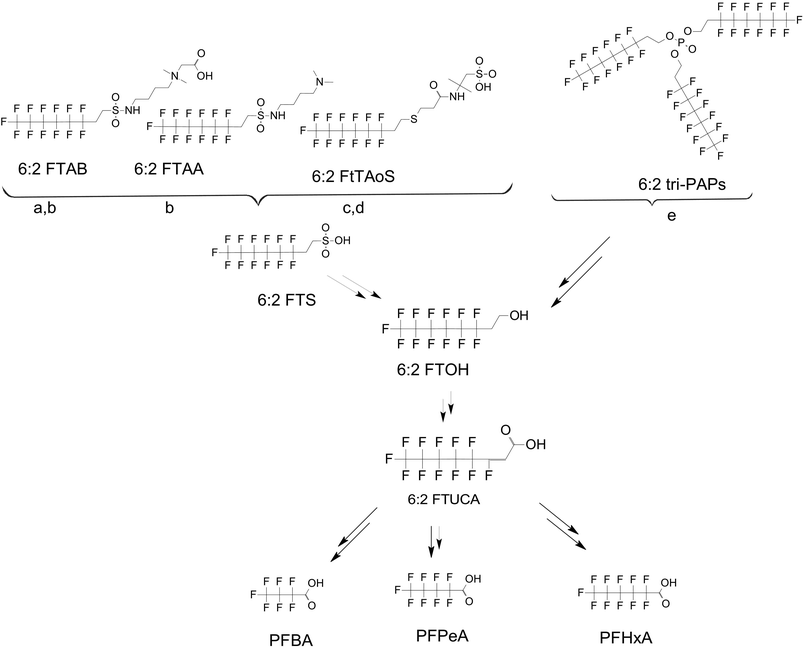 | ||
| Fig. 2 Transformation of 6:2 FTS and precursors a;66 b;70 c;72 d;71 e73 double arrows indicate multiple transformation steps. | ||
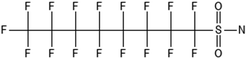
3.3 ECF precursor degradation
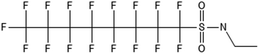
Unlike FT compounds, ECF compounds have a fully fluorinated carbon chain making them significantly more resistant to biological transformation. They have only been observed to undergo transformation of the functional head group and not defluorination (Fig. 3). Of these functional head transformations, the most studied pathway starts with N-ethyl perfluorooctane sulfonamidoethanol (EtFOSE). A common daughter product produced from EtFOSE is N-ethylperfluoro-1-octanesulfonamide (EtFOSA), the main ingredient in Sulframid, an ant killer frequently used in Brazil to reduce leaf cutting ant populations on agricultural lands.110 The application of this insecticide has been linked to high levels of PFOS (a potential daughter product of EtFOSA) in Brazilian surface water.78,110 In addition to ant killer, these sulfonamide-ECF compounds are common ingredients in AFFF formulations, especially in many 3M formulations from 1988–1998, lending an even greater relevance to understanding their biotransformation pathways for impacted soils and groundwaters.35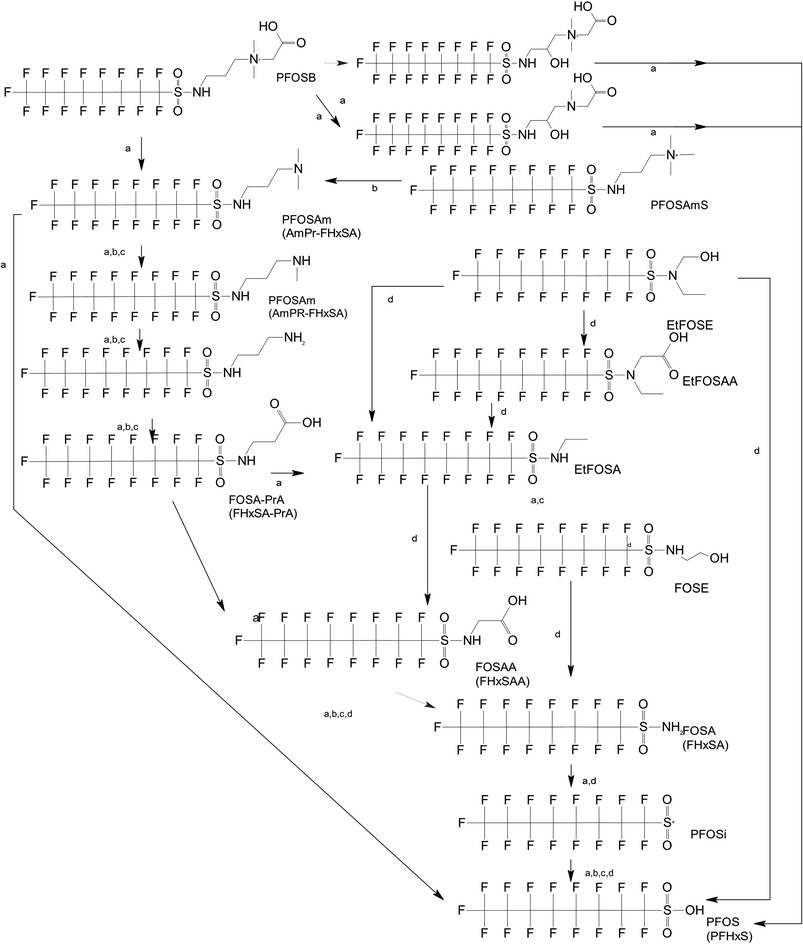 | ||
| Fig. 3 ECF Pathway. a;83 b;76 c;112 d111 names in parentheses indicate C6 homologues observed in ref. 84. | ||
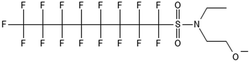
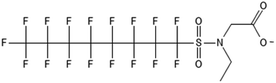
Biotransformation pathways have been proposed for several of the larger sulfonamide ECF compounds shown in Fig. 3. Many of these pathways converge at EtFOSA and then proceed to FOSAA, FOSA, and finally, PFOS. Cook et al. recently reported that the C6 homologue to the C8 sulfonamide PFOSAm, AmPr-FHxSA, transformed to PFHxS in aerobic soil enrichments amended with carbon substrates, including diethyl gycol monobutyl ether (DGBE), benzene, toluene, ethylbenzene, and xylene (BTEX), acetate, and methane.84 AmPr-FHxSA was observed to follow a similar transformation path as the C8 compounds, but with fewer identified intermediates. Demethylation, acetylation, and hydroxylation are all proposed enzymatic mechanisms for transformation of these compounds. Interestingly, several of these studies have proposed a direct transformation step from larger precursors (EtFOSE, PFOSAm, PFOSB intermediates formed by hydroxylation and demethylation) to PFOS, and this has subsequently been supported with modeling.80,111
The biotransformation pathways that result in terminal PFAAs have been of particular interest because of the regulatory focus on PFOA and PFOS specifically, and the high concentrations of these compounds found at contaminated sites.113 When looking at the transformation pathways, it is worth noting that only the linear isomers are shown here but there are typically two isomers of EtFOSE, and likely the other larger precursors as well, one linear (n) and one branched (br), and therefore all of the transformation products also have two isomers: linear isomers degrade to linear transformation products and branched to branched products.80 It was initially thought that the branched isomers would biotransform faster; however, several studies have reported the opposite: the linear EtFOSE isomer and transformation products, with the exception of perfluorooctanesulfonamide (FOSA), are more readily transformed by bacteria.78,80 This is opposite of the metabolic preference in high-order organisms such as fish.114 Although progress has been made over the past decade, the biotransformation pathways of ECF compounds need significantly more study. We need a better understanding of transformation reactions to terminal PFAAs and knowledge concerning reactions that may result in defluorination.
