Open Access Article
Open Access ArticleMulti-endpoint assessments for in vitro nano-bio interactions and uptake of biogenic phosphorus nanomaterials using HEK293 cells†
Ayushi
Priyam
 ab,
Luis O. B.
Afonso
b,
Aaron G.
Schultz
ab,
Luis O. B.
Afonso
b,
Aaron G.
Schultz
 b,
Amit Kumar
Dinda
c and
Pushplata Prasad
Singh
*ab
b,
Amit Kumar
Dinda
c and
Pushplata Prasad
Singh
*ab
aNational Centre of Excellence for Advanced Research in Agricultural Nanotechnology, TERI – Deakin Nanobiotechnology Centre, Sustainable Agriculture Division, The Energy and Resources Institute (TERI), DS Block, India Habitat Centre, Lodhi Road, New Delhi, 110003, India. E-mail: pushplata.singh@teri.res.in
bSchool of Life and Environmental Sciences, Deakin University, Geelong, Victoria 3217, Australia
cDepartment of Pathology, All India Institute of Medical Sciences, Ansari Nagar, New Delhi, 110029, India
First published on 11th March 2023
Abstract
Phosphorus (P)-based nanomaterials (NMs) are being explored as nanofertilisers due to higher uptake in plants, enhanced nutrient use efficiency, and minimal leaching. However, their colloidal behaviour in biologically relevant media and corresponding in vitro nano-bio interactions have been understudied topics for the evaluation of their potential cytotoxicity. Using HEK293, in this study, we investigated the interactions and any associated cytotoxicity of four different nanohydroxyapatites (nHAPs: (i) biogenic platelet-shaped, chemically synthesized (ii) nanorods, (iii) spherical, (iv) nanoneedles) and nanophosphorus (spherical biogenic nP derived from rock phosphate). The NM's interaction with cell culture media was studied by dynamic light scattering, transmission electron microscopy, and elemental and protein corona characterisation. The investigation on acute effects such as mitochondrial and lysosomal activities, membrane disintegrity, ROS generation, and cell viability was performed by following NIH guidelines. In addition to the dose-dependent approach, the cytotoxic effects of NM on changing the initial cell seeding density were also monitored. Our results suggest that physicochemically different P-based NMs interact differently with cell culture media resulting in NM transformation, minimal P release, and protein corona formation. Lipoprotein and albumins were the major components of the hard corona. No LC50 values were attained for any of the cytotoxic endpoints up to 1000 µg mL−1. Fluorescence and electron microscopy confirmed the uptake of NMs in the cells. This is the first report on the nano-bio interactions of differently shaped and sized P-based NMs using colloidal chemistry and two independent approaches-variable NM dose and variable cell densities. The findings suggest low cytotoxicity and hence the potential suitability of using the biogenic P-based NMs in agriculture as nanofertilisers.
Environmental significanceThe use of phosphorus (P) – based nanomaterials (NMs) as fertilisers to increase agricultural productivity is being investigated. It is therefore critical to investigate the colloidal behaviour of P-based nanofertilisers in biological media relevant to human physiology while assessing their cytocompatibility. This results in dynamic changes in NM behaviour in terms of protein corona acquisition, change in size, and P release. Bridging such changes with detailed cytological effects of NMs remains an understudied topic particularly for biosynthesised NMs. This study reports the NMs interaction with cell culture media, cellular uptake, and acute effects of P-based NMs on human cells using multi-end point assays. The study provided evidence of nano-safety that supports the recommendation of environmentally relevant concentrations and dosages of P-based nanofertilisers. |
1 Introduction
Nanomaterials (NMs) have been explored for their various applications in medicine and agriculture. Considering the depleting phosphorus (P) levels in agricultural soils across the globe, P-based NMs are proposed to be used as nanofertilisers.1 Lab-scale research on P-based NMs has shown promising fertilising effects.2–6 However, before agricultural applications, it is important to understand risks from the perspective of human health considering occupation safety and end-user applications. As compared to conventional fertilisers, these nanofertilisers are easily taken up by the plants and aid in enhancing nutrient use efficiency.7 There are several reports on the beneficial effects of P-based nanofertilisers on the overall quality and yield of agricultural crops.6,8,9 These nanofertilisers are reported to be either applied via soil6,8,10 or foliar doses.11,12 In either application mode, farmers and field workers will be primarily exposed to the nanofertilisers. This direct exposure may happen with the concentration ranges (10–1000 µg mL−1) that are used for different crops.6,8,10–12P-based NMs such as nanohydroxyapatites (nHAPs) have been evaluated multiple times to show in vitro biocompatibility, especially in osteogenic and related cell lines.13 This had relevance when nHAPs were to be explored for biomedical purposes such as bone grafting and tissue engineering. For such applications, majorly surface-functionalized nHAP and its composites with other materials such as chitosan,14 caprolactone,15 carbon nanotube,16 gelatin,17 and elements like europium,18 yttrium19 and strontium19 were used. Further, for nHAPs without any co-substitutions or modifications, there are some reports showing the toxicity of nHAP to BEAS-2B (lung cell line),20 RAW264.7 (mouse macrophage),21 catfish B (3B11) and T (28s 3) immune cells,22 L929 (murine fibroblasts),23 and primary rat osteoblasts.13 There are other reports that have shown size and morphology-dependent inflammatory responses and inhibition in macrophages.24–26 This prior background information suggests that investigation on cytocompatibility of nHAPs is largely dependent on their properties and type of cells. Although bulk HAP is a bone material and hence biocompatible, at the nanoscale the physicochemical properties can change significantly. Thus, nanoscale HAP will be different from bulk HAP, which may influence its biological behaviour.
Previously, research on cytocompatibility was focussed on chemically synthesized nHAPs. Such synthesis routes use chemicals that are potent toxicants or irritants and release hazardous effluents.27 It is now gradually been established that green or biosynthesized NMs have lesser synthesis-associated toxicity due to the elimination of harsh chemicals.28,29 Methods of biosynthesis have also been applied to develop P-based NMs.23,30,31
Biocompatibility of such biogenic P-based NMs remains a research gap. Additionally, little has been explored for biogenic P-based NMs in terms of phosphorus release and nano-bio interactions, especially their colloidal behaviour in the presence of serum-supplemented cell culture media (CCM). This kind of colloidal study in the biologically relevant media is of key importance to understanding the physicochemical mechanism of NM interaction with cells by estimating the influence of media components on pristine NMs. It has been explored previously that the attachment of biomolecules to NM surfaces changes the physicochemical identity of NMs. This interferes with the stability of NM and the colloidal dispersion state. A few researchers have shown interactions of bovine serum albumin with nHAPs,32,33 yet their interactions with serum proteins and CCM are not investigated much34 and there is a gap. Further, for thorough nano-bio interactions, evidence on the behaviour of P-based NMs with increasing/decreasing cell densities is lacking.
In our previous studies, we have reported the biological synthesis, characterisation, and toxicity of nHAP and nP in C. elegans and zebrafish.30,35–37 In the present study, the aim was to evaluate the in vitro nano-bio interactions using a set of complementary techniques to characterize phosphorus release from NMs and protein corona formation, uptake of biologically synthesized P-based NMs, and their effects on HEK293. It is known that once the NMs reach systemic circulation, they are cleared from the body by kidneys,38 making it critical to evaluate NM-induced renal toxicity. For Ca–P-based NMs, the release of Ca can form Randall plaques on renal epithelial cells that may further result in kidney stones.39,40 The study also involves a comparison of biologically synthesized NMs with chemically synthesized analogues. All these P-based NMs differ in size and shape, thus biological impacts of NMs with different physicochemical properties are highlighted. This is the first report where a thorough investigation of colloidal interactions of biogenic nHAPs and nP has been done to understand their behaviour in a dose-dependent manner as well as with variable cell densities. In addition, using different techniques, the results are the first standalone observations of the effects, uptake, and localisation of nP in human cells.
2 Methods
2.1 Materials
P-based nanomaterials: biologically synthesized nanohydroxyapatite (nHAP_B) and nanophosphorus (nP) were used as previously synthesized in our lab.35,41 Chemically synthesized nanohydroxyapatite, (nHAP_C) (previously synthesized in our lab),41 rock phosphate (RP) (bulk material used to synthesize nanophosphorus), and commercially available controls from Sigma Aldrich (nHAP_Sigma, USA) and SRL (nHAP_SRL, India) were used as standards for comparison in all physicochemical characterization.41 Fetal Bovine Serum (FBS) was purchased from Gibco (Massachusetts, USA). Cell culture media with 10% FBS was prepared using DMEM provided by Lonza (Basel, Switzerland). Heat-inactivated FBS (HiMedia) was added to the media after filter sterilization. The proteins were used without further purification. Autoclaved, sterile water (Millipore, Milli-Q water with a conductivity of 18.2 MΩ cm) was used wherever required.2.2 Physicochemical characterisation of P-based NMs and P release from nHAP_B and nP
The investigated NMs were characterized in detail by a combination of physicochemical techniques. NM stock suspensions (1 mg mL−1) prepared in in vitro cell culture media were sonicated for 30 minutes using a bath sonicator (25 kHz) before the preparation of test solutions. The test solutions (suspensions) were vortexed for 30 s before initiating characterization experiments. The hydrodynamic diameter and zeta (ζ) potential of NMs (1 mg mL−1) in in vitro media were assessed using Zetasizer Nano ZS (Malvern Instruments, UK) equipped with a 633 nm laser. The NMs were also characterized for their interactions with serum proteins and colloidal behaviour using UV-vis (Shimadzu, Japan), FTIR (Nicolet 6700, Thermofisher Scientific, USA), and dynamic light scattering after dispersal in cell culture media containing 10% fetal bovine serum (FBS). The shape and size of NMs in in vitro cell culture media were determined using transmission electron microscopy analysis (200 kV, Tecnai G2 30-U twin microscope, FEI, USA).The release of P from the biogenic nHAP and nP was calculated by using the previously published dialysis method.42 The dialysis membranes (14k MWCO and pore size = 3.9 nm) were washed and activated. The NMs suspensions (1 mg mL−1) were prepared in DI water and serum-supplemented cell culture media (CCM) and were added to the activated dialysis membrane. These dialysis bags were added to the containers with DI water and incubated at 37 °C and 5% CO2. Samples were then taken at 0, 2, 4, 6, 8, and 24 h to measure the P-release from the NMs by measuring the P contents using flame photometry. The initial content of P was 23.22% and 0.89% in nHAP and nP, respectively.
2.3 Protein corona study on nHAPs and nP in serum-supplemented media
Here a comparative analysis of P-based NMs–protein interactions with the application of cell culture media supplemented with FBS was performed to investigate the formation of the hard and soft protein corona. Briefly, the NMs at 1 mg mL−1 were incubated with 500 µL cell culture media + 10% FBS for 1 h at 37 °C under constant agitation. These were then separated from the supernatant by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf for 30 min. The NMs were washed with 1× PBS in three centrifugation steps at 15
000 rcf for 30 min. The NMs were washed with 1× PBS in three centrifugation steps at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf for 30 min. The pellet obtained after copious washing was analysed for protein attachment as the hard corona. The washes were kept for soft corona assessment. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), a protein quantification assay using Bradford's reagent,43 and fast protein liquid chromatography (FPLC, GE AKTA, Germany) were used to analyse the protein corona.
000 rcf for 30 min. The pellet obtained after copious washing was analysed for protein attachment as the hard corona. The washes were kept for soft corona assessment. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), a protein quantification assay using Bradford's reagent,43 and fast protein liquid chromatography (FPLC, GE AKTA, Germany) were used to analyse the protein corona.
2.4 Cell culture
Commercially available HEK293 was procured from National Centre for Cell Science, Pune, India. The cells were cultured with DMEM + 10% FBS, supplemented with 1% glutamine, and 1% penicillin–streptomycin. The cells were incubated at 37 °C and 5% CO2. The culture medium was changed every 3–4 days until the cells reached confluence. The adherent cells were then detached with a solution of 0.05% trypsin and 0.53 mM EDTA and the cells were then passaged.2.5 Cellular metabolic activity assessment using MTT and neutral red uptake assays
A total of 1 × 104 cells were seeded in 96 well plates before the exposure to nHAP and nP. At a confluent stage of 70–80%, the cells were exposed to increasing concentrations (1.56, 3.12, 6.25, 12.5, 25, 50, 100, 200, 500, and 1000 µg mL−1) of nHAP_B, nHAP_C, nHAP_Sigma, nHAP_SRL, nP, and RP. The time of exposure was kept at 24 h to study the acute effects of NMs. The MTT assay was carried out according to the protocol described by NCL, NIH, USA,44 and a neutral red uptake assay was performed as described by Borenfreund and Puerner.45 All the samples, along with positive (0.1% (v/v) Triton X), negative (no treatment, only cells), and media controls (no treatment, no cells) were run in triplicates. Absorbances were measured at 570 nm and 540 nm for MTT and neutral red assays, respectively, with a reference wavelength of 680 nm.2.6 Cell membrane integrity assessment using LDH and trypan blue assays
The effects of NMs on cell membrane integrity were estimated using lactate dehydrogenase release and trypan blue dye inclusion assays. The cells were seeded and exposed to NMs as described in the section above. The LDH assay was carried out according to the protocol described by NCL, NIH, USA44 and the trypan blue dye inclusion assay was performed as described by Uliasz and Hewett.46 All the samples, positive (0.1% (v/v) Triton X), negative (no treatment, only cells), and media controls (no treatment, no cells) were run in triplicate. Absorbances were measured at 490 nm and 590 nm for LDH and trypan assays, respectively, with a reference wavelength of 680 nm.2.7 Effect of NMs interaction on HEK293 with decreasing cell seeding density – an assessment by MTT for cellular metabolic activity
To analyze the nano-bio interactions w.r.t. variable cell densities, three different initial cell densities were chosen. A total of 1 × 104, 1 × 103, and 1 × 102 cells were seeded in separate 96 well plates before the exposure to nHAP and nP. The evaluation of mitochondrial activity was performed in a similar manner as stated above in Section 2.4.2.8 Intracellular oxidative stress assessment using DCFH_DA enzymatic assays
Oxidative stress due to exposure to NMs was determined using the DCFH_DA assay. The analysis was performed by following the protocol described by NCL, NIH, USA.47,48 For the DCFH_DA assay, briefly, the cells were seeded at a density of 1 × 105 cells per well and cultured for 24 h at 37 °C in a 12-well plate. After incubation, the media was discarded, and the cells were treated with different concentrations of NMs (50 and 500 µg mL−1) for 24 h. Post-exposure, the media was discarded and 20 µM of DCFDA solution was added to the wells with fresh media. The test plate was taken at 3 h post DCFDA addition and read at ex. 485 nm and em. 530 nm. The fold change in ROS activity was reported with respect to the negative control (untreated cells) and 300 µM of H2O2 was taken as the positive control.2.9 Uptake of fluorescent-tagged nanohydroxyapatite particles
FITC-tagged nHAP_B was used to see the time-dependent uptake of particles under a fluorescence microscope. The cells were seeded in a 35 mm culture dish at a seeding density of 0.3 × 106 cells. After 75–80% confluence these were exposed to 100 µg mL−1 of FITC-tagged nHAP_B. At different time points (2, 4, 6, 8, and 24 h), the exposure solution (media + NMs) was aspirated completely, and the cells were gently washed three times with 1× DPBS. Cells were fixed and after fixation imaged immediately under a fluorescence microscope (EVOS M5000 imaging system, ThermoFisher Scientific, USA) at ex. 488 nm and em. 516 nm. Untreated cells were also viewed under a fluorescence microscope to rule out autofluorescence.2.10 TEM visualization of cells exposed to biogenic P-based NMs
For tracking cellular uptake and localization, TEM analysis was performed after cells were exposed to 100 µg mL−1 of biogenic nHAP_B and nP. The cells were washed twice with 1× DPBS and detached. The cells were gently centrifuged at 800 rcf and washed with 1× DPBS. Cells were pre- and post-fixed with 2.5% glutaraldehyde solution for 30 minutes and 1% osmium tetroxide for 2 h, respectively. Post fixation, the samples were sequentially dehydrated using alcohol gradient and acetone. After dehydration, the samples were cast in blocks and infiltrated with epoxy resin. Samples blocks were sectioned using microtomy. Thin sections of 60–100 nm were placed on carbon-coated copper grids. Samples were stained, and after that, the resin sample block was trimmed and thin-sectioned at a thickness of 60–100 nm, followed by the collection on carbon-coated copper grids. The grids were stained primarily with 1% uranyl acetate and 2% lead citrate was used as a secondary stain. TEM (Tecnai G2 F20 S-TWIN, US) coupled with energy-dispersive X-ray analysis (EDX) was used to capture images and to evaluate the presence of particles inside the cells.2.11 Acridine orange/ethidium bromide dual staining for cell viability
Cells seeded on a coverslip at a density of 1 × 105 cells per well were cultured for 24 h at 37 °C in a 12-well plate. After incubation, the media was discarded, and the cells were treated with different concentrations of NMs (100 and 1000 µg mL−1) for 24 h. Untreated cells were taken as a negative control and the treatment with 300 µM of H2O2 was taken as a positive control. Cells were washed twice with 1× DPBS, fixed, and stained using dual fluorescent staining solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 100 µg mL−1 of each acridine orange and ethidium bromide) for 5 min. Excess stain was removed by washing the cells with 1× DPBS. Cells were observed under a fluorescence microscope with an automated cell counter (Cytation 5, Biotek, Agilent Technologies, USA). A total of 3715 cells as determined by the automated cell counter were analysed in each case.
1 mixture of 100 µg mL−1 of each acridine orange and ethidium bromide) for 5 min. Excess stain was removed by washing the cells with 1× DPBS. Cells were observed under a fluorescence microscope with an automated cell counter (Cytation 5, Biotek, Agilent Technologies, USA). A total of 3715 cells as determined by the automated cell counter were analysed in each case.
2.12 Statistical analyses
For each of the NM (nHAPs and nP), we characterized dose-effect relationships. All statistical analyses were performed using GraphPad Prism software, Version 9 (GraphPad Software Inc., San Diego, CA, USA). Prior to statistical analysis, the normal distribution of residuals was checked with the Shapiro–Wilk test and QQ plots. Results were subjected to analysis of variance for significance followed by Dunnett post hoc multiple comparison tests, to identify the treatment groups with significant differences in mean values.3 Results
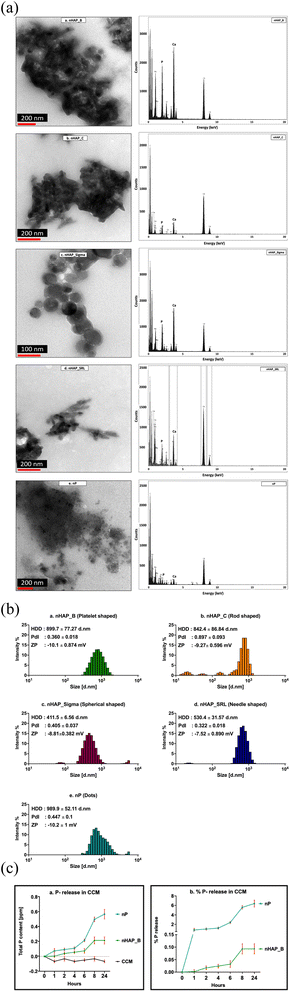
3.1 Physicochemical characterization of P-based NMs in cell culture media
The TEM micrographs (Fig. 1A) show the particles along with the media components such as proteins. The EDX results suggest that the Ca/P ratio was changed from 1.58, 1.79, 1.55, 1.62, and 3.69 to 1.6, 1.57, 1.52, 1.53, and 1.07 for nHAP_B, nHAP_C, nHAP_Sigma, nHAP_SRL, and nP, respectively. The primary sizes of NMs at a concentration of 1 mg mL−1 in CCM from TEM were 37.86 ± 8.8 nm for nHAP_B; L: 87.69 ± 14.5 nm and W: 25.97 ± 5.7 nm for nHAP_C; 43.12 ± 13.02 nm for nHAP_Sigma; L: 84.04 ± 16.76 nm and W: 18.42 ± 3.76 nm for nHAP_SRL; 18.41 ± 9.69 nm for nP. The histograms for the primary sizes obtained from TEM are presented in ESI Fig. 1.† As reported previously,30 the hydrodynamic sizes of the pristine NMs in de-ionised water were 325 ± 37.1 nm, 756.2 ± 28.8 nm, 874.3 ± 51.5 nm, 892.8 ± 21.1 nm, and 798.5 ± 23.4 nm, respectively, for nHAP_B, nHAP_C, nHAP_Sigma, nHAP_SRL, and nP. Upon interaction with 10% serum-supplemented CCM (Fig. 1B), the sizes were changed to 899.7 ± 77.27 nm, 842.4 ± 86.84 nm, 411.5 ± 6.56 nm, 530.4 ± 31.57 nm, 989.9 ± 52.11 nm for nHAP_B, nHAP_C, nHAP_Sigma, nHAP_SRL, and nP, respectively. In contrast, when the NMs were dispersed only in the CCM without serum, the hydrodynamic sizes were found to be 959.5 ± 115.47 nm, 1122.8 ± 373.05 nm, 441.4 ± 92.09 nm, 794.83 ± 231 nm, and 1201.5 ± 256.34 nm for nHAP_B, nHAP_C, nHAP_Sigma, nHAP_SRL, and nP, respectively. Correspondingly, the zeta potential values were also changed.It is hypothesized that once these NMs disperse within the CCM, they will interact with the cells either via colloidal dispersion or by sedimentation in the cell culture vessel. For each of the NM tested, a theoretical analysis of the concentration-dependent particle numbers corresponding to either colloidal dispersion or sedimentation was carried out and is presented in ESI Tables 1–3.† It can be observed that for P-based NMs, the number of particles interacting with cells via colloidal dispersion is more than by sedimentation at any tested concentration from 1.5–1000 µg mL−1.