3.4 PFOS and PFOA biotransformation
Most of the current literature focuses on the biotransformation of precursors to terminal end-products in environmental samples and microbial consortia. However, specific strains of bacteria have also been reported to defluorinate perfluorinated moieties such as PFOA and PFOS.90,92,115,116 The first strain to be identified as capable of breaking the carbon–fluorine bond in PFAS was Pseudomonas fluorescens D2, which was able to use the recalcitrant polyfluorinated H-PFOS as a source of sulfur; however, it was unable to transform PFOS itself (i.e., fully saturated with fluorine).116 Alpha oxidation was theorized to be the transformation mechanism of H-PFOS.116 The researchers studying P. fluorescens D2 were also the first to propose a link between sulfur metabolism and defluorination.116A few bacteria have been reported to be capable of using PFOS and/or PFOA as substrates resulting in degradation of these PFAAs. Such strains were isolated from contaminated material such as soil from old factory sites or activated sludge from wastewater treatment plants with high PFAS concentrations. These environmental sources were used as the original innocula followed by a series of isolations using PFOS as the substrate to isolate Pseudomonas aeruginosa strain HJ4, Ensifer adhaerens strain M1, and Pseudomonas plecoglossicida strain 2.4-D.90,96,115 Only one strain (Pseudomonas parafulva strain YAB1) to-date has been isolated using PFOA instead of PFOS.92 YAB1 was subsequently put through a series of gene shuffling and re-isolation procedures to modify the strain to have improved degradation efficiency.93 Although these papers show promising results, they are the only ones to report organisms able to grow solely on PFOA and/or PFOS.
A strain not isolated originally on PFOS or PFOA, but showing evidence of defluorination of both of these PFAAs is Acidimicrobium A6. This bacterium was originally isolated based on its capability to grow using the feammox reaction, oxidizing ammonium (NH4+) to nitrite (NO2−) while simultaneously reducing ferric iron (Fe(III)) to ferrous iron (Fe(II)).46,91,117 The degradation of PFOA and PFOS observed with A6 is thought to be co-metabolic (see next section), similar to its degradation of other contaminants such as perchloroethylene (PCE).91 In order to confirm defluorination, higher concentrations of PFAS were used than would commonly be found in the environment (i.e., 0.24 mM and 0.2 mM for PFOA and PFOS respectively).91 It is unclear yet whether this organism will be relevant for in situ treatment of PFAS. However, recent work by this group has shown that PFOA in biosolids could be partially defluorinated when incubated with both Fe(III) and strain A6.46 The enzymatic mechanism for defluorination of PFOA and PFOS has yet to be elucidated, however a model was published linking defluorination to ammonia oxidation.118 It is also worth noting that PFOS desulfurization was theorized to have occurred as well as defluorination, but the source of sulfate in samples could not be positively identified as originating from PFOS/PFOA.91
In addition to isolating individual strains, three studies have reported the transformation of PFOS and PFOA in mixed culture or in combination with other technologies.85–87 In these studies, a decrease in PFAA concentration in mixed communities was observed, however, evidence for microbial transformation was not confirmed. Other possible mechanisms for the observed mass reduction could be sorption or, in the case of Beskoski et al., the use of dimethyl sulfoxide (DMSO) as a solvent for PFOA, as DMSO has recently been shown to degrade PFOA at low temperatures.15,86
4. Co-metabolic degradation of PFAS
The majority of studies seem to implicate a co-metabolic degradation mechanism for the biotransformation of PFAS. This section discusses co-metabolism in the context of bioremediation and evaluates its role in the observed biotransformation of PFAS.4.1 What is co-metabolism?
Co-metabolic reactions are most commonly catalyzed by “promiscuous” enzymes that have binding sites with broad substrate specificity such that they perform both growth-linked and non-growth-linked reactions.119 Historically, many different oxygenase enzymes (e.g., methane monooxygenase, ammonia monooxygenase, toluene dioxygenase) have been observed to co-metabolize a range of xenobiotic compounds, which differ based on the organism/enzyme.During co-metabolic transformation of a pollutant, microbial growth is typically dependent on the primary growth substrate (e.g., methane or propane) rather than the contaminant. As such, this process can sometimes be applied as a bioremediation strategy to treat contaminants that are present at low environmental concentrations (e.g., μg L−1) but above relevant regulatory standards or toxicity thresholds.
For this reason, co-metabolism research has recently focused on a variety of contaminants that occur at low concentrations in environmental matrices, such as 1,4-dioxane (stabilizer in chlorinated solvents), n-nitrosodimethylamine (disinfection and liquid rocket fuel byproduct), 1,2,3-trichloropropane (solvent and pesticide manufacturing byproduct), and a broad array of pharmaceuticals, estrogens, and personal care products found in wastewater.120–122 Often, these compounds are present in the environment at concentrations too low to support microbial growth causing co-metabolic processes to be the primary path of removal. Since these and other recalcitrant, and often halogenated, compounds are broken down via co-metabolic pathways, the possibility to use co-metabolism to remediate PFAS should be explored.
In an effort to find useful patterns among the studies that have provided insight for how PFAS are biotransformed (Table 5), several trends are evident and mapped here: (I) defluorination has been observed only through co-metabolic processes, (II) larger precursors with complex functional groups have been observed to transform potentially through both metabolic and co-metabolic pathways, and (III) desulfonation appears to be a driving factor for the metabolic transformation of precursors in sulfur-limited environments.
| Source | PFAS | Notes on Co-metabolism in study |
|---|---|---|
| 62 | 8:2 FTOH | Replenishment of organic carbon enhanced microbial mineralization of multiple –CF2– groups in the fluorocarbon chain of 14C-8:2 FTOH. |
| 102 | 4,6,8:2 FTOH | Presence of formate increased FTOH transformation 6-fold, cannot grow on just FTOH |
| 99 | 8:2 FTOH | Ethanol, octanol, 1,4 dioxane used as carriers and carbons sources, could not grow on just FTOH |
| 100 | 6:2 FTOH | Supports co-metabolism, all strains studied used other carbon sources |
| 99 | 8:2 FTOH | Used 1,4, dioxane, ethanol and octanol. FTOH could not support growth by itself, concluded it is a co-metabolic process |
| 68 | 6:2 FTOH | Defluorination observed and alkane monooxygenases implicated as possible co-metabolic enzymes responsible |
| 73 | 6:2 FTOH | Showed dependency on co-substrate supporting co-metabolic mechanism |
| 72 | 6:2 FtTAoS | Found that BTEX amendments increased transformation of PFAS |
| 123 | HPFOS | Used glucose/acetate to partially degrade H-PFOS |
| 124 | 6:2 FTSA | Different carbon sources used and defluorination observed, haloacid dehalogenase implicated for defluorination activity |
| 70 | 6:2 FTAB and 6:2 FTSA | Used acetate as the carbon source, observed defluorination after desulfonation in media where PFAS were the only sulfur source. |
| 91 | PFOA | Feammox, NH4+ and Fe(III) needed for defluorination |
| 95 | PFCAs | Unable to isolate defluorinating organism, TCE as substrate |
| 73 | PAPs | Used same organisms as.100 The enzyme inducers resulted in less biotransformation of PAPs implying initial transformation is metabolic. |
| 84 | AmPr-FHxSA | Studied soil enrichments ability to transform AmPr-FHxSA with different carbon substrates, methane, acetate, DGBE, BTEX, methane and acetate produced highest amounts of transformation products |
Expanding on each of these three claims, first there is no convincing evidence of defluorination proceeding metabolically. Yu et al. observed defluorination and theorized a Dehalobacter strain may be responsible but were unable to identify a specific organism or show growth on PFAS.94 The same holds true for the feammox process as it requires the primary substrates of ammonium (or hydrogen) as the electron donor and ferric iron as the electron acceptor in order to observe PFOA and PFOS defluorination.46,91 Defluorination by the FTOH one-carbon-removal pathway, observed in activated sludge and aerobic soils, was shown to be dependent on primary substrates in pure culture studies, additionally supporting co-metabolic defluorination.101,102
Second, the functional groups of PFAS have been observed to transform both metabolically and co-metabolically with the latter appearing more commonly in the literature.72,100,124 Instances of metabolic transformation appear to be limited to very large functional groups such as 6:2 PAPs, where the initial transformation step is metabolically mediated and consecutive transformations appear to switch back to either co-metabolism, where the organism is not able to grow on the compound, or to metabolic desulfonation as described below73
Third, desulfonation appears to be the main mechanism for metabolic transformation of sulfur-containing precursors under sulfur-limiting conditions (Fig. 1). Several studies have observed that 6:2 FTSA is desulfonated only when there is no other source of sulfur, strongly suggesting that 6:2 FTSA is used as the sole sulfur source for some bacterial strains.70,124 In addition to precursor transformation, the sulfur cycle has also been linked to PFOS desulfonation in Antarctic ocean communities, further strengthening the importance of sulfur cycling to PFAS transformation.125 Overall, metabolic sulfur use and co-metabolism both appear to play a large role in PFAS biotransformation, and studies are ongoing to isolate the relevant enzymes and mechanisms, so these transformation reactions can be more fully understood. Some of these studies are reviewed in the subsequent sections.