Further, on examining the release kinetics of P (Fig. 1C) from biologically synthesized NMs dispersed in serum-supplemented cell culture media at 1000 µg mL−1, it was found that for nHAP_B, the maximum release was 0.093 ± 0.035% at 24 h and for nP, it was 6.38 ± 1.24% at 24 h.
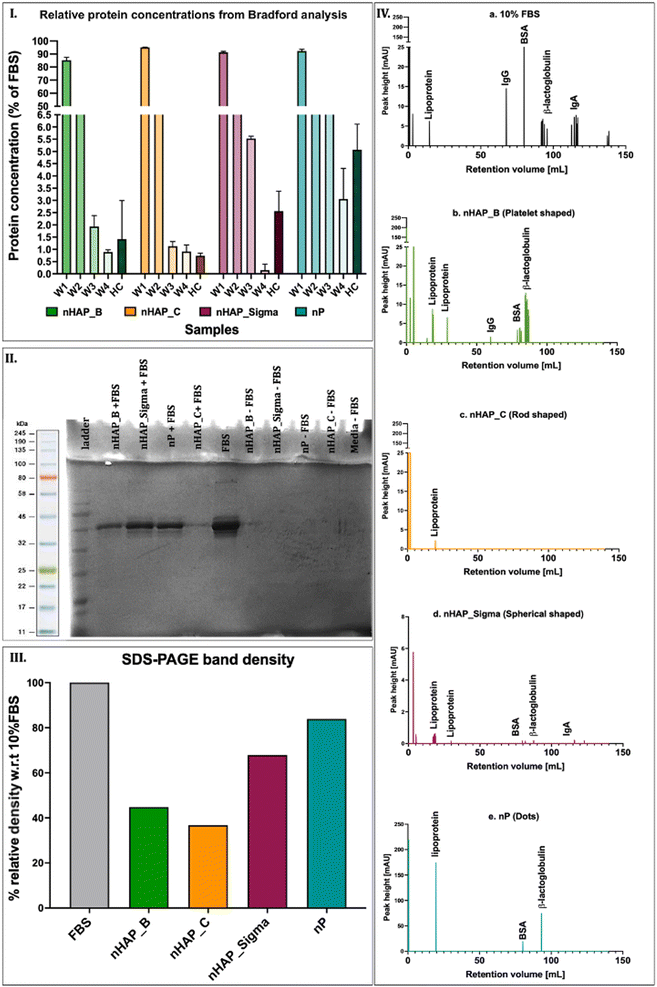
3.2 Evaluation of protein corona formation on P-based NMs
The protein content in the corona was determined in different washes (W) and in hard corona (HC) by Bradford's method. After 4 washes (W1, W2, W3, and W4), the protein content in HC was found in the order of nHAP_Sigma > nP > nHAP_B > nHAP_C (Fig. 2(I)). The SDS-PAGE results show distinct bands of HC for NM samples (Fig. 2(II)) and the quantified band intensities (Fig. 2(III)) corroborated with the protein concentration trend observed by Bradford's method. Complete protein profiles are shown by FPLC graphs for the HC samples (Fig. 2(IV)). Peaks for lipoprotein, IgG, IgA, BSA, and β-lactoglobulin were found in 10% FBS. Of these, lipoprotein was present in HC of all the NMs, IgG was present in HC of nHAP_B, IgA was present in HC of nHAP_Sigma, BSA, and β-lactoglobulin was present in HC of nHAP_B, nHAP_Sigma, and nP.3.3 Effect of NMs on cellular metabolic activities
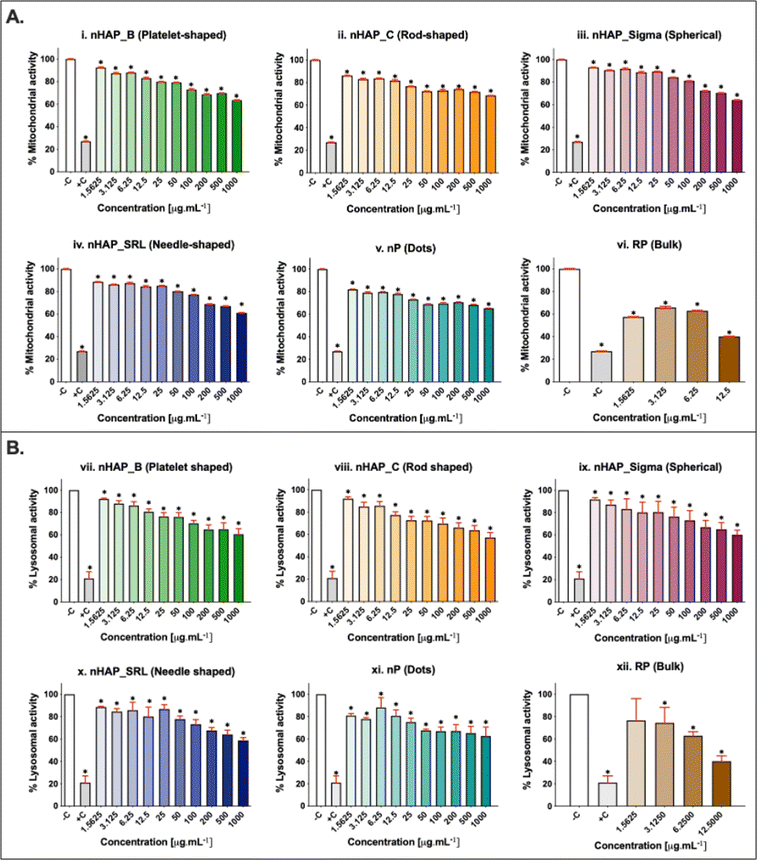
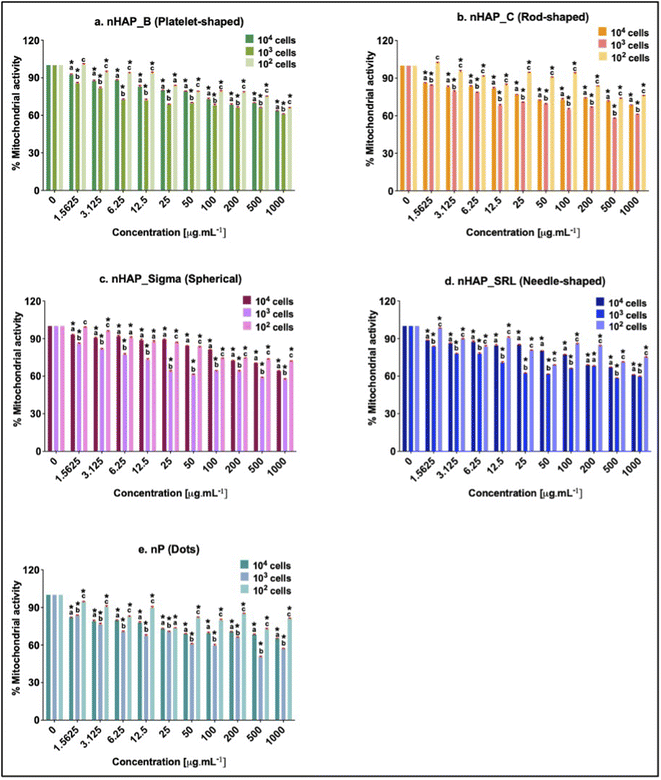
A dose-dependent decrease in cellular mitochondrial activity was seen for all NMs (Fig. 3A). The corresponding linear regression graphs and equations are presented in ESI Fig. 2A.† Nearly 20% decrease in mitochondrial activity was observed for different treatments for the environmentally relevant concentration of 100 µg mL−1. The minimum cellular mitochondrial activities were noted at the highest tested concentration of 1000 µg mL−1. A neutral red uptake assay was performed to assess the lysosomal activity on exposure to NMs and a significant (p < 0.001) dose-dependent decrease was seen (Fig. 3B) with a minimum activity at the highest tested concentration of 1000 µg mL−1. The linear regression graphs and equations for the neutral red uptake assay are presented in ESI Fig. 2B.† Similar to the mitochondrial activity, the lysosomal activity was also decreased by ∼20% on exposure to an environmentally relevant concentration of 100 µg mL−1. The rod-shaped nHAP_C caused a maximum decrease in cellular metabolic activities followed by the needle-shaped nHAP_SRL, platelet-shaped nHAP_B, and spherical nHAP_Sigma. For both assays, none of the NMs had LC50 at any tested concentrations. On the other hand, for bulk RP, LC50 was attained at much lower tested concentrations.3.4 Effect of NMs on cellular membrane integrity
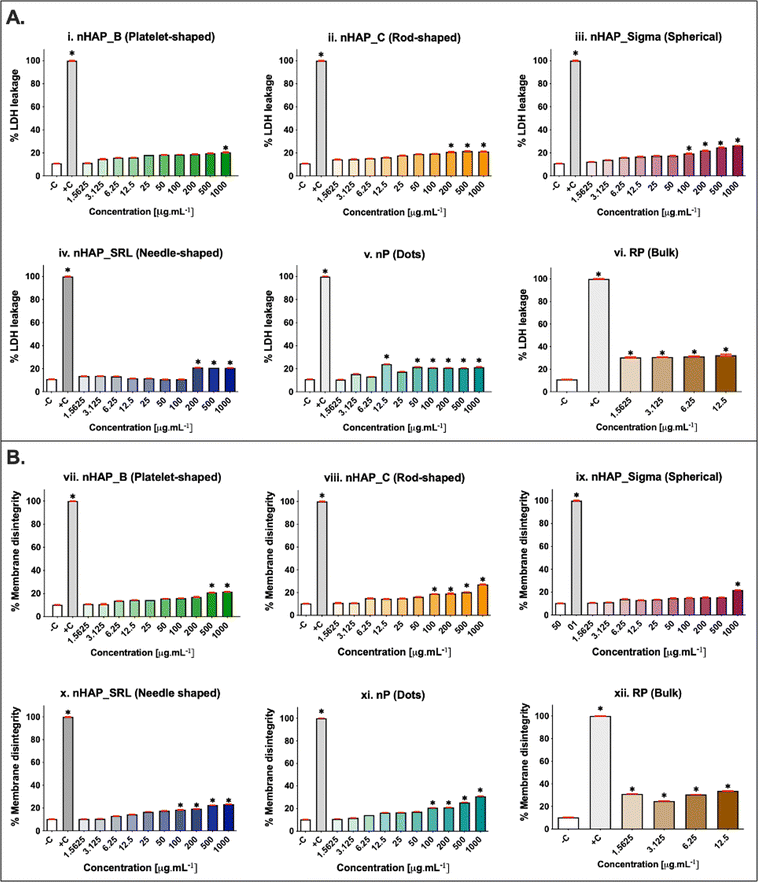
The membrane integrity was evaluated at a range of concentrations from 1.5 to 1000 µg mL−1. A dose-dependent increase in LDH leakage (Fig. 4A) was seen for all NMs. The maximum values were noted at the highest tested concentration of 1000 µg mL−1. The percentage of LDH leakage was <20% in all cases of exposure. The corresponding linear regression graphs and equations are presented in ESI Fig. 3A.† Trypan blue dye inclusion assay was performed to assess the membrane disintegrity on exposure to NMs and a dose-dependent increase was seen (Fig. 4B). On comparing with 0.1% triton-positive control (100% membrane dis-integrity), it was found that the highest exposure concentration of 1000 µg mL−1 resulted in maximum %membrane dis-integrity. The membrane disintegrity was <20% for all tested concentrations of NMs except at the highest concentrations for rod-shaped nHAP_C and nP. For both assays, none of the NMs had LC50 at any tested concentrations. The linear regression graphs and equations for the trypan blue assay are presented in ESI Fig. 3B.†3.5 Effects of NMs exposure on cellular activity for different initial seeding densities
Initial cell seeding densities of 104, 103, and 102 were used for testing the concentration range from 1.5 to 1000 µg mL−1. For nHAP_B, nHAP_C, and nP, minimum mitochondrial activity was seen in the case of 103 cells and the highest was in the case of 102 cells (Fig. 5). Similar observations were made for nHAP_Sigma and nHAP_SRL with some overlaps between the activity of 102 and 104 cells. However, in neither case, LC50 was seen for any of the NMs at any of concentrations. The minimum % metabolic activity, in any case, was found at the highest tested dose of 1000 µg mL−1. These values were statistically significant (p < 0.001) as compared to the untreated control cells.3.6 ROS generation monitored on exposure to NMs
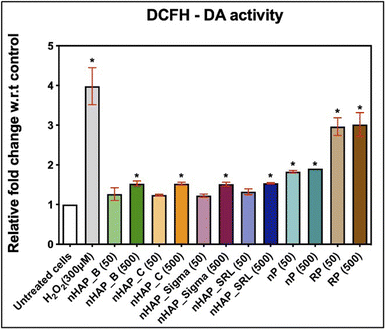
Intracellular ROS levels were monitored after the cells were exposed to 50 and 500 µg mL−1 of NMs from 2′,7′-dichlorofluorescein diacetate (DCFH-DA) oxidation. It was found that after 3 h of monitoring, results were significant for 500 µg mL−1 of NMs, as compared to the untreated control. The fold changes w.r.t. untreated control were 1.5 for nHAPs and 1.9 for nP at 500 µg mL−1 and at 3 h estimation (Fig. 6). These fold changes were significant (p < 0.001) for nP from evaluation for both 50 and 500 µg mL−1. Exposure to both concentrations of RP has significant (p < 0.001) DCF levels. This suggests that on getting converted to nanoform from RP to nP, the oxidative stress due to the exposure decreases.3.7 Time-dependent uptake of FITC-tagged nHAP_B