4.2 Potentially responsible enzymes
There have been several reviews on the enzymes potentially responsible for PFAS biotransformation reactions and more generally cleavage of the carbon–fluorine bond.27,126,127 Current research has focused on two main areas: co-metabolic and metabolic degradation using oxygenase enzymes and reductive dehalogenation using dehalogenase enzymes which are typically dependent on reduced corrinoid cofactors.In addition to oxygenase enzymes carrying out co-metabolic reactions, they have also been linked to metabolic degradation via desulfonation.55,68,109 In particular, the alkanesulfonate monooxygenase system has been upregulated in desulfonation of 6:2 FTSA and 6:2 FTAB with strains Gordonia sp. strain NB4-1Y and Rhodococcus jostii RHA.55,68,109 Within the class of alkanesulfonate monooxygenases, nitrilotriacetate monooxygenases and flavin- dependent monooxygenases (FMN) have been implicated for carrying out the desulfonation of PFAS.55,57 Additionally the alkB gene, which codes for oxygenases, was found to be a marker correlated with the PFAS transformation potential of systems.73 There have been a number of studies that have theorized on the role of oxygenase enzymes but none have explicitly studied them.62,70,90
5. Conclusions and recommendations for future work
Over the past two decades understanding of microbial degradation of PFAS has progressed from the conclusion that most PFAS were biologically inert to the idea that biologically mediated biotransformation and even biodefluorination are possible. While progress has been made in documenting PFAS biotransformation and understanding processes already occuring at contaminated sites, the potential for bioremediation to serve as a viable technology for treating PFAS in the environment remains unclear. There are many data gaps that must be filled to better understand this potential. Importantly, with few exceptions, the observed biotransformation reactions of PFAS to date do not result in the complete defluorination and/or detoxification of the parent molecules. In many instances, precursor compounds are transformed to terminal PFAAs, which are likely to persist based upon current knowledge. As noted in the introduction to this review, the microbial conversion of precursors to PFAAs could be part of an overall remedial strategy at sites as these compounds tend to be more readily extractable from groundwater and other environments.One of the more critical needs is to both reproduce and develop a clearer understanding of PFAS defluorination, as reported for strain A6 and a handful of other organisms and microbial enrichments. The development of in situ bioremediation strategies for PFAS destruction and detoxification depends largely on the capacity of bacteria (or other organisms including archaea and fungi) to defluorinate PFAS. Isolating a variety of organisms that carry out these reactions under different geochemical conditions, and then understanding the reactions themselves, is likely the key to determining whether bioremediation can be a viable strategy for PFAS treatment as it is now for so many other contaminants of concern. When defluorination reactions are better understood, studies can then be conducted to determine potential approaches to enhance this activity at contaminated sites (e.g., adding amendments, changing REDOX, bioaugmentation) and to assess the true potential for this process. Defluorination may also be influenced by both the relatively low concentrations of PFAS at most sites (i.e., limiting growth-linked reactions but perhaps supporting co-metabolic ones) and the diverse array of PFAS often present, as compounds may undergo different reactions. Once these reactions are more clearly understood, there is also the potential for genetically engineering strains specifically for PFAS defluorination.
Author contributions
Jessica LaFond: Writing-original draft. Kayleigh Millerick: Writing-review and editing. Paul Hatzinger: SERDP funding, writing- review and editing. Jennifer Guelfo: Writing-review and editing. Andrew Jackson: Conceptualization, Writing- review and editing.Conflicts of interest
The authors declare no conflict of interest associated with this review.Acknowledgements
The authors acknowledge the financial support from the Strategic Environmental Research and Development Program (SERDP) project number ER22-3312, and J. LaFond acknowledges funding support from the National Science Foundation Graduate Research Fellowships Program (NSF GRFP).References
- E. Kissa, Fluorinated Surfactants and Repellents, Marcel Bekker, Inc., NY, 2nd edn, 2001, p. 640 Search PubMed.
- K. M. Hinnant, S. L. Giles, A. W. Snow, J. P. Farley, J. W. Fleming and R. Ananth, An Analytically Defined Fire-Suppressing Foam Formulation for Evaluation of Fluorosurfactant Replacement, J. Surfactants Deterg., 2018, 21(5), 711–722, DOI:10.1002/jsde.12166.
- J. Glüge, M. Scheringer, I. T. Cousins, J. C. Dewitt, G. Goldenman and D. Herzke, et al., An overview of the uses of per- And polyfluoroalkyl substances (PFAS), Environ. Sci.: Processes Impacts, 2020, 22(12), 2345–2373, 10.1039/D0EM00291G.
- R. C. Buck, J. Franklin, U. Berger, J. M. Conder, I. T. Cousins and P. De Voogt, et al., Perfluoroalkyl and polyfluoroalkyl substances in the environment: Terminology, classification, and origins, Integr. Environ. Assess. Manage., 2011, 7(4), 513–541, DOI:10.1002/ieam.258.
- Z. R. Hopkins, M. Sun, J. C. DeWitt and D. R. U. Knappe, et al., Recently Detected Drinking Water Contaminants: GenX and Other Per- and Polyfluoroalkyl Ether Acids, J.-Am. Water Works Assoc., 2018, 110(7), 13–28, DOI:10.1002/awwa.1073.
- M. E. Mcguire, C. Schaefer, T. Richards, W. J. Backe, J. A. Field and E. Houtz, et al., Evidence of Remediation-Induced Alteration of Subsurface Poly- and Per fluoroalkyl Substance Distribution at a Former Fire Fighter Training Area, Environ. Sci. Technol., 2014, 48(12), 6644–6652, DOI:10.1021/es5006187.
- N. Yamashita, S. Taniyasu, G. Petrick, S. Wei, T. Gamo and P. K. S. Lam, et al., Perfluorinated acids as novel chemical tracers of global circulation of ocean waters, Chemosphere, 2008, 70(7), 1247–1255, DOI:10.1016/j.chemosphere.2007.07.079.
- N. Yamashita, K. Kannan, S. Taniyasu, Y. Horii, G. Petrick and T. Gamo, A global survey of perfluorinated acids in oceans Related papers, Mar. Pollut. Bull., 2005, 51, 658–668, DOI:10.1016/j.marpolbul.2005.04.026.
- J. A. Faust, PFAS on atmospheric aerosol particles: a review, Environ. Sci.: Processes Impacts, 2023, 35, 133–150, 10.1039/D2EM00002D.
- Stockholm Convention on Persistent Organic Pollutants Texts and Annexes, Stockholm Convention on Persistent organic Pollutants, May 2001, Stockholm, Sweden, UN Environment Programme, 2021 Search PubMed.