Exposures to FITC_nHAP (100 µg mL−1) in HEK293 were analysed in a time-dependent manner, and the data are shown in Fig. 7. Fig. 7(i) shows the control untreated cells. Fig. 7(ii)–(vii) show the time-dependent uptake at 2 h, 4 h, 6 h, 8 h, and 24 h. It can be suggested that the apparent uptake of P-based NMs starts after 4 h of treatment.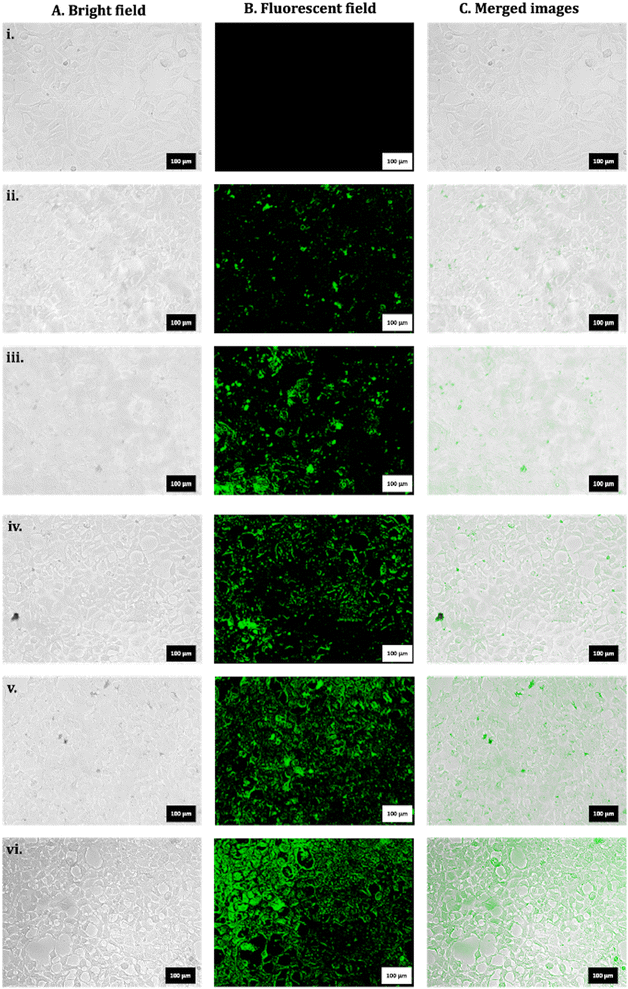 | ||
| Fig. 7 Representative microscopic images for HEK293 for (i) untreated control. Exposure to FITC_nHAP_B at (ii) 2 h, (iii) 4 h, (iv) 6 h, (v) 8 h, and (vii) 24 h (scale: 100 µm). | ||
3.8 TEM analysis of cellular uptake and localization
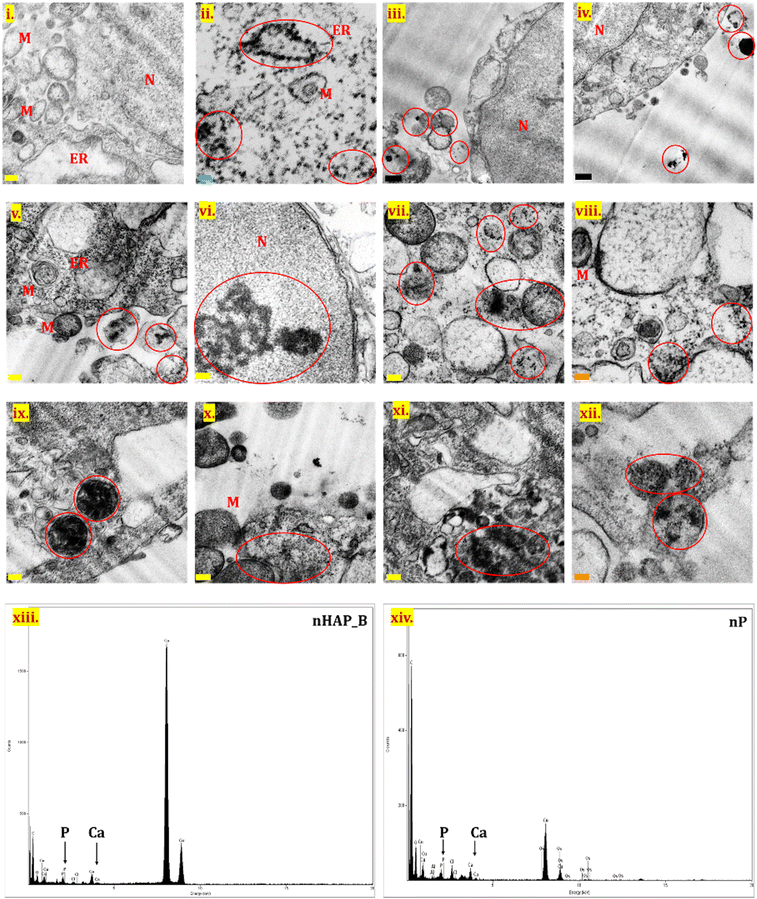
TEM analysis was performed for the untreated control cells (Fig. 8(i)) and for cells after exposure to nHAP_B and nP (100 µg mL−1) by processing samples immediately after NMs exposure (Fig. 8(iii) and (iv)) and after 24 h of exposure (Fig. 8(ii) and (v)–(xii)). No significant cellular internalization of NMs was seen for samples processed immediately after the exposure. NM internalization was seen in the case of both nHAP_B and nP after 24 h of exposure, contrary to the untreated control. NMs were seen interacting with prominent cell organelles such as cytoplasm, nucleus, mitochondria, endoplasmic reticulum, endosomes, lysosomes, and the membranes of different cellular organelles. nHAP_B aggregates were seen in the nucleus of the cell, while particles were also present in subcellular organelles. nP was more in the aggregated form inside the cells as compared to nHAP_B. Fig. 8(xiii) and (xiv) show EDX for nHAP_B and nP, respectively. indicating the presence of P and Ca.3.9 Live/dead cell assessment using acridine orange/ethidium bromide dual staining
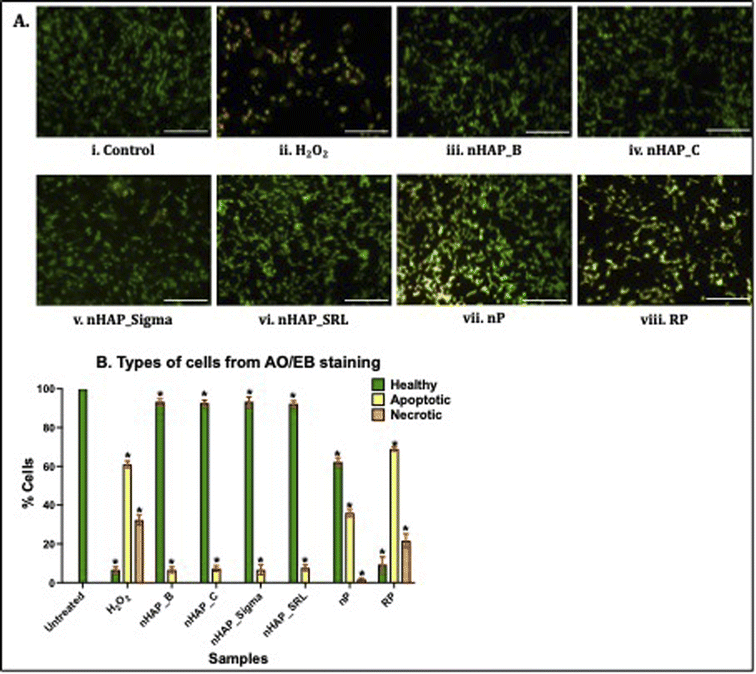
To investigate cell viability and apoptosis, dual staining using acridine orange and ethidium bromide was completed and 3715 cells in each case were auto-analysed using an automated cell counter (built-in function of Cytation 5, Biotek, Agilent Technologies, USA). In the untreated control, 100% of the HEK 293 cells appeared healthy (Fig. 9A(i) and B). Exposure of the cells to the positive control, H2O2, resulted in an increased number of apoptotic (61 ± 1.73%) and necrotic cells (32.33 ± 2.52%; Fig. 9A(ii) and B). Further, as shown in Fig. 9A(iii)–(viii) and B, healthy cells were observed for nHAPs. The exposure of cells to nP also caused an increase in apoptotic cells (36 ± 2%) compared to the control (Fig. 9A(vii) and B). Exposure of the cells to the bulk control, RP, was also observed to cause an increase in both apoptotic (69 ± 1%) and necrotic cells (21.67 ± 3.51%). All these values were statistically significant (p < 0.01) w.r.t. to untreated control cells.4 Discussion
The P-based NMs are proposed for agricultural applications as P-supplementing nanofertilisers. For occupational health and safety in response to the nanofertiliser exposure, it is critical to assess the potentially toxic effects of NMs on humans using the human cell lines or other model systems at their predicted application concentrations as fertilisers (up to 25 µg mL−1)49,50 and higher concentrations (25–1000 µg mL−1). Farmers and field workers are most likely to be exposed to nanofertilisers when applying it either via the soil or foliar spray in a direct exposure concentration range of 10–1000 µg mL−1.6,8,10–12 Dermal, inhalation, and ocular routes may be the primary ways of NM internalisation in the human body.51 After internalisation, renal clearance of the NMs that have reached the systemic circulation is an expected and possible elimination route from the human body.38 For this reason, kidneys may be one of the most important secondary target organs, and NM-induced renal toxicity should be evaluated. Further, excess Ca from HAP crystallites on the surface of renal epithelial cells is known to induce the formation of Randall plaques and even kidney stones.39,40 Due to the diversity of P-based NMs, especially their physicochemical properties that influence their characteristics and behaviour, many nano-bio interactions are yet to be investigated. Therefore, further studies are required to provide a comprehensive and critical evaluation of the toxicity of NMs in the renal system. In addition to providing insights into the effects of P-based NMs on nephrotoxicity, HEK293 cells, being epithelial cells, can also be used to assess the toxicity of P-based NMs on epithelial cells. Previously, the effects of P-based NMs, especially nHAP have been tested on mammalian cell lines and some results have shown toxicity20–23 and inflammatory22–26 responses of rod- and needle-shaped nHAPs. Recently, a study on rod-shaped mesoporous silica-coated nHAP reported that the NM was toxic to MC3T3-E1 cells on doses above 10 µg mL−1.52 However, a thorough investigation of the physicochemical modifications and protein corona formation remains a gap to understand the cytological behaviour of differently shaped P-based NMs.In this study, we investigated the in vitro nano-bio interactions of multiple variants of nHAPs and nP, which differed in physicochemical properties and origin as described previously.35,41 Four differently shaped nHAPs namely, nHAP_B (platelet-shaped), nHAP_C (rod-shaped), nHAP_Sigma (spherical), and nHAP_SRL (needle-shaped), were used in this study. Another P-based NM used in this study, nP (dots) was derived from rock phosphate. A systematic and thorough impact of these NMs was carried out in HEK293. In the serum-supplemented CCM, the hydrodynamic behaviour, physicochemical properties, P release, and protein absorption on the NM surface were investigated. Cell viability, cellular mitochondrial and lysosomal activity, membrane disintegrity, oxidative stress, uptake, and localization of these NMs in the cell and cellular organelles were studied. The effects of these NMs were also tested on the cellular metabolic activity at varying seeding densities.
The addition of the P-based NMs to serum-supplemented media resulted in increased hydrodynamic diameters for all nHAPs. The Z-average size of nHAP_C was the highest followed by nHAP_SRL, nHAP_B, and nHAP_Sigma. It is worth noting here that nHAP_C and nHAP_SRL are rods- and needle-shaped, respectively, and thus have higher sizes than nHAP_Sigma (spherical) or nHAP_B (platelet-shaped). From their TEM images, it was apparent that the media formed a film surrounding the surface of NMs. It can be suggested that the serum components form a capping over these NMs. The negative zeta potential values for all the NMs were decreased upon interaction with FBS. This could be due to the absorption of positively charged amine groups on the surface of NMs. A similar observation was made by Casals et al. (2010) for Au NMs.53 Similar observations were also made for nP, where the hydrodynamic diameter increased after dispersal in the serum-supplemented media. It was also discussed recently by Zhao et al. (2021) that serum protein can be deposited more on particles with smaller diameters.54 This agrees with the TEM profile obtained for nP where the pristine diameter was ∼10 nm (ref. 55) and upon interaction with serum-supplemented media, a large visible matrix was observed with small-embedded particles of nP (∼18 nm).
In this study, the investigations on protein corona revealed that the protein adsorption from the CCM onto the surfaces of P-based NMs was dependent on their inherent shapes and physicochemical properties. The proteins on hard corona were profiled in the FPLC analysis. These were then identified by referring to the peaks reported previously for a similar analysis.56 The main proteins identified in the hard corona were lipoprotein, β-lactoglobulin, albumin (BSA), and immunoglobulins (IgG and IgA). The presence of the protein corona on the NMs could affect the uptake and intracellular trafficking of the NMs.57,58 It has been shown previously that phagocytosis can be influenced by immunoglobulins in alveolar macrophages,59,60 and lipoproteins61 have been shown to affect the intracellular transport and localisation of the NMs in the rat epithelial cells62 and bone marrow-derived macrophages.63
Several interesting findings were observed when HEK293 cells were exposed to P-based NMs in a dose-dependent manner. It was also observed that the P-release from the biologically synthesised NMs was slow and minimal over the maximum exposure time of 24 h. Thus, the effects on the cells were dominantly due to interaction with NMs only. Exposure of HEK293 to nP derived from RP significantly decreased the mitochondrial and lysosomal activity, enhanced membrane disintegrity, and increased ROS and cellular apoptosis. However, these effects were further increased when the cells were exposed to the bulk RP at the same doses. The elemental composition of RP might be the reason for its severe cytotoxicity. Fagiya and colleagues64 have previously shown that RP is a heterogeneous mix of different elements: 20–28% P2O5, 43.9% O, 34.1% Ca, 13.6% P, 3.57% F, 1.43% Fe, 1.26% Si, and 2.14% other heavy elements such as Pb, Zn, and Al.64 It is however interesting to observe from this study that the nano-formulation from RP is comparatively safer than the bulk material. The cellular effects of nP can also be correlated with its protein corona. It has been shown by Levak et al. (2017) that increased serum albumin attachment to the NMs and NM aggregates reduces the bioavailability of NMs and hence results in comparatively higher cell viability.65 From the early investigations on protein corona, it was found that biologically synthesized nP had more protein content, as compared to the bulk RP and had a prominent BSA peak. Consequently, it was shown that at the highest concentration tested, it had improved mitochondrial and lysosomal activity, ROS activity, and cell viability, as compared to RP. When considering the cytotoxic effects of nP, oxidative stress may be a key cause. Buzea et al. have suggested that the toxicity of NMs is primarily because of the size of the particles and them being smaller than organelles.66 Several studies have already confirmed that NMs may induce inflammation67 or apoptosis by oxidative stress.68,69 Further, similar to findings in mouse alveolar macrophage cells (RAW264.1) by Mortensen et al. (2013)70 for SiO2 NMs and Xia et al. (2006)71 for polystyrene NMs, a plausible explanation about the nP-mediated ROS generation and toxicity can be linked with the protein corona. According to these reports, spherical NMs adsorb amino and carboxylic groups from the protein. The number of amino groups adsorbed on the surface of NMs dictates the cellular ROS generation in a direct proportion.