- EPA, Biden-Harris Administration Proposes First-Ever National Standard to Protect Communities from PFAS in Drinking Water, 2023, http://epa.gov Search PubMed.
- M. Trojanowicz, Removal of persistent organic pollutants (POPs) from waters and wastewaters by the use of ionizing radiation, Sci. Total Environ., 2020, 718, 134425, DOI:10.1016/j.scitotenv.2019.134425.
- R. Mahinroosta and L. Senevirathna, A review of the emerging treatment technologies for PFAS contaminated soils, J. Environ. Manage., 2020, 255, 109896, DOI:10.1016/j.jenvman.2019.109896.
- C. D. Vecitis, Y. Wang, J. Cheng, H. Park, B. T. Mader and M. R. Hoffmann, Sonochemical degradation of perfluorooctanesulfonate in aqueous film-forming foams, Environ. Sci. Technol., 2010, 44(1), 432–438, DOI:10.1021/es902444r.
- B. Trang, Y. Li, X. S. Xue, M. Ateia, K. N. Houk and W. R. Dichtel, Low-temperature mineralization of perfluorocarboxylic acids, Science, 2022, 377(6608), 839–845, DOI:10.1126/science.abm8868.
- H. Sharifan, M. Bagheri, D. Wang, J. G. Burken, C. P. Higgins and Y. Liang, et al., Fate and transport of per- and poly fluoroalkyl substances (PFASs) in the vadose zone, Sci. Total Environ., 2021, 771, 145427, DOI:10.1016/j.scitotenv.2021.145427.
- K. Zhang, J. Huang, G. Yu, Q. Zhang, S. Deng and B. Wang, Destruction of Perfluorooctane Sulfonate (PFOS) and Perfluorooctanoic Acid (PFOA) by Ball Milling, Environ. Sci. Technol., 2013, 47(12), 6471–6477, DOI:10.1021/es400346n.
- N. Merino, Y. Qu, R. A. Deeb, E. L. Hawley, M. R. Hoffmann and S. Mahendra, Degradation and Removal Methods for Perfluoroalkyl and Polyfluoroalkyl Substances in Water, Environ. Eng. Sci., 2016, 33(9), 615–649, DOI:10.1089/ees.2016.0233.
- R. K. Singh, S. Fernando, S. F. Baygi, N. Multari, S. M. Thagard and T. M. Holsen, Breakdown Products from Perfluorinated Alkyl Substances (PFAS) Degradation in a Plasma-Based Water Treatment Process, Environ. Sci. Technol., 2019, 53(5), 2731–2738, DOI:10.1021/acs.est.8b07031.
- M. J. Krause, E. Thoma, E. Sahle-Damesessie, B. Crone, A. Whitehill and E. Shields, et al., Supercritical Water Oxidation as an Innovative Technology for PFAS Destruction, J. Environ. Eng., 2022, 148(2), 1–8, DOI:10.1061/(ASCE)EE.1943-7870.0001957.
- M. Alexander, Biodegradation and Bioremediation, Gulf Professional Publishing, NY, 2nd edn, 1999 Search PubMed.
- M. Shojaei, N. Kumar, S. Chaobol, K. Wu, M. Crimi and J. Guelfo, Enhanced Recovery of Per- And Polyfluoroalkyl Substances (PFASs) from Impacted Soils Using Heat Activated Persulfate, Environ. Sci. Technol., 2021, 55(14), 9805–9816, DOI:10.1021/acs.est.0c08069.
- J. Liu and S. Mejia Avendaño, Microbial degradation of polyfluoroalkyl chemicals in the environment: A review, Environ. Interact., 2013, 61, 98–114, DOI:10.1016/j.envint.2013.08.022.
- C. M. Butt, D. C. G. Muir and S. A. Mabury, Critical Review Biotransformation Pathways of Fluorotelomer-Based Polyfluoroalkyl Substances: A Review Setac, Environ. Toxicol. Chem., 2014, 33, 243–267, DOI:10.1002/etc.2407.
- Z. Zhang, D. Sarkar, J. K. Biswas and R. Datta, Biodegradation of per- and polyfluoroalkyl substances (PFAS): A review, Bioresour. Technol., 2022, 344 DOI:10.1016/j.biortech.2021.126223.
- Y. J. Choi, D. E. Helbling, J. Liu, C. I. Olivares and C. P. Higgins, Microbial biotransformation of aqueous film-forming foam derived polyfluoroalkyl substances, Sci. Total Environ., 2022, 824, DOI:10.1016/j.scitotenv.2022.153711.
- L. P. Wackett, Nothing lasts forever: understanding microbial biodegradation of polyfluorinated compounds and perfluorinated alkyl substances, Microb. Biotechnol., 2021, 15(3), 773–792, DOI:10.1111/1751-7915.13928.
- A. Naumann, J. Alesio, M. Poonia and G. D. Bothun, PFAS fluidize synthetic and bacterial lipid monolayers based on hydrophobicity and lipid charge, J. Environ. Chem. Eng., 2022, 10(2), 107351, DOI:10.1016/j.jece.2022.107351.
- A. Presentato, S. Lampis, A. Vantini, F. Manea, F. Daprà and S. Zuccoli, et al., On the ability of perfluorohexane sulfonate (PFHxS) bioaccumulation by two Pseudomonas sp. strains isolated from PFAS-contaminated environmental matrices, Microorganisms, 2020, 8(1), 1–14, DOI:10.3390/microorganisms8010092.
- N. J. M. Fitzgerald, M. F. Simcik and P. J. Novak, Perfluoroalkyl Substances Increase the Membrane Permeability and Quorum Sensing Response in Aliivibrio Fischeri, Environ. Sci. Technol. Lett., 2018, 5(1), 26–31, DOI:10.1021/acs.estlett.7b00518.
- N. J. M. Fitzgerald, A. Wargenau, C. Sorenson, J. Pedersen, N. Tufenkji and P. J. Novak, et al., Partitioning and Accumulation of Perfluoroalkyl Substances in Model Lipid Bilayers and Bacteria, Environ. Sci. Technol., 2018, 52(18), 10433–10440, DOI:10.1021/acs.est.8b02912.
- G. Liu, S. Zhang, K. Yang, L. Zhu and D. Lin, Toxicity of per fluorooctane sulfonate and per fluorooctanoic acid to Escherichia coli : Membrane disruption , oxidative stress, and DNA damage induced cell inactivation and/or death, Environ. Pollut., 2016, 214, 806–815, DOI:10.1016/j.envpol.2016.04.089.
- T. S. Weathers and C. P. Higgins, Enhanced Biofilm Production by a Toluene-Degrading Rhodococcus Observed after Exposure to Perfluoroalkyl Acids, Environ. Sci. Technol., 2015, 49(9), 5458–5466, DOI:10.1021/es5060034.
- W. Chen, D. Yuan, M. Shan, Z. Yang and C. Liu, Single and combined effects of amino polystyrene and perfluorooctane sulfonate on hydrogen-producing thermophilic bacteria and the interaction mechanisms, Sci Total Environ, 2020, 703, 135015, DOI:10.1016/j.scitotenv.2019.135015.
- K. A. Barzen-hanson, S. C. Roberts, S. Choyke, K. Oetjen, A. Mcalees and N. Riddell, et al., Discovery of 40 Classes of Per- and Polyfluoroalkyl Substances in Historical Aqueous Film-Forming Foams (AFFFs) and AFFF-Impacted Groundwater, Environ. Sci. Technol., 2017, 51(4), 2047–2057, DOI:10.1021/acs.est.6b05843.
- K. K. Jefferson, What drives bacteria to produce a biofilm?, FEMS Microbiol. Lett., 2004, 236, 163–173, DOI:10.1111/j.1574-6968.2004.tb09643.x.