Exposure of the HEK293 cells to each of the nHAPs also caused significantly lower mitochondrial and lysosomal activities at the highest NM doses. Interestingly, while the cellular mitochondrial and lysosomal activities were decreased in a dose-dependent manner, the disintegrity of the membrane and ROS generation were not highly disturbed. Also, as confirmed by the dual staining with acridine orange/ethidium bromide for the live-dead cells assessments, the overall cell viability was not decreased when cells were exposed to nHAPs. Some of the previous studies have demonstrated that the nHAPs can cause increased cytoplasmic Ca loading, which is likely the cause of compromised organelle activities where the increased cellular Ca trigger and modulate mitochondrial and lysosomal dysfunctions.72–76 Zhang et al. (2017), have reported that 40 nm wide rod-shaped nHAP caused cytotoxicity in MC3T3-E1 cells after increasing the intracellular Ca levels.72 In another report, Jin et al. (2017) have shown that rod-shaped nHAPs (50 nm in width) enter the MC3T3 cells via an energy-dependent macro-pinocytosis pathway and co-localise mainly in lysosomes and mitochondria causing sub-lethal toxicity.73 Interestingly, in more recent studies, rod-shaped nHAP particles were found to be non-toxic to MC3T3-E1 cells.52,77 Cui et al. (2016) have provided evidence that nHAP is readily taken up by the human gastric MCG80-3 cells and dissolves in lysosomal acidic environments, which in turn increases Ca ion levels in the cells and causes an imbalance in the intracellular Ca-dependent homeostasis.74 Wang et al. have verified that the presence of nHAP changes the serum electrolyte ion balance in MC3T3 cells and Sprague Dawley rats, and decreases potassium ion concentrations, which is negatively correlated with calcium ion concentration in serum resulting in decreased cellular activities.75 Sun et al. (2020) have also shown that the endocytotic ability of renal epithelial cells for calcium phosphates (nano- or micro-forms) exhibits size dependence. They provided evidence that nHAP is endocytosed into cells at a greater rate than micron-sized calcium phosphate.76 Contrary to their results, where nHAP was more cytotoxic than the micron-sized HAP form of calcium phosphate, the results from the current study show that the behaviour and toxicity of nHAP were similar to the bulk form of calcium phosphate. This further suggests the importance of considering the physicochemical properties of nHAPs to determine their cellular behaviour and in turn, can indirectly reflect the importance of NM synthesis routes on governing the physicochemical properties of NMs. Overall, in this study, the cell viability was observed to be higher in the case of biologically synthesized nHAP_B, as compared to chemically synthesized nHAP_C. It is also important to highlight the importance nHAP shape has on cell viability, as platelet-shaped nHAP_B and spherical nHAP_Sigma had higher cellular viability, as compared to the rod- and needle-shaped nHAP_C and nHAP_SRL. Similar to our observations, in a recent study, it was reported that green synthesised spherical like nHAP was more cyto-compatible than the rod-shaped nHAP in L929 fibroblasts.23
An interesting new approach to further investigate the nano-bio interactions of P-based NMs was to assess the cellular mitochondrial activity by decreasing the initial cell seeding density by a factor of 10. It has been previously hypothesized that NMs can interact with cells either via colloidal diffusion or via gravitational settling,78–80 and thus, it becomes curious to test whether cell density influences such interactions any further. A new approach to further investigate the nano-bio interactions of P-based NMs was, therefore, conducted that assessed the cellular mitochondrial activity of the cells seeded at different densities and exposed to P-based NMs. The results obtained from this study indicate that initial cell density may play a potential role to identify the biological behaviour of NMs. With a lower cell count (density) the proportion of NM per cell increases, which may result in a more profound effect of the NMs on cells. Our results show that for the minimum cell density used in this experiment, the comparative mitochondrial activity was higher, as compared to the effects of the NMs for higher cell density. This suggests that cell density could be an important factor in the cellular uptake and toxicity of NMs. Future experiments could be conducted on this using more complex cell cultures (e.g., cell spheres) and other bioassays and molecular approaches to further investigate the effects of cell density and culture methods on NM toxicity.
The uptake of the biogenic P-based NMs in HEK293 cells was confirmed using FITC-tagged nHAP_B and fluorescence microscopy, as well as from TEM for both nP and nHAP_B. Interestingly, nHAP_B was observed to enter the cells in a time-dependent manner. Further analysis with TEM confirmed the migration of these NMs into the cytosol and cellular organelles. The EDX spectra also confirmed the presence of P and Ca from the area imaged for NMs. Similar internalisation and EDX patterns were observed by Zhang et al. (2017) for nHAP in MC3T3-E1 cells.72 The cellular uptake of the metallic NMs has also been previously confirmed using TEM. In one of the studies with Ag NMs, uptake in murine melanoma B16 cells was shown where the particles localise to intracellular vesicles and late endosomes, and clathrin-mediated internalisation was suggested.81 In another study on Ag NMs and human hepatocyte carcinoma HepG2, the authors reported localisation of particles in endosomes and lysosomes after being taken up by micropinocytosis and clathrin-mediated endocytosis.82 A study with Au NMs in RAW267.1 cells has shown the localisation of NMs in the perinuclear region and vesicles and has reported the internalisation via clathrin-mediated and dynamin-dependent endocytosis.83 In another study demonstrating the uptake of Fe NMs in human ovarian cancer (SKOV-3) and RAW 264.7 cells, it was shown that the NMs localise in endosomes and lysosomes suggesting the possible uptake by endocytosis.84 Another study found the time-dependent uptake of ZnO NMs in human mesenchymal stem cells after repetitive treatments and the localisation into the nucleus and mitochondria.85 Considering these previous studies, and the results from this study where P-based NMs were located around the lysosomes and the other membrane-bound organelles, it can be hypothesized that their internalisation would have been either via adsorptive or receptor-mediated endocytosis. However, it will be interesting to further investigate if the internalisation of P-based NMs in human cells is through multiple pathways or a single energy-dependent pathway.
The impacts of exposure to P-based NMs on acute cellular behaviour are significant. In a nutshell, for P-based NMs to be used potentially as nanofertilisers, it is important to know the specific safe doses for manufacturers and end users to handle these NMs. These doses are dependent on the physicochemical properties of nHAPs and nP. Although, some sub-lethal effects including mitochondrial and lysosomal activities were observed at the higher NM doses and were more evident after exposure to nP and RP compared to nHAPs. For environmentally relevant concentration of 100 µg mL−1, the safer NMs follow the order as nHAP_B ≈ nHAP_Sigma > nHAP_SRL > nHAP_C > nP. However, there were no LC50 values obtained in either of the assays for any of the P-based NMs up to 1000 µg mL−1. This highest tested concentration is hypothesized to be significantly higher than the environmentally relevant concentration for NM bioactivity, providing further support for these P-based NMs being further advanced for use as safe nanofertilisers for agriculture. Findings such as the transformation of NMs from the pristine state, NM colloidal interactions, and the effects on cellular metabolic activities, as discussed above, affirm that the effects of NMs on mammalian cells are due to multiple mechanisms and physicochemical factors. This further supports the aim of our study where the emphasis is to understand the nano-bio interactions with varying physicochemical properties of P-based NMs.
5 Conclusions
In this study, in vitro nano-bio interactions of five physicochemically different P-based NMs were evaluated by changes in size, P content, colloidal interactions, protein corona estimation, and effects on HEK293 cells. The colloidal interactions and protein hard corona composition were shape-dependent. Biogenic nHAP and nP were shown to be taken up by the cells and localise intracellularly in time-dependent manner. None of the variants of P-based NMs had LC50 up to 1000 µg mL−1 when cell viability, mitochondrial and lysosomal activity; and membrane disintegrity were evaluated. This highest tested concentration is hypothesised to be significantly higher than the environmentally relevant concentration for NM bioactivity, suggesting that the tested P-based NMs can be considered safe as nano-agromaterials. At the environmentally relevant dose of 100 µg mL−1 platelet (biogenic) and spherical-shaped nHAPs are safer as compared to the rod (chemically synthesised) and needle-shaped NMs. This study aims to provide experimental data on safer doses for the new generation of biogenic P-based NMs to be used as nanofertilisers to supplement P in agricultural soils.List of abbreviations
| nHAP_B | Nanohydroxyapatite biologically synthesized |
| nHAP_C| | Nanohydroxyapatite chemically synthesized |
| nHAP_Sigma | Nanohydroxyapatite procured from Sigma-Aldrich |
| nHAP_SRL | Nanohydroxyapatite procured from SRL Pvt Ltd |
| nP | Nanophosphorus |
| RP | Rock phosphate |
| CCM | Cell culture media |
| DPBS | Dulbecco's phosphate buffer saline |
| FBS | Fetal bovine serum |
| TEM | Transmission electron microscopy |
| DLS | Dynamic light scattering |
| FITC | Fluorescein isothiocynate |
Ethics for approval
All experiments were approved by Deakin University Human Research Ethics Committee (DUHREC reference no. 2021-095), Australia.Consent for publication
All the authors have read and approved the manuscript for publication.Data availability
The datasets used in the present study are available from the corresponding author upon reasonable request.Author contributions
Conceiving and designing the experiments: AP, AKD, PPS. Wet-lab work and data compilation: AP. Data analyses and manuscript writing: AP and PPS. Critical inputs and finalization of the manuscript: AP, LOA, AGS, AKD, and PPS.Conflicts of interest
The authors declare the following conflict of interest: nanophosphorus (nP) is the nanomaterial developed at TERI-Deakin Nanobiotechnology Centre (TDNBC), Gurgaon, India, and the synthesis and characterization of nP as the intellectual property of TDNBC will be filed with the Indian patent office.Acknowledgements
The research activities are supported by the Department of Biotechnology, India (grant no. BT/NNT/28/SP30280). The research activities of the authors are supported by the TERI-Deakin Nanobiotechnology Centre, Gurugram, India, and Deakin University, Victoria, Australia. The authors also acknowledge Mr Chandrakant Tripathi and Ms Deep Rajani (TDNBC, India) for TEM sample preparation and analysis. AP is supported by the Deakin University postgraduate scholarship.References
- R. Liu and R. Lal, Synthetic apatite nanoparticles as a phosphorus fertilizer for soybean (Glycine max), Sci. Rep., 2014, 4(1), 1–6 Search PubMed.