- J. Wingender, T. R. Neu and H.-C. F. Hans-Curt, Microbial Extracellular Polymeric Substances, 1999, pp. 1–19, DOI:10.1007/978-3-642-60147-7_1.
- T. S. Weathers, K. Harding-Marjanovic, C. P. Higgins, L. Alvarez-Cohen and J. O. Sharp, Perfluoroalkyl Acids Inhibit Reductive Dechlorination of Trichloroethene by Repressing Dehalococcoides, Environ. Sci. Technol., 2015, 50(1), 240–248, DOI:10.1021/acs.est.5b04854.
- B. Xu, S. Liu, J. L. Zhou, C. Zheng, J. Weifeng and B. Chen, et al., PFAS and their substitutes in groundwater: Occurrence, transformation and remediation, J. Hazard Mater., 2021, 412, 125159, DOI:10.1016/j.jhazmat.2021.125159.
- M. L. Butzen, J. T. Wilkinson, S. R. McGuinness, S. Amezquita, G. F. Peaslee and J. B. Fein, Sorption and desorption behavior of PFOS and PFOA onto a Gram-positive and a Gram-negative bacterial species measured using particle-induced gamma-ray emission (PIGE) spectroscopy, Chem. Geol., 2020, 552, 119778, DOI:10.1016/j.chemgeo.2020.119778.
- J. Ji, L. Peng, M. M. Redina, T. Gao, A. Khan and P. Liu, et al., Perfluorooctane sulfonate decreases the performance of a sequencing batch reactor system and changes the sludge microbial community, Chemosphere, 2021, 279, 130596, DOI:10.1016/j.chemosphere.2021.130596.
- X. Yu, F. Nishimura and T. Hidaka, Impact of Long-Term Perfluorooctanoic Acid (PFOA) Exposure on Activated Sludge Process, Water, Air, Soil Pollut., 2018, 229(4), 134, DOI:10.1007/s11270-018-3760-y.
- T. Yin, N. H. Tran, C. Huiting, Y. He and K. Y. H. Gin, Biotransformation of polyfluoroalkyl substances by microbial consortia from constructed wetlands under aerobic and anoxic conditions, Chemosphere, 2019, 233, 101–109, DOI:10.1016/j.chemosphere.2019.05.227.
- N. J. M. Fitzgerald, H. R. Temme, M. F. Simcik and P. J. Novak, Aqueous film forming foam and associated perfluoroalkyl substances inhibit methane production and Co-contaminant degradation in an anaerobic microbial community, Environ. Sci. Process Impacts, 2019, 21(11), 1915–1925, 10.1039/C9EM00241C.
- V. Ochoa-Herrera, J. A. Field and A. Luna-Velasco, Microbial toxicity and biodegradability of perfluorooctane sulfonate (PFOS) and shorter chain perfluoroalkyl and polyfluoroalkyl substances (PFASs), Environ. Sci.: Processes Impacts, 2016, 18, 1236–1246, 10.1039/C6EM00366D.
- S. Huang, M. Sima, Y. Long, C. Messenger and P. R. Jaffé, Anaerobic degradation of perfluorooctanoic acid (PFOA) in biosolids by Acidimicrobium sp. strain A6, J. Hazard Mater., 2022, 424, 127699, DOI:10.1016/j.jhazmat.2021.127699.
- E. Cerro-Galvez, J. L. Roscales, B. Jimenez, M. M. Sala, J. Dachs and M. Vila-costa, Microbial responses to per fluoroalkyl substances and per fuorooctanesulfonate (PFOS) desulfurization in the Antarctic marine environment, Water Res., 2020, 171, 115434, DOI:10.1016/j.watres.2019.115434.
- D. Zhang, W. Zhang and Y. Liang, Bacterial community in a freshwater pond responding to the presence of perfluorooctanoic acid (PFOA), Environ. Technol., 2020, 41(27), 3646–3656, DOI:10.1080/09593330.2019.1616828.
- Y. Sun, T. Wang, X. Peng, P. Wang and Y. Lu, Bacterial community compositions in sediment polluted by perfluoroalkyl acids (PFAAs) using Illumina high-throughput sequencing, Environ. Sci. Pollut. Res., 2016, 10556–10565, DOI:10.1007/s11356-016-6055-0.
- H. Hamid, L. Y. Li and J. R. Grace, Aerobic biotransformation of fluorotelomer compounds in landfill leachate-sediment, Sci. Total Environ., 2020, 713, 136547, DOI:10.1016/j.scitotenv.2020.136547.
- B. Lu, J. Qian, F. He, P. Wang, Y. He and S. Tang, et al., Effects of long-term perfluorooctane sulfonate (PFOS) exposure on activated sludge performance, composition, and its microbial community, Environ. Pollut., 2022, 295, 118684, DOI:10.1016/j.envpol.2021.118684.
- Y. Ke, J. Chen, X. Hu, T. Tong, J. Huang and S. Xie, Emerging perfluoroalkyl substance impacts soil microbial community and ammonia oxidation, Environ. Pollut., 2020, 257, 113615, DOI:10.1016/j.envpol.2019.113615.
- M. Zhang, K. Yamada, S. Bourguet, J. Guelfo and E. M. Suuberg, Vapor Pressure of Nine Perfluoroalkyl Substances (PFASs) Determined Using the Knudsen Effusion Method, J. Chem. Eng. Data, 2020, 65(5), 2332–2342, DOI:10.1021/acs.jced.9b00922.
- D. Q. Zhang, M. Wang, Q. He, X. Niu and Y. Liang, Distribution of perfluoroalkyl substances (PFASs) in aquatic plant-based systems: From soil adsorption and plant uptake to effects on microbial community, Environ. Pollut., 2020, 257, 113575, DOI:10.1016/j.envpol.2019.113575.
- E. M. Bottos, E. Y. AL-shabib, D. M. J. Shaw, B. M. McAmmond, A. Sharma and D. M. Suchan, et al., Transcriptomic response of Gordonia sp. strain NB4-1Y when provided with 6:2 fluorotelomer sulfonamidoalkyl betaine or 6:2 fluorotelomer sulfonate as sole sulfur source, Biodegradation, 2020, 31(4–6), 407–422, DOI:10.1007/s10532-020-09917-8.
- I. Wagner-Döbler and H. Biebl, Environmental biology of the marine Roseobacter lineage, Annu. Rev. Microbiol., 2006, 60, 255–280, DOI:10.1146/annurev.micro.60.080805.142115.
- D. M. O'Carroll, T. C. Jeffries, M. J. Lee, S. Thao, A. Yeung and S. Wallace, et al., Developing a roadmap to determine per- and polyfluoroalkyl substances-microbial population interactions, Sci. Total Environ., 2020, 712, 135994, DOI:10.1016/j.scitotenv.2019.135994.
- C. J. Newell, W. H. DiGuiseppi, D. P. Cassidy, C. E. Divine, J. M. Fenstermacher and N. W. Hagelin, et al., PFAS Experts Symposium 2: PFAS Remediation research – Evolution from past to present, current efforts, and potential futures, Remediation, 2022, 32(1–2), 65–73, DOI:10.1002/rem.21705.
- M. H. Russell, W. R. Berti, B. Szostek and R. C. Buck, Investigation of the Biodegradation Potential of a Fluoroacrylate Polymer Product in Aerobic Soils, Environ. Sci. Technol., 2008, 42(3), 800–807, DOI:10.1021/es0710499.
- J. W. Washington, J. J. Ellington, T. M. Jenkins, J. J. Evans, H. Yoo and S. C. Hafner, Comment on “Degradability of an acrylate-linked, fluorotelomer polymer in soil”, Environ. Sci. Technol., 2009, 44(2), 848, DOI:10.1021/es9002668.
- C. C. Lange, J. K. Lundberg, P. A. Services and S. S. Division, Aerobic Biodegradation ofN-EtFOSE Alcohol Solutions, For 3M Company, 2000 Search PubMed.
- P. W. Folsom, L. M. Sulecki, V. Capka, W. R. Berti and J. T. Gannon, Fluorotelomer Alcohol Biodegradation-Direct Evidence that Perfluorinated Carbon Chains Breakdown, Environ. Sci. Technol., 2005, 39(19), 7516–7528, DOI:10.1021/es0506760.
- K. Dasu and L. S. Lee, Aerobic biodegradation of toluene-2,4-di(8:2 fluorotelomer urethane) and hexamethylene-1,6-di(8:2 fluorotelomer urethane) monomers in soils, Chemosphere, 2016, 144, 2482–2488, DOI:10.1016/j.chemosphere.2015.11.021.
- L. A. Royer, L. S. Lee, M. H. Russell, L. F. Nies and R. F. Turco, Microbial transformation of 8:2 fluorotelomer acrylate and methacrylate in aerobic soils, Chemosphere, 2015, 129, 54–61, DOI:10.1016/j.chemosphere.2014.09.077.
- K. C. Harding-Marjanovic, E. F. Houtz, S. Yi, J. A. Field, D. L. Sedlak and L. Alvarez-Cohen, Aerobic biotransformation of fluorotelomer thioether amido sulfonate (Lodyne) in AFFF-amended microcosms, Environ. Sci. Technol., 2015, 49(13), 7666–7674, DOI:10.1021/acs.est.5b01219.
- L. A. D’Agostino and S. A. Mabury, Aerobic biodegradation of 2 fluorotelomer sulfonamide–based aqueous film–forming foam components produces perfluoroalkyl carboxylates, Environ. Toxicol. Chem., 2017, 36(8), 2012–2021, DOI:10.1002/etc.3750.
- M. Sun, J. Cui, J. Guo, Z. Zhai, P. Zuo and J. Zhang, Fluorochemicals biodegradation as a potential source of trifluoroacetic acid (TFA) to the environment, Chemosphere, 2020, 254, 126894, DOI:10.1016/j.chemosphere.2020.126894.
- S. H. Yang, Y. Shi, M. Strynar and K. H. Chu, Desulfonation and defluorination of 6:2 fluorotelomer sulfonic acid (6:2 FTSA) by Rhodococcus jostii RHA1: Carbon and sulfur sources, enzymes, and pathways, J. Hazard Mater., 2022, 423, 127052, DOI:10.1016/j.jhazmat.2021.127052.
- S. Zhang, X. Lu, N. Wang and R. C. Buck, Biotransformation potential of 6:2 fluorotelomer sulfonate (6:2 FTSA) in aerobic and anaerobic sediment, Chemosphere, 2016, 154, 224–230, DOI:10.1016/j.chemosphere.2016.03.062.
- D. M. J. Shaw, G. Munoz, E. M. Bottos, S. V. Duy, S. Sauvé and J. Liu, et al., Degradation and defluorination of 6:2 fluorotelomer sulfonamidoalkyl betaine and 6:2 fluorotelomer sulfonate by Gordonia sp. strain NB4-1Y under sulfur-limiting conditions, Sci. Total Environ., 2019, 647, 690–698, DOI:10.1016/j.scitotenv.2018.08.012.
- S. Yi, K. C. Harding-marjanovic, E. F. Houtz, Y. Gao, J. E. Lawrence and R. V. Nichiporuk, et al., Biotransformation of AFFF Component 6:2 Fluorotelomer Thioether Amido Sulfonate Generates 6:2 Fluorotelomer Thioether Carboxylate under Sulfate-Reducing Conditions, Environ. Sci. Technol. Lett., 2018, 5(5), 283–288, DOI:10.1021/acs.estlett.8b00148.
- C. I. Olivares, S. Yi, E. K. Cook, Y. J. Choi, R. Montagnolli and A. Byrne, et al., Aerobic BTEX biodegradation increases yield of perfluoroalkyl carboxylic acids from biotransformation of a polyfluoroalkyl surfactant, 6:2 FtTAoS, Environ. Sci. Process Impacts, 2022, 24, 439–446, 10.1039/D1EM00494H.
- M. Lewis, M. H. Kim, E. J. Liu, N. Wang and K. H. Chu, Biotransformation of 6:2 polyfluoroalkyl phosphates (6:2 PAPs): Effects of degradative bacteria and co-substrates, J. Hazard. Mater., 2016, 320, 479–486, DOI:10.1016/j.jhazmat.2016.08.036.
- R. Li, G. Munoz, Y. Liu, S. Sauvé, S. Ghoshal and J. Liu, Transformation of novel polyfluoroalkyl substances (PFASs) as co-contaminants during biopile remediation of petroleum hydrocarbons, J. Hazard. Mater., 2019, 362, 140–147, DOI:10.1016/j.jhazmat.2018.09.021.
- S. Yi, K. C. Harding-Marjanovic, E. F. Houtz, E. Antell, C. Olivares and R. V. Nichiporuk, et al., Biotransformation of 6:2 Fluorotelomer Thioether Amido Sulfonate in Aqueous Film-Forming Foams under Nitrate-Reducing Conditions, Environ. Sci. Technol., 2022, 56(15), 10646–10655, DOI:10.1021/acs.est.2c00063.
- S. Mejia Avendaño, S. V. Duy and J. Liu, Generation of Per fluoroalkyl Acids from Aerobic Biotransformation of Quaternary Ammonium Poly fluoroalkyl Surfactants, Environ. Sci. Technol., 2016, 50(18), 9923–9932, DOI:10.1021/acs.est.6b00140.
- S. Mejia Avendaño and J. Liu, Production of PFOS from aerobic soil biotransformation of two perfluoroalkyl sulfonamide derivatives, Chemosphere, 2015, 119, 1084–1090, DOI:10.1016/j.chemosphere.2014.09.059.
- I. Zabaleta, E. Bizkarguenaga, D. B. O. Nunoo, L. Schultes, J. Leonel and A. Prieto, et al., Biodegradation and Uptake of the Pesticide Sulfluramid in a Soil − Carrot Mesocosm, Environ. Sci. Technol., 2018, 52(5), 2603–2611, DOI:10.1021/acs.est.7b03876.
- L. Zhang, L. S. Lee, J. Niu and J. Liu, Kinetic analysis of aerobic biotransformation pathways of a perfluorooctane sulfonate (PFOS) precursor in distinctly different soils, Environ. Pollut., 2017, 229, 159–167, DOI:10.1016/j.envpol.2017.05.074.
- J. Liu, G. Zhong, W. Li and S. Mejia Avendaño, Isomer-specific biotransformation of perfluoroalkyl sulfonamide compounds in aerobic soil, Sci. Total Environ., 2019, 651, 766–774, DOI:10.1016/j.scitotenv.2018.09.214.
- C. C. Lange, Anaerobic biotransformation of N-methyl perfluorobutanesulfonamido ethanol and N-ethyl perfluorooctanesulfonamido ethanol, Environ. Toxicol. Chem., 2018, 37(3), 768–779, DOI:10.1002/etc.4014.
- H. Chen, M. Liu, G. Munoz, S. V. Duy, S. Sauvé and Y. Yao, et al., Fast Generation of Perfluoroalkyl Acids from Polyfluoroalkyl Amine Oxides in Aerobic Soils, Environ. Sci. Technol. Lett., 2020, 7(10), 714–720, DOI:10.1021/acs.estlett.0c00543.
- M. Liu, G. Munoz, S. Vo Duy, S. Sauvé and J. Liu, Stability of Nitrogen-Containing Polyfluoroalkyl Substances in Aerobic Soils, Environ. Sci. Technol., 2021, 55(8), 4698–4708, DOI:10.1021/acs.est.0c05811.