- L. Xiong, et al., Bioavailability and movement of hydroxyapatite nanoparticles (HA-NPs) applied as a phosphorus fertiliser in soils, Environ. Sci.: Nano, 2018, 5(12), 2888–2898 RSC.
- D. Montalvo, M. J. McLaughlin and F. Degryse, Efficacy of hydroxyapatite nanoparticles as phosphorus fertilizer in andisols and oxisols, Soil Sci. Soc. Am. J., 2015, 79(2), 551–558 CrossRef CAS.
- M. B. Taşkın, et al., Effect of synthetic nano-hydroxyapatite as an alternative phosphorus source on growth and phosphorus nutrition of lettuce (Lactuca sativa L.) plant, J. Plant Nutr., 2018, 41(9), 1148–1154 CrossRef.
- A. Priyam, et al., Fertilizing benefits of biogenic Phosphorous nanonutrients on Solanum lycopersicum in soils with variable pH, Heliyon, 2022, e09144 CrossRef CAS PubMed.
- A. Priyam, et al., Uptake and Benefits of Biogenic Phosphorus Nanomaterials Applied via Fertigation to Japonica Rice (Taipei 309) in Low-and High-Calcareous Soil Conditions, ACS Agricultural Science & Technology, 2022 Search PubMed.
- N. Basavegowda and K.-H. Baek, Current and future perspectives on the use of nanofertilizers for sustainable agriculture: the case of phosphorus nanofertilizer, 3 Biotech, 2021, 11(7), 1–21 CrossRef PubMed.
- A. Priyam, et al., Fertilizing benefits of biogenic phosphorous nanonutrients on Solanum lycopersicum in soils with variable pH, Heliyon, 2022, 8(3), e09144 CrossRef CAS PubMed.
- A. E. Szameitat, et al., Unravelling the interactions between nano-hydroxyapatite and the roots of phosphorus deficient barley plants, Environ. Sci.: Nano, 2021, 8(2), 444–459 RSC.
- H. P. G. Reis, et al., Role of Slow-Release Phosphate Nanofertilizers in Forage Nutrition and Phosphorus Lability, ACS Agricultural Science & Technology, 2022 Search PubMed.
- Q. Li, et al., Effects of phosphorus ensembled nanomaterials on nutrient uptake and distribution in Glycine Max L. under simulated precipitation, Agronomy, 2021, 11(6), 1086 CrossRef CAS.
- E. P. Pérez‐Álvarez, et al., Towards a more sustainable viticulture: foliar application of N‐doped calcium phosphate nanoparticles on Tempranillo grapes, J. Sci. Food Agric., 2021, 101(4), 1307–1313 CrossRef PubMed.
- Z. Xu, et al., Effects of four types of hydroxyapatite nanoparticles with different nanocrystal morphologies and sizes on apoptosis in rat osteoblasts, J. Appl. Toxicol., 2012, 32(6), 429–435 CrossRef CAS PubMed.
- H.-T. Lu, et al., Development of genipin-crosslinked and fucoidan-adsorbed nano-hydroxyapatite/hydroxypropyl chitosan composite scaffolds for bone tissue engineering, Int. J. Biol. Macromol., 2019, 128, 973–984 CrossRef CAS PubMed.
- T. M. Andrade, et al., In vitro and in vivo evaluation of rotary-jet-spun poly (ε-caprolactone) with high loading of nano-hydroxyapatite, J. Mater. Sci.: Mater. Med., 2019, 30(2), 19 CrossRef PubMed.
- N. M. Martinelli, et al., In vitro osteogenesis stimulation via nano-hydroxyapatite/carbon nanotube thin films on biomedical stainless steel, Materials, 2018, 11(9), 1555 CrossRef PubMed.
- J. C. Forero, et al., Development of useful biomaterial for bone tissue engineering by incorporating nano-copper-zinc alloy (nCuZn) in chitosan/gelatin/nano-hydroxyapatite (Ch/G/nHAp) scaffold, Materials, 2017, 10(10), 1177 CrossRef PubMed.
- S. P. Victor, et al., Europium doped calcium deficient hydroxyapatite as theranostic nanoplatforms: effect of structure and aspect ratio, ACS Biomater. Sci. Eng., 2017, 3(12), 3588–3595 CrossRef CAS PubMed.
- S. Lakshmanaperumal and C. Mahendran, Structural, dielectric, cytocompatibility, and in vitro bioactivity studies of yttrium and strontium co-substituted nano-hydroxyapatite by sol–gel method, J. Sol-Gel Sci. Technol., 2018, 88(2), 296–308 CrossRef.
- X. Zhao, et al., In vitro assessment of cellular responses to rod-shaped hydroxyapatite nanoparticles of varying lengths and surface areas, Nanotoxicology, 2011, 5(2), 182–194 CrossRef CAS PubMed.
- J. Scheel, et al., Exposure of the murine RAW 264.7 macrophage cell line to hydroxyapatite dispersions of various composition and morphology: assessment of cytotoxicity, activation and stress response, Toxicol. in Vitro, 2009, 23(3), 531–538 CrossRef CAS PubMed.
- X. Zhao, et al., Evaluating the toxicity of hydroxyapatite nanoparticles in catfish cells and zebrafish embryos, Small, 2013, 9(9–10), 1734–1741 CrossRef CAS PubMed.
- A. E. M. Aguilar, et al., Green synthesis of nano hydroxyapatite: morphology variation and its effect on cytotoxicity against fibroblast, Mater. Lett., 2021, 284, 129013 CrossRef.
- K.-W. Lee, et al., Physical properties and cellular responses to crosslinkable poly (propylene fumarate)/hydroxyapatite nanocomposites, Biomaterials, 2008, 29(19), 2839–2848 CrossRef CAS PubMed.
- Z. R. Huang, et al., Development and evaluation of lipid nanoparticles for camptothecin delivery: a comparison of solid lipid nanoparticles, nanostructured lipid carriers, and lipid emulsion, Acta Pharmacol. Sin., 2008, 29(9), 1094–1102 CrossRef CAS PubMed.
- T. T. Ding and J. Sun, Effect of Nanometer Hydroxyapatite Particles on Rat Macrophage at mRNA Level, in Key Engineering Materials, Trans Tech Publ, 2007 Search PubMed.
- T. J. Park, K. G. Lee and S. Y. Lee, Advances in microbial biosynthesis of metal nanoparticles, Appl. Microbiol. Biotechnol., 2016, 100(2), 521–534 CrossRef CAS PubMed.
- H. Duan, D. Wang and Y. Li, Green chemistry for nanoparticle synthesis, Chem. Soc. Rev., 2015, 44(16), 5778–5792 RSC.
- R. K. Das, et al., Biological synthesis of metallic nanoparticles: plants, animals and microbial aspects, Nanotechnol. Environ. Eng., 2017, 2(1), 1–21 CrossRef CAS.
- A. Priyam, et al., A new method for biological synthesis of agriculturally relevant nanohydroxyapatite with elucidated effects on soil bacteria, Sci. Rep., 2019, 9, 1 CrossRef CAS PubMed.
- F. Sharifianjazi, et al., Biocompatibility and mechanical properties of pigeon bone waste extracted natural nano-hydroxyapatite for bone tissue engineering, Mater. Sci. Eng., B, 2021, 264, 114950 CrossRef CAS.
- R. N. Rial, et al., Structural and kinetic visualization of the protein corona on bioceramic nanoparticles, Langmuir, 2018, 34(7), 2471–2480 CrossRef CAS PubMed.
- F. Catalano, et al., Towards the control of the biological identity of nanobiomaterials: Impact of the structure of 011-0 surface terminations of nanohydroxyapatite on the conformation of adsorbed proteins, Colloids Surf., B, 2020, 188, 110780 CrossRef CAS PubMed.
- Y. Zhu, et al., Protein corona of magnetic hydroxyapatite scaffold improves cell proliferation via activation of mitogen-activated protein kinase signaling pathway, ACS Nano, 2017, 11(4), 3690–3704 CrossRef CAS PubMed.
- A. Priyam, et al., Investigation into the trophic transfer and acute toxicity of phosphorus-based nano-agromaterials in Caenorhabditis elegans, NanoImpact, 2021, 23, 100327 CrossRef CAS PubMed.
- A. Priyam, et al., Exposure to biogenic phosphorus nano-agromaterials promotes early hatching and causes no acute toxicity in zebrafish embryos, Environ. Sci.: Nano, 2022,(9), 1364–1380 RSC.
- A. Priyam, et al., Abiotic factors and aging alter the physicochemical characteristics and toxicity of Phosphorus nanomaterials to zebrafish embryos, NanoImpact, 2022, 25, 100387 CrossRef CAS PubMed.