- E. K. Cook, C. I. Olivares, E. H. Antell, S. Yi, A. Nickerson and Y. J. Choi, et al., Biological and Chemical Transformation of the Six-Carbon Polyfluoroalkyl Substance N-Dimethyl Ammonio Propyl Perfluorohexane Sulfonamide (AmPr-FHxSA), Environ. Sci. Technol., 2022, 56(22), 15478–15488, DOI:10.1021/acs.est.2c00261.
- R. Ding, Y. Wu, F. Yang, X. Xiao, Y. Li and X. Tian, et al., Degradation of low-concentration perfluorooctanoic acid via a microbial-based synergistic method : assessment of the feasibility and functional microorganisms, J. Hazard. Mater., 2021, 416, 125857, DOI:10.1016/j.jhazmat.2021.125857.
- V. P. Beskoski, A. Yamamoto, T. Nakano, K. Yamamoto, C. Matsumura and M. Motegi, et al., Defluorination of per fluoroalkyl acids is followed by production of mono fluorinated fatty acids, Sci. Total Environ., 2018, 636, 355–359, DOI:10.1016/j.scitotenv.2018.04.243.
- A. Chiavola, C. D. Marcantonio, M. R. Boni, S. Biagioli, A. Frugis and G. Cecchini, PFOA and PFOS Removal Processes in Activated Sludge Reactor at Laboratory Scale, Front. Water Energy, 2020, 375–377, DOI:10.1007/978-3-030-13068-8_94.
- J. R. Lang, B. M. Allred, G. F. Peaslee, J. A. Field and M. A. Barlaz, Release of Per- and Poly fluoroalkyl Substances (PFASs) from Carpet and Clothing in Model Anaerobic Land fi ll Reactors, Environ Sci Technol, 2016, 50, 5024–5032, DOI:10.1021/acs.est.5b06237.
- A. Nickerson, A. C. Maizel, C. I. Olivares, C. E. Schaefer and C. P. Higgins, Simulating Impacts of Biosparging on Release and Transformation of Poly- And Perfluorinated Alkyl Substances from Aqueous Film-Forming Foam-Impacted Soil, Environ. Sci. Technol., 2021, 55(23), 15744–15753, DOI:10.1021/acs.est.1c03448.
- S. P. Chetverikov, D. A. Sharipov, T. Y. Korshunova and O. N. Loginov, Degradation of perfluorooctanyl sulfonate by strain Pseudomonas plecoglossicida 2.4-D, Appl. Biochem. Microbiol., 2017, 53(5), 533–538, DOI:10.1134/S0003683817050027.
- S. Huang and P. R. Jaffé, Defluorination of Perfluorooctanoic Acid (PFOA) and Perfluorooctane Sulfonate (PFOS) by Acidimicrobium sp. Strain A6, Environ Sci Technol, 2019, 53(19), 11410–11419, DOI:10.1021/acs.est.9b04047.
- L. B. Yi, L. Y. Chai, Y. Xie, Q. J. Peng and Q. Z. Peng, Isolation, identification, and degradation performance of a PFOA-degrading strain, Genet. Mol. Res., 2016, 15(2), 1–12, DOI:10.4238/gmr.15028043.
- L. Yi, Q. Peng, D. Liu, L. Zhou, C. Tang and Y. Zhou, et al., Enhanced degradation of perfluorooctanoic acid by a genome shuffling-modified Pseudomonas parafulva YAB-1, Environ. Technol., 2018, 40, 3153–3161, DOI:10.1080/09593330.2018.1466918.
- Y. Yu, S. Che, C. Ren, B. Jin, Z. Tian and J. Liu, et al., Microbial Defluorination of Unsaturated Per- and Polyfluorinated Carboxylic Acids under Anaerobic and Aerobic Conditions: A Structure Specificity Study, Environ. Sci. Technol., 2022, 56(8), 4894–4904, DOI:10.1021/acs.est.1c05509.
- Y. Yu, K. Zhang, Z. Li, C. Ren, J. Chen and Y. H. Lin, et al., Microbial Cleavage of C-F Bonds in Two C6Per- And Polyfluorinated Compounds via Reductive Defluorination, Environ. Sci. Technol., 2020, 54(22), 14393–14402, DOI:10.1021/acs.est.0c04483.
- S. P. Chetverikov and O. N. Loginov, A New Ensifer adhaerens Strain M1 is Capable of Transformation of Perfluorocarboxylic Acids, Microbiol., 2019, 88(1), 115–117, DOI:10.1134/S0026261718060085.
- L. Zhao, P. W. Folsom, B. W. Wolstenholme, H. Sun, N. Wang and R. C. Buck, 6:2 Fluorotelomer alcohol biotransformation in an aerobic river sediment system, Chemosphere, 2013, 90(2), 203–209, DOI:10.1016/j.chemosphere.2012.06.035.
- J. Liu, N. Wang, B. Szostek, R. C. Buck, P. K. Panciroli and P. W. Folsom, et al., 6-2 Fluorotelomer alcohol aerobic biodegradation in soil and mixed bacterial culture, Chemosphere, 2010, 78(4), 437–444, DOI:10.1016/j.chemosphere.2009.10.044.
- J. Liu, L. S. Lee, L. F. Nies, C. H. Nakatsu and R. F. Turco, Biotransformation of 8:2 fluorotelomer alcohol in soil and by soil bacteria isolates, Environ. Sci. Technol., 2007, 41(23), 8024–8030, DOI:10.1021/es0708722.
- M. H. Kim, N. Wang and K. H. Chu, 6:2 Fluorotelomer alcohol (6:2 FTOH) biodegradation by multiple microbial species under different physiological conditions, Appl. Microbiol. Biotechnol., 2013, 98(4), 1831–1840, DOI:10.1007/s00253-013-5131-3.
- N. Wang, B. Szostek, R. C. Buck, P. W. Folsom, L. M. Sulecki and J. T. Gannon, 8-2 Fluorotelomer alcohol aerobic soil biodegradation: Pathways, metabolites, and metabolite yields, Chemosphere, 2009, 75(8), 1089–1096, DOI:10.1016/j.chemosphere.2009.01.033.
- M. H. Kim, N. Wang, T. Mcdonald and K. Chu, Biodefluorination and Biotransformation of Fluorotelomer Alcohols by Two Alkane-Degrading Pseudomonas Strains, 2012, 109(12), 3041–3048, DOI:10.1002/bit.24561.
- N. Wang, R. C. Buck, B. Szostek, L. M. Sulecki and B. W. Wolstenholme, 5:3 Polyfluorinated acid aerobic biotransformation in activated sludge via novel one-carbon removal pathways, Chemosphere, 2012, 87(5), 527–534, DOI:10.1016/j.chemosphere.2011.12.056.
- J. Liu, N. Wang, R. C. Buck, B. W. Wolstenholme, P. W. Folsom and L. M. Sulecki, et al., Aerobic biodegradation of [14C] 6:2 fluorotelomer alcohol in a flow-through soil incubation system, Chemosphere, 2010, 80(7), 716–723, DOI:10.1016/j.chemosphere.2010.05.027.
- N. H. Tran and K. Y. H. Gin, Occurrence and removal of pharmaceuticals, hormones, personal care products, and endocrine disrupters in a full-scale water reclamation plant, Sci. Total Environ., 2017 DOI:10.1016/j.scitotenv.2017.05.097.
- N. Wang, B. Szostek, P. W. Folsom, L. M. Sulecki, V. Capka and R. C. Buck, et al., Aerobic Biotransformation of 14 C-Labeled 8-2 Telomer B Alcohol by Activated Sludge from a Domestic Sewage Treatment Plant, Environ. Sci. Technol., 2005, 39(2), 531–538, DOI:10.1021/es049466y.
- M. J. A. Dinglasan, Y. Ye, E. A. Edwards and S. A. Mabury, Fluorotelomer alcohol biodegradation yields poly- and perfluorinated acids, Environ. Sci. Technol., 2004, 38(10), 2857–2864, DOI:10.1021/es0350177.
- N. Wang, J. Liu, R. C. Buck, S. H. Korzeniowski, B. W. Wolstenholme and P. W. Folsom, et al., 6:2 Fluorotelomer sulfonate aerobic biotransformation in activated sludge of waste water treatment plants, Chemosphere, 2011, 82(6), 853–858, DOI:10.1016/j.chemosphere.2010.11.003.
- J. D. Van Hamme, E. M. Bottos, N. J. Bilbey and S. E. Brewer, Genomic and proteomic characterization of Gordonia sp. NB4-1Y in relation to 6 : 2 fluorotelomer sulfonate biodegradation, Microbiol., 2013, 159(8), 1618–1628, DOI:10.1099/mic.0.068932-0.
- J. Gilljam, J. Leonel, I. T. Cousins and J. P. Benskin, Is Ongoing Sul fluramid Use in South America a Significant Source of Perfluorooctanesulfonate (PFOS)? Production Inventories, Environmental Fate, and Local Occurrence, Environ. Sci. Technol, 2016, 653–659, DOI:10.1021/acs.est.5b04544.
- L. Zhang, L. S. Lee, J. Niu and J. Liu, Kinetic analysis of aerobic biotransformation pathways of a perfluorooctane sulfonate (PFOS) precursor in distinctly different soils, Environ. Pollut., 2017, 229, 159–167, DOI:10.1016/j.envpol.2017.05.074.
- H. Chen, M. Liu, G. Munoz, S. V. Duy, S. Sauvé and Y. Yao, et al., Fast Generation of Perfluoroalkyl Acids from Polyfluoroalkyl Amine Oxides in Aerobic Soils, Environ. Sci. Technol. Lett., 2020, 7(10), 714–720 CrossRef CAS , https://doi.org/10.1021/acs.estlett.0c00543.
- ITRC, 8.2.2.1 Toxic Substances Control Act (TSCA), PFAS — Per- and Polyfluoroalkyl Substances, 2020, available from: https://pfas-1.itrcweb.org/8-basis-of-regulations/#8_2 Search PubMed.
- H. Peng, S. Zhang, J. Sun, Z. Zhang, J. P. Giesy and J. Hu, Isomer-Specific Accumulation of perfluorooctanesulfonate from (N-ethyl per fluorooctanesulfonamido)ethanol-based Phosphate Diester in Japanese Medaka (Oryzias latipes), Environ. Sci. Technol., 2014, 48(2), 1058–1066, DOI:10.1021/es404867w.
- B. G. Kwon, H.-J. Lim, S.-H. Na, B.-I. Choi, D.-S. Shin and S.-Y. Chung, Comment on “Biodegradation of perfluorooctanesulfonate (PFOS) as an emerging contaminant, Chemosphere, 2014, 109, 221–225, DOI:10.1016/j.chemosphere.2014.01.072.
- B. D. Key, R. D. Howell and C. S. Criddle, Defluorination of organofluorine sulfur compounds by Pseudomonas sp. strain D2, Environ. Sci. Technol., 1998, 32(15), 2283–2287, DOI:10.1021/es9800129.
- S. Huang and P. R. Jaffé, Isolation and characterization of an ammonium-oxidizing iron reducer: Acidimicrobiaceae sp. A6, PLoS One, 2018, 13(4), 8–10, DOI:10.1371/journal.pone.0194007.
- M. W. Sima, S. Huang and P. R. Jaffé, Modeling the kinetics of perfluorooctanoic and perfluorooctane sulfonic acid biodegradation by Acidimicrobium sp. Strain A6 during the feammox process, J. Hazard Mater., 2023, 448, 130903, DOI:10.1016/j.jhazmat.2023.130903.
- L. P. Wackett, Questioning our perceptions about evolution of biodegradative enzymes, Curr. Opin. Microbiol., 2009, 12(3), 244–251, DOI:10.1016/j.mib.2009.05.001.
- K. Fischer and M. Majewsky, Cometabolic degradation of organic wastewater micropollutants by activated sludge and sludge-inherent microorganisms, Appl. Microbiol. Biotechnol., 2014, 98(15), 6583–6597, DOI:10.1007/s00253-014-5826-0.
- P. B. Hatzinger and D. R. Lippincott, Field demonstration of N-Nitrosodimethylamine (NDMA) treatment in groundwater using propane biosparging, Water. Res., 2019, 164, 114923, DOI:10.1016/j.watres.2019.114923.
- H. M. Rolston, M. R. Hyman and L. Semprini, Aerobic cometabolism of 1,4-dioxane by isobutane-utilizing microorganisms including Rhodococcus rhodochrous strain 21198 in aquifer microcosms: experimental and modeling study, Sci. Total Environ., 2019, 694, 133688 CrossRef CAS PubMed , https://doi.org/10.1016/j.scitotenv.2019.133688.
- B. D. Key, RoD. Howell and C. S. Criddle, Defluorination of Organofluorine Sulfur Compounds by Pseudomonas Sp. Strain D2, Environ. Sci. Technol., 1998, 32, 2283–2287 CrossRef CAS , https://doi.org/10.1021/es9800129.
- S. H. Yang, Y. Shi, M. Strynar and K. H. Chu, Desulfonation and defluorination of 6:2 fluorotelomer sulfonic acid (6:2 FTSA) by Rhodococcus jostii RHA1: Carbon and sulfur sources, enzymes, and pathways, J. Hazard Mater., 2022, 423, 127052, DOI:10.1016/j.jhazmat.2021.127052.
- E. Cerro-gálvez, C. Marrasé, J. M. Gasol, J. Dachs and M. Vila-costa, Modulation of microbial growth and enzymatic activities in the marine environment due to exposure to organic contaminants of emerging concern and hydrocarbons, Sci. Total Environ., 2019, 678, 486–498, DOI:10.1016/j.scitotenv.2019.04.361.
- Y. Wang and A. Liu, Carbon-fluorine bond cleavage mediated by metalloenzymes, Chem. Soc. Rev., 2020, 49(14), 4906–4925, 10.1039/C9CS00740G.
- M. Kiel and K. H. Engesser, The biodegradation vs. biotransformation of fluorosubstituted aromatics, Appl. Microbiol Biotechnol., 2015, 99(18), 7433–7464, DOI:10.1007/s00253-015-6817-5.
- Z. Xiao, W. Jiang, D. Chen and Y. Xu, Bioremediation of typical chlorinated hydrocarbons by microbial reductive dechlorination and its key players: A review, Ecotoxicol. Environ. Saf., 2020, 110925, DOI:10.1016/j.ecoenv.2020.110925.
- J. S. C. Liou, B. Szostek, C. M. DeRito and E. L. Madsen, Investigating the biodegradability of perfluorooctanoic acid, Chemosphere, 2010, 80(2), 176–183, DOI:10.1016/j.chemosphere.2010.03.009.
- J. Dolfing, Thermodynamic Considerations for Dehalogentation, Microbial Processes and Environmental Applications, Kluwer Academic Publishers, NY, 2003, pp. 89–114 Search PubMed.
- J. Liu, D. J. Van Hoomissen, T. Liu, A. Maizel, X. Huo and S. R. Fernández, et al., Reductive Defluorination of Branched Per- and Polyfluoroalkyl Substances with Cobalt Complex Catalysts, Environ. Sci. Technol. Lett., 2018, 5(5), 289–294, DOI:10.1021/acs.estlett.8b00122.
- Z. Sun, D. Geng, C. Zhang, J. Chen, X. Zhou and Y. Zhang, et al., Vitamin B12 (CoII) initiates the reductive defluorination of branched perfluorooctane sulfonate (br-PFOS) in the presence of sulfide, Chem. Eng. J., 2021, 423, 130149, DOI:10.1016/j.cej.2021.130149.
| This journal is © The Royal Society of Chemistry 2023 |