- H. S. Choi, et al., Renal clearance of quantum dots, Nat. Biotechnol., 2007, 25(10), 1165–1170 CrossRef CAS PubMed.
- C. Verrier, et al., Topography, composition and structure of incipient Randall plaque at the nanoscale level, J. Urol., 2016, 196(5), 1566–1574 CrossRef PubMed.
- H.-G. Tiselius, The role of calcium phosphate in the development of Randall's plaques, Urolithiasis, 2013, 41(5), 369–377 CrossRef CAS PubMed.
- A. Priyam, et al., A new method for biological synthesis of agriculturally relevant nanohydroxyapatite with elucidated effects on soil bacteria, Sci. Rep., 2019, 9(1), 1–14 CrossRef CAS PubMed.
- A. G. Schultz, et al., Silver nanoparticles inhibit sodium uptake in juvenile rainbow trout (Oncorhynchus mykiss), Environ. Sci. Technol., 2012, 46(18), 10295–10301 CrossRef CAS PubMed.
- N. J. Kruger, The Bradford method for protein quantitation, in The protein protocols handbook, Springer, 2009, pp. 17–24 Search PubMed.
- A. P. Stern ST and T. P. Potter, NCL Method GTA-1: LLC-PK1 Kidney Cytotoxicity Assay, 2020, available from: https://ncl.cancer.gov/resources/assay-cascade-protocols Search PubMed.
- E. Borenfreund and J. A. Puerner, Toxicity determined in vitro by morphological alterations and neutral red absorption, Toxicol. Lett., 1985, 24(2–3), 119–124 CrossRef CAS PubMed.
- T. F. Uliasz and S. J. Hewett, A microtiter trypan blue absorbance assay for the quantitative determination of excitotoxic neuronal injury in cell culture, J. Neurosci. Methods, 2000, 100(1–2), 157–163 CrossRef CAS PubMed.
- Z. B. Stern ST, NCL Method GTA-7: Hepatocyte Primary ROS Assay, 2010, available from: https://ncl.cancer.gov/resources/assay-cascade-protocols Search PubMed.
- P. T. Stern ST and B. Neun, NCL Method GTA-4: Hep G2 Hepatocyte Lipid Peroxidation Assay, 2010, available from: https://ncl.cancer.gov/resources/assay-cascade-protocols Search PubMed.
- J. Zhao, et al., Engineered nanomaterials in the environment: Are they safe?, Crit. Rev. Environ. Sci. Technol., 2020, 1–36 Search PubMed.
- M. Simonin, et al., Toxicity of TiO2 nanoparticles on soil nitrification at environmentally relevant concentrations: Lack of classical dose–response relationships, Nanotoxicology, 2017, 11(2), 247–255 CrossRef CAS PubMed.
- J. B. Raftis and M. R. Miller, Nanoparticle translocation and multi-organ toxicity: a particularly small problem, Nano Today, 2019, 26, 8–12 CrossRef CAS PubMed.
- S. Jia, et al., A novel vertical aligned mesoporous silica coated nanohydroxyapatite particle as efficient dexamethasone carrier for potential application in osteogenesis, Biomed. Mater., 2021, 16(3), 035030 CrossRef CAS PubMed.
- E. Casals, et al., Time evolution of the nanoparticle protein corona, ACS Nano, 2010, 4(7), 3623–3632 CrossRef CAS PubMed.
- Z. Zhao, et al., Identification and interaction mechanism of protein corona on silver nanoparticles with different sizes and the cellular responses, J. Hazard. Mater., 2021, 414, 125582 CrossRef CAS PubMed.
- A. Priyam, et al., Investigation into the trophic transfer and acute toxicity of phosphorus-based nano-agromaterials in Caenorhabditis elegans, NanoImpact, 2021, 100327 CrossRef CAS PubMed.
- G. Healthcare, Size exclusion chromatography: Sephacryl™ High Resolution resins HiPrep™ Sephacryl HR columns, 2016, p. 8 Search PubMed.
- S. Schöttler, et al., Protein adsorption is required for stealth effect of poly (ethylene glycol)-and poly (phosphoester)-coated nanocarriers, Nat. Nanotechnol., 2016, 11(4), 372–377 CrossRef PubMed.
- A. E. Nel, et al., Understanding biophysicochemical interactions at the nano-bio interface, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS PubMed.
- T. Göppert and R. Müller, Adsorption kinetics of plasma proteins on solid lipid nanoparticles for drug targeting, Int. J. Pharm., 2005, 302(1–2), 172–186 CrossRef PubMed.
- P. Camner, et al., Experimental and calculated parameters on particle phagocytosis by alveolar macrophages, J. Appl. Physiol., 2002, 92(6), 2608–2616 CrossRef PubMed.
- M. Lundqvist, et al., Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 CrossRef CAS PubMed.
- H. R. Kim, et al., Low-density lipoprotein receptor-mediated endocytosis of PEGylated nanoparticles in rat brain endothelial cells, Cell. Mol. Life Sci., 2007, 64(3), 356–364 CrossRef CAS PubMed.
- V. Di Francesco, et al., Modulating Lipoprotein Transcellular Transport and Atherosclerotic Plaque Formation in ApoE–/–Mice via Nanoformulated Lipid–Methotrexate Conjugates, ACS Appl. Mater. Interfaces, 2020, 12(34), 37943–37956 CrossRef CAS PubMed.
- A. O. Fayiga and O. Nwoke, Phosphate rock: origin, importance, environmental impacts, and future roles, Environ. Rev., 2016, 24(4), 403–415 CrossRef CAS.
- M. Levak, et al., Effect of protein corona on silver nanoparticle stabilization and ion release kinetics in artificial seawater, Environ. Sci. Technol., 2017, 51(3), 1259–1266 CrossRef CAS PubMed.
- C. Buzea, I. I. Pacheco and K. Robbie, Nanomaterials and nanoparticles: sources and toxicity, Biointerphases, 2007, 2(4), MR17–MR71 CrossRef PubMed.
- R. Stevenson, et al., Nanoparticles and Inflammation, Sci. World J., 2011, 11, 1300–1312 CrossRef CAS PubMed.
- G. Oberdörster, E. Oberdörster and J. Oberdörster, Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles, Environ. Health Perspect., 2005, 113(7), 823–839 CrossRef PubMed.
- H. N. Sun, G. Z. Yan and H. Y. Zhou, Nanoparticles-Induced Oxidative Stress, in Bioactivity of Engineered Nanoparticles, ed. B. Yan, H. Zhou and J. L. GardeaTorresdey, 2017. pp. 63–79 Search PubMed.
- N. P. Mortensen, et al., Dynamic development of the protein corona on silica nanoparticles: composition and role in toxicity, Nanoscale, 2013, 5(14), 6372–6380 RSC.
- T. Xia, et al., Comparison of the abilities of ambient and manufactured nanoparticles to induce cellular toxicity according to an oxidative stress paradigm, Nano Lett., 2006, 6(8), 1794–1807 CrossRef CAS PubMed.
- H. Zhang, et al., Cellular internalization of rod-like nano hydroxyapatite particles and their size and dose-dependent effects on pre-osteoblasts, J. Mater. Chem. B, 2017, 5(6), 1205–1217 RSC.
- Y. Jin, et al., Oxidative stress-induced apoptosis of osteoblastic MC3T3-E1 cells by hydroxyapatite nanoparticles through lysosomal and mitochondrial pathways, RSC Adv., 2017, 7(21), 13010–13018 RSC.
- X. Cui, et al., Correlation of particle properties with cytotoxicity and cellular uptake of hydroxyapatite nanoparticles in human gastric cancer cells, Mater. Sci. Eng., C, 2016, 67, 453–460 CrossRef CAS PubMed.
- L. Wang, et al., Nano-hydroxyapatite particles induce apoptosis on MC3T3-E1 cells and tissue cells in SD rats, Nanoscale, 2012, 4(9), 2894–2899 RSC.
- X.-Y. Sun, et al., Size-dependent cytotoxicity of hydroxyapatite crystals on renal epithelial cells, Int. J. Nanomed., 2020, 15, 5043 CrossRef CAS PubMed.
- R.-M. Kavasi, et al., In vitro biocompatibility assessment of nano-hydroxyapatite, Nanomaterials, 2021, 11(5), 1152 CrossRef CAS PubMed.
- J. G. Teeguarden, et al., Particokinetics in vitro: dosimetry considerations for in vitro nanoparticle toxicity assessments, Toxicol. Sci., 2007, 95(2), 300–312 CrossRef CAS PubMed.
- K. Wittmaack, Excessive delivery of nanostructured matter to submersed cells caused by rapid gravitational settling, ACS Nano, 2011, 5(5), 3766–3778 CrossRef CAS PubMed.
- H. Kato, et al., Dispersion characteristics of various metal oxide secondary nanoparticles in culture medium for in vitro toxicology assessment, Toxicol. in Vitro, 2010, 24(3), 1009–1018 CrossRef CAS PubMed.
- M. Wu, et al., Size-dependent cellular uptake and localization profiles of silver nanoparticles, Int. J. Nanomed., 2019, 14, 4247 CrossRef CAS PubMed.
- L. B. Ahmed, et al., Impact of surface functionalization on the uptake mechanism and toxicity effects of silver nanoparticles in HepG2 cells, Food Chem. Toxicol., 2017, 107, 349–361 CrossRef PubMed.
- X. Xie, et al., The effect of shape on cellular uptake of gold nanoparticles in the forms of stars, rods, and triangles, Sci. Rep., 2017, 7(1), 1–9 CrossRef PubMed.
- Q. Feng, et al., Uptake, distribution, clearance, and toxicity of iron oxide nanoparticles with different sizes and coatings, Sci. Rep., 2018, 8(1), 1–13 CAS.
- P. Ickrath, et al., Time-dependent toxic and genotoxic effects of zinc oxide nanoparticles after long-term and repetitive exposure to human mesenchymal stem cells, Int. J. Environ. Res. Public Health, 2017, 14(12), 1590 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2va00318j |
| This journal is © The Royal Society of Chemistry 2023 |